Abstract
Acinetobacter baumannii has emerged as a notorious multidrug-resistant pathogen, and development of novel control measures is of the utmost importance. Understanding the factors that play a role in drug resistance may contribute to the identification of novel therapeutic targets. Pili are essential for A. baumannii adherence to and biofilm formation on abiotic surfaces as well as virulence. In the present study, we found that biofilm formation was significantly induced in an imipenem-resistant (Impr) strain treated with a subinhibitory concentration of antibiotic compared to that in an untreated control and an imipenem-susceptible (Imps) isolate. Using microarray and quantitative PCR analyses, we observed that several genes responsible for the synthesis of type IV pili were significantly upregulated in the Impr but not in the Imps isolate. Notably, this finding is corroborated by an increase in the motility of the Impr strain. Our results suggest that the ability to overproduce colonization factors in response to imipenem treatment confers biological advantage to A. baumannii and may contribute to clinical success.
INTRODUCTION
Acinetobacter baumannii is one of the most important agents of multidrug-resistant (MDR) hospital infections, and it has been associated with worldwide mortality rates as high as 43.4% (bloodstream infections) and 60% (community-acquired pneumonia) (1, 2). Current treatment of A. baumannii infections involves the use of β-lactams (i.e., imipenem), aminoglycosides, polymyxins, tigecycline, and tetracyclines either alone or in combination (1). However, with the increasing incidence of MDR A. baumannii, options for effective chemotherapies have become very limited, and the lack of new antimicrobials in clinical trials raises concern among practitioners and scientists. Current efforts to develop new antibiotics have been focusing on the characterization of virulence factors, such as pili and biofilms (3, 4), whereby the rationale is based on the observation of a correlation between virulence and drug resistance (5).
Of note, bacterial pili are composed of proteins that confer adhesive properties or that serve as a scaffold for the deposit of molecules that mediate bacterial interaction with the environment. In addition, these organelles facilitate DNA uptake and transfer, play a critical role in motility, and can act as receptors for bacteriophages (6–12). In fact, the A. baumannii Csu pili were found to be essential for attachment to plastic and for biofilm formation (13, 14). Bacterial biofilms are arrangements in which participating cells are morphologically, metabolically, and physiologically distinct from their planktonic counterparts (11, 15, 16). These structures represent a defense mechanism employed for bacterial survival in harsh conditions and during infections of the host, conferring greater resistance against disinfectants and antimicrobials than that exhibited by planktonic cells (17–20). Additionally, A. baumannii biofilms play an important role in immune evasion (21–23), contributing to the extension of the pathogen's survival and to infectivity in clinical settings (4). In this study, we investigated the effect of a subinhibitory concentration of imipenem in an imipenem-resistant (Impr) A. baumannii strain and observed an increase in pilus synthesis, biofilm formation, and motility, which correlate with the upregulation of the pilin genes.
MATERIALS AND METHODS
Bacterial strains and MIC determination.
AB08-5110S is an A. baumannii clinical isolate susceptible to imipenem (Imps). The imipenem-resistant strain AB08-5110R (Impr) was generated by exposing the Imps strain to 16 successive in vitro passages (1:1 dilution in equal proportions with fresh medium at 37°C and 180 rpm agitation) in Mueller-Hinton (MH) broth under subinhibitory concentrations of imipenem (16 mg/liter). The Impr strain was streaked onto heart infusion (HI) agar (Oxoid) after the 16th passage, and a single colony from each plate was subcultured at 37°C and 180 rpm agitation and was used to determine the MIC of imipenem using the broth dilution method. MICs were confirmed by two independent replicates of the Etest (Oxoid), and Escherichia coli ATCC 25922 was used as the control strain. The MIC results were interpreted according to 2010 CLSI guidelines. Finally, pulsed-field gel electrophoresis (PFGE) was employed for genetic profiling and for the confirmation of the two strains as noncontaminants, and bacterial strains were stored at −70°C in a brain heart infusion (BHI) medium containing 20% glycerol.
Growth curves.
The growth kinetics of our strains were determined in the absence and in the presence of sub-MIC (16 and 1 mg/liter), MIC (32 and 2 mg/liter), and 2× MIC (64 and 4 mg/liter) of imipenem for Impr and Imps strains, respectively. Briefly, cultures in the stationary phase were used to inoculate 5-ml aliquots of MH broth to an initial optical density at 600 nm (OD600) of 0.01. These cultures were then incubated at 37°C and 180 rpm agitation. When bacterial growth reached an OD600 of 0.35 (early log phase, chosen based on the literature), the cultures were supplemented with sub-MIC, MIC, and 2× MIC of imipenem (Merck). Cultures were then incubated at 37°C under constant shaking, and their turbidity values (OD600) were estimated every 30 min. This assay was repeated at least three times for each strain. GraphPad Prism was used for the determination of growth curves and statistical significance (t test).
TEM analysis.
A. baumannii Impr and Imps strains were streaked onto HI agar and were grown overnight at 37°C. Three colonies from each plate were then inoculated separately into 10 ml of MH broth, and the cultures were incubated overnight at 37°C and 150 rpm agitation. Overnight cultures were used for a 1:100 inoculation (OD600, 0.05) and were incubated at 37°C and 180 rpm agitation until an OD600 of 0.35 was reached, and then imipenem was added as previously mentioned. Bacterial cultures were further incubated for 3 to 4 h and were harvested for preparation prior to transmission electron microscopy (TEM). Untreated bacterial cultures were used as controls. For pilus observation, whole-cell mounts were prepared using noncentrifuged bacterial cells suspended in growth medium. One drop of the bacterial suspension was placed onto a 50-nm-thick Formvar film that was previously placed onto a copper index TEM grid (Marivac). The cell suspension was allowed to air dry, and after 5 to 10 min, the remaining solution was removed using filter paper. The samples were rinsed with 2 ml of HEPES buffer (pH 6.8) followed by 1 ml of deionized water and were negatively stained by placing a drop of phosphotungstic acid (PTA) onto the samples for 1 to 2 min. The mixture was removed by pipetting, the excess liquid was removed with filter paper, and the samples were immediately examined using a Hitachi H-7100 TEM (Hitachi High-Technologies Corporation).
Biofilm assay.
Biofilm formation was assessed and quantified by slightly modifying the protocol described by Cevahir et al. (24). A. baumannii strains were grown overnight in LB broth at 37°C. The next day, the bacterial cultures were diluted to 1 × 106 CFU/ml, and 10 μl was used to inoculate sterile 96-well Costar flat bottom polystyrene plates (Corning) containing 200 μl of fresh LB broth. At 24 h postinoculation at 37°C, the 96 wells were washed 3 times with 200 μl of sterile phosphate-buffered saline (PBS) and then allowed to air dry. Biofilms were stained with crystal violet (1%, wt/vol), and optical density (OD590) was measured using a microplate reader. Each assay was performed in triplicate. The following values were assigned for biofilm determination: an OD595 of <1, no biofilm; an OD595 of ≥1 but of <2, weak biofilm; an OD595 of ≥ 2 but of <3, average biofilm; and an OD595 of ≥3, strong biofilm. Of note, LB broth was used as the milieu to assess biofilm formation as a rich medium and to create consistency in relation to other publications that correlate antibiotic resistance to increased biofilm formation in A. baumannii. Therefore, although MH broth was employed in other assays, we believe that upregulation of the target genes would be observed irrespective of the nutritional conditions, as it appeared to be a response to supplementation with a sub-MIC of imipenem.
Microarray and qRT-PCR.
Overnight cultures were diluted 1:100 in MH broth medium and were incubated at 37°C under 180 rpm until they reached early mid-log phase (OD600, 0.35 to 0.4); then a sub-MIC of imipenem was added, and the Impr and Imps cultures were incubated until late log phase. Untreated cultures were used as controls. For gene expression analyses, RNA was extracted using the TRI reagent (Molecular Research Center) according to the manufacturer's protocol and was purified twice using an RNeasy Mini RNA isolation kit (Qiagen). RNA samples were evaluated for their purity and concentration using the Implen NanoPhotometer (Implen GmbH), and integrity was evaluated using microfluidic capillary electrophoresis (Agilent 2100 Bioanalyzer). Samples with an RNA integrity number (RIN) of ≥8 were used for microarray and quantitative reverse transcription-PCR (qRT-PCR) analyses. Subsequently, 8 × 15K custom genomic microarrays, representing each A. baumannii AYE coding sequence, were developed using the Agilent eArray package (Agilent Technologies). At least four 60-mer DNA oligonucleotides were incorporated into the chip design, and pilus gene expression was determined as part of the A. baumannii transcriptome. For cDNA synthesis and microarray hybridization, approximately 200 ng of each total RNA was polyadenylated and reverse transcribed using the T7 oligonucleotide (deoxyribosylthymine [dT]) primer. Following cDNA synthesis, the cRNA was transcribed and labeled using the Quick-Amp Labeling kit (Agilent Technologies) according to the manufacturer's protocol. The labeled cRNA was then purified using the RNeasy minikit (Qiagen) according to the manufacturer's instructions and was fragmented and hybridized to the custom-designed 8 × 15K one-color gene expression microarray slide for 16 h at 65°C. After washing, slides were scanned using an Agilent G2565CA microarray scanner (Agilent Technologies) at a 5-μm scanning resolution. The microarray data were extracted using Feature Extraction software v10.1 and were normalized and analyzed using GeneSpring GX 10.0 analysis platform software (Agilent Technologies). Signal intensity values were adjusted to the minimal intensity (0.05), and data were normalized to the 50th percentile per chip. Differential gene expression was assessed using a volcano plot and was determined as a ≥2-fold change with a false discovery rate of 0.05. Unpaired t test analysis was used to determine statistical significance. Finally, quantitative RT-PCR was employed to confirm the expression levels of the type IV pilus (TFP) genes pilN, pilV, pilT, pilQ, pilM, pilW, pilP, pilJ, pilO, pliL, and pilY and a type IV pilin protein gene (Table 1; see also Table S1 in the supplemental material). Bacterial cDNA was synthesized using 200 ng of total RNA, which was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad Laboratories). Gene sequences were retrieved from GenBank, and gene-specific primers were designed using the Primer Express software v3.0 (Applied Biosystems), which produces 50- to 150-bp amplicons. Primer pair efficiency was determined by carrying out RT-PCR on serial dilutions of cDNA, and valid pairs presented efficiency of amplification between 90% and 100% with a minimum R2 of 0.98. A SYBR green PCR master mix was used following the manufacture's protocol (Applied Biosystems), and experiments were performed in triplicate using the StepOnePlus real-time PCR system (Applied Biosystems). A final volume of 20 μl was adopted for each reaction mixture, which contained 10 μl Power SYBR green master mix (250 nM for each primer), 7 μl nuclease-free water (NFW), and 5-ng cDNA template (0.05-ng template for 16S rRNA). The quantitative PCR (qPCR) thermal cycling parameters were as follows: 10 min at 95°C for polymerase activation followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The specificity of the amplicons was verified by melting curve analysis (60°C to 95°C) with a heating rate of 0.3°C per 15 s. The experiments were performed in triplicate. No-template and no-reverse transcription controls were included. The relative quantitation was determined by the ΔΔCT method after normalizing it to the endogenous gene. Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software). A P value of <0.05 was considered significant.
TABLE 1.
A. baumannii Impr gene dysregulation determined by microarray and confirmed by qPCR
| Gene name | ORF no. (ATCC 17978) | Fold changea |
Biological role | |
|---|---|---|---|---|
| Microarray | qPCR | |||
| pilM | A1S_3195 | 2.9 | 7.5 | Required for cell surface expression of type IV pili and associated with twitching motility |
| pilN | A1S_3194 | 3.9 | 11.1 | |
| pilO | A1S_3193 | 2.6 | 4.8 | |
| pilV | AS1_3882 | 3.8 | 3.9 | Required for type IV pili adherence by promoting the functional display of PilC (the adhesion protein) |
| pilT | A1S_0897 | 3.8 | 5.6 | Type IV pilin assembly protein required for twitching motility |
| pilQ | A1S_3191 | 3.2 | 5.7 | Plays a central role in extruding the pilus fibers through the outer membrane |
| pilW | A1S_3168 | 2.9 | 5.9 | Type IV pilin assembly proteins required for twitching motility; plays an essential role in PilQ stability |
| pilP | A1S_3192 | 2.8 | 5.1 | |
| pilJ | A1S_2812 | 2.7 | 3.8 | Regulator of twitching motility by controlling pilus extension and retraction |
| pilL | A1S_2811 | 2.8 | 6.0 | Type IV pilus hybrid sense kinase/receptor regulator |
| pilY | A1S_3167 | 2.1 | 3.4 | Type IV fimbriae biogenesis protein tip-associated adhesin |
| Type IV pili | AS1_3883 | 2.7 | 5.3 | Type IV pilin structural subunit |
Induced by a sub-MIC of imipenem (16 mg/liter).
Motility assay.
Twitching motility was investigated as previously described (25). Briefly, Imps and Impr overnight cultures were stab inoculated through 1% Mueller-Hinton (MH) agar plates and were incubated at 37°C overnight. Of note, cultures and plates were employed with and without a sub-MIC of imipenem, and twitching motility was measured at the agar-plate interface. Motile strains were defined as those exhibiting a zone of >10 mm around the site of inoculation, and assays were performed in triplicate. Statistical analyses were performed using GraphPad Prism 6.
RESULTS
An imipenem-resistant A. baumannii strain survives during antibiotic treatment.
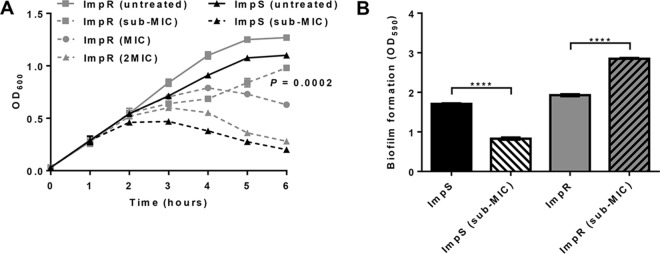
The MIC values of imipenem for A. baumannii Impr and Imps strains were found to be 32 mg/liter and 2 mg/liter, respectively. In addition, in vitro subculturing of the two strains under different concentrations of the antibiotic (sub-MIC, MIC, and 2× MIC) revealed various growth rates, whereby the A. baumannii Impr strain sustained normal growth at a sub-MIC (0.5-fold) (Fig. 1A). Furthermore, transmission electron microscopy (TEM) demonstrated that only minor morphological changes occurred in Impr under that concentration, which is contrary to the cell damage observed when the Imps strain was exposed to the same condition (see Fig. S1 in the supplemental material).
FIG 1.
Imipenem-resistant A. baumannii is biologically adapted to survive antibiotic treatment. (A) Growth of Imps and Impr strains under different concentrations of imipenem was assessed every hour over a 6-h period. Survival of the resistant strain under sub-MIC treatment was significantly increased compared to that of the Imps strain (four independent experiments; P = 0.0002, two-way ANOVA). (B) The mean biofilm formation per strain was determined (OD590, mean for 3 biological replicates. ****, P < 0.001, t test).
Biofilms are complex structures, often polymicrobial communities constituted by proteins, polysaccharides, and other molecules. One of the main factors influencing biofilm formation is the synthesis of bacterial surface components and cell appendages (i.e., pili, flagella, adhesins) (26, 27). Different Acinetobacter pili were found to be essential for (i) biofilm formation on medically relevant abiotic surfaces, such as polystyrene (11), (ii) adherence to biotic and abiotic surfaces (28), and (iii) twitching motility (29). Therefore, considering the critical role of pili in A. baumannii biofilm formation and the fact that Impr pilus synthesis is not affected by a subinhibitory concentration of imipenem, we decided to assess the impact of the antibiotic treatment on Impr biofilm production. Interestingly, we found that its exposure to a sub-MIC of imipenem significantly induced biofilm synthesis compared to that in Imps and uninduced Impr isolates (Fig. 1B).
Pilus gene expression is altered in response to a subinhibitory concentration of imipenem.
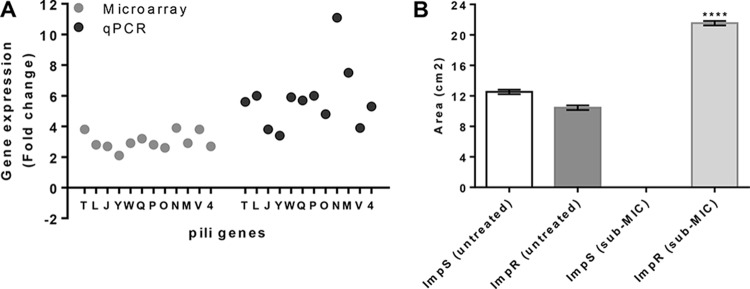
The results of our biofilm formation assay and TEM analysis suggest that the Impr strain is resistant to the cell-damaging effects caused by a subinhibitory concentration of imipenem, as would be expected from a resistant isolate. Accordingly, biofilms have been reported to prevent damage that directly kills bacterial cells (30). This was shown to be due, in part, to drug diffusion issues caused by altered biofilm architecture, as observed in resistant mutants and persister cells (31). Since biofilms are known to be dependent on pilus synthesis, we employed microarrays to investigate whether the expression of pilin-related genes would contribute to an explanation of our results. Microarray experiments were performed separately for each sub-MIC-treated A. baumannii strain and were compared to those of the respective untreated controls. In the Impr strain, 91 open reading frames (ORFs) were downregulated and 182 were upregulated, including 12 pilus-associated genes, one biofilm synthesis protein sequence (ABAYE1394, 2.77-fold), and an alginate biosynthesis regulatory protein gene (algR, ABAYE3509, 2.25-fold) (Table 1; see also Table S2 in the supplemental material). Of note, the AlgR response regulator is also required for the production of type IV pili in Pseudomonas aeruginosa, and biofilms in alginate-overproducing isolates present a highly structured architecture that responds for resistance to antibiotics and to the immune response of cystic fibrosis patients (32). Among the 12 pilus genes upregulated by >2-fold in the Impr strain are pilN, pilV, pilT, pilQ, pilM, pilW, pilP, pilJ, pilO, pliL, pilY, and a type IV pilin protein gene (Table 1), whereas the same sequences were unaffected in the Imps strain (see Table S3 in the supplemental material). In addition, the overexpression of the pil genes by 3-fold or more was confirmed by quantitative RT-PCR (Fig. 2A). The gene expression results are in agreement with our microscopy data and also contribute to an explanation of the increased biofilm formation exhibited by the Impr strain when treated with a sub-MIC of imipenem (Fig. 1B and 2A). Finally, a twitching motility assay was performed to confirm the influence of the treatment on the expression of the Impr pilus-related genes. Note that twitching motility was chosen as it is a well-characterized trait of A. baumannii, which is powered by the extension and retraction of type IV pili. As expected, Impr cells showed increased twitching motility at the agar-plate interface (above 10 mm), while it was inhibited in Imps cells (Fig. 2B; see also Fig. S2 in the supplemental material). These findings are corroborated by the reduced cell viability exhibited by the Imps strain when exposed to a sub-MIC of imipenem (Fig. 1A). In addition, the elevated motility of the Impr strain appears to be a consequence of the strain's ability to survive treatment with a sub-MIC of imipenem (Fig. 1A and 2B), which correlates to the upregulation of the pilus-related genes (Fig. 2A) and other phenotypes (Fig. 1B; see also Fig. S1 in the supplemental material). Of note, motility was not affected among untreated strains grown on MH medium. In summary, the data presented herein demonstrate that our Impr strain survives treatment with a sub-MIC of imipenem, a phenotype that correlates with increased biofilm synthesis, motility, and expression of pilus-associated genes.
FIG 2.
Differential gene expression explains Impr resistance to imipenem and overproduction of virulence factors during exposure to sub-MIC. (A) pil genes (letters) and the type IV pilus (4) gene expression were assessed by microarray (gray) and qPCR (black) analyses and were found to be significantly increased in the Impr strain upon treatment compared with those of Imps. Results are normalized and presented relative to the uninduced strains and are expressed as mean ± SEM for at least 3 biological replicates (P ≤ 0.001, t test). (B) Twitching motility was increased in the Impr strain upon exposure to imipenem. The area of the colonies shown in Fig. S2A to D in the supplemental material was quantified by direct measurement and was expressed in square centimeters, as mean ± SEM for at least 3 biological replicates (****, P < 0.0001, t test). Twitching motility was determined during antibiotic treatment with a sub-MIC of imipenem overnight on 1% agar medium at 37°C.
DISCUSSION
A. baumannii is recognized as a multidrug-resistant organism (1, 2), and an understanding of how environmental stress factors influence the expression of its virulence genes is expected to contribute to the development of novel directed therapies (5). Despite recent efforts, more studies are necessary to elucidate the correlation between A. baumannii virulence and antibiotic resistance (33). In A. baumannii, pili are essential for attachment to and colonization of biotic and abiotic surfaces, biofilm formation, and infection (11–13, 29). Bacteria in biofilms may exhibit increased antibiotic resistance due to cooperation among multiple microbial species, which confers numerous advantages, such as passive resistance, enlarged gene pool with more efficient DNA sharing, quorum-sensing systems, and other synergies (27). Imipenem is one of the drugs most commonly used against A. baumannii infections worldwide (1, 2, 34). Bearing this in mind, we attempted to contribute to the elucidation of the relationship between the synthesis of A. baumannii colonization factors and resistance to imipenem. To achieve this, we assessed phenotype alteration and differential gene expression in imipenem-resistant and imipenem-susceptible strains that were induced by their exposure to a subinhibitory concentration of the antibiotic.
Growth kinetics shows that the A. baumannii Impr strain exhibited sustained growth when exposed to a sub-MIC (Fig. 1A) and a significantly extended lag phase in relation to the susceptible cells when treated with the MIC (Fig. 1A). Notably, a recent study found that exposure of uropathogenic E. coli (UPEC) strains to sub-MICs of ciprofloxacin increased the expression of adhesive determinants, including peripherally located pili, compared to those of unexposed strains (35). Using TEM analysis, we observed that our A. baumannii Impr strain underwent significantly fewer morphological changes and retained most pili in response to treatment with a value of less than or equal to the MIC of imipenem. In contrast, Imps cells grown under all treatments were severely damaged (see Fig. S1 in the supplemental material). In addition, it is known that exposure of MDR A. baumannii to sub-MICs of imipenem increases biofilm formation and stimulates other virulence mechanisms (36). Likewise, sub-MICs of this antibiotic were found to boost the production of P. aeruginosa biofilm and alginate (37). In fact, our microscopy and biofilm quantification results demonstrate a strong correlation between the maintenance of pili and increased biofilm formation when the Impr strain is exposed to a subinhibitory concentration of imipenem. Taken together, our findings agree with the literature, suggesting that (i) biofilm-producing organisms present an altered growth rate upon exposure to antibiotics (38) and (ii) pilus synthesis has a direct impact on biofilm formation (13, 39, 40), although Impr mutant strains remain to be assessed regarding their phenotype during imipenem treatment.
Different types of pili, including type IV pili (TFP), have been found in Gram-negative bacteria such as Pseudomonas aeruginosa, Neisseria gonorrhoeae, and A. baumannii (7, 8, 29, 41–43). In these pathogens, pili are known to mediate a range of functions, from binding to abiotic and biotic surfaces to participation in biofilm formation, twitching motility, virulence, and persistence in the environment. Furthermore, pili have been found to facilitate DNA uptake and transfer between bacterial cells (29, 44–46). However, the role of pili in antibiotic resistance needs to be further investigated. Therefore, using transcriptomic approaches, we assessed the expression of pili genes in both Impr and Imps strains in response to supplementation of the cultures with a sub-MIC of imipenem. Microarray analysis showed significant upregulation of the A. baumannii Impr pil and type IV pilin genes (Table 1). In contrast, none of these genes was affected in imipenem-treated Imps cells. The pilin proteins encoded by the aforementioned genes are involved in TFP biogenesis and function (29, 41, 47–51). In fact, PilM, PilN, PilO, and PilP are required for assembly of the pilin subunits (PilA) into the pilus and constitute the core mechanism necessary for mechanical functioning (41). In the present study, their corresponding genes were upregulated between 2.6- and 3.9-fold upon treatment and were corroborated by quantitative RT-PCR (Table 1 and Fig. 2A). Therefore, we concluded that exposure of A. baumannii Impr to a sub-MIC of imipenem influenced its biology by enhancing the expression of genes critical for pilus synthesis and biofilm formation. Although the experiments performed do not allow us to directly confirm that A. baumannii colonization factors play a direct role in drug resistance, this statement may be true, as biofilms can act as physical barriers to antibiotic molecules (30, 31). Notably, A. baumannii comparative genomics showed that drug-resistant strains possess a higher catabolic capacity than sensitive ones due to the presence of virulence factors known to play a key role in infection, biofilm formation, iron uptake, quorum sensing, and gene expression (52). Furthermore, pilin-encoding genes (i.e., pil family) were found in antibiotic-resistant A. baumannii but are absent from the sensitive strain A. baumannii SDF (52, 53). To substantiate our molecular data, we performed twitching motility assays and observed that Impr strain motility was increased under exposure to a sub-MIC of imipenem. Our findings are in agreement with previous reports, whereby growth of bacterial pathogens under sub-MICs of tobramycin, tetracycline, norfloxacin, or cefodizime led to an increase in pilus synthesis, biofilm formation, motility, type III secretion system, and toxin production as a result of gene upregulation (54–56).
In addition, it is plausible that a subinhibitory concentration of imipenem also affected alternative stress response pathways, leading to the gene dysregulation that influenced biofilm formation and other noninvestigated mechanisms that contributed to the in situ survival of the Impr strain. This second hypothesis is supported by the upregulation of several transcriptional regulators (TetR, LysR, MerR, AlgR; 2.25- to 36.36-fold) and the downregulation of the ClpXP protease (2.7-fold), which is necessary for the stabilization of the Spx redox sensor and the transcriptional activator by proteolysis (32) (see Table S2 in the supplemental material). Finally, three genes corresponding to efflux systems were upregulated up to 3-fold when the Impr strain was grown under a sub-MIC of imipenem and may have participated in its survival.
Taken together, our findings help to explain the biology of an imipenem-resistant A. baumannii strain and suggest the possibility that multiple attributes may play a role in its survival during suboptimal chemotherapy. In addition, the molecular mechanism responsible for the overexpression of the aforementioned pilin genes is under investigation and is expected to shed light on the rational design of directed therapies against this troublesome pathogen. In summary, our results suggest that the pilus biosynthetic machinery may be an amenable target for the control of A. baumannii.
Supplementary Material
ACKNOWLEDGMENTS
We thank the University of Malaya Medical Centre for providing us with bacterial isolates and the Tropical Infectious Disease Research and Education Center (TIDREC) for supporting this study.
G. N. Dhabaan, S. AbuBakar, and H. Hassan designed the experiments, and H. Hassan supervised the research. G. N. Dhabaan and G. M. Cerqueira wrote the paper. G. N. Dhabaan, S. AbuBakar, H. Hassan, M. Al-Haroni, and G. M. Cerqueira revised the paper. G. N. Dhabaan and S. P. Pang performed the experiments. G. N. Dhabaan did the data analysis. All authors read and approved the final manuscript.
The authors declare that they have no competing interest.
Funding Statement
This study was funded by the vote FS189-2008B, FRGS grants FP026/2010B and FP022-2010A, and Research University Grant number RG025-09HTM.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01696-15.
REFERENCES
- 1.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 3.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. 2008. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol 6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abbott I, Cerqueira GM, Bhuiyan S, Peleg AY. 2013. Carbapenem resistance in Acinetobacter baumannii: laboratory challenges, mechanistic insights and therapeutic strategies. Expert Rev Anti Infect Ther 11:395–409. doi: 10.1586/eri.13.21. [DOI] [PubMed] [Google Scholar]
- 5.Ohlsen K, Ziebuhr W, Koller KP, Hell W, Wichelhaus TA, Hacker J. 1998. Effects of subinhibitory concentrations of antibiotics on alpha-toxin (hla) gene expression of methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother 42:2817–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sauer FG, Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. 2000. Bacterial pili: molecular mechanisms of pathogenesis. Curr Opin Microbiol 3:65–72. doi: 10.1016/S1369-5274(99)00053-3. [DOI] [PubMed] [Google Scholar]
- 7.Proft T, Baker EN. 2009. Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell Mol Life Sci 66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tobe T, Sasakawa C. 2002. Species-specific cell adhesion of enteropathogenic Escherichia coli is mediated by type IV bundle-forming pili. Cell Microbiol 4:29–42. doi: 10.1046/j.1462-5822.2002.00167.x. [DOI] [PubMed] [Google Scholar]
- 9.Karaolis DK, Somara S, Maneval DR Jr, Johnson JA, Kaper JB. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 10.Averhoff B, Friedrich A. 2003. Type IV pili-related natural transformation systems: DNA transport in mesophilic and thermophilic bacteria. Arch Microbiol 180:385–393. doi: 10.1007/s00203-003-0616-6. [DOI] [PubMed] [Google Scholar]
- 11.Gaddy JA, Actis LA. 2009. Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol 4:273–278. doi: 10.2217/fmb.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerqueira GM, Kostoulias X, Khoo C, Aibinu I, Qu Y, Traven A, Peleg AY. 2014. A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J Infect Dis 210:46–55. doi: 10.1093/infdis/jiu024. [DOI] [PubMed] [Google Scholar]
- 13.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 14.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. 2008. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154:3398–3409. doi: 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 15.Espinal P, Marti S, Vila J. 2012. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J Hosp Infect 80:56–60. doi: 10.1016/j.jhin.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Bano J, Marti S, Soto S, Fernandez-Cuenca F, Cisneros JM, Pachon J, Pascual A, Martinez-Martinez L, McQueary C, Actis LA, Vila J, Spanish Group for the Study of Nosocomial Infections (GEIH). 2008. Biofilm formation in Acinetobacter baumannii: associated features and clinical implications. Clin Microbiol Infect 14:276–278. doi: 10.1111/j.1469-0691.2007.01916.x. [DOI] [PubMed] [Google Scholar]
- 17.Olsen I. 2015. Biofilm-specific antibiotic tolerance and resistance. Eur J Clin Microbiol Infect Dis 34:877–886. doi: 10.1007/s10096-015-2323-z. [DOI] [PubMed] [Google Scholar]
- 18.Hoiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Nucleo E, Steffanoni L, Fugazza G, Migliavacca R, Giacobone E, Navarra A, Pagani L, Landini P. 2009. Growth in glucose-based medium and exposure to subinhibitory concentrations of imipenem induce biofilm formation in a multidrug-resistant clinical isolate of Acinetobacter baumannii. BMC Microbiol 9:270. doi: 10.1186/1471-2180-9-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao RS, Karthika RU, Singh SP, Shashikala P, Kanungo R, Jayachandran S, Prashanth K. 2008. Correlation between biofilm production and multiple drug resistance in imipenem resistant clinical isolates of Acinetobacter baumannii. Indian J Med Microbiol 26:333–337. doi: 10.4103/0255-0857.43566. [DOI] [PubMed] [Google Scholar]
- 21.Wand ME, Bock LJ, Turton JF, Nugent PG, Sutton JM. 2012. Acinetobacter baumannii virulence is enhanced in Galleria mellonella following biofilm adaptation. J Med Microbiol 61:470–477. doi: 10.1099/jmm.0.037523-0. [DOI] [PubMed] [Google Scholar]
- 22.Brossard KA, Campagnari AA. 2012. The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect Immun 80:228–233. doi: 10.1128/IAI.05913-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Breij A, Dijkshoorn L, Lagendijk E, van der Meer J, Koster A, Bloemberg G, Wolterbeek R, van den Broek P, Nibbering P. 2010. Do biofilm formation and interactions with human cells explain the clinical success of Acinetobacter baumannii? PLoS One 5:e10732. doi: 10.1371/journal.pone.0010732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cevahir N, Demir M, Kaleli I, Gurbuz M, Tikvesli S. 2008. Evaluation of biofilm production, gelatinase activity, and mannose-resistant hemagglutination in Acinetobacter baumannii strains. J Microbiol Immunol Infect 41:513–518. [PubMed] [Google Scholar]
- 25.Eijkelkamp BA, Stroeher UH, Hassan KA, Papadimitrious MS, Paulsen IT, Brown MH. 2011. Adherence and motility characteristics of clinical Acinetobacter baumannii isolates. FEMS Microbiol Lett 323:44–51. doi: 10.1111/j.1574-6968.2011.02362.x. [DOI] [PubMed] [Google Scholar]
- 26.Ghannoum M, O'Toole GA. 2004. Microbial biofilms. ASM Press, Washington, DC. [Google Scholar]
- 27.Wolcott R, Costerton JW, Raoult D, Cutler SJ. 2013. The polymicrobial nature of biofilm infection. Clin Microbiol Infect 19:107–112. doi: 10.1111/j.1469-0691.2012.04001.x. [DOI] [PubMed] [Google Scholar]
- 28.Gohl O, Friedrich A, Hoppert M, Averhoff B. 2006. The thin pili of Acinetobacter sp. strain BD413 mediate adhesion to biotic and abiotic surfaces. Appl Environ Microbiol 72:1394–1401. doi: 10.1128/AEM.72.2.1394-1401.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harding CM, Tracy EN, Carruthers MD, Rather PN, Actis LA, Munson RS Jr. 2013. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 4:e00360-13. doi: 10.1128/mBio.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirby AE, Garner K, Levin BR. 2012. The relative contributions of physical structure and cell density to the antibiotic susceptibility of bacteria in biofilms. Antimicrob Agents Chemother 56:2967–2975. doi: 10.1128/AAC.06480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lebeaux D, Ghigo JM, Beloin C. 2014. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang YW, Sussman M, Liu D, Poxton I, Schwartzman J. 2014. Molecular medical microbiology. Elsevier Science, Philadelphia, PA. [Google Scholar]
- 33.Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie JM. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karageorgopoulos DE, Falagas ME. 2008. Current control and treatment of multidrug-resistant Acinetobacter baumannii infections. Lancet Infect Dis 8:751–762. doi: 10.1016/S1473-3099(08)70279-2. [DOI] [PubMed] [Google Scholar]
- 35.Balague C, Fernandez L, Perez J, Grau R. 2003. Effect of ciprofloxacin on adhesive properties of non-P mannose-resistant uropathogenic Escherichia coli isolates. J Antimicrob Chemother 51:401–404. doi: 10.1093/jac/dkg048. [DOI] [PubMed] [Google Scholar]
- 36.Nucleo E, Steffanoni L, Fugazza G, Migliavacca R, Giacobone E, Navarra A, Pagani L, Landini P. 2009. Growth in glucose-based medium and exposure to subinhibitory concentrations of imipenem induce biofilm formation in a multidrug-resistant clinical isolate of Acinetobacter baumannii. BMC Microbiol 9:270. doi: 10.1186/1471-2180-9-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bagge N, Schuster M, Hentzer M, Ciofu O, Givskov M, Greenberg EP, Hoiby N. 2004. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and beta-lactamase and alginate production. Antimicrob Agents Chemother 48:1175–1187. doi: 10.1128/AAC.48.4.1175-1187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan RM, Kuang Z, Hao Y, Lau GW. 2014. Type IV pilus of Pseudomonas aeruginosa confers resistance to antimicrobial activities of the pulmonary surfactant protein-A. J Innate Immun 6:227–239. doi: 10.1159/000354304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo LM, Wu LJ, Xiao YL, Zhao D, Chen ZX, Kang M, Zhang Q, Xie Y. 2015. Enhancing pili assembly and biofilm formation in Acinetobacter baumannii ATCC19606 using non-native acyl-homoserine lactones. BMC Microbiol 15:62. doi: 10.1186/s12866-015-0397-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ayers M, Sampaleanu LM, Tammam S, Koo J, Harvey H, Howell PL, Burrows LL. 2009. PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J Mol Biol 394:128–142. doi: 10.1016/j.jmb.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 42.Drake SL, Koomey M. 1995. The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol Microbiol 18:975–986. doi: 10.1111/j.1365-2958.1995.18050975.x. [DOI] [PubMed] [Google Scholar]
- 43.Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, Fernandez R, Schoolnik GK. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 44.Leighton TL, Buensuceso R, Howell PL, Burrows LL. 2015. Biogenesis of Pseudomonas aeruginosa type IV pili and regulation of their function. Environ Microbiol 17:4148–4163. doi: 10.1111/1462-2920.12849. [DOI] [PubMed] [Google Scholar]
- 45.Melville S, Craig L. 2013. Type IV pili in Gram-positive bacteria. Microbiol Mol Biol Rev 77:323–341. doi: 10.1128/MMBR.00063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burrows LL. 2012. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol 66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 47.Bulyha I, Schmidt C, Lenz P, Jakovljevic V, Hone A, Maier B, Hoppert M, Sogaard-Andersen L. 2009. Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins. Mol Microbiol 74:691–706. doi: 10.1111/j.1365-2958.2009.06891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forsberg A, Guina T. 2007. Type II secretion and type IV pili of Francisella. Ann N Y Acad Sci 1105:187–201. doi: 10.1196/annals.1409.016. [DOI] [PubMed] [Google Scholar]
- 49.Salomonsson EN, Forslund AL, Forsberg A. 2011. Type IV pili in Francisella—a virulence trait in an intracellular pathogen. Front Microbiol 2:29. doi: 10.3389/fmicb.2011.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Craig L, Li J. 2008. Type IV pili: paradoxes in form and function. Curr Opin Struct Biol 18:267–277. doi: 10.1016/j.sbi.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carbonnelle E, Helaine S, Nassif X, Pelicic V. 2006. A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol Microbiol 61:1510–1522. doi: 10.1111/j.1365-2958.2006.05341.x. [DOI] [PubMed] [Google Scholar]
- 52.Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, Oztas S, Poulain J, Segurens B, Robert C, Abergel C, Claverie JM, Raoult D, Medigue C, Weissenbach J, Cruveiller S. 2008. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hacker J, Ott M, Hof H. 1993. Effects of low, subinhibitory concentrations of antibiotics on expression of a virulence gene cluster of pathogenic Escherichia coli by using a wild-type gene fusion. Int J Antimicrob Agents 2:263–270. doi: 10.1016/0924-8579(93)90060-I. [DOI] [PubMed] [Google Scholar]
- 55.Braga PC, Sasso MD, Sala MT. 2000. Sub-MIC concentrations of cefodizime interfere with various factors affecting bacterial virulence. J Antimicrob Chemother 45:15–25. doi: 10.1093/jac/45.1.15. [DOI] [PubMed] [Google Scholar]
- 56.Linares JF, Gustafsson I, Baquero F, Martinez JL. 2006. Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci U S A 103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.