Abstract
Infection elicits CD4+ memory T lymphocytes that participate in protective immunity. Although memory cells are the progeny of naïve T cells, it is unclear that all naïve cells from a polyclonal repertoire have memory cell potential. Using a single cell adoptive transfer and spleen biopsy method, we found that in mice, essentially all microbe-specific naïve cells produced memory cells during infection. Different clonal memory cell populations had different B cell or macrophage helper compositions that matched effector cell populations generated much earlier in the response. Thus, each microbe-specific naïve CD4+ T cell produces a distinctive ratio of effector cell types early in the immune response that is maintained as some cells in the clonal population become memory cells.
Infection of vertebrates elicits CD4+ memory T lymphocytes that participate in protective immunity (1, 2). The process begins when major histocompatibility complex class II (MHCII)-bound microbial peptides are displayed on host cells and recognized by T cell receptors (TCRs) on a few naïve CD4+ T cells from a vast repertoire. These cells proliferate and differentiate into distinct types of effector cells that help B cells or macrophages eliminate the infection (3). About 90% of the cells then disappear, leaving a population of long-lived memory cells. Some naïve CD4+ T cells have been reported to make terminally differentiated effector cells while others, perhaps those with the most avid TCRs (4, 5), make memory cells (6). In contrast, other studies of one TCR showed a single naïve cell can make both effector and memory cells (7, 8). Thus, the contribution of all naïve T cells in a polyclonal repertoire to the memory cell pool is unclear.
We addressed this issue by determining the fates of many single cells from the repertoire of naïve CD4+ T cells specific for an MHCII (I-Ab)-bound peptide (LLOp) from the listeriolysin O protein of Listeria monocytogenes. Using I-Ab tetramer-based cell enrichment and flow cytometry (9, 10), we confirmed that uninfected C57BL/6 (B6) mice contained about 80 CD4+ CD44low LLOp:I-Ab tetramer-binding naïve cells (Fig. 1A) (10). Seven days after intravenous infection with the attenuated ActA-deficient L. monocytogenes strain (referred to hereafter as L. monocytogenes), these cells proliferated to produce about 200,000 CD44high effector cells (Fig. 1A) consisting of CXCR5− PD-1low T helper 1 (Th1) macrophage helpers (10), and 2 kinds of B cell helpers: CXCR5+ PD-1− T follicular helper cells (Tfh), and CXCR5++ PD-1+ germinal center Tfh cells (GC-Tfh) (11) (Fig. 1B). The number of LLOp:I-Ab tetramer-binding cells then fell about 10-fold 21 days after infection (10) and the surviving cells were CD44high memory cells of 2 types: CXCR5− PD-1− Th1 effector memory cells and CXCR5+ PD-1− Tfh-like central memory cells (Fig. 1B) (10). Thus, acute systemic infection with L. monocytogenes caused a naïve T cell population to generate at least 2 long-lived memory cell populations that resemble earlier populations of effector cells.
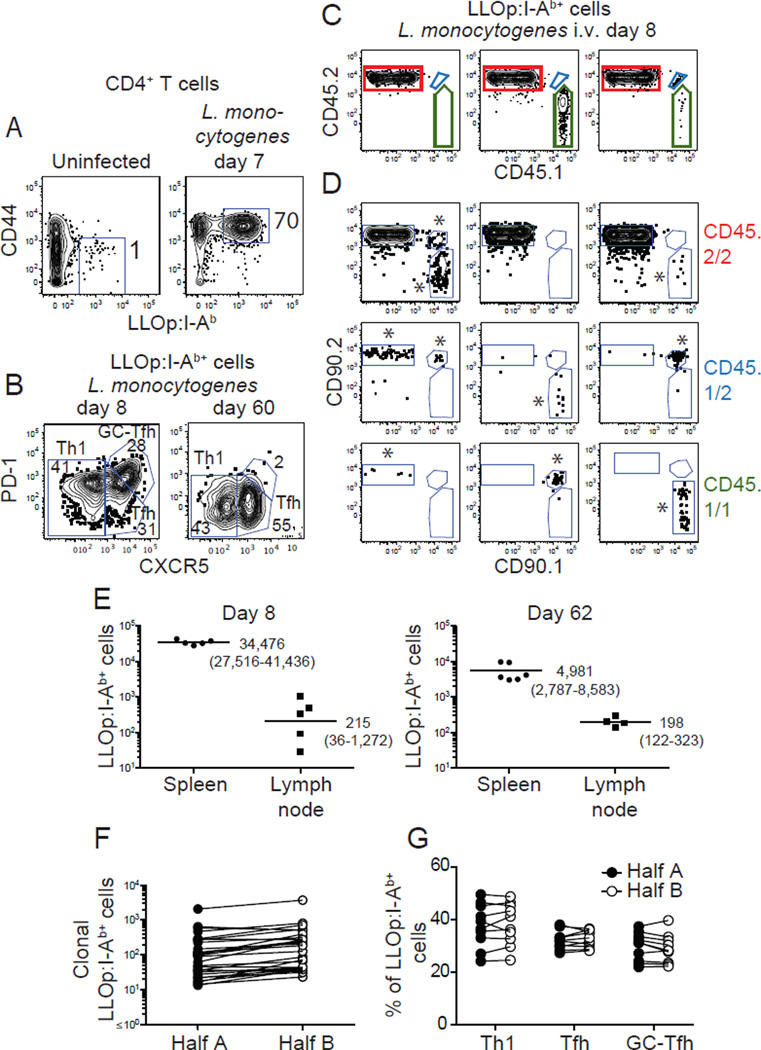
Fig. 1. Identification of clonal LLOp:I-Ab tetramer-binding T cells.
(A) LLOp:I-Ab tetramer-enriched CD4+ T cells from an uninfected (left panel) or a day 7 L. monocytogenes-infected B6 mouse with gates on CD44low or CD44high tetramer-binding cells. (B) Gates to identify LLOp:I-Ab tetramer-binding CXCR5− PD-1− Th1, CXCR5+ PD-1− Tfh, and CXCR5++ PD-1+ GC-Tfh effector cells (left panel) or memory cells (right panel). (C) CD45.1 versus CD45.2 expression by LLOp:I-Ab tetramer-binding cells from day 8 L. monocytogenes-infected B6 mice that received 7x105 CD4+ T cells from 8 unique CD45 and CD90 congenic strains. (D) CD90.1 versus CD90.2 expression on cells expressing CD45.2/2 (red), CD45.1/2 (blue), or CD45.1/1 (green) identified as in (C) from 9 different mice. Cells of recipient origin were CD45.2/2 CD90.2/2. Donor-derived populations considered to be genuine (at least 5 events) are indicated with asterisks. (E) LLOp:I-Ab tetramer-binding CD4+ T cells in the spleens or lymph nodes of B6 mice after L. monocytogenes infection. Each dot is a value from a single mouse. Geometric mean values are shown with 95% confidence intervals in parentheses. (F) Number of donor-derived LLOp:I-Ab tetramer-binding cells derived from single naïve cells in each half of the spleen 7 days after L. monocytogenes infection. Lines connect values for the same clonal population. (G) Frequencies of Th1, Tfh, and GC-Tfh cells (as defined in (B)) in clonal LLOp:I-Ab tetramer-binding cell populations in halves of the same spleens. In (F) and (G), lines connect populations derived from the same naive cell. Representative data from single experiments are shown in (A-C). Pooled results from 2 independent experiments are shown in (E-G).
A limiting dilution adoptive transfer strategy was then used to study the progeny of single naïve cells (12). CD4+ T cells from 8 different uninfected congenic strains expressing various combinations of CD45.1, CD45.2, CD90.1, or CD90.2 were transferred together into B6 mice (CD45.2/2 CD90.2/2) at a number expected to contain on average less than one LLOp:I-Ab tetramer-binding naïve T cell from each donor population. Recipient mice were then infected with L. monocytogenes and 8 days later, LLOp:I-Ab tetramer-enriched cells were stained with fluorochrome-labeled CD45.1, CD45.2, CD90.1, and CD90.2 antibodies and analyzed by flow cytometry. Cells expressing CD45.1 and/or CD45.2 were identified in the LLOp:I-Ab tetramer-binding population (Fig. 1C), and then cells expressing CD90.1 and/or CD90.2 were identified in those populations (Fig. 1D). This strategy identified 9 different LLOp:I-Ab tetramer-binding effector cell populations (Fig. 1D), one derived from the naïve cells of the recipients (CD45.2/2 CD90.2/2) and 8 others from single cells from one of the donor populations (Fig. 1D). Importantly, each of the 8 donor cell-derived populations was detected in only 20–75% of the recipient mice and thus had a 83–98% chance of being derived from a single naïve cell (13). The earlier finding that all the cells from donor-derived populations like these had the identical Tcrb-VDJ sequence supports this contention (12).
The approach was then modified so that clonal effector and memory cells could be analyzed in the spleen from the same animal. This was accomplished by surgical removal of part of the spleen for analysis of effector cells, followed by the other part several months later for analysis of memory cells. This strategy was possible because greater than 95% of secondary lymphoid organ-resident LLOp:I-Ab tetramer-binding CD4+ effector and memory T cells were in the spleen on days 8 and 62 after infection (Fig. 1E). In addition, limiting dilution transfer experiments revealed that clonal populations were in both halves of the spleen on day 8 after infection (Fig. 1F) and the clonal cells in each half had similar Th1, Tfh, and GC-Tfh ratios (Fig 1G).
This strategy was then used to track the progeny of single naïve CD4+ T cells. Examples for 2 different recipient mice, each containing 2 donor-derived clonal populations (CD45.1/2 and CD45.1/1 for mouse 1 and CD90.1/2 and CD90.1/1 for mouse 2), are shown in Fig. 2A. All 4 of these clonal effector cell populations generated memory cells 60 days after infection. In this experiment, 73 different clonal effector cell populations ranging from 30–6,000 cells and averaging about 500 cells were detected in 31 mice on day 8 after infection (Fig. 2B). Sixty-seven of these populations (92%) yielded detectable memory cells on day 60–62. All but one of the clonal memory cell populations were smaller than their effector cell predecessor populations and on average had the same 15% survival rate as polyclonal cells of recipient origin but were more variable in this regard (Fig. 2C). Although large effector cell populations tended to have lower survival rates as suggested in other studies (14), this weak trend was not statistically significant (Fig. 2D). On the contrary, the 6 populations that did not produce detectable memory cells all contained less than 120 cells on day 8. Although this result may indicate that naïve cells that produce few effector cells are less likely to make memory cells, it is possible that memory cells were produced from these small populations but fell below the limit of detection of the assay. Essentially identical results were obtained in a second set of experiments in which 87% (41 of 47) of day 8 clonal effector cell populations produced memory cells on day 30 after infection (Fig. S1). Thus, while clonal effector cell populations undergo contraction, almost all produce memory cells.
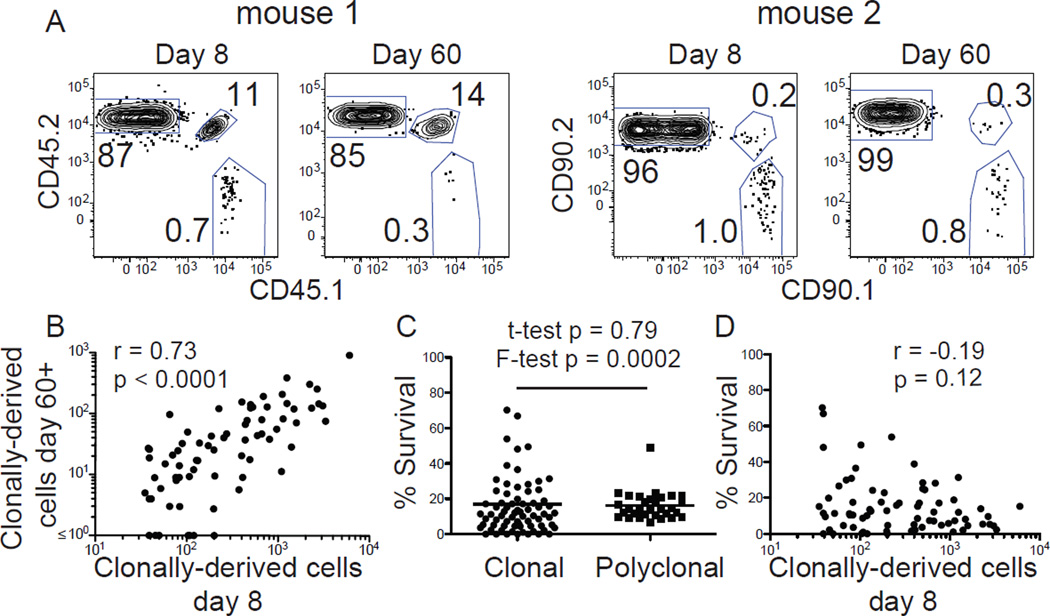
Fig. 2. Nearly all clonal effector cell populations produce memory cell populations.
(A) LLOp:I-Ab tetramer-binding CD4+ T cells from two different B6 mice that received 7x105 CD4+ T cells from 8 unique CD45 and CD90 congenic strains, identified by partial splenectomy and tetramer-based cell enrichment 8 days (left panel) and 60 days after L. monocytogenes infection (right panel). Each mouse contained a recipient-derived polyclonal (CD45.2/2 for mouse 1 or CD90.2/2 for mouse 2) and 2 donor-derived clonal populations (mouse 1 - CD45.1/2 and CD45.1/1, mouse 2 – CD90.1/2 and CD90.1/1) (B) Number of cells produced by different single naïve cells at day 8 and 60 or 62 after infection. (C) Percent survival between day 8 and 60 or 62 after infection for different donor-derived clonal or recipient-derived polyclonal effector cell populations. Note that one clone, with a calculated percent survival of 149% is not shown. (D) Number of effector cells produced by different single naïve cells at day 8 versus the percentage that survived on day 60 or 62. Statistical values in (B) and (D) were calculated with the Spearman correlation test and in (C) with Student’s t-test for unpaired samples or an F-test of equality of variances. Pooled results from 2 independent experiments are shown.
The phenotypes of clonal LLOp:I-Ab tetramer-binding T cell populations were also assessed. Different clonal populations had different numbers and percentages of CXCR5− Th1, CXCR5+ PD-1− Tfh and CXCR5++ PD-1+ GC-Tfh effector cells on day 8 (Fig. 3A and S1) as previously described (12). The number of cells in each population decreased by day 60–62 but the surviving memory populations had about the same percentage of CXCR5− and CXCR5+ cells as the parent effector cell populations (Fig. 3A-C). However, CXCR5++ PD-1+ cells, which were present in many effector populations, were absent in memory cell populations, while CXCR5+ PD-1− cells were found at elevated frequencies (Figure 3A, C, and D). Adoptive transfer experiments demonstrated that all tetramer-binding and most tetramer-negative CXCR5++ PD-1+ effector cells become CXCR5+ PD-1− memory cells (Fig. S2) as previously reported by others (15, 16). Thus, after contracting, memory cells retain the phenotype of their effector cell predecessors except that CXCR5++ PD-1+ cells lose PD-1.
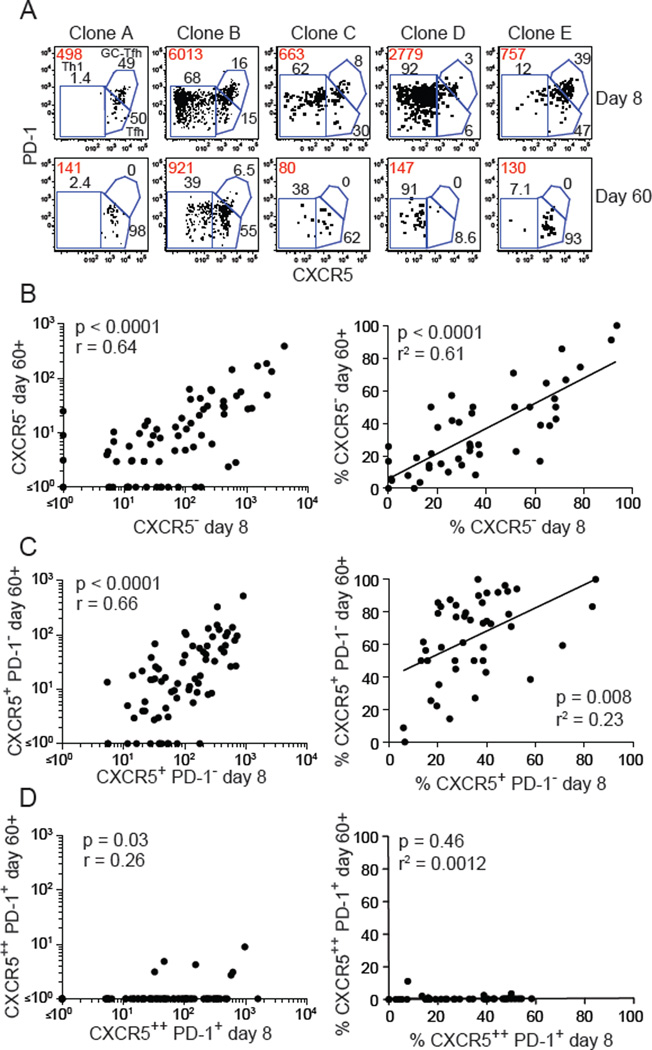
Fig. 3. Memory cells retain the CXCR5− and CXCR5+ phenotype of effector cells.
(A) Identification of Th1, Tfh, and GC-Tfh effector cells in different clonal populations of CD44high LLOp:I-Ab tetramer-binding cells on day 8 and day 60 after infection. Red values indicate the absolute number of cells in each population and black values indicate the percentages of each subset. (B-D, left panels) Numbers of CXCR5− (B), CXCR5+ PD-1− (C), or CXCR5++ PD-1+ (D) cells in clonal populations at day 8 and 60 or 62 after infection. Statistical values were calculated with the Spearman correlation test. (B-D, right panels) Percentages of cells in LLOp:I-Ab tetramer-binding clonal populations that were CXCR5− (B), CXCR5+ (C), or CXCR5++ PD-1+ (D) on day 8 plotted versus the percentages of those subpopulations 60–62 days after infection. Each dot represents a clonal population. Only populations in which 5 or more events were recovered at day 60 or 62 were included to optimize the meaningfulness of the percentage values. Statistical values and trend lines from linear regression analyses are shown. Each dot represents a single clonal population. Pooled results from 2 independent experiments are shown.
We then examined the recall responses of clonal memory cell populations. Limiting numbers of CD4+ T cells from uninfected congenic strains were transferred into B6 mice, which were then infected with L. monocytogenes. Sixty-five donor-derived clonal LLOp:I-Ab tetramer-binding memory cell populations ranging from 6–1,010 cells and averaging about 40 cells were detected by partial splenectomy and cell enrichment 60 days later (Fig. 4A and B). Clonal memory cell populations expanded about 15-fold 7 days after another L. monocytogenes infection (Fig. 4A and B) to produce an average of about 600 effector cells (Fig. 4C), which tended to have the same CXCR5 phenotype as their memory cell predecessors (Fig. 4C and D). Thus, clonal memory cell populations expanded during a recall infection, albeit less well than naïve cells, and produced effector cell subsets like themselves.
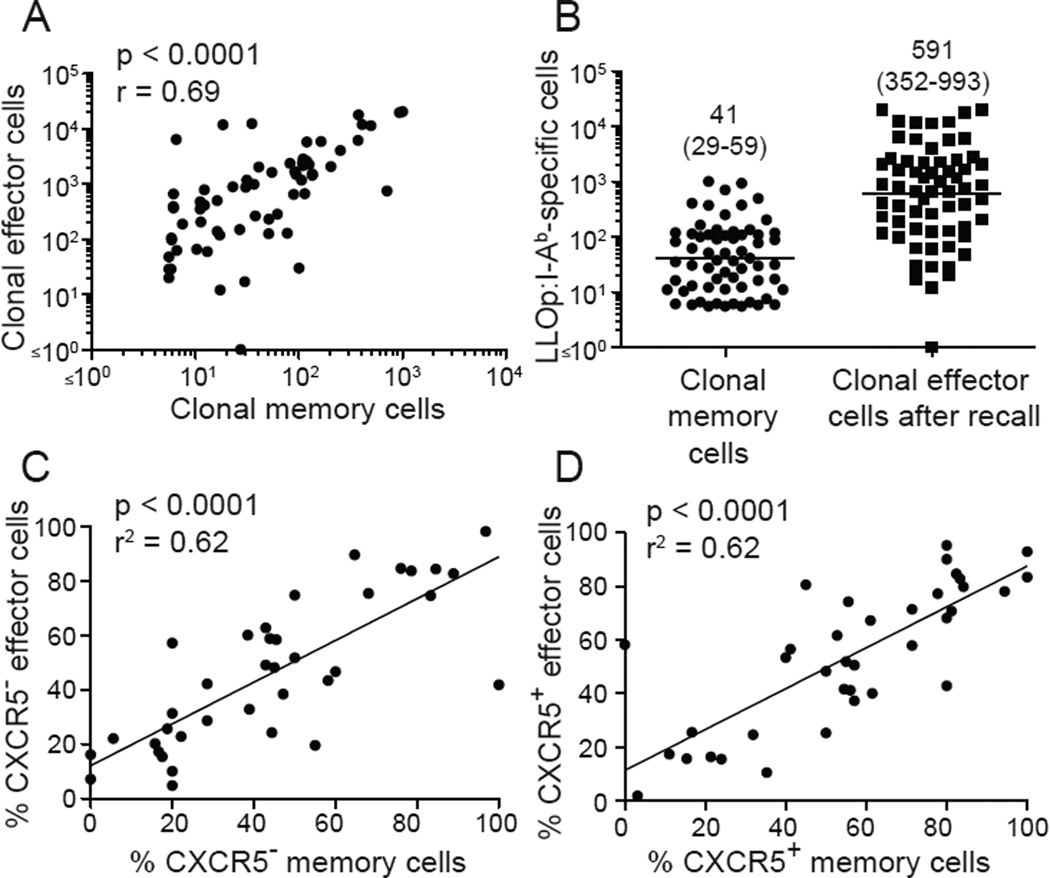
Fig. 4. Memory cells produce effector cells with a similar phenotype.
(A) Numbers of LLOp:I-Ab tetramer-binding memory cells in 65 clonal populations plotted versus the number of effector cells produced from those populations 7 days after a second infection. (B) Numbers of LLOp:I-Ab tetramer-binding memory cells in 65 clonal populations before (left column) or 7 days after a second infection (right column). Geometric mean values with 95% confidence intervals in parentheses are shown. (C-D) Percentages of CXCR5− or CXCR5+ memory cells in individual clonal populations plotted versus the percentages of CXCR5− or CXCR5+ effector cells generated from those populations after secondary infection. Statistical values and trend lines from linear regression analyses are shown. Pooled results from 3 independent experiments are shown.
These results indicate that essentially all naïve T cells in a polyclonal repertoire that respond to a bacterial p:MHCII ligand by producing effector cells also produce memory cells. Although we have evidence that the tetramer used here detects many of the T cells with TCRs specific for this ligand (17), it should be noted that some p:MHCII-specific T cells do not bind to the relevant tetramer, presumably due to expression of low affinity TCRs (18). Thus, although all naïve CD4+ T cells with TCRs that bind p:MHCII tetramer can become memory cells, it remains to be seen whether this conclusion applies to any p:MHCII-specific cells that do not.
Although the nature of the TCR signal experienced by individual naïve clones could influence their effector cell fate (12), the present results suggest that all TCRs can support memory cell formation although some may be better at it than others (4–6). The observation that a clonal memory cell population was very likely to have the same helper cell subset ratio as its predecessor population is consistent with each effector cell in the population having the same chance of becoming a memory cell. This model is consistent with studies of CD8+ T cells indicating that memory cells arise from the effector cell pool by a TCR-independent stochastic process (14, 19–21).
Our finding that a clonal CD4+ memory T cell population tended to produce an effector cell population with the same subset composition after recall suggests that a CXCR5− memory cell produces CXCR5− effector cells and a CXCR5+ memory cell produces CXCR5+ effector cells. Thus, although others (15) and we (10) found that bulk CXCR5+ memory cells can produce CXCR5− and CXCR5+ effector cells, the current results fit the suggestion that both CXCR5+ and CXCR5− cells are relatively lineage-committed (15). An advantage of this process is that the helper cell subset diversity of the effector cell pool is carried into the memory cell pool and retained thereafter.
Supplementary Material
Acknowledgments
We thank S. Jameson and D. Masopust for critical discussions and reading the manuscript, and J. Walter for help with mouse breeding and screening. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. This work was funded by grants to M.K.J. (R01 AI039614), B.T.F. (R01 AI106791), and N.J.T. (F32 AI107995).
References and Notes
- 1.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 2.Jameson SC, Masopust D. Diversity in T cell memory: an embarrassment of riches. Immunity. 2009;31:859. doi: 10.1016/j.immuni.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pepper M, Jenkins MK. Origins of CD4(+) effector and central memory T cells. Nat. Immunol. 2011;131:467. doi: 10.1038/ni.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim C, Wilson T, Fischer KF, Williams MA. Sustained interactions between T cell receptors and antigens promote the differentiation of CD4(+) memory T cells. Immunity. 2013;39:508. doi: 10.1016/j.immuni.2013.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams MA, Ravkov EV, Bevan MJ. Rapid culling of the CD4+ T cell repertoire in the transition from effector to memory. Immunity. 2008;28:533. doi: 10.1016/j.immuni.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savage PA, Boniface JJ, Davis MM. A kinetic basis for T cell receptor repertoire selection during an immune response. Immunity. 1999;10:485. doi: 10.1016/s1074-7613(00)80048-5. [DOI] [PubMed] [Google Scholar]
- 7.Stemberger C, et al. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27:985. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 8.Gerlach C, et al. One naive T cell, multiple fates in CD8+ T cell differentiation. J. Exp. Med. 2010;207:1235. doi: 10.1084/jem.20091175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moon JJ, et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing signals from the bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crotty S. Follicular helper CD4 T cells (TFH) Annu. Rev. Immunol. 2011;29:621. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 12.Tubo NJ, et al. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during Infection. Cell. 2013;153:785. doi: 10.1016/j.cell.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taswell C. Limiting dilution assays for the determination of immunocompetent cell frequencies. I. Data analysis. J. Immunol. 1981;126:1614. [PubMed] [Google Scholar]
- 14.Buchholz VR, et al. Disparate individual fates compose robust CD8+ T cell immunity. Science. 2013;340:630. doi: 10.1126/science.1235454. [DOI] [PubMed] [Google Scholar]
- 15.Hale JS, et al. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 2013;38:805. doi: 10.1016/j.immuni.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luthje K, et al. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat. Immunol. 2012;13:491. doi: 10.1038/ni.2261. [DOI] [PubMed] [Google Scholar]
- 17.Nelson RW, et al. T cell receptor cross-reactivity between similar foreign and self peptides influences naive cell population size and autoimmunity. Immunity. 2015;42:95. doi: 10.1016/j.immuni.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez RJ, Evavold BD. Lower affinity T cells are critical components and active participants of the immune response. Front. immunol. 2015;6:468. doi: 10.3389/fimmu.2015.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blattman JN, Sourdive DJ, Murali-Krishna K, Ahmed R, Altman JD. Evolution of the T cell repertoire during primary, memory, and recall responses to viral infection. J. Immunol. 2000;165:6081. doi: 10.4049/jimmunol.165.11.6081. [DOI] [PubMed] [Google Scholar]
- 20.Gerlach C, et al. Heterogeneous differentiation patterns of individual CD8+ T cells. Science. 2013;340:635. doi: 10.1126/science.1235487. [DOI] [PubMed] [Google Scholar]
- 21.Graef P, et al. Serial transfer of single-cell-derived immunocompetence reveals stemness of CD8(+) central memory T cells. Immunity. 2014;41:116. doi: 10.1016/j.immuni.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 22.Ertelt JM, et al. Selective priming and expansion of antigen-specific Foxp3- CD4+ T cells during Listeria monocytogenes infection. J. Immunol. 2009;182:3032. doi: 10.4049/jimmunol.0803402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.