The Fe-S binding monothiol GrxS15 is localized to mitochondria; its absence slows growth and respiration and increases arsenic susceptibility.
Abstract
Glutaredoxins (Grxs) are small proteins that function as oxidoreductases with roles in deglutathionylation of proteins, reduction of antioxidants, and assembly of iron-sulfur (Fe-S) cluster-containing enzymes. Which of the 33 Grxs in Arabidopsis (Arabidopsis thaliana) perform roles in Fe-S assembly in mitochondria is unknown. We have examined in detail the function of the monothiol GrxS15 in plants. Our results show its exclusive mitochondrial localization, and we are concluding it is the major or only Grx in this subcellular location. Recombinant GrxS15 has a very low deglutathionylation and dehydroascorbate reductase activity, but it binds a Fe-S cluster. Partially removing GrxS15 from mitochondria slowed whole plant growth and respiration. Native GrxS15 is shown to be especially important for lipoic acid-dependent enzymes in mitochondria, highlighting a putative role in the transfer of Fe-S clusters in this process. The enhanced effect of the toxin arsenic on the growth of GrxS15 knockdown plants compared to wild type highlights the role of mitochondrial glutaredoxin Fe-S-binding in whole plant growth and toxin tolerance.
Glutaredoxins (Grxs) are small proteins that function as oxidoreductases. Their classical function is the deglutathionylation of proteins. This involves the glutathione (GSH)-dependent reduction of mixed disulfides between protein thiols and GSH with glutathione reductase as a regeneration partner. Glutathionylation is thought to be a mechanism to protect sensitive thiols during oxidative stress and to act as a component of the redox signaling network by modulating activities of the affected proteins depending on the GSH/GSSG ratio (Ströher and Millar, 2012). Deglutathionylation can happen spontaneously or can be catalyzed by Grx. The reaction can be catalyzed using a dithiol or monothiol mechanism. Furthermore, Grx itself can reduce protein disulfides as well as the low molecular mass antioxidant dehydroascorbate (DHA).
In 2002, research on Grxs steered in a new direction, when yeast mitochondrial monothiol Grx5 was hypothesized to be involved in Fe-S cluster synthesis or repair (Rodríguez-Manzaneque et al., 2002). Soon after, the human dithiol Grx2 was proven to bind a [2Fe-2S] cluster, resulting in the formation of a Grx homodimer (Lillig et al., 2005). Since then, many more Grxs have been shown to possess the ability to bind Fe-S clusters. Based on protein sequence, Grx in eukaryotic photosynthetic organisms can be grouped into several classes (Couturier et al., 2009). Class I include Grx with the most classical functions, Class II contains only monothiol Grx and some bind a Fe-S cluster, but have limited and/or lower rates of classical Grx activities, while class III Grx are involved in higher plant-specific processes (Ströher and Millar, 2012). Most class I dithiol Grx homodimers are inactive in regards to classical Grx functions. For human Grx2, it was shown that the cluster is released under oxidative stress and the protein is then activated. In contrast, the Fe-S clusters of most class II monothiol Grxs, including human Grx5, are unstable and easily lost. This loss is a prerequisite for their function in biogenesis of Fe-S proteins, likely receiving the cluster from scaffold proteins and transferring it to recipient apoproteins (Bandyopadhyay et al., 2008). The most detailed information currently available about Grxs and iron comes from Grxs residing in the cytosol in yeast cells. Saccharomyces cerevisiae Grx3 and Grx4 are involved in the iron-dependent inhibition of two iron-responsive transcription factors, Aft1 and Aft2 (activators of ferrous transport). The molecular mechanism was revealed recently and involves the subsequent transfer of a Fe-S cluster from the Grx3 homodimer via the heterodimer between the BolA-like protein Fe repressor of activation-2 (Fra2) and Grx3 to Aft2 (Poor et al., 2014). In plant mitochondria, it is unclear if and how Grxs play a role in the Fe-S cluster assembly pathway.
Uncovering the precise role of different Grxs is very complex. Grxs are ubiquitous and their sequence is highly conserved. Family sizes vary from 5 to 35, with the larger numbers in higher plants due to the appearance of class III Grxs (Ziemann et al., 2009). Further complicating the discovery is the significant functional specification and different target protein preference and their location in different cellular organelles. Not only does evidence exist that proteins are glutathionylated in plant organelles in vivo (Dixon et al., 2005; Michelet et al., 2005; Zaffagnini et al., 2007; Leferink et al., 2009; Palmieri et al., 2010; Zaffagnini et al., 2012), raising the question if and which Grxs are present to remove this modification and regenerate modified proteins, but also essential Fe-S-cluster proteins and all components of an Fe-S cluster pathway would be required in organelles.
Arabidopsis (Arabidopsis thaliana) mitochondrial GrxS15 has appeared in a number of other studies as a protein of interest (Chew et al., 2003; Herald et al., 2003; Bandyopadhyay et al., 2008) and was even the focus of one study, which proposed that it is located in the chloroplast (Cheng, 2008). When combined together, these reports provide a confusing picture of the localization and function of this Grx and its roles in plant growth and stress tolerance. Cheng (2008) showed that GrxS15 (Grx4 was the nomenclature of the time due to high similarity to E. coli Grx4 but this was revised by Rouhier et al. [2004]) could complement the yeast grx5 strain, restoring its iron accumulation and, furthermore, proposed a role related to iron in chloroplasts. In contrast, a study published at the same time showed that GrxS15 did not bind Fe when compared to other tested monothiol Grxs (Bandyopadhyay et al., 2008). Using yeast two-hybrid analysis and a bimolecular fluorescence complementation assay in planta, monothiol Grx including GrxS15 were shown to interact and form heterodimers with BolA family members in the same respective compartment (Couturier et al., 2014). Except for GrxS15, it was shown that these heterodimers bind a labile Fe-S cluster (Dhalleine et al., 2014). Thus, both homo- and heterodimers could act as scaffold and/or carrier proteins during the maturation of Fe-S cluster proteins. Interestingly due to the absence of BolA proteins from the cytosol, the heterodimers are limited to organelles and might perform organelle specific reactions (Dhalleine et al., 2014).
Here, we have undertaken a comprehensive analysis to search for the full range of plant mitochondrial Grxs, and we have re-examined in detail the function of GrxS15 in plants by investigating its subcellular location, the impact of its removal, and its likely role in whole plant functions. We conclude that GrxS15 is the major, if not the only, Grx in mitochondria and that it plays a role somewhat analogous to the human Grx5, based on the impact on Fe-S-dependent processes in mitochondria. This has clear and measurable consequences for lipoic acid (LA)-dependent enzymes and whole plant tolerance to arsenic that target these processes.
RESULTS
Subcellular Localization of Grxs in Plant Mitochondria Identifies Only GrxS15
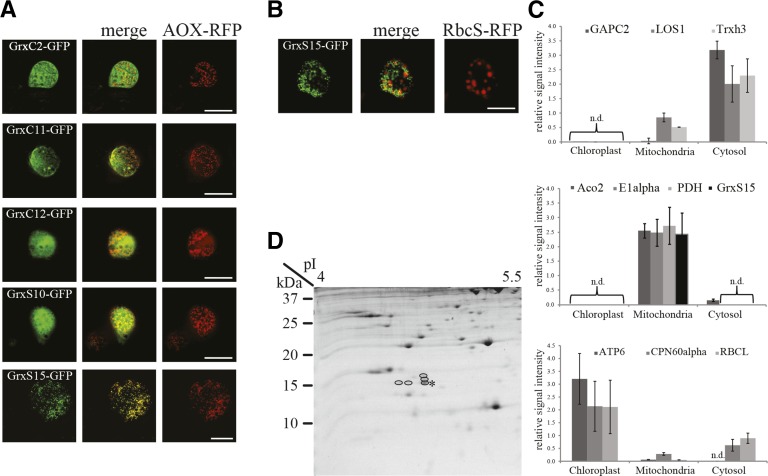
The subcellular localization of Grxs was analyzed using several different techniques. First, a targeted approach using chimeric fusions to GFP was performed. Prediction of the subcellular localization of mitochondrial Grxs was performed in silico using SUBA3 (suba3.plantenergy.uwa.edu.au). Of the 33 family members, five candidates were predicted by two or more predictors to be targeted to mitochondria and were selected to be analyzed experimentally for their subcellular localization, namely GrxC2, GrxC11, GrxC12, GrxS10, and GrxS15. GFP was fused separately to the N- and C-terminal of the full-length sequence of each Grx. Alternative oxidase, fused to red fluorescent protein (RFP), was used as a marker for mitochondrial subcellular localization. None of the N-terminal fusions showed fluorescence in cellular organelles (data not shown). The C-terminal fusion constructs showed that only fluorescence of GrxS15-GFP coincided with the fluorescence of the mitochondrial marker (Fig. 1A). All other tested Grxs showed a diffused location. Therefore, it can be concluded that under our conditions, of the Grxs tested, only GrxS15 is located in mitochondria. This confirms the results reported elsewhere using GFP and mass spectrometry (Chew et al., 2003; Herald et al., 2003; Klodmann et al., 2011; Taylor et al., 2011; Nikolovski et al., 2012). To test the claims by Cheng (2008), who presented a chloroplast location in tobacco leaves, an additional experiment was performed using the small subunit of Rubisco fused to RFP as a marker for chloroplast localization. There was no overlap with the signal emitted by the chloroplast marker for GrxS15-GFP (Fig. 1B). None of the three most recent large-scale chloroplast proteomes published could identify GrxS15 (Zybailov et al., 2008; Ferro et al., 2010; Huang et al., 2013a). To confirm this, targeted multiple reaction monitoring (MRM) assays for GrxS15 along with cytosol, chloroplast, and mitochondria peptide markers (Aebersold et al., 2013) were used (Fig. 1C). The cytosol marker peptides were detected primarily in the cytosol fraction with lower signals in the mitochondria fraction and no detection in the chloroplast fraction. Mitochondrial marker peptides were only detectable in mitochondria fractions with the exception of aconitase 2 peptides, which showed a low signal in the cytosol fraction. The majority of the chloroplast marker peptides were detected in chloroplasts, with lower signals in the cytosol fraction and even lower signal in the mitochondrial fraction. Peptides for GrxS15 were exclusively found in the mitochondria samples, closely resembling the distribution of the other mitochondrial marker peptides assayed. It can be concluded that there is no evidence of dual-targeting to the mitochondria and the chloroplast. Isolated mitochondrial proteins from an Arabidopsis cell culture line were separated using 2D-Tricine gel electrophoresis in an attempt to directly target a search for additional mitochondrial Grxs based on their small molecular mass (Fig. 1D). The 50 definable spots in the 10 to 20 kD region were analyzed for their protein identity using mass spectrometry. The only Grx identified was GrxS15. In fact, several protein spots in close proximity to the 15 kD marker and pI 4.5 were all identified with high confidence to be GrxS15 (Fig. 1D). The multiple spots likely represent post-translation modifications of this single protein.
Figure 1.
Subcellular localization of Grx. A, Expression of Grx-GFP fusion constructs in Arabidopsis cell suspension culture. GFP signal (left), mitochondrial marker AOX-RFP (right), and the merged image (middle). Bar = 20 µm. B, Expression of GrxS15-GFP fusion construct in Arabidopsis cell suspension culture. GFP signal (left), plastid marker RbcS-RFP (right), and the merged image (middle). Bar = 20 µm. C, MRM analysis using specific peptides for GrxS15 and marker proteins for cytosol (top), mitochondria (middle), and chloroplast (bottom) in cell fractions from 2-week-old wild-type leaves. Top: GAPC2 (At1g13440), LOS1 (At1g56070), Trxh3 (At5g42980). Middle: Aco2 (At2g05710), E1α (At1g59900), PDH (At1g24180), GrxS15 (At3g15660). Bottom: ATP6 (AtCg00480), CPN60α (At2g28000), RBCL (AtCg00490). D, Mitochondrial protein of green tissue separated on a 2D Tricine gel with a 4–7 pI strip as the first dimension. Spots identified as GrxS15 are circled. The unmodified form is highlighted with a star.
Molecular Characterization of the GrxS15 Mutants
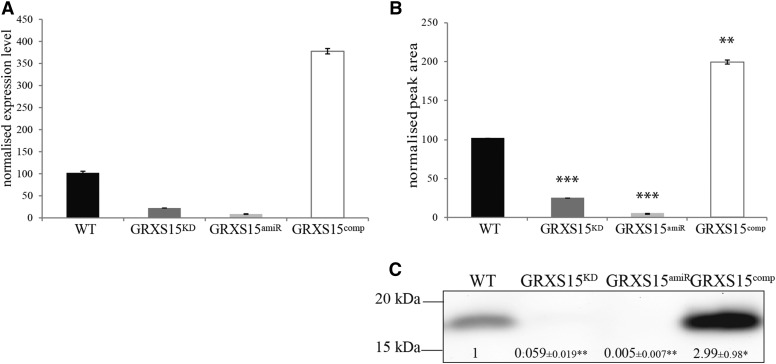
To analyze the in vivo function of GrxS15, mutant lines were studied. The only available T-DNA insertion line was Salk_112767, which is the same line studied by Cheng (2008), where it was named grx4-1. The insertion was recorded in the 3′-UTR of the gene. An independent line, GRXS15amiR, was generated in this study by artificial microRNA technology and Agrobacterium-mediated transformation. Furthermore, a complemented line, GRXS15comp, was generated using Agrobacterium-mediated transformation of Salk_112767. For both additional lines, seeds were screened for homozygous plants, and subsequently, the transcript level of GrxS15 in all four lines was analyzed using qPCR (Fig. 2A). For Salk_112767, the transcript level was reduced to 22% of the wild-type level; in GRXS15amiR, the GrxS15 transcript was < 10% of wild type, whereas in GRXS15comp, it was increased to nearly 350% compared to wild type (Fig. 2A). The GrxS15 protein content was analyzed using two independent techniques. First, antibodies were raised against Arabidopsis GrxS15, and western blot analysis indicated a reduction to < 6% and < 1% of the GrxS15 protein amount in Salk_112767 and GRXS15amiR compared to wild type (Fig. 2C; Supplemental Fig. S2), whereas the GrxS15 protein amount increased to ∼ 300% of wild type in GRXS15comp (Fig. 2C; Supplemental Fig. S2). Secondly, MRM assays for two specific peptides for GrxS15 showed a significant decrease in abundance in Salk_112767 and GRXS15amiR (23% and 3% compared to wild type) and significantly increased in abundance in GRXS15comp (197% compared to wild type; Fig. 2B). Statistical analysis showed that Salk_112767 and GRXS15amiR are also significantly different from each other (Student’s t test, P = 0.0002). Because MRM assays in general are considered to have a better linear range (Aebersold et al., 2013), it is highly likely that the latter result more accurately reflects the GrxS15 protein content in the two mutants. Based on these transcript and protein data, the Salk_112767 can be considered a knockdown of GrxS15, rather than a knockout. From here on, we refer to it as GRXS15KD to link this line to the new Grx nomenclature (Rouhier et al., 2004) and to identify it as a knockdown.
Figure 2.
GRXS15KD T-DNA insertion knockdown and phenotype. A, Relative expression level determined by qPCR of GrxS15 transcript in wild type, GRXS15KD, GRXS15amiR, and GRXS15comp. Shown is average ± SD, n = 3. B, MRM analysis using two specific peptides for GrxS15 of whole tissue mitochondrial protein in wild type, GRXS15KD, GRXS15amiR, and GRXS15comp. Peak area was normalized to the total MS2scan and standardized to wild type. Shown is median ± sem, n = 3. Student’s t test has been performed, ** indicates P < 0.001, *** indicates P < 0.0001. C, Western blot analysis with primary antibody raised against GrxS15 using isolated mitochondria from whole tissue of wild type, GRXS15KD, GRXS15amiR, and GRXS15comp. Shown is an exemplary image with values obtained from densitometry analysis of all replicates. Median and SEM are displayed, n = 4. Student’s t test has been performed, * indicates P < 0.1, ** indicates P < 0.0001.
Analysis of the GRXS15KD Mitochondrial Proteome Reveals Specific Changes in Iron-Dependent and LA-Conjugated Proteins
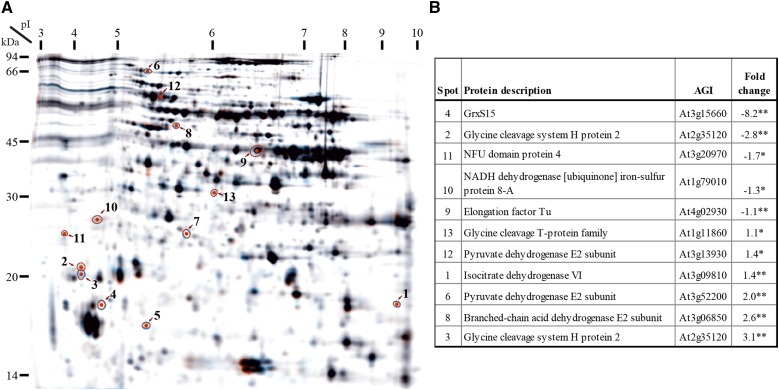
To determine the function of GrxS15, differences in protein abundance in isolated mitochondria from wild type and GRXS15KD plants were analyzed using differential in-gel electrophoresis (DIGE; Fig. 3A). The majority of proteins were unaltered in abundance in GRXS15KD, 13 protein spots were significantly up or down in abundance compared to wild type, of which 11 were identified by mass spectrometry (Fig. 3B). The spot that changed most dramatically was GrxS15 (−8.2-fold). Other spots that decreased in abundance were the NFU domain protein 4 and the NADH dehydrogenase iron-sulfur protein 8-A (−1.7-fold and −1.3-fold). The former is a scaffold for Fe-S cluster assembly of mitochondrial Fe-S proteins (Léon et al., 2003), and the latter is one of the 49 subunits of complex I in the mitochondrial respiratory chain in Arabidopsis, which is known to bind two Fe-S clusters per subunit (Schmidt-Bleek et al., 1997; Klodmann et al., 2010). The spots for other well-known Fe-S cluster proteins, e.g. Aconitase 2 and Aconitase 3 and the 24kD and 75kD subunits of Complex I, were visible on the DIGE, but the abundance did not change significantly in GRXS15KD compared to wild type. The abundance of Elongation factor Tu was slightly but significantly decreased in GRXS15KD (−1.1-fold). This protein is involved in binding aminoacyl-tRNA to the ribosomes during protein biosynthesis. It has been identified as a thioredoxin target before (Balmer et al., 2004; Yoshida et al., 2013) and has been shown to have intramolecular disulphide bonds under oxidative conditions (Winger et al., 2007). The abundance of the spot corresponding to Gly cleavage H protein 2 was decreased 2.8-fold, but the same protein was identified in a protein spot with a slightly lower apparent molecular mass, which showed a 3.1-fold increase in abundance. This is potentially caused by a protein modification that changes the mobility of this protein in the gels from GRXS15KD. H protein is one of four proteins of the Gly decarboxylase (GDC) complex where it plays an essential role acting as a mobile substrate via its lipoate group covalently bound by a Lys (Douce et al., 2001). There are three isoforms of GDC H protein in Arabidopsis. H protein 1 and H protein 3 are the most abundant forms and share 94% sequence identity, while H protein 2 is lower in abundance, shows ∼ 65% sequence identity to the other two, and migrates differently on 2D gels (Lee et al., 2008). Also, the T protein of the GDC complex was identified with increased abundance (1.1-fold) in the mutant compared to wild type, as was the catalytic subunit 6 of isocitrate dehydrogenase with 1.4-fold greater abundance compared to wild type. Other spots that increased in abundance (2-fold and 2.6-fold) were identified as proteins that belong to the pyruvate dehydrogenase (PDC), and the 2 branched-chain acid dehydrogenase (BCODC) complexes. Notably, of these complexes, only protein subunits that contained the cofactor LA were identified: two isoforms of pyruvate dehydrogenase E2 subunit (dihydrolipoyl acetyltransferase) and one isoform of the 2 branched-chain acid dehydrogenase E2 subunits (dihydrolipoyl acyltransferase). In all cases, the LA is used as a carrier for the substrate, thereby coupling the active sites of the E1 and the E3 subunit (Taylor et al., 2004). Comparison of the spot location of the two pyruvate dehydrogenase E2 subunits to other DIGE experiments performed in our lab clearly showed slightly faster migration and a shift toward the cathode compared to the reported major spot location (Lee et al., 2008), which is in line with a gain of a Lys charge due to the lacking LA cofactor. Due to the extreme pI of the H protein, the pI shift is negligible and only the faster migration can be seen in this case.
Figure 3.
Mitochondrial proteome response to GrxS15 reduction. A, DIGE 2D PAGE. Displayed is the merged image of three dyes. Spots which changed significantly between the mitochondrial proteome isolated from green tissue of wild type and GRXS15KD are circled and numbered. B, Table lists the identity of the proteins with their agi number and fold change when compared to wild type. The numbers refer to the image in A. A student’s t test was performed, ** indicates P < 0.05, and * indicates P < 0.1.
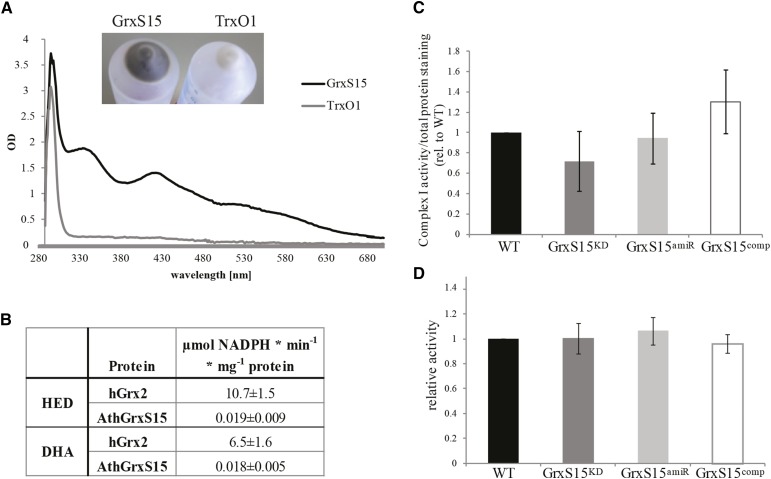
To interpret the abundance of changes in the identified iron-dependent protein set, we needed to reassess the previous claim that GrxS15 does not bind iron (Bandyopadhyay et al., 2008). Bandyopadhyay et al. (2008) measured iron binding in heterologously expressed GrxS15 alongside other monothiol Grxs, however did not take into account that several amino acids close to the GrxS15 active site are encoded by rare codons. Considering this, we used a Rosetta gami strain of E. coli, optimized for rare tRNA codon genes, to heterologously express GrxS15. Mitochondrial antioxidant protein, thioredoxin O1 (TrxO1), was used as a negative control. The E. coli suspension showed a strong brownish color only when expressing GrxS15 (Fig. 4A, insert), and when bound to the metal-chelate column material by the His-tag, the color became more obvious. Upon aerobic purification, the color was lost rapidly, but it could be stabilized in the presence of low concentrations of GSH and DTT. Spectrophotometric analysis of reduced, purified recombinant GrxS15 (but not TrxO1) showed characteristic absorbance peaks centered around 330 nm, 415 nm, and 460 nm, with a broad shoulder around 550 nm (Fig. 4A), which is characteristic for biological [2Fe-2S]2+ clusters (Dailey et al., 1994). Measurement of total complexed iron content revealed 0.78 ± 0.09 iron molecules per GrxS15 molecule for recombinant GrxS15 and no detectable Fe associated with TrxO1 (n = 3). These findings are close to values measured for other class II Grxs (Picciocchi et al., 2007; Bandyopadhyay et al., 2008; Johansson et al., 2011) and are consistent with recombinant GrxS15 binding a [2Fe-2S] cluster in a dimeric formation. To test for classical Grx activities, first, the 2-hydroxyethyl disulfide (HED) assay with commercially available human Grx2 as a control was used to test for deglutathionylation activity. The results showed that GrxS15, after removal of the Fe-S cluster, has an activity around 560 times lower than human Grx2 (Fig. 4B). Second, the ability of GrxS15 to reduce dehydroascorbate was tested, again using human Grx2 as a control. GrxS15 showed a very low activity, in fact, 360 times lower than human Grx2 (Fig. 4B). It can be concluded that GrxS15 has deglutathionylation and antioxidant reduction activity in mitochondria, but while thermodynamically possible, they are kinetically very limited compared to human Grx2. To further investigate the impact of lowering GrxS15 levels on major mitochondrial Fe-S cluster proteins, the activity of Complex I and aconitase were analyzed in isolated mitochondria of wild type, GRXS15KD, GRXS15amiR, and GRXS15comp. No statistically significant change in activity could be observed for Complex I (Fig. 4C, P > 0.2, n = 5) or aconitase (Fig. 4D, P > 0.5, n = 5).
Figure 4.
Fe-S cluster binding and activity of GrxS15. A, Spectrophotometric analysis of Fe-S cluster bound by purified heterologous expressed GrxS15 and another mitochondrial antioxidant protein thioredoxin O1 at 2.5 mg/mL. Insert shows the color of the E. coli pellet after heterologous expression of GrxS15 and TrxO1. n = 3, exemplary spectrum shown. B, Classical Grx activity assay was performed for GrxS15 and human Grx2 as comparison. HED and DHA assay were performed. n = 3; averages and sd were calculated. C, Densitometry quantification and statistical analysis of Complex I activity staining following separation of mitochondria isolated from whole tissue by Blue-Native PAGE, calculated as the ratio (complex I activity/total protein) relative to wild type. n = 5; shown is median and SEM, A student’s t test was performed, and no difference was detected. D, Aconitase activity assay using mitochondria isolated from whole tissue, calculated as the ratio of activity compared to wild type. n = 5, shown is median and SEM, A student’s t test was performed, and no difference was detected. The average wild-type aconitase activity was 94 nmol NADPH min−1 mg−1 mitochondrial protein.
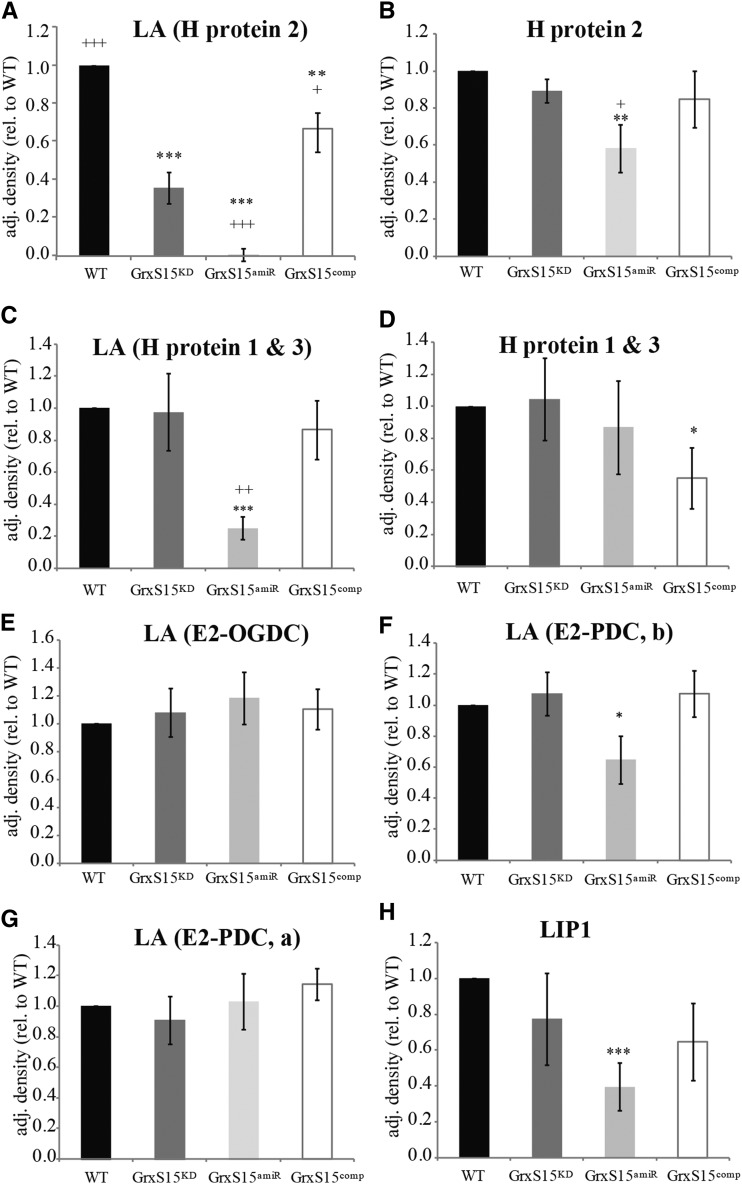
The DIGE result led us to further examine the lipoamide-containing subproteome using antibodies raised against the lipoic acid (LA) moiety (Fig. 5, A, C, and E–G; Supplemental Fig. S3A). A range of LA-containing proteins are located in plant mitochondria. Five major bands were detected after separating the protein extract on 1D-SDS-PAGE (Supplemental Fig. S3A). This pattern is in agreement with other reports and identifications published by our laboratory (Taylor et al., 2002; Taylor et al., 2004; Lee et al., 2008). No significant change could be observed for LA-bound OGDC-E2 1 and PDC-E2 1 (Fig. 5, E and G), but a significant decrease could be detected for LA-bound PDC-E2 2 and 3 (Fig. 5F) and for LA bound to H proteins 1 and 3 in GRXS15amiR compared to wild type (Fig. 5C). Both these H proteins have similar properties and consequently they are detected as one protein band. A significant decrease of LA bound to H protein 2 was observed for GRXS15KD and GRXS15amiR, however the strongest reduction was seen in GRXS15amiR (1% compared to wild type) (Fig. 5A). In GRXS15comp, the LA level is significantly raised above GRXS15KD and is restored toward the wild-type level (Fig. 5A). To confirm that the reduction in signal was due to a change in bound LA and not H protein itself, we performed western blot analysis using anti-H protein antibodies (Fig. 5, B and D; Supplemental Fig. S3B). H protein 1 and 3 was slightly reduced in GRXS15comp, whereas H protein 2 showed a slight reduction in protein level in GRXS15amiR. These results on H protein 2 confirm the DIGE results described earlier. The much greater reduction in the proportion of LA-bound protein led us to conclude that decreasing GrxS15 abundance leads to a selective decrease even among LA-bound proteins. H protein is mainly lipoylated via the mitochondrial KAS-Lip2-Lip1 pathway, while PDC and OGDC retain their lipoylation to much higher levels in the absence of mtKAS (Ewald et al., 2007). Lip1 is the mitochondrial lipoyl synthase that catalyzes the last step in the lipoylation pathway. Activity measurement of this enzyme has not been achieved in plants, but a western blot analysis using anti-Lip1 revealed lower abundance of this protein in GrxS15amiR (Fig. 5H).
Figure 5.
Impact of GrxS15 reduction on LA containing enzymes. Densitometry analysis of western blots of mitochondria isolated from whole tissue from WT, GRXS15KD, GRXS15amiR and GRXS15comp using antibodies against LA (A, C, E, F, G), H protein (B, D), and Lip1 (H). n = 4–6; shown is median and SEM; student’s t test was separately performed for all datasets against wild type, * indicates P < 0.1, ** indicates P < 0.05, and *** indicates P < 0.01, and against GRXS15KD, + indicates P < 0.1, ++ indicates P < 0.05, and +++ indicates P < 0.01.
Knockdown of GrxS15 Disrupts Plant Root Growth
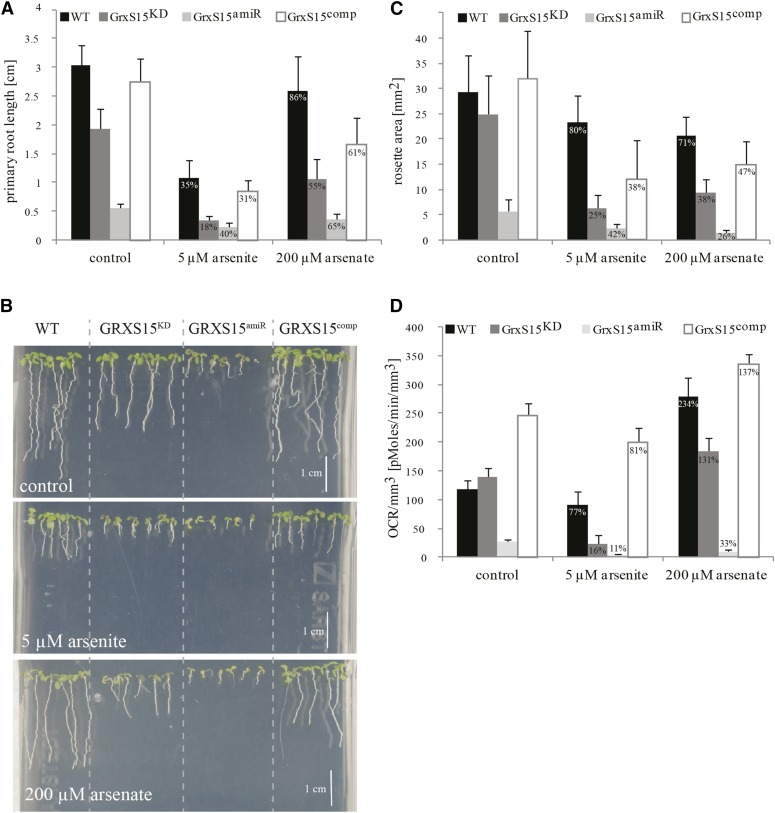
Phenotypic analysis of these lines grown on soil showed that GRXS15KD and GRXS15comp looked like the wild type, whereas the GRXS15amiR showed much reduced growth under long day growth conditions (Supplemental Fig. S1A). When grown on MS plates, horizontally and vertically, a difference in phenotype was apparent. The primary root length was significantly reduced in GRXS15KD and even more in GRXS15amiR to 63% and 18% of wild type, respectively (Fig. 6, A and B, top). However, GRXS15comp showed a recovery to 90% of the wild-type root length. A similar result for GRXS15KD alias Salk_112767 was reported before (Cheng, 2008). The rosette area was analyzed for plants grown on MS plates in short day growth conditions for 11 d. GRXS15comp plants showed wild-type-like growth, but both GRXS15KD and GRXS15amiR showed a significantly reduced rosette area, 85% and 19% of the wild type, respectively (Fig. 6C; Supplemental Fig. S1B). As the root length was affected, we measured root tip respiration, and while no significant differences were detected between wild type and GRXS15KD, GRXS15amiR showed a significantly lower respiration rate (22% compared to wild type), and GRXS15comp showed respiratory rates significantly higher than wild type (207% of wild type; Fig. 6D).
Figure 6.
Effect of Arsenic on Grx15 mutant lines, root and leaf growth, and root respiration. A, Quantification of the primary root length of GrxS15 mutant lines, grown for 9 d on vertical MS agar plates as seen in B. Percentages shown are calculated with the value measured under control conditions set as 100% for the individual line. A two-way ANOVA was performed: n = 27–30 F(6/332) = 44.13, P < 0.0005, see Supplemental Figure S6. B, Root length phenotype of 9 d old GrxS15 mutant seedlings compared to wild type on vertical MS agar plates supplemented with 5 µM arsenite and 200 mm arsenate compared to control plates. Exemplary images are shown. C, Rosette area measurements of GrxS15 mutant lines compared to wild type, measured after 11 d of growth on horizontal agar MS plates. Percentages shown are calculated with the value measured under control conditions set as 100% for the individual line. A two-way ANOVA was performed: n = 17–20, F(6/216) = 9.61, P < 0.0005, see Supplemental Figure S5. D, Oxygen consumption rate (OCR) of root tips of GrxS15 mutant lines compared to wild type when grown for 10 d on MS agar plates supplemented with arsenate and arsenite compared to control plates as in B. Percentages shown are calculated with the value measured under control conditions set as 100% for the individual line. A two-way ANOVA was performed: n = 15-39, F(6/261)=3.19, P = 0.005, see Supplemental Figure S7.
Treatment of GRXS15KD and GRXS15amiR with Arsenic Selectively Inhibited Plant Growth
A number of reports warranted the analysis of an arsenic-dependent stress phenotype. First, Grxs have been shown to be involved in arsenic metabolism, possibly via a deglutathionylation step of arsenate reductase (Mukhopadhyay et al., 2000; Martin et al., 2001); second, arsenic is known to target LA-dependent processes (Peters et al., 1946; Bergquist et al., 2009); and third, Pteris vittata L. glutaredoxin (Grx) Pv5–6 (PtGrx5) has been shown to increase arsenic tolerance in transgenic Arabidopsis plants (Sundaram et al., 2009). The root lengths of mutants were measured in response to different arsenic concentrations (Supplemental Fig. S4). For all further experiments, only 5 µM arsenite and 200 µM arsenate were used, as these concentrations caused the largest significant difference between GRXS15KD and wild type (Supplemental Fig. S4, A and B). The rosette area of plants grown on vertical plates for 11 d was measured to analyze the effect of arsenic stress on the leaves. For both chemicals, the complemented line showed partial restoration of the wild-type rosette area (Fig. 6C). In the following description, we use percentages to describe the behavior in response to the stress treatments. The values measured under control conditions in the individual lines are set to 100%, and the values measured after the stress treatments were calculated accordingly. In comparison to plants grown under control conditions, arsenite stress affected the rosette area of GRXS15KD more strongly than the other lines. The rosette area of GRXS15KD was reduced to 25%, whereas the GRXS15amiR was only reduced to 42%. However, the arsenate treatment affected GRXS15KD and GRXS15amiR by a similar magnitude, leading to a reduction to 38% and 26% of their growth under control conditions (Fig. 6C). A two-way ANOVA confirmed a significant interaction between the individual genotype and the treatments influencing the size of the rosette areas, F(6/216)= 9.61, P < 0.0005 (Supplemental Fig. S5).
After 9 d of treatment, 5 µM arsenite significantly reduced wild-type root length to 35% compared to growth under control conditions (Fig. 6, A and B). GRXS15comp showed a similar behavior. The primary root lengths of GRXS15KD were more affected compared to GRXS15amiR by this treatment. The root length of GRXS15KD was reduced to about 18% of GRXS15KD grown on control plates, whereas the GRXS15amiR was only reduced to 40%. Supplementing the MS media with 200 µM arsenate had a smaller impact on the primary root growth (Fig. 6, A and B). The arsenate treatment affected GRXS15KD slightly more than GRXS15amiR, leading to a reduction to 55% and 65% of their growth under control conditions. The highly significant interaction between genotype and treatment, affecting the root length, was confirmed by a two-way ANOVA, F(6/332) = 44.13, P < 0.0005 (Supplemental Fig. S6).
Respiration was measured in the root tips of plants grown on MS plates and normalized to the root volume used for the measurement (Fig. 6D). Treatment with 5 µM arsenite or 200 µM arsenate reduced respiration in GRXS15amiR to almost the point below detection, whereas wild type and GRXS15comp were quite tolerant to these treatments. A significant reduction in respiration in GRXS15KD was observed after treatment with 5 µM arsenite, but not 200 µM arsenate (Fig. 6D), with rates of 16% and 131% compared to the respiration rate of GrxS15KD under control condition. A two-way ANOVA revealed a significant interaction between genotype and the treatment on the respiration rates, F(6/261) = 3.19, P = 0.005 (Supplemental Fig. S7).
Based on root length assays, Cheng (2008) reported that in plants, a loss of GrxS15 led to oxidative stress sensitivity following H2O2, but not diamide treatment. In order to determine the specificity of the arsenic phenotype, a wider range of stress treatments was performed here. After 10 d of growth on vertical MS agar plates supplemented with diamide, H2O2, NaCl, and Paraquat showed no enhanced significant differences in the root phenotype of GRXS15KD compared to its growth under control conditions (Supplemental Fig. S4). It can be concluded that any observable effect is likely to be very subtle compared to the arsenic sensitivity, possibly highlighting a specificity in GrxS15 function.
DISCUSSION
Grxs in Plant Mitochondria
In mitochondria, Grxs of at least two types can be expected: (1) a Grx which could be involved in the mitochondrial Fe-S cluster pathway and (2) a Grx which can deglutathionylate target proteins, as experimental evidence has shown that deglutathionylation occurs in plant mitochondria (Dixon et al., 2005; Leferink et al., 2009; Palmieri et al., 2010). In yeast and humans, two Grxs are located in mitochondria and in both organisms: one has been described to perform deglutathionylation reactions (Lillig et al., 2005; Pedrajas et al., 2010), whereas the other has a labile Fe-S cluster and is involved in the maturation of Fe-S cluster proteins (Picciocchi et al., 2007; Johansson et al., 2011). In addition, except for dehydroascorbate reductase, all other enzymes for the functional ascorbate/GSH cycle are located in mitochondria, and it was hypothesized that a mitochondrial Grx might fulfill this function (Chew et al., 2003). Based on the results presented here, in Arabidopsis, GrxS15 is exclusively localized in mitochondria and the only one in this organelle. GrxS15 is a class II Grx and has been tested in the classical Grx activity assays. It has been shown to be able to perform deglutathionylation and DHA reduction, but very inefficiently compared to a class I Grx. This is in agreement with findings for other monothiol Fe-S cluster binding Grxs in class II (Couturier et al., 2011). Mitochondrial location, Fe-S cluster binding, and absence of oxidoreductase activity of GrxS15 have also been described in a very recent report (Moseler et al., 2015) that was published during the revision of this report. While these results uncover the involvement of GrxS15 in the Fe-S cluster machinery, it seemingly leaves the plant mitochondria with no Grx that can perform deglutathionylation effectively. The identification of other proteins that could catalyze this reaction has to be investigated further.
A Role of GrxS15 in Fe-S Biogenesis in Plant Mitochondria
Monothiol Grxs have previously been shown to be involved in the assembly and/or transfer of Fe-S clusters to apoproteins (Bandyopadhyay et al., 2008; Mühlenhoff et al., 2010; Mapolelo et al., 2013) or are proposed to be involved in the protection of Fe-S proteins (Knuesting et al., 2015). In plants, approximately 100 proteins contain Fe-S clusters (Kessler and Papenbrock, 2005; Balk and Pilon, 2011), many with essential functions in electron transfer or in catalytic cycle themselves. Mitochondria are one of the main sites of iron cluster biogenesis and harbor its own Fe-S cluster pathway, which is also required for a functional Fe-S cluster biogenesis in the cytosol (Bernard et al., 2013). Together with other proteins, e.g. BolA proteins, monothiol Grxs are proposed to be involved in the last step of the Fe-S cluster maturation pathway, transferring the Fe-S cluster to apoproteins (Balk and Schaedler, 2014). So far for plants, the involvement of a monothiol Grx, such as GrxS15, has only been speculated, mainly as the presence of a Fe-S cluster could not be shown. The data presented here reveals that recombinant GrxS15 indeed binds a Fe-S cluster and, moreover, supports the presence of a [2Fe-2S] type cluster similar to other class II Grxs. Together with our phenotypic analysis of GrxS15 knockdown plants, this supports the proposed role of GRXS15 in Fe-S dependent processes in plant mitochondria and reveals a possible mechanism.
Carrier proteins, such as members of the BolA family, have been shown to interact with monothiol Grxs, and together, they are involved in the Fe-S cluster machinery in the cytosol and mitochondria (Kumánovics et al., 2008; Poor et al., 2014). This interaction has recently been confirmed for GrxS15 and BolA4 in Arabidopsis mitochondria (Couturier et al., 2014). Interestingly, for human and yeast mitochondria, it has been described earlier that there are Fe-S cluster maturation pathways specific for particular target proteins (Cameron et al., 2011; Lill et al., 2012). BolA is essential for the transfer to only some of the apoproteins; its absence negatively affects transfer of Fe-S clusters to mitochondrial proteins, such as lipoyl synthase and complex I, but not to aconitase (Cameron et al., 2011). Plant BolA could play a similar role. In the knockdown lines GRXS15KD and/or GRXS15amiR, proteins that require LA as cofactor show reduced abundance of the holoprotein, which could reflect limited LA synthesis, which depends on the Fe-S containing protein lipoyl synthase, but no effect on abundance or activity of other classes of Fe-S cluster proteins such as aconitase (Fig. 4D) and complex I (Fig. 4C) that may not assemble via BolA/GrxS15 in plants. The novel finding that GrxS15 binds a Fe-S cluster in our study, very recently confirmed by Moseler et al. (2015), and the published finding that it interacts with BolA4 (Couturier et al., 2014), could explain this phenotype of the GRXS15KD. The interaction with BolA could provide a mechanism for Grx to target specific recipient apoproteins to transfer its Fe-S cluster, including the mitochondrial lipoyl synthase. In Arabidopsis, five NFU proteins are described: two have been shown to be in mitochondria (NFU4 and -5) and three in chloroplasts (NFU1, -2, and -3). The chloroplastic NFU2 assembles a [2Fe-2S] and a [4F-4S] cluster, and specific transfer of the former cluster to GrxS16 (but not GrxS14) and the latter cluster to adenosine 5′-phosphosulfate reductase has been shown in vitro (Gao et al., 2013). A similar scenario can be hypothesized for mitochondria, and in line with this a reduction in GrxS15 protein amount would reduce the demand for NFU4. As it is known that reduction of mitochondrial NFU specifically affects maturation of lipoyl synthase in humans (Cameron et al., 2011), this further strengthens our hypothesis of the involvement of GrxS15 in the maturation of lipoyl synthase. Recent investigation of the deficient Gly cleavage enzyme activity in human patients suffering from nonketotic hyperglycinemia identified mutations in three genes for human mitochondrial proteins, lipoate synthase, BOLA3, and GLRX5, further highlighting the tight association of the full functionality in these genes to GDC function via lipoylation (Baker et al., 2014). In addition to target protein-specific pathways, thresholds of the involved Fe-S carriers might also play an important role. Complete absence of GrxS15 was recently shown to be embryo-lethal (Moseler et al., 2015); our data show that ∼ 5% of the wild-type level is enough for survival, albeit with a smaller growth phenotype, while ∼ 20% of the wild-type level allows wild-type-like growth under control conditions (Fig. 6). This may well be linked to degrees of efficiency in Fe-S cluster transfer to specific target proteins. Complementing a GrxS15 KO with a mutated protein variant with less efficient Fe-S cluster binding led to a reduced cellular aconitase activity (Moseler et al., 2015). The absence of this phenomenon using our knockdown genotype set could be explained by the fact that the Fe-S cluster transfer to aconitase is still sufficient at ∼ 5% GrxS15 abundance, or that the mutated protein variant introduces a new dysfunction in transfer to aconitase in the absence of any wildtype GrxS15 protein.
The GrxS15 Role in Root Growth
GrxS15 shows especially high transcript expression in roots (Brady et al., 2007), in particular in the meristematic zone, which is essential for growth. Protein levels seem to follow suit and have been shown to be highest in roots and cell culture cells compared to other plant tissues and organs (Baerenfaller et al., 2008). Especially in the root meristem, more cellular energy is required to fuel active cell division. Consequently, loss of mitochondrial proteins often leads to a short root phenotype, sometimes in combination with reduced shoot growth, leading to an overall small plant. This has been shown for multiple mitochondrial proteins (Morgan et al., 2008; Han et al., 2010; van der Merwe et al., 2010; Solheim et al., 2012; Huang et al., 2013b). In addition, reduced amounts of proteins involved in mitochondrial antioxidant networks tend to also affect the root system more than the general growth of the plant, (Finkemeier et al., 2005; Miller et al., 2007; Morgan et al., 2008). Analysis of root length in the GrxS15 mutant lines showed a reduction of primary root length in GRXS15KD and GRXS15amiR compared to wild-type. This phenotype was more severe in the latter, and largely recovered in the complemented line GRXS15comp. Hence, we conclude that GrxS15 protein is required for efficient energy supply in the roots, which affects root growth. Respiration is one of the key processes for energy supply of the cell. Analysis of the mutants showed that even though under control conditions respiration in GRXS15KD was unchanged, its rate dropped significantly in the more severely reduced line GRXS15amiR. It can be concluded that the reduction of GrxS15 has a negative effect on mitochondrial respiration and, once it reaches a certain threshold, leads to a short root phenotype and impaired respiration. Based on findings here, it can be hypothesized that this could be a consequence of the incomplete LA moiety loading of important TCA cycle enzymes, most prominently the H protein (Fig. 5, B and C). It can also be speculated that once a critical GrxS15 protein threshold is reached, other Fe-S proteins in the electron transport chain could be affected too. Such a hierarchy for Fe-S incorporation has already been proposed in yeast.
GrxS15’s Role in Arsenic Stress Tolerance
Arsenic treatment led to an enhancement of the short root phenotype in the GrxS15 mutant lines. Arsenic (As) is an ubiquitous but nonessential metalloid and is an environmental concern because of its presence in drinking water and its concentration in plant tissues, which have negative impacts on plant yield and human health (Finnegan and Chen, 2012). Arsenic exists in different oxidation states, e.g. arsenate (AsV), which is an analog for inorganic phosphate and replaces it in important biochemical reactions, and arsenite (AsIII), which attacks thiol groups and can bind up to three SH groups at a time (Ida et al., 2014).
Roots are particularly involved in As reception as they are usually the first point of contact and consequently show reduced elongation and branching (Abercrombie et al., 2008). One possible reason for this could be that all reserves and energy are used to produce additional molecules for detoxification (Finnegan and Chen, 2012). Interestingly, the meristematic region in the root is also where GrxS15 is expressed the highest in planta (Brady et al., 2007). Being able to analyze the two knockdown lines with varying protein amount has been essential to understand the importance of GrxS15 under steady-state conditions, as well as under stress conditions. Overall, these results show that the GRXS15amiR line is already so severely affected by the stronger reduction of the GrxS15 protein that further exogenous stresses do not show an additional effect. GRXS15KD appears to cope under optimal conditions, but as soon as a specific exogenous stress is applied, it shows a stronger stress phenotype than wild type. Especially these two lines in combination have enabled us to analyze the in planta function of GrxS15 in more detail. There are multiple mechanisms to explain the role of mitochondrial GrxS15 in this arsenic stress tolerance. The first plant Grx was shown to be involved in arsenic metabolism was Pteris vittata L. glutaredoxin (Grx) Pv5–6 (PtGrx5) (Sundaram et al., 2008). PtGrx5 increases arsenic tolerance when overexpressed in Arabidopsis plants. The exact mechanism is not defined but might involve PtGrx5’s glutathionylation activity (Sundaram et al., 2009). In contrast, recombinant GrxS15 did not show any deglutathionylation activity. Instead, GrxS15 could be involved in direct or indirect regeneration of low molecular antioxidants upon arsenic stress, as synthesis of ascorbate, GSH, and phytochelatin is increased throughout the plant, particularly in roots (Bernard et al., 2009; Han et al., 2010; Zhou et al., 2011; Rouault, 2012). Class I Grx have been shown to regenerate DHA (Lundberg et al., 2001; Zaffagnini et al., 2008; Gao et al., 2010; Couturier et al., 2011; Riondet et al., 2012; Zaffagnini et al., 2012). However, GrxS15 has a very low DHA reductase activity and is, as other class II Grxs such as yeast Grx5, not able to catalyze this reaction effectively enough to be its main function (Tamarit et al., 2003). Previously, the function of Grx in arsenic tolerance has been linked to interconversion of the two inorganic forms, AsV and AsIII, catalyzed by arsenate reductase possibly involving a deglutathionylation step (Ellis et al., 2006; Duan et al., 2007). a much higher dose of arsenate suggests that arsenite is what is directly affecting the GrxS15 mutant lines. This would exclude a disrupted arsenate reductase catalytic cycle and point toward involvement of thiol group-containing proteins and cofactors such as LA (Bergquist et al., 2009). Not only do mitochondrial and plastid dehydrogenase complexes rely on this cofactor for their function, LA is a known target for AsIII and Lip1 is reduced in abundance in the more severe line GRXS15amiR and is known to be a Fe-S containing protein (Miller et al., 2000; Cronan, 2014), so insufficient Fe-S cluster loading might explain both the lower LA-bound targets and the apparent instability of this protein in severe knockdown of GrxS15. It can be speculated that the Fe-S cluster loading in this line might be affected to such an extent that under a LA-demanding condition (such as exposure to arsenite), the limitation of the enzyme becomes even obvious. While there are reports of in vitro assays of recombinant Lip1 from E. coli (Miller et al., 2000; Cronan, 2014), its low activity have prevented direct measurements in plant tissues (Yasuno and Wada, 1998, 2002) that would be needed to test this hypothesis of mechanism. Here, we showed that recombinant GrxS15 binds iron, and hence, its native form, like class II Grxs in other organisms, may be involved in Fe-S transfer to lipoyl synthase (Cameron et al., 2011). By this or another mechanism, plants with less GrxS15 clearly have a lower proportion of protein with bound LA. The finding that GrxS15 binds a Fe-S cluster together with proteome and phenotypical analysis presented here would suggest that GrxS15-deficient plants are more vulnerable to arsenic stress indirectly through the role of GrxS15 in iron-cluster transfer.
MATERIALS AND METHODS
Plant Lines
Seeds of Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (wild type) and T-DNA insertion line (Salk_112767) were obtained from ABRC (http://abrc.osu.edu). Standard PCR based methods were used to screen for homozygous insertion using the primers listed in Supplemental Table S1. Seeds from the same batch without insertion were used as wild-type plants in this study. They can be considered the background and closest relative to the GrxS15 line. The insertion is reported to be in the 3′UTR of GrxS15 (Cheng, 2008).
Complementation of GrxS15
The complementation of the GrxS15 mutant (Salk_112767) line was conducted by cloning the full length At3g15660 cDNA into the GATEWAY pDONR vector 207 (Thermo Fisher Scientific), verified by DNA sequencing. Subsequently, the construct was recombined into the modified binary vector pMDC43 (Curtis and Grossniklaus, 2003). This construct was introduced into Agrobacterium tumefaciens, which was then used to transform homozygous GRXS15KD using a modified floral dip method (Pracharoenwattana et al., 2005). Transformed plants were selected by germinating seedlings on agar plates containing 25 mg l−1 hygromycin. This line was named GRXS15comp.
Construction of the amiRNA Line
The silencing target sequence (TACATAATAACTTCCACTCGC, last exon of the GrxS15 coding sequence) was selected using the Web MicroRNA Designer tool (WMD3; wmd3.weigelworld.org; Ossowski et al., 2008). The primers were designed according to the instructions of the WMD3 tool, with the exception of gateway tails according to Andersen et al. (2008) (Supplemental Table S1). Cloning of the construct was done as described in the standard protocol from the WMD3 Web site. The resulting fragment and corresponding construct was treated as described above for the complementation. This line was named GRXS15amiR.
Plant Materials and Growth Conditions
Surface sterilized Arabidopsis seeds of wild-type cv Columbia, GRXS15KD, GRXS15amiR, and GRXS15comp were sown onto agar plates (1/2 strength MS medium, 2 mm MES, 1% [w/v] Suc, 1% [w/v] agar, pH 5.7). Plates were stratified for 3 d before they were moved and vertically placed in a growth chamber with short day growth conditions (8 h light/16 h dark), an irradiance of 250 µmol m−2 s−1, a relative humidity of 75%, and a temperature cycle of 22°C day/17°C night.
For growth on soil, the seeds were stratified on wet filter paper for 3 d before they were placed onto soil mixture containing compost, perlite, and vermiculite in a ratio of 3:1:1 in trays and grown under short day condition as described above.
The hydroponic seedling cultures were prepared and maintained as described in Lee et al. (2008) with slight modifications. Briefly, surface sterilized seeds were carefully laid on a stainless steel wire mesh platform, which was previously layered with 1% (w/v) agarose in a round transparent plastic vessel containing 300 mL of liquid medium (1/4 strength MS medium without vitamins, 1/4 strength Gamborg B5 vitamin solution, 2 mM MES, 1% [w/v] Suc, pH 5.8). Arabidopsis plants were grown under a 16-h/8-h light/dark period with a light intensity of 200 μmol m−2 s−1 at 22°C for 3 weeks.
For the liquid seedling culture, seeds were surface sterilized and added to a round transparent plastic vessel containing 80 ml of liquid medium (1/2 strength Murashige and Skoog medium without vitamins, 1/4 strength Gamborg B5 vitamin solution, 2 mm MES, 2.5% (w/v) Suc, pH 5.8). Plants were grown for 18 d under a 16/8-h light/dark period with light intensity of 200 μmol m−2 s−1 at 22°C on a rotary shaker at 130 rpm. The heterotrophic cell culture was maintained under the same conditions as described previously (Lee et al., 2008).
GFP Analysis
The full-length coding sequences of GrxC2, GrxC11, GrxC12, GrxS10, and GrxS15 were cloned as both N- and C-terminal GFP fusions using Gateway technology, first into pDonr207 (Thermo Fisher Scientific) and from there into pDest/pGem/CGFP (Carrie et al., 2009) under the control of the 35S CaMV promoter. Marker proteins for mitochondria and chloroplast were selected, then biolistic transformation, visualization, and image analysis were performed as described before (Carrie et al., 2009).
Isolation of Mitochondrial Proteins
Isolation of mitochondria from heterotrophic cell suspension culture, hydroponic seedling culture, and liquid culture was carried out as previously described (Taylor et al., 2014). Leaf tissue was used for the isolation of mitochondria for DIGE analysis and the whole seedling was used for the isolation of mitochondria for Complex I and aconitase activities and all western blot analyses. Samples were used directly for assays or frozen at −80°C for further analysis. Total protein quantification was performed as previously described (Law et al., 2012).
Isolation of Cytosol and Chloroplast Fractions
Cytosol and chloroplast fractions from 14 d wild-type plants grown in the hydroponic seedling culture system described above were prepared as described previously (Estavillo et al., 2014).
2D Tricine SDS Page
Isolated mitochondrial proteins from hydroponically grown wild type were separated by 2D-tricine PAGE. A total of 1.3 mg of protein was acetone precipitated and resolubilized in lysis buffer (7 m urea, 2 m thiourea, 4% [w/v] CHAPS, 40 mm Tris base, pH 8.5) and separated on IEF strips (pH 4–7, 24 cm, GE Healthcare, http://www.gelifesciences.com) according to the manufacturer’s instructions. The strips were rinsed in 1× gel buffer, equilibrated in 10 mm DTT and 125 mm IAA, and mounted on top of a tricine gel, with the following specifications: 16% T, 6% C separating gel was overlaid with a 10% T, 3% C spacer gel and a 4% T, 3% C stacking gel (Schägger, 2006). The strip was covered with 1% agarose in cathode buffer. Running conditions were 30 V until the sample entered the separating gel followed by constant 50 V for the rest of the run. Staining was performed overnight with colloidal Coomassie (G-250). Spots of interest were picked manually from the gel and further analyzed by mass spectrometry.
DIGE
DIGE 2D-PAGE using isolated mitochondrial proteins from hydroponically grown wild type and GRXS15KD leaves from three independent experiments was performed according to (Eubel et al., 2007). Fluorescent protein spots were visualized on a Typhoon laser scanner (GE Healthcare), and image comparison was done using the Delta-2D software package (Decodon, https://www.decodon.com). Matched spots of interest were picked manually from the preparative gel and further analyzed by mass spectrometry.
Mass Spectrometry
Spots were manually excised for identification. Excised gel plugs were destained and digested with trypsin as previously described (Grassl et al., 2010). Tryptic peptides were extracted in 70% ACN/0.2% formic acid, dried under vacuum, and stored at −80°C. Samples were resuspended in 5% ACN and 0.1% formic acid before MS analysis using Liquid Chromatography connected to an accurate-mass Quadrupole Time-of-Flight MS (Agilent 6510, http://www.agilent.com) equipped with a Chip Cube ion source as described previously (Grassl et al., 2012). MS/MS spectra analysis and database searching was performed as described (Lee et al., 2008).
Blue Native Page Complex I Activity Staining
Isolated mitochondria of wild type, GRXS15KD, GRXS15amiR, and GRXS15comp seedlings were separated using blue native PAGE as described previously (Eubel et al., 2005). The gel was rinsed in MilliQ water. The gel was scanned for overall protein staining, and the image was used later for the normalization. For complex I activity staining, the BN-native gel was incubated in the following staining medium: 0.1 m Tris-HCl (pH 7.4), 0.2 mm NADH, 0.2% (w/v) nitro-blue tetrazolium (Sabar et al., 2005). After 15 min, the reaction was stopped by the addition of fixing solution (40% methanol and 10% acetic acid). The gel was scanned and analyzed using Image Studio Lite (http://www.licor.com/).
Heterologous Expression of GrxS15 and TrxO1, Spectrometry, and Activity Assays for Grx
Mature proteins of GrxS15 and TrxO1, e.g. without putative mitochondrial transit peptide, were amplified using the primer combination in Supplemental Table S1. Gateway cloning into the GATEWAY pDONR vector 207 (Thermo Fisher Scientific) and subsequently into pETG-10K (a gift from A. Geerlof and EMBL laboratories, http://www.embl.de) was achieved and products were confirmed by sequencing. After transformation into Rosetta gami E. coli cells, protein expression was induced by addition of 0.4 mm IPTG at 30°C for 5 h. At the same time, 2 mm Cys, 0.2 mg/mL ferrous sulfate, 0.2 mg/mL ferric citrate, and/or 0.2 mg/mL ferric ammonium citrate was added as described elsewhere (Moseler et al., 2015). Cells were harvested and prepared for the purification using Ni-NTA agarose according to the manufacturer (Qiagen, www.qiagen.com), except that 10 mm GSH and 3 mm DTT was added to all buffers Subsequently, protein concentrations were determined as described previously (Law et al., 2012) and absorbance spectra 280 to 700nm were recorded with elution buffer as blank. For activity measurements GrxS15 was treated with 5 mm EDTA and desalted using Zeba chromatography columns (Thermo Fisher Scientific) and 50 mm NaH2PO4 (pH 8.0), 300 mm NaCl, 5% (w/v) glycerol, 5 mm GSH. HED and DHA assays were performed as described in (Holmgren and Lu, 2010). Human Grx2 (G6673, Sigma-Aldrich, http://www.sigmaaldrich.com) and GrxS15 were added in different concentrations (5 nM, 10 nM, 20 nm and 100 nM) to the mixture. Fe content in recombinant proteins were analyzed as described previously (Fish, 1988).
Aconitase Activity Assay
Aconitase activity was determined based on a modified method (MacDougall and ap Rees, 1991). Isolated mitochondria of wild type, GRXS15KD, GRXS15amiR, and GRXS15comp were used in the aconitase assay containing the following: 80 mm HEPES-NaOH (pH 7.5), 0.5 mm NADP, 0.5 mm MnCl2, 2 units NADP-isocitrate dehydrogenase, and 0.05% (v/v) Triton X-100, mM cis-aconitate. The rate change at A340 nm was monitored.
Quantitative PCR
qPCR analysis was performed on leaves from approximately 4-week-old Arabidopsis plants grown on soil. The material was collected in biological triplicates and snap-frozen under liquid nitrogen. Total RNA isolations were performed using the RNeasy plant mini kit (Qiagen) according to the manufacturer’s instructions. DNase treatment was performed using the Turbo DNA-free (AMBION, Thermo Fisher Scientific) according to the manufacturer’s instructions, except a classical ethanol precipitation rather than the Inactivation reagent was used to stop the reaction. Briefly, 70% ethanol was added to the pellet, prior to centrifugation at 15,000 rpm for 15 mins. The supernatant was removed and the pellet was dried for 5 mins at 37°C before it was resuspended in RNase-free water. One µg of total RNA was used for the cDNA synthesis using the SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. Transcript levels were analyzed using the LightCycler 480 SYBR Green I Master (Roche, http://www.roche-australia.com) and the LightCycler 480. Absolute transcript abundance measurements, determined by standard curves for each gene, were normalized to YLS8 (At5g08290), Actin2 (At3g18780), and Clatherin (At5g46630), which were analyzed as constitutive controls. qPCR primer sequences are listed in Supplemental Table S1.
Western Blotting
Anti-GrxS15 antibodies were commercially raised in rabbit, against amino acids Ser-43-Val-57 (SDSDTHDDFKPTQKV), specific to GrxS15 (Auspep, http://www.auspep.com.au). Recombinant proteins and isolated mitochondria from hydroponically grown plants (see above) were used to test the specificity of the anti-GrxS15 antibody. Anti-H protein (AS05 074, www.agrisera.com) and anti-LA (ab58724, www.abcam.com) are commercially available antibodies, and the anti-LIP1 was a kind donation from H. Wada (Yasuno and Wada, 1998). Western blot analysis was carried out against 30 μg of mitochondrial proteins for anti-GrxS15, anti-LA, and anti-LIP1, and 15 µg for anti-H protein, separated on Any kD Criterion TGX Gels, 18 well (Bio-Rad, http://www.bio-rad.com), and transferred to PVDF membrane (www.merckmillipore.com) for anti-GrxS15 or Hybond ECL membrane (http://www.gelifesciences.com) for anti-H protein, anti-LA, and anti-LIP1 using a ECL semidry blotter. Blots were probed with the following antibody combinations at specific concentrations. Detection of GrxS15, primary antibody, anti-GrxS15 1/10000; secondary antibody, HRP-conjugated goat anti-rabbit (1/20000; Thermo Fisher Scientific); detection of LA: primary antibody anti-LA 1/1000 (Abcam, http://www.abcam.com); secondary antibody, HRP-conjugated goat antirabbit (1/20000); detection of H protein, primary antibody anti-H protein 1/5000; secondary antibody, HRP-conjugated goat antirabbit (1/20000; Thermo Fisher Scientific). For all blots, ECL Prime substrate (GE Healthcare) was used, and chemiluminescence was visualized using an ImageQuant RT ECL Imager. For a loading control, total protein on the membrane was stained using Ponceau S or amido black (Sigma Aldrich) according to the manufacturer’s instructions. The images were analyzed using Image Studio Lite (http://www.licor.com/).
MRM Assay
Protein extracts from trypsin digests of either chloroplasts, mitochondria or cytosol of wild type or isolated mitochondria from whole tissue of wild type, GRXS15KD, GRXS15amiR, and GRXS15comp were analyzed as described previously (Huang et al., 2013b). Based on previous MS-Q-ToF experiments, parent ions and transitions were selected (Supplemental Table S2). A triple-quadrupole mass spectrometer (LC-QQQ-MS, Agilent Technologies 6430) was run in positive ion mode, and for each transition, the fragmentor was set to 130 and dwell time was 5 ms. A MS2Scan was performed (m/z 100 to 1000), and the resulting chromatogram was used for normalization. After normalization, the sum of all detected peptides per protein per replicate was calculated. For the standardization, this value was divided by the average of total signal for the protein. Finally, median and SEM were calculated.
MS Agar Plates
For plate growth, surface sterilized seeds were sown on 1/2 Murashige and Skoog plates containing 1% agar, 1% Suc, 1.8 mm MES with a pH at 5.8 adjusted by KOH. Arsenite and Arsenate stock solutions were prepared and filter-sterilized prior to usage. Arsenite was added in a final concentration of 5 µM, and arsenate was added in a final concentration of 200 µM. Seeds were stratified for 2 d at 4°C in the dark. Subsequently, plates were transferred to long-day controlled growth conditions as described above. For rosette area analysis, the plates were set in horizontal position, and for root length analysis as well as oxygen consumption rate measurements, the plates were set in a vertical position. Analysis was done with ImageJ, and the plugin rosette tracker (De Vylder et al., 2012) was used for the rosette area analysis.
Oxygen Consumption Rate Measurements
After 10 d of growth, 3 mm of the root tip were cut for respiration assay. The 96-well sensor cartridge was hydrated in 200 µL per well XF Calibrant Solution (Seahorse Bioscience, http://www.seahorsebio.com). To prevent the root tips from floating, 1 µL of 2.5% (v/v) Leukosan adhesive in 0.25% (w/v) agarose was pipetted onto the center of the well bottom of an additional 96-well utility plate. In each well, two root tips were placed on top of the adhesive mixture at the bottom of the well, and eight replicates for each treatment were measured. The adhesive mixture set after a few minutes, and the wells were filled with 200 µL of respiration buffer (10 mm HEPES, 10 mm MES, and 2 mm CaCl2, pH 7.2). Six cycles of mixing (2 min), waiting (3 min), and measurement (5 min) were run, and the oxygen consumption rate of the root tips was recorded by Seahorse XF Acquisition and Analysis Software (Version 1.3; Seahorse Bioscience).
Statistics
Data were analyzed using student’s t test or two-way ANOVA and Tukey's Honestly Significant Difference Method using SPSS version 21 as indicated.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL under the following accession numbers: GrxC2 (At5g40370) GrxC11 (At3g62950), GrxC12 (At2g47870), GrxS10 (At3g21460), GrxS15 (At3g15660), TrxO1 (At2g35010), human Grx2 (AF290514), Aconitase 2 (At4g26970), Aconitase 3 (At2g05710), Complex I 24kDa(At4g02580), Complex I 75kDa (At5g37510), H protein 1 (At2g35370), H protein 2 (At2g35120), H protein 3 (At1g32470), PDC-E2 1 (At3g52200), PDC-E2 2 (At3g13930), PDC-E2 3 (At1g54220), PDC-E2 4 (At3g25860), OGDC-E2 1 (At5g55070), Lip1 (At2g20860), GAPC2 (At1g13440), LOS1 (At1g56070), Trxh3 (At5g42980), E1α (At1g59900), PDH (At1g24180), ATP6 (AtCg00480), CPN60α (At2g28000), RBCL (AtCg00490).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypic analysis of WT, GrxS15KD, GrxS15amiR, and GrxS15comp.
Supplemental Figure S2. Western blot analysis of mitochondria using specific antibodies raised against GrxS15, including Ponceau staining.
Supplemental Figure S3. Western blot analysis of mitochondria using specific antibodies raised against lipoic acid and H protein, including Ponceau staining.
Supplemental Figure S4. Analysis of primary root length of WT and GRXS15KD.
Supplemental Figure S5. A two-way ANOVA with Tukey post hoc test for the rosette area data set.
Supplemental Figure S6. A two-way ANOVA with Tukey post hoc test for the root length data set.
Supplemental Figure S7. A two-way ANOVA with Tukey post hoc test for the root tip respiration data set.
Supplemental Table S1. PCR primers used in this study.
Supplemental Table S2. Transitions of Marker Peptides Monitored by MRMs.
Supplementary Material
Acknowledgments
The Lip1 antibodies were a kind donation from H. Wada, University of Tokyo. We would like to thank S. Tanz for advice on the GFP experiments.
Glossary
- GSH
glutathione
- DHA
dehydroascorbate
- LA
lipoic acid
- MRM
multiple reaction monitoring
- DIGE
differential in-gel electrophoresis
- GDC
Gly decarboxylase
- HED
2-hydroxyethyl disulfide
Footnotes
Articles can be viewed without a subscription.
References
- Abercrombie JM, Halfhill MD, Ranjan P, Rao MR, Saxton AM, Yuan JS, Stewart CN Jr (2008) Transcriptional responses of Arabidopsis thaliana plants to As (V) stress. BMC Plant Biol 8: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebersold R, Burlingame AL, Bradshaw RA (2013) Western blots versus selected reaction monitoring assays: time to turn the tables? Mol Cell Proteomics 12: 2381–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SU, Buechel S, Zhao Z, Ljung K, Novák O, Busch W, Schuster C, Lohmann JU (2008) Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 20: 88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Baker PR II, Friederich MW, Swanson MA, Shaikh T, Bhattacharya K, Scharer GH, Aicher J, Creadon-Swindell G, Geiger E, MacLean KN, Lee WT, Deshpande C, et al. (2014) Variant non ketotic hyperglycinemia is caused by mutations in LIAS, BOLA3 and the novel gene GLRX5. Brain 137: 366–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J, Pilon M (2011) Ancient and essential: the assembly of iron-sulfur clusters in plants. Trends Plant Sci 16: 218–226 [DOI] [PubMed] [Google Scholar]
- Balk J, Schaedler TA (2014) Iron cofactor assembly in plants. Annu Rev Plant Biol 65: 125–153 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Tanaka CK, Hurkman WJ, Gelhaye E, Rouhier N, Jacquot JP, Manieri W, Schürmann P, Droux M, Buchanan BB (2004) Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc Natl Acad Sci USA 101: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, Huynh BH, Herrero E, Jacquot JP, Johnson MK, Rouhier N (2008) Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J 27: 1122–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergquist ER, Fischer RJ, Sugden KD, Martin BD (2009) Inhibition by methylated organo-arsenicals of the respiratory 2-oxo-acid dehydrogenases. J Organomet Chem 694: 973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard DG, Cheng Y, Zhao Y, Balk J (2009) An allelic mutant series of ATM3 reveals its key role in the biogenesis of cytosolic iron-sulfur proteins in Arabidopsis. Plant Physiol 151: 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard DG, Netz DJ, Lagny TJ, Pierik AJ, Balk J (2013) Requirements of the cytosolic iron-sulfur cluster assembly pathway in Arabidopsis. Philos Trans R Soc Lond B Biol Sci 368: 20120259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Cameron JM, Janer A, Levandovskiy V, Mackay N, Rouault TA, Tong WH, Ogilvie I, Shoubridge EA, Robinson BH (2011) Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes. Am J Hum Genet 89: 486–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Kühn K, Murcha MW, Duncan O, Small ID, O’Toole N, Whelan J (2009) Approaches to defining dual-targeted proteins in Arabidopsis. Plant J 57: 1128–1139 [DOI] [PubMed] [Google Scholar]
- Cheng NH. (2008) AtGRX4, an Arabidopsis chloroplastic monothiol glutaredoxin, is able to suppress yeast grx5 mutant phenotypes and respond to oxidative stress. FEBS Lett 582: 848–854 [DOI] [PubMed] [Google Scholar]
- Chew O, Whelan J, Millar AH (2003) Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J Biol Chem 278: 46869–46877 [DOI] [PubMed] [Google Scholar]
- Couturier J, Wu HC, Dhalleine T, Pégeot H, Sudre D, Gualberto JM, Jacquot JP, Gaymard F, Vignols F, Rouhier N (2014) Monothiol glutaredoxin-BolA interactions: redox control of Arabidopsis thaliana BolA2 and SufE1. Mol Plant 7: 187–205 [DOI] [PubMed] [Google Scholar]
- Couturier J, Ströher E, Albetel AN, Roret T, Muthuramalingam M, Tarrago L, Seidel T, Tsan P, Jacquot JP, Johnson MK, Dietz KJ, Didierjean C, et al. (2011) Arabidopsis chloroplastic glutaredoxin C5 as a model to explore molecular determinants for iron-sulfur cluster binding into glutaredoxins. J Biol Chem 286: 27515–27527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J, Jacquot JP, Rouhier N (2009) Evolution and diversity of glutaredoxins in photosynthetic organisms. Cell Mol Life Sci 66: 2539–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE. (2014) Biotin and Lipoic Acid: Synthesis, Attachment and Regulation. Ecosal Plus 6: 10.1128/ecosalplus.ESP-0001-2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey HA, Finnegan MG, Johnson MK (1994) Human ferrochelatase is an iron-sulfur protein. Biochemistry 33: 403–407 [DOI] [PubMed] [Google Scholar]
- De Vylder J, Vandenbussche F, Hu Y, Philips W, Van Der Straeten D (2012) Rosette tracker: an open source image analysis tool for automatic quantification of genotype effects. Plant Physiol 160: 1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalleine T, Rouhier N, Couturier J (2014) Putative roles of glutaredoxin-BolA holo-heterodimers in plants. Plant Signal Behav 9: e28564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Skipsey M, Grundy NM, Edwards R (2005) Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol 138: 2233–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R, Bourguignon J, Neuburger M, Rébeillé F (2001) The glycine decarboxylase system: a fascinating complex. Trends Plant Sci 6: 167–176 [DOI] [PubMed] [Google Scholar]
- Duan GL, Zhou Y, Tong YP, Mukhopadhyay R, Rosen BP, Zhu YG (2007) A CDC25 homologue from rice functions as an arsenate reductase. New Phytol 174: 311–321 [DOI] [PubMed] [Google Scholar]
- Ellis DR, Gumaelius L, Indriolo E, Pickering IJ, Banks JA, Salt DE (2006) A novel arsenate reductase from the arsenic hyperaccumulating fern Pteris vittata. Plant Physiol 141: 1544–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo GM, Verhertbruggen Y, Scheller HV, Pogson BJ, Heazlewood JL, Ito J (2014) Isolation of the plant cytosolic fraction for proteomic analysis. Methods Mol Biol 1072: 453–467 [DOI] [PubMed] [Google Scholar]
- Eubel H, Braun HP, Millar AH (2005) Blue-native PAGE in plants: a tool in analysis of protein-protein interactions. Plant Methods 1: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H, Lee CP, Kuo J, Meyer EH, Taylor NL, Millar AH (2007) Free-flow electrophoresis for purification of plant mitochondria by surface charge. Plant J 52: 583–594 [DOI] [PubMed] [Google Scholar]
- Ewald R, Kolukisaoglu U, Bauwe U, Mikkat S, Bauwe H (2007) Mitochondrial protein lipoylation does not exclusively depend on the mtKAS pathway of de novo fatty acid synthesis in Arabidopsis. Plant Physiol 145: 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M, Brugière S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, Kieffer-Jaquinod S, Bruley C, et al. (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics 9: 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I, Goodman M, Lamkemeyer P, Kandlbinder A, Sweetlove LJ, Dietz KJ (2005) The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J Biol Chem 280: 12168–12180 [DOI] [PubMed] [Google Scholar]
- Finnegan PM, Chen W (2012) Arsenic toxicity: the effects on plant metabolism. Front Physiol 3: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish WW. (1988) Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol 158: 357–364 [DOI] [PubMed] [Google Scholar]
- Gao H, Subramanian S, Couturier J, Naik SG, Kim SK, Leustek T, Knaff DB, Wu HC, Vignols F, Huynh BH, Rouhier N, Johnson MK (2013) Arabidopsis thaliana Nfu2 accommodates [2Fe-2S] or [4Fe-4S] clusters and is competent for in vitro maturation of chloroplast [2Fe-2S] and [4Fe-4S] cluster-containing proteins. Biochemistry 52: 6633–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XH, Zaffagnini M, Bedhomme M, Michelet L, Cassier-Chauvat C, Decottignies P, Lemaire SD (2010) Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii: kinetics and specificity in deglutathionylation reactions. FEBS Lett 584: 2242–2248 [DOI] [PubMed] [Google Scholar]
- Grassl J, Pružinská A, Hörtensteiner S, Taylor NL, Millar AH (2012) Early events in plastid protein degradation in stay-green Arabidopsis reveal differential regulation beyond the retention of LHCII and chlorophyll. J Proteome Res 11: 5443–5452 [DOI] [PubMed] [Google Scholar]
- Grassl J, Scaife C, Polden J, Daly CN, Iacovella MG, Dunn MJ, Clyne RK (2010) Analysis of the budding yeast pH 4-7 proteome in meiosis. Proteomics 10: 506–519 [DOI] [PubMed] [Google Scholar]
- Han L, Qin G, Kang D, Chen Z, Gu H, Qu LJ (2010) A nuclear-encoded mitochondrial gene AtCIB22 is essential for plant development in Arabidopsis. J Genet Genomics 37: 667–683 [DOI] [PubMed] [Google Scholar]
- Herald VL, Heazlewood JL, Day DA, Millar AH (2003) Proteomic identification of divalent metal cation binding proteins in plant mitochondria. FEBS Lett 537: 96–100 [DOI] [PubMed] [Google Scholar]
- Holmgren A., and Lu J. (2010). Glutaredoxin catalysis and function in redox regulation. Mary Ann Liebert, Inc. Publications, New York. [Google Scholar]
- Huang M, Friso G, Nishimura K, Qu X, Olinares PD, Majeran W, Sun Q, van Wijk KJ (2013a) Construction of plastid reference proteomes for maize and Arabidopsis and evaluation of their orthologous relationships; the concept of orthoproteomics. J Proteome Res 12: 491–504 [DOI] [PubMed] [Google Scholar]
- Huang S, Taylor NL, Ströher E, Fenske R, Millar AH (2013b) Succinate dehydrogenase assembly factor 2 is needed for assembly and activity of mitochondrial complex II and for normal root elongation in Arabidopsis. Plant J 73: 429–441 [DOI] [PubMed] [Google Scholar]
- Ida T, Sawa T, Ihara H, Tsuchiya Y, Watanabe Y, Kumagai Y, Suematsu M, Motohashi H, Fujii S, Matsunaga T, Yamamoto M, Ono K, Devarie-Baez NO, Xian M, Fukuto JM, Akaike T (2014) Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc Natl Acad Sci USA 111: 7606–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson C, Roos AK, Montano SJ, Sengupta R, Filippakopoulos P, Guo K, von Delft F, Holmgren A, Oppermann U, Kavanagh KL (2011) The crystal structure of human GLRX5: iron-sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem J 433: 303–311 [DOI] [PubMed] [Google Scholar]
- Kessler D, Papenbrock J (2005) Iron-sulfur cluster biosynthesis in photosynthetic organisms. Photosynth Res 86: 391–407 [DOI] [PubMed] [Google Scholar]
- Klodmann J, Senkler M, Rode C, Braun HP (2011) Defining the protein complex proteome of plant mitochondria. Plant Physiol 157: 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klodmann J, Sunderhaus S, Nimtz M, Jänsch L, Braun HP (2010) Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell 22: 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesting J, Riondet C, Maria C, Kruse I, Bécuwe N, König N, Berndt C, Tourrette S, Guilleminot-Montoya J, Herrero E, Gaymard F, Balk J, et al. (2015) Arabidopsis glutaredoxin S17 and its partner, the nuclear factor Y subunit C11/negative cofactor 2α, contribute to maintenance of the shoot apical meristem under long-day photoperiod. Plant Physiol 167: 1643–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumánovics A, Chen OS, Li L, Bagley D, Adkins EM, Lin H, Dingra NN, Outten CE, Keller G, Winge D, Ward DM, Kaplan J (2008) Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J Biol Chem 283: 10276–10286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law SR, Narsai R, Taylor NL, Delannoy E, Carrie C, Giraud E, Millar AH, Small I, Whelan J (2012) Nucleotide and RNA metabolism prime translational initiation in the earliest events of mitochondrial biogenesis during Arabidopsis germination. Plant Physiol 158: 1610–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CP, Eubel H, O’Toole N, Millar AH (2008) Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol Cell Proteomics 7: 1297–1316 [DOI] [PubMed] [Google Scholar]
- Leferink NGH, van Duijn E, Barendregt A, Heck AJR, van Berkel WJH (2009) Galactonolactone dehydrogenase requires a redox-sensitive thiol for optimal production of vitamin C. Plant Physiol 150: 596–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon S, Touraine B, Ribot C, Briat JF, Lobréaux S (2003) Iron-sulphur cluster assembly in plants: distinct NFU proteins in mitochondria and plastids from Arabidopsis thaliana. Biochem J 371: 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Hoffmann B, Molik S, Pierik AJ, Rietzschel N, Stehling O, Uzarska MA, Webert H, Wilbrecht C, Mühlenhoff U (2012) The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta 1823: 1491–1508 [DOI] [PubMed] [Google Scholar]
- Lillig CH, Berndt C, Vergnolle O, Lönn ME, Hudemann C, Bill E, Holmgren A (2005) Characterization of human glutaredoxin 2 as iron-sulfur protein: a possible role as redox sensor. Proc Natl Acad Sci USA 102: 8168–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg M, Johansson C, Chandra J, Enoksson M, Jacobsson G, Ljung J, Johansson M, Holmgren A (2001) Cloning and expression of a novel human glutaredoxin (Grx2) with mitochondrial and nuclear isoforms. J Biol Chem 276: 26269–26275 [DOI] [PubMed] [Google Scholar]
- MacDougall AJ, ap Rees T (1991) Control of the Krebs Cycle in Arum Spadix. J Plant Phys 131: 683–690 [Google Scholar]
- Mapolelo DT, Zhang B, Randeniya S, Albetel AN, Li H, Couturier J, Outten CE, Rouhier N, Johnson MK (2013) Monothiol glutaredoxins and A-type proteins: partners in Fe-S cluster trafficking. Dalton Trans 42: 3107–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P, DeMel S, Shi J, Gladysheva T, Gatti DL, Rosen BP, Edwards BFP (2001) Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme. Structure 9: 1071–1081 [DOI] [PubMed] [Google Scholar]
- Michelet L, Zaffagnini M, Marchand C, Collin V, Decottignies P, Tsan P, Lancelin JM, Trost P, Miginiac-Maslow M, Noctor G, Lemaire SD (2005) Glutathionylation of chloroplast thioredoxin f is a redox signaling mechanism in plants. Proc Natl Acad Sci USA 102: 16478–16483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Rizhsky L, Hegie A, Koussevitzky S, Mittler R (2007) Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol 144: 1777–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Busby RW, Jordan SW, Cheek J, Henshaw TF, Ashley GW, Broderick JB, Cronan JE Jr., Marletta MA (2000) Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein. Biochemistry 39: 15166–15178 [DOI] [PubMed] [Google Scholar]
- Morgan MJ, Lehmann M, Schwarzländer M, Baxter CJ, Sienkiewicz-Porzucek A, Williams TC, Schauer N, Fernie AR, Fricker MD, Ratcliffe RG, Sweetlove LJ, Finkemeier I (2008) Decrease in manganese superoxide dismutase leads to reduced root growth and affects tricarboxylic acid cycle flux and mitochondrial redox homeostasis. Plant Physiol 147: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseler A, Aller I, Wagner S, Nietzel T, Przybyla-Toscano J, Mühlenhoff U, Lill R, Berndt C, Rouhier N, Schwarzländer M, Meyer AJ (2015) The mitochondrial monothiol glutaredoxin S15 is essential for iron-sulfur protein maturation in Arabidopsis thaliana. Proc Natl Acad Sci USA 112: 13735–13740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenhoff U, Molik S, Godoy JR, Uzarska MA, Richter N, Seubert A, Zhang Y, Stubbe J, Pierrel F, Herrero E, Lillig CH, Lill R (2010) Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab 12: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Shi J, Rosen BP (2000) Purification and characterization of ACR2p, the Saccharomyces cerevisiae arsenate reductase. J Biol Chem 275: 21149–21157 [DOI] [PubMed] [Google Scholar]
- Nikolovski N, Rubtsov D, Segura MP, Miles GP, Stevens TJ, Dunkley TP, Munro S, Lilley KS, Dupree P (2012) Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol 160: 1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Palmieri MC, Lindermayr C, Bauwe H, Steinhauser C, Durner J (2010) Regulation of plant glycine decarboxylase by s-nitrosylation and glutathionylation. Plant Physiol 152: 1514–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrajas J.R., Padilla C.A., McDonagh B. Barcena J.A. (2010). Glutaredoxin participates in the reduction of peroxides by the mitochondrial 1-CYS peroxiredoxin in Saccharomyces cerevisiae. Antioxid Redox Signal 13, 249–258. [DOI] [PubMed] [Google Scholar]
- Peters RA, Sinclair HM, Thompson RHS (1946) An analysis of the inhibition of pyruvate oxidation by arsenicals in relation to the enzyme theory of vesication. Biochem J 40: 516–524 [PMC free article] [PubMed] [Google Scholar]
- Picciocchi A, Saguez C, Boussac A, Cassier-Chauvat C, Chauvat F (2007) CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry 46: 15018–15026 [DOI] [PubMed] [Google Scholar]
- Poor CB, Wegner SV, Li H, Dlouhy AC, Schuermann JP, Sanishvili R, Hinshaw JR, Riggs-Gelasco PJ, Outten CE, He C (2014) Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2. Proc Natl Acad Sci USA 111: 4043–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM (2005) Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 17: 2037–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riondet C, Desouris JP, Montoya JG, Chartier Y, Meyer Y, Reichheld JP (2012) A dicotyledon-specific glutaredoxin GRXC1 family with dimer-dependent redox regulation is functionally redundant with GRXC2. Plant Cell Environ 35: 360–373 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Manzaneque MT, Tamarit J, Bellí G, Ros J, Herrero E (2002) Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell 13: 1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA. (2012) Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease. Dis Model Mech 5: 155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Jacquot JP (2004) Plant glutaredoxins: still mysterious reducing systems. Cell Mol Life Sci 61: 1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabar M, Balk J, Leaver CJ (2005) Histochemical staining and quantification of plant mitochondrial respiratory chain complexes using blue-native polyacrylamide gel electrophoresis. Plant J 44: 893–901 [DOI] [PubMed] [Google Scholar]
- Schägger H. (2006) Tricine-SDS-PAGE. Nat Protoc 1: 16–22 [DOI] [PubMed] [Google Scholar]
- Schmidt-Bleek K, Heiser V, Thieck O, Brennicke A, Grohmann L (1997) The 28.5-kDa iron-sulfur protein of mitochondrial complex I is encoded in the nucleus in plants. Mol Gen Genet 253: 448–454 [DOI] [PubMed] [Google Scholar]
- Solheim C, Li L, Hatzopoulos P, Millar AH (2012) Loss of Lon1 in Arabidopsis changes the mitochondrial proteome leading to altered metabolite profiles and growth retardation without an accumulation of oxidative damage. Plant Physiol 160: 1187–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ströher E, Millar AH (2012) The biological roles of glutaredoxins. Biochem J 446: 333–348 [DOI] [PubMed] [Google Scholar]
- Sundaram S, Rathinasabapathi B, Ma LQ, Rosen BP (2008) An arsenate-activated glutaredoxin from the arsenic hyperaccumulator fern Pteris vittata L. regulates intracellular arsenite. J Biol Chem 283: 6095–6101 [DOI] [PubMed] [Google Scholar]
- Sundaram S, Wu S, Ma LQ, Rathinasabapathi B (2009) Expression of a Pteris vittata glutaredoxin PvGRX5 in transgenic Arabidopsis thaliana increases plant arsenic tolerance and decreases arsenic accumulation in the leaves. Plant Cell Environ 32: 851–858 [DOI] [PubMed] [Google Scholar]
- Tamarit J, Belli G, Cabiscol E, Herrero E, Ros J (2003) Biochemical characterization of yeast mitochondrial Grx5 monothiol glutaredoxin. J Biol Chem 278: 25745–25751 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Day DA, Millar AH (2002) Environmental stress causes oxidative damage to plant mitochondria leading to inhibition of glycine decarboxylase. J Biol Chem 277: 42663–42668 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Millar AH (2011) The Arabidopsis thaliana 2-D gel mitochondrial proteome: Refining the value of reference maps for assessing protein abundance, contaminants and post-translational modifications. Proteomics 11: 1720–1733 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Ströher E, Millar AH (2014) Arabidopsis organelle isolation and characterization. Methods Mol Biol 1062: 551–572 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Day DA, Millar AH (2004) Lipoic acid-dependent oxidative catabolism of alpha-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol 134: 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe MJ, Osorio S, Araújo WL, Balbo I, Nunes-Nesi A, Maximova E, Carrari F, Bunik VI, Persson S, Fernie AR (2010) Tricarboxylic acid cycle activity regulates tomato root growth via effects on secondary cell wall production. Plant Physiol 153: 611–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger AM, Taylor NL, Heazlewood JL, Day DA, Millar AH (2007) Identification of intra- and intermolecular disulphide bonding in the plant mitochondrial proteome by diagonal gel electrophoresis. Proteomics 7: 4158–4170 [DOI] [PubMed] [Google Scholar]
- Yasuno R, Wada H (1998) Biosynthesis of lipoic acid in Arabidopsis: cloning and characterization of the cDNA for lipoic acid synthase. Plant Physiol 118: 935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuno R, Wada H (2002) The biosynthetic pathway for lipoic acid is present in plastids and mitochondria in Arabidopsis thaliana. FEBS Lett 517: 110–114 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Noguchi K, Motohashi K, Hisabori T (2013) Systematic exploration of thioredoxin target proteins in plant mitochondria. Plant Cell Physiol 54: 875–892 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Bedhomme M, Lemaire SD, Trost P (2012) The emerging roles of protein glutathionylation in chloroplasts. Plant Sci 185-186: 86–96 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Michelet L, Massot V, Trost P, Lemaire SD (2008) Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. J Biol Chem 283: 8868–8876 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Michelet L, Marchand C, Sparla F, Decottignies P, Le Maréchal P, Miginiac-Maslow M, Noctor G, Trost P, Lemaire SD (2007) The thioredoxin-independent isoform of chloroplastic glyceraldehyde-3-phosphate dehydrogenase is selectively regulated by glutathionylation. FEBS J 274: 212–226 [DOI] [PubMed] [Google Scholar]
- Zhou X, Li Q, Chen X, Liu J, Zhang Q, Liu Y, Liu K, Xu J (2011) The Arabidopsis RETARDED ROOT GROWTH gene encodes a mitochondria-localized protein that is required for cell division in the root meristem. Plant Physiol 157: 1793–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemann M, Bhave M, Zachgo S (2009) Origin and diversification of land plant CC-type glutaredoxins. Genome Biol Evol 1: 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q. van Wijk K.J. (2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3, e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.