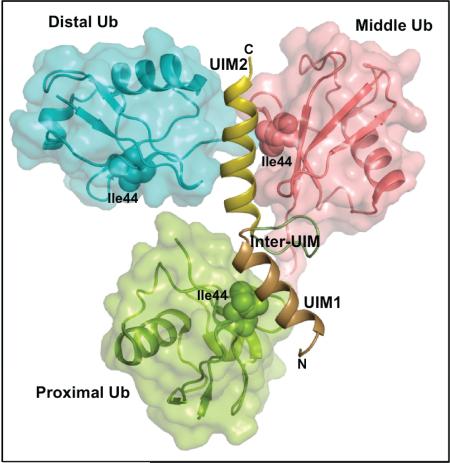
Abstract
Lys48-linked ubiquitin chains act as the main targeting signals for protein degradation by the proteasome. Here we report selective binding of AIRAPL, a protein that associates with the proteasome upon exposure to arsenite, to Lys48-linked tri-ubiquitin chains. AIRAPL comprises two ubiquitin-interacting motifs in tandem (tUIMs) that are linked through a flexible inter-UIM region. In the complex crystal structure UIM1 binds the proximal ubiquitin, whereas UIM2 (the double-sided UIM) binds non-symmetrically to the middle and distal ubiquitin moieties on either side of the helix. Specificity of AIRAPL for Lys48-linked ubiquitin chains is determined by UIM2, and the flexible inter-UIM linker increases avidity by placing the two UIMs in an orientation that facilitates binding of the third ubiquitin to UIM1. Unlike middle and proximal ubiquitins, distal ubiquitin binds UIM2 through a novel surface, which leaves the Ile44 hydrophobic patch accessible for binding to the proteasomal ubiquitin receptors.
Graphical Abstract
Introduction
Toxic effects of protein misfolding (proteotoxicity) are balanced by the cellular folding capacity and protein degradation machinery. As such, both cellular abilities are dynamically regulated to accommodate for fluctuations in the amount of misfolded proteins that occur in response to cellular and environmental changes. The ubiquitin-proteasome pathway (UPP) is the major cellular machinery for regulated proteolysis and its specificity is provided by versatility of the ubiquitin system (Kravtsova-Ivantsiv and Ciechanover, 2012, Schwartz and Ciechanover, 2009).
Ubiquitin is a small protein modifier with 76 amino acid residues, which regulates a wide range of biological functions. Ubiquitin signals are generated through covalent attachment of the C-terminal glycine of ubiquitin to a specific lysine residue in a protein substrate through a process known as ubiquitylation. Versatility of the ubiquitin system is accomplished by the ability of ubiquitin to interact with substrates as a single moiety (mono-ubiquitin) or different types of poly-ubiquitin chains. Poly-ubiquitin chains are typically linked via any of the seven lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 and Lys63) or the N-terminal methionine residue of ubiquitin (Kulathu and Komander, 2012).
Among various poly-ubiquitin chains, Lys48-linked chains are known as the canonical protein turnover signals (Chau et al., 1989, Thrower et al., 2000). However, other poly-ubiquitin chains are also shown to be involved in proteasomal degradation (Xu, 2009, Kravtsova-Ivantsiv and Ciechanover, 2012). For example, Lys11-linked ubiquitin chains regulate the cell cycle progression by promoting the degradation of mitotic proteins in the 26S proteasome (Matsumoto et al., 2010, Budhavarapu et al., 2012). Met1-linked (also called linear) ubiquitin chains have also been reported to target the eukaryotic replication clamp PCNA for proteasomal degradation (Zhao and Ulrich, 2010).
Ubiquitin signals are decoded by the specialized modules in proteins known as ubiquitin-binding domains (UBDs) (Husnjak and Dikic, 2012). They recognize and non-covalently bind ubiquitylated proteins or free ubiquitin molecules. UBDs are structurally and functionally diverse and are often selective for a specific type of ubiquitin species, thereby providing specificity for ubiquitin-signaling pathways (Dikic et al., 2009, Ikeda et al., 2010, Rahighi and Dikic, 2012). To date, 20 different families of UBDs are characterized (Hicke et al., 2005). Ubiquitin-interacting motifs (UIMs) represent a family of UBDs that were initially identified in the S5a/Rpn10 protein at the regulatory subunit of the proteasome (Young et al., 1998). A UIM is characterized as a stretch of ~ 20 amino acid residues forming an amphipathic α-helical structure (Hofmann and Falquet, 2001, Fisher et al., 2003). Typically, a single UIM has only a modest affinity for binding to ubiquitin molecules (0.1-1 mM), which may not be sufficient to generate efficient signaling effects in the context of living cells (Fisher et al., 2003). Therefore, UIMs are often found in tandem repetitions (tUIMs), double-sided UIMs, or in combination with UBDs of other types, resulting in multivalent ubiquitin-binding (Husnjak and Dikic). Tandem UIMs of Vps27 and the double-sided UIM of Hrs, a component of the endosomal sorting machinery are examples of UBDs recognizing multiple mono-ubiquitylated proteins (Onishi et al., 2007, Swanson et al., 2003, Hirano et al., 2006), while tUIMs in Rap80 and the combination of UIM and VHS (Vps27/Hrs/Stam) domains in STAM2 result in a selective binding to Lys63-linked di-ubiquitin chains in a cooperative manner (Sato et al., 2009) (Lange et al., 2012).
Although several studies have revealed structural basis for the recognition of ubiquitin chains by UBDs, to date structural data explaining selectivity toward a specific ubiquitin chain type are limited to di-ubiquitins of a subset of linkages. Hence, there are no structural explanations available for cases such as Lys48-linked ubiquitin signaling to proteasomal degradation where chains longer than di-ubiquitins are found to be more relevant in terms of the ubiquitin chain length and signaling efficiency (Thrower et al., 2000, Sakata et al., 2012).
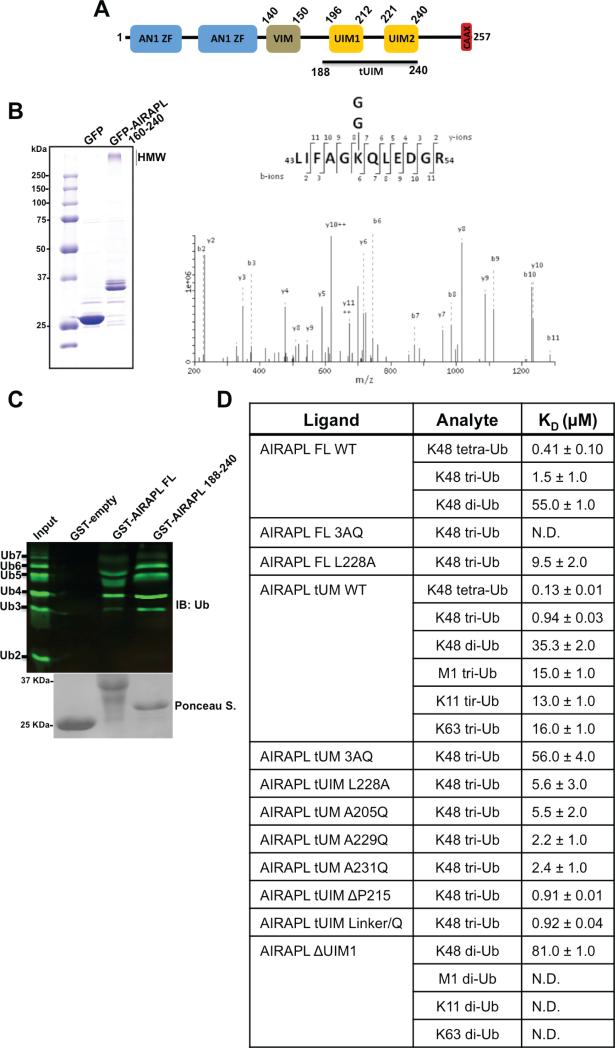
AIRAPL (arsenite-inducible proteasomal 19S regulatory particle-associated protein-like) is a ubiquitin-binding protein that resides on the cytosolic surface of the ER membrane (Yun et al., 2008). Also, it is a P97 (VCP/Cdc48) adaptor (Glinka et al., 2013) and impairment of aip-1 (a C. elegans homologue of AIRAPL) is reported to reduce longevity and accelerate protein aggregation in two model disease animals (poly Q and beta amyloid peptide (Hassan et al., 2009, Yun et al., 2008)). The AIRAPL polypeptide is composed of two AN1 Zinc finger (ZF) domains, a valosin-containing protein-interacting motif (VIM), two UIMs in tandem (tUIMs), and a C-terminal CAAX box (Fig. 1A). In this report we demonstrate specificity of AIRAPL for Lys48-linked tri-ubiquitin chains triggered by intrinsic selectivity of its double-sided UIM (UIM2) for Lys48 linkage and a flexible inter-UIM linker that results in an overall conformation which confers simultaneous binding of tUIMs to three consecutive ubiquitin moieties in a Lys48-linked chain.
Figure 1. AIRAPL is an ubiquitin-binding protein and is selective for Lys48-linked ubiquitin chains.
(A) Schematic domain organization of mouse AIRAPL protein. AN1 ZF, AN1-type zinc finger; VIM, valosin-containing protein (VCP)-interacting motif; UIM, ubiquitin-interacting motif; CAAX box, a sequence of cysaliphatic-aliphatic-undefined amino acids. (B) Left: high molecular weight (HMW) polyubiquitin chains co-purified with GFP tagged-AIRAPL (residues 160-240) were subjected to liquid chromatography-tandem mass spectrometry (LCMS/MS). GFP alone was used as a negative control for binding to ubiquitin. Right: the MS/MS spectrum of the Lys48-linkage GG-containing peptide 43-54 derived from Ub with a precursor ion mass of [M+2H] 2+ 731.0 Da was obtained from cells expressing Flag-AIRAPL. The b/y fragment ions are indicated. (C) Binding of GST-tagged AIRAPL FL and tUIM to Lys48-linked (2-7)-ubiquitin chains was analyzed by immunoblotting using anti-ubiquitin antibody. Loading of GST-tagged proteins was determined by Ponceau S staining. (See also Fig. S1) (D) Binding affinity (KD or equilibrium constants) of AIRAPL FL, tUIM and ΔUIM1, WT (wild type) or mutants, for various ubiquitin chain types and lengths measured by surface plasmon resonance (SPR). For each measurement, His-tagged AIRAPL was immobilized on NiHC1000m chip surface and ubiquitin chains were loaded over the chip. Each measurement was done at least two times and data was analyzed using Scrubber 2 (see also Fig. S2). FL, full-length; tUIM, a construct of AIRAPL comprising tandem UIMs (residues 188-240); ΔUIM1, tUIM construct lacking UIM1; ND, not detected.
Results
AIRAPL is selective for Lys48-linked poly-ubiquitin chains
In order to test the selectivity of AIRAPL for ubiquitin chains we first used ubiquitin chain linkage-specific antibodies (Newton et al., 2012). Immunoprecipitation of AIRAPL from HEK 293T cells and the use of ubiquitin linkage-specific antibodies indicated the interaction of AIRAPL with Lys48-linked poly-ubiquitin chains, while Lys11- and Lys63-linked ubiquitin chains were absent in the AIRAPL complex (Fig. S1). High molecular weight (HMW) polyubiquitin chains co-purified with AIRAPL (Fig. 1B, left) were then evaluated by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The results showed that Lys48 was the only GG-modified peptide identified and all the other lysines or the N-terminal methionine were excluded as possible ubiquitylation sites (Fig. 1B, right).
To verify the minimum length of ubiquitin chains required for binding to AIRAPL, we performed in vitro pull-down binding assays and surface plasmon resonance (SPR) measurements using AIRAPL full-length (FL) and a construct encompassing tUIMs (residues 188-240 referred to as tUIM, hereafter, Fig. 1A). Our in vitro pull-down assays of a mixture of Lys48-linked [2-7] ubiquitin chains with GST-tagged AIRAPL tUIM and FL showed binding of both proteins to tri- or longer ubiquitin chains, while binding to mono- or di-ubiquitins was not detected (Fig. 1C). Also, the SPR results indicated that AIRAPL tUIM and FL bind diubiquitin chains with KD of 35 and 55 μM, respectively, which is ~35 times lower than their affinity for tri-ubiquitin chains (1-1.5 μM) (Fig. 1D, Fig. S2A-F). Although AIRAPL tUIM and FL show stronger binding to tetra-ubiquitin chains (0.13-0.41 μM), we chose tri-ubiquitins as the shortest chains that bind AIRAPL with reasonably high affinity and used them for further structural studies. It has to be noted that in these measurements, AIRAPL FL binds ubiquitin chains with consistently lower affinity than tUIM. The reason might be that the ubiquitin-binding sites in the isolated tUIM construct are more accessible than in FL, which is a multi-domain protein (Fig. 1A). Indeed, structural details of FL AIRAPL are yet unknown.
To further investigate the ubiquitin-binding specificity of AIRAPL, we performed SPR measurements using tUIM and Lys48-, Lys63-, Lys11- and Met1-linked tri-ubiquitin chains. The data demonstrated that tUIM binds Lys48-linked chains with a KD of ~1μM while it binds Lys63-, Lys11-, and Met1-linked chains with at least 13 times lower affinity (KD ranging from 13 to 16 μM), thereby confirming the in vitro selectivity of AIRAPL towards Lys48-linked ubiquitin chains (Fig. 1D, Fig. S2B, S2G-I).
Crystal structure of the AIRAPL tUIM /Lys48-linked tri-ubiquitin chain complex
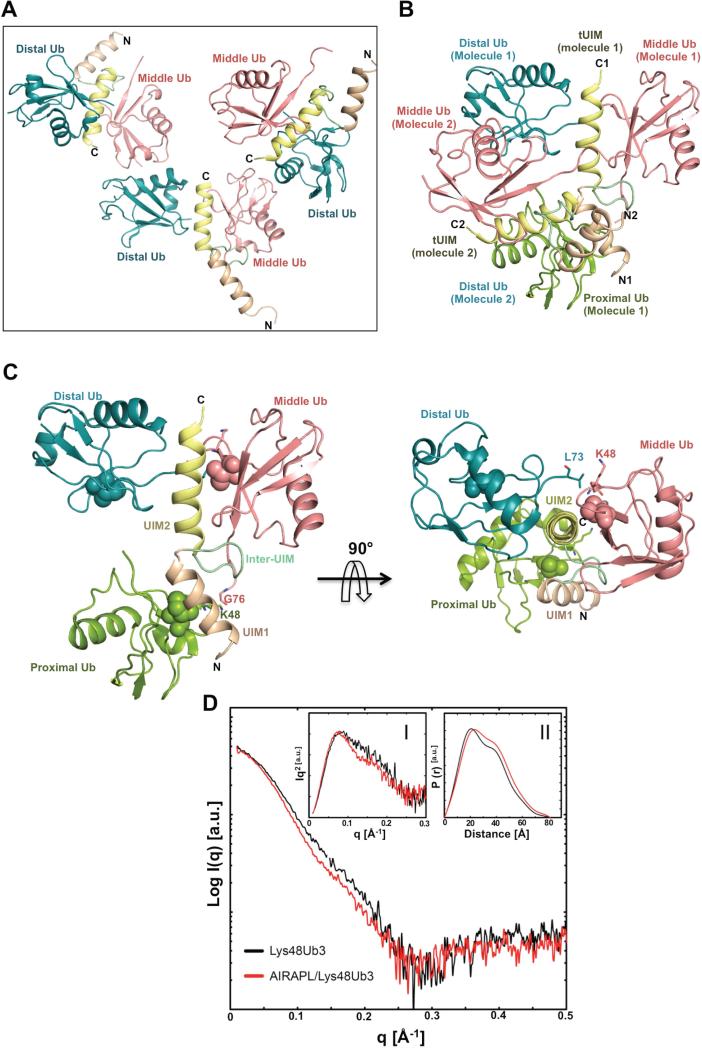
To obtain further insight into the selective recognition of ubiquitin chains by AIRAPL, we carried out co-crystallization of tUIM with Lys48-linked tri- or tetra-ubiquitin chains. However we only obtained co-crystals with tri-ubiquitin chains that appeared after 120 days and diffracted up to 3.0 Å resolution (Table 1). The crystals belonged to the P31 space group and each asymmetric unit contained three molecules. Each molecule was, unexpectedly, composed of a tUIM bound to two ubiquitin moieties (Fig. 2A). This might be due to the cleavage of iso-peptide bonds in the ubiquitin chains by the time that co-crystals formed. But, intriguingly, one of the two ubiquitin moieties from a symmetry-related molecule in the crystal was found to bind to the N-terminal UIM (UIM1) (Fig. 2B, Fig. S3A), which in effect made the entire complex comprising of one tUIM and three ubiquitin moieties that can be linked through Lys48 (Fig. 2C). The two UIMs of AIRAPL formed α-helical structures linked by a loop-structured inter-UIM region (Fig. S3B). Apart from some minor deviations in the structure of inter-UIMs, the three complex molecules in the asymmetric unit of the crystal adopted similar conformations and could be well superimposed (~ 0.8 Å, Fig. S3C).
Table 1.
The X-ray data collection and refinement statistics
| AIRAPL tUIM / Lys48Ub3 | |
|---|---|
| Data collection | |
| Beamline | PF-BL17A |
| Space group | P31 |
| Cell dimensions | |
| a, b, c (Å) | 90.9, 90.9, 61.0 |
| α, β, γ (°) | 90.0, 90.0, 120. 0 |
| Wavelength (Å) | 1.00 |
| Resolution (Å) | 50.0-3.0 (3.2-3.0) |
| CC (½) | 0.97 (0.38) |
| Rmerge | 0.16 (0.76) |
| I/σI | 6.8 (2.5) |
| Completeness (%) | 99.4 (99.7) |
| Redundancy | 2.9 (2.9) |
| Refinement | |
| Resolution (Å) | 50.0-3.0 |
| No. reflections | 11211 |
| Rwork/Rfreeb | 20.1-25.5 |
| No. atoms | |
| Protein | 4421 |
| Water | 15 |
| B-factors | |
| Protein (average) | 48.3 |
| AIRAPL tUIM | 49.1 |
| Ubiquitins | 46.9 |
| Water | 39.2 |
| R.m.s. deviation | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.171 |
Figure 2. Structure of the AIRAPL tUIM in complex with a Lys48-linked tri-ubiquitin chain.
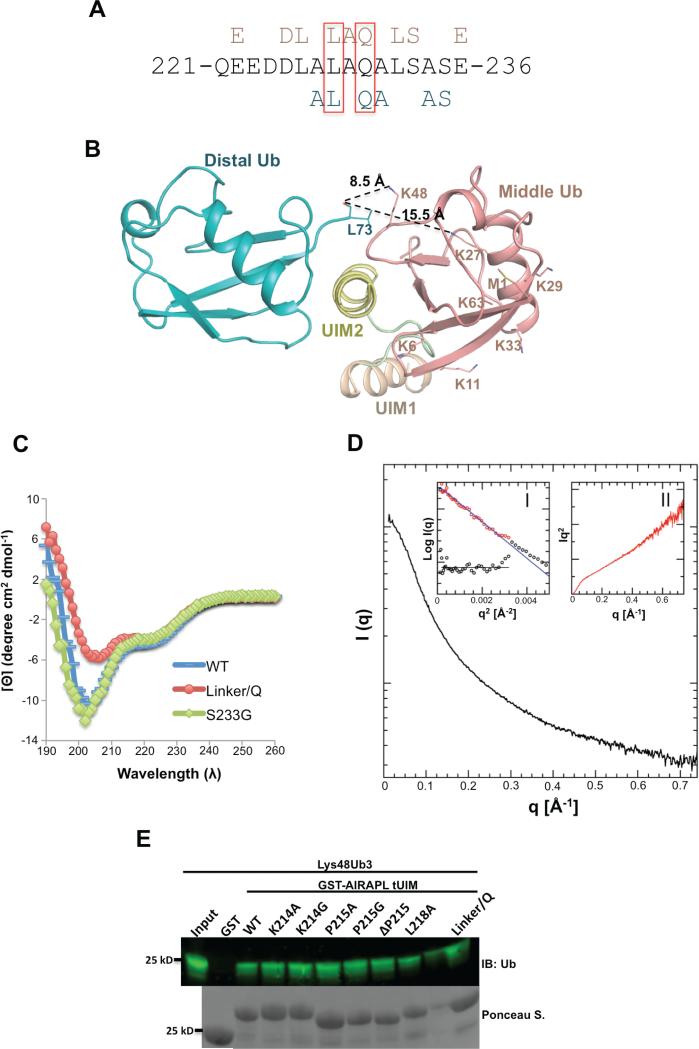
(A) There are three complex molecules in each asymmetric unit (AU) that each complex is composed of one tUIM and two ubiquitin moieties (distal and middle) (See also Fig. S3a-c). (B) Two complex molecules from neighboring asymmetric units (molecules 1 & 2) where distal ubiquitin of molecule 2 acts as the proximal ubiquitin for molecule 1. N1, C1 and N2, C2 indicate N- and C-term of tUIMs from molecules 1 and 2, respectively. (C) The overall structure of tUIM/ Lys48-linked tri-ubiquitin in two orthogonal views. UIM1 and UIM2 are shown in light orange and yellow, respectively. The inter-UIM loop is colored in peach. The distal, middle and proximal ubiquitins are shown in blue, pink and green. The spheres represent Ile44 from each ubiquitin moiety. The most C-terminal residues from distal and middle ubiquitins and Lys48 residue of middle and proximal ubiquitins are indicated as sticks. (D) SAXS intensity of Lys48-linked tri-ubiquitins alone (black) and in complex with AIRAPL tUIM (red). Kratky plots and P(r) functions of the data were shown in Inset I and II, respectively. Inset I: Tails of Kratky plots of both curves gradually converged to a baseline at higher q range, suggesting well-folded multi-domain conformations. Inset II: a significant peak shift was observed in P(r) function indicating that the complex has a more globular shape than Lys48-linked tri-ubiquitins alone (See also Fig. S3D).
In each AIRAPL/tri-ubiquitin complex, we refer to the ubiquitin moiety with a free C-terminal tail as proximal, and the two other ubiquitins are sequentially termed as middle and distal. In all three complex molecules in the asymmetric unit, the most C-terminal residue of the distal ubiquitins observed in the electron density map is Leu73, suggesting the flexibility of the C-terminal tail (a.a. 73-76) of distal ubiquitin, which is not sufficiently stabilized by interacting with AIRAPL. In a similar manner, no direct interaction is detected between the C-terminal tail of the middle ubiquitin and tUIMs. In only one of the three complexes in the asymmetric unit, the C-terminal tail of the middle ubiquitin could be fully modeled in the electron density map.
SAXS study of the AIRAPL tUIM/ Ly48-linked tri-ubiquitin chains complex in solution
To study the interactions of AIRAPL with Lys48-linked ubiquitin and stoichiometry of their complex in solution, we carried out in-line size exclusion chromatography- small angle x-ray scattering (SEC-SAXS) experiments on Lys48-linked tri-ubiquitin alone and in a mixture with AIRAPL tUIM. The ubiquitin chains were pre-mixed with tUIM in a molar ratio of 1:5 and a Superdex 75 3.2/300 column (GE Healthcare) was employed to separate the tUIM/Lys48-linked tri-ubiquitin complex from excess tUIM. The results indicated different SAXS profiles for free Lys48-linked tri-ubiquitin and the complex form. Tails of Kratky plots of both curves (Fig 2D, Inset I) gradually converged to a baseline at higher q range, suggesting well-folded multi-domain conformations. Both SAXS profiles displayed similar Rg and Dmax values (Fig. S3D) however interestingly a significant peak shift was observed in P(r) function (Fig 2D, Inset II), indicating that the complex is more globular in shape than Lys48-linked ubiquitin chains alone. Moreover, molecular weights (MW) were estimated from SAXS profiles using Porod volumes (Petoukhov et al.). While the Mw of Lys48-linked ubiquitin chains was estimated to be 22.3kDa, which is in reasonably good agreement with their theoretical Mw (25.5kDa), the estimated MW of the complex was 30.1kDa, implying a 1:1 stoichiometry in solution, in which a Lys48-linked triubiquitin binds one AIRAPL tUIM (Fig. S3D).
Mapping the interactions between tUIM and ubiquitin molecules
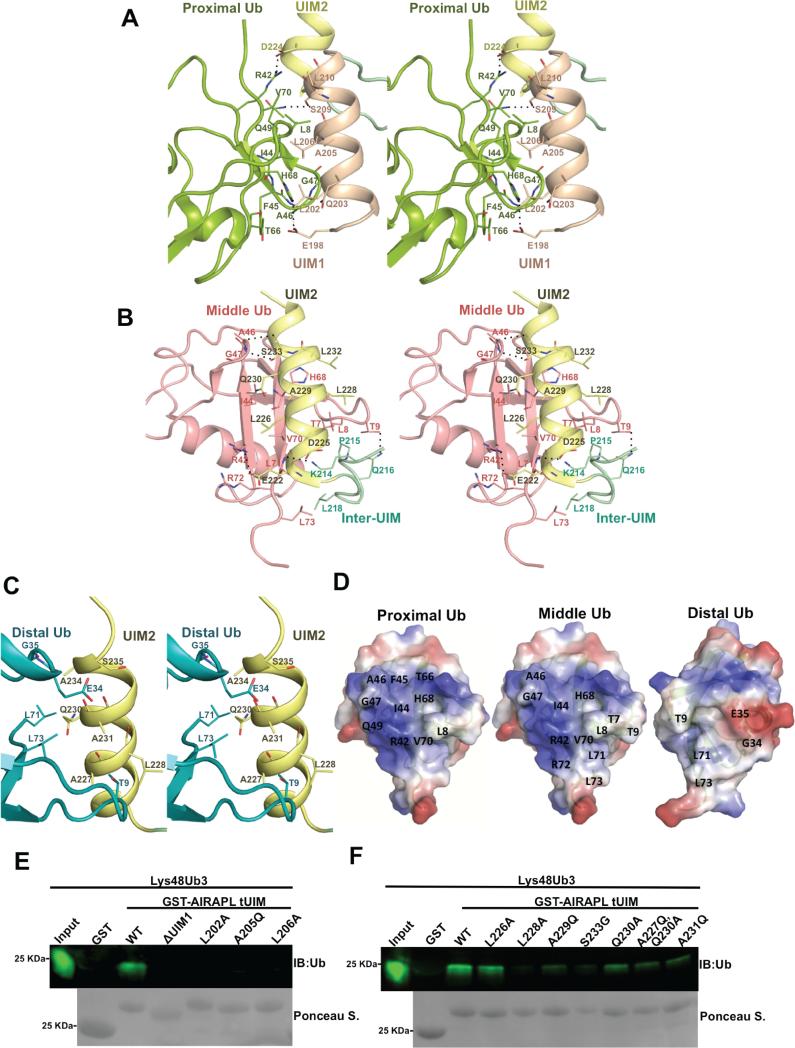
In the crystal structure, the proximal ubiquitin is inverted (turned upside down) compared with the orientation of ubiquitins involved in conventional ubiquitin/UIM interactions (Fig. S4A). In spite of that, the proximal ubiquitin still uses the hydrophobic patch centered on Ile44 to interact with UIM1 residues ranging from Glu198 to Leu210 as well as Asp224 of the UIM2 (Fig. 3A). Although the interactions are mostly hydrophobic, there are salt bridges and hydrogen bonds formed between His68/Glu198, Arg42/Asp224 and Gln49/Ser209 of proximal ubiquitin/tUIM (Fig. 3A).
Figure 3. Characterization of the interactions between AIRAPL tUIM and ubiquitin moieties.
Stereo-views of the interactions between (A) UIM1 and proximal ubiquitin, (B) UIM2/inter-UIM and middle ubiquitin, (C) UIM2 and distal ubiquitin. The color codes are similar to Fig. 2b. Hydrogen bonds and salt bridges are indicated with dashed lines. (D) Surface representation of the interacting residues from the proximal, middle and distal ubiquitins. The surfaces are colored according to their electrostatic surface potential (blue, positive; red, negative). (E) Binding of the GST-tagged AIRAPL FL, tUIM WT and mutants to Lys48-linked tri-ubiquitin chains was analyzed by immunoblotting using anti-ubiquitin antibody. Loading of GST-tagged proteins was determined by Ponceau S staining (See also Fig. S4).
AIRAPL UIM2 (residues 221-235) binds non-symmetrically to the middle and distal ubiquitins (Fig. 2C). While the middle ubiquitin binds AIRAPL through the Ile44 hydrophobic patch, distal ubiquitin uses a novel surface including Thr9, Glu34, Gly35 and the C-terminal tail including Leu71 and Leu73 for the interactions (Fig. 3B-D). The C-terminal tail of distal ubiquitin wraps around the UIM2 α-helix to reach Lys48 of the middle ubiquitin. The middle ubiquitin is the main binding partner for the tUIM in a tri-ubiquitin chain as judged by the extent of interactions and buried surface area on ubiquitin molecules (727 Å2 compared with 453 Å2 and 315 Å2 on proximal and distal ubiquitins, respectively). Distal and middle ubiquitin-binding surfaces on the UIM2 are highly conserved among AIRAPL proteins from various species (Fig. S4B). Interactions between distal and middle ubiquitins and UIM2 are mainly hydrophobic; however, from the middle ubiquitin Arg42 forms a salt bridge with Glu222, and backbone amino groups of Leu71 and Gly47 make hydrogen bonds with Asp225 and Ser233, respectively. Furthermore, backbone amino group of Ala46 makes hydrogen bond with the backbone carbonyl group of Ser233 (Fig. 3B). In addition to UIM2, middle ubiquitin contacts Lys214, Pro215, Gln216 and Leu218 residues of the inter-UIM loop region through Leu8, Thr7, Thr9, and Leu71 (Fig. 3B).
Based on the specific interactions observed in the complex crystal structure, we performed SPR binding assays as well as in vitro pull-down experiments using various mutants of either FL or tUIM AIRAPL and examined their binding to Lys48-linked tri-ubiquitin chains (Fig. 1D, 3E). The SPR results indicate 2-6 times decrease in the binding affinity as a result of single mutations including A205Q, L228A, A229Q and A231Q. However, either tUIM or FL AIRAPL bearing 3AQ (A205/229/231/Q) mutation has drastically lower affinity for binding to ubiquitin chains. While tUIM 3AQ mutant binds with ~60 times lower affinity than tUIM WT, no binding was detected for FL 3AQ and Lys48-linked triubiquitins (Fig. 1D, Fig. S2J-P). Also, as demonstrated by the in vitro GST pull-down experiments, mutations in any of the ubiquitin-binding sites on AIRAPL tUIM reduces binding to Lys48-linked tri-ubiquitins (Fig. 3E,F).
UIM2 defines specificity of AIRAPL for Lys48-linked ubiquitin chains
As illustrated by the crystal structure, distal and middle ubiquitins of a Lys48-linked tri-ubiquitin bind AIRAPL UIM2 in a non-symmetric manner, meaning that the two ubiquitins employ completely different surfaces for binding to either side of the UIM2 α-helix (Fig. 2C, 4A, Fig.S5A). This is unlike the previously reported structure of Hrs double-sided UIM that binds two independent mono-ubiquitin moieties with identical binding modes (Fig. S5B). The non-symmetric binding mode appears to make the linkage between distal and middle ubiquitins possible only through Lys48, as the second closest Lys residue is ~8 Å away from the carbonyl group of Leu73 (the most C-terminal residue observed in the electron density map) (Fig. 4B). This observation prompted us to test if AIRAPL UIM2 is selective for a specific ubiquitin chain type. Thus we performed SPR experiments using a tUIM construct lacking residues 196-213 (ΔUIM1) and various di-ubiquitin chains. The results indicated that ΔUIM1 binds Lys48-linked di-ubiquitins with KD of 81 μM, but interestingly no binding was detected for ΔUIM1 and Lys63-, Lys11-, or Met1-linked diubiquitins (Fig. 1D, Fig. S2S-V), thereby suggesting intrinsic selectivity of the AIRAPL UIM2 for Lys48-linked ubiquitin chains.
Figure 4. The roles of AIRAPL UIM2 and inter-UIM linker in defining specificity towards Lys48-linked ubiquitin chains.
(A) UIM2 sequence comprising amino acid residues 221-236. Residues from UIM2 that interact with the middle (top, pink) and/or distal (below, blue) ubiquitins are indicated. Leu228 and Gln230 that are involved in the interactions with both distal and middle ubiquitins are marked by red boxes. (See also Fig. S5a) (B) Orientation of the distal and middle ubiquitins exclusively allows Lys48 linkage binding. The last C-terminal residues (74-76) of distal ubiquitins are not visible in the electron density map. In spite of that, Lys48 is the closest lysine residue from middle ubiquitin to the main chain carbonyl group of Leu73 (distance: 8.5 Å). The second closest lysine (Lys27) is 15.5 Å away from Leu73 (See also Fig. S5b-d) (C) Circular Dichroism (CD) measurements of AIRAPL tUIM WT and two mutants (S233G and linker/Q). The data is reported as mean residue ellipticities in the range of λ: 190-260 nm and indicate ~32% helical content for tUIM WT and S233G, and 25% for the linker/Q mutant. In linker/Q mutant all residues of the linter/UIM linker are substituted with glutamine. (D) SAXS intensity of AIRAPL tUIM in solution. Inset I: the Guinier analysis of the data exhibits good linearity within the fit region (red circles) with unbiased residuals (depicted enlarged by a factor of two as purple circles distributed around a straight line in the lower left corner). Inset II: the Kratky representation of the SAXS data monotonically increases in the high q-region and does not show a pronounced peak at lower q indicative of an extended, flexible structure. (E) Binding of GST-tagged tUIM WT and inter-UIM linker mutants to Lys48-linked tri-ubiquitin chains was analyzed by immunoblotting using anti-ubiquitin antibody. Loading of GST-tagged proteins was determined by Ponceau S staining.
We also searched for putative double-sided UIMs and found MEKK1 (MAPK/ERK kinase kinase 1) as a possible Lys48 linkage-selective protein based on conservation of residues on the distal ubiquitin-binding site (Fig. S5C). The LC-MS/MS analysis of HMW poly-ubiquitin chains co-purified with a MEKK1 UIM domain identified only Lys48-linked chains, suggesting the Lys48-linkage specificity of MEKK1 double-sided UIM (Fig. S5D).
The flexible inter-UIM facilitates binding of AIRAPL to Lys48-linked triubiquitins
The AIRAPL inter-UIM linker comprises 8 amino acid residues (213-220), and adopts a highly flexible structure resulting in flexibility of the overall structure of tUIM. Circular dichroism (CD) data indicate helical content of ~32 % for the tUIM region (Fig. 4C). Thus, to gain better understanding of the inter-UIM linker structure in solution, we carried out SAXS analysis of AIRAPL tUIM. The SAXS results confirmed the flexible nature of the overall tUIM structure. While the linearity of the Guinier plot clearly confirms the absence of larger aggregates or intermolecular repulsion and yields a radius of gyration (Rg) of 22.6 Å for the AIRAPL tUIM, the Kratky representation of the SAXS profile exhibits no pronounced peak and shows an increasing behavior in the high q-region indicative of a rather extended and disordered conformation (Fig. 4D). On the contrary, in the complex crystal structure the inter-UIM linker forms a well-defined loop structure, which implies stabilization of the linker region by binding of tUIM to ubiquitin chains. Importance of the inter-UIM structure in the linkage-specific ubiquitin chain recognition has been previously demonstrated for the RAP80 tUIMs, where the inter-UIM forms a continuous helical structure and results in a selective binding to Lys63-linked di-ubiquitins (Sims and Cohen, 2009, Sato et al., 2009). Therefore, to elucidate the role of AIRAPL inter-UIM linker in ubiquitin-binding, we made various mutants of the linker region including single residue mutations (K214A, K214/G, P215A, and L218A), single residue deletion (ΔP215), and substitution of all linker residues with Gln (linker/Q) and tested for binding to Lys48-linked tri-ubiquitins. The linker/Q mutant was made to have an inter-UIM linker with totally different amino acid composition and potentially different structure compared with the WT protein. As indicated by our CD measurements, tUIM linker/Q has 25% helical content which is lower than tUIM WT (helical content ~32%) (Fig. 4C). Interestingly, none of the abovementioned mutations affected binding of the tUIM to Lys48-linked tri-ubiqiuitin chains, significantly (Fig. 1D, 4E, Fig. S2Q-R). These data imply that amino acid content and length of the inter-UIM linker region are not determining factors for the ubiquitin chain linkage specificity of AIRAPL. However, although the inter-UIM linker is not involved in determining specificity, its flexible nature works towards increasing avidity. While UIM2 defines the selectivity and binds exclusively to Lys48-linked ubiquitin chains (Fig. 1D), its binding is rather weak and requires the inter-UIM linker to place UIM1 in a proper orientation that would facilitate binding of the third ubiquitin in the chain to UIM1.
Importance of the Lys48-linked tri-ubiquitin binding by AIRAPL in vivo
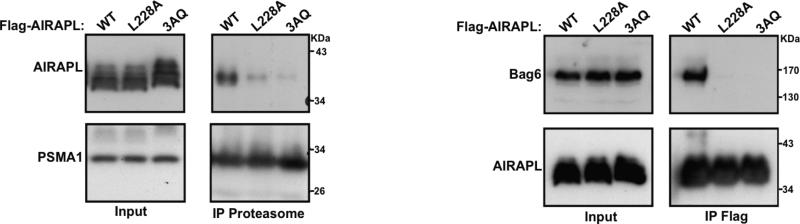
We have previously shown that AIRAPL binding to the 26S proteasome is dependent on its binding to poly-ubiquitin chains (Glinka et al., 2013, Yun et al., 2008). Additionally, Bag6 is a HSP70 co-chaperone involved in various ribosomal and proteasomal quality control processes (Lee and Ye, 2013) and has been recently shown to bind AIRAPL in a UIM-dependent manner (Glinka et al., 2013). Therefore, to investigate the biological relevance of our structural data, we evaluated the ability of AIRAPL to bind proteasome and Bag6 using L228A and the 3AQ (A205/229/231Q) mutants. Indeed, the proteasome-and Bag6- binding ability of both AIRAPL 3AQ and L228A mutants were substantially disrupted (Fig. 5), implying the role of Lys48-linked ubiquitin chain- binding by tUIM in association of AIRAPL with Bag6 and the proteasome. The ubiquitin-binding ability of FL AIRAPL is abolished in 3AQ but the L228A mutant still binds Lys48-linked tri-ubiquitins with a KD of ~ 10 μM (Fig. 1D). Leu228 is located on UIM2, which is responsible for the ubiquitin linkage specificity and is involved in binding to both distal and middle ubiquitins (Fig. S5A). Therefore, we hypothesize that the L228A mutation affects ubiquitin chain linkage selectivity resulting in less efficient signaling and association with the proteasome and Bag6.
Figure 5. The Lys48-linked ubiquitin-binding by AIRAPL tUIM is required for its association with the 26S proteasome (left) and Bag6 (right).
Left-HEK293T cells transiently transfected with Flag-AIRAPL were treated with arsenite and subjected to proteasome affinity purification. Levels of AIRAPL, and 20S proteasome subunit PSMA1 were detected by immunoblotting. Right-HEK293T cells transiently transfected with Flag-AIRAPL were directly subjected to Flag affinity purification, AIRAPL and endogenous Bag6 levels were detected by immunoblotting (See also Fig. S6).
Discussion
The cooperative binding of multiple UBDs to ubiquitin(s) and/or direct recognition of the ubiquitin chain linker regions by UBDs are known as the main factors that determine selectivity towards a specific type of ubiquitin chains (Kulathu et al., 2009, Rahighi et al., 2009, Zhang et al., 2009, Sato et al., 2011). In this study we demonstrated the structural basis for recognition of Lys48-linked tri-ubiquitin chains by the AIRAPL protein. Novelty of the AIRAPL tUIM structure over other, so far identified, UIM domains is the unique architecture of the tUIM which combines a single and a double-sided UIM (UIM2), providing three distinct ubiquitin-binding sites along the AIRAPL polypeptide chain. UIM2 defines selectivity for Lys48 ubiquitin linkage and binds non-symmetrically to two ubiquitin moieties (distal and middle ubiquitins) on either side of the UIM. Moreover, flexibility of the inter-UIM region positions the two UIMs in an orientation, which allows for binding of the third ubiquitin to UIM1 and thereby increases the binding avidity. However, AIRAPL does not recognize the isopeptide bonds between either the proximal-middle or middle-distal ubiquitins, a feature that has been observed in several other UBD/ubiquitin chain structures (Zhang et al., 2009, Kulathu et al., 2009, Sato et al., 2009, Sato et al., 2011).
Among the three ubiquitin moieties in a Lys48-linked ubiquitin chain, distal ubiquitin has the smallest extent of interactions with AIRAPL; nonetheless, this particular mode of binding combined with binding of the middle ubiquitin to the other side of UIM2 helix defines the Lys48 linkage selectivity (Fig. 1D and 4B). Although UIM2 is highly specific for Lys48 linkage and exclusively binds Lys48-linked ubiquitins, AIRAPL tUIM which encompasses both UIM1 and UIM2 binds tri-ubiquitin chains of other linkage types (Met1, Lys11 and Lys63) with a KD of ~13-16 μM (Fig. 1D). This is due to the flexible structure of the inter-UIM linker that results in non-specific interactions with other types of ubiquitin chains than Lys48-linked, albeit with significantly lower affinity.
AIRAPL is a p97 adaptor that has been demonstrated to bind Bag6 and the proteasome (Glinka et al., 2013, Yun et al., 2008) and is involved in the proteasomal processing of pre-emptive quality control substrates (Braunstein et al., 2015). Functional impairment of AIRAPL homologues is linked to the acceleration of ageing and protein aggregation (Yun et al., 2008). While mutations in AIRAPL that eliminate binding to p97 do not impair its ability to bind Bag6 and ubiquitin (Yun et al., 2008), we demonstrate that specific binding of AIRAPL to Lys48-linked ubiquitin chains is required for its interaction with both Bag6 and the proteasome (Fig. 5) as well as pre-emptive quality control substrates that are designated for proteasomal degradation (Braunstein et al., 2015). Therefore, the selective binding of AIRAPL to Lys48-linked poly-ubiquitin chains is not surprising as these chains are considered the canonical targeting signals for the proteasome (Ciechanover and Stanhill, 2014).
Recent findings regarding the architecture of the 19S regulatory particle have enabled modeling of the recruitment and initial degradation steps of ubiquitylated substrates in the proteasome (Beck et al., 2012, da Fonseca et al., 2012, Lander et al., 2012). Proteasomal ubiquitin receptors including Rpn10 and Rpn13 are suggested to cooperate in binding to the ubiquitylated substrates and their localization at the 19S regulatory particle indicates a ~100 Å distance between the two proteins which is in agreement with the required minimal length of tetra-ubiquitin chain for the substrate recruitment (Beck et al., 2012, da Fonseca et al., 2012, Lander et al., 2012, Thrower et al., 2000). Ubiquitin-binding domains of both Rpn10 (UIM) and Rpn13 (Pru) were shown to bind the Ile44 surface of a Lys48-linked chain (Schreiner et al., 2008, Zhang et al., 2009). In a model based on the crystal structure of AIRAPL tUIM/Lys48-linked triubiquitin, one of the ubiquitin receptors (Rpn10 or Rpn13) at the proteasome 19S regulatory particle recognizes distal ubiquitin and the chain is elongated by two ubiquitins from the proximal site to bind the other ubiquitin receptor (Fig. S6). Although the process through which AIRAPL associates with the proteasome complex is not yet elucidated, high affinity binding of AIRAPL to the Lys48-linked tri- and tetra-ubiquitins, and the fact that the Ile44 surface on the distal ubiquitin is still available for binding to the ubiquitin receptors are in line with the structural requirements for recruiting a Lys48-linked ubiquitylated substrate to the proteasome and passing it onto the proteasome without its disengagement from AIRAPL.
Experimental procedures
Mass spectrometry
The analysis of AIRAPL and MEKK1 (mitogen-activated protein kinase kinase binding) poly-ubiquitin linkage types by tandem mass spectrometry was performed essentially as described (Sobott et al., 2009). In brief, excised gel bands were subjected to in-gel digestion with trypsin using chloroacetamide for the alkylation step to minimize false positive identification of G-G adducts on lysine side chains. For the analysis of digested protein material, liquid chromatography was performed using an Ultimate 3000 nano-HPLC system (Thermo/Dionex) coupled to a 3D high capacity ion trap mass spectrometer (HCTplus, Bruker Daltonics) utilizing 10 m ID distal coated SilicaTips (New Objective) in positive nano-electrospray ionization (ESI) mode. Up to 4 precursor ions were selected per cycle with active exclusion (0.5 min) in CID mode excluding singly charged ions. CID fragmentation was achieved using helium gas and a 30-200 % collision energy sweep with amplitude 1.0 (ions are ejected from the trap as soon as they fragment). Raw LC-MS/MS data were processed and Mascot-compatible files created using DataAnalysis 3.4 software (Bruker Daltonics). Searches were performed using Mascot software (version 2.2) and the SwissProt database (release 54.0, 07/2007, number of entries 276256) with the following parameters: 2+ and 3+ ions, peptide tolerance 1.2 Da, 13C=1, fragment tolerance 0.6 Da, missed cleavages: 2, instrument type: ESI-TRAP (for CID) and ETD-TRAP, respectively, for the two separate data sets. The G-G tag on lysine residues (+114.1 Da) was included as a variable modification. The interpretation and presentation of MS/MS data was performed according to published guidelines (Taylor and Goodlett, 2005). Individual MS/MS spectra for peptides with a Mascot Mowse score lower than 40 were inspected manually and only included if a series of at least 4 continuous fragment ions was observed.
Crystallization, data collection, and structure determination
Co-crystals of AIRAPL/Lys48-linked tri-ubiquitin grew in sitting drops containing 20% (w/v) polyethylene glycol 3350, 0.02 M calcium chloride, 0.02 M cadmium chloride, and 0.02 M cobalt chloride in the reservoir solution. Crystals appeared after 120 days and tested for x-ray diffraction using 20 % glycerol in reservoir solution as the cryoprotectant. X-ray diffraction data was collected to 3.0 Å resolution at 100K at the beamline BL-17A of Photon Factory, KEK (Tsukuba, Japan) and processed using iMOSFLM (Leslie, 2007). Examination of the data with POINTLESS (Evans, 2006, Evans, 2011) and CHECK from the CCP4 package (Collaborative Computational Project, Number 4, 1994) indicated the space group as P31 bearing merohedral twinning with alpha (twinning fraction) equal to 0.3 and a twin operator of –h, h+k, −l. The structure was solved using mono-ubiquitin (PDB: 1UBQ) and a truncated single α-helix from the Rap80 (PDB: 3A1Q) structures as search models for molecular replacement in MOLREP. The model was further built using COOT (Emsley and Cowtan, 2004) and refined by the application of amplitude-based twin refinement in REFMAC5 (Murshudov et al., 2011, Murshudov et al., 1997). Data collection and refinement statistics are summarized in Table 1, and the atomic coordinates and structure factors are deposited to the PDB database under accession code 4XKH. All structure figures were prepared in PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.5 Schrödinger, LLC.).
Surface Plasmon Resonance (SPR) binding assays
The SPR experiments were performed using the BiOptix 404pi instrument (BiOptix, Boulder, Colorado). His-tagged AIRAPL FL, tUIM or ΔUIM1 were immobilized on the Xantech NiNTA (NiHC1000m, BiOptix) sensor chip. Running buffer contained PBS supplemented with 50 μM EDTA, 200 mM NaCl, and 0.005 % Tween-20. After immobilization of the ligand, 20 μl of 2 mg/ml BSA in running buffer was injected to reduce non-specific interaction of ubiquitin chains with surface of the chip. Each experiment was repeated at least two times and results were analyzed using Scrubber 2 (BioLogic software).
Small Angle X-ray Scattering
SAXS experiments were performed at the Bio-SAXS beamline BL4-2 at Stanford Synchrotron Radiation Light source (SSRL) (Igor L. Smolsky, 2007). Data were collected using a Rayonix MX225-HE CCD detector (Rayonix, Evanston, IL) with a 1.1 m sample-to-detector distance and beam energy of 11 keV (wavelength, λ = 1.127 Å). SAXS data were measured in the range of 0.01 Å−1 ≤ q ≤ 0.7 Å−1 (q = 4πsin(θ)/λ with 2θ being the scattering angle). The q scale was calibrated with silver behenate powder and water scattering intensity was used for absolute intensity scaling. Online SEC-SAXS experiments were performed with Lys48-linked ubiquitins and its complex with AIRAPL in a buffer containing 50mM Tris-HCl (pH=8.0), 150mM NaCl and 5mM DTT (as a radical scavenger to minimize radiation damage). Complex form was prepared in advance by mixing Lys48-linked ubiquitins and AIRAPL at a molar ratio of 1:5. A 50ul of ~4 mg/ml sample (concentration of Lys48-linked ubiquitins in solution) was applied into a Superdex 75 3.2/300 column with a flow rate of 0.05 ml/min. The details of SEC-SAXS procedure at BL4-2 have been previously described (Edwards et al., Matsui et al., 2014). Equilibrium SAXS data of the AIRAPL were collected in 50 mM Tris-HCl buffer (pH=8.0) containing 150 mM NaCl. Concentration series were measured from 0.25 mg/ml to 8.2 mg/ml in order to detect (and eliminate) concentration-dependent intermolecular interactions. The SAXS data were taken in series of twelve two-second exposures. These images were then analyzed for possible effects of radiation damage, normalized according to the transmitted intensity and averaged using the program SasTool (http://ssrl.slac.stanford.edu/~saxs/analysis/sastool.htm). The scaled and averaged buffer curve was then subtracted from the averaged protein curve. A single SAXS profile was produced by merging the low and high concentration data. The SAXS profiles of SEC-SAXS were generated by averaging 10 frames (1sec exposure per frame) around the main peak. No concentration dependence and radiation damage was observed over the elution. The P(r) and Porod volume were determined by using the program GNOM (Svergun, 1992).
Circular Dichroism (CD) measurements
The CD experiments were performed using a Jasco J-815 (Jasco Analytical Instruments) with 1.0 nm bandwidth and scanning speed of 50 nm/min. Each measurement was repeated 3 times and “mean residue ellipticity” values were calculated as described in reference 54 (Kelly et al., 2005). Helical contents of the proteins were determined using absorption at λ: 208 and λ: 222 as in (Colin J. Barrow, 1992): ([θ]208 − 4000 / −33,000 − 4000) × 100 and ([θ]222 − 3000 / −36,000 − 3000) × 100.
In vitro GST pull-down assays
GST-tagged proteins immobilized on the GS4B beads (GE Healthcare) were incubated with 1 μg of ubiquitin chains in PBS buffer containing 0.5 % Triton-X100 for 2 hours at 4°C. The beads were then washed 3 times with the same buffer and boiled with Laemmli buffer for 1 minute. Western blots were performed using PVDF membrane and anti-ubiquitin antibody (P4D1, Santa Cruz Biotechnology) and detected by Odyssey infrared imaging system (LI-COR Biosciences).
Cell culture and immunopurifications
HEK293T cells were grown in 10% FCS DMEM medium. Cell lysis was performed in TNH buffer (20 mM HEPES, pH 7.9; 100 mM NaCl; 1% Triton X-100; 1 mM EDTA; 1.5 mM MgCl2; 20 mM NEM and protease inhibitors) and lysate clarified at 20,000xg for 10 minutes. For endogenous AIRAPL polyubiquitin linkage immunoblots, immunopurifications of AIRAPL using a polyclonal AIRAPL antibody were performed (Yun et al., 2008). Purified material was resolved on SDS-PAGE and analyzed for linkage specificity byusing Ub linkage specific antibodies (Newton et al., 2012). For proteasomal interactions with the indicated AIRAPL mutants, Flag-AIRAPL mammalian expression vectors were transiently transfected in HEK293T cells. Untreated cell lysates were Flag affinity purified to detect Bag6 interactions. Alternatively, cells were arsenite treated (0.5 mM 30 min), proteasomes were affinity purified by using a PSMA1 antibody, as previously described (Yun et al., 2008) and AIRAPL interaction evaluated by Flag immunoblot. For mass-spectrometry ubiquitin chain analysis, ectopic expression of Flag-AIRAPL, its UIM domain or MEKK1 fused to Flag-GFP was performed in HEK293T cells. Flag affinity purification followed by peptide elution (M2, Sigma Aldrich) was performed and resolved on SDS-PAGE. HMW parts of the Coomassie stained gels were analyzed by mass-spectrometry.
Supplementary Material
Highlights.
Crystal structure of tandem UIMs of AIRAPL in complex with Lys48Ub3 was determined.
AIRAPL contains two ubiquitin-interacting motifs connected via a flexible linker.
UIM2 is double-sided and determines the specificity for Lys48-linked ubiquitins.
Inter-UIM linker increases avidity by facilitating binding of ubiquitin to UIM1.
Acknowledgments
We would like to thank staff of the Photon Factory BL-17A for their support with X-ray data collection, R. Sertchook for initial analysis of AIRAPL tUIM configuration, M. Edri and V. Bronner from Bio-Rad Haifa Protein Technologies for initial SPR experiments, F. Jabbarpour and V. Bhadkamkar for their help in the preparation of proteins for binding assays, and K. Husnjak, J. Hermann, and J. Tan for critical reading of the manuscript. The research was supported by the Israeli Science Foundation (ISF) grant 438/14 to A.S., and S.R. was a recipient of the JSPS Fellowship for Research in Japan (Long-Term) and Katherine McCormick Advanced Postdoctoral Fellowship at Stanford. S.W. is a Stanford Bio-X Affiliated Faculty and part of this work was conducted at the James H. Clark Center, the hub for Stanford Bio-X. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health National Institute of General Medical Sciences (including P41GM103393).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
S.W., A.S. and S.R. designed the research. S.R. performed the crystallography, CD and in vitro binding assays, I.B. purified proteins for MS and in vivo experiments. N.T. and B.K. performed MS, T.M. and T.M.W. performed SAXS experiments. M.K. and R.K. provided expertise and feedback. S.R, A.S., S.W, T.M and T.M.W. wrote the paper. All authors reviewed and contributed to the manuscript.
Accession numbers
Atomic coordinates and structure factors of the AIRAPL tUIMs/ Lys48-linked triubiquitin complex structure are deposited in the Protein Data Bank, under accession code 4XKH.
Supplemental Information
Supplemental information includes five figures, experimental procedures and references.
References
- BECK F, UNVERDORBEN P, BOHN S, SCHWEITZER A, PFEIFER G, SAKATA E, NICKELL S, PLITZKO JM, VILLA E, BAUMEISTER W, FORSTER F. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci U S A. 2012;109:14870–5. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRAUNSTEIN I, ZACH L, ALLAN S, KALIES KU, STANHILL A. Proteasomal degradation of pre-emptive quality control (pQC) substrates is mediated by aN AIRAPL-p97 complex. Mol Biol Cell. 2015 doi: 10.1091/mbc.E15-02-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUDHAVARAPU VN, WHITE ED, MAHANIC CS, CHEN L, LIN FT, LIN WC. Regulation of E2F1 by APC/C Cdh1 via K11 linkage-specific ubiquitin chain formation. Cell Cycle. 2012;11:2030–8. doi: 10.4161/cc.20643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAU V, TOBIAS JW, BACHMAIR A, MARRIOTT D, ECKER DJ, GONDA DK, VARSHAVSKY A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–83. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- CIECHANOVER A, STANHILL A. The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim Biophys Acta. 2014;1843:86–96. doi: 10.1016/j.bbamcr.2013.07.007. [DOI] [PubMed] [Google Scholar]
- COLIN J, BARROW AY, KENNY PETERTM, ZAGORSKI MICHAELG. Solution conformations and aggregational properties of synthetic amyloid beta-peptides of Alzheimer's disease. J. Mol. Biol. 1992;225:1075–1093. doi: 10.1016/0022-2836(92)90106-t. [DOI] [PubMed] [Google Scholar]
- DA FONSECA PC, HE J, MORRIS EP. Molecular model of the human 26S proteasome. Mol Cell. 2012;46:54–66. doi: 10.1016/j.molcel.2012.03.026. [DOI] [PubMed] [Google Scholar]
- DIKIC I, WAKATSUKI S, WALTERS KJ. Ubiquitin-binding domains -from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–71. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EDWARDS AL, MATSUI T, WEISS TM, KHOSLA C. Architectures of Whole-Module and Bimodular Proteins from the 6-Deoxyerythronolide B Synthase. Journal of Molecular Biology. 2014;426:2229–2245. doi: 10.1016/j.jmb.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMSLEY P, COWTAN K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- EVANS P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- EVANS PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67:282–92. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FISHER RD, WANG B, ALAM SL, HIGGINSON DS, ROBINSON H, SUNDQUIST WI, HILL CP. Structure and ubiquitin binding of the ubiquitin-interacting motif. J Biol Chem. 2003;278:28976–84. doi: 10.1074/jbc.M302596200. [DOI] [PubMed] [Google Scholar]
- GLINKA T, ALTER J, BRAUNSTEIN I, TZACH L, WEI SHENG C, GEIFMAN S, EDELMANN M, KESSLER B, STANHILL A. Signal-peptide mediated translocation is regulated by a P97-AIRAPL complex. Biochem J. 2013 doi: 10.1042/BJ20130710. [DOI] [PubMed] [Google Scholar]
- HASSAN WM, MERIN DA, FONTE V, LINK CD. AIP-1 ameliorates beta-amyloid peptide toxicity in a Caenorhabditis elegans Alzheimer's disease model. Hum Mol Genet. 2009;18:2739–47. doi: 10.1093/hmg/ddp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HICKE L, SCHUBERT HL, HILL CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–21. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- HIRANO S, KAWASAKI M, URA H, KATO R, RAIBORG C, STENMARK H, WAKATSUKI S. Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nat Struct Mol Biol. 2006;13:272–7. doi: 10.1038/nsmb1051. [DOI] [PubMed] [Google Scholar]
- HOFMANN K, FALQUET L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci. 2001;26:347–50. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- HUSNJAK K, DIKIC I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- SMOLSKY IGORL, P. L., NIEBUHR MARC, ITO KAZUKI, WEISS THOMASM, TSURUTA HIRO. Biological small-angle X-ray scattering facility at the Stanford Synchrotron Radiation Laboratory. Journal of Applied Crystallography. 2007;40(Supplement):s453–s458. [Google Scholar]
- IKEDA F, CROSETTO N, DIKIC I. What determines the specificity and outcomes of ubiquitin signaling? Cell. 2010;143:677–81. doi: 10.1016/j.cell.2010.10.026. [DOI] [PubMed] [Google Scholar]
- KELLY SM, JESS TJ, PRICE NC. How to study proteins by circular dichroism. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- KRAVTSOVA-IVANTSIV Y, CIECHANOVER A. Non-canonical ubiquitin-based signals for proteasomal degradation. J Cell Sci. 2012;125:539–48. doi: 10.1242/jcs.093567. [DOI] [PubMed] [Google Scholar]
- KULATHU Y, AKUTSU M, BREMM A, HOFMANN K, KOMANDER D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol. 2009;16:1328–30. doi: 10.1038/nsmb.1731. [DOI] [PubMed] [Google Scholar]
- KULATHU Y, KOMANDER D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13:508–23. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- LANDER GC, ESTRIN E, MATYSKIELA ME, BASHORE C, NOGALES E, MARTIN A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–91. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANGE A, CASTANEDA C, HOELLER D, LANCELIN JM, FUSHMAN D, WALKER O. Evidence for cooperative and domain-specific binding of the signal transducing adaptor molecule 2 (STAM2) to Lys63-linked diubiquitin. J Biol Chem. 2012;287:18687–99. doi: 10.1074/jbc.M111.324954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE JG, YE Y. Bag6/Bat3/Scythe: a novel chaperone activity with diverse regulatory functions in protein biogenesis and degradation. Bioessays. 2013;35:377–85. doi: 10.1002/bies.201200159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LESLIE AGW, POWELL HR. Processing diffraction data with mosflm . Evolving Methods for Macromolecular Crystallography. 2007;245:41–51. [Google Scholar]
- MATSUI T, GU SY, LAM KH, CARTER LG, RUMMEL A, MATHEWS II, JIN RS. Structural Basis of the pH-Dependent Assembly of a Botulinum Neurotoxin Complex. Journal of Molecular Biology. 2014;426:3773–3782. doi: 10.1016/j.jmb.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATSUMOTO ML, WICKLIFFE KE, DONG KC, YU C, BOSANAC I, BUSTOS D, PHU L, KIRKPATRICK DS, HYMOWITZ SG, RAPE M, KELLEY RF, DIXIT VM. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–84. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- MURSHUDOV GN, SKUBAK P, LEBEDEV AA, PANNU NS, STEINER RA, NICHOLLS RA, WINN MD, LONG F, VAGIN AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MURSHUDOV GN, VAGIN AA, DODSON EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- NEWTON K, MATSUMOTO ML, FERRANDO RE, WICKLIFFE KE, RAPE M, KELLEY RF, DIXIT VM. Using linkage-specific monoclonal antibodies to analyze cellular ubiquitylation. Methods Mol Biol. 2012;832:185–96. doi: 10.1007/978-1-61779-474-2_13. [DOI] [PubMed] [Google Scholar]
- ONISHI KH, BAILLARGEON R, LESLIE AM. 15-month-old infants detect violations in pretend scenarios. Acta Psychol (Amst) 2007;124:106–28. doi: 10.1016/j.actpsy.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETOUKHOV MV, FRANKE D, SHKUMATOV AV, TRIA G, KIKHNEY AG, GAJDA M, GORBA C, MERTENS HDT, KONAREV PV, SVERGUN DI. New developments in the ATSAS program package for small-angle scattering data analysis. Journal of Applied Crystallography. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAHIGHI S, DIKIC I. Selectivity of the ubiquitin-binding modules. FEBS Lett. 2012;586:2705–10. doi: 10.1016/j.febslet.2012.04.053. [DOI] [PubMed] [Google Scholar]
- RAHIGHI S, IKEDA F, KAWASAKI M, AKUTSU M, SUZUKI N, KATO R, KENSCHE T, UEJIMA T, BLOOR S, KOMANDER D, RANDOW F, WAKATSUKI S, DIKIC I. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- SAKATA E, BOHN S, MIHALACHE O, KISS P, BECK F, NAGY I, NICKELL S, TANAKA K, SAEKI Y, FORSTER F, BAUMEISTER W. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc Natl Acad Sci U S A. 2012;109:1479–84. doi: 10.1073/pnas.1119394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SATO Y, FUJITA H, YOSHIKAWA A, YAMASHITA M, YAMAGATA A, KAISER SE, IWAI K, FUKAI S. Specific recognition of linear ubiquitin chains by the Npl4 zinc finger (NZF) domain of the HOIL-1L subunit of the linear ubiquitin chain assembly complex. Proc Natl Acad Sci U S A. 2011;108:20520–5. doi: 10.1073/pnas.1109088108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SATO Y, YOSHIKAWA A, MIMURA H, YAMASHITA M, YAMAGATA A, FUKAI S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 2009;28:2461–8. doi: 10.1038/emboj.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHREINER P, CHEN X, HUSNJAK K, RANDLES L, ZHANG N, ELSASSER S, FINLEY D, DIKIC I, WALTERS KJ, GROLL M. Ubiquitin docking at the proteasome through a novel pleckstrinhomology domain interaction. Nature. 2008;453:548–52. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHWARTZ AL, CIECHANOVER A. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol. 2009;49:73–96. doi: 10.1146/annurev.pharmtox.051208.165340. [DOI] [PubMed] [Google Scholar]
- SIMS JJ, COHEN RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell. 2009;33:775–83. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOBOTT F, WATT SJ, SMITH J, EDELMANN MJ, KRAMER HB, KESSLER BM. Comparison of CID versus ETD based MS/MS fragmentation for the analysis of protein ubiquitination. J Am Soc Mass Spectrom. 2009;20:1652–9. doi: 10.1016/j.jasms.2009.04.023. [DOI] [PubMed] [Google Scholar]
- SVERGUN DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- SWANSON KA, KANG RS, STAMENOVA SD, HICKE L, RADHAKRISHNAN I. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 2003;22:4597–606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAYLOR GK, GOODLETT DR. Rules governing protein identification by mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:3420. doi: 10.1002/rcm.2225. [DOI] [PubMed] [Google Scholar]
- THROWER JS, HOFFMAN L, RECHSTEINER M, PICKART CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XU P. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–45. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOUNG P, DEVERAUX Q, BEAL RE, PICKART CM, RECHSTEINER M. Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J Biol Chem. 1998;273:5461–7. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- YUN C, STANHILL A, YANG Y, ZHANG Y, HAYNES CM, XU CF, NEUBERT TA, MOR A, PHILIPS MR, RON D. Proteasomal adaptation to environmental stress links resistance to proteotoxicity with longevity in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2008;105:7094–9. doi: 10.1073/pnas.0707025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG N, WANG Q, EHLINGER A, RANDLES L, LARY JW, KANG Y, HARIRINIA A, STORASKA AJ, COLE JL, FUSHMAN D, WALTERS KJ. Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol Cell. 2009;35:280–90. doi: 10.1016/j.molcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO S, ULRICH HD. Distinct consequences of posttranslational modification by linear versus K63-linked polyubiquitin chains. Proc Natl Acad Sci U S A. 2010;107:7704–9. doi: 10.1073/pnas.0908764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.