A wheat microRNA promotes heading by negatively regulating expression of homologs of the clock gene TOC1.
Abstract
Timing of flowering is not only an interesting topic in developmental biology, but it also plays a significant role in agriculture for its effects on the maturation time of seed. The hexaploid wheat (Triticum aestivum) is one of the most important crop species whose flowering time, i.e. heading time, greatly influences yield. However, it remains unclear whether and how microRNAs regulate heading time in it. In our current study, we identified the tae-miR408 in wheat and its targets in vivo, including Triticum aestivum TIMING OF CAB EXPRESSION-A1 (TaTOC-A1), TaTOC-B1, and TaTOC-D1. The tae-miR408 levels were reciprocal to those of TaTOC1s under long-day and short-day conditions. Wheat plants with a knockdown of TaTOC1s via RNA interference and overexpression of tae-miR408 showed early-heading phenotype. Furthermore, TaTOC1s expression was down-regulated by the tae-miR408 in the hexaploid wheat. In addition, other important agronomic traits in wheat, such as plant height and flag leaf angle, were regulated by both tae-miR408 and TaTOC1s. Thus, our results suggested that the tae-miR408 functions in the wheat heading time by mediating TaTOC1s expression, and the study provides important new information on the mechanism underlying heading time regulation in wheat.
The correct timing of the switch from vegetative to reproductive growth, namely the floral transition, is critical for reproductive success in flowering plants (Huijser and Schmid, 2011; Yamaguchi and Abe, 2012). This transition is controlled by genetic, epigenetic, and environmental factors (Kim et al., 2009; Srikanth and Schmid, 2011; Andrés and Coupland, 2012; Gu et al., 2013; Spanudakis and Jackson, 2014; Hong and Jackson, 2015; Teotia and Tang, 2015). In Arabidopsis (Arabidopsis thaliana), five genetically defined pathways, including the vernalization pathway, the autonomous pathway, the gibberellin pathway, the photoperiod pathway, and the age pathway, regulate the floral transition and are mediated by endogenous and exogenous signals (Srikanth and Schmid, 2011; Teotia and Tang, 2015). These signaling pathways converge on the flowering integrator genes, e.g. FLOWERING LOCUS T (FT; Kobayashi et al., 1999). The FT protein is a mobile florigen that moves from the leaves into the phloem and travels to the shoot apical meristem for the induction of the phase transition (Corbesier et al., 2007). The change in day length or photoperiod is a key cue for the seasonal control of the floral transition. CONSTANS (CO) is a key regulator in the photoperiod pathway of Arabidopsis (Putterill et al., 1995). Under the control of the circadian clock, the expression of CO shows a biphasic diurnal expression profile (Suárez-López et al., 2001). The CO protein as a transcriptional activator is stabilized by light and induces the expression of FT in the leaf under long-day (LD, 16 h light/8 h dark) conditions (Kobayashi et al., 1999). Plants synchronize the timings of their floral transition to seasonal changes via the interactions among circadian-clock-regulated components (Niwa et al., 2007; Johansson and Staiger, 2015; Song et al., 2015). The circadian clock measures the day-length change through an input system and then regulates the transcriptional activities that ultimately control the floral transition pathways (McClung, 2001).
TIMING OF CAB EXPRESSION 1 (TOC1), also known as PSEUDO-RESPONSE REGULATOR 1 (PRR1), is a key component of the plant circadian clock (Somers et al., 1998). Its function is involved in the clock’s evening loop, whereby it directly represses the transcription of morning loop genes LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1; Alabadí et al., 2001). The expression of TOC1 at both the transcriptional and translational levels shows a circadian change, even under constant light or dark conditions (Strayer et al., 2000; Más et al., 2003b). The mRNA of TOC1 starts to accumulate in the morning, and its level reaches a peak in the late day (Matsushika et al., 2000; Strayer et al., 2000). An increase or decrease of TOC1 expression could result in the alteration of normal circadian rhythm in Arabidopsis (Makino et al., 2002; Más et al., 2003a). Thus, the maintenance and regulation of TOC1 rhythmic expression is essential for proper functioning of the circadian clock (Más, 2008; McClung and Gutiérrez, 2010). Mutations of TOC1 in different genetic backgrounds produce earlier flowering under short-day (SD, 10 h light/14 h dark) conditions, but slightly later flowering or with little effect on flowering under LD conditions (Somers et al., 1998; Niwa et al., 2007). Genetic analysis has suggested that the CCA1/LHY-TOC1 circadian clock is closely linked to a CO-FT flowering pathway (Niwa et al., 2007).
MicroRNAs (miRNAs) are small noncoding RNAs of 21 to 24 nucleotides with a wide distribution in animals and plants (Bartel, 2009). They act as crucial ubiquitous regulators of gene expression at the transcriptional, posttranscriptional, and translational levels by repressing gene translation or degrading target mRNAs in most eukaryotic genomes (Mallory and Vaucheret, 2004; Chellappan et al., 2010; Khraiwesh et al., 2010). In plants, miRNAs play essential roles in various biological processes, such as the floral transition during the plant growth and development (Aukerman and Sakai 2003; Palatnik et al., 2003; Rubio-Somoza and Weigel 2011). Several miRNA families are involved in the pathways controlling flowering as inhibitors or promoters of the floral transition (Zhou and Wang, 2013; Spanudakis and Jackson, 2014). The miR156 and miR172 are two main factors that control the flowering time in the plant aging pathway (Huijser and Schmid, 2011; Yamaguchi and Abe, 2012; Wang, 2014). Other miRNA families including miR159, miR399, and OsmiR393 have also been shown to function in the control of flowering time (Achard et al., 2004; Kim et al., 2011; Xia et al., 2012). These miRNA-regulated pathways that control plant flowering time are themselves regulated by environmental factors, e.g. photoperiod and temperature (Teotia and Tang, 2015). Thus, miRNAs are important regulators of the floral transition in plants.
Wheat (Triticum aestivum), one of the most important commercial crops worldwide, is defined as an LD plant species (Thomas and Vince-Prue, 1997). A number of miRNA families have now been identified in wheat, and about twenty miRNA families are conserved in diverse plant species (Yao et al., 2007; Feng et al., 2014; Han et al., 2014; Kumar et al., 2014; Sun et al., 2014). Of these, miR408 contains 21 nucleotides and has been annotated in more than 30 plant species to date, indicating a high level of conservation in different plant species (Axtell and Bowman, 2008; Kozomara and Griffiths-Jones, 2011). The expression of miR408 is responsive to various environmental factors, e.g. copper, light, boron, water deficit, and salinity (Yamasaki et al., 2007; Trindade et al., 2010; Ozhuner et al., 2013; Mutum et al., 2013; Feng et al., 2013; Jovanović et al., 2014; Zhang et al., 2014; Hajyzadeh et al., 2015). Previous studies have indicated that plantacyanin, laccases, AFG1, and TaCLP1 are genuine targets of miR408 (Wang et al., 2004; Zhang et al., 2006; Abdel-Ghany and Pilon, 2008; Feng et al., 2013; Ozhuner et al., 2013). Moreover, the overexpression of miR408 in Arabidopsis promotes vegetative development (Zhang and Li, 2013). In wheat, TaCLP1 encodes a chemocyanin-like protein target of tae-miR408 (Feng et al., 2013). Previous functional analysis has indicated that tae-miR408 regulates the resistance of host plants to abiotic stresses and stripe rust (Feng et al., 2013). However, little is currently known about the function of tae-miR408 in the plant development of wheat.
In our current study, the functions of the Triticum aestivum TIMING OF CAB EXPRESSION 1 (TaTOC1) genes in the hexaploid wheat were analyzed. We performed knockdowns of the TaTOC1s in wheat by RNA interference (RNAi) that resulted in an early-heading time. We further found that TaTOC1 expression was down-regulated by tae-miR408 in the hexaploid wheat. Expression analyses revealed that tae-miR408 was highly expressed in leaves, although its transcripts could be detected in various other organs in wheat. Moreover, the expression patterns of tae-miR408 were found to have a rhythmic manner under both LD and SD conditions. Overexpression of tae-miR408 promoted the heading time of transgenic wheat. As expected, the expression levels of the TaTOC1 genes were down-regulated in tae-miR408 transgenic plants. Thus, our current results suggest that tae-miR408 functions in the wheat heading time process by mediating TaTOC1s expression. Our present findings provide important new information for our understanding of the mechanism underlying heading time regulation of wheat.
RESULTS
Identification of TaTOC1 Genes in the Hexaploid Wheat
TOC1 is a key regulator of the circadian clock and flowering time in Arabidopsis (Somers et al., 1998; Niwa et al., 2007). To determine the functions of the TOC1 genes in wheat, three cDNAs with high similarity (TaTOC-A1, TaTOC-B1, and TaTOC-D1; Supplemental Table S1) were identified from the leaf tissues of the hexaploid wheat (Triticum aestivum L. cv. Lunxuan987) by reverse transcription PCR (RT-PCR). These genes harbor a 1551-bp, 1551-bp, and 1563-bp open reading frame, respectively. They showed high homology with three wheat TOC1 genes: Traes_6AL_A0A31AA9F (99.7%), Traes_6BL_ED40C8806 (99.9%), and Traes_6DL_C215BACFD (99.8%), deposited in the gramene database (http://www.gramene.org). The deduced amino acid sequences of their proteins were found to contain the pseudo-receiver domain of about 120 amino acids at its N terminus and the short CCT motif of about 50 amino acids at the C terminus (Supplemental Fig. S1). Further dendrogram analysis indicated that the three TaTOC1 proteins belong to a PRR1 cluster and are closest to the rice OsPRR1s (Supplemental Fig. S2). Thus, we confirmed that three TaTOC1 genes had been identified from the hexaploid wheat genomes.
Analysis of TaTOC1 Genes Expression Patterns
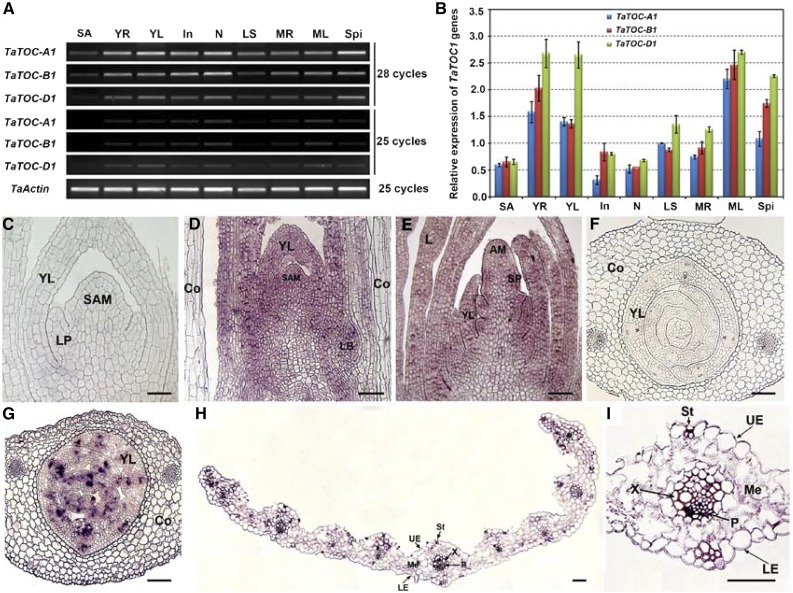
We further analyzed the expression patterns of the TaTOC1 genes in various organs of wheat by RT-PCR and quantitative RT-PCR (qRT-PCR). As shown in Figure 1A and B, these three genes showed similar expression patterns and were expressed at different levels in all of the tissues examined, including shoot apices, young roots, and young leaves at the Zadoks decimal growth stage (Z) 1.5, as well as in nodes, internodes, leaf sheaths, mature roots, mature leaves, and young spikelets at Z3.5 (staging according to Zadoks et al., 1974). Predominant expression was evident in young roots, young leaves, and mature leaves. In situ hybridization analysis revealed that strong signals for three genes localized in the vegetative and reproductive meristems and young leaves (Fig. 1, D, E, and G) and in some cells in the vascular bundles and guard cells of the mature leaf (Fig. 1, H–I).
Figure 1.
Expression patterns of TaTOC-A1, TaTOC-B1, and TaTOC-D1 in different wheat organs. A, Total RNAs were isolated from various organs of hexaploid wheat at ZT12 under 12-h-light/12-h-dark conditions. Expression of TaTOC-A1, TaTOC-B1, and TaTOC-D1 expression was analyzed by RT-PCR. B, Real-time qRT-PCR was performed using homolog-specific primers of TaTOC-A1, TaTOC-B1, and TaTOC-D1 under highly stringent conditions. The TaActin gene was used as an endogenous control. The data are the means from three replicates. C–I, In situ localization of the TaTOC1 mRNAs in the vegetative shoot apices, inflorescence primordia, and leaves of wheat. C-E, Longitudinal sections of the vegetative shoot apex and an inflorescence primordium. F and G, Transverse sections of the leaf primordia and young leaves in a seedling at day 1 after germination. H, Transverse section of young leaf from seedlings grown for 14 d. I, An enlarged view of the leaf middle vein in H. D, E, H, and I, Leaf sections that were hybridized with a TaTOC1 antisense mRNA probe. C and F, Controls hybridized with a sense probe. AM, apical meristem; Co, coleoptile; In, internodes; L, leaf; LB, lateral bud; LE, lower epidermal; LP, leaf primordia; LS, leaf sheaths; MR, mature roots; Me, mesophyl; ML, mature leaves; N, nodes; P, phloem; SA, shoot apices; SAM, shoot apical meristem; Spi, spikelets; UE, upper epidermal; St, stoma; X, xylem; YL, young leaf; YR, young root. Bars in C–E, 200 μm; in F and G, 350 μm; in H and I, 15 μm.
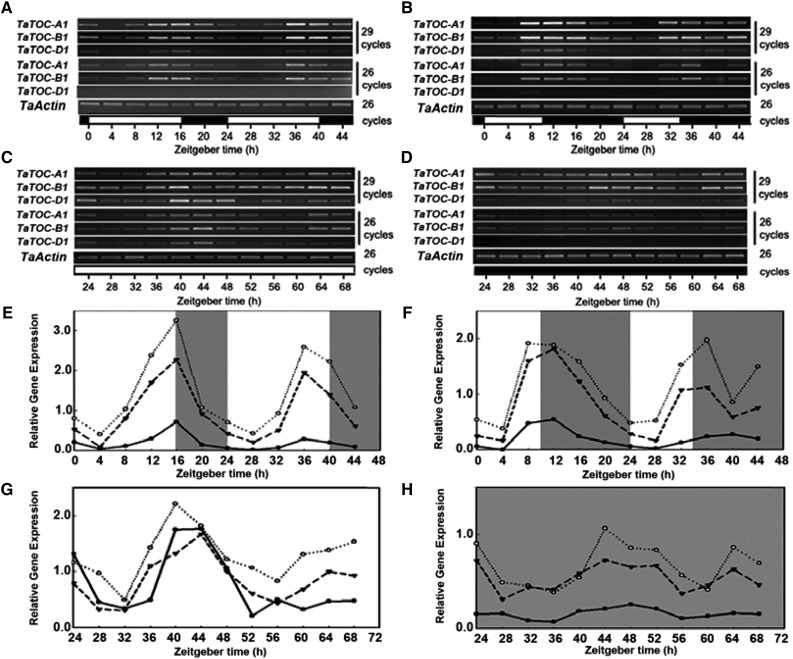
As a major component of the circadian clock, TOC1 in Arabidopsis is itself regulated in a circadian manner and participates in a feedback loop to control its own expression (Strayer et al., 2000). Our present analyses revealed that expression of TaTOC-A1, TaTOC-B1, and TaTOC-D1 expression peaked in the evening and showed diurnal patterns under LD and SD conditions (Fig. 2, A, B, E, and F). To test whether the rhythmic cycling of TaTOC1 transcript levels in leaves is controlled by a circadian clock, wheat seedlings grown under standard light (SL, 12 h light/12 h dark) conditions were transferred to either continuous light or continuous dark, and the TaTOC-A1, TaTOC-B1, and TaTOC-D1 transcript levels were then examined every 4 h over a period of 48 h. The transcript levels of these genes continued to cycle under both sets of conditions (Fig. 2, C, D, G, and H), but the gene transcript levels were greatly reduced in continuous dark compared with those in continuous light, suggesting that light plays an important role in the maintenance of the TaTOC-A1, TaTOC-B1, and TaTOC-D1 mRNA concentrations. Thus, our data indicated that the TaTOC1 genes are circadian clock controlled.
Figure 2.
Daily oscillation of TaTOC-A1, TaTOC-B1, and TaTOC-D1 mRNA abundance in wheat leaves under different conditions. Leaves of two-week-old wheat seedlings grown under LD (16L/8D) (A and E) and SD (10L/14D) (B and F) were harvested at ZT0 and at 4-h intervals thereafter. The seedlings grown under SL (12L/12D) conditions were transferred to continuous light (C and G) and continuous dark (D and H) conditions, respectively. One day later, leaves were collected at 6:00 a.m. (ZT24) and at 4-h intervals thereafter. Both RT-PCR (A–D) and qRT-PCR (E–H) analyses were performed using homolog-specific primers of TaTOC-A1, TaTOC-B1, and TaTOC-D1 under highly stringent conditions. The TaActin gene was used as an endogenous control. Data are the means from three replicates. The white and black boxes at the bottom of each graph in A–D represent light and dark periods, respectively. The white and gray areas in E–H represent light and dark periods, respectively.
Wheat Heading Time Is Promoted by TaTOC1s Knockdown
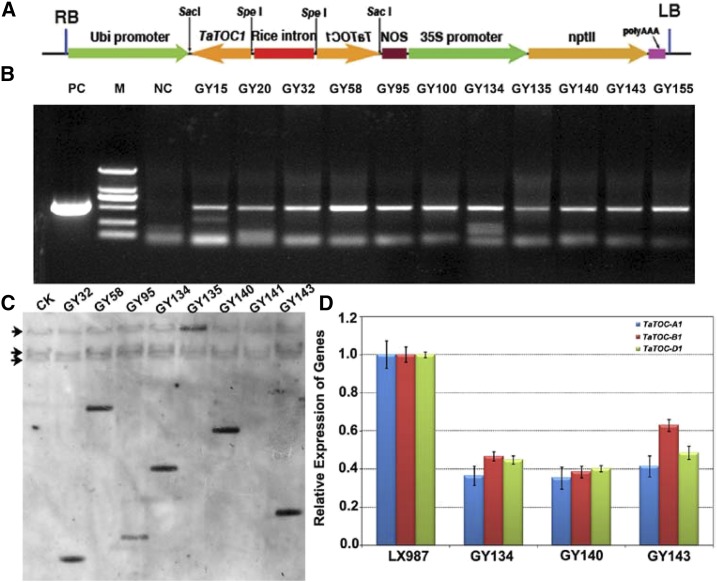
To determine the function of the three TaTOC1 genes in the hexaploid wheat, we used an RNAi strategy to knockdown these genes in wheat (Fig. 3A). Eleven independent kanamycin-resistant transgenic wheat lines were generated through the Agrobacterium-mediated transformation of wheat immature embryos (Fig. 3B). Southern-blot analysis confirmed that the Pubi:TaTOC1-RNAi construct was integrated into the genomes of six of these transgenic lines at one copy in diverse patterns into the genomes of the six transgenic lines (Fig. 3C). The transcript levels of the three TaTOC1 genes in these transgenic lines were significantly reduced compared with ones in wild type (cv Lunxuan 987; Fig. 3D). Thus, these kanamycin-resistant plants were verified as TaTOC1s silenced transgenic wheat lines. Homozygous plants from three T5 transgenic lines (GY134, GY140, and GY143) were chosen for further analysis.
Figure 3.
Identification of TaTOC1-RNAi transgenic wheat plants. A, Schematic representation of the TaTOC1-RNAi construct used for wheat transformation. The vector backbone was derived from the pZP211 plasmid. The cassettes consisted of the Ubi (the maize ubiqutin) promoter, the sense and antisense regions from a wheat TaTOC1 cDNA, rice intron (rice SBEI intron 9), NOS (a terminator of the nopaline synthase gene), the 35S (Cauliflower Mosaic Virus promoter) promoter, and nptII (neomycin phospho transferase II). B, PCR detection of kanamycin-resistant regenerated lines. M, DL2000 DNA marker; NC, negative control; PC, positive control. C, Southern-blot assay of the eight kanamycin-resistant regenerated lines. Genomic DNA was digested with BamHI and then hybridized to an amplified fragment specific for TaTOC1. D, qRT-PCR analyses of the expression of TaTOC-A1, TaTOC-B1, and TaTOC-D1 in three T5 RNAi transgenic lines. Bars indicate sem.
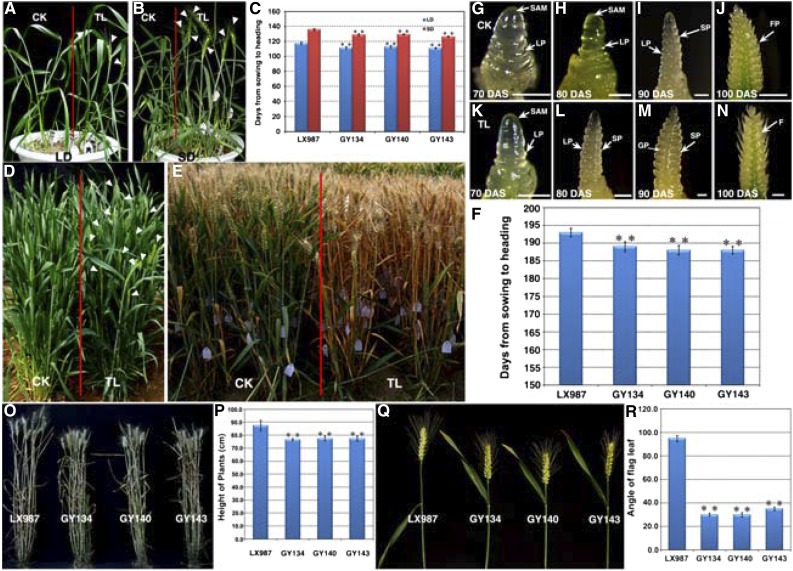
The GY134, GY140, and GY143 transgenic wheat plants showed an early-heading phenotype under both LD and SD conditions in a growth chamber (Fig. 4, A and B). The heading time of these transgenic plants was shortened by 4 to 9 d compared with that of the wild-type plants (Fig. 4C). These three transgenic lines also displayed earlier heading time than the wild-type plants when cultivated in a field (Fig. 4, D and E). Through the emergence of half-length ears as an indicator of the heading time, we found that the transgenic plants headed, on average, about 5 d earlier than the wild-type plants (P < 0.001; Fig. 4F). To further dissect the effects of TaTOC1 genes silencing on the heading time in wheat, we examined developmental changes in the apex. As shown in Figures 4G to 4N, the floral transition of transgenic plants started earlier than that of wild type. At 70 d after sowing, the apex of TaTOC1-RNAi transgenic plant was more extended than that of control (Fig. 4, G and K). Spike primordia emerged between the leaf ridge on the elongated meristem of transgenic plant at 80 d after sowing (Fig. 4L), indicating the transition from vegetative to reproductive state. By contrast, the status of apex in control is still at single ridge stage (Fig. 4I). Thus, flowering initiation and heading time were promoted in these transgenic wheat plants.
Figure 4.
Phenotype analysis of TaTOC1-RNAi transgenic wheat. A and B, TaTOC1-RNAi transgenic wheat (TL) and control (CK) plants were grown in a growth chamber under LD (A) and SD (B) conditions. The white arrowheads indicate the heading spikelet. C, Heading time in the TL and CK plants, represented by the period (days) from sowing to heading when one-half of the spike had emerged. All plants were grown under LD and SD conditions, respectively. Data are indicated as the means ± SEM of 20 plants. D, TL and CK wheat grown under natural conditions in a field. The white arrowheads indicate the heading spikelet. E, TL plants mature more quickly than CK plants under natural conditions. F, Heading times of TL and CK plants grown under natural conditions. Data are the mean ± SEM of 100 plants. G-N, Comparison of spike development between TL andCK plants. DAS, days after sowing; F, floret; FP, floret primordium; GP, glume primordium; LP, leaf primordium; SP, spikelet primordium; SAM, shoot apical meristem. Bars in G-N, 100 μm. O and P, Comparison of the TL and CK plant heights. Q and R, Comparison of the TL and CK flag leaf angle. Statistically significant differences were determined based on three replicate analyses (t test, **P < 0.01). The plants in O–R were grown in the experimental field under natural conditions. Fifty plants per replicate were sampled and analyzed in P and R.
Other important agronomic traits, such as plant height and flag leaf angle, were also clearly altered in the GY134, GY140, and GY143 plants (Fig. 4, O-R). The plant height in cereal crops, which is a critical aspect of plant architecture, is directly linked to the harvest index and yield potential. The height of the transgenic wheat in our current study showed a clear reduction (Fig. 4O), which our statistical analyses revealed to be nearly 10 cm below the controls (P < 0.01; Figure 4P). The flag leaf angle is positively and significantly correlated with the photosynthetic ability of a plant and also with the translocation of assimilation products that influence grain yield (Khaliq et al., 2008; Biswal and Kohli, 2013). Our data further showed that the flag leaf angles of the transgenic lines were significantly decreased (Fig. 5, Q and R). These results revealed that the TaTOC1 genes also function in other developmental traits in wheat.
Figure 5.
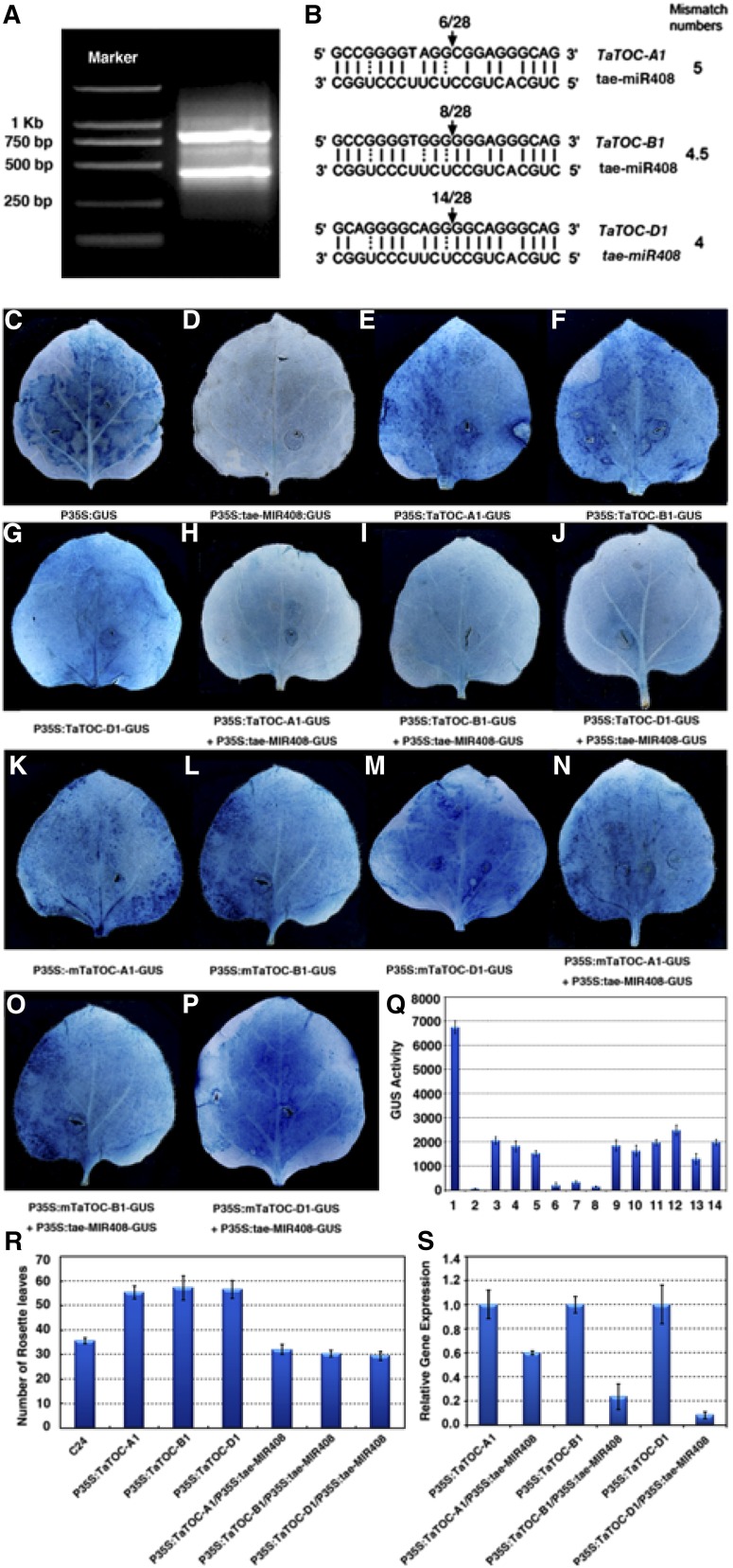
Identification and validation of TaTOC1s cleavage by tae-miR408. A, Two bands were detected by Hi-TAIL-PCR. DL2000 DNA markers were used. B, Predication and validation of tae-miR408 targets. Arrows indicate the cleavage sites detected by 5ʹ RACE. C–Q, Cotransformation of tobacco leaves with TaTOC-A1, TaTOC-B1, TaTOC-D1, and tae-MIR408. The recombinant vectors were transferred into tobacco leaves via the Agrobacterium strain GV3101. GUS phenotypes were observed by histochemical staining. C, The pBI121 vector was used as positive control. D, P35S:tae-MIR408-GUS. E, P35S:TaTOC-A1-GUS. F, P35S:TaTOC-B1-GUS. G, P35S:TaTOC-D1-GUS. H, A mixture of P35:tae-MIR408-GUS and P35S:TaTOC-A1-GUS. I, A mixture of P35S:tae-MIR408-GUS and P35S:TaTOC-B1-GUS. J, A mixture of P35:tae-MIR408-GUS and P35S:TaTOC-D1-GUS. K, P35S:mTaTOC-A1-GUS. L, P35S:mTaTOC-B1-GUS. M, P35S:mTaTOC-D1-GUS. N, A mixture of P35S:tae-MIR408-GUS and P35S:mTaTOC-A1-GUS. O, A mixture of P35S:tae-MIR408-GUS and P35S:mTaTOC-B1-GUS. P, A mixture of P35:tae-MIR408-GUS and P35S:mTaTOC-D1-GUS. Q, Quantitative analysis using a fluorospectrophotometer of GUS enzyme activity in leaves inoculated with different recombinant vectors. The numbers from 1 to 14 represent different constructs or construct combinations, respectively. 1, P35S:GUS; 2, P35S:tae-MIR408-GUS; 3, P35S:TaTOC-A1-GUS; 4, P35S:TaTOC-B1-GUS; 5, P35S:TaTOC-D1-GUS; 6, P35:tae-MIR408-GUS+P35S:TaTOC-A1-GUS; 7, P35S:tae-MIR408-GUS+P35S:TaTOC-B1-GUS; 8, P35:tae-MIR408-GUS+P35S:TaTOC-D1-GUS; 9, P35S:mTaTOC-A1-GUS; 10, P35S:mTaTOC-B1-GUS; 11, P35S:mTaTOC-D1-GUS; 12, P35S:tae-MIR408-GUS+P35S:mTaTOC-A1-GUS; 13, P35S:tae-MIR408-GUS+P35S:mTaTOC-B1-GUS; 14, P35:tae-MIR408-GUS+P35S:mTaTOC-D1-GUS. Data are the mean ± SEM of 10 plants. R, Flowering time analysis of transgenic Arabidopsis overexpressing TaTOC-A1, TaTOC-B1, TaTOC-D1, tae-MIR408, or different vector combinations. The number of rosette leaves was used to indicate flowering time. Wild-type Arabidopsis (cv C24) was used as a control. Data are the indicated as mean ± SEM of 20 plants. S, The relative expression levels of TaTOC-A1, TaTOC-B1, and TaTOC-D1, detected as overexpression of a single gene or different gene combinations, respectively.
TaTOC1s Are Targets of tae-miR408
Although the draft genome sequences of wheat and its progenitors have been acquired previously via shotgun sequencing (Brenchley et al., 2012; Jia et al., 2013; Ling et al., 2013; Chapman et al., 2015), the sequencing data for the hexaploid wheat are still fragmentary and incomplete. To elucidate the upstream-regulatory sequences of the three TaTOC1 genes, high-efficiency thermal asymmetric interlaced PCR (hiTAIL-PCR; Liu and Chen, 2007) was performed. We identified a number of potential regulatory motifs at about 1 kb upstream of the TaTOC1 genes (Supplemental Table S2). These included ACE, G-box, and Sp1 motifs belonging to light responsive elements, and an EE motif that is involved in circadian rhythmicity (Lescot et al., 2002). Interestingly, two differently sized TaTOC1 transcripts were identified (Fig. 5A), suggesting that the three TaTOC1 genes might be targeted by miRNAs. Through our analysis of the microRNA database (http://microrna.sanger.ac.uk), we found that tae-miR408 matches the TaTOC-D1, TaTOC-B1, and TaTOC-A1 cDNAs. Normally, all functional miRNA targets in plants have up to five mismatches to miRNA (Schwab et al., 2005; Liu et al., 2014). In our current analysis, the mismatch number within the predicted complementary region of tae-miR408 with TaTOC-D1, TaTOC-B1, and TaTOC-A1 was found to be 4, 4.5, and 5 bases, respectively, when counting G:U pairs as 0.5 mismatches (Fig. 5B). This suggested that the three TaTOC1 genes transcripts might be cleaved by tae-miR408. To investigate whether the target relationship between miR408 and TOC1 was ubiquitous in plants, we extended our analysis to Arabidopsis, soybeans, rice, barley, Brachypodium, and maize. Sequence alignments revealed that the putative miR408/TOC1 duplexes in most of these species carried several large bulge loops and had more than five mismatch numbers (Supplemental Fig. S3). By contrast, the five-mismatch bases were identified within the predicted complementary region of HvmiR408 with HvTOC1 in barley.
To confirm that TaTOC1 genes were targets of tae-miR408, a modified 5ʹ end RACE (5ʹ RACE) was carried out. The results showed that TaTOC-A1, TaTOC-B1, and TaTOC-D1 mRNA could be cleaved in vivo at the site between bases 10 and 11 within the region of these transcripts that is complementary to tae-miR408 (Fig. 5B), which was identified previously as a canonical miRNA cleavage site (Peters and Meister, 2007; Höck and Meister, 2008). To further validate the cleavage function of tae-miR408 with respect to TaTOC1 mRNAs, transient assays were performed in tobacco leaves. The leaves transformed with the pBI121 vector harboring the GUS reporter gene were used as controls, and the GUS signal was observed by histochemical staining (Fig. 5, C–P). Leaves inoculated with P35S:TaTOC-A1-GUS, P35S:TaTOC-B1-GUS, or P35S:TaTOC-D1-GUS showed a similar phenotype to the control (Fig. 5, E–G). In contrast, the leaves inoculated with P35S:tae-MIR408-GUS showed no GUS signal (Fig. 5D). When the leaves inoculated with both P35S:tae-MIR408-GUS and either P35S:TaTOC-A1-GUS, P35S:TaTOC-B1-GUS, or P35S:TaTOC-D1-GUS, the GUS signal was very weak compared with the leaves inoculated with the corresponding expression vectors for the TaTOC1 genes (Fig. 5, E–G and H–J). A previous study reported that mutation of miRNA target sites could suppress miRNAs function and block the cleavage of the target gene (Wu et al., 2013). We thus mutated the cleavage sites targeted by tae-miR408 in TaTOC-A1, TaTOC-B1, and TaTOC-D1, respectively (referred to as mTaTOC-A1, mTaTOC-B1, and mTaTOC-D1; Supplemental Fig. S4). The leaves inoculated with P35S:mTaTOC-A1-GUS, P35S:mTaTOC-B1-GUS, or P35S:mTaTOC-D1-GUS showed strong GUS signals (Fig. 5, K–M). However, when the P35S:tae-MIR408-GUS construct was cotransformed with P35S:mTaTOC-A1-GUS, P35S:mTaTOC-B1-GUS, or P35S:mTaTOC-D1-GUS, the GUS signals were comparable to the controls (Fig. 5, N–P). Next, the enzyme activity of GUS in leaves inoculated with the Agrobacterium strain GV3101 containing different recombinant vectors was measured using a fluorospectrophotometer (Fig. 5Q). The enzyme activity in leaves of different treatments was consistent with the GUS signal level in leaves of those corresponding treatments.
Overexpression of TaTOC-A1, TaTOC-B1, or TaTOC-D1 in wild-type Arabidopsis caused a later-flowering phenotype (Fig. 5R). When the P35S:TaTOC-A1, P35S:TaTOC-B1, and P35S:TaTOC-D1 constructs were separately cotransformed with the P35S: tae-MIR408 vector, all of the plants harboring tae-miR408 and the TaTOC1s showed marginally early flowering compared with the control plants (Fig. 5R). The transcript levels of the TaTOC1 genes in these plants were reduced compared with those overexpressing TaTOC1s (Fig. 5S). In Arabidopsis, AtTOC1 cannot be targeted by miR408 for limited sequence matching (Supplemental Fig. S3). Consistently, the ectopic expression of tae-miR408 in Arabidopsis did not result in any obvious alteration in the expression level of AtTOC1 (Supplemental Fig. S5). These results indicate that the TaTOC1s are targeted by tae-miR408.
Characterization and Expression Pattern of tae-miR408 in Wheat
Previously, tae-miR408 was predicted to be a 21-nucleotide miRNA in wheat (Yao et al., 2007) and to be involved in regulating the resistance of host plants to abiotic stresses and stripe rust by targeting a putative plantacyanin protein gene (TaCLP1; Feng et al., 2013). However, the fundamental function of tae-miR408 in wheat development remains unknown. The primary miRNA of tae-miR408 (GenBank No. BE419354) was identified by Yao et al. (2007), and this gene is located on chromosome 7BL (referred to as tae-MIR408b). By further analysis of the published wheat genome sequences of wheat in the NCBI (www.ncbi.nlm.nih.gov), the other two tae-MIR408 genes, located on chromosome 7AL and 7DL, respectively, were identified and referred to as tae-MIR408a and tae-MIR408c. Three tae-MIR408 genes were isolated from wheat (cv. Lunxuan987) using RT-PCR and confirmed the amplified fragment by sequencing (Supplemental Fig. S6A). The secondary structures of the three tae-miR408 precursor sequences were predicted by MFold web server (Zuker, 2003) to form a hairpin loop with mature miR408 in its stem region, a characteristic of miRNA precursors (Ambros et al., 2003; Supplemental Fig. S6, B–D). These three tae-MIR408 genes could produce the same mature miRNA (tae-miR408). Phylogenetic analysis of miR408 in 31 plant species further showed that tae-miR408 is closer to its homologs in monocots, such as rice and B. distachyon, than to those in dicots (Supplemental Fig. S6E).
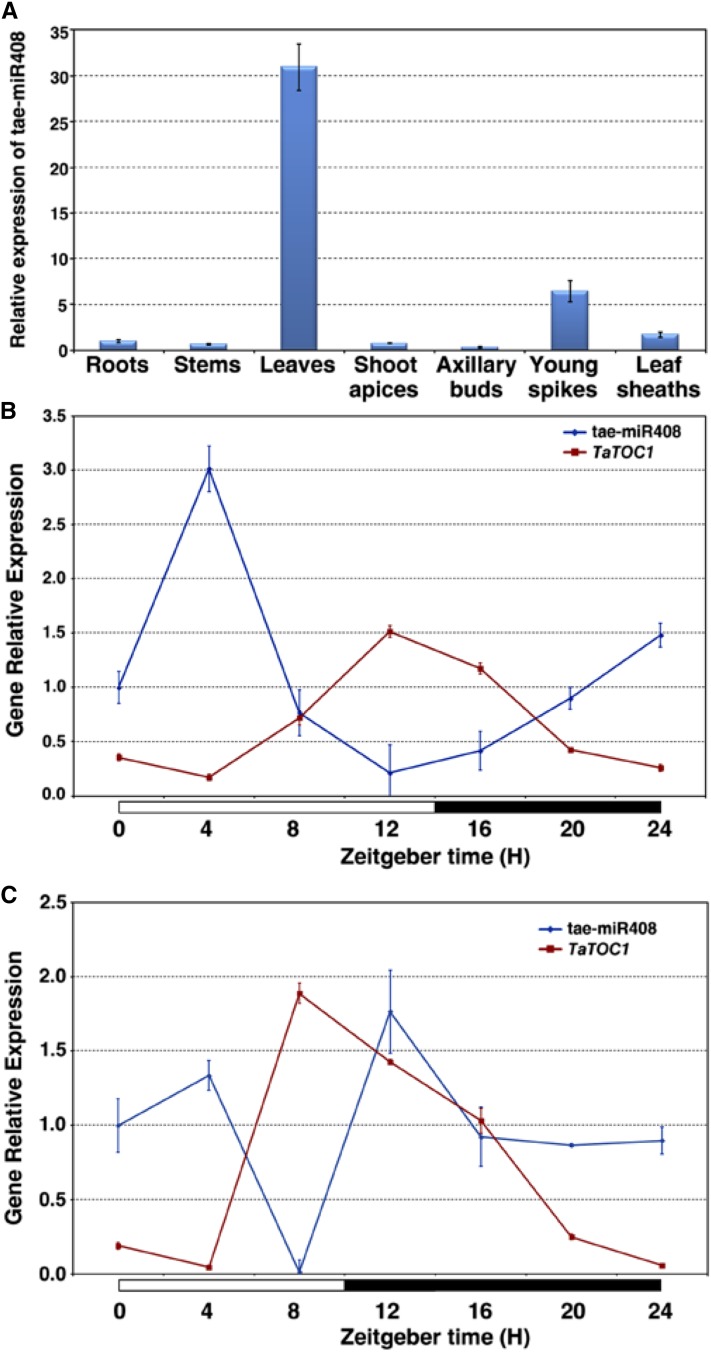
The relative levels of tae-miR408 transcripts were analyzed by qRT-PCR in various organs of wheat. As shown in Figure 6A, tae-miR408 is ubiquitously expressed in wheat, but higher transcript levels could be detected in leaves (Fig. 6A). To determine whether tae-miR408 expression was day-length dependent, we detected its accumulation in leaves under LD and SD conditions. Under both conditions, tae-miR408 was found to be expressed in different rhythmic patterns (Fig. 6, B and C). The expression levels of tae-miR408 reached a peak at about 4 h after dawn and fell to the lowest level toward the evening under LD conditions (Fig. 6B). In contrast, this expression peaked at 4 h and 12 h after dawn, and its lowest level was at 8 h after dawn under SD conditions (Fig. 6C). Interestingly, the expression levels of tae-miR408 under both sets of conditions are reciprocal to those of the TaTOC1s, indicating that the TaTOC1 genes could be cleaved in vivo by tae-miR408.
Figure 6.
Expression Pattern of tae-miR408. A, qRT-PCR analysis of tae-miR408 expression in various wheat organs, including the roots, stems, shoots apices, axillary buds, young spikes, and leaf sheaths. B and C, Diurnal expression patterns of tae-miR408 and TaTOC1s under LD (B) and SD (C) conditions. The white and black boxes at the bottom of each graph in B and C represent light and dark periods, respectively.
Tae-miR408 Regulates the Heading Time in Transgenic Wheat
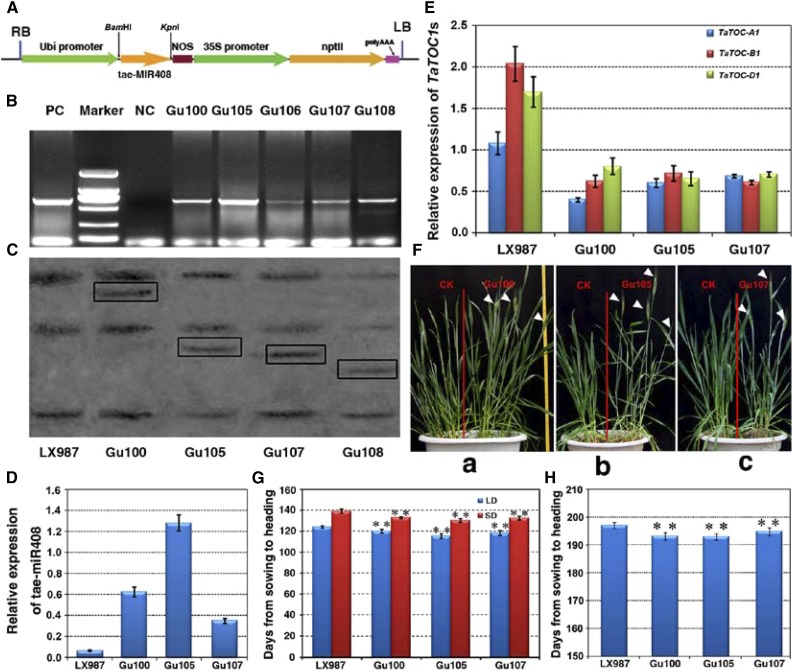
To further elucidate the function of tae-miR408, we constructed a vector carrying the tae-MIR408b gene driven by the maize ubiquitin promoter (Fig. 7A) and transformed this construct into hexaploid wheat (cv. Lunxuan 987) through the Agrobacterium-mediated transformation of immature embryos. Both PCR and southern blotting showed that the tae-MIR408 gene had integrated into the genomes of the transgenic plants at one copy (Fig. 7, B and C). The transcript abundance of tae-miR408 in the T3 transgenic lines (Gu100, Gu105, and Gu107) was significantly increased compared with wild-type plants (Fig. 7D). As the targets of tae-miR408, the expression levels of TaTOC-A1, TaTOC-B1, and TaTOC-D1 were also markedly reduced in the transgenic wheat overexpressing tae-miR408 (Fig. 7E; Supplemental Fig. S7). Furthermore, phenotypic analysis showed the heading time of these transgenic lines was earlier than that of the controls when cultivated in a growth chamber (Fig. 7F). Under both LD and SD conditions, tae-miR408 transgenic plants showed earlier heading by about 5 to 9 d compared with the controls (Fig. 7G). Moreover, the heading time of transgenic wheat was also shorter than that of controls under field growth conditions (Fig. 7H). The heading time in tae-miR408 transgenic plants was found to be very similar to those in the TaTOC1-RNAi transgenic plants. Thus, these findings confirm that tae-miR408 regulates the heading time of wheat.
Figure 7.
Characterization of transgenic wheat overexpressing tae-miR408. A, Schematic representation of the tae-miR408 overexpressing construct for wheat transformation. The vector backbone was derived from the pZP211 plasmid. The cassettes consisted of the Ubi promoter, the tae-MIR408b gene, NOS, the 35S promoter, and the resistance gene nptII. B, PCR detection of kanamycin-resistant regenerated lines. Marker, DL2000 DNA; NC, negative control; PC, positive control. C, Southern-blot analysis of the four kanamycin-resistant regenerated lines (Gu100, Gu105, Gu107, Gu108). Genomic DNA was digested with HindIII and then hybridized to an amplified fragment specific for the tae-MIR408b. D, qRT-PCR analyses of the expression of tae-miR408 in three T3 transgenic lines (20 plants per line). Data are the mean as mean ± SEM. E, Relative expression levels of TaTOC-A1, TaTOC-B1, and TaTOC-D1 in tae-miR408 overexpressing transgenic plants. Data are the mean ± SEM. F, Comparison of phenotypes between the transgenic lines (Gu100, Gu105, Gu107) and CK plants grown in a growth chamber. The white arrowheads indicate the heading spikelets. G, Heading times in plants overexpressing tae-miR408 represented by the period (days) from sowing to the emergence of one-half of the spike emerged. Both the T3 transgenic lines and controls were grown under LD and SD conditions, respectively. Data are the mean ± SEM of 20 plants. (t test, ** = P < 0.01). H, Heading times in plants overexpressing tae-miR408 was represented by days from sowing to heading when one-half of spike emerged. Both T3 transgenic lines and controls were grown in the experimental field under natural conditions. Data are the mean ± SEM of 100 plants (t test, **P < 0.01).
Since TaTOC1 genes expression is regulated by tae-miR408, we speculated that this miRNA might affect other developmental traits in transgenic plants. As expected, other traits, such as plant height and flag leaf angle, were also quite severely altered in transgenic plants compared with those of wild-type plants (Supplemental Fig. S8). Thus, the altered traits in tae-miR408 transgenic plants were found to be very similar to those in TaTOC1-RNAi transgenic wheat, suggesting that these traits are mediated by tae-miR408 through the regulation of TaTOC1s function.
The tae-miR408 Mediated Down-Regulation of TaTOC1s Caused Early Heading by Activation of TaFT1 Gene
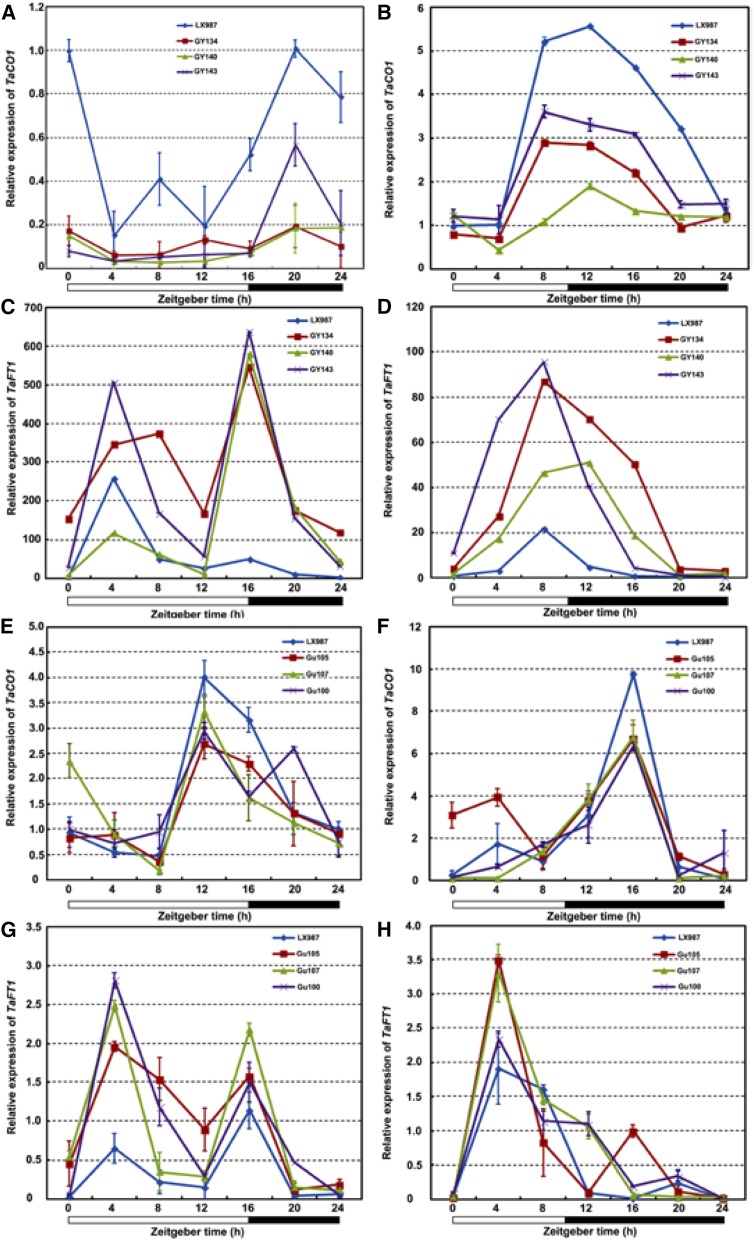
The flowering time gene CO acts downstream of circadian clock in Arabidopsis (An et al., 2004), and its ortholog in wheat, TaCO1, also plays key roles in flowering (Shimada et al., 2009). FT encodes a mobile protein that moves from leaves via the phloem to the shoot apical meristem to promote flowering (Corbesier et al., 2007). TaFT1 (VRN3) was identified as an ortholog of FT in wheat (Yan et al., 2006). Under either LD or SD conditions, the transcript levels of both TaCO1 and TaFT1 were significantly altered, although their expression patterns in the TaTOC1-RNAi and tae-miR408-OE transgenic plants were similar to those in the wild type (Fig. 8). The expression of TaFT1 was substantially upregulated, whereas that of TaCO1 was significantly reduced in the transgenic plants (Fig. 8). These results suggest that tae-miR408 functions in the heading time of wheat by down-regulating TaTOC1s and TaFT1 in wheat.
Figure 8.
Expression of TaCO1 and TaFT1 in TaTOC1-RNAi (A–D) and tae-miR408-OE (E–H) transgenic wheat plants. The white and black boxes at the bottom of each graph represent light and dark periods, respectively. Data are the mean ± SEM.
DISCUSSION
The plant circadian clock integrates environmental signals with internal cues to coordinate diverse physiological outputs (Adams and Carré, 2011; Kinmonth-Schultz et al., 2013). Many plants synchronize their flowering time with seasonal changes by means of circadian clock to improve the chances of reproductive success (Song et al., 2015). Wheat is a LD crop and is distributed widely throughout the world because of the adaptability of its flowering time to different environmental conditions (Worland and Snape, 2001). This adaptability might be facilitated by the complex genome in hexaploid wheat, i.e. the large genome size, a complex structure of homologous genomes, and the highly repetitive genomic sequences, implying that molecular mechanisms controlling the flowering time in wheat are also complex. Indeed, we show from our current analyses that the TaTOC1-mediated flowering time is regulated by an epigenetic factor.
In our present study, we identified three genes, TaTOC-A1, TaTOC-B1, and TaTOC-D1, from the hexaploid wheat. The transcript levels of these TaTOC1 genes were clearly reduced in entrained plants shifted to continuous dark, during which time the rhythmic expression pattern also became indistinct (Fig. 2). A similar phenomenon was also observed for the OsTOC1 gene of rice under continuous dark conditions (Murakami et al., 2005). In contrast, neither the transcript level nor the rhythmic pattern of Arabidopsis TOC1 was reported to show any obvious change in continuous dark (Strayer et al., 2000). Moreover, our previous study found that the TaGI1 transcript levels are greatly reduced in continuous dark compared with continuous light (Zhao et al., 2005). These data suggest that light may be an important factor in the maintenance of circadian expression and clock gene transcript levels in monocots.
The TOC1 transcriptional factor regulates thousands of potential targets in Arabidopsis (Gendron et al., 2012). Some clock-regulated biological processes, such as the flowering time, response to stimulus, metabolic process, and development, were found in previous studies to be enriched for potential target genes of TOC1 (Gendron et al., 2012; Huang et al., 2012), implying its complex and dynamic function as a regulator of oscillation. Our present results indicate that the TOC1 homologs in wheat play a role not only in circadian clock controlled flowering time but also in other developmental traits, such as plant height and flag leaf angle. Master regulators of plant height, Rht-B1 and Rht-D1, were reported to have a significant effect on the heading time of wheat (Wilhelm et al., 2013). It is likely therefore that the TaTOC1-mediated developmental traits may involve their downstream genes functions, and the divergence of the mechanisms responsible for these phenotypes might have occurred during the evolution of wheat and Arabidopsis.
Recently, miR408 was identified as a conserved microRNA family across many species (Trindade et al., 2010; Mutum et al., 2013; Jovanović et al., 2014; Thiebaut et al., 2014; Wu et al., 2014; Zhang et al., 2014; Alaba et al., 2015; Boke et al., 2015; Hajyzadeh et al., 2015). The expression of this miRNA is regulated by environmental factors such as light, nutrition, and abiotic stress, and it plays multiple roles in plant development and in plant resistance to biotic and abiotic stresses. In Arabidopsis, miR408 is regulated by the HY5-SPL7 module and functions in the coordinated response to light and copper (Zhang et al., 2014), suggesting that it responds to light signals in leaves. In accordance with its multiple functions, miR408 in plants targets a number of genes (Trindade et al., 2010; Mutum et al., 2013; Jovanović et al., 2014; Thiebaut et al., 2014; Wu et al., 2014; Alaba et al., 2015; Boke et al., 2015; Hajyzadeh et al., 2015). In our current study, we identified tae-miR408 from hexaploid wheat and characterized its expression and biological functions. The diurnal expression underlying tae-miR408 accumulation is sensitive to day lengths (Fig. 6). As the targets of tae-miR408, the TaTOC1 genes in wheat also showed rhythmic expression controlled by photoperiods and circadian clocks (Fig. 2). Moreover, the expression of tae-miR408 and TaTOC1s clearly showed anticyclical patterns in wheat leaves. Thus, tae-miR408 might play a functional role in wheat leaves in the sensing of day length changes through the mediation of TaTOC1s expression.
Our present results indicated that tae-miR408 directly targets the TaTOC-A1, TaTOC-B1, and TaTOC-D1 genes in wheat. Our bioinformatics analysis showed that the cleavage site may exist in the TOC1 mRNA that would be targeted by miR408 in barley, but not in rice, Brachypodim, maize, Arabidopsis, or soybeans (Supplemental Fig. S4). Moreover, overexpression of tae-miR408 in Arabidopsis showed no obvious effects on the expression of TOC1 (Supplemental Fig. S5). So, this cleavage mechanism may exist in a portion of the grass species.
In Arabidopsis, at least three miRNA families are involved in flowering time control: miR172, miR156, and miR159. In the aging pathway, miR172 and miR156 antagonistically regulate reproductive competency. The nature of the aging pathway is dependent on the sequential activity of two miRNA families (Zhu and Helliwell, 2011; Yamaguchi and Abe, 2012). The miR159 molecule functions in flowering time through the gibberellin pathway (Yamaguchi and Abe, 2012). In our present analyses, we showed that the flowering time was regulated by miR408 in wheat and that the altered traits in tae-miR408 transgenic wheat are very similar to those in the TaTOC1-RNAi transgenic lines, suggesting that the tae-miR408-mediated development of these traits might occur through the regulation of TaTOC1s activity.
The circadian clock plays important roles in controlling the flowering time in plants (Mouradov et al., 2002). In commercial wheat cultivation, an optimal timing of anthesis is desirable, because it is helpful for wheat to avoid high temperatures during its critical developmental stage. Overexpression of tae-miR408 led to earlier wheat heading by nearly 1 week due to the resultant down-regulation of TaTOC1 genes expression. This might be helpful for genetic manipulation of heading time in commercially important crops. Our further analysis revealed that TaFT1, the key gene involved in flowering time, was upregulated in both tae-miR408 overexpressing and TaTOC1s knockdown transgenic wheat, while the expression of TaCO1 was reduced. This finding seems to conflict with the idea that the function activation cascade from CO to FT is conserved in Arabidopsis and barley (Turner et al., 2005). However, Shaw et al. (2012) have reported that there is a feedback effect that reduces TaCO1 expression once TaFT1 is activated to prevent a runaway flowering effect. This provides a reasonable explanation for the gene expression profile of the three TaTOC1 genes in wheat that we identified in our current study. Taken together, our present data suggest that the tae-miR408 regulation of wheat heading time is dependent on its TaTOC1s mRNA cleavage activity and enhances the expression of TaFT1.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wheat (Triticum aestivum L. cv. Lunxuan987) was grown in a field at the experimental station of Shandong Agricultural University (Taian, Shandong, China). Immature seeds were collected at 13-14 d after pollination, and the immature embryos of 1.0-1.2 mm in size were harvested for transformation. Three T5 TaTOC1-RNAi lines (GY134, GY140, and GY143) and three T3 tae-miR408 OE lines (Gu100, Gu105, and Gu107) were selected for further analysis. These lines are single-copy-integrated transgenic plants and genetically stable. For phenotype examination, wheat seedlings after germination were also vernalized at 4°C for 4 weeks and grown in a growth chamber under LD (16 h light/8 h dark) and SD (10 h light/14 h dark) conditions at 25 to 26°C. Transgenic wheat and control plants were also grown in the experimental field under natural conditions. For gene expression analysis in different organs of wheat plants, the plants were grown under SL (12 h light/12 h dark) conditions for 15 d (about Z1.5) and 90 d (about Z3.5), respectively. The apices, young roots, and young leaves were harvested from 15-d-old seedlings. Other tissues, including internodes, nodes, leaf sheaths, mature roots, mature leaves, and spikelets, were sampled from 90-d-old plants. For rhythmic expression analysis, wheat seedlings were grown under various light-dark cycle conditions (LD, SD, and SL) for 14 d. Leaves from LD or SD plants were collected at Zeitgeber time 0 (ZT0, light on 6:00 am) and at 4-h intervals thereafter over a 44-h period. Seedlings grown in SL for 14 d were transferred to continuous light and continuous dark in the morning (ZT0), respectively. Collection of leaf materials were started at 6:00 am (ZT24) on the fifteenth day, and sampling was carried out every 4 h over a 48-h period. All plant tissues were immediately frozen in liquid nitrogen and stored at -80°C until used.
Wild-type Arabidopsis (Arabidopsis thaliana; ecotype C24) seeds were surface-sterilized and kept for 2 d at 4°C in the dark and then transferred to 16-h-light/8-h-dark conditions at 22°C. One week after germination, the seedlings were transplanted to vermiculite and later into soil in growth chambers at 20-22°C under LD or SD conditions. Plants with six rosette leaves were harvested for gene expression analysis. For the cotransformation assay, Nicotiana benthamiana plants were cultivated in a growth chamber under LD conditions at 26°C.
DNA Isolation and RNA Extraction
Genomic DNA for each sample was isolated from the wheat leaves using the CTAB method (Saghai-Maroof et al., 1984). Total RNA was extracted from the plants tissues using the Trizol reagent (Ambion, Cat. No. 15596-026) and treated with RNase-free DNaseI (NEB, Cat. No. M0303S) for purification.
RT-PCR and qRT-PCR Analysis
Two micrograms (μg) of RNA per sample was used to synthesize first-strand cDNAs using the Superscript II First-Strand Synthesis Kit (Invitrogen, Cat. No. 11904-018). RT-PCR was used to analyze the transcript levels of TaTOC-A1, TaTOC-B1, and TaTOC-D1 in various wheat organs and the expression patterns of these genes under different day-length conditions. The wheat TaActin gene was used as internal references controls to normalize the expression of the genes of interest.
The qRT-PCR amplifications were performed with each cDNA dilution using a SYBR Green Master mix with Chromo4 in accordance with as described in the manufacturer’s protocol (Bio-Rad Laboratories). The amplification efficiency for each primer pair was determined by a dilution series of cDNA samples with Ct value related to the logarithm of the dilution factor and the slope of the line of best fit used to measure reaction efficiency [efficiency = -1+10(-1/slope)]. The RNA accumulation for the genes in each sample was normalized to that of TaActin, and the measurements were performed using three biological replicates. The comparative CT method, the means, and the standard deviations were used to calculate and analyze the results.
The primers used in the RT-PCR and qRT-PCR reactions are listed in Supplemental Table S3.
Constructs and Plant Transformation
The 463-bp TaTOC1s specific fragment was amplified with a specific primer pair (TaTRNi-F and TaTRNi-R; Supplemental Table S3). The amplicons were then double digested with each of two restriction enzyme combinations (SpeI and SacI, KpnI and BamHI) and fused in both sense and antisense orientations to flank the 478-bp rice SBEI gene intron 9 contained within the pTCK303-RNAi vector. This recombinant DNA was then digested with SacI and ligated into the pZP211 vector to generate the Pubi:TaTOC1-RNAi construct. To generate the tae-miR408-overexpressing transformation vector, the sequence encompassing the 227-bp tae-MIR408b stem-loop sequence was amplified with the primers tae-MIR408bF1 and tae-MIR408bR1 (Supplemental Table S3). The amplifications were double digested with BamHI and KpnI and cloned downstream of the ubiquitin promoter in the pZP211 vector (designated as Pubi:tae-MIR408b).
The full-length cDNAs of TaTOC-A1, TaTOC-B1, and TaTOC-D1 were amplified with specific primers (TaTOC-A1F and TaTOC-A1R, TaTOC-B1F and TaTOC-B1R, TaTOC-D1F and TaTOC-D1R, respectively) and cloned into the pMD18-T vector (TaKaRa, Cat. No. D101A). The antisense inserts were selected, digested with XbaI and SpeI, and ligated into the pCAM-HA-SDIR vector. The gene of interest was controlled by the CaMV35S promoter, and these vectors were designated as P35S:TaTOC-A1, P35S:TaTOC-B1, P35S:TaTOC-D1, respectively. The 227-bp tae-MIR408b fragment was amplificated with the primer pair tae-MIR408b-F2 and tae-MIR408b-R2. The amplicons were cloned into the pMD18-T vector, and the recombined DNAs in the antisense orientation were digested with XbaI and SalI and subcloned into the pBI121 vector. The new recombinant vector was denoted P35S:tae-MIR408b.
The Pubi:TaTOC1-RNAi and Pubi:tae-MIR408 plasmids were transformed into the hexaploid wheat (cv. lunxuan987) via Agrobacterium-mediated transformation as described by Tao et al. (2011). The TaTOC1 knockdown (referred to as TaTOC1-Ri) transgene was detected by a positive PCR product (478 bp) amplified with specific to pTCK303-D, located in the rice intron region and TaTRNi-R, in the TaTOC1 coding region. The presence of the tae-miR408 overexpression transgene was also verified by a PCR using the primers pTCK303-F, located in the rice intron region, and tae-miR408-F that corresponds to a region in the tae-MIR408b. Transgenic Arabidopsis plants harboring P35S:TaTOC-A1, P35S:TaTOC-B1, P35S:TaTOC-D1, P35S:tae-MIR408 were separately generated using the floral dipping method (Clough and Bent, 1998).
In Situ Hybridization Analysis
The shoot apexes, young inflorescences, and mature leaves were harvested from the wheat plants under analysis and fixed in formaldehyde-acetic acid overnight at 4°C. Following dehydration, the fixed tissues were embedded in Paraplast (Sigma-Aldrich) and sectioned at 8 μm. Inserts for antisense and sense RNA probes for the 436-bp specific cDNAs of TaTOC-A1, TaTOC-B1, and TaTOC-D1 were inserted into the pGEM-T easy vector (Promega, Cat. No. A1360) and synthesized in vitro with digoxigenin-UTP using SP6 and T7 RNA polymerases (digoxigenin RNA labeling kit; Boehringer Mannheim). The probes were used in subsequent hybridizations according to the detailed procedures, as previously described by Zhao et al. (2006). The DNA templates for these probes were amplified with the TaTRNi-F and TaTRNi-R primers (Supplemental Table S3).
Southern Blotting
Genomic DNA from hexaploid wheat was isolated from young leaves with the DNeasy Plant Mini Kit (QIAGEN, Cat. No. 69106) and digested with BamHI and HindIII. Fifteen micrograms of this digested genomic DNA were loaded per lane onto 0.8% (w/v) agarose gels and separated by electrophoresis. The resolved genomic DNA was then transferred to the positively charged nylon membranes (Hybond-N+, Amersham Pharmacia Biotech) using a model 785 vacuum blotter system (Bio-Rad). Gene-specific probes were synthesized from 436-bp (for TaTOC1) and 227-bp (for tae-MIR408) fragments, respectively, using a DIG high primer DNA labeling and detection starter kit II (Roche, Cat. No. 11 585 614 910). The probes were purified using a high pure PCR product purification kit (Roche, Cat. No. 11 732 668 001). The DNA blots were prehybridized at 42°C for 1 h in DIG easy hyb granule and then hybridized to denatured DIG-labeled probes for 20 h under the same conditions and buffer used for the prehybridization. The blots were then washed twice with 2× SSC and 0.1% (w/v) SDS for 15 min each and washed twice with 1× SSC and 0.1% (w/v) SDS for 15 min each. Immunological detection of the probes was carried out in accordance with the manufacturer’s instructions for DIG high primer DNA labeling and detection starter kit II.
hiTAIL-PCR and 5′ RACE Assays
To identify the upstream-regulatory sequences of three TaTOC1 genes, hiTAIL-PCR was carried out with TP-0, TP-1, and TP-2 primers specific for these genes and an abridged universal amplification primer (LAD1-4) according to the method of Liu and Chen (2007). The cDNAs were synthesized with the GSP1 primer by using M-MuLV reverse transcriptase (NEB, Cat. M0253S) and purified with DNA clean-up system (Promega, Cat. A7280). The primer pair AAP and GSP2 was used for subsequent amplification. The amplified products were diluted 40-fold and used as the template for the second round of nested-PCR using the AUAP and GSP3 primers. After these reactions, the 5ʹ RACE PCR products were separated in a 2.5% agarose gel and visualized ethidium bromide staining. The DNA fragments were gel purified and subcloned using the TOPO TA cloning Kit (Invitrogen) for sequencing. All of the primers used in this analysis are listed in Supplemental Table S3.
GUS Staining
For tobacco leaf cotransformation assays of the tae-miR408 and TaTOC1 genes, the tae-MIR408 and the full-length TaTOC1 genes containing a wild-type or mutated cleavage site for tae-MIR408 were amplified by PCR using the appropriate primers and then were cloned into T-simple vectors. After validation by sequencing, the correct clones were digested with XbaI and SmaI and inserted downstream of CaMV35S promoter in the pBI121 vector. The primers used in this assay are listed in Supplemental Table S3. The different recombinant vectors thereby generated were transferred into tobacco leaves using the method of Feng et al. (2013). Histochemical GUS staining and quantitative detection of GUS were carried out as described by Jefferson et al. (1987). All assays were performed for three independent biological replicates. Leaves were photographed using an Olympus BH-2 microscope equipped with an Olympus DP12 digital camera.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers TaActin (AF326781), TaCO1 (AB094487), TaFT1 (DQ890165), pri-tae-miR408b (BE419354), tae-MIR408a (KU712264), tae-MIR408c (KU712266), TaTOC-A1 (KU712261), TaTOC-B1 (KU712262), TaTOC-D1 (KU712263).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of the amino acid sequences of TaTOC-A1, TaTOC-B1, TaTOC-D1, and TOC1.
Supplemental Figure S2. Phylogenetic tree of deduced amino acid sequences of PRR in some plant species.
Supplemental Figure S3. Predication of miR408 Targets in a number of plant species.
Supplemental Figure S4. Mutation in TaTOC-A1, TaTOC-B1, and TaTOC-D1.
Supplemental Figure S5. Expression of AtTOC1 in transgenic Arabidopsis overexpressing tae-miR408 (tae-miR408-OE).
Supplemental Figure S6. Identification of tae-miR408 and phylogenetic analysis among miR408s in various species.
Supplemental Figure S7. Expression of TaTOC-A1 (A and D), TaTOC-B1 (B and E), and TaTOC-D1 (C and F) in transgenic wheat overexpressing tae-miR408.
Supplemental Figure S8. Comparison of the plant heights (A and C) and flage leaf angle (B and D) between the transgenic wheat overexpressing tae-miR408 (TL) and controls (CK).
Supplemental Table S1. Comparison of cDNA sequence identity (%) of TaTOC-A1, TaTOC-B1, and TaTOC-D1
Supplemental Table S2. Main regulatory motifs found within the promoter sequences of the three TaTOC1 genes.
Supplemental Table S3. Primers used in this study.
Supplementary Material
Acknowledgments
We thank all persons who generously provided plant vectors for use in this study: Qi Xie (Chinese Academy of Sciences) and Kang Chong (Chinese Academy of Sciences). We also thank the ABRC for providing the Arabidopsis mutant seeds.
Glossary
- hiTAIL-PCR
high-efficiency thermal asymmetric interlaced PCR
- LD
long-day
- miRNA
microRNA
- 5ʹ RACE
5ʹ end RACE
- qRT-PCR
quantitative RT-PCR
- RNAi
RNA interference
- RT-PCR
reverse transcription PCR
- SD
short-day
- SL
standard light
- Z
Zadoks decimal growth stage
- ZT
Zeitgeber time
Footnotes
This work was funded by the National 973 Research Program of China (2014CB138102), the National Natural Science Foundation of China (31171475), and the National 863 Project of China (2006AA10Z150).
Articles can be viewed without a subscription.
References
- Abdel-Ghany SE, Pilon M (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem 283: 15932–15945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P, Herr A, Baulcombe DC, Harberd NP (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365 [DOI] [PubMed] [Google Scholar]
- Adams S, Carré IA (2011) Downstream of the plant circadian clock: output pathways for the control of physiology and development. Essays Biochem 49: 53–69 [DOI] [PubMed] [Google Scholar]
- Alaba S, Piszczalka P, Pietrykowska H, Pacak AM, Sierocka I, Nuc PW, Singh K, Plewka P, Sulkowska A, Jarmolowski A, Karlowski WM, Szweykowska-Kulinska Z (2015) The liverwort Pellia endiviifolia shares microtranscriptomic traits that are common to green algae and land plants. New Phytol 206: 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, et al. (2003) A uniform system for microRNA annotation. RNA 9: 277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, Coupland G (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Bowman JL (2008) Evolution of plant microRNAs and their targets. Trends Plant Sci 13: 343–349 [DOI] [PubMed] [Google Scholar]
- Bartel DP. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal AK, Kohli A (2013) Cereal flag leaf adaptations for grain yield under drought: knowledge status and gaps. Mol Breed 31: 749–766 [Google Scholar]
- Boke H, Ozhuner E, Turktas M, Parmaksiz I, Ozcan S, Unver T (2015) Regulation of the alkaloid biosynthesis by miRNA in opium poppy. Plant Biotechnol J 13: 409–420 [DOI] [PubMed] [Google Scholar]
- Brenchley R, Spannagl M, Pfeifer M, Barker GL, D’Amore R, Allen AM, McKenzie N, Kramer M, Kerhornou A, Bolser D, Kay S, Waite D, et al. (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491: 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JA, Mascher M, Buluç A, Barry K, Georganas E, Session A, Strnadova V, Jenkins J, Sehgal S, Oliker L, Schmutz J, Yelick KA, et al. (2015) A whole-genome shotgun approach for assembling and anchoring the hexaploid bread wheat genome. Genome Biol 16: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan P, Xia J, Zhou X, Gao S, Zhang X, Coutino G, Vazquez F, Zhang W, Jin H (2010) siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res 38: 6883–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Feng H, Zhang Q, Wang Q, Wang X, Liu J, Li M, Huang L, Kang Z (2013) Target of tae-miR408, a chemocyanin-like protein gene (TaCLP1), plays positive roles in wheat response to high-salinity, heavy cupric stress and stripe rust. Plant Mol Biol 83: 433–443 [DOI] [PubMed] [Google Scholar]
- Feng H, Duan X, Zhang Q, Li X, Wang B, Huang L, Wang X, Kang Z (2014) The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol Plant Pathol 15: 284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Wang Y, He Y (2013) Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biol 11: e1001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajyzadeh M, Turktas M, Khawar KM, Unver T (2015) miR408 overexpression causes increased drought tolerance in chickpea. Gene 555: 186–193 [DOI] [PubMed] [Google Scholar]
- Han R, Jian C, Lv J, Yan Y, Chi Q, Li Z, Wang Q, Zhang J, Liu X, Zhao H (2014) Identification and characterization of microRNAs in the flag leaf and developing seed of wheat (Triticum aestivum L.). BMC Genomics 15: 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höck J, Meister G (2008) The Argonaute protein family. Genome Biol 9: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Jackson S (2015) Floral induction and flower formation--the role and potential applications of miRNAs. Plant Biotechnol J 13: 282–292 [DOI] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X, Jing R, Zhang C, et al. ; International Wheat Genome Sequencing Consortium (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496: 91–95 [DOI] [PubMed] [Google Scholar]
- Johansson M, Staiger D (2015) Time to flower: interplay between photoperiod and the circadian clock. J Exp Bot 66: 719–730 [DOI] [PubMed] [Google Scholar]
- Jovanović Ž, Stanisavljević N, Mikić A, Radović S, Maksimović V (2014) Water deficit down-regulates miR398 and miR408 in pea (Pisum sativum L.). Plant Physiol Biochem 83: 26–31 [DOI] [PubMed] [Google Scholar]
- Khaliq I, Ishad A, Ahsan M (2008) Awns and flag leaf contribution towards grain yield in spring wheat (Triticum aestivum L.). Cereal Res Commun 36: 65–76 [Google Scholar]
- Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W (2010) Transcriptional control of gene expression by microRNAs. Cell 140: 111–122 [DOI] [PubMed] [Google Scholar]
- Kim DH, Doyle MR, Sung S, Amasino RM (2009) Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol 25: 277–299 [DOI] [PubMed] [Google Scholar]
- Kim W, Ahn HJ, Chiou TJ, Ahn JH (2011) The role of the miR399-PHO2 module in the regulation of flowering time in response to different ambient temperatures in Arabidopsis thaliana. Mol Cells 32: 83–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinmonth-Schultz HA, Golembeski GS, Imaizumi T (2013) Circadian clock-regulated physiological outputs: dynamic responses in nature. Semin Cell Dev Biol 24: 407–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Singh D, Kanodia P, Prabhu KV, Kumar M, Mukhopadhyay K (2014) Discovery of novel leaf rust responsive microRNAs in wheat and prediction of their target genes. J Nucleic Acids 2014: 570176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H-Q, Zhao S, Liu D, Wang J, Sun H, Zhang C, Fan H, Li D, Dong L, Tao Y, Gao C, Wu H, et al. (2013) Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496: 87–90 [DOI] [PubMed] [Google Scholar]
- Liu Q, Wang F, Axtell MJ (2014) Analysis of complementarity requirements for plant microRNA targeting using a Nicotiana benthamiana quantitative transient assay. Plant Cell 26: 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Chen Y (2007) High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43: 649–650 [DOI] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T (2002) The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol 43: 58–69 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Vaucheret H (2004) MicroRNAs: something important between the genes. Curr Opin Plant Biol 7: 120–125 [DOI] [PubMed] [Google Scholar]
- Más P. (2008) Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends Cell Biol 18: 273–281 [DOI] [PubMed] [Google Scholar]
- Más P, Kim WY, Somers DE, Kay SA (2003a) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Más P, Alabadí D, Yanovsky MJ, Oyama T, Kay SA (2003b) Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T (2000) Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol 41: 1002–1012 [DOI] [PubMed] [Google Scholar]
- McClung CR. (2001) Circadian rhythms in plants. Annu Rev Plant Physiol Plant Mol Biol 52: 139–162 [DOI] [PubMed] [Google Scholar]
- McClung CR, Gutiérrez RA (2010) Network news: prime time for systems biology of the plant circadian clock. Curr Opin Genet Dev 20: 588–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14(Suppl): S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Matsushika A, Ashikari M, Yamashino T, Mizuno T (2005) Circadian-associated rice pseudo response regulators (OsPRRs): insight into the control of flowering time. Biosci Biotechnol Biochem 69: 410–414 [DOI] [PubMed] [Google Scholar]
- Mutum RD, Balyan SC, Kansal S, Agarwal P, Kumar S, Kumar M, Raghuvanshi S (2013) Evolution of variety-specific regulatory schema for expression of osa-miR408 in indica rice varieties under drought stress. FEBS J 280: 1717–1730 [DOI] [PubMed] [Google Scholar]
- Niwa Y, Ito S, Nakamichi N, Mizoguchi T, Niinuma K, Yamashino T, Mizuno T (2007) Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol 48: 925–937 [DOI] [PubMed] [Google Scholar]
- Ozhuner E, Eldem V, Ipek A, Okay S, Sakcali S, Zhang B, Boke H, Unver T (2013) Boron stress responsive microRNAs and their targets in barley. PLoS One 8: e59543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Peters L, Meister G (2007) Argonaute proteins: mediators of RNA silencing. Mol Cell 26: 611–623 [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I, Weigel D (2011) MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16: 258–264 [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81: 8014–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Shaw LM, Turner AS, Laurie DA (2012) The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum). Plant J 71: 71–84 [DOI] [PubMed] [Google Scholar]
- Shimada S, Ogawa T, Kitagawa S, Suzuki T, Ikari C, Shitsukawa N, Abe T, Kawahigashi H, Kikuchi R, Handa H, Murai K (2009) A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. Plant J 58: 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Webb AA, Pearson M, Kay SA (1998) The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125: 485–494 [DOI] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T (2015) Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol 66: 441–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanudakis E, Jackson S (2014) The role of microRNAs in the control of flowering time. J Exp Bot 65: 365–380 [DOI] [PubMed] [Google Scholar]
- Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sun F, Guo G, Du J, Guo W, Peng H, Ni Z, Sun Q, Yao Y (2014) Whole-genome discovery of miRNAs and their targets in wheat (Triticum aestivum L.). BMC Plant Biol 14: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L-L, Yin G-X, Du L-P, Shi Z-Y, She M-Y, Xu H-J, Ye X-G (2011) Improvement of plant regeneration from immature embryos of wheat infected by Agrobacterium tumefacien. Agric Sci China 10: 317–326 [Google Scholar]
- Teotia S, Tang G (2015) To bloom or not to bloom: role of microRNAs in plant flowering. Mol Plant 8: 359–377 [DOI] [PubMed] [Google Scholar]
- Thiebaut F, Rojas CA, Grativol C, Motta MR, Vieira T, Regulski M, Martienssen RA, Farinelli L, Hemerly AS, Ferreira PC (2014) Genome-wide identification of microRNA and siRNA responsive to endophytic beneficial diazotrophic bacteria in maize. BMC Genomics 15: 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue D (1997) Photoperiodism in Plants, Ed 2 Academic Press, London, pp 355-365 [Google Scholar]
- Trindade I, Capitão C, Dalmay T, Fevereiro MP, Santos DM (2010) miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 231: 705–716 [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Wang JW. (2014) Regulation of flowering time by the miR156-mediated age pathway. J Exp Bot 65: 4723–4730 [DOI] [PubMed] [Google Scholar]
- Wang X-J, Reyes JL, Chua N-H, Gaasterland T (2004) Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol 5: R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm EP, Boulton MI, Al-Kaff N, Balfourier F, Bordes J, Greenland AJ, Powell W, Mackay IJ (2013) Rht-1 and Ppd-D1 associations with height, GA sensitivity, and days to heading in a worldwide bread wheat collection. Theor Appl Genet 126: 2233–2243 [DOI] [PubMed] [Google Scholar]
- Worland T, Snape J (2001) Genetic basis of worldwide wheat varietal improvement. The World Wheat Book: A History of Wheat Breeding. Paris, Lavoisier Publishing, pp 61-67 [Google Scholar]
- Wu F, Shu J, Jin W (2014) Identification and validation of miRNAs associated with the resistance of maize (Zea mays L.) to Exserohilum turcicum. PLoS One 9: e87251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Liu D, Wu J, Zhang R, Qin Z, Liu D, Li A, Fu D, Zhai W, Mao L (2013) Regulation of FLOWERING LOCUS T by a microRNA in Brachypodium distachyon. Plant Cell 25: 4363–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K, Wang R, Ou X, Fang Z, Tian C, Duan J, Wang Y, Zhang M (2012) OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One 7: e30039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Abe M (2012) Regulation of reproductive development by non-coding RNA in Arabidopsis: to flower or not to flower. J Plant Res 125: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M (2007) Regulation of copper homeostasis by micro-RNA in Arabidopsis. J Biol Chem 282: 16369–16378 [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103: 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q (2007) Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biol 8: R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14: 415–421 [Google Scholar]
- Zhang B, Pan X, Anderson TA (2006) Identification of 188 conserved maize microRNAs and their targets. FEBS Lett 580: 3753–3762 [DOI] [PubMed] [Google Scholar]
- Zhang H, Li L (2013) SQUAMOSA promoter binding protein-like7 regulated microRNA408 is required for vegetative development in Arabidopsis. Plant J 74: 98–109 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhao X, Li J, Cai H, Deng XW, Li L (2014) MicroRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated response to light and copper. Plant Cell 26: 4933–4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XY, Cheng ZJ, Zhang XS (2006) Overexpression of TaMADS1, a SEPALLATA-like gene in wheat, causes early flowering and the abnormal development of floral organs in Arabidopsis. Planta 223: 698–707 [DOI] [PubMed] [Google Scholar]
- Zhao XY, Liu MS, Li JR, Guan CM, Zhang XS (2005) The wheat TaGI1, involved in photoperiodic flowering, encodes an Arabidopsis GI ortholog. Plant Mol Biol 58: 53–64 [DOI] [PubMed] [Google Scholar]
- Zhou CM, Wang JW (2013) Regulation of flowering time by microRNAs. J Genet Genomics 40: 211–215 [DOI] [PubMed] [Google Scholar]
- Zhu QH, Helliwell CA (2011) Regulation of flowering time and floral patterning by miR172. J Exp Bot 62: 487–495 [DOI] [PubMed] [Google Scholar]
- Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.