Abstract
The process of protein complex dissociation remains to be understood at the atomic level of detail. Computers now allow microsecond timescale molecular-dynamics simulations, which make the visualization of such processes possible. Here, we investigated the dissociation process of the EphA2-SHIP2 SAM-SAM domain heterodimer complex using unrestrained all-atom molecular-dynamics simulations. Previous studies on this system have shown that alternate configurations are sampled, that their interconversion can be fast, and that the complex is dynamic by nature. Starting from different NMR-derived structures, mutants were designed to stabilize a subset of configurations by swapping ion pairs across the protein-protein interface. We focused on two mutants, K956D/D1235K and R957D/D1223R, with attenuated binding affinity compared with the wild-type proteins. In contrast to calculations on the wild-type complexes, the majority of simulations of these mutants showed protein dissociation within 2.4 μs. During the separation process, we observed domain rotation and pivoting as well as a translation and simultaneous rolling, typically to alternate and weaker binding interfaces. Several unsuccessful recapturing attempts occurred once the domains were moderately separated. An analysis of protein solvation suggests that the dissociation process correlates with a progressive loss of protein-protein contacts. Furthermore, an evaluation of internal protein dynamics using quasi-harmonic and order parameter analyses indicates that changes in protein internal motions are expected to contribute significantly to the thermodynamics of protein dissociation. Considering protein association as the reverse of the separation process, the initial role of charged/polar interactions is emphasized, followed by changes in protein and solvent dynamics. The trajectories show that protein separation does not follow a single distinct pathway, but suggest that the mechanism of dissociation is common in that it initially involves transitions to surfaces with fewer, less favorable contacts compared with those seen in the fully formed complex.
Introduction
How protein-protein complexes form and dissociate is as yet incompletely understood, as are the details of certain protein interactions, especially if these interactions are dynamic. For example, it is still difficult to correctly predict protein-protein interactions that involve conformational fluctuations by means of ab initio docking or simulation methods. This challenge is illustrated by the observation that the solvent-accessible surface area that is buried upon protein complex formation is typically small, accounting for only ∼8% of the total surface area of both proteins (1). What, then, makes the surfaces that are involved in the predominant interactions special? Many protein-complex structures have been analyzed (e.g., (1, 2, 3, 4, 5)) and it has become clear that fluctuations of the proteins as well as changes in their solvation are critical for protein complex formation and dissociation (6, 7, 8, 9). Accordingly, the recent incorporation of protein flexibility into protein-protein docking calculations has led to an increased accuracy of predictions (10, 11, 12). However, to date, there have been very few calculations of protein-protein association (or dissociation) processes at the all-atom level of detail. Recently, Chong and Ham (13, 14) studied the association of amyloid-β peptides, and Blöchliger et al. (15) investigated PDZ-peptide dissociation/association without biasing restraints. Similarly, Ahmad and colleagues simulated the association of a SH3 domain with a peptide starting 13 all-atom simulations with an extended peptide conformation at different peptide-protein separation and orientations (16). The few all atom studies on protein-protein association have, by computational necessity, used a more limited set of starting configurations in unrestrained simulations (17) or umbrella sampling to characterize the free energy surface of protein complex formation (18). By contrast, dissociation simulations have protein complexes as starting structures and, here, we present an all-atom simulation of a protein-protein dissociation process between two five-helix bundle proteins, carried out without any restraints to separate the proteins.
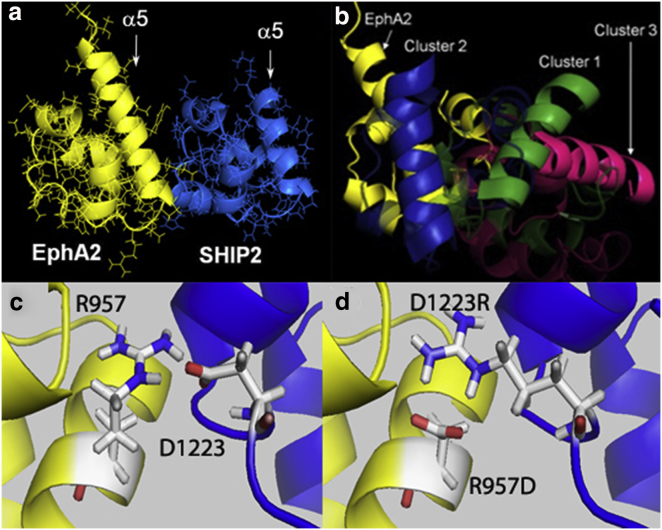
Many signal transduction processes involve adaptor protein domains whose binding helps to localize enzymes near their substrates and/or cause a conformational change that activates their targets (19). EphA2 is a transmembrane receptor tyrosine kinase (RTK) with key roles not only in cell migration during developmental processes but also in cancer metastasis (20). In contrast to other RTKs, Eph receptors possess a C-terminal sterile α motif (SAM) domain. SAM domains are α-helix bundles (Fig. 1 a) that are found in more than 200 human proteins and are used to form dimers, if not higher-order oligomers, of proteins (21). The SAM domain of EphA2 interacts with adaptor proteins such as the SAM domain of the enzyme SHIP2. We previously reported a refined NMR structure of the EphA2-SHIP2 SAM-SAM heterodimer complex (22). Although 80% of the complexes populated one configuration, denoted cluster 1, two additional clusters of structures were observed. These configurations (clusters 1–3) have different orientations of the SAM domains relative to each other (Fig. 1, a and b) while maintaining essentially the same interaction surfaces. To examine whether transitions between these configurations occur on a nanosecond–microsecond timescale, as implied by the fast exchange behavior seen in solution NMR spectroscopy, we carried out duplicate trajectories initiated with each of the three NMR-derived clusters. The simulations were extended to 2.4 μs on the Anton supercomputer, which is highly optimized for molecular-dynamics (MD) simulations (23). The trajectories sampled structures that were closely similar to clusters 1–3, as previously described (24). Importantly, degenerate and temporary cross-interface ion-pair interactions enabled transitions between different configurations, with average lifetimes of 50–150 ns (24).
Figure 1.
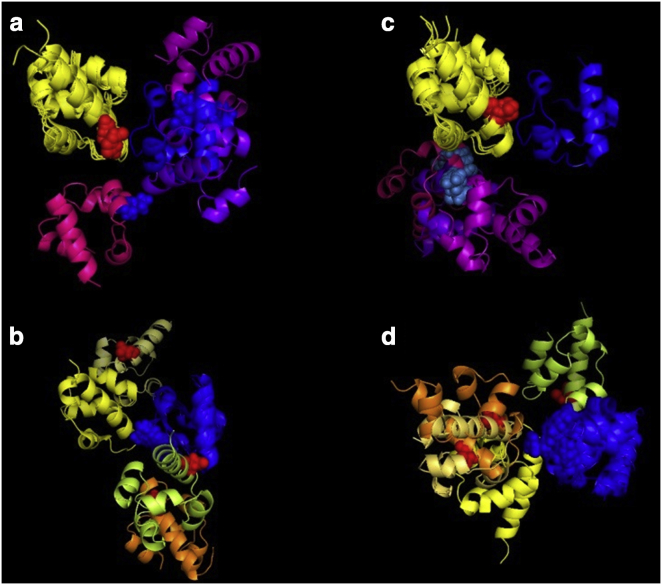
Depiction of the SAM-SAM complex, in multiconfigurational geometry, and a swap mutation. (a) EphA2-SHIP2 SAM-SAM heterodimer as a ribbon diagram; helix 5 of each domain is marked. (b) Complex viewed from the side, with transparent shading to emphasize the different orientations of the domains, described by the angle between helix 5 of EphA2 (yellow in back) to helix 5 of each SHIP2 domain orientation in front (green, blue, and red). (c and d) WT (c) and model (d) for the EphA2 R957D/SHIP2 D1223R swap mutant, based on the cluster 2 configuration.
Protein interactions are frequently validated by the use of site-directed mutagenesis. As the EphA2-SHIP2 SAM-SAM complex samples multiple conformers, swap mutations across the EphA2/SHIP2 protein-protein interface (e.g., Fig. 1, c and d) are expected to stabilize only a subset of the configurations, thereby shifting the relative populations of structures in the ensemble. Here, we describe the behavior of two such swap mutants, EphA2 K956D/SHIP2 D1235K (swap-mutant 1) and EphA2 R957D/SHIP2 D1223R (swap-mutant 2). In most of the simulations, we observed an eventual dissociation of the two SAM domains. The simulations offer an opportunity to analyze the dissociation process and the interactions involved at the all-atom level with full residue flexibility. The trajectories indicate that there is no single distinct pathway for the proteins to dissociate, and instead the separation process proceeds through a common mechanism involving low-contact interfaces and configurations. We also examine the changes in protein dynamics and solvation, which are both thought to contribute significantly to the separation process. The pattern of the changes in protein dynamics points to a regional relaxation of structures at and near the predominant binding interface. Away from this region, the changes suggest a dynamic allosteric feature in SAM domains. We discuss the generality of these observations also with respect to the reverse process, protein association.
Materials and Methods
Experimental methods
The human EphA2 and SHIP2 SAM domains (residues 897–976 and 1192–1258, respectively) were expressed in E. coli as wild-type (WT) and mutant proteins, and then purified as described previously (22). Surface plasmon resonance (SPR) was carried out on a Biacore T100 instrument with proteins in 10 mM HEPES at pH 7.4, 150 mM NaCl, 1 mM TCEP-HCl, 0.005% surfactant P-20 at 25°C (25) (see Supporting Materials and Methods in the Supporting Material for added details and discussion; Table S1; Figs. S3 and S4).
Simulation methods
Simulations were performed with swap-mutant 1 (K956D/D1235K) and swap-mutant 2 (R957D/D1223R) EphA2-SHIP2 SAM-SAM complexes. The double-mutant structures were built using VMD (26) based on the three WT complex configurations that were previously determined from solution NMR restraints (22) and were the starting structures of extensive simulations reported previously (24) (clusters 1–3 were the lowest-energy structures of the configurations). The simulation protocols, which we previously described (24, 27), were employed with the all-atom CHARMM C36 potential function (28). The complexes were centered in a 90 × 70 × 70 Å3 box and solvated by ∼16,500 explicit waters (TIP3P) (29). Not all configurations were well compatible with the swap mutations, and some swaps separated within 20 ns of the NAMD run (26) that followed the initial energy minimization and equilibration steps. In total, we built six systems based on both kinds of swap mutants and starting from three different initial cluster configurations. We performed at least three runs on each system with different random numbers in NAMD simulations to ensure that at least one stable complex structure would be ready before we continued with the Anton simulations (23). All simulations were run at a constant pressure of 1 atm and temperature of 300 K, using periodic boundary conditions with a 12 Å cutoff and the particle-mesh Ewald method for treating long-range electrostatics. The six Anton simulations (three for each swap mutant) were run for 2.4 μs or until the proteins separated (i.e., distance between centers of mass >70 Å). Coordinates were saved every 50 ps in the ordinary run. For more detailed analyses, the trajectories were restarted at a time point 10–50 ns before protein separation occurred and were run with an increased saving frequency of every 2 ps. These detailed trajectories with increased saving frequencies are termed zoomed-in trajectories.
Analysis
Details of the simulation analysis, including standard hydrogen-bond identification, surface and interaction energy calculations are reported in the Supporting Material (Table S2, a–c). The solvation free energy (ΔGs) was calculated for structures at 0.2 ns intervals using the PBEQ program (30). The protein residues were classified according to their location at the predominant interacting surface, at secondary binding surfaces near it, or far away from these surfaces (residues are listed in Table S3). We estimated protein entropy from the simulations using two different methods. In the first method, we used a quasi-harmonic (Q-entropy) approach (31) implemented in the Wordom program (32) to analyze representative segments of a 2.4 μs WT trajectory (these simulations were reported in our previous study (24)) and of trajectories after SAM domain separation (Table S4). In the second method, we estimated protein entropy differences for these residue groups due to changes in the orientational bond motions of main- and side-chain groups using the Lipari-Szabo order parameter S2, following the approach of Yang and Kay (33). The parameter S2 reflects the amplitude of bond fluctuations on the picosecond–nanosecond timescale, and we calculated it together with the correlation times using CHARMM over the same trajectory intervals used for the quasi-harmonic approach (Table S5).
Results
Simulation and experimental studies show that the mutant complexes have altered binding affinities. Simulations were started from each of the three WT starting configurations (Fig. 1 b), with the mutant side chains modeled as described in Materials and Methods. Swap-mutant 1, K956D/D1235K, is most compatible with the cluster 1 configuration, and indeed this is populated to a greater extent than in the WT simulation (Fig. S1 i). For swap-mutant 2, R957D/D1223R, cluster 2 is slightly more populated relative to cluster 1 and the WT simulations (Fig. S2 i). The altered populations of the bound states, as well as further swap mutants, will be discussed elsewhere (S.B. and M.B., unpublished data). As shown for the WT simulations (24), interconversion between the clusters can occur on the 100 ns timescale. Thus, the three starting configurations can interconvert and are not expected to lead to distinct trajectories. Importantly, whereas all simulations on the WT protein were stable over 2.4 μs (24), five of six simulations of the mutant complexes showed domain-domain separation after 350–1100 ns of Anton simulation time, with swap-mutant 2 dissociating earlier than swap-mutant 1.
An analysis of the equilibrium dissociation constants (KD) by SPR measurements demonstrates that the mutants have higher KD values (20 and 106 μM for swap-mutants 1 and 2, respectively) compared with the WT complex (2.2 μM), suggesting weaker binding (Table S1; Fig. S3). Therefore, a greater dissociation of the mutant complexes in the MD simulations, even on the microsecond timescale, is to be expected (see discussion in Supporting Materials and Methods).
Analysis of events in dissociation trajectories
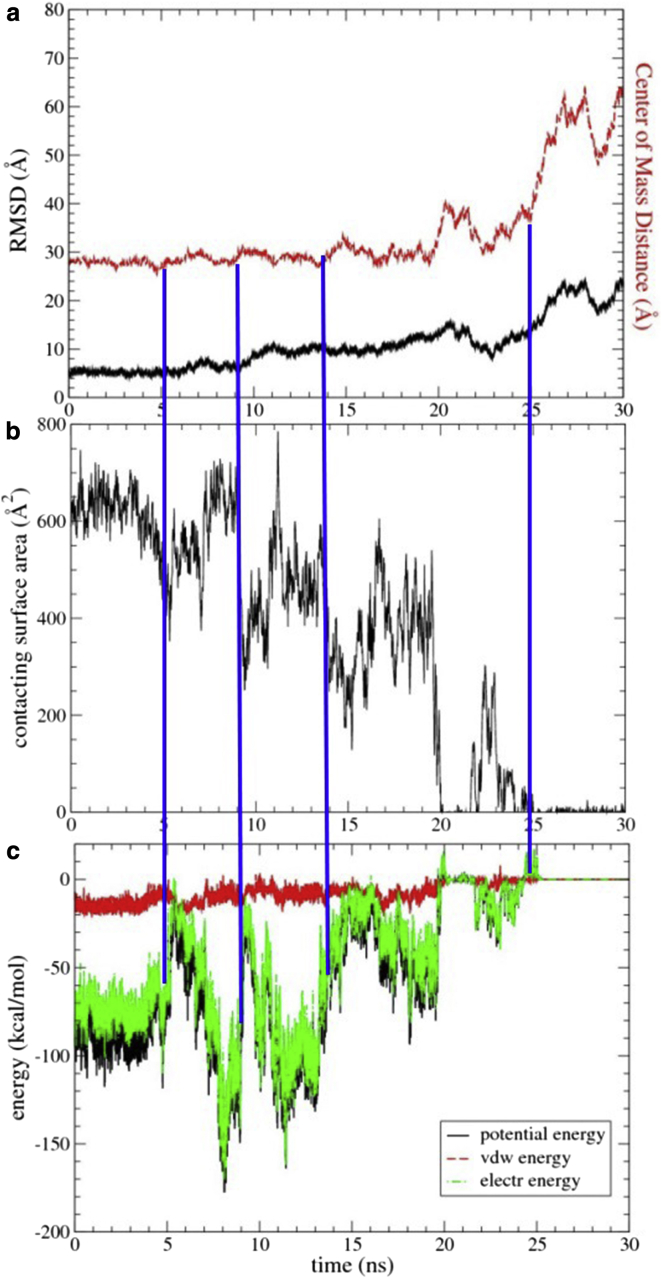
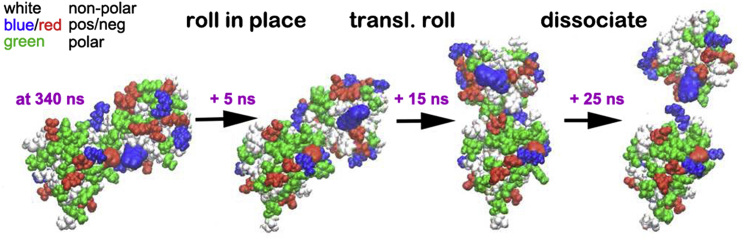
The initial Anton simulations did not sample the dissociation process in enough detail for some of the analysis; therefore, we repeated segments of the swap-mutant 2 trajectories with an increased saving frequency for coordinate frames. An example of such a zoomed-in trajectory, showing SAM domain separation, is depicted in Figs. 2 and 3, as well as in Movie S1 (see Figs. S6 and S8 for the two other trajectories). Fig. S6 shows the results from the simulation started from cluster 1. Just before the final separation, one SAM domain is flipped and moved so that only a fraction of the original interfaces (which are in contact at the start of the simulation) interact. The cluster 3-started simulation also moves to a structure that is flipped (shown in Fig. S8 for the zoomed-in portion). At the start of the zoomed-in trajectories, only the cluster 2-started simulation (shown in Fig. 2) is relatively close to the starting structure, but is shifted, accounting for the loss of buried area. In a movie of the latter trajectory (Movie S1), we observe a rolling-in-place and then a rolling-around (or translation and rolling) event without significant separation (at 340 ns +5 and +9 ns, respectively; see Fig. 3). For the first event, the identity of the EphA2 residues in contact remains substantially unchanged; however, for the second event (translational and simultaneous rolling), the EphA2 protein transitions to a different interface, whereas the SHIP2 interface is preserved. A breathing transition (based on protein separation) then occurs at this new interface (at +12–15 ns). The transitions lead to substantial (if mostly transient) losses of buried surface area and of potential interaction energy (Fig. 2, b and c). A certain amount of recovery in buried surface area and energy is seen after the transition events; however, at +20 ns there is a sudden but brief separation, followed by a recapture event before the final separation occurs at +25 ns (Figs. 2, 3, and 4; Movie S1). Similar observations are made in the other trajectories. In general, the pathways to protein dissociation appear to involve diverse transitions to states that bury much less overall surface area. For example, in the case of a simulation started with a cluster 2 structure, there is a marked loss of nonpolar surface area, whereas the polar surface area is relatively constant right up to the point of initial separation (Figs. 2 b and 4 b). In other simulations (Figs. S6–S9) the reverse is true (i.e., an earlier loss of polar surface area), suggesting a diversity of separation mechanisms.
Figure 2.
The protein-protein dissociation process encompasses several events. Results from an analysis of a zoomed-in region (after 340 ns) of the swap-mutant 2 trajectory, started with the cluster 2 configuration, are shown. (a) RMSD (data in black, left y axis) and center-of-mass separation (data in red, right axis). (b) Total solvent-accessible surface area buried in complex (BSA). (c) Interprotein electrostatic and van der Waals (VDW) potential interaction energies. Blue lines indicate significant transitions. To see this figure in color, go online.
Figure 3.
Structures of the SAM-SAM complex at points before protein separation in the zoomed-in portion of the swap-mutant 2 trajectory (see also Fig. 2). Time points (after 340 ns of Anton simulation) and events are indicated. The mutant residues are shown as 2× VDW spheres for easier localization.
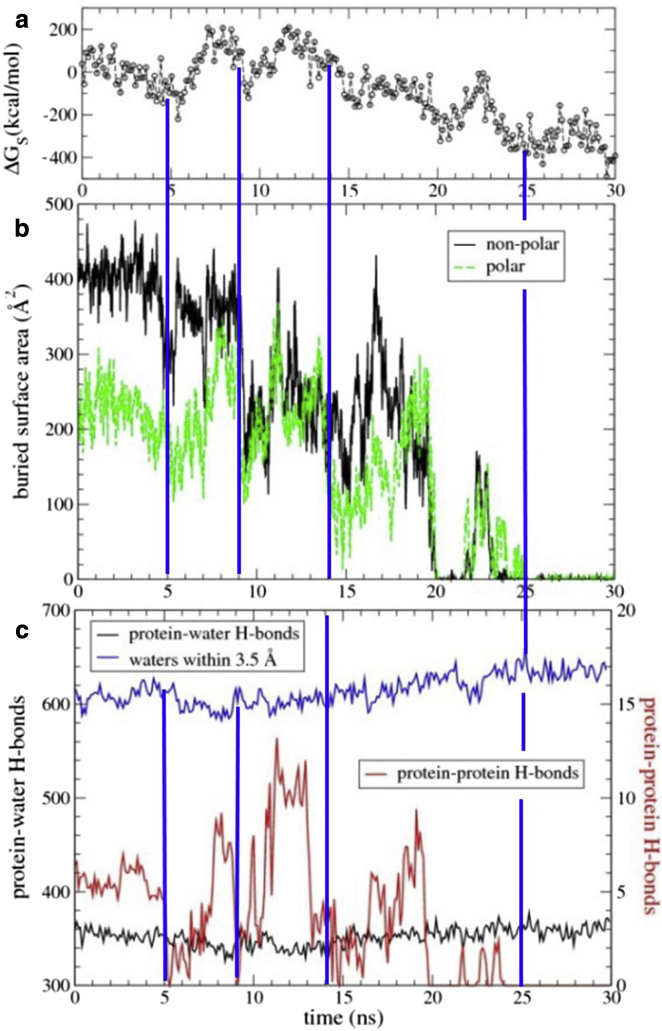
Figure 4.
Protein solvation increases during dissociation of the cluster 2-started trajectory of swap-mutant 2 (Fig. 2). (a) Change in solvation free energy (ΔGs). (b) Polar and nonpolar BSA. (c) Total number of protein-solvent hydrogen bonds and water-protein contacts within 3.5 Å of the protein surface (left). Number of interprotein hydrogen bonds (right y-axis). To see this figure in color, go online.
The potentials plotted in Fig. 2 c represent the unscreened forces experienced by the proteins with respect to each other. An estimate of solvent free energy, ΔGs, can be calculated with the use of a continuum model (30). Fig. 4 a shows that thermodynamically favorable decreases in ΔGs accompany the rolling-in-place and rolling-around transitions as well as the breathing motion at ∼+14 ns and initial separation at +20 ns. Typically, the change in ΔGs mirrors the drop in buried surface area, which precedes larger changes in the protein’s separation and/or root mean-square deviation (RMSD). Intriguingly, solvation can follow changes in the separation of the centers of mass with a slight delay. However, a different order of events is seen in two other trajectories (Figs. S6–S9). In one (Fig. S9, a and b), changes in buried surface areas and in ΔGs are complete by +9 ns, the domains are slightly separated, and the proteins still hang onto each other until ∼+11 ns (Fig. S8 a). In the other trajectory (Fig. S6), the proteins are only loosely associated at the start of the zoomed-in trajectory and fully dissociate at +21 ns, but the change in ΔGs seems to lag behind, only reaching a plateau at +25 ns.
Changes in protein solvation and hydrogen bonding
Since the change in ΔGs was highly correlated with changes in the solvent-accessible surface area, we examined hydrogen bonding and water molecules present between and around the proteins. The number of protein-solvent hydrogen bonds remains relatively constant (see Fig. 4 c and Table S2) because even before separation occurs, the majority of protein donor and acceptor groups are already involved in bifurcated hydrogen bonding with the solvent. The number of additional hydrogen bonds that form upon protein separation is small (0–20 relative to 350 bonds) and corresponds roughly to the number of interprotein hydrogen bonds that are lost upon dissociation (5–12 bonds). In addition to the polar surface area, the nonpolar surface is also exposed to solvent upon protein separation. Consequently, the number of contacts to waters surrounding the protein increases by ∼30. Again, this is a small number relative to the 600 waters that surround the protein complex generally. Remarkably, such an increase is seen consistently across all the simulations that show protein separation.
Changes in protein dynamics upon dissociation
We analyzed the motions at the predominant binding surface (clusters 1–3) compared with other surfaces of the protein complex, both before and after separation occurred (see Materials and Methods for details). We used two methods to analyze the change in dynamics. First, we employed a quasi-harmonic entropy calculation. Fluctuations in the proteins vary across the structures with averages of 1.16 to 1.39 cal mol−1 atom−1 for atoms at the predominant interface to 1.25 to 1.47 cal mol−1 atom−1 for atoms at the sites away from the interface (Table S4). This shows that noninterface atoms are more dynamic than atoms located at interfaces, which is consistent with their involvement in protein-protein interactions. After dissociation of the proteins, the main- and side-chain EphA2 atoms at the predominant interface show a considerable increase in dynamics (13% and 20%, respectively), whereas the SHIP2 predominant interface atoms show a much smaller increase (3% and 7%). Other changes occur at the less populated interface and noninterface sites for EphA2 atoms (+1% to +6%). The fluctuations of noninterface SHIP2 side chains actually decrease (−6%). We also analyzed the changes in dynamics by calculating the autocorrelation functions of bond fluctuations as well as the amplitudes of main- and side-chain N-H, C-H, and C=O bond motions (see S2 analysis in Materials and Methods; results are shown in Table S5). A comparison of the motions in the complex and the free proteins reveals a picture that is very similar to that obtained by the Q-entropy analysis. Again, main and side chains at the EphA2 predominant interface, as well as side chains at the SHIP2 predominant interface, are affected the most; the magnitude of the changes is also similar to that indicated by the Q-entropy analysis. More modest changes are seen at the low-population interfaces and finally at the noninterface region. However, as discussed below, it is also clear that the changes are long range and that the SHIP2 SAM domain, in particular, displays an allosteric behavior.
Discussion
The observation of protein dissociation in the majority of microsecond MD simulations of two mutation-destabilized EphA2-SHIP2 SAM-SAM complexes allowed us, for the first time to our knowledge, to examine the changes in protein domain interactions and dynamics that accompany a protein dissociation process at the all-atom level.
A general mechanism of protein dissociation, but no common separation pathway
The energy landscape of proteins is typically complicated, and simulations that are initiated away from experimentally derived starting structures (or from unstable structures) can easily become trapped, as recently described (e.g., (24, 34)). The simulation results suggest that the energy landscape of the SAM domain heterodimer complex was significantly perturbed by the mutations. NMR studies also confirm the altered ensembles of configurations (S.B. and M.B., unpublished data). In this report, we focus on features that characterize the dissociation process, such as possible pathways, if not events. In an example of a dissociation trajectory (Movie S1; Figs. 2, 3, and 4), we observe several transitions without a significant initial separation. In the early events, only contacts on one interface are changed substantially at a time, but just before separation occurs, generally neither of the surfaces involved corresponds to the one in the original complex. For the WT SAM-SAM complexes (as well as for the mutant proteins initially in configurations 1–3), a common interacting surface predominates and utilizes regions of the proteins that are the most dense in hydrogen-bond acceptor/donor as well as charged groups (characterized in our previous study (24)). In contrast, separation events generally occur from less populated, less dense, and thus low-affinity surfaces (Fig. 5 and see below). This is true for all of the trajectories of both swap mutants that show separation and for the three zoomed-in trajectories of the R957D/D1223R mutant. This process is consistent with the observation that the configurational transitions of the complex toward such interfaces lead to a progressive loss of buried surface area and of potential interaction energy (Fig. 2, b and c). As noted in other reports, binding affinity usually scales with the number of contacts between proteins and the area of the interacting surfaces. This measure is used by protein structure analysis programs to characterize interactions; for example, the program PISA uses it to discriminate between protein interactions and crystal contacts (35).
Figure 5.
Highly diverse structures of the mutant complexes just before protein separation occurs. Complexes are shown 5 ns before the final separation in two views superimposed on the initial WT complex structure (cluster 2 configuration; EphA2 in yellow and SHIP2 in blue). (a–d) One view is superimposed on the EphA2 domain (a and c) and the other is superimposed on the SHIP2 SAM domain (b and d). (a and b) For the three K956D/D1235K swap-mutant 1 separation trajectories, pink, violet, and light blue indicate SHIP2 SAM domain positions and orientations from trajectories started with clusters 1–3, respectively. Orange, pale yellow, and lemon indicate EphA2 SAM domains from the same trajectories. Mutant residue atoms are shown as VDW spheres (colored according to residue charge: red for positively charged and blue for negatively charged). (c and d) Separation trajectories for the R957D/D1223K swap-mutant 2 are depicted using the same color scheme as above; (d) shows a rotated view (rotated by +90° around the x axis relative to b) for clarity.
The mechanism for transitions within a protein-protein complex is similar to that seen in the WT protein complex. Transitions involve degenerate sets of hydrogen-bonding and salt-bridge interactions in a monkey-bar-type mechanism (i.e., intermediary contacts are formed or persist to facilitate the transitions) (24). Transitions to a low-affinity surface involve a pivoting event, where several persistent contacts form a ridge and then one domain rotates around this anchor point. Although the general mechanism of these transitions, including the overall progressive loss of interface area and contacts, is common to the separation trajectories, the detailed events are significantly different. For example, a consideration of the buried nonpolar surface area shows that in one trajectory the nonpolar residues become gradually exposed (Fig. 4 c, swap-mutant 2 started from cluster 2), whereas in another, there is a sudden change (Fig. S7; swap-mutant 2 started from cluster 1). An examination of the structures 5 ns before the final separation in the eight dissociation trajectories (shown in Fig. 5) supports the view that there is no common dissociation pathway or event at the individual residue interaction level of detail.
Although the number of separation trajectories that could be run for this study does not provide a high level of statistics, the observations nevertheless suggest that there likely is no common pathway that underlies the protein dissociation process. It is possible that a collection of pathways would be discernible with a larger number (by 1–2 orders of magnitude) of trajectories. However, we conclude that despite the absence of a single predominant dissociation pathway, dissociation invariably proceeds through a number of weakly bound states.
A tipping point for dissociation and implications for protein association
An intriguing question is, at what point in time does a complex become unstable, resulting in imminent protein separation? An analysis of the buried solvent-accessible surface area suggests that as early as tens to hundreds of nanoseconds before dissociation, the protein complexes assume a configuration that predestines dissociation. Specifically, the surface area buried between the SAM-SAM interfaces (400–600 Å2) in the three zoomed-in trajectories suggests that these structures are much less stable than the WT or mutant proteins near the start of the trajectories (burying 800–1100 Å2; Table S2). Buried protein surface area can be lost substantially or completely, but only for relatively short periods of time (on the order of a few nanoseconds or less). Also, before final separation occurs, the buried surface area is frequently reduced to 200–400 Å2. Once separation occurs, several of the simulations show a brief recapture event (again for a few nanoseconds) with this level of area burial. We also notice that the nonpolar and polar buried surface areas for the unstable complexes are nearly equal, whereas in persistent complexes this ratio is closer to 2:1 (Fig. 4 b; see also Table S2). Thus, in accord with previous studies on protein complexes (1, 2, 3), we find that a minimum surface area needs to be buried for a stable complex, and a major part of this area tends to be nonpolar.
Viewed in reverse, the dissociation process may be considered as a series of protein-protein association events. From this perspective, a complex that forms due to protein recapture would correspond to an encounter state. Although overall this would be beneficial, a large fraction of such states would dissociate (8, 36). Simulations of protein association at the all-atom level will be needed to examine in detail the earliest interactions that form. However, the time-reversed trajectories of protein dissociation are consistent with the view (based on experimental as well as rigid-body simulation studies) that longer-range electrostatic interactions play an important role in the protein association process (8, 13, 36, 37). Specifically, as shown in Figs. 2, 3, and 4, electrostatic interactions diminish late in dissociation/form early in association, several protein-protein hydrogen bonds also diminish late/form early, and substantial nonpolar surface area is lost early upon protein separation/is buried late upon complex formation. Polar and nonpolar contacts are predicted to initially increase equally as the native interfaces come into contact upon initial protein association. However, the predominant interfaces in the native complex are associated with a patch of nonpolar residues that only closely interact once these interfaces are in contact late in the protein association process. It is now appreciated that the events of protein complex formation are substantially different from the process of protein folding, during which an early hydrophobic collapse and short-range hydrogen-bonding interactions are dominant (38, 39, 40).
Why are the mutant protein complexes less stable?
We examined all of the dissociation trajectories for evidence that the mutated residues actively contribute to the dissociation process. In one simulation, the complex experiences a dynamic and directional push toward separation (see Fig. S10), but this appears to be fortuitous rather than general. Fig. 5 shows the structures before their separation. Except for the one simulation just mentioned, it is clear that the swap residues (which are in close proximity in at least some of the starting configurations) are well separated before dissociation occurs. In fact, mostly the mutant side chains point away from the other domain and are no longer involved in the protein complex. This suggests that complex destabilization is an early event that serves to enable the transitions to the weaker binding/nonbinding interfaces, consistent with the general process proposed above (see also Fig. 5). In accord with changes in KD, changes in the association and dissociation kinetics are indicated by SPR (see Figs. S2 and S3; however, as discussed in the Supporting Materials and Methods, the surface techniques give kinetics that are orders of magnitude different from the simulation and NMR results). Further experimental studies of the SAM-SAM complex dissociation kinetics, using other techniques, are needed before conclusions can be drawn. Destabilization of the SAM-SAM complex can also be achieved by a nonoptimal interaction between the mutated side chains across the interface in the initial SAM heterodimer configuration or by local clashes with neighboring side chains on the same interface. This is a complex issue that needs to be further explored both computationally and experimentally, given that the complexes are dynamic. Nevertheless, several of the starting configurations were stable and the proteins did not dissociate immediately; rather, they began to dissociate after 350–1100 ns, a time that traditionally would be considered a very lengthy equilibration for all-atom MD simulations.
Changes in internal protein dynamics are in part local and in part point to allosteric pathways within SAM domains
A simple analysis in terms of an implicit solvation model (Figs. 4, S7, and S9) reveals a varied behavior in the changes of solvation that occur during the preseparation transitions. Again, this supports our conclusion that there is no predominant dissociation pathway. Once separation occurs, ΔGs is lower (more favorable) for the unbound proteins than for the protein complex in all simulations (Table S2). Both protein-water solvation enthalpy and entropy contribute to this change, but quantitative contributions are difficult to estimate without much more extensive and explicit models (e.g., (13, 41)). Similar to what has been observed in other systems, polar side chains are at least partly solvated in the complex (42, 43, 44). Notably, in our analysis of transitions in the WT SAM-SAM complex, we only found one side-chain residue (a tryptophan in the SHIP2 SAM domain at the center of the interface) that was temporarily fully desolvated (24). Our results show accordingly that the solvation of polar groups is essentially unchanged upon protein dissociation. However, the number of water molecules close to the protein increases, corresponding to the area that is exposed upon protein dissociation. Although this additional solvation requires an overall ordering of water molecules (and is not a thermodynamically favorable event in terms of entropy), it could be offset by the additional enthalpy of protein-water interactions (see discussion in Supporting Materials and Methods). In addition, water bound at the polar protein-protein interface in the complex becomes more dynamic as the proteins dissociate (e.g., (4, 41)). Generally, water is able to move quickly to the newly uncovered interfaces and is unlikely to have a limiting kinetic influence on the dissociation process (6, 8). As discussed elsewhere (e.g., (45)), substantial protein fluctuations are an order or two slower than solvent motions. The dynamics of hydrogen-bonded waters is expected to mirror the fluctuations of protein side chains, which are more rigid even at the rim of the interface. Water-mediated structures and water dynamics clearly play an important role in protein association processes as recently described in publications of the Ham and the Helms laboratories (14, 17). Compared with the dynamics of water, the internal dynamics of proteins can be more straightforwardly evaluated. A change in protein dynamics provides a deeper insight into the process of dissociation.
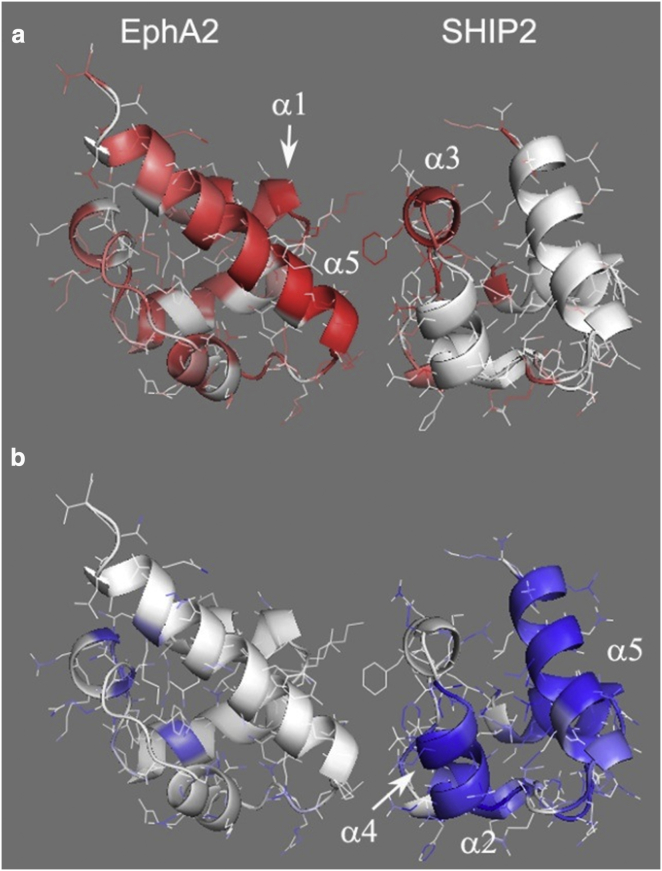
We used two methods to evaluate changes in protein entropy. The first method compared the fluctuations in the proteins in the bound complex with those of their dissociated states. The second one compared the relative change in entropy for each domain between their free and bound states during the protein dissociation process, and gave consistent results. A broad equivalence between different methods of entropy evaluation has been reported (46, 47, 48). Similar to what has been reported in other studies, we found it difficult to obtain an exact accounting of the different contributions and typically had to make adjustments due to nonconsidered terms (see Supporting Materials and Methods). However, it is clear that the entropy changes due to altered protein dynamics alone account for a significant proportion of the estimated entropy differences (up to 50% in terms of magnitude). Since the changes were consistent by themselves, we examined the distribution of the entropy differences at the residue level in both proteins. The order parameter (S2)-derived dynamics changes were mapped to a representative structure of the complex (shown in Fig. 6). Our analyses point to a substantial increase in protein fluctuations in parts of the SHIP2 SAM and EphA2 SAM interface regions (especially the latter).
Figure 6.
(a and b) Increased dynamics at the EphA2-SHIP2 interface (a) and decreased motions in wide regions of SHIP2 (b) are observed upon dissociation. The residues are colored from white to red for positive changes in the S2-derived entropy value multiplied by the temperature (a), and white to blue for negative changes (b) (in both, the color range is given by a value of ln (1) to ln (376)). The sum of the average changes in dynamics for the residue bonds that were analyzed is projected onto the main chain of the complex, shown in ribbon and line representation.
Both analyses reveal a small decrease in the dynamics for parts of the SHIP2 domain away from the interface region. Numerically, the change in dynamics for the groups at the predominant interface of EphA2 and SHIP2 outweighs the changes at the SHIP2 noninterface surface, even after we consider a >3-fold higher number of bonds and correspondingly larger protein surface area for the latter. Thus, changes that occur far away from the interaction interfaces are more modest than those observed at the predominant interaction interface itself. Similar dynamic allostery has been characterized in other systems (48, 49, 50), and the importance of the protein interaction rim region, as well as the noninteracting regions, is currently a topic of investigation in the field of protein-protein interactions (e.g., (51, 52, 53)). Thus, for our system, it is likely that the binding event on one interface may be used to alter the binding of proteins or ligands at other SAM domain interfaces via a mechanism that may be exploited for possible signaling functions.
In summary, WT EphA2-SHIP2 SAM-SAM complexes are stable in all-atom microsecond MD simulations. By contrast, the majority of simulations of two cross-interface ion-pair mutants show protein dissociation on this timescale. Before protein dissociation occurs, several transitions (especially in-place domain rotations, pivoting with translational movement and rolling), as well as transient separation and recapture events, are seen. It appears that such transitions are critical and likely obligatory events in the dissociation process, as they serve to move the complex structure away from its preferred binding interface to interfaces with weaker and less extensive contacts. The mutated residues appear to facilitate this initial process. Assuming that protein association is the reverse of the protein dissociation process, our findings make predictions about the nature of interactions in encounter complexes and bound intermediates. Although protein solvation plays a large role, another major contribution to the dissociation process is altered fluctuations, mostly by increased side- and main-chain dynamics in the separated proteins. Based upon the pattern of changes in dynamics, we conclude that SAM domains could be allosteric proteins.
Author Contributions
L.Z. carried out and analyzed the MD simulations. S.B. purified proteins and carried out SPR experiments and data analysis. M.B. oversaw the project and carried out additional analysis. L.Z., S.B., and M.B. wrote the manuscript.
Acknowledgments
This work was funded by grants to M.B. (NIH R01GM112491) and postdoctoral fellowships to L.Z. (NIH T32DK007470) and S.B. (American Heart Association 13POST17110051). Use of the Anton supercomputer at the Pittsburgh Supercomputing Center is supported by an NIH award (RC2GM093307) to Carnegie Mellon University. Part of this work was carried out on the Lonestar supercomputer under XEDES grant MCB120047, and at the High Performance Computing Cluster at Case Western Reserve University.
Editor: Michael Feig.
Footnotes
Liqun Zhang’s present address is Chemical Engineering Department, Tennessee Technological University, Cookeville, Tennessee.
Supporting Materials and Methods, ten figures, five tables, and one movie are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)00043-6.
Supporting Material
The trajectory is centered on the initial orientation of EphA2 SAM. Residues are colored as follows: white, nonpolar side chains; green, hydrophilic side chains; blue and red, positively and negatively charged side chains, respectively. The mutated residues EphA2 R957D and SHIP2 D1223R are shown as 2× van der Waals radii spheres. The interaction exists at the start of the movie but is broken by the transitions. Eventually the mutated SHIP2 residue D1223R participates in a repulsive event with EphA2 arginines (see Fig. S10).
References
- 1.Lo Conte L., Chothia C., Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 2.Jones S., Thornton J.M. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kortemme T., Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl. Acad. Sci. USA. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keskin O., Ma B., Nussinov R. Hot regions in protein–protein interactions: the organization and contribution of structurally conserved hot spot residues. J. Mol. Biol. 2005;345:1281–1294. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 5.Davis F.P., Sali A. PIBASE: a comprehensive database of structurally defined protein interfaces. Bioinformatics. 2005;21:1901–1907. doi: 10.1093/bioinformatics/bti277. [DOI] [PubMed] [Google Scholar]
- 6.Camacho C.J., Kimura S.R., Vajda S. Kinetics of desolvation-mediated protein-protein binding. Biophys. J. 2000;78:1094–1105. doi: 10.1016/S0006-3495(00)76668-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spaar A., Dammer C., Helms V. Diffusional encounter of barnase and barstar. Biophys. J. 2006;90:1913–1924. doi: 10.1529/biophysj.105.075507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schreiber G., Haran G., Zhou H.X. Fundamental aspects of protein-protein association kinetics. Chem. Rev. 2009;109:839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khare S.D., Fleishman S.J. Emerging themes in the computational design of novel enzymes and protein-protein interfaces. FEBS Lett. 2013;587:1147–1154. doi: 10.1016/j.febslet.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Bonvin A.M. Flexible protein-protein docking. Curr. Opin. Struct. Biol. 2006;16:194–200. doi: 10.1016/j.sbi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Zacharias M. Accounting for conformational changes during protein-protein docking. Curr. Opin. Struct. Biol. 2010;20:180–186. doi: 10.1016/j.sbi.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Torchala M., Moal I.H., Bates P.A. SwarmDock: a server for flexible protein-protein docking. Bioinformatics. 2013;29:807–809. doi: 10.1093/bioinformatics/btt038. [DOI] [PubMed] [Google Scholar]
- 13.Chong S.H., Ham S. Impact of chemical heterogeneity on protein self-assembly in water. Proc. Natl. Acad. Sci. USA. 2012;109:7636–7641. doi: 10.1073/pnas.1120646109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chong S.H., Ham S. Distinct role of hydration water in protein misfolding and aggregation revealed by fluctuating thermodynamics analysis. Acc. Chem. Res. 2015;48:956–965. doi: 10.1021/acs.accounts.5b00032. [DOI] [PubMed] [Google Scholar]
- 15.Blöchliger N., Xu M., Caflisch A. Peptide binding to a PDZ domain by electrostatic steering via nonnative salt bridges. Biophys. J. 2015;108:2362–2370. doi: 10.1016/j.bpj.2015.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad M., Gu W., Helms V. Mechanism of fast peptide recognition by SH3 domains. Angew. Chem. Int. Ed. Engl. 2008;47:7626–7630. doi: 10.1002/anie.200801856. [DOI] [PubMed] [Google Scholar]
- 17.Ahmad M., Gu W., Helms V. Adhesive water networks facilitate binding of protein interfaces. Nat. Commun. 2011;2:261. doi: 10.1038/ncomms1258. [DOI] [PubMed] [Google Scholar]
- 18.Gumbart J.C., Roux B., Chipot C. Standard binding free energies from computer simulations: What is the best strategy? J. Chem. Theory Comput. 2013;9:794–802. doi: 10.1021/ct3008099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hynes N.E., Ingham P.W., Pawson T. Signalling change: signal transduction through the decades. Nat. Rev. Mol. Cell Biol. 2013;14:393–398. doi: 10.1038/nrm3581. [DOI] [PubMed] [Google Scholar]
- 20.Pasquale E.B. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat. Rev. Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiao F., Bowie J.U. The many faces of SAM. Sci. STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 22.Lee H.J., Hota P.K., Buck M. NMR structure of a heterodimeric SAM:SAM complex: characterization and manipulation of EphA2 binding reveal new cellular functions of SHIP2. Structure. 2012;20:41–55. doi: 10.1016/j.str.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw D.E., Deneroff M.M., Wang S.C. Anton, a special-purpose machine for molecular dynamics simulation. Commun. ACM. 2008;51:91–97. [Google Scholar]
- 24.Zhang L., Buck M. Molecular simulations of a dynamic protein complex: role of salt-bridges and polar interactions in configurational transitions. Biophys. J. 2013;105:2412–2417. doi: 10.1016/j.bpj.2013.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myszka D.G. Improving biosensor analysis. J. Mol. Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buck M., Bouguet-Bonnet S., MacKerell A.D., Jr. Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophys. J. 2006;90:L36–L38. doi: 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorgensen W.L., Madura J.D. Solvation and conformation of methanol in water. J. Am. Chem. Soc. 1983;105:1407–1413. [Google Scholar]
- 30.Jo S., Vargyas M., Im W. PBEQ-Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Res. 2008;36:W270–W275. doi: 10.1093/nar/gkn314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andricioaei I., Karplus M. On the calculation of entropy from covariance matrices of the atomic fluctuations. J. Chem. Phys. 2001;115:6289. [Google Scholar]
- 32.Seeber M., Cecchini M., Caflisch A. Wordom: a program for efficient analysis of molecular dynamics simulations. Bioinformatics. 2007;23:2625–2627. doi: 10.1093/bioinformatics/btm378. [DOI] [PubMed] [Google Scholar]
- 33.Yang D., Kay L.E. Contributions to conformational entropy arising from bond vector fluctuations measured from NMR-derived order parameters: application to protein folding. J. Mol. Biol. 1996;263:369–382. doi: 10.1006/jmbi.1996.0581. [DOI] [PubMed] [Google Scholar]
- 34.Zeiske T., Stafford K.A., Palmer A.G., 3rd Starting-structure dependence of nanosecond timescale intersubstate transitions and reproducibility of MD-derived order parameters. Proteins. 2013;81:499–509. doi: 10.1002/prot.24209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krissinel E. Crystal contacts as nature’s docking solutions. J. Comput. Chem. 2010;31:133–143. doi: 10.1002/jcc.21303. [DOI] [PubMed] [Google Scholar]
- 36.Cohen M., Reichmann D., Schreiber G. Similar chemistry, but different bond preferences in inter versus intra-protein interactions. Proteins. 2008;72:741–753. doi: 10.1002/prot.21960. [DOI] [PubMed] [Google Scholar]
- 37.Long D., Brüschweiler R. Directional selection precedes conformational selection in ubiquitin-UIM binding. Angew. Chem. Int. Ed. Engl. 2013;52:3709–3711. doi: 10.1002/anie.201208683. [DOI] [PubMed] [Google Scholar]
- 38.Xu D., Tsai C.J., Nussinov R. Hydrogen bonds and salt bridges across protein-protein interfaces. Protein Eng. 1997;10:999–1012. doi: 10.1093/protein/10.9.999. [DOI] [PubMed] [Google Scholar]
- 39.Fitzpatrick A.W., Knowles T.P., Dobson C.M. Inversion of the balance between hydrophobic and hydrogen bonding interactions in protein folding and aggregation. PLOS Comput. Biol. 2011;7:e1002169. doi: 10.1371/journal.pcbi.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scanu S., Foerster J.M., Ubbink M. Role of hydrophobic interactions in the encounter complex formation of the plastocyanin and cytochrome f complex revealed by paramagnetic NMR spectroscopy. J. Am. Chem. Soc. 2013;135:7681–7692. doi: 10.1021/ja4015452. [DOI] [PubMed] [Google Scholar]
- 41.Li Z., Lazaridis T. Water at biomolecular binding interfaces. Phys. Chem. Chem. Phys. 2007;9:573–581. doi: 10.1039/b612449f. [DOI] [PubMed] [Google Scholar]
- 42.Rodier F., Bahadur R.P., Janin J. Hydration of protein-protein interfaces. Proteins. 2005;60:36–45. doi: 10.1002/prot.20478. [DOI] [PubMed] [Google Scholar]
- 43.Ubbink M. The courtship of proteins: understanding the encounter complex. FEBS Lett. 2009;583:1060–1066. doi: 10.1016/j.febslet.2009.02.046. [DOI] [PubMed] [Google Scholar]
- 44.Chang C.-E.A., McLaughlin W.A., McCammon J.A. Entropic contributions and the influence of the hydrophobic environment in promiscuous protein-protein association. Proc. Natl. Acad. Sci. USA. 2008;105:7456–7461. doi: 10.1073/pnas.0800452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamaneh M.B., Zhang L., Buck M. A direct coupling between global and internal motions in a single domain protein? MD investigation of extreme scenarios. Biophys. J. 2011;101:196–204. doi: 10.1016/j.bpj.2011.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kasinath V., Sharp K.A., Wand A.J. Microscopic insights into the NMR relaxation-based protein conformational entropy meter. J. Am. Chem. Soc. 2013;135:15092–15100. doi: 10.1021/ja405200u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trbovic N., Cho J.H., Palmer A.G., 3rd Protein side-chain dynamics and residual conformational entropy. J. Am. Chem. Soc. 2009;131:615–622. doi: 10.1021/ja806475k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Popovych N., Sun S., Kalodimos C.G. Dynamically driven protein allostery. Nat. Struct. Mol. Biol. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai C.J., Del Sol A., Nussinov R. Protein allostery, signal transmission and dynamics: a classification scheme of allosteric mechanisms. Mol. Biosyst. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bouguet-Bonnet S., Buck M. Compensatory and long-range changes in picosecond-nanosecond main-chain dynamics upon complex formation: 15N relaxation analysis of the free and bound states of the ubiquitin-like domain of human plexin-B1 and the small GTPase Rac1. J. Mol. Biol. 2008;377:1474–1487. doi: 10.1016/j.jmb.2008.01.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kastritis P.L., Rodrigues J.P., Bonvin A.M. Proteins feel more than they see: fine-tuning of binding affinity by properties of the non-interacting surface. J. Mol. Biol. 2014;426:2632–2652. doi: 10.1016/j.jmb.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 52.Visscher K.M., Kastritis P.L., Bonvin A.M. Non-interacting surface solvation and dynamics in protein-protein interactions. Proteins. 2015;83:445–458. doi: 10.1002/prot.24741. [DOI] [PubMed] [Google Scholar]
- 53.David A., Sternberg M.J. The contribution of missense mutations in core and rim residues of protein-protein interfaces to human disease. J. Mol. Biol. 2015;427:2886–2898. doi: 10.1016/j.jmb.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The trajectory is centered on the initial orientation of EphA2 SAM. Residues are colored as follows: white, nonpolar side chains; green, hydrophilic side chains; blue and red, positively and negatively charged side chains, respectively. The mutated residues EphA2 R957D and SHIP2 D1223R are shown as 2× van der Waals radii spheres. The interaction exists at the start of the movie but is broken by the transitions. Eventually the mutated SHIP2 residue D1223R participates in a repulsive event with EphA2 arginines (see Fig. S10).