Summary
Neuronal exocytosis is mediated by SNARE proteins, which assemble into a highly stable four helical bundle in a process that is not well-understood. In the present study, EPR spectroscopy was used to examine how the t-SNAREs syntaxin and SNAP25 assemble in the presence and absence of the regulatory protein Munc18-1. Syntaxin and SNAP25 form a 2:1 complex, which is structurally heterogeneous and persists in the presence of excess SNAP25. Munc18-1 dissociates this 2:1 complex, but a 1:1 complex is retained where syntaxin is in a closed state. In the absence of an N-terminal fragment of syntaxin, Munc18-1 also stabilizes a 1:1 complex of sytaxin:SNAP25; however, syntaxin now samples an open state. These data demonstrate that the open-closed syntaxin equilibrium is shifted towards the open state when syntaxin and Munc18-1 are associated with SNAP-25, and the results indicate that a syntaxin:SNAP25:Munc18-1 complex is a likely starting point for SNARE assembly.
In the central nervous system, neurotransmitter is released into the synapse by a neuronal exocytosis event that fuses synaptic vesicles to the presynaptic plasma membrane. This fusion event is driven by three soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins: syntaxin-1a and SNAP25 in the plasma membrane (t-SNAREs), and synaptobrevin in the vesicle membrane (v-SNARE). The assembly of these SNAREs results in a highly stable four helical SNARE bundle that is believed to provide the energy required to drive vesicle and target membranes together and promote fusion (Jahn and Fasshauer, 2012; Jahn and Scheller, 2006; Rizo and Sudhof, 2012; Sudhof and Rothman, 2009). A number of other proteins are critical for this fusion process, including synaptotagmin-1, a vesicle associated membrane protein that acts as the calcium sensor for synchronous vesicle fusion (Chapman, 2008), and complexin, which appears to modulate the efficiency of synchronous fusion (Brose, 2008). Munc18-1, which is an SM protein, and Munc13, which is related to the CATCHR (Complex Associated with Tethering Containing Helical Rods) family of tethering complexes, are also required for fusion (Rizo and Sudhof, 2012). These two proteins may help assemble syntaxin and SNAP25 into an acceptor complex and facilitate binding of the v-SNARE synaptobrevin.
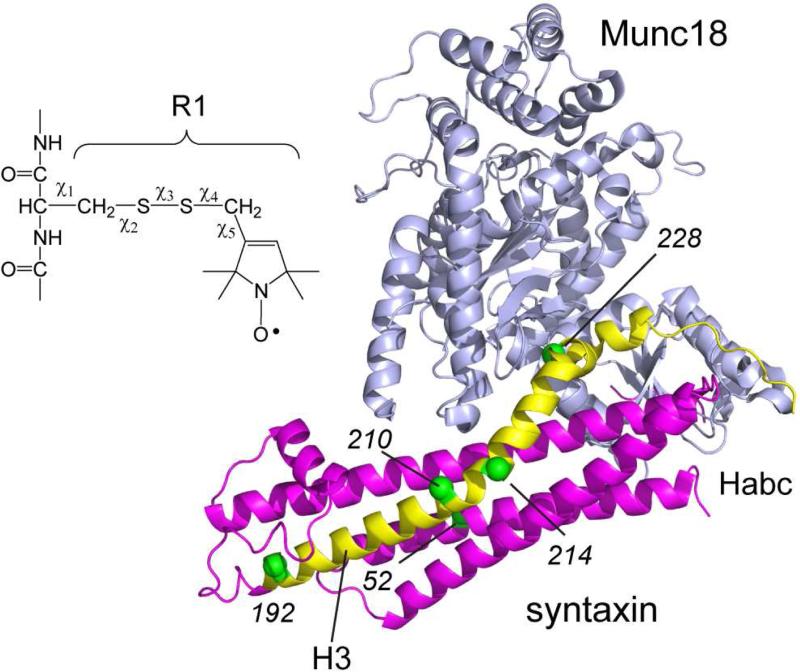
The precise role or roles of Munc18-1 have been difficult to define. While it is required for neuronal fusion, it appears to have both stimulatory and inhibitory activities towards fusion. When Munc18-1 binds to syntaxin, the regulatory Habc domain, which consists of a three-helical bundle, and the SNARE forming H3 motif, which contributes one helix to the assembled SNARE complex, are brought into close proximity as seen in Figure 1. This places syntaxin into a closed state where it is inhibited from either aggregating or assembling into the SNARE complex (Burkhardt et al., 2008). In this manner Munc18-1 appears to act as a chaperone for syntaxin (Yang et al., 2000). In addition to the binding interaction shown in Figure 1, a second interaction involves a different site on Munc18-1 interacting with the N-terminus of syntaxin. This second binding mode may allow Munc18-1 to remain associated to an intermediate or assembled SNARE complex, and when attached to the SNARE complex Munc18-1 may act to stimulate fusion (Rizo and Sudhof, 2012).
Figure 1.
Crystal structure of the complex between Munc18-1 (grey) and syntaxin. On syntaxin, the H3 segment is shown in yellow and Habc domain in red (PDB ID: 3C98) (Burkhardt et al., 2008). Derivatization of cysteine with the MTSL spin label produces the R1 side chain. The label is linked to the protein backbone through 5 rotatable bonds, although only a subset of the possible rotamers are typically seen. The α carbons of several sites to which the spin-labeled side chain R1 have been attached are shown as green spheres.
Neither the state of the t-SNAREs in the acceptor complex nor the configuration of the SNAREs in the fusion ready state is well-characterized. The sequence of events that lead to SNARE assembly is also not understood. If syntaxin and SNAP25 are mixed together, syntaxin assembles preferentially into a 2:1 (Syntaxin:SNAP25) complex that is slow to disassemble (Fasshauer et al., 1997; Stein et al., 2007). This is thought to be an off-pathway complex and it may explain why the kinetics of some in-vitro fusion experiments are slow. It has been proposed that Munc18-1 acts to dissociate syntaxin from SNAP25, resulting in a Munc18-1/syntaxin complex that is the starting point for fusion (Ma et al., 2013). From this starting point, Munc13 might act to open syntaxin and assemble the SNAREs, perhaps with the help of the protein machinery that disassembles the SNAREs, NSF and αSNAP. However, there are also indications that Munc18-1 may act with the syntaxin:SNAP25 complex to facilitate SNARE assembly (Rodkey et al., 2008; Shen et al., 2007).
In the present work, we engineered the spin-labeled side chain R1 (Figure 1) into sites in syntaxin and SNAP25 to characterize the interactions between syntaxin and SNAP25 in the presence and absence of Munc18-1. Dipolar interactions between spin labels using double electron-electron resonance (DEER) indicate the formation of a 2:1 syntaxin:SNAP25 complex. This complex is heterogeneous, which may result from different alignments of syntaxin in the complex. The addition of Munc18-1 dissociates this 2:1 complex; however, one syntaxin remains attached to SNAP25 in a closed configuration. Removal of the N-terminal segment of syntaxin does not alter the ability of Munc18-1 to dissociate the 2:1 complex, but it does promote an open state of syntaxin that is not seen in the absence of SNAP25. These results demonstrate that a syntaxin:SNAP25:Munc18-1 complex can be formed. The results also explain previous data on SNARE assembly and indicate that this t-SNARE complex is a likely starting point for SNARE assembly.
Results
Syntaxin oligomerizes in the presence of SNAP25 and forms a complex that is structurally heterogeneous
EPR spectra provide information on the local structure and dynamics at the labeled site (Columbus and Hubbell, 2002; Fanucci and Cafiso, 2006; Hubbell et al., 1998), and pulse methods such as DEER yield distances and distance distributions between pairs of spin labels (Jeschke, 2012). Using continuous wave (CW) and pulse EPR, we examined the formation and configuration of the sytaxin:SNAP25 complex using a soluble syntaxin fragment (1-262) containing the H3 (SNARE forming) and Habc (regulatory) motifs (Figure 1).
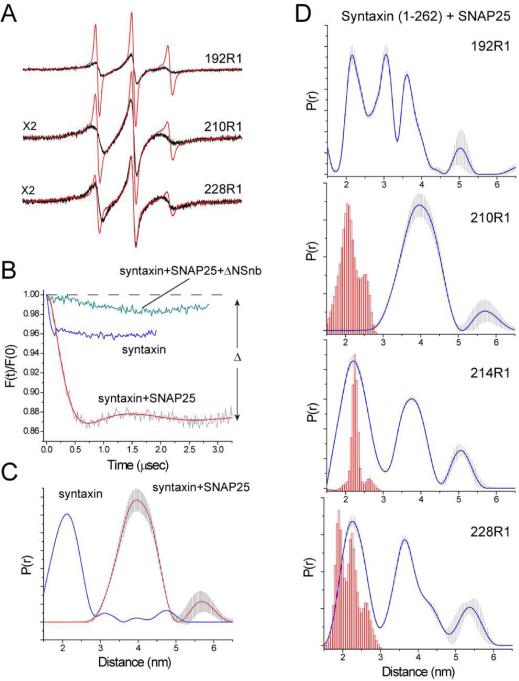
Shown in Figure 2A are EPR spectra obtained from three sites labeled within the H3 (SNARE forming) motif of syntaxin (labeled sites are indicated in Figure 1). These EPR spectra are dominated by a motional component with a short correlation time, consistent with an unstructured protein segment. In some spectra, particularly 228R1, the spectra are multicomponent and as described previously this likely reflects conformational exchange (Dawidowski and Cafiso, 2013) and/or the formation of a syntaxin dimer (Margittai et al., 2001). When SNAP25 is added to syntaxin, the correlation time for labels at these sites increases and the lines broaden. These changes are consistent with the induction of secondary structure and the formation of a complex between syntaxin and SNAP25. Syntaxin and SNAP25 are known to assemble into a 2:1 (syntaxin:SNAP25) complex, where the second syntaxin replaces synaptobrevin in a 4-helix bundle resembling the core SNARE complex (Xiao et al., 2001). The EPR spectra in Figure 2A are generally consistent with those reported previously using a shorter fragment of syntaxin lacking the Habc domain (Margittai et al., 2001, Zhang et al., 2002).
Figure 2.
EPR data showing the formation of the Syntaxin:SNAP25 2:1 complex. (A) Normalized X-band EPR spectra from several sites along the H3 motif of syntaxin in the absence (red trace) and presence (black trace) of SNAP25. EPR spectra were obtained for 20 μM syntaxin in the presence of 160 μM SNAP25 and are 100 Gauss scans. (B) Background corrected DEER data obtained for 210R1 in solution at low ionic strength (blue trace), 20 μM syntaxin in the presence of 120 μM SNAP25 (red trace) or in the presence of synaptobrevin (49-96) to form the artificial ΔN synaptobrevin acceptor complex (green trace). (C) Distance distributions for syntaxin 210R1 in solution at low ionic strength (blue trace) and syntaxin 210R1 in the presence of SNAP25 (red trace). (D) Distance distributions obtained from single labeled sites on syntaxin in the Syntaxin:SNAP25 complex. The red histograms represent distance predictions (see Experimental) based upon the core SNARE complex (Sutton et al., 1998) using rotamer libraries appropriate for exposed helical sites (Warshaviak et al., 2011). The grey error range in the distribution indicates the range of distance distributions obtained by varying the background form factor that produce fits that are within 15% of the best fit.
The formation of this complex is also observed by pulse EPR. Shown in Figure 2B are background corrected double electron-electron resonance (DEER) data for syntaxin labeled at site 210 (210R1). If there were no oligomerization, a single spin-labeled site would yield a spin-echo (F(t)/F(0)) of constant amplitude with no time-dependent oscillation or decay. However, syntaxin alone is known to dimerize in a manner that is both concentration and ionic strength dependent (Chen et al., 2008; Lerman et al., 2000; Margittai et al., 2001), and as seen Figure 2B syntaxin 210R1 yields a small dipolar evolution at low ionic strength that represents about 4% of the amplitude of the total spin echo. The signal change or modulation depth (Δ) is dependent upon the number of interacting spins, and for syntaxin 210R1 Δ is approximately 4 fold smaller than it would be for an efficiently double-labeled protein under similar experimental conditions. If SNAP25 is added to syntaxin 210R1, the modulation depth dramatically increases indicating the formation of the 2:1 complex so that two syntaxin H3 segments are interacting. The corresponding distance distributions in the absence and presence of SNAP25 are shown in Figure 2C and they indicate that the syntaxin dimer in solution is different than the dimer associated to SNAP25. Three other sites were labeled along the H3 syntaxin segment and also yield strong dipolar interactions, where the distance distributions for all four sites are shown in Figure 2D along with distance predictions for the R1 side chain (see Methods). These predictions assume that the 2:1 (Syntaxin:SNAP25) complex is identical to the 4-helical core SNARE complex (Sutton et al., 1998), where the second syntaxin occupies the same position as synaptobrevin.
As seen in Figure 2D, the distributions obtained from the syntaxin:SNAP25 complex are broad and heterogeneous. Except for site 210, there is a short distance component that matches the prediction based upon the core SNARE complex. However, each of these sites exhibits longer distances that differ significantly from the prediction. The assembled SNAREs have been reported to oligomerize (Rickman et al., 2005), and the heterogeneity seen in Figure 2d might be due to some oligomerization of the 2:1 complex. We performed two experiments to test for oligomerization. First, self-association of the 2:1 complex should be sensitive to concentration, as it is for syntaxin in solution. However, this structural heterogeneity was not altered when the concentration was varied by an order of magnitude. Second, sucrose acts as an osmolyte and it is expected to modulate aggregation and conformational states that are in equilibrium (Lopez et al., 2009). However, the distributions in Figure 2D were not affected by the addition of 30% w/v sucrose. These results suggest that the structure itself is heterogeneous, perhaps with the second syntaxin assembling into the synaptobrevin binding site in multiple ways. Taken together, the data in Figure 2D indicate that while a structure resembling the core SNARE complex is formed, additional structural forms are present in this 2:1 t-SNARE complex.
It has been reported that an artificial acceptor complex can be formed by the addition of a short synaptobrevin segment (49-96 or ΔN synaptobrevin) to the syntaxin/SNAP25 complex (Stein et al., 2007). This fragment both dissociates the 2:1 complex to form 1:1 syntaxin:SNAP25 complex, and it promotes much more efficient membrane fusion. Pulse EPR is consistent with this finding. As seen in Figure 2B, addition of the ΔN synaptobrevin fragment to the syntaxin:SNAP25 complex virtually eliminates the dipolar signal from 210R1 indicating the dissociation of the 2:1 complex.
SNAP25 gains secondary structure and folds in the presence of syntaxin
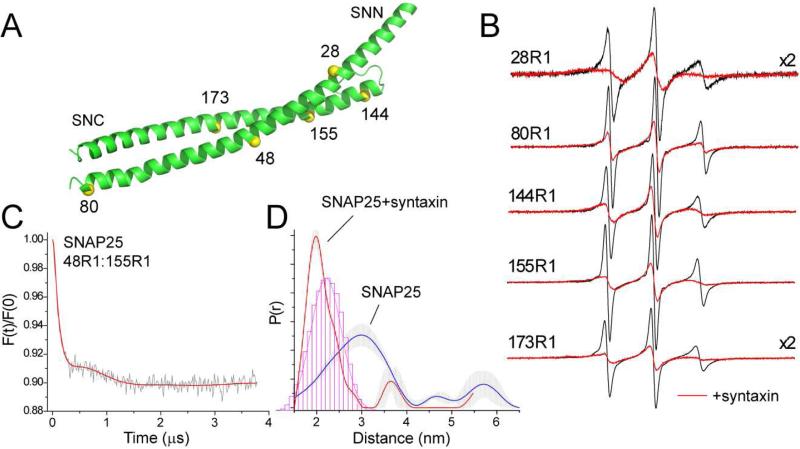
Formation of the syntaxin:SNAP25 complex may also be observed by labeling SNAP25. EPR spectra for the R1 label at 5 positions in SNAP25 are shown in Figure 3B. Except for site 28, all these sites are outward facing sites in the assembled core SNARE complex. In the absence of syntaxin these spectra have correlation times of approximately 0.5 ns, indicating that these segments are disordered and highly dynamic. Addition of syntaxin produces an increase in the label correlation time and a dramatic change in the EPR spectra. These results are similar to those obtained previously using a shorter fragment of syntaxin (Margittai et al., 2001). The spectra in Figure 3B indicate the induction of helical structure in SNAP25 upon the addition of syntaxin. The exception is 28R1 where the EPR spectrum shows evidence for tertiary contact of the label, consistent with the position of site 28 within the four-helix SNARE complex (Sutton et al., 1998).
Figure 3.
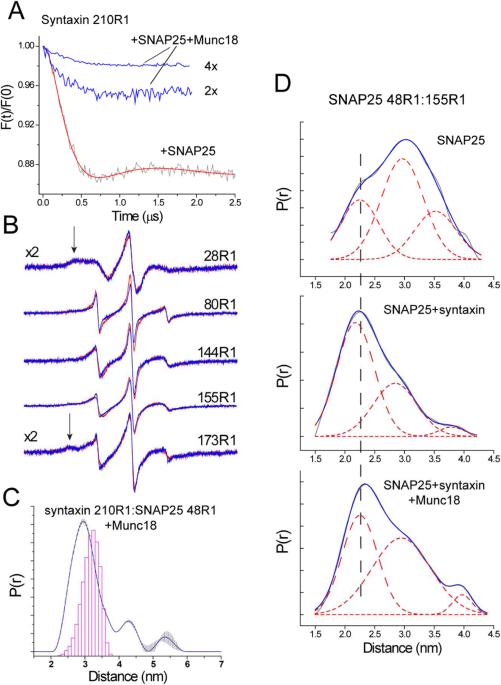
Syntaxin folds SNAP25. (A) Spin labels were placed at several positions on the N and C-terminal SNARE forming motifs of SNAP25 (SNN and SNC, respectively). (B) X-band EPR spectra of single SNAP25 labels in the absence (black trace) and presence (red trace) of syntaxin (1-262). Labeled SNAP25 is present at concentrations of 40 to 60 μM and syntaxin is present at 60 to 90 μM. (C) Distances measured between the SNN and SNC segments of SNAP25. Background corrected DEER data for 50 μM SNAP25 48R1:155R1 in the presence of 5 fold excess unlabeled SNAP25. (D) Distance distributions obtained between the SNN and SNC segments in the absence (red trace) and presence (blue trace) of syntaxin (1-262). For the syntaxin sample, labeled SNAP25 is present at 10 μM with 30 M syntaxin and 30 μM unlabeled SNAP25. The histogram (red) represents the distances expected if SNAP25 has the same configuration that it does in the core SNARE complex.
Using pulse EPR, distances were measured between 48R1 in the N-terminal SNARE-forming segment and 155R1 in the C-terminal SNARE-forming segment of SNAP25. Single labeled SNAP25 in solution produced a small dipolar signal, and excess unlabeled SNAP25 was added to the double-labeled SNAP25 to prevent any contributions due to an intermolecular dipolar coupling. The dipolar evolution obtained is shown in Figure 3C for SNAP25 alone, and the distance distributions are shown in Figure 3D in the absence and presence of syntaxin. In the absence of syntaxin, a broad distribution spanning over 20 Å is observed, where the most populated distance is approximately 30 Å. This is consistent with the spectra in Figure 3B, which indicate that SNAP25 alone is highly dynamic and largely unstructured. Addition of syntaxin dramatically narrows this distribution and produces a shorter main distance of about 20 Å. This is close to the distance expected between spin labels for SNAP25 that is assembled into the core SNARE complex. This result suggests that the two helices of SNAP25 are folded in a similar manner to that in the four helix SNARE bundle when associated with syntaxin.
Syntaxin is in an open state when bound to SNAP25, but can be closed by the addition of Munc18-1
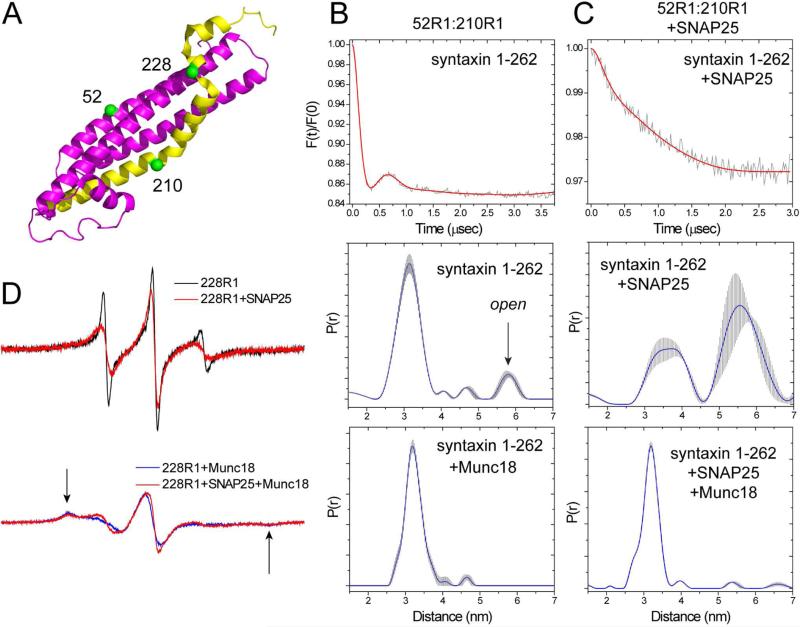
Munc18-1 has a high affinity for syntaxin, and when bound to syntaxin it acts to bring the regulatory (Habc) domain into contact with the H3 domain and close the syntaxin structure (Burkhardt et al., 2008). We determined whether Munc18-1 could close syntaxin when bound to SNAP25 using a double spin label variant where labels were placed at positions 210 in the H3 segment and 52 in the Habc segment of syntaxin (Figure 4A). Shown in Figure 4B are DEER data obtained previously for syntaxin 52R1:210R1 (Dawidowski and Cafiso, 2013). Syntaxin (1-262) is predominantly closed in solution, as indicated by the major peak around 30 Å, and there is a small population that represents an open state (middle panel, Figure 4B)). Adding Munc18-1 narrows the distance distribution and eliminates the longer distance (bottom panel, Figure 4B). Shown in Figure 4C are the DEER signals obtained for syntaxin 52R1:210R1 when bound to SNAP25 in the 2:1 complex. In this case, syntaxin 52R1:210R1 was diluted with unlabeled syntaxin to reduce the intermolecular dipolar interaction that would arise from 210R1 (as seen in Figure 2B). In contrast to syntaxin alone in in solution, the distance distribution across the H3 to the Habc domain is very broad, indicating that syntaxin is in an open configuration when bound to SNAP25. Subsequent addition of Munc18-1 dramatically narrows the distance distribution and yields a distance virtually identical to the distribution seen for syntaxin bound to Munc18-1 in solution (compare bottom panels, Figures 4B, C). Direct contact between syntaxin and Munc18-1 could be confirmed by examining EPR spectra from site 228. This site interacts in a pocket on Munc18-1 and the spin label becomes immobilized upon Munc18-1 association (Dawidowski and Cafiso, 2013). As shown in Figure 4D, the EPR spectra from syntaxin 228R1 show evidence for strong tertiary contact in the Munc18-1 bound state, regardless of whether Munc18-1 is added to syntaxin in solution or syntaxin bound to SNAP25. Thus, for syntaxin that is bound to SNAP25 in a 2:1 complex, addition of Munc18-1 binds and promotes the closed configuration of syntaxin. It should be noted that the modulation depths for the DEER data in the presence of Munc18-1 indicate that the conversion to the closed state is virtually complete either in the absence or presence of SNAP25.
Figure 4.
Determining whether syntaxin is open or closed when bound to SNAP25 or Munc18-1. (A) a pair of labels was placed at sites 52 and 210 to monitor the open and closed states of syntaxin, and the single label at position 228 is sensitive to Munc18-1 binding. (B) Background corrected DEER trace for syntaxin 52R1:210R1 (top), corresponding distance distribution (middle), and (bottom) distance distribution in the presence of Munc18-1 (data in (B) were published previously (Dawidowski and Cafiso, 2013)). (C) Background corrected DEER data for syntaxin 52R1:210R1 in the presence of SNAP25 (top), the corresponding distance distribution (middle), and the distribution obtained upon the addition of excess Munc18-1 (bottom). In both cases, syntaxin is at a concentration of 5 μM with 3 fold excess unlabeled syntaxin and 150 μM SNAP25. (D) The EPR spectra from syntaxin 228R1 are sensitive to Munc18-1 binding. Top traces: spectra of 40 μM syntaxin 228R1 in the absence (black) and presence (red) of excess SNAP25 (160 μM); bottom traces: EPR spectra of 40 μM syntaxin 228R1 following the addition of 60 μM Munc18-1 (black), and 40 μM syntaxin 228R1 when bound to 160 μM SNAP25 following the addition of 60 μM Munc18-1 (red).
Munc18-1 eliminates the syntaxin oligomer but does not completely dissociate syntaxin from SNAP25
From earlier HMQC spectra of selectively labeled syntaxin, it was concluded that Munc18-1 completely dissociated syntaxin from SNAP25 (Ma et al., 2013). Here we performed several measurements to test this conclusion. Shown in Figure 5A is a comparison of DEER signals from syntaxin 210R1 when bound to SNAP25 in the absence and presence of excess Munc18-1. When Munc18-1 is added, the modulation depth in the dipolar evolution signal is significantly attenuated, and dipolar interactions are lost between spins at site 210 in syntaxin. This indicates that Munc18-1 eliminates the syntaxin oligomer. This loss in oligomerization could either be due to a total dissociation of the Munc18-1-bound syntaxin from SNAP25 or the formation of a 1:1:1 complex of syntaxin, SNAP25 and Munc18-1.
Figure 5.
Syntaxin remains associated to SNAP25 following the addition of Munc18-1. (A) Background corrected DEER data obtained for 20 μM syntaxin 210R1 in the presence of 120 μM SNAP25 (red trace) indicates the formation of the 2:1 Syntaxin:SNAP25 complex; addition of Munc18-1 in 2 and 4 fold excess over syntaxin dissociates the 2:1 complex. (B) EPR spectra from sites on SNAP25 (40 μM) when bound to syntaxin (60 μM) in the absence (red) and presence (blue) of Munc18-1 (120 μM). The arrows indicate components due to tertiary contact of the label, which results in incompletely averaged hyperfine interactions. (C) Distance distribution obtained between syntaxin 210R1 and SNAP25 48R1 (both at 40 μM in the presence of 80 μM Munc18-1. (D) Distance distributions obtained for SNAP25 48R1/155R1 in solution (top), bound to syntaxin (1-262) (middle), and bound to syntaxin and Munc18-1 (bottom). Shown in each figure is the distribution giving the best fit to the DEER data (grey trace), and a fit to this distribution using 3 Gaussians (solid red trace and three dashed traces). Samples contain 50 μM SNAP25, 50 μM syntaxin (1-262) and 90 μM Munc18-1.
A number of observations demonstrate that Munc18-1 does not completely dissociate syntaxin from SNAP25, but that one syntaxin remains bound to SNAP25 in a closed state. First, as seen in Figure 5B, the EPR lineshapes along SNAP25 are not affected by the addition of Munc18-1 and are virtually superimposable with the spectra obtained in the presence of syntaxin. These spectra are highly sensitive to backbone dynamics and secondary structure and indicate that the ordering that takes place in SNAP25 upon syntaxin binding is retained after Munc18-1 binding. Second, the primary distance that is observed when measuring across the N- and C-terminal domains of SNAP25 does not resemble the syntaxin-free state of SNAP25 (Figure 5D). Although the distribution broadens slightly upon the addition of Munc18-1, the primary distance near 23 Å is close to that seen for the SNAP25-syntaxin complex. These data suggest that the two helices of SNAP25 are folded, but slightly more separated than they are in the 2:1 syntaxin:SNAP25 complex. Finally, pulse EPR indicates that spin labels in the H3 motif interact with spin labels in SNAP25 in the presence of Munc18-1 (Figure 5C), and that the distance distribution is close to that expected based upon the crystal structure of the core SNARE complex. Thus, Munc18-1 when added to syntaxin bound to SNAP25 dissociates the 2:1 complex, but preserves a 1:1 syntaxin:SNAP25 complex where syntaxin is in a closed state (recall Figure 4C). Thus, the result suggests that a syntaxin:Munc18-1 complex favors the SNAP25 bound state.
It should be noted that the data in Figure 4A were taken with excess Munc18-1, and that addition of 2 fold excess Munc18-1 was less effective at dissociating the 2:1 complex than 4-fold excess Munc18-1. This is in contrast to similar measurements made for syntaxin (1-262) alone in solution, where stoichiometric addition of Munc18-1 to syntaxin was sufficient to close the structure and dissociate the syntaxin oligomer (Dawidowski and Cafiso, 2013). As a result, the nM affinity observed between Munc18-1 and syntaxin does not appear to hold for syntaxin bound to SNAP25.
The N-terminus of syntaxin is required to achieve a fully closed state of syntaxin in complex with Munc18-1 and SNAP25
In addition to contacting the H3 and Habc motifs, Munc18-1 also interacts with an N-terminal segment of syntaxin (sites 2-9) at a different site (Figure 6A). This interaction accounts for about 30% of the Syntaxin:Munc18-1 binding energy (Burkhardt et al., 2008), and is believed to be important in the attachment of Munc18-1 to a SNARE complex (Rizo and Sudhof, 2012). The precise role of the N-terminus of syntaxin is not known, but it has been reported to mediate the stimulatory activity of Munc18-1 and to play a role in the initiation of SNARE assembly.
Figure 6.
EPR measurements on an N-terminal truncated syntaxin mutant. (A) An N-terminal segment of syntaxin (residues 2-9) is resolved in the crystal structure of the Syntaxin-Munc18-1 complex (PDB ID: 3C98).(Burkhardt et al., 2008) (B) Background corrected DEER data for syntaxin (27-262) 210R1 (40 μM) bound to SNAP25 (160 μM) in the absence (black trace) and presence (red trace) of Munc18-1 (80 μM). (C) Top: background corrected DEER data, and bottom: distance distribution obtained from syntaxin (27-262) 52R1:210R1 (40 μM) bound to SNAP25 (160 μM) with Munc18-1 (80 μM). Both closed and open states in syntaxin (27-262) are present. (D) Previously published data in the absence of SNAP-25 for syntaxin (27-262) shown for comparison (Dawidowski and Cafiso, 2013). Top: background corrected DEER data, and bottom: distance distribution obtained from syntaxin (27-262) 52R1:210R1 (40 μM) bound to Munc18-1 (40 μM) in the absence of SNAP-25. Without SNAP25 no open state is observed.
In order to determine whether the N-terminal segment might modulate that state of syntaxin when bound to SNAP25, we compared the behavior of a shortened version of syntaxin lacking the N-terminus, syntaxin (27-262) or ΔN syntaxin. Shown in Figure 6B are DEER traces obtained for a single spin-labeled ΔN syntaxin (ΔN syntaxin 210R1). When bound to SNAP25, the DEER traces indicate the presence of the 2:1 complex, and yield a trace that is similar to that obtained for syntaxin (1-262) (Figure 2B). Upon the addition of Munc18-1, the modulation depth is dramatically attenuated and the 2:1 complex appears to dissociate, as was observed for syntaxin (1-262) (Figure 5A). However, in contrast to syntaxin (1-262), ΔN syntaxin is not completely closed in the presence of SNAP25 when Munc18-1 is bound. Shown in Figure 6C (top) is the background subtracted DEER trace in the presence of Munc18-1 and the distribution (bottom) obtained from these data. In contrast to syntaxin (1-262), a significant open population can be seen in the distribution, and this represents approximately 40% of the ΔN syntaxin. When SNAP25 is absent, ΔN syntaxin behaves differently. Data obtained previously for ΔN syntaxin in the absence of SNAP25 are shown in Figure 6D, and in this case, Munc18-1 completely closes ΔN syntaxin.
As indicated above, removal of the syntaxin N-terminus is reported to reduce the affinity of Munc18-1 to syntaxin. It is possible that removal of the N-terminus also reduced the affinity of Munc18-1 for syntaxin when bound to SNAP25, and that the appearance of an open state in Figure 6C is due to a large fraction of syntaxin not associated to Munc18-1. However, other data suggest this is not the case. First, addition of Munc18-1 to the ΔN syntaxin:SNAP25 complex suppresses most of the modulation in the DEER signal (Figure 6B), indicating that greater than 80% of the 2:1 syntaxin:SNAP25 complex is dissociated. Second, the syntaxin dimer yields a distance at 40 Å (Figure 2C), and this component would be a dominant peak if Munc18-1 were not bound. In Figure 6C, this distance component is minor and corresponds to less than 8% of the distribution. As a result, ΔN syntaxin must be nearly completely bound to Munc18-1 under the conditions of this experiment. Taken together, the data imply that syntaxin assumes an alternate configuration from that seen in the crystal structure when bound to SNAP25 and Munc18-1 in the absence of its N-terminus, such that the open-closed equilibrium is shifted towards the open state.
Discussion
The sequence of events leading to SNARE assembly and the triggering of neuronal exocytosis is presently poorly understood. In the work carried out here, site-directed spin labeling was used to examine the complex formed between syntaxin and SNAP25 to determine the effect of Munc18-1 on the configuration of syntaxin and its association with SNAP25. It has been proposed that a 1:1 complex of syntaxin and Munc18-1 acts as the starting point for the assembly of the SNARE complex, and that Munc18-1 acts to dissociate syntaxin from its initial non-productive association with SNAP25 (Ma et al., 2013). In experiments presented here, Munc18-1 is found to dissociate syntaxin from the 2:1 Syntaxin:SNAP25 complex; however, one syntaxin remains associated with SNAP25. This conclusion is based upon EPR spectra and DEER data from SNAP25 showing that this t-SNARE does not resemble free SNAP25, and pulse EPR measurements showing a direct interaction between SNAP25 and syntaxin in the presence of Munc18-1 (Figure 5). The result suggests that while Munc18-1 functions to prevent the formation of a non-productive 2:1 complex, a syntaxin:SNAP25:Munc18-1 complex is formed that might function as a starting point for SNARE assembly. Assembly is likely to require other regulatory proteins, such as Munc13, that act to open the Habc domain of syntaxin and permit binding of synaptobrevin (Ma et al., 2011).
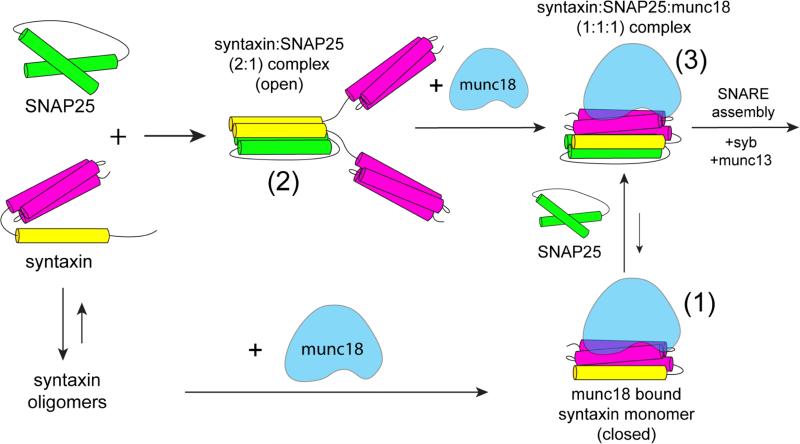
Shown in Figure 7 is a summary of some of the states that are found for syntaxin. Syntaxin readily oligomerizes in solution, and Munc18-1 will bind syntaxin ti dissociate these oligomers yielding a 1:1 syntaxin:Munc18-1 complex (form (1) in Figure 7). In the presence of SNAP25, syntaxin will bind to SNAP25 to form a 2:1 syntaxin:SNAP25 complex (form (2)). In the 2:1 Syntaxin:SNAP25 complex, syntaxin is in an open configuration with the Habc domain dissociated from each H3 motif (Figure 4). This is expected, since the four helix bundle may not allow each Habc domain to closely approach its corresponding H3 motif. The addition of Munc18-1 to the 2:1 complex dissociates one syntaxin from the complex and produces a sytaxin:SNAP25:Munc18-1 (1:1:1) complex (form (3)). As seen in Figure 4C, syntaxin is now in a closed configuration, resembling the 1:1 Munc18-1:syntaxin complex seen in the absence of SNAP25 (Chen et al., 2008; Dawidowski and Cafiso, 2013; Margittai et al., 2003). Apparently, this complex ((3) in Figure 7) is favored over the dissociated syntaxin:Munc18-1 complex ((1) in Figure 7), suggesting that the regulation of SNARE assembly takes place when one syntaxin is associated withSNAP25.
Figure 7.
Syntaxin (1-262) exists in several states in the presence of SNAP25 and Munc18-1. In solution, syntaxin readily forms oligomers in a manner that is dependent upon ionic strength and syntaxin concentration. When Munc18-1 is added, the oligomer is dissociated and syntaxin forms a 1:1 complex with Munc18-1 (form (1)) where syntaxin is in a closed configuration. If SNAP25 is added in excess to syntaxin (1-262), syntaxin preferentially forms a 2:1 syntaxin:SNAP25 complex (form (2)). Addition of Munc18-1 dissociates this 2:1 complex, and produces a 1:1:1 syntaxin:SNAP25:Munc18-1 complex where syntaxin is in a closed configuration (form (3)). The equilibrium between complexes (1) and (3) favors form (3). Form (3) is a likely starting point for SNARE assembly, because syntaxin appears to be more easily opened in this state.
An interesting observation made here is that removal of the syntaxin N-terminus alters the ability of Munc18-1 to drive syntaxin to a closed conformation. For syntaxin (1-262), Munc18-1 closes the structure whether SNAP25 is absent or present (Figures 4B, C). However, a significant open population remains for ΔN syntaxin in complex with Munc18-1 when bound to SNAP25 (Figure 6C), but not when ΔN syntaxin is bound to Munc18-1 in the absence of SNAP25 (Figure 6D). Thus, both the N-terminal segment of syntaxin and the binding of syntaxin to SNAP25 may alter the open-closed equilibrum of syntaxin in the presence of Munc18-1. These data provide an explanation for earlier observations on the role of the N-terminus of syntaxin in SNARE assembly (Burkhardt et al., 2008). When added to syntaxin (1-262) along with SNAP25 and synaptobrevin, Munc18-1 was found to block formation of the SNARE complex. However, SNARE assembly could proceed when the shortened ΔN syntaxin was used. An interesting conclusion that can be drawn from these observations is that SNARE assembly is more likely to take place for the syntaxin-Munc18-1 complex when it is associated with SNAP-25 than it is for the syntaxin-Munc18-1 complex alone, because it is easier to shift the open-closed equilibrium towards the open state when Munc18-1 and syntaxin are associated with SNAP25.
Another observation made here indicates that syntaxin association to SNAP25 alters its interaction with Munc18-1. As shown in Figure 4A, an excess of Munc18-1 is required to dissociate the 2:1 complex and close syntaxin when bound to SNAP25. This was an unexpected observation, since the SNAREs are present at concentrations in the tens of μM range, and the reported Munc18-1-syntaxin affinity is in the nM range (Burkhardt et al., 2008). Indeed, in the absence of SNAP25 we found in a previous study that stoichiometric additions of Munc18-1 were sufficient to eliminate the oligomerization of syntaxin in solution and close syntaxin (Dawidowski and Cafiso, 2013). We have not attempted to carry out measurements of the binding affinity between Munc18-1 and syntaxin in this ternary system, but given the concentrations of protein used for EPR, we expect that the affinity of Munc18-1 for syntaxin might be shifted to the μM range in the presence of SNAP25.
Several previous EPR studies have examined the configuration of the binary 2:1 syntaxin:SNAP25 complex (Margittai et al., 2001; Xiao et al., 2001; Zhang et al., 2002). Although there are indications from these studies that segments of this complex are disordered, the results indicate that the syntaxin:SNAP25 complex is a 4-helix bundle resembling the core SNARE complex where the position of synaptobrevin is occupied by a second syntaxin. The results presented here are generally consistent with this conclusion, however they indicate that there is considerable heterogeneity in the structure that may be due to syntaxin assuming one of multiple positions in the synaptobrevin binding site. Synaptobrevin is an R SNARE containing arginine at layer zero in the SNARE complex (Fasshauer et al., 1998) (syntaxin and SNAP25 are Q-SNAREs containing glutamine at the zero layer), and it is possible that the second syntaxin may not be able to align properly in the synaptobrevin binding site because it lacks the appropriate charged residue at the layer zero. It should be noted that there are a number of differences between these previous studies and the present study. In the present study, data were obtained from a longer syntaxin construct (1-262) and the use of pulse EPR allowed for a longer distance range with better resolution of the distributions than did previous continuous wave studies.
Munc18-1 is generally thought to have at least two binding modes to SNAREs, one which involves an interaction with syntaxin to place it into a closed configuration, and a second binding mode to fully assembled SNARE complexes, that involves interactions with the syntaxin N-terminus (Khvotchev et al., 2007). The latter interaction is thought to be the primary binding mode for most SM (Sec1/Munc18-1-like) proteins (Dulubova et al., 2007). The data in Figure 6 suggest that the Munc18-1 is able to bind to syntaxin when it is associated with SNAP25 in the absence of the N-terminal interaction, and that this interaction places syntaxin is a different configuration than that seen for full-length syntaxin. This result indicates that there is a third binding mode for Munc18-1 where syntaxin samples an open state. Indeed, alternate binding modes of Munc18-1 to syntaxin have been observed by high-resolution NMR (Liang et al., 2013) and neutron diffraction (Christie et al., 2012) There is evidence that Munc18-1 functions as a template for SNARE assembly to bring together the three neuronal SNAREs (Parisotto et al., 2014; Shen et al., 2015; Shen et al., 2007). Conceivably, Munc18-1 when associated with syntaxin in its open state may function in this template mode.
In conclusion, the data presented here provide clear evidence that syntaxin will associate with SNAP25 while bound to Munc18-1. The data suggest that one role for Munc18-1 may be to prevent the formation of a non-productive 2:1 syntaxin:SNAP25 complex, and simultaneously stabilize a 1:1 syntaxin:SNAP25 complex. Although this 1:1 complex is not competent by itself to assemble into the 4-helical SNARE bundle, the open-closed equilibrium of syntaxin appears to be shifted towards the open state when syntaxin is bound to SNAP25. This implies that syntaxin is more easily opened when associated with SNAP25, and that a syntaxin:Munc18-1:SNAP25 complex is a likely starting point for SNARE assembly.
Experimental Procedures
Protein mutagenesis, expression, and purification
Plasmid constructs (pET28a) for soluble syntaxin (residues 1-262), full-length syntaxin (residues 1-288), SNAP25 (residues 1-206), C-terminal half of soluble synaptobrevin (ΔN synaptobrevin; residues 49-96) and Munc18-1 were generously provided by Reinhard Jahn and Dirk Fasshauer (Max Planck Institute for Biophysical Chemistry, Gottingen, Germany). The soluble syntaxin construct without the N-terminus (ΔN syntaxin; residues 27-262) was prepared from soluble syntaxin using two-step PCR to delete the first 26 amino acids. All genetic constructs were derived from Rattus Norvegicus. Cysteine mutations were introduced into syntaxin and SNAP25 using QuickChange PCR (Agilent Technologies, Wilmington, DE) to allow attachment of the spin labeled side chain R1 (Figure 1). Soluble and full-length Syntaxin and Munc18-1 were expressed as described previously (Dawidowski and Cafiso, 2013). The same protocol was also used to express ΔN syntaxin, SNAP25 and the ΔN synaptobrevin.
Soluble syntaxin (1-262) and Munc18-1 were purified using an NiNTA resin (Biorad, Hercules, CA) using a previously established protocol (Dawidowski and Cafiso, 2013). This procedure was also used to purify ΔN syntaxin (27-262), SNAP25 and ΔN synaptobrevin and in each case the protein attached His-tag was removed by thrombin cleavage. Labeling of all syntaxin variants and SNAP25 was performed as described previously for syntaxin (Dawidowski and Cafiso, 2013), except that the MTSL label (1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl methanethiosulfonate) was purchased from Santa Cruz Biotechnology (Dallas, TX).
Sample preparation and EPR measurements
For all samples, protein aliquots of purified spin-labeled or unlabeled protein were mixed in the specified molar ratios and concentrated by ultrafiltration (Ultracel 30K; EMD Millipore); see figure legends for details on protein concentrations and ratios used. Unless otherwise specified, samples were run in a buffer containing 139 mM KCl, 12 mM NaCl, 20 mM MOPS, pH 7.4. For syntaxin data recorded at low ionic strength, samples contained only buffer. For pulse EPR measurements, approximately 10% deuterated glycerol (Cambridge Isotope laboratories, Andover, MA) was added to the final protein mixtures before freezing.
Continuous wave (CW) and four pulse double electron-electron resonance (DEER) EPR measurements were performed and analyzed as described previously (Dawidowski and Cafiso, 2013). For DEER, samples were placed into quartz capillaries (2.0 mm i.d. and 2.4 mm o.d.) and frozen using a dry-ice isopropanol bath. Pulse experiments were carried out at Q-band using a Bruker Elexsys E580 spectrometer and EN5107D2 dielectric resonator (Bruker Biospin, Billerica, MA). A standard four-pulse DEER sequence was used where 16-ns π/2 and 32-ns π observe pulses were separated by a 32-ns π pump pulse (Pannier et al., 2000). The pump frequency was set to the center maximum of the nitroxide spectrum and the observe frequency was set 60 MHz lower. The dipolar evolution data were processed and distance distributions determined using Tikhonov regularization incorporated into the DeerAnalysis2013 software package (Jeschke et al., 2006). This program contains a validation routine that was used to assess the error produced by background subtraction upon the distance distributions. At each distance in the distribution, ranges were plotted that represent fits that are within 15% RMSD of the best fit.
The experimental distance distributions obtained by DEER were compared with the predictions based upon a crystal structure or molecular model using the program MMM (Multiscale Modeling of Macromolecular systems) (Polyhach et al., 2011). In this program, we employed a rotamer library based upon density functional theory that matches the experimental rotamers found by crystallography of spin-labeled proteins (Warshaviak et al., 2011). Structures were visualized using the PyMOL Molecular Graphics System, Version 1.7.4 (Schrödinger, LLC).
Highlights.
Munc18 stabilizes a 1:1 syntaxin:SNAP25 complex
Syntaxin is in a closed state when associated with SNAP25 in the presence of Munc18
The syntaxin N-terminus regulates open and closed states in t-SNAREs
The syntaxin:SNAP25:Munc18 complex is a starting point for SNARE assembly
Acknowledgements
This work was supported by NIH grant NIGMS GM072694. We would like to thank Prof. Reinhard Jahn, Prof. Lukas Tamm and members of their research groups for helpful discussions during the course of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
DD and DSC designed the experiments, DD conducted the experiments, DD and DSC wrote the manuscript.
Syntaxin and SNAP25 poised for fusion
Neuronal fusion is driven by SNARE proteins that assemble and bring vesicle and target membranes together. EPR spectroscopy was used to show that Munc18 stabilizes a 1:1 syntaxin:SNAP25 t-SNARE complex and that this complex is a likely starting point for fusion.
References
- Brose N. For better or for worse: complexins regulate SNARE function and vesicle fusion. Traffic. 2008;9:1403–1413. doi: 10.1111/j.1600-0854.2008.00758.x. [DOI] [PubMed] [Google Scholar]
- Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 2008;27:923–933. doi: 10.1038/emboj.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman ER. How does synaptotagmin trigger neurotransmitter release? Annu Rev Biochem. 2008;77:615–641. doi: 10.1146/annurev.biochem.77.062005.101135. [DOI] [PubMed] [Google Scholar]
- Chen X, Lu J, Dulubova I, Rizo J. NMR analysis of the closed conformation of syntaxin-1. J Biomol NMR. 2008;41:43–54. doi: 10.1007/s10858-008-9239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie MP, Whitten AE, King GJ, Hu SH, Jarrott RJ, Chen KE, Duff AP, Callow P, Collins BM, James DE, et al. Low-resolution solution structures of Munc18:Syntaxin protein complexes indicate an open binding mode driven by the Syntaxin N-peptide. Proc Natl Acad Sci U S A. 2012;109:9816–9821. doi: 10.1073/pnas.1116975109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Columbus L, Hubbell WL. A new spin on protein dynamics. Trends Biochem Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- Dawidowski D, Cafiso DS. Allosteric control of syntaxin 1a by Munc18-1: characterization of the open and closed conformations of syntaxin. Biophys J. 2013;104:1585–1594. doi: 10.1016/j.bpj.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Khvotchev M, Liu S, Huryeva I, Sudhof TC, Rizo J. Munc18-1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci U S A. 2007;104:2697–2702. doi: 10.1073/pnas.0611318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanucci GE, Cafiso DS. Recent advances and applications of site-directed spin labeling. Curr Opin Struct Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Otto H, Eliason WK, Jahn R, Brunger AT. Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J Biol Chem. 1997;272:28036–28041. doi: 10.1074/jbc.272.44.28036. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci U S A. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490:201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. DeerAnalysis2006 - a comprehensive software package for analyzing pulsed ELDOR data. Applied Magnetic Resonance. 2006;30:473–498. [Google Scholar]
- Khvotchev M, Dulubova I, Sun J, Dai H, Rizo J, Sudhof TC. Dual modes of Munc18-1/SNARE interactions are coupled by functionally critical binding to syntaxin-1 N terminus. J Neurosci. 2007;27:12147–12155. doi: 10.1523/JNEUROSCI.3655-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman JC, Robblee J, Fairman R, Hughson FM. Structural analysis of the neuronal SNARE protein syntaxin-1A. Biochemistry. 2000;39:8470–8479. doi: 10.1021/bi0003994. [DOI] [PubMed] [Google Scholar]
- Liang B, Kiessling V, Tamm LK. Prefusion structure of syntaxin-1A suggests pathway for folding into neuronal trans-SNARE complex fusion intermediate. Proc Natl Acad Sci U S A. 2013;110:19384–19389. doi: 10.1073/pnas.1314699110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CJ, Fleissner MR, Guo Z, Kusnetzow AK, Hubbell WL. Osmolyte perturbation reveals conformational equilibria in spin-labeled proteins. Protein Sci. 2009;18:1637–1652. doi: 10.1002/pro.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Li W, Xu Y, Rizo J. Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat Struct Mol Biol. 2011;18:542–549. doi: 10.1038/nsmb.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339:421–425. doi: 10.1126/science.1230473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margittai M, Fasshauer D, Pabst S, Jahn R, Langen R. Homo- and heterooligomeric SNARE complexes studied by site-directed spin labeling. J Biol Chem. 2001;276:13169–13177. doi: 10.1074/jbc.M010653200. [DOI] [PubMed] [Google Scholar]
- Margittai M, Widengren J, Schweinberger E, Schroder GF, Felekyan S, Haustein E, Konig M, Fasshauer D, Grubmuller H, Jahn R, et al. Single-molecule fluorescence resonance energy transfer reveals a dynamic equilibrium between closed and open conformations of syntaxin 1. Proc Natl Acad Sci U S A. 2003;100:15516–15521. doi: 10.1073/pnas.2331232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- Parisotto D, Pfau M, Scheutzow A, Wild K, Mayer MP, Malsam J, Sinning I, Sollner TH. An extended helical conformation in domain 3a of Munc18-1 provides a template for SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex assembly. J Biol Chem. 2014;289:9639–9650. doi: 10.1074/jbc.M113.514273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyhach Y, Bordignon E, Jeschke G. Rotamer libraries of spin labelled cysteines for protein studies. Phys Chem Chem Phys. 2011;13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- Rickman C, Hu K, Carroll J, Davletov B. Self-assembly of SNARE fusion proteins into star-shaped oligomers. Biochem J. 2005;388:75–79. doi: 10.1042/BJ20041818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Sudhof TC. The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices--guilty as charged? Annu Rev Cell Dev Biol. 2012;28:279–308. doi: 10.1146/annurev-cellbio-101011-155818. [DOI] [PubMed] [Google Scholar]
- Rodkey TL, Liu S, Barry M, McNew JA. Munc18a scaffolds SNARE assembly to promote membrane fusion. Mol Biol Cell. 2008;19:5422–5434. doi: 10.1091/mbc.E08-05-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Rathore SS, Yu H, Gulbranson DR, Hua R, Zhang C, Schoppa NE, Shen J. The trans-SNARE-regulating function of Munc18-1 is essential to synaptic exocytosis. Nat Commun. 2015;6:8852. doi: 10.1038/ncomms9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- Stein A, Radhakrishnan A, Riedel D, Fasshauer D, Jahn R. Synaptotagmin activates membrane fusion through a Ca2+-dependent trans interaction with phospholipids. Nat Struct Mol Biol. 2007;14:904–911. doi: 10.1038/nsmb1305. [DOI] [PubMed] [Google Scholar]
- Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Warshaviak DT, Serbulea L, Houk KN, Hubbell WL. Conformational analysis of a nitroxide side chain in an alpha-helix with density functional theory. J Phys Chem B. 2011;115:397–405. doi: 10.1021/jp108871m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Poirier MA, Bennett MK, Shin YK. The neuronal t-SNARE complex is a parallel four-helix bundle. Nat Struct Biol. 2001;8:308–311. doi: 10.1038/86174. [DOI] [PubMed] [Google Scholar]
- Yang B, Steegmaier M, Gonzalez LC, Jr., Scheller RH. nSec1 binds a closed conformation of syntaxin1A. J Cell Biol. 2000;148:247–252. doi: 10.1083/jcb.148.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Chen Y, Kweon DH, Kim CS, Shin YK. The four-helix bundle of the neuronal target membrane SNARE complex is neither disordered in the middle nor uncoiled at the C-terminal region. J Biol Chem. 2002;277:24294–24298. doi: 10.1074/jbc.M201200200. [DOI] [PubMed] [Google Scholar]