Abstract
Pseudomonas aeruginosa, a major cause of nosocomial and chronic infections, is considered a paradigm of antimicrobial resistance development. However, the evolutionary trajectories of antimicrobial resistance and the impact of mutator phenotypes remain mostly unexplored. Therefore, whole-genome sequencing (WGS) was performed in lineages of wild-type and mutator (ΔmutS) strains exposed to increasing concentrations of relevant antipseudomonal agents. WGS provided a privileged perspective of the dramatic effect of mutator phenotypes on the accumulation of random mutations, most of which were transitions, as expected. Moreover, a frameshift mutagenic signature, consistent with error-prone DNA polymerase activity as a consequence of SOS system induction, was also seen. This effect was evidenced for all antibiotics tested, but it was higher for fluoroquinolones than for cephalosporins or carbapenems. Analysis of genotype versus phenotype confirmed expected resistance evolution trajectories but also revealed new pathways. Classical mechanisms included multiple mutations leading to AmpC overexpression (ceftazidime), quinolone resistance-determining region (QRDR) mutations (ciprofloxacin), oprD inactivation (meropenem), and efflux pump overexpression (ciprofloxacin and meropenem). Groundbreaking findings included gain-of-function mutations leading to the structural modification of AmpC (ceftazidime), novel DNA gyrase (GyrA) modification (ciprofloxacin), and the alteration of the β-lactam binding site of penicillin-binding protein 3 (PBP3) (meropenem). A further striking finding was seen in the evolution of meropenem resistance, selecting for specific extremely large (>250 kb) genomic deletions providing a growth advantage in the presence of the antibiotic. Finally, fitness and virulence varied within and across evolved antibiotic-resistant populations, but mutator lineages showed a lower biological cost for some antibiotics.
INTRODUCTION
Pseudomonas aeruginosa, a ubiquitous microorganism, is one of the most relevant pathogens causing human opportunistic infections (1). Due to its impressive metabolic plasticity and versatility, P. aeruginosa is capable of infecting/colonizing a wide range of ecological niches, including aquatic and soil habitats, animals, and plants (2). P. aeruginosa is one of the most frequent and severe causes of acute nosocomial infections, particularly affecting immunocompromised patients or those admitted to the intensive care unit (ICU) (3). Likewise, P. aeruginosa is the most frequent and severe driver of chronic respiratory infections in patients suffering from cystic fibrosis (CF) or other chronic underlying diseases (4). The versatility and adaptability conferred by the large proportion (>8%) of regulatory genes encoded in its large genome (>6 Mb), its remarkable repertoire of virulence determinants, and its outstanding capacity to evade the activity of antimicrobial treatments make P. aeruginosa one of the most feared bacterial pathogens (1, 2, 5). The emergence of mutator (or hypermutable) variants, which are particularly prevalent in chronic infections, further increases adaptability and antimicrobial resistance development (6, 7).
P. aeruginosa is genetically equipped with outstanding intrinsic antibiotic resistance machinery (5, 8, 9). The production of an inducible AmpC cephalosporinase, the constitutive or inducible expression of efflux pumps, and the reduced permeability of its outer membrane are thought to be those mechanisms having a greater impact on the basal lower susceptibility of P. aeruginosa to antibiotics compared with that of other Gram-negative pathogens. Moreover, the complexity of and concern about antimicrobial resistance are much further enhanced by the extraordinary capacity of this pathogen for developing resistance to all available antibiotics through the selection of mutations in chromosomal genes leading to the upregulation of AmpC or efflux pumps, the inactivation of the carbapenem porin OprD, or the modification of the drug target (DNA topoisomerases) in the case of fluoroquinolones. In addition to these well-studied canonical resistance mechanisms, recent whole-genome screening mutant libraries reveal a plethora of genes, collectively known as the resistome, which have an impact on antimicrobial susceptibility, including many with central metabolic functions (5, 10–13). However, transposon mutagenesis-based approaches are biased by the type of introduced genetic event, not allowing the study of gain-of-function/modify-function mutations in essential and nonessential genes. Other potential limitations of these approaches are the absence of competition between emerging variants and that the obtained information is restricted to single genetic events.
Thus, evolutionary trajectories of antimicrobial resistance and fitness in P. aeruginosa remain mostly unexplored. Likewise, current knowledge on the impact of mutator phenotypes on bacterial evolution under antibiotic pressure is still very limited. In order to fulfill this gap, we followed a whole-genome sequencing (WGS) approach to decipher the evolutionary pathways leading to high-level resistance to three of the most relevant classes of antipseudomonal agents, including cephalosporins, fluoroquinolones, and carbapenems, under wild-type and high mutation supply rates. For this purpose, the evolution of antibiotic resistance and its fitness consequences were studied in populations of wild-type P. aeruginosa PAO1 and its mutS DNA mismatch repair-deficient derivative P. aeruginosa PAOMS exposed to increasing concentrations of ceftazidime, ciprofloxacin, or meropenem for approximately 70 generations.
MATERIALS AND METHODS
Bacterial strains.
The parent strains used included the wild-type reference strain P. aeruginosa PAO1 (14) and its mismatch repair-deficient (ΔmutS) mutator derivative (PAOMS) (7). In a previous study (15), PAO1 and PAOMS strains were subjected to stepwise exposure to increasing concentrations (0.5 times to 64 times the MIC) of ceftazidime (MIC, 1 mg/liter), ciprofloxacin (MIC, 0.125 mg/liter), or meropenem (MIC, 0.5 mg/liter) for 7 days. Each day, tubes from the highest antibiotic concentration showing growth were diluted (1:1,000) into fresh medium containing concentrations up to 64 times the MIC. The same procedure was repeated for seven consecutive days, allowing a total of approximately 70 generations of growth. Three independent isolates from each of three independent experiments per strain (PAO1 or PAOMS) and antibiotic (ceftazidime, ciprofloxacin, meropenem, or none) were purified for characterization in this work through WGS. Thus, a total of 26 lineages were sequenced, including original PAO1 and PAOMS stocks from our laboratory used as the template.
Whole-genome sequencing.
Total DNA was isolated by the use of a commercial capture system (High Pure PCR template preparation kit; Roche Diagnostics), and indexed paired-end libraries were generated by using a commercial kit (Nextera XT DNA library preparation kit; Illumina). All samples were then sequenced with a MiSeq desktop sequencer cartridge (MiSeq reagent kit version 3; Illumina). Raw data yielded a total of 6.5 Gb of information, from which 83.4% was >Q30. The reads were demultiplexed by sample, obtaining a mean coverage of 34× (range, 15× to 49×), and then were aligned against the P. aeruginosa PAO1 genome (RefSeq accession no. NC_002516.2) using Burrows-Wheeler Aligner (BWA). Variants were called using Genome Analysis Toolkit (GATK) and then annotated and curated manually. The Pseudomonas Genome Database was used for gene function analysis (14, 16). dN/dS, the ratio of the rate of nonsynonymous substitutions (dN) to the rate of synonymous substitutions (dS), was calculated as a measure of the selection pressure acting on the protein-coding genome, as previously described, assuming that 25% of all single-nucleotide polymorphisms (SNPs) result in synonymous changes (17). dN/dS is expected to be >1 if natural selection promotes changes in protein sequences and <1 if natural selection suppresses changes.
Susceptibility testing.
The MICs of ceftolozane-tazobactam, ceftazidime, cefepime, piperacillin-tazobactam, aztreonam, imipenem, meropenem, ciprofloxacin, ticarcillin, tobramycin, amikacin, and colistin were determined by broth microdilution using customized Sensititre plates (reference FRCNRP; Thermo Fisher Scientific).
Characterization of resistance mechanisms.
The expression of the genes encoding the chromosomal β-lactamase AmpC (ampC) and four P. aeruginosa efflux pumps, MexAB-OprM (mexB), MexCD-OprJ (mexD), MexEF-OprN (mexF), and MexXY-OprM (mexY), were determined from late-log-phase Luria-Bertani (LB) broth cultures at 37°C and 180 rpm by real-time reverse transcription-PCR (RT-PCR) with an Illumina Eco real-time PCR system, as previously described (18). When needed, genes involved in the regulation of resistance mechanisms, including AmpC (dacB, ampD, and ampR), mexAB-OprM (mexR or nalB, nalC, and nalD), mexCD-OprJ (nfxB), mexEF-OprN (mexS, mexT and mvaT), and MexXY-OprM (mexZ and PA5471) were sequenced. PCR amplification (primers listed in Table S1 in the supplemental material) was performed on whole-DNA extracts (DNeasy tissue kit; Qiagen, Hilden, Germany), and two independent PCR products for each isolate and gene were sequenced on both strands. Likewise, the quinolone resistance-determining regions (QRDR) of gyrA, gyrB, parC, and parE were sequenced in ciprofloxacin-resistant mutants (19). Outer membrane protein (OMP) profiles were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue (18). The obtained OprD profiles were compared with those of PAO1 and its OprD-deficient mutant. oprD sequences were also determined in all carbapenem-resistant mutants.
PBP assays.
Membranes containing the penicillin-binding proteins (PBPs) of each P. aeruginosa strain were obtained, according to previously described protocols (20, 21). Briefly, 500 ml of late-log-phase (optical density at 600 nm [OD600], 1) Luria-Bertani (LB) (Sigma-Aldrich, St. Louis, MO) cultures were collected by centrifugation (4,400 × g for 10 min) and then washed and suspended in 50 ml of 20 mM KH2PO4 and 140 mM NaCl (pH 7.5) (buffer A). Cells were then sonicated using a Digital Sonifier unit model S-450D (Branson Ultrasonics Corporation, Danbury, CT) at 20 W for three 30-s bursts (while immersed in an ice bath) and centrifuged at 12,000 × g for 10 min. Membranes containing the PBPs were isolated from the supernatant through two steps of ultracentrifugation at 150,000 × g and 4°C for 1 h using an Optima L-XP series preparative ultracentrifuge (Beckman Coulter, Inc., Palo Alto, CA) and suspension in buffer A. Total protein content was measured through the Bradford method using the Quick Start Bradford protein assay kit, with bovine serum albumin as a standard (Bio-Rad Laboratories, Hercules, CA), according to the manufacturer's instructions. For the analysis of the PBP profiles, membrane fractions were adjusted to 1 mg/ml, and 10 μl of the adjusted preparations was labeled with 25 μM penicillin (Bocillin FL) (20) and subsequently separated through 10% gel SDS-polyacrylamide gel electrophoresis. Labeled PBPs were visualized (excitation at 488 nm and emission at 530 nm) using a Bio-Rad FX Pro molecular imager (Bio-Rad Laboratories, Hercules, CA). In order to evaluate reproducibility and consistency, the profiles were determined in three independent occasions for each of the strains. For the determination of 50% inhibitory concentrations (IC50), previously described protocols were used (21, 22). Briefly, 20 μl (final volume) of PBPs containing solution was incubated (30 min at 37°C) in the presence of increasing concentrations of ceftazidime or meropenem (range of concentrations tested, 0.0156 to 2 μg/ml) and were afterwards labeled with a 25 μM concentration of penicillin (Bocillin FL). The reaction mixtures were then denatured with 20 μl each of SDS denaturing solution at 100°C for 3 min. The PBPs were then separated through 10% SDS-polyacrylamide gel electrophoresis. The protein gels were rinsed in water immediately after electrophoresis. Labeled PBPs were visualized using a Bio-Rad FX Pro molecular imager, and the IC50s of ceftazidime and meropenem were determined from triplicate independent experiments using the Quantity One software (Bio-Rad Laboratories) and compared using the Student's t test. P values of <0.05 were considered statistically significant.
Characterization of large genomic deletions selected upon meropenem exposure.
PCR amplification of external, internal, and immediate flanking genes of each deletion was performed on whole-DNA extracts (High Pure PCR template preparation kit; Roche Diagnostics) for each mutant, using primers listed in Table S1 in the supplemental material. The obtained PCR products were then fully sequenced.
Pyomelanin production.
For quantification of the spontaneous emergence of dark brown colonies, denoting the hyperproduction of pyomelanin, PAO1 was grown in triplicate at 37°C in LB broth overnight, and serial dilutions were plated in LB agar containing or not containing 1 mg/liter meropenem. After 48 h, approximately 1,000 randomly selected colonies of those growing were streaked into a fresh LB agar plate, and the proportion of colonies showing pyomelanin hyperproduction was scored after 24 h of incubation.
In vitro competition experiments and growth rates.
In vitro competition experiments between each of the resistant mutants and a gentamicin-tagged (att intergenic neutral chromosomal locus) wild-type PAO1 were performed (23, 24). Exponentially growing cells were mixed in a 1:1 ratio and diluted in 0.9% saline solution. Approximately 103 cells from each of the mixtures were inoculated into eight 10-ml LB broth flasks and grown at 37°C and 180 rpm for 16 to 18 h, corresponding to approximately 20 generations. Serial 10-fold dilutions were plated in duplicate onto LB agar supplemented with 15 μg/ml gentamicin to enumerate wild-type CFU, and LB agar supplemented with 10 times the wild-type MIC of ceftazidime (10 μg/ml), ciprofloxacin (1.25 μg/ml), or meropenem (5 μg/ml) was plated to enumerate the CFU of the corresponding resistant mutants. The competition index (CI) was defined as the mutant-to-wild-type ratio. For performing the growth curves of PAO1 and meropenem-resistant derivatives, overnight cultures were adjusted at an initial OD600 of 0.01 and incubated at 37°C and 180 rpm in Mueller-Hinton broth (MHB) and MHB with subinhibitory (0.25 μg/ml) and high (32 μg/ml) concentrations of meropenem. The OD600 was then monitored every hour for 10 h, and curves were generated by plotting the mean ± standard deviation (SD) values of the results from three independent experiments.
Caenorhabditis elegans killing assay.
The assay for studying bacterial killing of C. elegans was performed as described previously (25). Briefly, a fresh culture of each bacterial strain to be tested was layered onto a 55-mm-diameter plate containing 5 ml of potato dextrose agar. After spreading of the bacterial culture, the plates were incubated at 37°C for 24 h to form bacterial lawns. The bacterial plates were kept overnight, and 5 worms per plate were poured on top of these bacterial lawns. The plates were incubated at 24°C and scored to detect the presence of living worms at 0 h, 24 h, 72 h, and 168 h. The nematodes were examined at 20× and 40× magnifications, and a worm was considered dead if it did not move spontaneously. Five replicate independent experiments per bacterial strain were performed and the means ± SD recorded.
RESULTS AND DISCUSSION
Numbers and nature of mutations arising upon exposure to different antimicrobial agents in wild-type and mutator (ΔmutS) strains.
As shown in Table 1, PAO1 derivatives grown for 7 days (approximately 70 generations) in the absence of antibiotics acquired 1 to 2 mutations (0.014 to 0.028 mutations per genome per generation), which included transitions, transversions, and frameshifts. On the other hand, PAO1 lineages grown in the presence of antibiotics acquired 3 to 6 mutations; the excess mutations of antibiotic-exposed lineages correlated well with the number of acquired resistance mutations (see below) and provided a clear signature of positive selection (nonsynonymous-to-synonymous mutations ratio [dN/dS], >1). However, significant differences in the total number of mutations were not documented among PAO1 lineages evolved in the presence of the three different antibiotics. Mutations acquired in the presence of antibiotics also included transitions, transversions, and frameshifts, but the presence of very large deletions (>250 kb) in two of the three PAO1 lineages exposed to meropenem is noteworthy.
TABLE 1.
Number and nature of mutations documented in wild-type (PAO1) and mutator (PAOMS) strain derivatives exposed to increasing concentrations of different antibiotics for 7 days
| Strain-antibiotica | No. of SNPs (silent + intergenic) |
No. of indels (intergenic) | No. of frameshifts (intergenic) | Total no. of mutations | Mutations/genome/generationb | dN/dSc | |
|---|---|---|---|---|---|---|---|
| Transition | Transversion | ||||||
| PAO1.1-none | 1 (1 + 0) | 1 | 2 | 0.028 | —f | ||
| PAO1.2-none | 1 (0 + 1) | 1 | 0.014 | ||||
| PAO1.3-none | 1 | 1 | 0.014 | ||||
| PAOMS.1-none | 40 (13 + 2) | 2 (1) | 42 | 0.60 | 0.64 | ||
| PAOMS.2-none | 41 (10 + 5) | 2 | 43 | 0.61 | 0.87 | ||
| PAOMS.3-none | 48 (13 + 2) | 2 (2) | 50 | 0.71 | 0.85 | ||
| PAO1.1-CAZ | 2 | 1 | 3 | 0.042 | >1g | ||
| PAO1.2-CAZ | 1 | 2 (0 + 1) | 3 | 0.042 | >1 | ||
| PAO1.3-CAZ | 1 | 2 | 1 | 4 | 0.056 | >1 | |
| PAOMS.1-CAZ | 46 (11 + 6) | 5 (2) | 51 | 0.73 | 0.88 | ||
| PAOMS.2-CAZ | 45 (13 + 3) | 4 (2) | 49 | 0.7 | 0.74 | ||
| PAOMS.3-CAZ | 41 (9 + 4) | 6 (1) | 47 | 0.67 | 1.04 | ||
| PAO1.1-CIP | 3 | 3 | 0.042 | >1 | |||
| PAO1.2-CIP | 1 | 3 (0 + 1) | 2 | 6 | 0.084 | >1 | |
| PAO1.3-CIP | 3 | 1 | 4 | 0.056 | >1 | ||
| PAOMS.1-CIP | 54 (11 + 5) | 1 | 12 (1) | 67 | 0.96 | 1.15 | |
| PAOMS.2-CIP | 47 (16 + 5) | 2 | 8 (2) | 57 | 0.81 | 0.58 | |
| PAOMS.3-CIP | 47 (10 + 5) | 9 (2) | 56 | 0.80 | 1.07 | ||
| PAO1.1-MER | 3 | 1d | 4 | 0.056 | >1 | ||
| PAO1.2-MER | 3 | 3 | 6 | 0.084 | >1 | ||
| PAO1.3-MER | 1 | 1 | 1e | 2 | 5 | 0.07 | >1 |
| PAOMS.1-MER | 48 (15 + 7) | 7 | 55 | 0.79 | 0.58 | ||
| PAOMS.2-MER | 45 (14 + 3) | 5 (2) | 50 | 0.71 | 0.67 | ||
| PAOMS.3-MER | 37 (12 + 3) | 5 (1) | 42 | 0.60 | 0.61 | ||
CAZ, ceftazidime; CIP, ciprofloxacin; MER, meropenem.
The estimated number of generations for the 7-day experiments was 70.
dN/dS, the ratio of the rate of nonsynonymous substitutions (dN) to the rate of synonymous substitutions (dS) was calculated as a measure of the selection pressure acting on protein-coding genome, as previously described, assuming that 25% of all SNPs result in synonymous changes. dN/dS is expected to be greater than 1 if natural selection promotes changes in protein sequences and less than 1 if natural selection suppresses changes.
Deletion of 258,659 base pairs from position 2183460.
Deletion of 338,658 base pairs from position 2092676.
dS/dN cannot be determined for total numbers of mutations of <3.
All mutations were nonsynonymous.
In sharp contrast, PAOMS derivatives grown in the absence of antibiotics displayed 42 to 50 mutations (0.6 to 0.7 mutations per genome per generation), and most of them (40 to 48 mutations) were transitions, as expected (26). PAOMS lineages grown in the presence of antibiotics acquired 42 to 67 mutations. Thus, globally, the numbers of total mutations did not differ significantly in lineages grown in the absence or presence of antibiotics. Moreover, dN/dS ratios of ≤1 (indicating negative to neutral selection) show that although found in a quite large number (see below), positively selected antibiotic resistance mutations are exceeded by random mutations that accumulate due to very high mutation rates. However, major differences in the total numbers of mutations were seen across the three antibiotics. Indeed, the lineages exposed to ciprofloxacin showed a higher number of mutations (57.7 ± 2.1 mutations) than those exposed to ceftazidime (49.0 ± 2.0 mutations) or meropenem (49 ± 6.5 mutations) (P = 0.015, t test). Among the different types of mutations, only frameshifts were significantly more frequent among ciprofloxacin-exposed derivatives (9.7 ± 2.1 frameshifts) than among ceftazidime (5 ± 1 frameshifts) or meropenem (5.7 ± 1.2 frameshifts) lineages (P = 0.004, t test). The results obtained for ciprofloxacin are thus consistent with its predicted mutagenic effect driven by SOS system induction, leading to error-prone DNA polymerase DinP overexpression (27, 28). Moreover, despite the number being lower than that for ciprofloxacin, the number of frameshift mutations acquired by ceftazidime- and meropenem-exposed lineages were significantly higher than those acquired by unexposed ones (2 ± 0 frameshifts, P = 0.03). Thus, these results are consistent with previous work showing that β-lactams can also exert a mutagenic effect through DinP induction (29, 30). The number of frameshift mutations tended also to be higher in PAO1 lineages exposed to antibiotics (particularly ciprofloxacin and meropenem), although the overall small numbers of mutations precluded any determination of statistical significance.
Correlation between genotype and antibiotic resistance phenotype: expected findings and new traits.
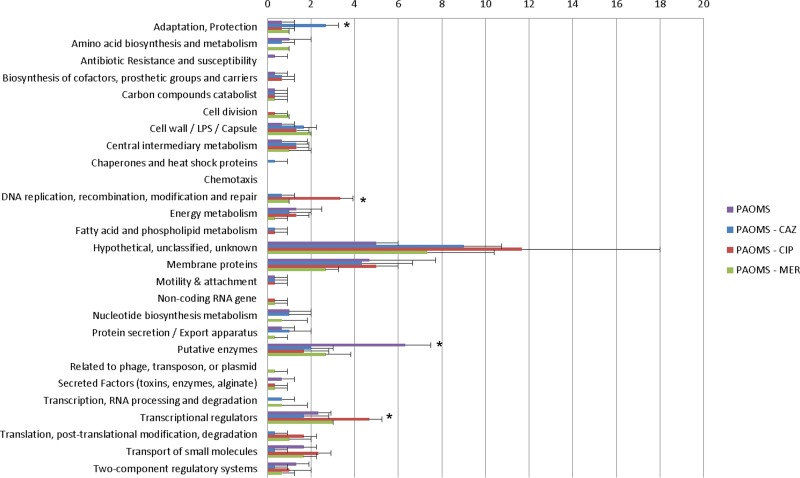
The complete list of mutations detected in the PAO1 and PAOMS lineages is available in Table S2 in the supplemental material, while Fig. 1 shows the number of nonsynonymous mutations by gene function category documented for PAOMS derivatives. According to gene function category, compared to the other antibiotics, ciprofloxacin-exposed lineages more frequently acquired mutations in transcriptional regulators and the DNA replication, modification, and repair categories, whereas ceftazidime-exposed lineages more frequently had mutations in genes from the adaptation and protection category. However, these differences correlated well with the documented specific resistance mutations (see below). On the other hand, intriguingly, unexposed lineages were more frequently mutated in putative enzyme categories than lineages exposed to any of the antibiotics.
FIG 1.
Number of nonsynonymous mutations by gene function category documented in PAOMS derivatives exposed for 7 days to increasing concentrations of ceftazidime (CAZ), ciprofloxacin (CIP), meropenem (MER), or control without antibiotics. The mean ± SD values obtained for three independent lineages are shown. Statistically significant differences (P < 0.05, Student's t test) for one condition respect to each of the other 3 are indicated with an asterisk.
In order to determine the correlation between genotype and antibiotic resistance phenotype, we performed a deep characterization of the evolved antibiotic resistance profiles. Table 2 shows the MICs for a comprehensive set of 10 antipseudomonal agents, and Table 3 shows the expression of resistance mechanisms and the correlation with the corresponding resistance mutations.
TABLE 2.
Susceptibility profiles of PAO1 and PAOMS lineages evolved in the presence of ceftazidime, meropenem, or ciprofloxacin in each of three experiments
| Strain | MIC (μg/ml) by antibiotic (CLSI susceptibility breakpoint)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TOL-TAZ | CAZ (≤8) | FEP (≤8) | PIP-TZ (≤16) | AZT (≤8) | IMI (≤4) | MER (≤4) | CIP (≤1) | TIC (≤16) | TOB (≤4) | AMK (≤16) | COL (≤2) | |
| PAO1 | 0.5 | 1 | 1 | 4 | 4 | 2 | 0.5 | ≤0.12 | 16 | 0.5 | ≤2 | 2 |
| PAOMS | 0.5 | 2 | 1 | 8 | 2 | 2 | 1 | ≤0.12 | 32 | 1 | 4 | 4 |
| PAO1.1-CAZ | 4 | 32 | 16 | 128 | 32 | 2 | 2 | ≤0.12 | 128 | 0.5 | ≤2 | 2 |
| PAO1.2-CAZ | 4 | 64 | 16 | 256 | 32 | 2 | 2 | ≤0.12 | 128 | 0.5 | ≤2 | 2 |
| PAO1.3-CAZ | 4 | 64 | 16 | 256 | 64 | 2 | 2 | ≤0.12 | 128 | 0.5 | ≤2 | 2 |
| PAOMS.1-CAZ | 16 | 64 | 16 | 128 | 64 | 2 | 1 | ≤0.12 | 128 | 1 | 4 | 4 |
| PAOMS.2-CAZ | 8 | 64 | 16 | 128 | 64 | 2 | 1 | ≤0.12 | 128 | 1 | 4 | 4 |
| PAOMS.3-CAZ | 8 | 64 | 32 | 256 | 64 | 2 | 4 | ≤0.12 | 128 | 0.5 | 4 | 2 |
| PAO1.1-CIP | ≤0.25 | ≤0.5 | 4 | ≤2 | ≤1 | ≤0.5 | ≤0.25 | >16 | ≤4 | ≤0.25 | ≤2 | 2 |
| PAO1.2-CIP | ≤0.25 | ≤0.5 | 4 | ≤2 | ≤1 | ≤0.5 | 0.5 | 8 | ≤4 | ≤0.25 | ≤2 | 2 |
| PAO1.3-CIP | 0.5 | 4 | 4 | 16 | 16 | 1 | 2 | >16 | 64 | 0.5 | ≤2 | 2 |
| PAOMS.1-CIP | 0.5 | ≤0.5 | 4 | ≤2 | ≤1 | 1 | 0.5 | >16 | ≤4 | 0.5 | 4 | 4 |
| PAOMS.2-CIP | ≤0.25 | ≤0.5 | 2 | ≤2 | ≤1 | 2 | 1 | >16 | ≤4 | 0.5 | 4 | 4 |
| PAOMS.3-CIP | 0.5 | ≤0.5 | 4 | ≤2 | ≤1 | 2 | 1 | >16 | 8 | 0.5 | 4 | 4 |
| PAO1.1-MER | 8 | 32 | 32 | 128 | 128 | 32 | >32 | 0.5 | 512 | ≤0.25 | ≤2 | ≤0.5 |
| PAO1.2-MER | 2 | 32 | 64 | 64 | 64 | 16 | >32 | 0.5 | 512 | 0.5 | ≤2 | 4 |
| PAO1.3-MER | 4 | 32 | 32 | 64 | 64 | 64 | >32 | 0.5 | 256 | ≤0.25 | ≤2 | ≤0.5 |
| PAOMS.1-MER | 1 | 4 | 16 | 8 | 64 | 32 | >32 | 0.5 | 512 | 0.5 | ≤2 | 2 |
| PAOMS.2-MER | 2 | 8 | 16 | 32 | 64 | 64 | >32 | 0.25 | 512 | 1 | 4 | 2 |
| PAOMS.3-MER | 1 | 16 | 16 | 64 | 32 | 32 | >32 | 0.5 | 512 | 1 | 4 | 4 |
TOL-TAZ, ceftolozane-tazobactam; CAZ, ceftazidime; FEP, cefepime; PIP-TZ, piperacillin-tazobactam; ATZ, aztreonam; IMI, imipenem; MER, meropenem; CIP, ciprofloxacin; TIC, ticarcillin; TOB, tobramycin; AMK, amikacin; COL, colistin.
TABLE 3.
Characterization of the resistance mechanisms of PAO1 and PAOMS lineages evolved in the presence of ceftazidime, meropenem, or ciprofloxacin in each of three experiments
| Straina | Relative mRNA expression level by geneb |
OprD lossc | Resistance mutation(s)d | ||||
|---|---|---|---|---|---|---|---|
| ampC | mexB | mexD | mexF | mexY | |||
| PAO1.1-CAZ | 350 ± 180 | <2 | <5 | dacB (W273X) + ampD (M1T) | |||
| PAO1.2-CAZ | 32 ± 5 | <2 | <5 | dacB (G407D) + ampD (V101G) | |||
| PAO1.3-CAZ | 68 ± 31 | <2 | <5 | dacB (nt307Δ4) + ampD (V101G) | |||
| PAOMS.1-CAZ | 244 ± 90 | <2 | <5 | dacB (G420D) + ampC (V239A) | |||
| PAOMS.2-CAZ | 673 ± 237 | <2 | <5 | dacB (G366D) + ampC (V239A) | |||
| PAOMS.3-CAZ | 254 ± 144 | <2 | <5 | ampR (D135N) | |||
| PAO1.1-CIP | <5 | <2 | 633 ± 62 | <5 | <5 | nfxB (W115X) + gyrA (T83I) + parC (S87L) | |
| PAO1.2-CIP | <5 | <2 | 690 ± 92 | <5 | <5 | nfxB (nt59Δ1) + gyrA (E153K) | |
| PAO1.3-CIP | <5 | 11.6 ± 4.3 | <5 | <5 | <5 | mexR (R83H) + gyrA (T83I) + parC (S87L) | |
| PAOMS.1-CIP | <5 | <2 | 656 ± 69 | <5 | <5 | nfxB (L172P) + gyrA (T83I) + parC (E91K) | |
| PAOMS.2-CIP | <5 | <2 | 652 ± 96 | <5 | <5 | nfxB (R42H) + gyrA (T83I) + parC (S87L) + dacB (A299T) | |
| PAOMS.3-CIP | <5 | <2 | 544 ± 317 | <5 | <5 | nfxB (L26P) + gyrA (T83I) + parC (S87L) | |
| PAO1.1-MER | <5 | 6.0 ± 0.8 | <5 | <5 | + | oprD (A150X) + mexR (T130P) | |
| PAO1.2-MER | <5 | 5.3 ± 2.0 | <5 | <5 | + | oprD (nt1329InsG) + mexR (R63H) | |
| PAO1.3-MER | <5 | 6.5 ± 1.4 | <5 | <5 | + | oprD (nt109Δ681) + mexR (nt307Δ1) | |
| PAOMS.1-MER | <5 | 7.7 ± 0.3 | <5 | <5 | + | oprD (W277X) + nalD (L91P) + mexT (F172I) | |
| PAOMS.2-MER | <5 | 21.9 ± 9.9 | <5 | <5 | + | oprD (W65X) + mexR (R91C) | |
| PAOMS.3-MER | <5 | 17.0 ± 1.0 | <5 | <5 | + | oprD (nt635InsG) + mexR (G101R) + mexT (A179V) | |
PAO1 or PAOMS ceftazidime (CAZ)-, ciprofloxacin (CIP)-, and meropenem (MER)-resistant mutant derivatives.
Relative ampC, mexB, mexD, mexF, and mexY expression levels compared to that of wild-type PAO1. Previously described breakpoints (18) were applied for defining overexpression: ampC, mexD, mexF, and mexY, positive >10-fold compared to wild-type PAO1, negative <5-fold; mexB, positive >3-fold compared to wild-type PAO1, negative <2-fold.
Lack of expression of OprD, as evidenced by the analysis of outer membrane proteins (OMPs) through SDS-PAGE.
Mutations in genes known to affect ampC, mexB, mexD, mexF, and mexY expression, oprD, and QRDR of gyrA, gyrB, parC, and parE.
Evolution of β-lactam resistance.
All mutants selected upon ceftazidime exposure showed high-level resistance to ceftazidime, cefepime, piperacillin-tazobactam, and aztreonam, caused by the overexpression of the chromosomal cephalosporinase AmpC, due to mutations in the peptidoglycan-recycling enzymes DacB (PBP4), AmpD, and AmpR. Cross-resistance to carbapenems, fluoroquinolones, aminoglycosides, or polymyxins was not detected in any of the ceftazidime-selected mutants, and none of them showed efflux pump overexpression. Intriguingly, despite the dramatic difference in the total number of mutations in favor of the ΔmutS strain, all PAO1 derivatives showed two mutations in ampC regulatory genes (ampD and dacB), while PAOMS ceftazidime-resistant evolved populations showed only one mutation (in ampD, dacB, or ampR). However, among many other mutations, two of the PAOMS derivatives showed a V239A substitution in AmpC (V213A, according to the mature protein), which was located in the Ω loop and shown very recently to confer high-level ceftazidime resistance among clinical strains (31). On the other hand, one of the multiple mutations detected in the third PAOMS derivative was tracked to galU, encoding a UDP-glucose pyrophosphorylase required for lipopolysaccharide (LPS) core synthesis (32); indeed, a recent analysis of transposon mutant libraries showed that the inactivation of galU increases ceftazidime and meropenem MICs (10, 11), while it reduces those of colistin (13), as observed for PAOMS.3-CAZ. Moreover, this mutant showed also a substitution (G476S) in the sensor/response regulator hybrid GacS, previously shown to be involved in virulence, biofilm formation, and antibiotic resistance (33).
Evolution of fluoroquinolone resistance.
All PAO1 and PAOMS mutants selected upon ciprofloxacin exposure showed QRDR mutations (gyrA with or without parC) and overexpressed efflux pumps (Table 3). Remarkably, while the mutations detected most frequently included the classical GyrA T83I and ParC S87L substitutions, one of the mutants showed a non-previously described substitution in GyrA (E153K). The specific effects of this new QRDR mutation in quinolone resistance are under investigation in our laboratory. Additionally, 5 of the 6 mutants overexpressed the efflux pump MexCD-OprJ, correlating with the presence of nfxB mutations and the expected hypersusceptibility to imipenem and tobramycin. In contrast, the sixth mutant overexpressed MexAB-OprM, correlating with a mexR mutation and the expected reduced β-lactam susceptibility. In addition to these classical fluoroquinolone resistance mechanisms, one of the PAO1 derivatives (PAO1.2) showed, intriguingly, a deletion of three nucleotides in pmrB. PmrAB is a two-component regulatory system mediating the transcription of the arnBCADTEF-pmrE operon involved in the addition of 4-amino-l-arabinose to phosphate groups within the lipid A and core oligosaccharide moieties of the lipopolysaccharide (LPS), which is linked to polymyxin resistance in clinical strains (34, 35). Colistin MICs were not enhanced in this mutant, but recent studies show the existence of regulatory pathways interconnecting LPS modifications with fluoroquinolone resistance (36). Also intriguingly, one of the PAOMS ciprofloxacin-resistant derivatives showed a dacB (PBP4) mutation. However, the strain did not show elevated β-lactam MICs or increased ampC expression, which might be related to recent findings showing that the overexpression of MexCD-OprJ impairs dacB mutation-mediated β-lactam resistance. Also possibly related to our findings, ciprofloxacin has been suggested to select β-lactam resistance driven by AmpC hyperproduction (37). Other interesting mutated genes detected among PAOMS derivatives include those coding for the global transcriptional regulators GacR and RetS or the single-stranded-DNA-specific exonuclease RecJ.
Evolution of carbapenem resistance.
All PAO1 and PAOMS mutants selected upon meropenem exposure contained oprD-inactivating mutations, which correlated with the loss of OprD expression in OMP analysis (see Fig. S1 in the supplemental material) and resistance to imipenem and meropenem. Moreover, all mutants showed mexR (most frequently) or nalD mutations, which correlated with MexAB-OprM overexpression and reduced susceptibility to all β-lactams and fluoroquinolones. Thus, meropenem exposure selected multidrug-resistant (MDR) profiles. Moreover, despite none of the mutants overexpressing MexEF-OprN, two of the meropenem-resistant mutator derivatives contained a mutation in mexT, a regulator of this efflux pump; interestingly, however, mexT has been shown to be a global transcriptional regulator, along with other genes, including oprD (38). Moreover, the lack of correlation with MexEF-OprN overexpression mutations might be related to the fact that PAO1 mexT contains an 8-bp insertion resulting in a nonfunctional regulator (14). Additionally, one of the PAOMS derivatives showed a substitution (N587S) in pbpA, encoding PBP2, a major target of carbapenems. Much more revealing and unexpected, all three PAOMS derivatives and one of the PAO1 derivatives showed mutations (each a different one: R504H, A539T, V465A, or F533L) in ftsI, encoding PBP3. All these residues are located close to the β-lactam binding site of the essential transpeptidase PBP3; of particular relevance, F533 has been demonstrated to play a key role in β-lactam recognition (39). Indeed, an analysis of PBP 50% inhibitory concentrations (IC50s) in two of these mutants, as an example, revealed a major and specific increase in meropenem PBP3 IC50s (Table 4). Therefore, these results indicate that PBP3 is a major unexpected target for meropenem resistance development that should be explored among clinical strains. Indeed, the positive selection of PBP3 mutations was documented in a very recent study monitoring the evolution of P. aeruginosa in the lungs of chronically infected cystic fibrosis patients (40), although potential links with meropenem exposure remain to be explored.
TABLE 4.
Ceftazidime and meropenem PBP3 IC50 in wild-type PAO1 and selected PBP3 mutants
| Strain | IC50 (mg/liter) ± SD for: |
|
|---|---|---|
| PBP3 CAZ | PBP3 MER | |
| PAO1 | 0.13 ± 0.08 | 0.028 ± 0.003 |
| PAOMS.1-MER (PBP3 A539T) | 0.23 ± 0.07 | 0.089 ± 0.04a |
| PAOMS.3-MER (PBP3 F533L) | 0.18 ± 0.05 | 0.074 ± 0.04a |
P < 0.05 compared to wild-type PAO1.
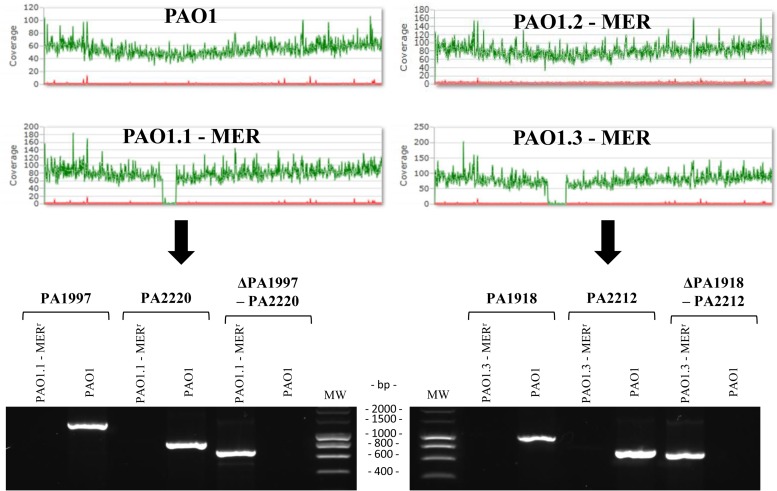
On the other hand, surprisingly, the two PAO1 derivatives that did not contain ftsI mutations showed an extremely large (around 5% of the genome) chromosomal deletion, with 258,659 nucleotides in PAO1.1-MER and 338,658 in PAO1.3-MER.
Meropenem selects extremely large deletions in specific areas of PAO1 genome.
Figure 2 shows the MiSeq reporter run plots evidencing the chromosomal deletions and the PCR mapping used for their confirmation. The complete representation of the deleted genome fragments is shown in Fig. S2 in the supplemental material. Analysis of the genomic regions showed that in both cases, deletions occurred between two inverted repeats, of 5 nucleotides in PAO1.1-MER and of 13 nucleotides in PAO1.3-MER. The deleted regions included 223 genes (PA1997 to PA220) for PAO1.1-MER and 295 genes (PA1918 to PA2212) for PAO1.3-MER. Therefore, interestingly, major overlapping (215 genes) of the deleted sequences in both strains was evidenced.
FIG 2.
MiSeq Reporter run plots and PCR confirmation of deleted regions. Upper panel, MiSeq Reporter run plots of P. aeruginosa reference strain PAO1 and the 3 derivative lineages evolved in the presence of meropenem for 7 days. Lower panel, agarose gel electrophoresis results of PCR products obtained to confirm the deleted regions in PAO1.1-MER and PAO1.3-MER. PCRs for genes within right and left margins of the deletions were positive for PAO1 only. On the other hand, PCR amplifications of DNA regions flanking the deletion yielded the expected size in PAO1.1-MER and PAO1.3-MER and were negative for PAO1. In all cases, PCR products were confirmed by sequencing. MW, molecular weight.
A first phenotypical characterization of PAO1.1-MER and PAO1.3-MER colonies revealed the presence of a dark-brown pigment (pyomelanin) in contrast to the greenish pigment produced by wild-type PAO1 (see Fig. S3 in the supplemental material). The hyperproduction of pyomelanin is known to be caused by the disruption of hmgA, which encodes a homogentisate-1,2-dioxygenase (41). Indeed, hmgA (PA2209) was deleted in both mutants (see Fig. S2 in the supplemental material). Using the dark-brown pigment as a marker, we further analyzed 10 colonies (2 from each of 5 independent experiments of 7 days of meropenem exposure), and pyomelanin hyperproduction was detected in 4 of the colonies from 3 independent experiments. PCR mapping confirmed the deletion of the hmgA gene in the context of large deletions that were similar, but not identical, to those of PAO1.1-MER or PAO1.3-MER. Moreover, the selection of pyomelanin hyperproduction by meropenem was also analyzed by plating overnight cultures of PAO1 in Mueller-Hinton agar plates with and without meropenem at 1 mg/liter (2 times the MIC). Indeed, analysis of around 1,000 colonies revealed that 1.3% of one-step 1 mg/liter meropenem-resistant mutants showed pyomelanin hyperproduction, while the frequency was <0.1% for colonies grown in meropenem-free medium. Altogether, our results show that meropenem exposure selects for a large chromosomal deletion in the hmgA region.
Interestingly, hmgA inactivation (including deletion) occurs relatively frequently in chronic infections and has been shown to increase oxidative stress resistance and persistence (41). However, hmgA inactivation does not directly affect antibiotic resistance; therefore, the conferred resistance phenotype (and the primary selection driver) should be conferred to another (or several) of the hundreds of deleted genes. Indeed, galU (PA2023) was deleted in both mutants, and as mentioned above, recent analysis of transposon mutant libraries has shown that the inactivation of galU increases meropenem resistance (10, 11). Additionally, the inactivation of galU should explain the marked colistin hypersusceptibility of both mutants (Table 2) (13), although the EraRS two-component regulator, which was also deleted, has recently been shown to be involved in colistin susceptibility (42). Moreover, both strains show a marked tobramycin hypersusceptibility, correlating with the deletion of genes encoding MexX and MexY, components of the inducible efflux pump responsible for intrinsic aminoglycoside resistance (43). Interestingly, this efflux pump uses the same outer membrane channel (OprM) as MexAB-OprM, which is involved in meropenem resistance and is overexpressed in all meropenem-exposed mutants (Table 3). Thus, in addition to galU inactivation, eliminating MexXY might also favor meropenem tolerance, reducing the competition of MexAB for OprM and therefore increasing the efficiency of meropenem efflux. Indeed, this type of antagonistic interaction between resistance mechanisms has been noted (44). However, in addition to galU and mexXY, many other deleted genes might eventually contribute to making possible the selection of these extraordinarily large deletions upon meropenem exposure.
Fitness and virulence vary within and across evolved antibiotic-resistant populations: impact of mutator phenotypes.
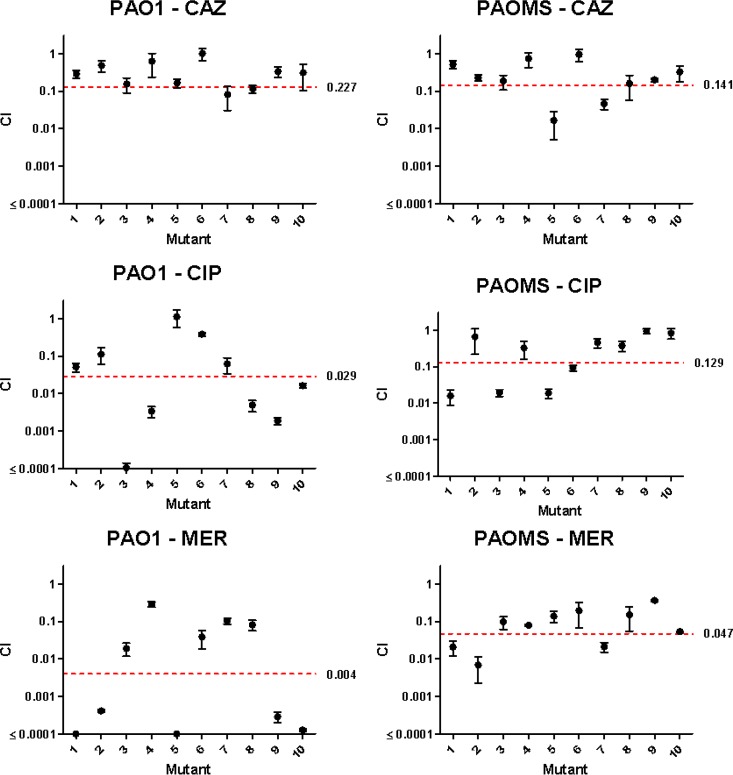
The fitness of the antibiotic-resistant derivatives was evaluated through the determination of the competition indexes (CIs) in a competitive growth assay with a wild-type strain, and the results are shown in Table 4. As can be observed, in the majority of the cases, a biological cost was evidenced in the evolved resistance strains, although the impact on fitness varied considerably, depending on the antibiotic and specific mutants analyzed. Overall, the impact on fitness was significantly (P = 0.02) lower for ceftazidime (median CI, 0.18) than for ciprofloxacin (CI, 0.02) and meropenem (CI, 0.016). As previously noted (45), fitness was slightly reduced in the mutator strain PAOMS (CI, 0.66). However, despite the dramatically higher number of mutations acquired, the fitness levels of the antibiotic-resistant derivatives from PAOMS (median CI, 0.02) were not significantly (P = 0.7) lower than those of PAO1 derivatives (CI, 0.05). Within strain and antibiotic, PAO1 ciprofloxacin-resistant derivatives showed the widest variability, with CIs ranging from 0.0001 to 1.12. PAO1.3-CIP (CIP, ciprofloxacin), showing no fitness cost (CI, 1.12), overexpressed MexAB-OprM, while the other two overexpressed MexCD-OprJ, which is known to be associated with a significant biological cost (44, 46). Moreover, strain PAO1.2-CIP, showing the lowest fitness (CI, 0.0001) among ciprofloxacin-resistant derivatives, showed the non-previously described substitution in GyrA (E153K). Although the specific impact of this mutation in fitness remains to be explored, the fact that it has not been detected in the hundreds of ciprofloxacin-resistant clinical strains sequenced so far might suggest that it is associated with a major fitness cost. Not surprisingly, the only other two mutants showing comparable extremely low CIs were the two PAO1 meropenem-resistant derivatives showing the large chromosomal deletions. Indeed, the deletion of such an extraordinarily high number of genes is expected to have a major impact on bacterial physiology. In fact, 27 of the genes deleted in one or both mutants are not represented in available saturated transposon mutant libraries, suggesting that they are essential for in vitro growth (47–49). However, our results indicate that although they may significantly compromise growth, they should be removed from the list of essential genes.
In order to confirm these findings, we evaluated the fitness of an extended collection, including 10 mutants (2 mutant CFU from each of 5 independent experiments) for each antibiotic series, and the results are shown in Fig. 3. As can be observed, the documented tendencies were largely confirmed. First, the fitness of the lineages that evolved in the presence of ceftazidime was higher than that of lineages exposed to ciprofloxacin and, particularly, meropenem. Second, for ciprofloxacin and meropenem but not for ceftazidime, the evolved lineages showed an extraordinary variability in fitness, with CIs ranging from <0.0001 (very major fitness cost) to 1 (no cost at all). This extraordinary variability of fitness occurred, however, in the lineages evolved from wild-type PAO1 but not in those from the mutator strain PAOMS. Moreover, the evolved mutator lineages showed statistically significant higher fitness than that of evolved wild-type lineages for ciprofloxacin (P = 0.04) and meropenem (P = 0.03); in contrast, no differences were observed for ceftazidime (P = 0.43). Altogether, these results indicate that for some antibiotics (such as ceftazidime), wild-type mutation supply rates provide a large enough pool of mutations (such as the multiple mutational targets for AmpC derepression), ensuring a stepwise high-level resistance development at a relatively low fitness cost. On the other hand, high-level resistance development to other antibiotics, such as ciprofloxacin or meropenem, under wild-type mutation supply rates determines major population bottlenecks in which the pool of available mutations is very limited, allowing the eventual selection of rare high-level resistant lineages even if they are associated with a major fitness cost. However, the mutator phenotype is expected to significantly increase the pool of total mutations, leading to natural selection not picking up those mutations associated with an excessive cost. Moreover, the high mutation rates should facilitate the selection of cost-compensatory mutations.
FIG 3.
Competition index (CI) values for 10 selected mutants (2 mutant CFU from each of 5 independent experiments) for each antibiotic series. The mean ± SD of 3 replicates are represented. The median CI values for the 10 mutants in each series are indicated by the red dashed line (with the value shown to the right).
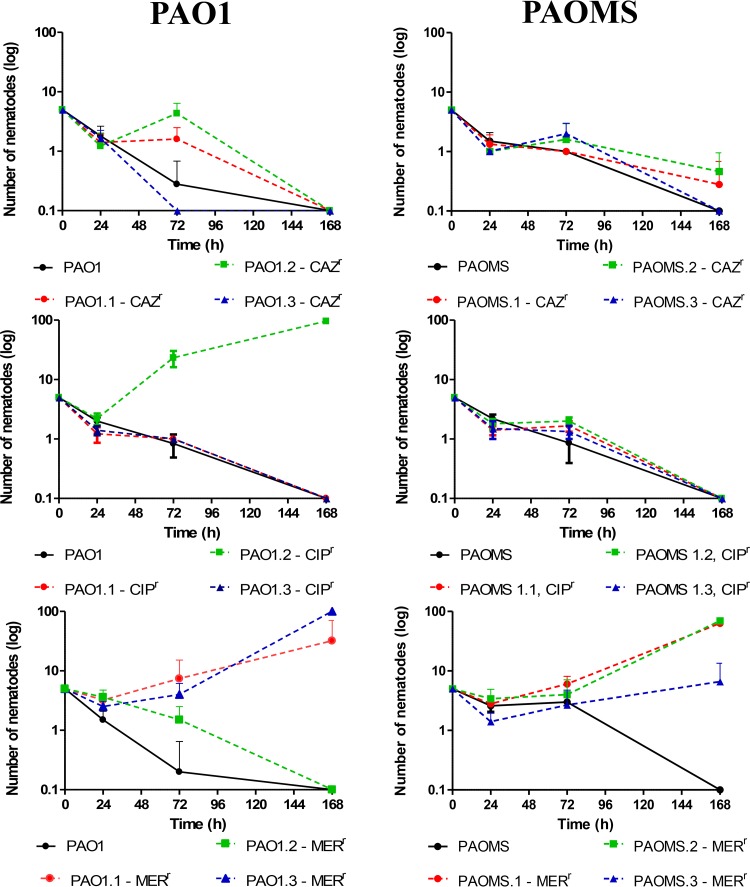
As a next step, we evaluated the pathogenicity of the evolved antibiotic-resistant derivatives. For this purpose, we monitored the interaction between each of the strains and the nematode C. elegans for 7 days, and the results are shown in Table 5 and Fig. 4. As can be observed, almost all PAO1 and PAOMS ceftazidime- or ciprofloxacin-resistant mutants conserved their pathogenic capacity, since they killed nearly all nematodes by day 7. The single exception was the PAO1 ciprofloxacin-resistant derivative containing the E153K substitution, which not only did not kill the nematode but allowed it to proliferate, increasing its population from 5 at the beginning of the experiments to nearly 100 by day 7. In contrast to derivatives that were resistant to ceftazidime and ciprofloxacin, all but one of the meropenem-resistant derivatives showed impaired pathogenicity, including the 3 PAOMS derivatives and the 2 PAO1 derivatives showing the chromosomal deletions.
TABLE 5.
Fitness and virulence of wild-type and mutator lineages evolved in the presence of different antibiotics
| Strain | CI | No. of surviving nematodes at 168 h (mean ± SD) |
|---|---|---|
| PAO1 | 1.34 ± 0.23 | <1 |
| PAOMS | 0.66 ± 0.12 | <1 |
| PAO1.1-CAZ | 0.29 ± 0.07 | <1 |
| PAO1.2-CAZ | 0.16 ± 0.07 | <1 |
| PAO1.3-CAZ | 0.16 ± 0.04 | <1 |
| PAOMS.1-CAZ | 0.52 ± 0.12 | <1 |
| PAOMS.2-CAZ | 0.19 ± 0.08 | <1 |
| PAOMS.3-CAZ | 0.02 ± 0.018 | <1 |
| PAO1.1-CIP | 0.05 ± 0.01 | <1 |
| PAO1.2-CIP | 0.0001 ± 0.00004 | 95.6 ± 9.8 |
| PAO1.3-CIP | 1.12 ± 0.55 | <1 |
| PAOMS.1-CIP | 0.02 ± 0.007 | <1 |
| PAOMS.2-CIP | 0.02 ± 0.004 | <1 |
| PAOMS.3-CIP | 0.02 ± 0.006 | <1 |
| PAO1.1-MER | 0.00002 ± 0.000003 | 32 ± 38.5 |
| PAO1.2-MER | 0.012 ± 0.008 | <1 |
| PAO1.3-MER | 0.0001 ± 0.00003 | >100 |
| PAOMS.1-MER | 0.02 ± 0.01 | 63 ± 50.8 |
| PAOMS.2-MER | 0.1 ± 0.04 | 69.5 ± 35.5 |
| PAOMS.3-MER | 0.14 ± 0.05 | 6.6 ± 6.8 |
FIG 4.
Virulence on C. elegans of PAO1 and PAOMS lineages evolved in the presence of different antibiotics. The mean ± SD numbers of surviving nematodes at 0, 24, 72, and 168 h are shown. CAZr, ceftazidime resistant; CIPr, ciprofloxacin resistant; MERr, meropenem resistant.
Large deletions in specific areas of PAO1 genome promote growth in the presence of carbapenems.
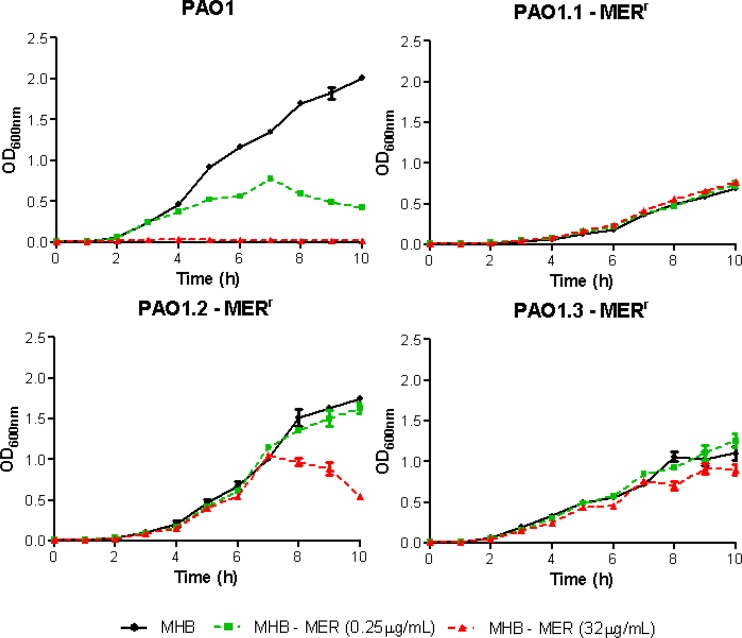
Considering the enormous fitness cost associated with the extraordinarily large deletions detected in the PAO1 chromosome, we wondered why these deletions should then be selected by meropenem exposure, considering the existence of alternative mutational pathways leading to high-level meropenem resistance. To solve this uncertainty, we analyzed the growth rates of PAO1 and the three meropenem-resistant derivatives in the absence of meropenem and in the presence of two concentration of this antibiotic: one subinhibitory (0.5 times the MIC [0.25 mg/liter]) for PAO1, and one high concentration (32 mg/liter) that was subinhibitory for the three high-level meropenem-resistant derivatives. As can be observed in Fig. 5, the growth rate was, as expected, significantly reduced for the two derivatives containing the deletion (PAO1.1-MER and PAO1.3-MER). On the other hand, even the low (0.25 mg/liter) subinhibitory concentration had a marked effect on the growth rate of PAO1, similar to the high subinhibitory concentration (32 mg/liter) for the PAO1.2-MER high-level meropenem-resistant derivative. In sharp contrast, growth rates were not affected even by the 32 mg/liter concentrations in the two mutants showing the deletion. Thus, the two lineages containing the deletion were extremely well adapted to grow in the presence of meropenem. Whether meropenem treatments could also select this type of extremely large deletion in vivo remains to be explored, but recent whole-genome analysis of adapted CF isolates shows that the evolution of P. aeruginosa in chronic infections frequently involves large genome reductions (50), and, as noted above, pyomelanin hyperproduction, a marker of these specific deletions, is frequent among CF isolates (41).
FIG 5.
Grow rates (OD600nm) of wild-type PAO1 and the 3 derivative lineages evolved in the presence of meropenem. Growth was monitored in the presence of 0, 0.25, and 32 μg/ml meropenem.
In summary, we have performed the first comparative analysis of the evolution of resistance to relevant antibiotics in wild-type and mismatch repair (MMR)-deficient mutator P. aeruginosa strains. WGS provided a privileged perspective of the dramatic effect of mutator phenotypes on the accumulation of random mutations, of which the vast majority were transitions, as expected. Much more revealing, WGS of antibiotic-exposed mutator lineages also evidenced a frameshift mutagenic signature, with a consistent error-prone DNA polymerase activity consequence of SOS system induction. This mutagenic signature was evidenced for all three antibiotics tested, but it was much higher for fluoroquinolones than for cephalosporins or carbapenems. The subsequent analysis of the correlation between genotype and antibiotic resistance phenotype confirmed expected resistance evolution trajectories but also revealed new major pathways. Classical pathways included multiple mutational targets (ampD, ampR, or dacB) leading to AmpC overexpression selected upon ceftazidime exposure, QRDR mutations selected by ciprofloxacin, or oprD mutational inactivation in the presence of meropenem. Likewise, mutational overexpression of efflux pumps was seen in all lineages evolved in the presence of ciprofloxacin and meropenem but not with ceftazidime. Less-expected pathways included the structural modification of AmpC and the mutation of the UDP-glucose pyrophosphorylase GalU, both conferring ceftazidime resistance, or the detection of a non-previously described GyrA mutation involved in fluoroquinolone resistance. However, the most striking findings were seen in the evolution of meropenem resistance, in which classical mechanisms were always accompanied by either the modification of the β-lactam binding site of PBP3 or extremely large genomic deletions providing a growth advantage in the presence of the antibiotic. Further studies are thus needed to determine whether the same evolutionary trajectories are seen in vivo and for strains with different genetic backgrounds. Finally, while fitness and virulence varied within and across evolved antibiotic-resistant populations, despite the dramatically higher number of mutations acquired, mutator lineages that evolved in the presence of ciprofloxacin and meropenem showed a lower biological cost. Thus, altogether, the provided data are a major advancement in the understanding of the evolution of antimicrobial resistance and fitness in wild-type and mutator bacterial populations.
Supplementary Material
Funding Statement
Ministerio de Economía y Competitividad (MINECO), Instituto de Salud Carlos III, cofinanced by European Regional Development Fund “A way to achieve Europe” ERDF, provided funding to Jesús Blázquez and Antonio Oliver through the Spanish Network for the Research in Infectious Diseases (grants RD06/0008 and RD12/0015) and grants PI12/00103 and PI15/00088.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02676-15.
REFERENCES
- 1.Gellatly SL, Hancock RE. 2013. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 2.Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW. 2011. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35:652–680. doi: 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- 3.Vincent JL. 2003. Nosocomial infections in adult intensive-care units. Lancet 61:2068–2077. [DOI] [PubMed] [Google Scholar]
- 4.Oliver A, Mena A, Macià MD. 2008. Evolution of Pseudomonas aeruginosa pathogenicity: from acute to chronic infections, p 433–444. In Baquero F, Nombela C, Cassell GH, Gutíerrez JA (ed), Evolutionary biology of bacterial and fungal pathogens. ASM Press, Washington, DC. [Google Scholar]
- 5.Breidenstein EB, de la Fuente-Núñez C, Hancock RE. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Oliver A, Cantón R, Campo P, Baquero F, Blázquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 7.Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, Perez JL, Oliver A. 2008. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J Bacteriol 190:7910–7917. doi: 10.1128/JB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvarez-Ortega C, Wiegand I, Olivares J, Hancock RE, Martínez JL. 2010. Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to beta-lactam antibiotics. Antimicrob Agents Chemother 54:4159–4167. doi: 10.1128/AAC.00257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dötsch A, Becker T, Pommerenke C, Magnowska Z, Jänsch L, Häussler S. 2009. Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:2522–2531. doi: 10.1128/AAC.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fajardo A, Martínez-Martín N, Mercadillo M, Galán JC, Ghysels B, Matthijs S, Cornelis P, Wiehlmann L, Tümmler B, Baquero F, Martínez JL. 2008. The neglected intrinsic resistome of bacterial pathogens. PLoS One 3:e1619. doi: 10.1371/journal.pone.0001619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández L, Alvarez-Ortega C, Wiegand I, Olivares J, Kocíncová D, Lam JS, Martínez JL, Hancock RE. 2013. Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:110–119. doi: 10.1128/AAC.01583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2001. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. [DOI] [PubMed] [Google Scholar]
- 15.Cabot G, Bruchmann S, Mulet X, Zamorano L, Moyà B, Juan C, Haussler S, Oliver A. 2014. Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple mutations leading to overexpression and structural modification of AmpC. Antimicrob Agents Chemother 58:3091–3099. doi: 10.1128/AAC.02462-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39:D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang L, Jelsbak L, Marvig RL, Damkiær S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Høiby N, Sommer MO, Molin S. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci U S A 108:7481–7486. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cabot G, Ocampo-Sosa AA, Tubau F, Macia MD, Rodríguez C, Moya B, Zamorano L, Suárez C, Peña C, Martínez-Martínez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI). 2011. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob Agents Chemother 55:1906–1911. doi: 10.1128/AAC.01645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabot G, Ocampo-Sosa AA, Domínguez MA, Gago JF, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI). 2012. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother 56:6349–6357. doi: 10.1128/AAC.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao G, Meier T, Kahi SD, Gee KR, Blaszczak LC. 1999. BOCILLIN FL, a sensitive and commercially available reagent for the detection of penicillin-binding proteins. Antimicrob Agents Chemother 43:1124–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moyá B, Zamorano L, Juan C, Ge Y, Oliver A. 2010. Affinity of the new cephalosporin CXA-101 to penicillin-binding proteins of Pseudomonas aeruginosa. Antimicrob Agents Chemother 54:3933–3937. doi: 10.1128/AAC.00296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moyà B, Beceiro A, Cabot G, Juan C, Zamorano L, Alberti S, Oliver A. 2012. Pan-β-lactam resistance development in Pseudomonas aeruginosa clinical strains: molecular mechanisms, penicillin-binding protein profiles, and binding affinities. Antimicrob Agents Chemother 56:4771–4778. doi: 10.1128/AAC.00680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moyà B, Juan C, Albertí S, Pérez JL, Oliver A. 2008. Benefit of having multiple ampD genes for acquiring beta-lactam resistance without losing fitness and virulence in Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:3694–3700. doi: 10.1128/AAC.00172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulet X, Cabot G, Ocampo-Sosa AA, Domínguez MA, Zamorano L, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI). 2013. Biological markers of Pseudomonas aeruginosa epidemic high-risk clones. Antimicrob Agents Chemother 57:5527–5535. doi: 10.1128/AAC.01481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navas A, Cobas G, Talavera M, Ayala JA, López JA, Martínez JL. 2007. Experimental validation of Haldane's hypothesis on the role of infection as an evolutionary force for metazoans. Proc Natl Acad Sci U S A 104:13728–13731. doi: 10.1073/pnas.0704497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horst JP, Wu TH, Marinus MG. 1999. Escherichia coli mutator genes. Trends Microbiol 7:29–36. doi: 10.1016/S0966-842X(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 27.Cirz RT, O'Neill BM, Hammond JA, Head SR, Romesberg FE. 2006. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J Bacteriol 188:7101–7110. doi: 10.1128/JB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SR, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damaged DNA. Proc Natl Acad Sci U S A 94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez-Capilla T, Baquero MR, Gómez-Gómez JM, Ionel A, Martín S, Blázquez J. 2005. SOS-independent induction of dinB transcription by beta-lactam mediated inhibition of cell wall synthesis in Escherichia coli. J Bacteriol 187:1515–1518. doi: 10.1128/JB.187.4.1515-1518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blázquez J, Gómez-Gómez JM, Oliver A, Juan C, Kapur V, Martín S. 2006. PBP3 inhibition elicits adaptive responses in Pseudomonas aeruginosa. Mol Microbiol 62:84–99. [DOI] [PubMed] [Google Scholar]
- 31.Berrazeg M, Jeannot K, Ntsogo Enguéné VY, Broutin I, Loeffert S, Fournier D, Plésiat P. 2015. Mutations in β-lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob Agents Chemother 59:6248–6255. doi: 10.1128/AAC.00825-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dean CR, Goldberg JB. 2002. Pseudomonas aeruginosa galU is required for a complete lipopolysaccharide core and repairs a secondary mutation in a PA103 (serogroup O11) wbpM mutant. FEMS Microbiol Lett 210:277–283. doi: 10.1111/j.1574-6968.2002.tb11193.x. [DOI] [PubMed] [Google Scholar]
- 33.Gooderham WJ, Hancock RE. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol Rev 33:279–294. [DOI] [PubMed] [Google Scholar]
- 34.Barrow K, Kwon DH. 2009. Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:5150–5154. doi: 10.1128/AAC.00893-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moskowitz SM, Brannon MK, Dasgupta N, Pier M, Sgambati N, Miller AK, Selgrade SE, Miller SI, Denton M, Conway SP, Johansen HK, Høiby N. 2012. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 56:1019–1030. doi: 10.1128/AAC.05829-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller C, Plésiat P, Jeannot K. 2011. A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother 55:1211–1221. doi: 10.1128/AAC.01252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolter DJ, Schmidtke AJ, Hanson ND, Lister PD. 2007. Increased expression of ampC in Pseudomonas aeruginosa mutants selected with ciprofloxacin. Antimicrob Agents Chemother 51:2997–3000. doi: 10.1128/AAC.00111-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochs MM, McCusker MP, Bains M, Hancock REW. 1999. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob Agents Chemother 43:1085–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han S, Zaniewski RP, Marr ES, Lacey BM, Tomaras AP, Evdokimov A, Miller JR, Shanmugasundaram V. 2010. Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 107:22002–22007. doi: 10.1073/pnas.1013092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diaz Caballero J, Clark ST, Coburn B, Zhang Y, Wang PW, Donaldson SL, Tullis DE, Yau YC, Waters VJ, Hwang DM, Guttman DS. 2015. Selective sweeps and parallel pathoadaptation drive Pseudomonas aeruginosa evolution in the cystic fibrosis lung. mBio 6(5):e00981-15. doi: 10.1128/mBio.00981-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodríguez-Rojas A, Mena A, Martín S, Borrell N, Oliver A, Blázquez J. 2009. Inactivation of the hmgA gene of Pseudomonas aeruginosa leads to pyomelanin hyperproduction, stress resistance and increased persistence in chronic lung infection. Microbiology 155:1050–1057. doi: 10.1099/mic.0.024745-0. [DOI] [PubMed] [Google Scholar]
- 42.Lee JY, Chung ES, Na IY, Kim H, Shin D, Ko KS. 2014. Development of colistin resistance in pmrA-, phoP-, parR- and cprR-inactivated mutants of Pseudomonas aeruginosa. J Antimicrob Chemother 69:2966–2971. doi: 10.1093/jac/dku238. [DOI] [PubMed] [Google Scholar]
- 43.Jeannot K, Sobel ML, El Garch F, Poole K, Plésiat P. 2005. Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J Bacteriol 187:5341–5346. doi: 10.1128/JB.187.15.5341-5346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mulet X, Moyà B, Juan C, Macià MD, Pérez JL, Blázquez J, Oliver A. 2011. Antagonistic interactions of Pseudomonas aeruginosa antibiotic resistance mechanisms in planktonic but not biofilm growth. Antimicrob Agents Chemother 55:4560–4568. doi: 10.1128/AAC.00519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mena A, Maciá MD, Borrell N, Moyà B, de Francisco T, Pérez JL, Oliver A. 2007. Inactivation of the mismatch repair system in Pseudomonas aeruginosa attenuates virulence but favors persistence of oropharyngeal colonization in cystic fibrosis mice. J Bacteriol 189:3665–3668. doi: 10.1128/JB.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martínez-Ramos I, Mulet X, Moyà B, Barbier M, Oliver A, Albertí S. 2014. Overexpression of MexCD-OprJ reduces Pseudomonas aeruginosa virulence by increasing its susceptibility to complement-mediated killing. Antimicrob Agents Chemother 58:2426–2429. doi: 10.1128/AAC.02012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Held K, Ramage E, Jacobs M, Gallagher L, Manoil C. 2012. Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J Bacteriol 194:6387–6389. doi: 10.1128/JB.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewenza S, Falsafi RK, Winsor G, Gooderham WJ, McPhee JB, Brinkman FS, Hancock RE. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res 15:583–589. doi: 10.1101/gr.3513905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rau MH, Marvig RL, Ehrlich GD, Molin S, Jelsbak L. 2012. Deletion and acquisition of genomic content during early stage adaptation of Pseudomonas aeruginosa to a human host environment. Environ Microbiol 14:2200–2211. doi: 10.1111/j.1462-2920.2012.02795.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.