Abstract
Tay-Sachs or Sandhoff disease result from mutations in either the evolutionarily related HEXA or HEXB genes encoding respectively, the α- or β-subunits of β-hexosaminidase A (HexA). Of the three Hex isozymes, only HexA can interact with its cofactor, the GM2 activator protein (GM2AP), and hydrolyze GM2 ganglioside. A major impediment to establishing gene or enzyme replacement therapy based on HexA is the need to synthesize both subunits. Thus, we combined the critical features of both α- and β-subunits into a single hybrid µ-subunit that contains the α-subunit active site, the stable β-subunit interface and unique areas in each subunit needed to interact with GM2AP. To facilitate intracellular analysis and the purification of the µ-homodimer (HexM), CRISPR-based genome editing was used to disrupt the HEXA and HEXB genes in a Human Embryonic Kidney 293 cell line stably expressing the µ-subunit. In association with GM2AP, HexM was shown to hydrolyze a fluorescent GM2 ganglioside derivative both in cellulo and in vitro. Gene transfer studies in both Tay-Sachs and Sandhoff mouse models demonstrated that HexM expression reduced brain GM2 ganglioside levels.
Introduction
The in vivo hydrolysis of GM2 ganglioside (GM2) requires the correct synthesis, folding and interaction of three gene products. Mutations in any of these genes can result in one of the three forms of the lysosomal storage disease, GM2 gangliosidosis. The HEXA and HEXB genes encode the ~60 kDa α- and β- subunits, respectively, of the heterodimeric β-hexosaminidase A (HexA) isozyme. Deficiencies in either the α- or β-subunit leads to Tay-Sachs (TSD) or Sandhoff disease (SD), respectively. These are autosomal recessive, progressive neurodegenerative disorders that account for the vast majority of GM2 gangliosidosis patients. However, it has been estimated that only ~10% of normal HexA activity is all that is needed to prevent GM2 storage.1
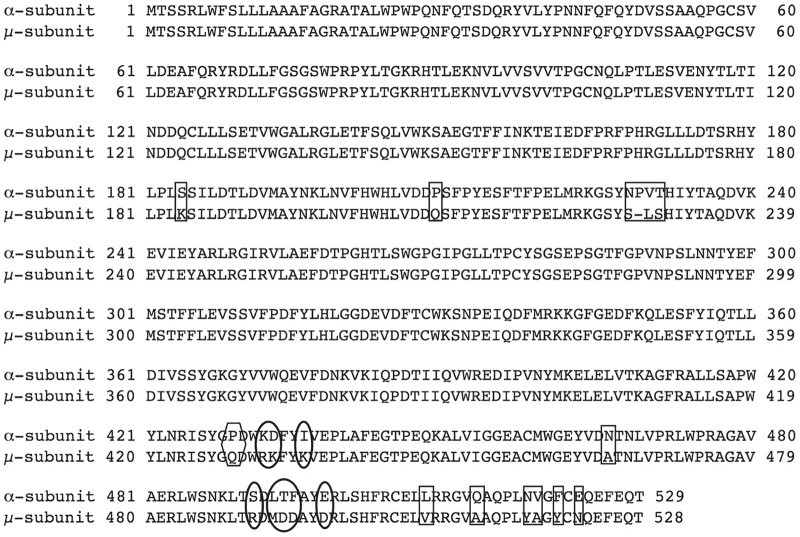
The HEXA and HEXB genes are evolutionarily related, with their protein products sharing ~60% sequence identity. While each subunit contains its own active site, residues from the neighboring subunit must stabilize the site in order for it to become functional.2–4 Thus, monomers are not active. In normal human cells, there are two major Hex isozymes, HexA (αβ) and HexB (ββ). An unstable isozyme is also detectable at very low levels in SD patient samples, HexS (αα). Due to sequence variations in each of the α- and β-subunit-subunit interfaces, the stability of the active dimeric forms of the isozymes vary markedly, i.e., the stability of HexB> HexA>> HexS. While all three isozymes can hydrolyze soluble, neutral substrates, e.g., 4-methylumbelliferyl 2-acetamido-2-deoxy-β-D-glucopyranoside (MUG), only the α-subunit-containing HexA and HexS isozymes can efficiently hydrolyze negatively charged substrates,5 such as MUG-6-sulfate6 (MUGS). The variation in substrate specificities is due to a positively charged binding pocket in the α-subunit, which is negatively charged in the β-subunit.2,3,7 This pocket is also responsible for binding the negatively charged sialic acid moiety in GM2.2,8
The third gene responsible for GM2 gangliosidosis, GM2A, encodes the GM2 activator protein (GM2AP).9 Mutations in the GM2A gene lead to the ultra-rare AB-variant form. GM2AP is a small, glycolipid-transport protein that removes a molecule of GM2 from the lysosomal membrane and presents it to HexA for hydrolysis (reviewed in refs. 10,11). Thus, HexA and the GM2AP-GM2 complex must interact in the lysosome to form a soluble, active quaternary complex with the GM2 substrate correctly positioned for hydrolysis by the active site of the α-subunit. This complex has been modeled.2 Consistent with the genetic and biochemical evidences that only HexA can efficiently turnover GM2, nonconserved protein patches, located in different areas of the α- and β-subunits, were predicted by the model (Figures 1 and 2) to be required for efficient interaction of the GM2AP-GM2 complex with HexA.2
Figure 1.
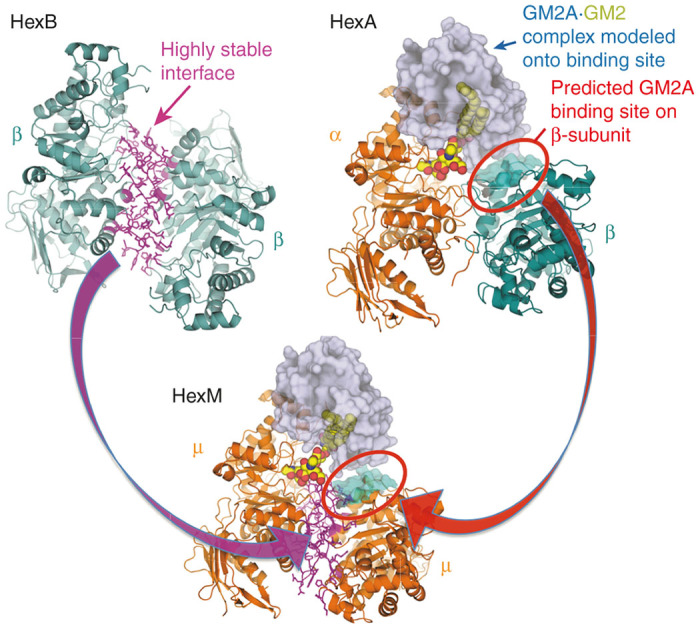
Model of the active HexM quaternary complex. The μ-subunit of HexM is derived from the α-subunit (orange) of human HexA, which was modified to include the stable homodimer interface (magenta) formed between the β-subunits (teal) of human HexB and a region from the β-subunit predicted to interact with GM2AP. The GM2AP (grey) bound to GM2 (spheres) was modeled onto HexA as described2,3 and this position was maintained when modeling the HexM quaternary complex.
Figure 2.
Substitutions that were made from the Hex β-subunit sequence into the human α-subunit to produce the μ-subunit of homodimeric HexM. Changes in; rectangles = β-like dimer interface; those in ovals = β-like GM2AP binding; and the one in a hexagon = both β-like interface and activator binding sites.
The heterodimeric nature of HexA is a major impediment to developing either gene therapy or enzyme replacement therapy (ERT) for TSD and SD. Previous gene therapy approaches for Tay-Sachs and Sandhoff have relied on delivery of a single Hex subunit, or a dual vector approach separately delivering both subunits.12–17 While these studies have generated promising results, they are not ideal. Our approach to overcoming this impediment, is to engineer a new artificial Hex subunit (µ) that contains the α-active site, regions on the surfaces of the α- and β-subunits that are needed to functionally interact with the GM2AP-GM2 complex in vivo, and the β-subunit interface needed to form a new stable µ-homodimer (HexM). The resulting size of the DNA-coding sequence for µ is well within the packaging capacity of the efficient self-complementary (sc)AAV.18 There have also been reports that intravenously administered AAV9 can cross the blood–brain barrier, wherein the efficiency of gene transfer to the CNS was greatly enhanced by the use of scAAV vectors.19–21
Results
Constructing HexM
We previously demonstrated that neither of two hybrid Hex subunit engineered by ourselves22 or others23 were able to hydrolyze GM2 in a GM2AP-dependent manner as homodimers.22 Both of these subunits were constructed by substituting regions of α-subunit sequence in the β-subunit, e.g., HexBMM.22 Thus, we engineered a new hybrid based on the human α-subunit. To promote rapid dimerization and stability, the dimerization interface of the α-subunit was exchanged with that of the β-subunit. Next, two small areas in the C-terminal 20% of the β-subunit, predicted to be necessary for GM2AP binding, were used to replace the aligned sequences in the α-subunit (Figures 1 and 2 and Supplementary Table S1). In the process of making this new construct, the nucleotide sequence was also codon-optimized for more efficient protein expression in mammalian cells (Supplementary Figure S1).
Establishing stably transfected HEXA-/-/HEXB-/- HEK-293 cells secreting HexM-His6
One of the problems we encountered in our previous attempt to engineer a β-subunit-based Hex hybrid subunit was the formation of heterodimers between our transfected hybrid and the endogenous normal α subunit in SD cells. These α-hybrid heterodimers were then capable of hydrolyzing a fluorescent GM2 derivative (NBD-GM2) in cellulo and produced false positive results.22 To address this problem, we used CRISPR-based genome editing24 to render both the HEXA and HEXB genes in clonal populations of HEK cells nonfunctional (HEKHexABKO). The double inactivation of HEXA/B genes was confirmed by: (i) direct sequencing or PCR analyses of exon1 and 11 of HEXA and exon 1 of HEXB (Supplementary Figure S2a), which were targeted by the guide RNAs we used; (ii) western blot analyses of the wild type (WT) versus the HEKHexABKO cell lysates (Supplementary Figure S2b); and (iii) Hex activity assays based on the fluorogenic substrates MUG and MUGS (Supplementary Figure S2c). These results confirmed the presence of deletions in both alleles of HEXA and HEXB (Supplementary Figure S2a) resulting in a reduction of 3 log orders in total MUGS and MUG activities. The very low levels of residual activity likely result from other neutral glycosidases, such as O-GlcNAcase.25
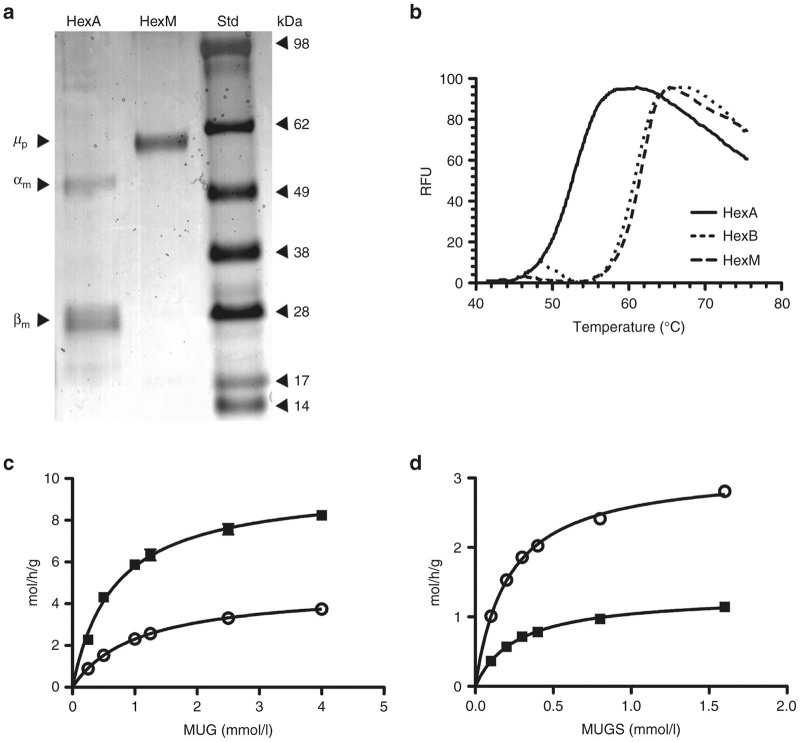
The HEK-HexABKO cells were next transfected with a construct encoding the µ-subunit with a C-terminal His6 tag. A clonal population of these cells, stably expressing high levels of HexM-His6 (HEK-HexABKO-HexMHis) activity, was selected for further study. Initially, it was confirmed that these live cells could hydrolyze NBD-GM2, which we26 have previously shown to be taken-up by cultured cells and transported to the lysosome where it is primarily hydrolyzed by HexA in a GM2AP-dependent manner (Figure 3a,b). These in cellulo data were confirmed in vitro by comparing the activities of purified HexA and HexM-His6 toward NBD-GM2 in the presence or absence of recombinant human GM2AP (rGM2AP). Our previously constructed β-base hybrid (HexBMM), in its enriched homodimeric form,22 was used as a negative control. These assays were performed using an equal number of MUGS units of each isozyme (Figure 3c,d). Interestingly, the resulting data suggest that both active sites in HexM are able to bind and hydrolyze a GM2:GM2AP complex simultaneously, given that they can do this with the smaller MUGS substrate (Figure 4c,d; Table 1, discussed below). Based on the crystal structures of HexA, HexB, and GM2AP, a model was constructed to predict whether HexM could simultaneously bind a GM2:GM2AP complex at each of its two active sites. The resulting model suggests this could occur without significant steric hindrance (data not shown).
Figure 3.
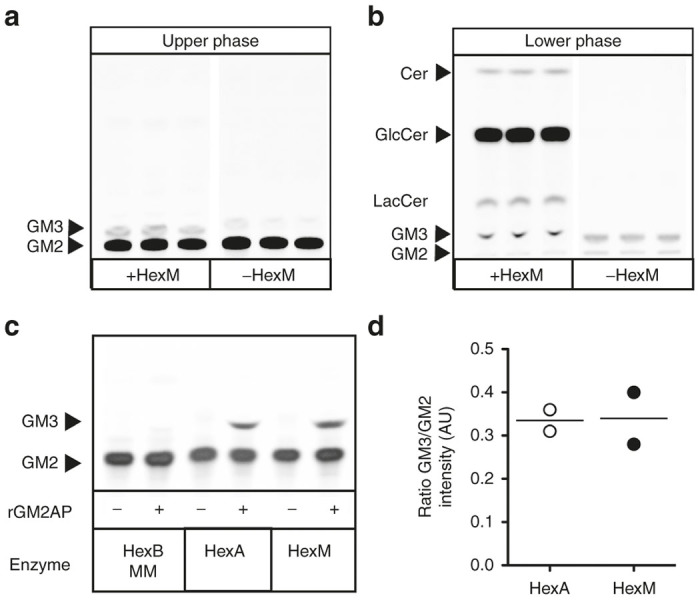
HexM is capable of hydrolyzing NBD-GM2 in a GM2AP-dependent manner. In cellulo hydrolysis of NBD-GM2 was accessed by HPTLC separation of the Folch-extracted NBD-glycolipids from HexM expressing (+HexM) and untransfected (-HexM) HEKHexABKO cells. The upper phase (a) contains acidic glycolipids (gangliosides) and the lower phase (b) contains primarily neutral glycolipids. Conduritol B epoxide (50 µmol/l), a nonreversible inhibitor of glucocerebrosidase, was also present in the medium to inhibit NBD-GlcCer degradation.26 (c) In vitro assay of NBD-GM2 hydrolysis by equal MUGS units of three human Hex isozymes; HexBMM,22 HexA and HexM; in the absence (−) or presence (+) of human rGM2AP. (d) Quantitative comparison of hydrolysis of NBD-GM2 to NBD-GM3 by HexA and HexM (n = 2) expressed in arbitrary units (AU) and as a ratio of band intensities corresponding to NBD-GM3 and NBD-GM2 in HPTLC shown in panel c.
Figure 4.
Characterization of the heat stability and kinetic properties of HexM as compared to other Hex isozymes. (a) Protein stained SDS-PAGE gel demonstrating the purity of secreted HexM after Ni-NTA chromatography. The secreted, purified HexM precursor polypeptide (µp) versus the major mature α- and β- polypeptides (αm, βm) of isolated placental HexA. Molecular weight standards (Std) are shown at the right. (b) Confirmation was obtained using differential scanning fluorimetry that the β-subunit interface was successfully transferred into the HexM protein structure. The Tm of HexA calculated from its heat denaturation curve (solid line) was significantly lower (~10 °C) than those calculated for HexB (dotted line) and HexM (dashed line), which were virtually identical. (c,d) Comparison of the kinetics of purified HexM versus HexA for artificial substrates. The Km and Vmax values of HexM (open circles) and HexA (filled squares) for the MUG (c) and MUGS (d) substrates were determined from the specific activities of each isozyme (y-axis) at various concentrations of each substrate (x-axis) and fitted to the Michaelis-Menten equation using Prism Graphpad.
Table 1. Comparison of Km and Vmax values of HexM toward a neutral and a negatively charged artificial substrate with those of other Hex isozymes.
| Hex | MUG Km (mmol/l) | MUG Vmaxa | MUGS Km (mmol/l) | MUGS Vmaxa | Reference |
|---|---|---|---|---|---|
| M (μμ)b | 1.10 ± 0.06 | 4.7 ± 0.1 | 0.21 ± 0.01 | 3.13 ± 0.03 | PRc |
| A (αβ) | 0.67 ± 0.06 | 9.7 ± 0.3 | 0.26 ± 0.02 | 1.32 ± 0.03 | PR |
| S (αα) | 1.5 ± 0.2 | 2.0 ± 0.1d | 0.3 ± 0.1 | 0.94 ± 0.04d | 7 |
| B (ββ) | 0.71 ± 0.1 | 14.5 ± 0.5 | NDe | NDe | 7 |
Moles (MU) h−1 g−1 (Hex).
Indicates a homodimer of our α-based hybrid subunit.
Present report.
These values may be low due to the presents of significant amounts of inactive HexS (an unstable isozyme).
Not determined because of insufficient activity toward MUGS (MUG/MUGS ~1/300).
Characterization of HexM
We have previously shown that a C-terminal His6-tag does not affect the kinetics of the precursor form of HexB, which is indistinguishable from the kinetics of the mature lysosomal form.27 Furthermore, we have shown that the His6-tag is removed once the enzyme enters the protease-rich environment of the lysosome,27 where proteolytic processing produces the mature, multi-polypeptide-containing subunits28 (reviewed in ref. 11). Thus, we purified the secreted precursor form of HexM-His6 using Ni-NTA resin (Figure 4a), and compared its melting temperature (Tm) to those of purified placental HexA and HexB, as determined using differential scanning fluorimetry29,30 (Figure 4b). The calculated Tm for HexB and HexM were virtually identical at 61 and 62 °C, respectively. In contrast, the Tm calculated for HexA was significantly lower at 53 °C. These data indicate that the β-subunit interface was successfully transferred to the µ-subunits of HexM.
Using the purified His6-tagged precursor form of HexM and the purified mature form of placental HexA,31 the KM and Vmax values for each isozyme were determined for both the MUG (Figure 4c) and MUGS (Figure 4d) substrates and compared to previously published data for HexB and HexS (Table 1). The KM and Vmax of the α- and β-subunits’ active sites are known to differ for the MUG and MUGS substrates.6 As reported in Table 1, HexA, HexM, and HexS have virtually the same KM for MUGS. This result is expected, since the β-active site in HexA (and HexB) has a very low affinity for negatively charged substrates, and thus would not contribute significantly to the KM determination for HexA. However, the Vmax of MUGS for HexM is ~2-fold that for HexA (Fig. 4d). This was also expected, as both active sites in HexM should be able to hydrolyze negatively charged MUGS.
Interestingly, the Vmax of HexA for MUG is ~2-fold higher than that of HexM. MUG can be hydrolyzed by either the α- or β-active site; however the β-site has a significantly high affinity (lower KM) and turnover rate (Vmax) for this substrate than does the α-site.6 Thus, the KM value of HexA for MUG is strongly influenced by its β-site while its Vmax value would be the average rate of hydrolysis for both subunits (Figure 4c). This is consistent with the observation that the Vmax of MUG for HexA, ~10 moles (MU) h−1 g−1 (Hex), is virtually equal to the average of those determined for HexB and HexM (Table 1).
Confirmation that HexM oligosaccharides contain M6P residues
Most soluble lysosomal enzymes, including Hex, depend on their Asn-linked oligosaccharides being specifically tagged in the ER/Golgi with mannose-6-phosphate residues (M6P) to both target them to lysosomes and allow their secreted forms to be “recaptured” by other cells.32,33 This could be advantageous for developing effective gene therapy. Although evidence that the recapture system in brain requires M6P-liganded enzyme is sparse, Kyttälä et al.34 have reported that by adding 5 mmol/l M6P along with recombinant aspartylglucosaminidase to the growth medium of aspartylglucosaminidase-deficient mouse primary neurons, a significant amount of the enzyme could be blocked from being endocytosed after a 24 hours incubation period.
In order to confirm that HexM also contains M6P residues, the 48-hour growth medium (conditioned medium) from HEK-HexABKO-HexMHis cells was collected and used to replace the medium in which an adult TSD patient fibroblast line35 was being grown. Lysates from these TSD cells, grown for 48 hours in conditioned medium produced a 40-fold increase in MUGS activity; whereas cells grown in conditioned medium containing M6P monosaccharides (5 mmol/l) produced only a 10-fold increase in specific activity (~75% reduction in endocytosis, Table 2).
Table 2. The effects of M6P on the internalization of HexM, secreted into the growth medium of transfected HEK (HEXA-/-, HEXB-/-) cells (conditioned medium), by adult TSD cells (5–10% residual MUGS activity).
Final concentration in the adult TSD cells’ growth medium.
MUGS, nmoles hydrolyzed * h−1 * mg−1 (total lysate protein) ± standard deviation (n = 3).
Not determined.
Intracranial administration of vectors expressing HexM in TSD mice reduces the level of accumulated GM2 ganglioside
To determine the functionality of HexM in lowering the GM2 levels in vivo, we performed intracranial injections of scAAV9 vectors containing either the HexM gene (HEXM), HEXA, or HEXBMM22 into the left hemisphere of 4-month-old Tay-Sachs hexa-/- mice. Two mice were injected with a mixture of one of the vectors and an AAV9-GFP vector at a dose of 109 vector genomes (vg) of each vector. Two additional mice were injected with the AAV9-GFP vector alone. Coronal sections from the injected brain regions were subjected to immuno-histochemistry (IHC) against GFP and GM2 (Figure 5). Extensive expression of GFP throughout the lateral cortex and striatum is apparent on the ipsilateral side, with scattered GFP-positive cells visible on the contralateral side (Figure 5a–d). This is consistent with axonal transport of AAV9 that has been reported.36 The Hex expression is expected to overlap with the areas of GFP expression, since identical AAV capsids, vector regulatory sequences, and doses were used for all vectors. GM2 accumulation is clearly visible at 5 months of age in these mice. As expected, in the case of sections from mice injected with GFP vector alone, there is no decrease in the level of GM2 accumulation (Figure 5e) on the ipsilateral side of the brain compared to the contralateral side. In the case of brain sections from mice injected with a mix of GFP and different Hex vectors, there is visible reduction in the GM2 levels on the injected side of the brain compared to the contralateral side of the same section (Figure 5f–h), and this reduction overlaps with the area of peak GFP expression (Figure 5b–d). To control for variability between mice, the comparison is mainly between the ipsilateral and contralateral sides of the same mouse. The extent of reduction in GM2 levels, apparent by the lighter DAB staining in these sections, is clear in the sections from all three HEX vector-treated mice. The HEXM vector (Figure 5h) was at least as efficient in reducing GM2 levels as the vector containing the HEXA gene (Figure 5g) and both of these were significantly more efficient than the vector containing the HEXBMM gene (Figure 5f).
Figure 5.
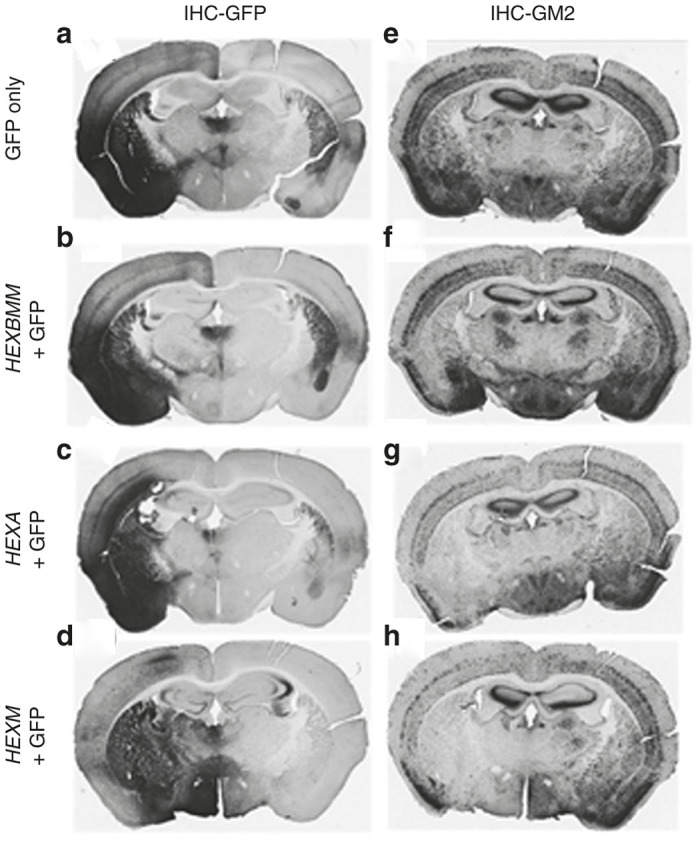
Reduction in GM2 ganglioside levels following intracranial injection of AAV9-Hex vectors. Four-month-old Tay-Sachs disease knock out mice received a single unilateral intracranial injection (left side) of either AAV9-GFP (109 vg) or a 2-vector mixture containing equal amounts (109 vg each) of AAV9-GFP and an AAV9-Hex vector, as designated on the left panel. Coronal sections of the brain were analyzed by immunohistochemistry (IHC) for green fluorescent protein (GFP) expression to indicate vector spread (a–d) and for GM2 ganglioside levels to assess the relative activity of Hex (e–h).
Reduced GM2 ganglioside accumulation in neonatal SD mice following the intravenous injection of a vector expressing HexM
Neonatal SD (hexb-/-) mice were injected intravenously with a scAAV9.47 vector containing a synthetic promoter to drive the expression of the HEXM gene. Eight-week postinjection the mice were sacrificed and GM2 levels (relative to those of GD1a) and enzyme activity toward MUGS and MUG (relative to total protein) from the mid-brain homogenate were determined. Although the Hex activity in the brains of the mice injected with the vector expressing the HexM µ-subunit was only slightly increased above those in mice injected with an empty vector, GM2 levels in these mice were significantly decreased (Figure 6, HexM versus vehicle). However, the GM2 levels in these mice were not reduced to the levels found in mice heterozygous for the hexb deletion mutation (Figure 6, HexM versus Het). This observation is likely due to a low level of gene transfer into the brain from the intravenous injection of the HEXM vector, which can also be inferred from the small increases in MUG and MUGS activity found in the same samples.
Figure 6.
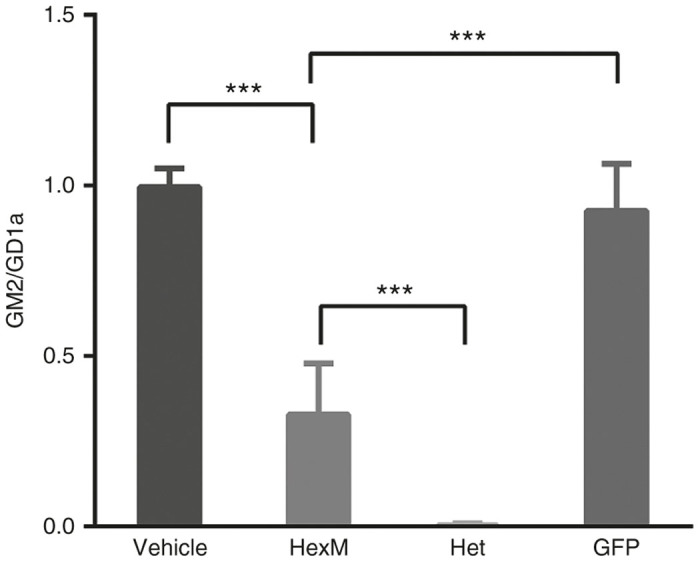
GM2 levels are significantly reduced in the brains of Sandhoff disease mice 8 weeks after neonatal, intravenous injection of a AAV9.47-HEXM vector. GM2 levels are expressed as a ratio of the densities of the GM2 versus GD1a spots from HPTLC plates (y-axis). Mice injected with either vehicle, AAV9.47-HEXM, or AAV9.47-GFP were compared to untreated heterozygous, hexb-/+, mice (Het). N = 6; ***P < 0.001.
Discussion
GM2 gangliosidosis (TSD and SD) presents challenges to developing both ERT and gene therapies in that the only Hex isozyme that turns over GM2 in vivo, HexA, is a heterodimer composed of similar but non-identical subunits. AAV are limited in their packaging capacity, i.e., ~4.5 kb for traditional single-strand AAV, and ~2.1 kb for the more efficient scAAV.18 The DNA coding sequences alone for both the α- and β-subunits of HexA total ~3.2 kb. Packaging only the deficient subunit is well within the size constraints of the AAV genome. However, overexpression of only one of the subunits would not lead to a similar level of expression and secretion of the missing heterodimeric HexA isozyme (to be recaptured by other non-infected cells), as the normal endogenous subunit would become a limiting factor. Much the same problem exists for ERT, which would require the mammalian cells producing the enzyme to be transfected with vectors encoding both subunits. Additionally, the secreted Hex from these cells would have to be purified and the Hex isozymes separated. Our approach to overcoming these challenges was to design a hybrid Hex subunit comprised of the desired functional areas of both the α- and β-subunits that could form stable homodimers and, using the GM2AP as a co-factor, turnover GM2 in vivo. We22 and others23 have previously attempted to accomplish this by modifying the stable β-subunit with α-sequences. However our previously published data demonstrated that neither homodimeric forms of these β-based hybrids could efficiently hydrolyze GM2 in a human rGM2AP dependent manner in vitro,22 e.g., HexBMM (Figure 3c).
The major problem in evaluating the ability of any homodimer of an engineered α-/β-hybrid subunit to hydrolyze GM2 in TSD/SD patient cells or animal models is the potential for the hybrid to form an active heterodimer with the remaining endogenous WT subunit, thereby masking its inactivity as a homodimer.22 There are two additional problems in testing gene therapy or ERT in the common mouse models. The first is that mice have a biological bypass system not found in humans, that allows them to hydrolyze GM2 through first converting it to its asialo derivative, GA2, which is then hydrolyze, although poorly, by mHexB37 in a mGM2AP-dependent manner.38 Thus the TSD mouse has only a mild, slowly progressing phenotype. This suggests that a significant increase in HexB or possibly even HexS activity particularly in the SD mouse model could produce a decrease in the level of stored GM2 and increased lifespans. On the other hand, human patients lack the lysosomal sialidase necessary to convert GM2 to GA2 and their hGM2AP is much more specific for the HexA isozyme than is the mGM2AP.39 This highlights another problem in evaluating ERT or gene transfer using human-based enzyme or expressed cDNA in animal models; i.e., the vagaries of the strength and specificities of the three sets of interspecies protein–protein interactions required to form the active quaternary complex and hydrolyze GM2. To minimize these problems, CRISPR-based genome editing was used to produce a (human) HEK cell line that does not express either the α- or β-subunits of HexA, HEKHexABKO (Supplementary Figure S2). We also ameliorated some of the potential problems in using a single mouse model to confirm the functionality of homodimeric HexM by demonstrating that AAV HEXM vectors can reduce GM2 levels in the brains of both TSD (Figure 5h) and SD (Figure 6) mice.
Considering that the MUGS Vmax for HexM is approximately twice that of HexA (Table 1) and that equal MUGS units of HexA and HexM hydrolyze, in a rGM2AP dependent manner, equal amounts of NDB-GM2 in vitro (Figure 3c,d), it appears that each of the active sites in HexM also have the ability to simultaneously bind and hydrolyze a GM2:GM2AP complex. This conclusion is further supported by molecular modeling, which indicates that two bound complexes could be accommodated in the HexM homodimer.
A promising observation for the potential application of ERT to TSD and SD is the reported ability of another lysosomal enzyme, sulphamidase, containing an Apolipoprotein B binding domain at its C-terminus and expressed in the liver of deficient (MPSIIIA) mice, to cross the BBB and correct the enzyme deficiency.40 The potential use of a similarly tagged version of HexM for ERT is attractive given the high level of expression we have observed in the HEKHexABKO cells (~1 mg of Hex M-His6 was purified from 1 l of media collected over time from 4× T75 flasks of cells), the observation that secreted HexM can be recaptured by TSD cell in a M6P dependent manner, and the extreme heat stability of the homodimer (Figure 4b).
The next steps in bringing HexM gene therapy or ERT into the clinic is a longer-term trial preferably in a larger animal model lacking the biological bypass pathway found in mice, such as the TSD sheep,41 prior to human trials. Future studies will also need to evaluate whether the incorporation of amino acid residues from the β-subunit into the α-subunit to create our hybrid µ-subunit result in new epitopes that could elicit an immune response
Materials and Methods
Designing the HexM subunit (μ)
The μ-subunit of HexM was engineered based on the α-subunit structure of human HexA. Firstly, the dimerization interface of the HexA α-subunit was identified from a PISA analysis (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html)42 of the HexA crystal structure.3 The identified interface was then replaced with residues comprising the more stable homodimeric β-subunit:β-subunit interface found in human HexB, which was identified by a PISA analysis of the human HexB2 crystal structure. Next, based on a model of human HexA bound to a GM2A/GM2 complex,2 two areas of residues near the C-terminus of the β-subunit, predicted to be necessary for GM2AP binding, were used to replace the aligned sequences in the α-subunit (Figures 1 and 2 and Supplementary Table S1). All structural analyses and modeling were carried out using PyMOL (Schrodinger, New York, NY (2010) The PyMOL Molecular Graphics System, Version 1.7.4.).
Generation of HEXA and HEXB knockout HEK293T cell line (HEKHexABKO) using CRISPR
Guide RNAs specifically targeting exon1 and exon 11 of HexA and exon 1 of HexB were chosen from the database of gRNAs targeting the human genome described in Mali et al.43 Oligonucleotides coding for the; (i) HexA gRNA targeting exon 1 (CGTCCTTTACGGTGTTTCGTCCTTTCCAC, HumanHexA-Ex1Rev and TAGCGCTGGTGTTTTAGAGCTAGAAATAGC, HumanHexA-Ex1For); (ii) HexA gRNA targeting exon 11 (TCCTATGGCCGGTGTTTCGTCCTTTCCAC, HumanHexA-Ex11Rev and TATACGGTTCGTTTTAGAGCTAGAAATAGC, HumanHexA-Ex11For); and (iii) oligonucleotides coding for the HexB gRNA targeting Exon 1 (AGTCAGCAGCGGTGTTTCGTCCTTTCCAC HuHexB-Ex1RevB and AGGTGGCGCGTTTTAGAGCTAGAAATAGC, HuHexB-Ex1For) were used to replace the guide RNA present in the Church gRNA expression vector described in Mali et al. https://www.addgene.org/41824/, Addgene, Cambridge, MA) via Round-the-horn site-directed mutagenesis http://openwetware.org/wiki/’Round-the-horn_site-directed_mutagenesis using Platinum Taq polymerase (Life Technologies, Grand Island, NY) in place of the Q5 polymerase in the Q5 Site-directed mutagenesis kit (New England Biolabs, Ipswich, MA). Insertion of the appropriate guide RNA sequence was verified by DNA sequencing (ACGT, Toronto, Canada). HEK 293T cells (100,000–250,000 cells in a six-well plate) were cotransfected overnight with HexA and HexB gRNA vectors (125 ng) together with the wild type Cas9 nuclease expression vector (125 ng) at 1:1:1:1 ratio, using lipofectamine LTX (Life Technologies). Following 3 days growth, single transfected cells were cloned by limiting cell dilution (ca. 0.5 cells/well) into eight 96-well tissue culture plates. After 2 weeks of growth, media were replaced with DMEM supplemented with 0.5% HSA and 100 mmol/l NH4Cl to facilitate secretion of Hex isoforms into the media, for an overnight incubation. Hex activities were evaluated in duplicate aliquots (10 µl) of the media in 384-well plates using either MUG (0.4 mmol/l) or MUGS (0.4 mmol/l) as the substrate. Potential clones “knocked out” for the HEXA and HEXB genes were identified as those wells, which contained cells and had background levels (i.e., medium only) of MUG and MUGS activity. Those clones were further expanded and the lack of HexA and HexB activity were reconfirmed using MUG and MUGS substrates relative to another lysosomal enzyme, β-galactosidase (bGal)44 (Supplementary Figure S2c). Absence of α- and β-subunits were confirmed by western blotting using a previously described rabbit polyclonal Ab against Hex A/B44 (Supplementary Figure S2b). HexA exon 11 and HexB exon 1 and surrounding introns from HEKHexABKO lines were amplified by PCR using primers shown in Supplementary Table S2. Resulting amplification products were either directly sequenced using the same primers as were used for PCR or in the case of HexB exon 1, the product was first cloned into PCR2.1 TOPO TA (Life Technologies) prior to sequencing (ACGT, Toronto, Canada). Characterization of HexA exon 1 and sequencing of HexA exon 11 were performed at TCAG (SickKids, Toronto, Canada, we are grateful to Dr. Peter Ray and Carol-Ann Ryan for their help). Overlapping sequences in the case of HexB exon 1 were deconvolved using PolyPeak Parser (http://yosttools.genetics.utah.edu/PolyPeakParser/).
Generation of stable HEKHexABKO line expressing His-tagged HexM
To generate HexM tagged at the C-terminus with poly Histidine (His6), the HexM cDNA was amplified with the following: Forward pigBHexHybAttFor2 (TACAAAAAAGCAGGCTCCaaagccaccatgacctcttctagactgtgg) and Reverse primers pigBHxHybHs6AttRev2 (ACAAGAAAGCTGGGTCTCAATGATGATGATGATGATGTCTACCCTCGATagtctgctcaaactcctggttg). The amplified product was cloned into gateway entry vector (pENTR201) using the BP reaction followed by transfer to the piggybac vector PB-T-RfA45 using the LR reaction, according to the manufacturers protocol (Life Technologies). Positive clones (PB-T-HexMHis) were verified by DNA sequencing of the HexM cDNA. The plasmid PB-T-HexMHis contained the His6 tagged HexM cDNA under the control of a tertracycline inducible promotor and flanked with the piggybac 5’ and 3’ terminal repeats, and incorporated the puromycin selectable marker. The HEKHexABKO line (ca. 250,000 cells in a six-well tissue culture plate) was co-transfected with PB-T-HexMHis, pBASE (coding for the piggybac transposase enabling transposon mediated genomic integration) and PB-RB (coding the reverse tetracycline transactivator) at an 8:1:1 weight ratio using lipofectamine LTX transfection reagent (Life Technologies). Followings, 3 days unselected growth, stable tranfectants were selected with Puromycin (2 µg/ml) and Blasticidin S (2 µg/ml), followed by single cell cloning in 96-well tissue culture plates. After 2 weeks growth, expression was induced following addition of 1 µmol/l tetracycline and 100 mmol/l NH4Cl to facilitate secretion of HexM. Positive clones were identified by the presence of HexM activity in their media monitored with the MUG substrate (1.6 mmol/l).
In the experiments aimed at confirming that secreted HexM could be recaptured by deficient cells (adult Tay-Sachs cells), the expression levels of HexM from transfected HEKHexABKO cells were high enough that the NH4Cl (used to promote the secretion of lysosomal enzymes) could be omitted during the generation of the “conditioned” medium. This eliminated the usual dialysis step needed to remove the NH4Cl from the conditioned medium before using it to replace the growth media, ± 5 mmol/l M6P, of the adult TSD fibroblasts for the 48 hours HexM uptake experiments.
Chemicals, Hex enzyme, and heat stability assays
Sources of the synthetic fluorogenic substrates, MUG and MUGS (Toronto Research Chemicals, Toronto, Canada), and their methods of use in the associated Hex isozyme assays, and the components that make-up the in cellulo and in vitro NBD-GM2 assays have been previously described.22 Kinetic analyses of the Hex isozymes using MUG or MUGS were performed as previously described.7 The method of evaluating the heat stability of the purified Hex isozymes by differential scanning fluorimetry using NanoOrange has also been previously described.29
Cell lines and tissue culture
An adult Tay-Sachs cell line (17662), homozygous for a αG269S missense mutation in exon 7 of HEXA,35 was used to demonstrate that secreted HexM could be recaptured in a MPR-dependent manner. HEK cells were obtained from The Hospital for Sick Children (Toronto, Canada) tissue culture facility. All cells were grown in α-minimal essential medium from Wisent (Canada) in the presence of 1% antibiotics (penicillin and streptomycin, Wisent, Montreal, Canada) and supplemented with 10% fetal bovine serum (FBS) (Wisent). The cell lines were grown at 37 °C in a humidified atmosphere with 5% CO2.
SDS-PAGE and western blots
Western blotting lysates (20 µg WT HEK or 40 µg HEKHexABKO of total protein) were subjected to SDS-PAGE on a 10% bis:acrylamide gel and transferred to nitrocellulose (Bio-Rad, Hercules, CA), processed and developed as previously.22,35 Silver (protein) staining of the SDS-Page gel was preformed as reported by Wray et al.46
Virus and plasmid preparation
Recombinant AAV vectors were generated as described47 using proprietary methods developed at the UNC Gene Therapy Center Vector Core facility (Chapel Hill, NC). The following scAAV9 or 9.47 vectors were produced and used for this study: AAV9-GFP, AAV9-HEXA (human), AAV9-HEXM; AAV9-HEXBMM. The HEXA, HEXM, and HEXB(MM) transgenes were codon-optimized for optimal protein expression in mammalian cells, using DNA2.0 (Menlo Park, CA). In order to provide similar transgene expression and allow scAAV packaging, all vectors used the same SV40 polyA and a proprietary, short synthetic promoter–intron regulatory sequence, provided by J. Keimel and M. Kaytor (New Hope Research Foundation, North Oaks, MN).
Experimental TSD and SD mouse models
The Tay-Sachs disease mouse (strain: B6;129S-hexa(-/-)) model developed by disruption of the hexa gene48,49 was obtained from The Jackson Laboratory and maintained in a 12-hour light–dark cycle with free access to food and water. All care and procedures were in accordance with the Guide for the Care and Use of Laboratory Animals and received prior approval by the University of North Carolina Institutional Animal Care and Usage Committee. The different AAV vectors were injected stereotaxically into the brain parenchyma of 4-month-old TSD hexa knock-out mice as previously described.50 The injections were placed between the cortex and striatum, using the following stereotaxic coordinates relative to bregma: rostral/caudal: +0.5 mm (toward rostral), left/right: left +3 mm, up/down: −4 mm. Two mice were injected with each vector combination. Each injection used a volume of 1 µl at a rate of 0.1 µl/minute, using a mixture containing 1 × 109 vg of Hex vector and 1 × 109 vg of GFP vector. After injection, the mice were allowed to recover, and at 4 weeks postinjection, the mice were sacrificed and transcardially perfused with PBS containing 1 U/ml heparin (Abraxis Pharmaceutic Products, Shaumburg, IL).
The SD mouse model (strain B6;129S4-Hexbtm1Rlp/J, hexb knock-out) was obtained from the Jackson Laboratory (Bar Harbor, ME). All animal experiments were carried out under protocols approved by the University Animal Care Committee at Queen’s University, Kingston, ON, Canada under Canadian Council on Animal Care regulations. The neonatal mice were intravenously injected with a AAV9-HEXM vector at day 0–1. They were sacrificed at 8 weeks and their mid-brains were collected for determining Hex activity as previously described51 and for GM2 analysis (described below).
Immuno-histochemical analysis (TSD mice)
The brains collected from injected TSD mice were fixed in 4% paraformaldehyde in PBS for 48 hours and then sectioned at 40 microns using a Leica vibrating microtome. One set of sections from the rostral half of the brain was immunostained for GFP, and another set was immunostained for GM2 gangliosides as described.48 Additionally, sections were treated with hydrogen peroxide to remove endogenous peroxidase activity prior to blocking. The primary antibodies were as follows: rabbit anti-GFP (1:1,000; Millipore, Billerica, MA; AB3080) or anti-GM2 (1:1,000; gift from Kyowa Hakko Kirin, Tokyo, Japan; KM966). Brain sections were digitized using a Scan-Scope slide scanner (Aperio Technologies, Vista, CA). Virtual slides were viewed using ImageScope software package (v. 10.0; Aperio Technologies). The Hex activity was assessed by the clearance of GM2 within the injected region compared to the contralateral brain hemisphere. The area of AAV transduction was visualized by staining for GFP.
GM2 ganglioside analysis (SD mice)
Gangliosides were extracted using modification of a method described earlier.52 Briefly, murine mid-brain sections were weighed and sonicated in 0.5 ml methanol at amplitude 15–20% for 3 × 10 second pulses with cooling on ice between pulses (Sonic Dismembrator Model 500, Fisher Scientific, Pittsburgh, PA). Samples were centrifuged for 15 minutes at 10,000 RPM at 4 °C and 0.4 ml of supernatant was removed for enzyme activity studies, the remainder was suspended (including pellet) up to a final volume of 1.5 ml methanol added to 2.5 ml chloroform. Acidic gangliosides were isolated using C18-E columns (Phenomenex, Torrence, CA) and eluted sequentially in 2 ml methanol, then in 2 ml 1:1 methanol: Chloroform mixture. The eluate was then evaporated under a stream of nitrogen, and the residue is resuspended in 1:1 methanol:chloroform and then applied to HPTLC Silica Gel 60 plate (Fisher Scientific) 10 µl at a time with drying between applications. Plates were developed with running buffer (chloroform:methanol:CaCl2 0.22%—55:45:10). After staining with orcinol (Sigma-Aldrich, St Louis, MO), and baking at 105 °C for 10 minutes, plates were immediately scanned for Spot Densitometric analysis (Image J software) using a mix of monosialoganglioside standards (Matreya LLC, Pleasant Gap, PA). The ganglioside ratios were presented as GM2:GD1a.
Acknowledgments
This research was funded by the New Hope Research Foundation http://newhoperesearch.org (J.S.W., S.J.G., and D.M.) and donations from the Uger Estate (M.B.T. and D.M.). Indirect support for S.J.G. was provided by Research to Prevent Blindness to the UNC Department of Ophthalmology. B.L.M. is supported by a Manitoba Research Chair (Research Manitoba). The authors thank Kyowa Hakko Kirin Co. for providing the KM966 antibody. B.L.M. and D.M. designed the primary structure of HexM, coordinated the overall project, and produced the first draft of the manuscript. M.B.T. produced the double K/O HEKs and with D.M. coordinated in vitro and in cellulo assays performed by S.Y. S.Y. also performed all the protein and kinetic analyses. The AAV vector constructs were generated by S.J.G. and AAV vectors were produced by S.K.-M. and S.J.G. The work on the TSD mice was done by S.K.-M. and directed by S.J.G. The work on the SD mice was accomplished by P.T. and directed by J.S.W. W.W. synthesized the NBD-GM2 used in the in cellulo assays. M.B.T., J.S.W., S.J.G., S.K.-M., B.L.M., and D.M. participated in writing the manuscript.
Footnotes
S.J.G. has received patent royalties from Asklepios Biopharma that are not directly related to these studies. A patent application has been filed on the sequence and potential uses of HexM, as well as the synthetic promoter used in these AAV vectors. The other authors declare no conflict of interest.
References
- Leinekugel, P, Michel, S, Conzelmann, E and Sandhoff, K (1992). Quantitative correlation between the residual activity of beta-hexosaminidase A and arylsulfatase A and the severity of the resulting lysosomal storage disease. Hum Genet 88: 513–523. [DOI] [PubMed] [Google Scholar]
- Mark, BL, Mahuran, DJ, Cherney, MM, Zhao, D, Knapp, S and James, MN (2003). Crystal structure of human beta-hexosaminidase B: understanding the molecular basis of Sandhoff and Tay-Sachs disease. J Mol Biol 327: 1093–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux, MJ, Mark, BL, Cherney, MM, Withers, SG, Mahuran, DJ and James, MN (2006). Crystallographic structure of human beta-hexosaminidase A: interpretation of Tay-Sachs mutations and loss of GM2 ganglioside hydrolysis. J Mol Biol 359: 913–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, T, Strater, N, Schuette, CG, Klingenstein, R, Sandhoff, K and Saenger, W (2003). The X-ray crystal structure of human beta-hexosaminidase B provides new insights into Sandhoff disease. J Mol Biol 328: 669–681. [DOI] [PubMed] [Google Scholar]
- Kresse, H, Fuchs, W, Glössl, J, Holtfrerich, D and Gilberg, W (1981). Liberation of N-acetylglucosamine-6-sulfate by human beta-N-acetylhexosaminidase A. J Biol Chem 256: 12926–12932. [PubMed] [Google Scholar]
- Kytzia, HJ and Sandhoff, K (1985). Evidence for two different active sites on human beta-hexosaminidase A. Interaction of GM2 activator protein with beta-hexosaminidase A. J Biol Chem 260: 7568–7572. [PubMed] [Google Scholar]
- Sharma, R, Deng, H, Leung, A and Mahuran, D (2001). Identification of the 6-sulfate binding site unique to alpha-subunit-containing isozymes of human beta-hexosaminidase. Biochemistry 40: 5440–5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, R, Bukovac, S, Callahan, J and Mahuran, D (2003). A single site in human beta-hexosaminidase A binds both 6-sulfate-groups on hexosamines and the sialic acid moiety of GM2 ganglioside. Biochim Biophys Acta 1637: 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, EM, Schwarzmann, G, Fürst, W and Sandhoff, K (1991). The human GM2 activator protein. A substrate specific cofactor of beta-hexosaminidase A. J Biol Chem 266: 1879–1887. [PubMed] [Google Scholar]
- Mahuran, DJ (1998). The GM2 activator protein, its roles as a co-factor in GM2 hydrolysis and as a general glycolipid transport protein. Biochim Biophys Acta 1393: 1–18. [DOI] [PubMed] [Google Scholar]
- Mahuran, DJ (1999). Biochemical consequences of mutations causing the GM2 gangliosidoses. Biochim Biophys Acta 1455: 105–138. [DOI] [PubMed] [Google Scholar]
- Bradbury, AM, Gray-Edwards, HL, Shirley, JL, McCurdy, VJ, Colaco, AN, Randle, AN et al. (2015). Biomarkers for disease progression and AAV therapeutic efficacy in feline Sandhoff disease. Exp Neurol 263: 102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachón-González, MB, Wang, SZ, Lynch, A, Ziegler, R, Cheng, SH and Cox, TM (2006). Effective gene therapy in an authentic model of Tay-Sachs-related diseases. Proc Natl Acad Sci USA 103: 10373–10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachón-González, MB, Wang, SZ, McNair, R, Bradley, J, Lunn, D, Ziegler, R et al. (2012). Gene transfer corrects acute GM2 gangliosidosis–potential therapeutic contribution of perivascular enzyme flow. Mol Ther 20: 1489–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachón-González, MB, Wang, SZ, Ziegler, R, Cheng, SH and Cox, TM (2014). Reversibility of neuropathology in Tay-Sachs-related diseases. Hum Mol Genet 23: 730–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti, JE, Mignon, A, Haase, G, Caillaud, C, McDonell, N, Kahn, A et al. (1999). Adenoviral gene therapy of the Tay-Sachs disease in hexosaminidase A-deficient knock-out mice. Hum Mol Genet 8: 831–838. [DOI] [PubMed] [Google Scholar]
- Sargeant, TJ, Wang, S, Bradley, J, Smith, NJ, Raha, AA, McNair, R et al. (2011). Adeno-associated virus-mediated expression of β-hexosaminidase prevents neuronal loss in the Sandhoff mouse brain. Hum Mol Genet 20: 4371–4380. [DOI] [PubMed] [Google Scholar]
- Gray, SJ (2013). Gene therapy and neurodevelopmental disorders. Neuropharmacology 68: 136–142. [DOI] [PubMed] [Google Scholar]
- Gray, SJ, Matagne, V, Bachaboina, L, Yadav, S, Ojeda, SR and Samulski, RJ (2011). Preclinical differences of intravascular AAV9 delivery to neurons and glia: a comparative study of adult mice and nonhuman primates. Mol Ther 19: 1058–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust, KD, Nurre, E, Montgomery, CL, Hernandez, A, Chan, CM and Kaspar, BK (2009). Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol 27: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaranch, L, Salegio, EA, San Sebastian, W, Kells, AP, Foust, KD, Bringas, JR et al. (2012). Adeno-associated virus serotype 9 transduction in the central nervous system of nonhuman primates. Hum Gene Ther 23: 382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinici, I, Yonekawa, S, Tkachyova, I, Gray, SJ, Samulski, RJ, Wakarchuk, W et al. (2013). In cellulo examination of a beta-alpha hybrid construct of beta-hexosaminidase A subunits, reported to interact with the GM2 activator protein and hydrolyze GM2 ganglioside. PLoS One 8: e57908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka, K, Tamura, T, Tsuji, D, Dohzono, Y, Kitakaze, K, Ohno, K et al. (2011). Therapeutic potential of intracerebroventricular replacement of modified human β-hexosaminidase B for GM2 gangliosidosis. Mol Ther 19: 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P, Yang, L, Esvelt, KM, Aach, J, Guell, M, DiCarlo, JE et al. (2013). RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley, MS, Whitworth, GE, Debowski, AW, Chin, D and Vocadlo, DJ (2005). O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J Biol Chem 280: 25313–25322. [DOI] [PubMed] [Google Scholar]
- Tropak, MB, Bukovac, SW, Rigat, BA, Yonekawa, S, Wakarchuk, W and Mahuran, DJ (2010). A sensitive fluorescence-based assay for monitoring GM2 ganglioside hydrolysis in live patient cells and their lysates. Glycobiology 20: 356–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y, Vocadlo, DJ, Leung, A, Withers, SG and Mahuran, D (2001). Characterization of the Glu and Asp residues in the active site of human beta-hexosaminidase B. Biochemistry 40: 2201–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasilik, A and Neufeld, EF (1980). Biosynthesis of lysosomal enzymes in fibroblasts. Synthesis as precursors of higher molecular weight. J Biol Chem 255: 4937–4945. [PubMed] [Google Scholar]
- Kornhaber, GJ, Tropak, MB, Maegawa, GH, Tuske, SJ, Coales, SJ, Mahuran, DJ et al. (2008). Isofagomine induced stabilization of glucocerebrosidase. Chembiochem 9: 2643–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard-Borger, ED, Tropak, MB, Yonekawa, S, Tysoe, C, Mahuran, DJ and Withers, SG (2012). Rapid assembly of a library of lipophilic iminosugars via the thiol-ene reaction yields promising pharmacological chaperones for the treatment of Gaucher disease. J Med Chem 55: 2737–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahuran, D and Lowden, JA (1980). The subunit and polypeptide structure of hexosaminidases from human placenta. Can J Biochem 58: 287–294. [DOI] [PubMed] [Google Scholar]
- Hasilik, A and Neufeld, EF (1980). Biosynthesis of lysosomal enzymes in fibroblasts. Phosphorylation of mannose residues. J Biol Chem 255: 4946–4950. [PubMed] [Google Scholar]
- Natowicz, MR, Chi, MM, Lowry, OH and Sly, WS (1979). Enzymatic identification of mannose 6-phosphate on the recognition marker for receptor-mediated pinocytosis of beta-glucuronidase by human fibroblasts. Proc Natl Acad Sci USA 76: 4322–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyttälä, A, Heinonen, O, Peltonen, L and Jalanko, A (1998). Expression and endocytosis of lysosomal aspartylglucosaminidase in mouse primary neurons. J Neurosci 18: 7750–7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropak, MB, Reid, SP, Guiral, M, Withers, SG and Mahuran, D (2004). Pharmacological enhancement of beta-hexosaminidase activity in fibroblasts from adult Tay-Sachs and Sandhoff Patients. J Biol Chem 279: 13478–13487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cearley, CN and Wolfe, JH (2006). Transduction characteristics of adeno-associated virus vectors expressing cap serotypes 7, 8, 9, and Rh10 in the mouse brain. Mol Ther 13: 528–537. [DOI] [PubMed] [Google Scholar]
- Sango, K, Yamanaka, S, Hoffmann, A, Okuda, Y, Grinberg, A, Westphal, H et al. (1995). Mouse models of Tay-Sachs and Sandhoff diseases differ in neurologic phenotype and ganglioside metabolism. Nat Genet 11: 170–176. [DOI] [PubMed] [Google Scholar]
- Liu, Y, Hoffmann, A, Grinberg, A, Westphal, H, McDonald, MP, Miller, KM et al. (1997). Mouse model of GM2 activator deficiency manifests cerebellar pathology and motor impairment. Proc Natl Acad Sci USA 94: 8138–8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuziuk, JA, Bertoni, C, Beccari, T, Orlacchio, A, Wu, YY, Li, SC et al. (1998). Specificity of mouse GM2 activator protein and beta-N-acetylhexosaminidases A and B. Similarities and differences with their human counterparts in the catabolism of GM2. J Biol Chem 273: 66–72. [DOI] [PubMed] [Google Scholar]
- Sorrentino, NC, D’Orsi, L, Sambri, I, Nusco, E, Monaco, C, Spampanato, C et al. (2013). A highly secreted sulphamidase engineered to cross the blood-brain barrier corrects brain lesions of mice with mucopolysaccharidoses type IIIA. EMBO Mol Med 5: 675–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, PA, Zeng, BJ, Porter, BF, Alroy, J, Horak, F, Horak, J et al. (2010). Tay-Sachs disease in Jacob sheep. Mol Genet Metab 101: 357–363. [DOI] [PubMed] [Google Scholar]
- Krissinel, E and Henrick, K (2007). Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797. [DOI] [PubMed] [Google Scholar]
- Mali, P, Esvelt, KM and Church, GM (2013). Cas9 as a versatile tool for engineering biology. Nat Methods 10: 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa, GH, Tropak, M, Buttner, J, Stockley, T, Kok, F, Clarke, JT et al. (2007). Pyrimethamine as a potential pharmacological chaperone for late-onset forms of GM2 gangliosidosis. J Biol Chem 282: 9150–9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z, Michael, IP, Zhou, D, Nagy, A and Rini, JM (2013). Simple piggyBac transposon-based mammalian cell expression system for inducible protein production. Proc Natl Acad Sci USA 110: 5004–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray, W, Boulikas, T, Wray, VP and Hancock, R (1981). Silver staining of proteins in polyacrylamide gels. Anal Biochem 118: 197–203. [DOI] [PubMed] [Google Scholar]
- Gray, SJ, Nagabhushan Kalburgi, S, McCown, TJ and Jude Samulski, R (2013). Global CNS gene delivery and evasion of anti-AAV-neutralizing antibodies by intrathecal AAV administration in non-human primates. Gene Ther 20: 450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phaneuf, D, Wakamatsu, N, Huang, JQ, Borowski, A, Peterson, AC, Fortunato, SR et al. (1996). Dramatically different phenotypes in mouse models of human Tay-Sachs and Sandhoff diseases. Hum Mol Genet 5: 1–14. [DOI] [PubMed] [Google Scholar]
- Taniike, M, Yamanaka, S, Proia, RL, Langaman, C, Bone-Turrentine, T and Suzuki, K (1995). Neuropathology of mice with targeted disruption of Hexa gene, a model of Tay-Sachs disease. Acta Neuropathol 89: 296–304. [DOI] [PubMed] [Google Scholar]
- Gray, SJ, Choi, VW, Asokan, A, Haberman, RA, McCown, TJ and Samulski, RJ (2011). Production of recombinant adeno-associated viral vectors and use in in vitro and in vivo administration. Curr Protoc Neurosci Chapter 4: Unit 4.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia, JS, Altaleb, N, Bello, A, Kruck, C, LaFave, MC, Varshney, GK et al. (2015). Long-term correction of Sandhoff disease following intravenous delivery of rAAV9 to mouse neonates. Mol Ther 23: 414–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyrantepe, V, Lema, P, Caqueret, A, Dridi, L, Bel Hadj, S, Carpentier, S et al. (2010). Mice doubly-deficient in lysosomal hexosaminidase A and neuraminidase 4 show epileptic crises and rapid neuronal loss. PLoS Genet 6: e1001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.