Abstract
Aim:
To determine the genetic basis and types of beta-lactamase encountered among enterobacterial isolates of wild pets from the animal exhibit.
Materials and Methods:
A total of 17 beta-lactamase-producing enterobacteria recovered from fecal samples of wild pet animals were analyzed for a selected beta-lactamase gene by polymerase chain reaction.
Results:
Molecular analysis identified one or more β-lactamase-encoding genes in 14 enterobacterial isolates as a single or gene combination. The most frequent extended-spectrum β-lactamases types were TEM and CTX-M, and the most common AmpC enzymes were CMY-2 and DHA types.
Conclusions:
The study is the first in Saudi Arabia, have established the presence of β-lactamase-encoding genes in the fecal isolates of wild pets.
Keywords: animal exhibit, extended-spectrum β-lactamases/AmpC beta-lactamase, fecal samples, polymerase chain reaction, Saudi Arabia
Introduction
Antibiotic-resistant bacteria are extremely important to human health. The production of ß-lactamases is the major mechanism of bacterial resistance to β-lactam antibiotics which are considered the most widely used class of antibiotics against both Gram-negative and Gram-positive bacteria. Resistance to this class of antimicrobial agents is therefore of immense clinical significance.
A major reason for resistance of Enterobacteriaceae to beta-lactam antibiotics is the production of extended-spectrum β-lactamases (ESBLs) and AmpC beta-lactamases, capable of inactivating the effects of broad-spectrum cephalosporins and penicillins [1]. Exposure to ESBL/AmpC-producing microorganisms can occur through any means, but the hospital has always been thought to be the greatest risk [2]. The occurrence of ESBL/AmpC-producing microorganisms is on the rise globally, with prevalence varying from country to country and within a country from institution to institution [3]. The genes that encode for these enzymes may be plasmid-borne or chromosomally located.
Wild animals provide a biological mechanism for the spread of antibiotic resistance genes [4]. Recently, a number of studies describing the occurrence of ESBL-resistant Escherichia coli in wildlife [5-14].
Data from the Arabian Peninsula, including Saudi Arabia, suggested that extended-spectrum and AmpC beta-lactam-resistant bacteria constitute a major problem in nosocomial and community-acquired infections [15,16]. However, there is scarce information on the occurrence and genetic characteristics of β-lactamase-producing bacteria in wild pet animals. Therefore, this study was carried to investigate the occurrence and distribution of beta-lactamase encoding genes within enterobacteria derived from wild pet animals in Saudi Arabia.
Materials and Methods
Ethical approval
The fecal samples were collected aseptically with adequate precautionary measures to minimize pain and/or discomfort to the animals and carried out in accordance with the Saudi animal welfare laws.
Bacterial strains
A total of 17 positive ESBL/AmpC enterobacterial isolates recovered from 75 fecal samples of wild animals at pet market, Taif, Western Saudi Arabia (5 rock hyrax, 4 Yemen Linnet, 3 common kestrel, 3 red foxes, 3 long-tailed finches, 2 caracal, 2 peacock, 1 rock dove, 1 hamadryas baboon, 1 orange-winged parrot, 1 Burmese python, 1 Hill Mynah, 1 African gray parrot, 1 common myna) were included. Wild animals are caught or bought for pet, shops, local breeder or traded (sometimes illegally). The enterobacterial isolates were 9 E. coli, and single isolates of Klebsiella pneumonia, Klebsiella oxytoca, Proteus mirabilis, Proteus vulgaris, Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii, and Citrobacter youngae. Isolates were identified and confirmed by commercially available biochemical test (API tests; bioMérieux). The ESBLs and AmpC beta-lactamase production were achieved by commercially available Etest (bioMérieux).
Molecular investigation
Rapid DNA preparation was performed by a boiling technique that includes heating at boiling of an overnight bacterial culture (200 µl) mixed with 800 µl of distilled water, followed by cooling, centrifugation and the supernatant was used as the DNA template for the polymerase chain reaction (PCR).
The presence of genes encoding TEM, SHV, OXA, CTX-M, CMY-2, and DHA type β-lactamases was studied by multiplex PCR using universal primers and conditions previously reported [17,18]. The PCR was conducted in a Thermal Cycler PXE-0.5 (THERMO; Electron Corporation) and the resulting PCR products were subjected to electrophoretic separation in 1.5% agarose gel. Visualization of amplicons was completed by staining with ethidium bromide (Sigma-Aldrich) (1 µg/ml) under UV transluminator and photographed. DNA bands of each amplicon were compared with 100-bp DNA mass marker (Figure-1a-c). Primers sequence and PCR condition are presented in Table-1.
Figure-1.
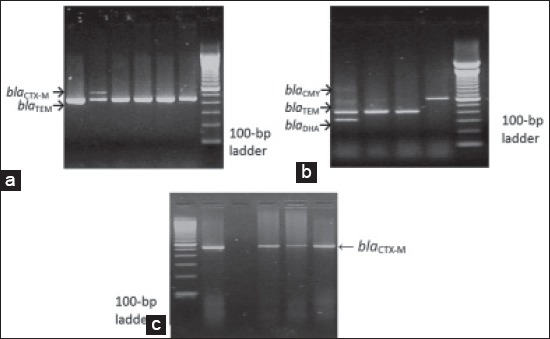
(a) The result of the multiplex polymerase chain reaction (PCR) amplification of the DNA target gene loci of 593-bp fragment DNA region coding for CTX-M; 431-bp fragment DNA region coding for TEM, (b) the result of the multiplex PCR amplification of the DNA target gene loci of: 695-bp fragment DNA region coding for CMY-2; 431-bp fragment DNA region coding for TEM; 314-bp fragment DNA region coding for DHA, (c) the result of the multiplex PCR amplification of the DNA target gene loci of 593-bp fragment DNA region coding for CTX-M.
Table-1.
Primers used in this study to detect betalactamase (bla) genes.
| Primer target | Primer name | Sequence (5’-3’) | Annealing temperature | Product size (bp) | Reference |
|---|---|---|---|---|---|
| TEM (blaTEM) | TEM-F TEM-R |
AGTGCTGCCATAACCATGAGTG CTGACTCCCC GTCGTGTAGATA |
61°C for 1 min | 431 | [18] |
| SHV (blaSHV) | SHV-F SHV-R |
GATGAACGCTTTCCCATGATG CGCTGTTATCGCTCATGGTAA |
61°C for 1 min | 214 | [18] |
| CTX-M (blaCTX-M) | CTX-M-F CTX-M-R |
ATGTGCAGYACCAGTAARGTKATGGC TGGGTRAARTARGTSACCAGAAYCAGCGG |
61°C for 1 min | 593 | [17] |
| OXA (blaOXA) | OXA-F OXA-R |
ACACAATACATATCAACTTCGC AGTGTGTTTAGAATGGTGATC |
61°C for 1 min | 813 | [17] |
| PampC (blaCMY-2) | CMY-F2 CMY-R2 |
AGCGATCCGGTCACGAAATA CCCGTTTTATGCACCCATGA |
61°C for 1 min | 695 | [18] |
| PampC (blaDHA) | DHA (F) DHA (R) |
GTGGTGGACAGCACCATTAAA CCTGCGGTATAGGTAGCCAGAT |
61°C for 1 min | 314 | [18] |
Results
PCR detection of ß-lactamase encoding genes
A total of 17 beta-lactamase positive enterobacterial strains recovered from the feces of wild pet animals were screened for beta-lactamase (bla)-encoding genes. The PCR screening identified the presence of the beta-lactamase genes encoding TEM, CTX-M, CMY-2, and DHA in 14 of them (Figure-1a-c). None of the isolates were reacted positively for blaOXA and blaSHV. No beta-lactamase genes were identified in the remaining three isolates.
Overall, variety of beta-lactamase genes were found within nine bacterial species isolated from various wild pets species. TEM enzyme was detected in nine isolates of beta-lactamase-producing, respectively, which included 4 isolates of E. coli and single isolate of E. aerogenes, P. mirabilis, C. youngae, and P. vulgaris (Table-2). The CTX-M enzyme was identified in five strains among of beta-lactamase-producing isolates, as a single isolate of E. coli, K. pneumonia, E. cloacae, K. oxytoca and C. freundii (Table-2). Both of CMY-2 and DHA, a plasmid-mediated AmpC beta-lactamases were detected in two different isolate of E. coli (Table-2).
Table-2.
Prevalence and multiplicity of β-Lactamase genes among ESBLs- positive fecal bacteria derived from wild pet animals in Saudi Arabia.
| Bacterial species | ESBL positive no | β-Lactamase- associated genes | |||||
|---|---|---|---|---|---|---|---|
| TEM | CTX-M | CMY-2 | CTX-M, TEM | TEM, DHA | Total | ||
| E. coli | 9 | 3 | 1 | 1 | - | 1 | 6 |
| K. pneumonia | 1 | - | - | - | 1 | - | 1 |
| P. mirabilis | 1 | 1 | - | - | - | - | 1 |
| E. cloacae | 1 | - | 1 | - | - | - | 1 |
| K. oxytoca | 1 | - | 1 | - | - | - | 1 |
| C. youngae | 1 | 1 | - | - | - | - | 1 |
| C. freundii | 1 | - | 1 | - | - | - | 1 |
| P. vulgaris | 1 | 1 | - | - | - | - | 1 |
| E. aerogenes | 1 | 1 | - | - | - | - | 1 |
| Total | 17 | 7 | 4 | 1 | 1 | 1 | 14 |
E. aerogenes=Enterobacter aerogenes, K. pneumonia=Klebsiella pneumonia, K. oxytoca=Klebsiella oxytoca, P. mirabilis=Proteus mirabilis, P. vulgaris=Proteus vulgaris, E. cloacae=Enterobacter cloacae, C. freundii=Citrobacter freundii, C. youngae=Citrobacter youngae, TEM=Temoneira, DHA=Dhahran, CTX-M=Cefotaxime – Munich, CMY=Cephamycinase, SHV=Sulfhydryl Variable, ESBL=Extended spectrum β-Lactamase
Distribution of bla genes
The ß-lactamase-producing isolates were distributed into two categories, the first harbored only one type of ß-lactamase encoding gene, the second harbored two types (Table-2). Twelve (12/17) of the total beta-lactamase-producing entrerobacteria were harboring only one beta-lactamase encoding gene, including five strains of E. coli and a single isolate of E. cloacae, K. oxytoca, C. youngae, P. vulgaris, C. freundii, P. mirabilis and E. aerogenes.
The blaTEM, a narrow-spectrum ß-lactamase was detected alone in 7 isolates; E. coli (3 isolates) and a single isolate of C. youngae, P. vulgaris, P. mirabilis and E. aerogenes. The blaCTX-M, an extended-spectrum ß-lactamase was detected alone in four isolates; single isolate of K. oxytoca from Yemen linnet feces, E. coli from common kestrel, E. cloacae from rock dove, and C. freundii from African gray parrot (Table-3). The plasmid-mediated ß-lactamases, blaCMY-2 and blaDHA were detected in two different E. coli isolates recovered from Arabian red fox and Hill Mynah, respectively.
Table-3.
Genotypic characteristics and occurrence of β-lactamases encoding genes in enterobacteria from wild pet animals.
| Isolate ID | Bacteria (no) | Animal species (scientific name) | bla gene | Betalactam resistance phenotype |
|---|---|---|---|---|
| RH-1 | E. coli (1) | Rock hyrax (Procavia capensis) | TEM | AMP, CEP |
| CK-3 | E. coli (1) | Common kestrel (Falco tinnuculus) | CTX-M | AMP, CEP, AZT, CXM, CTX, CAZ |
| HM-7 | E. coli (1) | Hill mynah (Gracula religosa) | TEM, DHA | AMC, AMP, CEP, AZT, CXM, CTX, CAZ, FOX |
| AF-17 | E. coli (1) | Arabian red fox (Vulpes vulpes) | CMY-2 | AMC, AMP, CEP, AZT, CXM, CTX, CAZ FEP, FOX |
| OP-22 | P. mirabilis (1) | Orange-winged Parrot (Amazona amazonica) | TEM | AMP, CEP |
| BP-19 | C. youngae (1) | Burmese python (Python molurus) | TEM | AMP, CEP |
| LF-27 | E. aerogene (1) | Long-tailed finches (Taeniopygia guttata) | TEM | AMP, CEP |
| RD-33 | E. cloacae (1) | Rock dove (Columba livia) | CTX-M | AMP, CEP, CXM, AZT, CAZ |
| PC-6 | P. vulgaris (1) | Peacock (Pavo cristatus) | TEM | AMP, CEP |
| BM-11 | K. pneumonia (1) | Baboon Monkey (Papio hamadryas) | TEM, CTX-M | AMP, CEP, CXM, AZT, CAZ |
| CA-31 | E. coli (1) | Caracal (Caracal caracal) | TEM | AMP, CEP |
| CM-29 | E. coli (1) | Common myna (Acridotheres tristis) | TEM | AMP, CEP |
| YL-8 | K. oxytoca (1) | Yemen linnet (Carduelis yemenensis) | CTX-M | AMP, CEP, AZT, CEF, CXM, CAZ |
| AP-13 | C. freundii (1) | African gray parrot (Psittacus erithacus) | CTX-M | AMP, CEP, AZT, CXM, CTX, CAZ |
E. aerogenes=Enterobacter aerogenes, K. pneumonia=Klebsiella pneumonia, K. oxytoca=Klebsiella oxytoca, P. mirabilis=Proteus mirabilis, P. vulgaris=Proteus vulgaris, E. cloacae=Enterobacter cloacae, C. freundii=Citrobacter freundii, C. youngae=Citrobacter youngae
A total of two (2/17) of the total beta-lactamase-producing isolates were harboring gene combinations of blaTEM and blaDHA in E. coli recovered from the feces of Hill Mynah and blaTEM and blaCTX-M in K. pneumonia delivered from the feces of baboon monkey.
Discussion
The resistance to beta-lactam and beta-lactamase inhibitors is of great clinical significance in several countries. Resistance to beta-lactam antibiotics is primarily mediated by beta-lactamases production. Many different β-lactamases have been described, but TEM, SHV, OXA, CMY-2, and CTX-M β-lactamases are currently regarded the most common among Enterobacteriaceae spp. [2].
Recently, many studies carried out in different countries describing the prevalence and characteristics of beta-lactamase gene harbored Enterobacteriaceae in wildlife free-living Canada geese in Georgia and North California [19], wild animals in Portugal [8,20], zoo animals in Japan [21], black-headed gulls in the Czech Republic [4] and wild birds and free-range poultry in Bangladesh [22]. Since there seem to be geographical variations in the occurrence of different ESBLs, we describe prevalence and characteristics of ESBL/AmpC-genotypes within enterobacterial isolates from wild pet animals presenting at live animal market in Taif, Western Saudi Arabia.
Prevalence of beta-lactamase genes
The beta-lactamase genes harboring enterobacterial isolates from wild pet animals were detected in 14 out of 17 isolates including six E. coli and single isolate of K. pneumonia, P. mirabilis, E. cloacae, K. oxytoca, C. youngae, C. freubdii, P. vulgaris, and E. aerogenes. The rate of bla genes in this study was consistent with that previously reported [8,20,21], whereas E. coli is the most prevalent and encountered bla genes among enterobacteria from wild animals.
Determination of the types of bla genes
In this study, PCR screening revealed detection of beta-lactamase encoding genes of TEM, CTX-M, CMY, and DHA. None of the isolates were positive for blaOXA and blaSHV. The remaining three isolates did not show any of the bla genes investigated. Similarly, previous studies also detected many β-lactamase-encoding genes in wild animals [12,14,20,21,22].
A TEM-β-lactamase is a narrow-spectrum beta-lactamase gene, which confers resistance against penicillin’s and first-generation cephalosporins [23]. In this study, blaTEM being detected in 7 isolates out of 17 enterobacteria-producing beta-lactamase as a sole mechanism of resistance to beta-lactams and all these isolates showed an ampicillin, cephalothin and or cefuroxime resistance phenotypes. TEM-β-lactamase has been previously detected in fecal isolates from magpies and wild rabbits from West Wales [24], free-living Canada geese in Georgia and North Carolina [19], wild animals in Portugal [20], Zoo animals in Japan [21], black-headed gulls in the Czech Republic [4], yellow-legged gulls in France [5], imported flamingos in Japan [25], gulls population in Sweden [12], migratory and resident population of rooks in Austria [26], seagulls and crows in Bangladesh [27].
Recently, there has been worldwide increase in the incidence of ESBLs [3]. In this study, blaCTX-M, an ESBL-encoding gene, was detected in five isolates of enterobacteria from feces of wild animals. The blaCTX-M has been previously identified in fecal bacteria from wild animals in Portugal [20], masked palm civet in Japan [21], imported flamingos in Japan [25], gulls in Sweden [12], migrating and resident population of rooks in Austria [26].
Furthermore, the plasmid-mediated AmpC genes (blaCMY-2 and blaDHA), were observed in two of strains of enterobacteria showed a typical AmpC-beta-lactamase resistance phenotype. The presence of AmpC β-lactamases have been found worldwide but are less common than ESBLs [28]. The information on the presence of AmpC producing Enterobacteriaceae in wildlife is scarce. Recently, the blaCMY has been reported previously from jaybird isolates of K. oxytoca in Japan [21], migrating and resident population of rooks in Austria [26]. The blaDHA was the first identified from clinical isolates of Salmonella enteritidis in Saudi Arabia [29]. Recently, in Magnolia, the blaDHA was detected in one E. coli from clinical sources [30].
Analysis of bla genes multiplicity among isolates
As in previous studies, bla-genes in this study were detected within enterobacteria from wild animals either as a single gene loci or as gene combination of two or more gene loci for beta-lactamases [13,21,26].
A comparative view of Arabian Gulf region and Saudi Arabia showed a high occurrence of ESBL-producing isolates harboring TEM, SHV, OXA, and CTX-M- β-lactamases from hospitals [16,31-34] and raw chicken [35].
Conclusions
It is of interest the detection of ESBL/AmpC-producing bacteria in wild animals at pet market. This is the first study, to our knowledge, of enterobacteria harboring β-lactamase genes in wild animals in Saudi Arabia. The fact that these animals often live in close contact with their owners and other people in market make the occurrence of transmission between them even more likely. More studies should be carried out in the future in order to track the variants and evolution of β-lactamase genes compared to those from human isolates.
Authors’ Contributions
SAH conceived, designed the study, drafted and revised the manuscript. MYS collected and analyzed samples. Both authors read and approved the final manuscript.
Acknowledgements
The authors are grateful for financial support of deanship of scientific research of Taif University, Saudi Arabia (project no. 1/435/3319).
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Bush K, Jacoby G.A, Medeiros A.A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 1995;39(6):1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford P.A. Extended spectrum β-lactamases in the 21st century:Characterization, epidemiology and the detection of this important resistance threat. Clin. Microbiol. Rev. 2001;14:933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paterson D.L, Bonomo R.A. Extended spectrum beta-lactamases:A clinical update. Clin. Microbiol. Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dolejska M, Cizek A, Literak I. High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from black-headed gulls in the Czech Republic. J. Appl. Microbiol. 2007;103:11–19. doi: 10.1111/j.1365-2672.2006.03241.x. [DOI] [PubMed] [Google Scholar]
- 5.Bonnedahl J, Drobni M, Gauthier-Clerc M, Hernandez J, Granholm S, Kayser Y, Melhus A, Kahlmeter G, Waldenstrom J, Johansson A, Olsen B. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the South of France. PLoS One. 2009;4:e5958. doi: 10.1371/journal.pone.0005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Literak I, Dolejska M, Janoszowska D, Hrusakova J, Meissner W, Rzyska H, Bzoma S, Cizek A. Antibiotic resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum beta-lactamase and QnrS, in water birds on the Baltic Sea Coast of Poland. Appl. Environ. Microbiol. 2010;76:8126–8134. doi: 10.1128/AEM.01446-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guenther S, Grobbel M, Beutlich J, Bethe A, Friedrich N.D, Goedecke A, Luebke-Becker A, Guerra B, Wieler L.H, Ewers C. CTX-M-15 type extended-spectrum beta-lactamases-producing Escherichia coli from wild birds in Germany. Environ. Microbiol. Rep. 2010;2:641–645. doi: 10.1111/j.1758-2229.2010.00148.x. [DOI] [PubMed] [Google Scholar]
- 8.Pinto L, Radhouani H, Coelho C, Martins da Costa P, Simoes R, Brandao R.M, Torres C, Igrejas G, Poeta P. Genetic detection of extended spectrum beta-lactamase-containing Escherichia coli isolates from birds of prey from Serra da Estrela Natural Reserve in Portugal. Appl. Environ. Microbiol. 2010;76:4118–4120. doi: 10.1128/AEM.02761-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radhouani H, Pinto L, Coelho C, Goncalves A, Sargo R, Torres C, Igrejas G, Poeta P. Detection of Escherichia coli harboring extended-spectrum beta-lactamases of the CTX-M classes in faecal samples of common buzzards (Buteo buteo) J. Antimicrob. Chemother. 2010;65:171–173. doi: 10.1093/jac/dkp403. [DOI] [PubMed] [Google Scholar]
- 10.Garmyn A, Haesebrouck F, Hellebuyck T, Smet A, Pasmans F, Butaye P, Martel A. Presence of extended spectrum beta-lactamase-producing Escherichia coli in wild geese. J. Antimicrob. Chemother. 2011;66:1643–1644. doi: 10.1093/jac/dkr148. [DOI] [PubMed] [Google Scholar]
- 11.Silva N, Igrejas G, Rodrigues P, Rodrigues T, Goncalves A, Felgar A.C, Pacheco R, Goncalves D, Cunha R, Poeta P. Molecular characterization of vancomycin-resistant Enterococci and extended-spectrum beta-lactamase-containing Escherichia coli isolates in wild birds from the Azores Archipelago. Avian Pathol. 2011;40:473–479. doi: 10.1080/03079457.2011.599061. [DOI] [PubMed] [Google Scholar]
- 12.Wallensten A, Hernandez J, Ardiles K, González-Acuña D, Drobni M, Olsen B. Extended spectrum beta-lactamases detected in Escherichia coli from gulls in Stockholm, Sweden Infect . Ecol. Epidemiol. 2011;1:7030. doi: 10.3402/iee.v1i0.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goncalves A, Igrejas G, Radhouani H, Estepa V, Alcaide E, Zorrilla I, Serra R, Torres C, Poeta P. Detection of extended-spectrum beta-lactamase-producing Escherichia coli isolates in faecal samples of Iberian lynx. Lett. Appl. Microbiol. 2012;54:73–77. doi: 10.1111/j.1472-765X.2011.03173.x. [DOI] [PubMed] [Google Scholar]
- 14.Radhouani H, Igrejas G, Goncalves A, Estepa V, Sargo R, Torres C, Poeta P. Molecular characterization of extended-spectrum beta lactamase-producing Escherichia coli isolated from red foxes in Portugal. Arch. Microbiol. 2013;195(2):141–144. doi: 10.1007/s00203-012-0853-7. [DOI] [PubMed] [Google Scholar]
- 15.Mokaddas E.M, Abdulla A.A, Shati S, Rotimi V.O. The technical aspects and clinical significance of detecting extended-spectrum beta-lactamase producing Enterobacteriaceae at a tertiary-care hospital in Kuwait. J. Chemother. 2008;20:445–451. doi: 10.1179/joc.2008.20.4.445. [DOI] [PubMed] [Google Scholar]
- 16.Al-Agamy M.H, Shibl A.M, Tawfic A.F. Prevalence and molecular characterization of extended-spectrum beta-lactamase producing Klebsiella pneumoniae in Riyadh, Saudi Arabia. Ann. Saudi Med. 2009;29:253–257. doi: 10.4103/0256-4947.55306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang H, Ataker F, Hedin G, Dornbusch K. Molecular epidemiology of extended-spectrum beta-lactamases among Escherichia coli isolates collected in a Swedish hospital and its associated health care facilities from 2001 to 2006. J Clin. Microbiol. 2008;46(2):707–712. doi: 10.1128/JCM.01943-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Jeon S, Rhie H, Lee B, Park M, Lee H, Lee J, Kim S. Rapid detection of extended spectrum β-Lactamase (ESBL) for Enterobacteriaceae by use of a multiplex PCR-based method. Infect. Chemother. 2009;41(3):181–184. [Google Scholar]
- 19.Cole D, Drum D.J, Stalknecht D.E, White D.G, Lee M.D, Ayers S, Sobsey M, Maurer J.J. Free-living Canada geese and antimicrobial resistance. Emerg. Infect. Dis. 2005;11:935–938. doi: 10.3201/eid1106.040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa D, Poeta P, Saenz Y, Vinue L, Rojo-Bezares B, Jouini A, Zarazaga M, Rodrigues J, Torres C. Detection of Escherichia coli harbouring extended-spectrum beta-lactamases of the CTX-M, TEM and SHV classes in faecal samples of wild animals in Portugal. J. Antimicrob. Chemother. 2006;58:1311–1312. doi: 10.1093/jac/dkl415. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed A.M, Motoi Y, Sato M, Maruyama A, Watanabe H, Fukumoto Y, Shimamoto T. Zoo animals as reservoirs of gram-negative bacteria harboring integrons and antimicrobial resistance genes. Appl. Environ. Microbiol. 2007;73:6686–6690. doi: 10.1128/AEM.01054-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasan B, Sandegren L, Melhus A, Drobni M, Hernandez J, Waldenstroem j, Alam M, Olsen B. Antimicrobial drug-resistant Escherichia coli in wild birds and free-range poultry, Bangladesh. Emerg. Infect. Dis. 2012;18(12):2055–2058. doi: 10.3201/eid1812.120513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livermore D.M, Woodford N. The β-lactamase threat in Enterobacteriaceae Pseudomonas and Acinetobacter. Trends Microbiol. 2006;14:413–420. doi: 10.1016/j.tim.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 24.Livermore D.M, Warner M, Hall L.M, Enne V.I, Projan S.J, Dunman P.M, Wooster S.L, Harrison G. Antibiotic resistance in bacteria from magpies (Pica pica) and rabbits (Oryctolagus cuniculus) from West Wales. Environ. Microbiol. 2001;3:658–661. doi: 10.1046/j.1462-2920.2001.00239.x. [DOI] [PubMed] [Google Scholar]
- 25.Sato M, Ahmed A.M, Noda A, Watanabe H, Fukumoto Y, Shimamoto T. Isolation and molecular characterization of multidrug-resistant Gram-negative bacteria from imported flamingos in Japan. Acta Vet. Scand. 2009;51:46–50. doi: 10.1186/1751-0147-51-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loncaric I, Stalder G.L, Mehinagic K, Rosengarten R, Hoelzl F, Knauer F, Walzer C. Comparison of ESBL –and AmpC producing Enterobacteriaceae and methicillin-resistant Staphylococcus aureus (MRSA) isolated from migratory and resident population of rooks (Corvus frugilegus) in Austria. PLoS ONE. 2013;8(12):e84048. doi: 10.1371/journal.pone.0084048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasan B. Antimicrobial resistance and production of extended spectrum beta-lactamases in Enterobacteriaceae from birds in Bangladesh. Uppsala: Acta Universitatis Upsaliensis; 2013. pp. 911–975. [Google Scholar]
- 28.Jacoby G.A. AmpC beta-lactamases. Clin. Microbiol. Rev. 2009;22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnaud G, Arlet G, Verdet C, Gaillot O, Lagrange P.H, Philippon A. Salmonella enteritidis: AmpC plasmid-mediated inducible-lactamase (DHA-1) with an ampR gene from Morganella morganii . Antimicrob. Agents Chemother. 1998;42:2352–2358. doi: 10.1128/aac.42.9.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaftandzieva A, Trajkovska-Dokic E, Panovski N. Prevalence and molecular characterization of extended spectrum beta-lactamases (ESBLs) producing Escherichia coli and Klebsiella pneumonia. Sec. Biol. Med. Sci. 2011;XXXII(2):129–141. [PubMed] [Google Scholar]
- 31.Rotimi V.O, Jamal W, Pal T, Sovenned A, Albert M.J. Emergence of CTX-M-15 type extended-spectrum beta-lactamase-producing Salmonella spp. in Kuwait and the United Arab Emirates. J. Med. Microbiol. 2008;57:881–886. doi: 10.1099/jmm.0.47509-0. [DOI] [PubMed] [Google Scholar]
- 32.Bindayna K.M, Khanfar H.S, Senok A.C, Botta G.A. Production of CTX-M genotype among ESBL-isolates in a tertiary hospital in Saudi Arabia. Saudi Med. J. 2010;31:859–863. [PubMed] [Google Scholar]
- 33.Alsultan A.A, Abulmagd E, Amin T.T. Extended beta-lactamse-producing Escherichia coli and Klebsiella pneumoniae in Al-Ahsa, Saudi Arabia:Antibiotic susceptibility and production of blaSHV and blaTEM. J. Infect. Dev. Ctries. 2013;7:1016–1019. doi: 10.3855/jidc.3764. [DOI] [PubMed] [Google Scholar]
- 34.Mohmid E.A, El-Sayed E.A, Abdel El-Haliem M.F. Molecular study on extended spectrum beta-lactam producing Gram-negative bacteria isolated from Ahmadi hospital in Kuwait. Afr. J. Biotechnol. 2013;12:5040–5035. [Google Scholar]
- 35.Altalhi A.D, Gherbawy Y.A, Hassan S.A. Antibiotic resistance in Escherichia coli isolated from retail raw chicken meat in Taif, Saudi Arabia. Foodborne Pathog. Dis. 2010;7:281–285. doi: 10.1089/fpd.2009.0365. [DOI] [PubMed] [Google Scholar]


