Abstract
Inappropriate activation of innate immune responses in intestinal epithelial cells underlies the pathophysiology of inflammatory disorders of the intestine. Here we examine the physiological effects of immune hyperactivation in the intestine of the nematode Caenorhabditis elegans. We previously identified an immunostimulatory xenobiotic that protects C. elegans from bacterial infection by inducing immune effector expression via the conserved p38 MAP kinase pathway, but was toxic to nematodes developing in the absence of pathogen. To investigate a possible connection between the toxicity and immunostimulatory properties of this xenobiotic, we conducted a forward genetic screen for C. elegans mutants that are resistant to the deleterious effects of the compound, and identified five toxicity suppressors. These strains contained hypomorphic mutations in each of the known components of the p38 MAP kinase cassette (tir-1, nsy-1, sek-1, and pmk-1), demonstrating that hyperstimulation of the p38 MAPK pathway is toxic to animals. To explore mechanisms of immune pathway regulation in C. elegans, we conducted another genetic screen for dominant activators of the p38 MAPK pathway, and identified a single allele that had a gain-of-function (gf) mutation in nsy-1, the MAP kinase kinase kinase that acts upstream of p38 MAPK pmk-1. The nsy-1(gf) allele caused hyperinduction of p38 MAPK PMK-1-dependent immune effectors, had greater levels of phosphorylated p38 MAPK, and was more resistant to killing by the bacterial pathogen Pseudomonas aeruginosa compared to wild-type controls. In addition, the nsy-1(gf) mutation was toxic to developing animals. Together, these data suggest that the activity of the MAPKKK NSY-1 is tightly regulated as part of a physiological mechanism to control p38 MAPK-mediated innate immune hyperactivation, and ensure cellular homeostasis in C. elegans.
Keywords: innate immunity, immune regulation, C. elegans genetics, host-pathogen interactions, genetics of immunity
Coordination of innate immune responses at mucosal surfaces is a critical determinant of cellular homeostasis in evolutionarily diverse organisms (Peterson and Artis 2014). Exaggerated or aberrantly triggered immune responses, for example, underlie the pathophysiology of inflammatory disorders of the human intestine (Xavier and Podolsky 2007). In flies, immune pathway hyperactivation also has negative physiological consequences and is subject to feedback control (Aggarwal and Silverman 2008).
To understand ancient mechanisms of pathogen detection and immune regulation, our group and others are examining innate immune responses in the nematode Caenorhabditis elegans (Irazoqui et al. 2010; Pukkila-Worley and Ausubel 2012; Cohen and Troemel 2015). As in other metazoans, nematodes coordinate inducible immune defenses from intestinal epithelial cells (IECs), which provide a critical barrier against ingested pathogens (Kim et al. 2002; Shivers et al. 2009; Pukkila-Worley et al. 2011). Studies from C. elegans and other diverse organisms have revealed that key innate immune signaling regulators are strongly conserved (Irazoqui et al. 2010; Pukkila-Worley and Ausubel 2012; Visvikis et al. 2014; Cohen and Troemel 2015). For example, the NSY-1-SEK-1-PMK-1 Mitogen Activated Protein Kinase (MAPK) pathway in C. elegans, which is orthologous to the mammalian ASK1-MKK3/6-p38 MAPK pathway, controls the induction of putative antimicrobial immune effectors, and is required in intestinal epithelial cells for nematodes to survive challenge from ingested pathogens (Kim et al. 2002; Troemel et al. 2006; Shivers et al. 2010; Pukkila-Worley et al. 2011). In mammals, the ASK1-MKK3/6-p38 signaling cassette is a central regulator of inflammatory cytokine production in response to pathogen detection at epithelial surfaces, and pathway activation is tightly regulated through negative regulatory circuits (Kyriakis and Avruch 2012). Misregulation of p38 signaling in IECs has been implicated in the pathogenesis of inflammatory bowel disease, cancer, autoimmune disorders, and immunodeficiency syndromes (Waetzig et al. 2002; Kyriakis and Avruch 2012). From an evolutionary perspective, it is therefore logical that mechanisms of immune homeostasis are selected for as part of a survival strategy, particularly for organisms such as bacterivorous nematodes that live in microbe-rich environments and must distinguish pathogens from potential food sources.
Here we focus on the p38 MAPK pathway in C. elegans, and examine the physiological consequences of innate immune hyperactivation in intestinal epithelial cells. From two different forward genetic screens, we present evidence that aberrant activation of p38 MAPK-mediated defenses in intestinal epithelial cells is deleterious to nematodes. We found that the toxicity associated with exogenous stimulation of the p38 MAPK-mediated immune responses can be suppressed through loss-of-function mutations in p38 MAPK pathway components. We also identify and characterize a gain-of-function allele of the MAPKKK nsy-1 that drives hyperactivation of the p38 MAPK PMK-1 pathway. Accordingly, the nsy-1(gf) allele is protective against bacterial infection, but the concomitant hyperactivation of innate immune effectors is deleterious to developing animals. These data suggest that the MAPKKK NSY-1 is negatively regulated as a part of mechanism to ensure immune homeostasis.
Materials and Methods
C. elegans and bacterial strains
C. elegans was grown and propagated on NGM plates with Escherichia coli OP50, as described (Brenner 1974). AU307, an N2 Bristol-derived strain carrying the agIs44 transgene (pF08G5.6::GFP::unc-54-3′UTR; pmyo-2::mCherry), which was outcrossed to the wild-type strain N2 five times (Pukkila-Worley et al. 2014), was used as the wild-type strain for these studies, unless otherwise noted. N2 was used as the wild-type strain for the studies presented in Figure 1. Previously isolated and characterized mutants used in this study were: nsy-1(ag3) II (Kim et al. 2002), tir-1(qd4) III (Shivers et al. 2009), xbp-1(zc12) III; zcIs4 V (Calfon et al. 2002), pmk-1(km25) IV (Kim et al. 2002), and zcIs4 V (Calfon et al. 2002). RPW1 nsy-1(ums1) II, RPW2 tir-1(ums2) III, RPW3 tir-1(ums3) III, RPW4 pmk-1(ums4) IV, and RPW5 sek-1(ums5) X were isolated in this study as described below. RPW43 agIs44; nsy-1(ums8) was also isolated in this study as described below, and outcrossed six times to the wild-type strain N2.
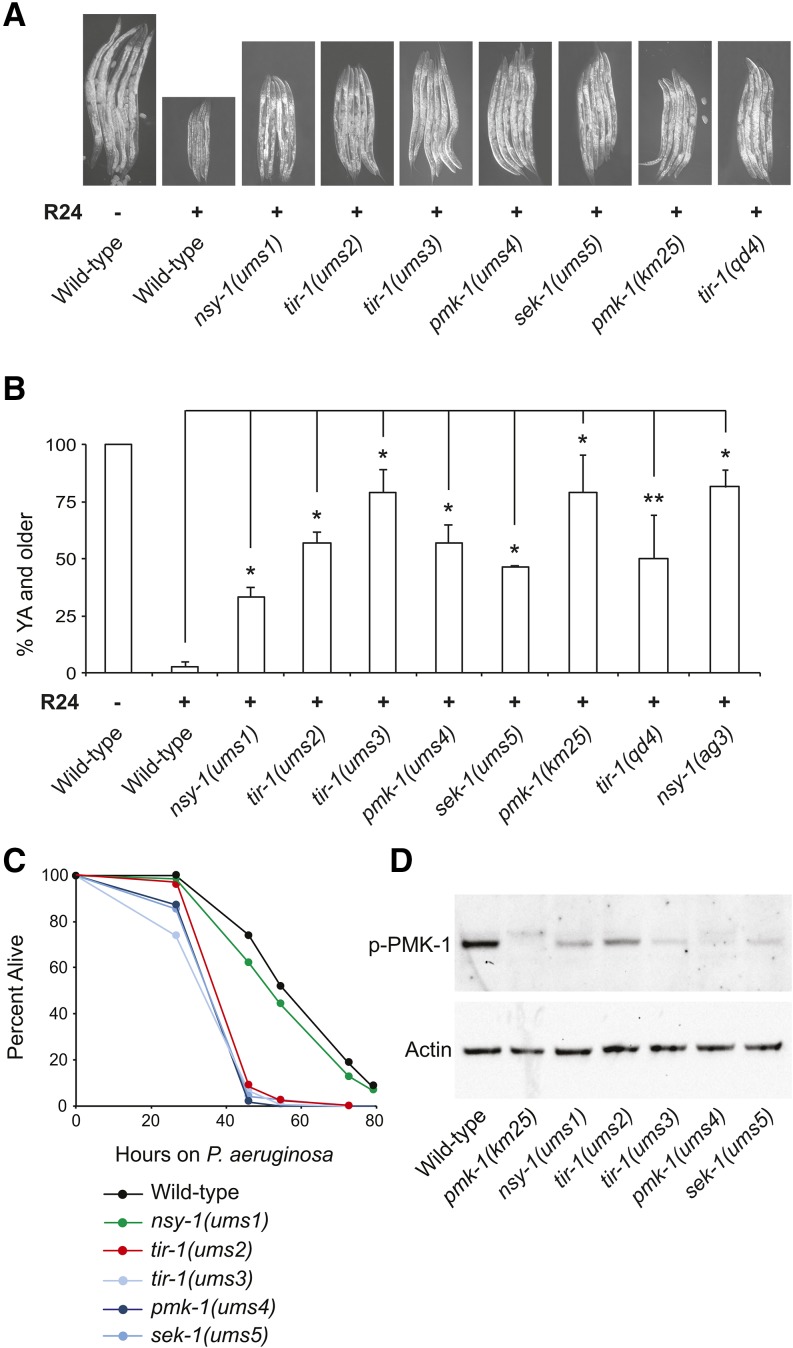
Figure 1.
p38 MAPK mutants have a Xenobiotic toxicity suppressor (Xts) phenotype. (A) Representative images of C. elegans mutants with an Xts phenotype photographed after 3 d of development at 20°C in the presence (+) or absence (–) of 140 μM R24. C. elegans N2 animals were used as the wild-type control strain. (B) Quantification of the percentage of animals that grew from the L1 to young adult (YA) stage for the experiment shown in (A). Data are the average of two technical replicates, with error bars giving the standard deviation between plates. The sample sizes for this experiment are: N2 (196), nsy-1(ums1) (171), tir-1(ums2) (193), tir-1(ums3) (220), pmk-1(ums4) (145), sek-1(ums5) (196), pmk-1(km25) (155), tir-1(qd4) (269) and nsy-1(ag3) (193). * P < 0.05, ** P = 0.07. The data are representative of multiple biological replicate experiments, which were conducted during the backcrossing of the ums mutants to wild-type animals. (C) A P. aeruginosa pathogenesis assay with C. elegans wild-type N2 and mutants with an Xts phenotype is shown. The difference in P. aeruginosa susceptibility between all mutants with an Xts phenotype and wild-type animals is significant (P < 0.001), except nsy-1(ums1) (P = n.s.). Sample sizes are: wild-type N2 (152), nsy-1(ums1) (140), tir-1(ums2) (151), tir-1(ums3) (150), pmk-1(ums4) (124), and sek-1(ums5) (139). (D) Immunoblot analysis of lysates from L4 larval stage animals of the indicated genotype using antibodies that recognize the doubly phosphorylated TGY motif of PMK-1 (p-PMK-1) and actin.
Isolation and identification of mutants with an Xts phenotype
Mutagenesis of wild-type N2 animals was performed with ethyl methane sulfonate (EMS, Sigma-Aldrich Co.) following standard methods (Jorgensen and Mango 2002). In two separate rounds of EMS screening, F2 animals from approximately 100,000 mutagenized haploid genomes were synchronized by hypochlorite treatment, and plated onto “slow kill” media (Tan et al. 1999) containing 140 μM of R24. We previously used 70 μM for our characterization of R24 immunostimulatory properties (Pukkila-Worley et al. 2012). The lower concentration also delayed the development of wild-type nematodes, but the phenotype was more pronounced at a concentration of 140 μM. Animals that reached at least the L4 larval stage after approximately 60 hr at 20° were singled to a new plate. Of the unmutagenized animals treated in parallel, 100% were at the L2 or L3 larval stage at this time point. Five mutants with the most penetrant Xts phenotypes after three rounds of retesting were identified using next-generation sequencing technology (Illumina, Inc.) following established methods (Sarin et al. 2008). Briefly, the progeny from individual F2 recombinant animals that had the mutant phenotype following a 1X backcross to N2 animals were pooled for ums3, ums4, and ums5 (40, 38, and 33 pooled recombinants, respectively). DNA was isolated from these samples, and from the original ums1 and ums2 mutants, using the Gentra Puregene Kit (Qiagen). Libraries for deep sequencing were prepared using the NEBNext DNA Library Prep Reagent Set and Oligos (New England Biolabs, Inc.), and sequenced using Illumina HiSeq 2500 following instructions from the manufacturer. Homozygous variants from the WS220 (ce10) C. elegans reference genome were identified using CloudMap (Minevich et al. 2012), and removed from further analysis if they were present in the unmutagenized parent wild-type N2 strain, the genome of which was also sequenced.
Isolation and identification of a constitutive activator of F08G5.6::GFP
EMS mutagenesis was performed on strain agIs44 as described above. Synchronized F1 progeny from approximately 170,000 mutagenized haploid genomes were screened for animals that constitutively express agIs44 GFP (green fluorescent protein) fluorescence using a dissecting microscope that is able to visualize GFP. A single mutant allele, ums8, was identified. To identify the ums8 mutation, DNA was isolated and sequenced using the methods described above. Pools of progeny from a total of 52 individual recombinants from a single outcross to wild-type N2, and from 43 individual recombinants that were outcrossed twice to N2, were sequenced. All recombinants selected for sequencing expressed agIs44 GFP fluorescence constitutively. Homozygous variants from the WS220 (ce10) C. elegans reference genome that were present in both the 1X and 2X backcross samples, but were not present in the parent agIs44 strain, were identified using CloudMap (Minevich et al. 2012).
RNAi of nsy-1
Two RNAi constructs were created using 731 bp and 1007 bp segments of nsy-1 coding region (bases 32,533 to 33,264, and 30,950 to 31,957 on cosmid F59A6, respectively), which were amplified by PCR and subcloned into the Fire vector pPD129.36, ligation number L4440 (referred to throughout the manuscript as L4440) to create plasmids pHC1 and pHC2, respectively. These plasmids were transformed into the RNAi bacterial feeder strain HT115, and RNAi experiments were carried out with these strains following established protocols (Timmons et al. 2001) using HT115 bacteria expressing the empty vector L4440 as the control. For all RNAi experiments, L4 larval stage animals of the indicated genotypes were picked to RNAi bacteria, and F1 progeny were used for subsequent studies, as described in more detail below.
nanoString nCounter gene expression and quantitative RT-PCR (qRT-PCR)
For the nanoString transcription profiling experiment, hypochlorite-synchronized L1 larval stage animals were added to NGM plates seeded with OP50. nsy-1(ums8) mutant animals were added to these plates approximately 24 hr before the nsy-1(ag3) and agIs44 animals, to allow the animals to reach the L4 larval stage at the time of harvest. The agIs44 animals were used as the wild-type control for this experiment. Worms were flash-frozen in an ethanol and dry ice bath, lysed in 0.5% SDS, 5% β-mercaptoethanol, 10 mM EDTA, 10 mM Tris-HCl pH 7.4., 0.5 mg/ml Proteinase K at 55° for 15 min using a previously described protocol (Ding et al. 2015), RNA was isolated using Tri-reagent (Sigma-Aldrich Co.), and 100 ng was analyzed by the nanoString nCounter Gene Expression System (nanoString Technologies, Inc.) using a “codeset” designed by nanoString that contained probes for 118 C. elegans genes (Supporting Information, Table S1). Probe hybridization, data acquisition and analysis were performed according to instructions from nanoString with the expression data from each sample normalized to the geometric mean of expression values for the control genes snb-1, ama-1, and act-1.
To determine the gene induction in nsy-1(ums8)/+ heterozygotes, RNA was isolated from 100 L4 larval stage agIs44 (+/+), nsy-1(ums8)/nsy-1(ums8) homozygotes, and nsy-1(ums8)/+ heterozygotes, the latter of which were the F1 progeny from a cross between agIs44 animals and nsy-1(ums8) homozygotes. RNA was isolated from three replicates using Trizol (Sigma-Aldrich Co.), treated with recombinant DNaseI (Ambion), reverse transcribed to cDNA using the Retro-script kit (Life Technologies), and analyzed using iQ SYBR Green detection (Bio-Rad Laboratories, Inc.) in duplicate 20-μl reactions on a CFX1000 machine (Bio-Rad Laboratories, Inc.) with previously published primers (Troemel et al. 2006; Pukkila-Worley et al. 2012). All values were normalized against the control gene snb-1. Fold change was calculated using the Pfaffl method (Pfaffl 2001). For the other qRT-PCR studies, four L4 larval stage animals of the indicated genotype were added to RNAi bacteria, and RNA was harvested from the mixed-stage F1 progeny from these animals. To ensure that animals would be approximately stage-matched at the time of harvest, nsy-1(ums8) mutants were allowed to lay their brood and develop at 20°, while agIs44 and nsy-1(ag3) animals were kept at 15°. RNA was isolated from three replicates using Tri-reagent (Sigma-Aldrich Co.), reversed transcribed into cDNA, and studied by qRT-PCR following the protocol described above.
Immunoblot analyses
C. elegans was prepared in the manner described for the nanoString experiment to ensure that stage-matched animals at the young L4 larval stage were studied in each condition. A previously described protocol (Ding et al. 2015) was adapted for the immunoblot analysis. Harvested animals were washed twice with M9 buffer, incubated in a roller at room temperature for 15 min to allow the nematode intestine to clear of bacteria, washed an additional time and flash-frozen in RIPA Buffer (Cell Signaling Technology, Inc.) using an ethanol and dry ice bath. Samples were lysed by sonication and centrifuged. Protein was quantified from the supernatant of each sample using Bradford Reagent (Bio-Rad Laboratories, Inc.). Laemmli buffer (Bio-Rad Laboratories, Inc.) was added to a concentration of 1X, and the total protein from each sample was resolved on NuPage 4–12% gels (Life Technologies), transferred to nitrocellulose membranes (Life Technologies), blocked with 5% powdered milk in TBST, and probed with a 1:2000 dilution of an antibody that recognizes the doubly phosphorylated TGY motif of PMK-1 (Promega Corporation). The blot was then stripped and reprobed with a 1:10,000 dilution of an anti-actin antibody (Thermo Fisher Scientific, Inc.). Horseradish peroxidase (HRP)-conjugated anti-rabbit, and anti-mouse IgG secondary antibodies (Thermo Fisher Scientific, Inc.) were used to detect the primary antibodies following the addition of ECL reagents (Thermo Fisher Scientific, Inc.), which were visualized using a Fujifilm LAS-400 luminescent image analyzer.
C. elegans bacterial infection and development assays
“Slow killing” P. aeruginosa infection assays were performed as previously described (Tan et al. 1999). A single colony of P. aeruginosa PA14 was inoculated into 3 ml of Luria-Bertani (LB) medium, and allowed to incubate at 37° for 14–15 hr; 10 μl of this culture was added to 35-mm tissue culture plates containing 4 ml of slow kill agar (0.35% peptone, 0.3% sodium chloride, 1.7% agar, 5 μg/ml cholesterol, 25 mM potassium phosphate, 1 mM magnesium sulfate, 1 mM calcium chloride). Plates were incubated for 24 hr at 37°, and 24 hr at 25°. At 1–2 hr before the start of the assay, 0.1 mg/ml 5-fluorodeoxyuridine (FUDR) was added to the medium to prevent progeny from hatching. A total of 40–50 animals at the young L4 larval stage were picked to each of three or four assay plates per experimental condition. C. elegans was prepared for the pathogenesis assays in the manner described above for the nanoString experiment to ensure that stage-matched nematodes were used for these experiments. Animals were scored as live or dead on a daily basis by gently touching them with a platinum wire. Worms that crawled onto the wall of the tissue culture plate were eliminated from the analysis. P. aeruginosa killing assays were conducted at 25°.
The development assay presented in Figure 1 was conducted by placing approximately 100 hypochlorite-synchronized L1 larval stage animals of the indicated genotype on “slow kill” medium plates (Tan et al. 1999) containing 140 μM R24 or the solvent control DMSO (1%), and monitoring development to the young adult stage for 3 d at 20° on two replicate plates per condition. For the development assays conducted with RNAi bacteria (Figure 4B), two L4 larval stage animals of the indicated genotype were allowed to lay their brood on RNAi bacteria at 15°. Plates were then transferred to 20° for 3 d. The stage of approximately 300 animals on each of three replicate plates per condition was recorded, and the percentage of animals at the L4 larval stage was reported. Representative animals from each condition in this experiment were photographed. For the experiments with the xbp-1(zc12) mutant (Figure S4), four animals were allowed to lay their brood for 8 hr at 20° in the presence or absence of 70 μM R24. C. elegans carrying the zcIs4 transgene was used as the control for these experiments.
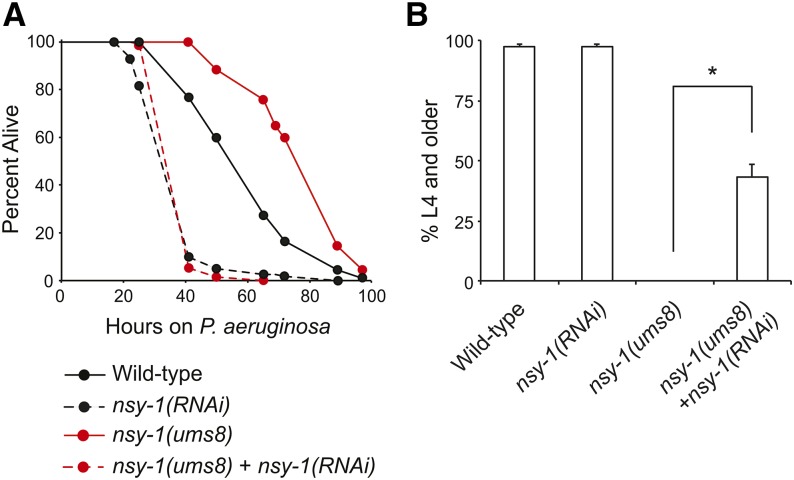
Figure 4.
Endogenous hyperactivation of p38 MAPK innate immune responses protects nematodes from bacterial infection and delays the development of wild-type nematodes. In both (A) and (B), wild-type animals were grown on RNAi bacteria expressing the empty vector L4440 (Wild-type), or a construct designed to knockdown nsy-1 [nsy-1(RNAi)]. nsy-1(ums8) animals, were grown in parallel on RNAi bacteria expressing the empty vector L4440 [nsy-1(ums8)], or a construct designed to knockdown nsy-1 [nsy-1(ums8) + nsy-1(RNAi)]. (A) P. aeruginosa pathogenesis assays were performed on RNAi-treated animals of the indicated genotypes. The difference in P. aeruginosa susceptibility between wild-type and nsy-1(ums8) animals is significant, as is the survival difference between nsy-1(ums8) and nsy-1(ums8) + nsy-1(RNAi) (P < 0.001). Data are representative of two biological replicates. The sample sizes for this experiment are: wild-type (115), nsy-1(RNAi) (122), nsy-1(ums8) (111), and nsy-1(ums8) + nsy-1(RNAi) (133) (B) The development of RNAi-treated animals of the indicated genotypes to the L4 larval stage or older was recorded. The data are the average of three plates, with error bars showing the standard deviation between plates. The sample sizes for this experiment are: wild-type + L4440 (1,200), wild-type + nsy-1(RNAi) (1179), nsy-1(ums8) + L4440 (956), and nsy-1(ums8) + nsy-1(RNAi) (1010). Data are representative of two biological replicates. * P < 0.001.
Microscopy
Nematodes were mounted onto 2% agar pads, paralyzed with levamisole (Sigma-Aldrich Co.), and photographed using a AXIO Imager Z1 microscope with a AxioCam HRm camera and Axiovision 4.6 software (Zeiss), or an Eclipse E400 with a DS-QilMc camera and NIS Elements Imaging Software (Nikon Corporation). Photographs were acquired using the same imaging conditions for a given experiment, and were processed in Photoshop (Adobe Systems, Inc.).
Statistical analyses and amino acid alignment
Differences in survival of C. elegans animals in the P. aeruginosa pathogenesis assays were determined with the log-rank test. Fold changes in the qRT-PCR analyses and differences in the development of animals to the indicated stage were compared using unpaired, two-tailed student t-tests. Amino acid alignment between human ASK1, the Drosophila Pk92B, and C. elegans NSY-1, was determined using ClustalW2 (Larkin et al. 2007).
Data availability
Strains are available upon request. Accession numbers for genes and gene products are given for the publically available database Wormbase (http://www.wormbase.org). The accession numbers for the principal genes mentioned in this paper are: C17H12.8, C32H11.12, F08G5.6, F35E12.5, F56D6.2, nsy-1 (F59A6.1), pmk-1 (B0218.3), sek-1 (R03G5.2), tir-1(F13B10.1), T24B8.5. Other accession numbers are given in Table S1.
Results and Discussion
Exogenous hyperactivation of the p38 MAP kinase pathway is toxic to developing C. elegans
The small molecule R24, also called RPW-24, acts upstream of the p38 MAPK pathway to cause the induction of putative antibacterial immune effectors in the intestine of C. elegans and, accordingly, protects nematodes from bacterial infection (Pukkila-Worley et al. 2012, 2014). In addition, exposure to R24 causes the induction of detoxification enzymes and delays the development of wild-type nematodes in the absence of pathogen, suggesting that this compound is toxic to C. elegans under normal laboratory growth conditions (Pukkila-Worley et al. 2012, 2014). To investigate the relationship between the immunostimulatory properties and the toxicity of this anti-infective xenobiotic, we screened nematodes derived from approximately 100,000 mutagenized haploid genomes for mutants that were able to develop faster in the presence of 140 μM R24 (Figure 1, A and B). We chose this concentration because the developmental delay phenotype was more pronounced than observed for 70 μM R24, the concentration we had used in our previous studies (Pukkila-Worley et al. 2012, 2014). For the screen, we required the selected mutants to be L4 larval stage or older at a time point when 100% of unmutagenized worms treated with R24 in parallel were at the L2 or L3 larval stage. Seven mutants with a xenobiotic toxicity suppressor (Xts) phenotype were identified, and we focused on the five mutants that had the most penetrant Xts phenotypes which we have called ums1, ums2, ums3, ums4 and ums5 (Figure 1, A and B). To identify the mutation in these strains that permitted improved development in the presence of R24, we used next-generation sequencing technology (Sarin et al. 2008; Minevich et al. 2012). For three mutants (ums3, ums4, and ums5), we sequenced DNA from pooled F2 recombinants that had the mutant phenotype following a backcross to wild-type N2 animals, and for two mutants (ums1 and ums2) we identified mutations after sequencing the original mutant strain. Interestingly, these five mutants each contained missense mutations in one of the four known components of the p38 MAPK signaling cassette (tir-1, nsy-1, sek-1, or pmk-1, Table 1).
Table 1. Xenobiotic toxicity suppressors.
| Gene | Allele | Mutation | Description, Mammalian Homolog |
|---|---|---|---|
| tir-1 | ums2 | L730F | TIR domain adaptor protein, SARM |
| tir-1 | ums3 | A723T | TIR domain adaptor protein, SARM |
| nsy-1 | ums1 | P874L | MAPKKK, ASK1 |
| sek-1 | ums5 | G199S | MAPKK, MKK3/6 |
| pmk-1 | ums4 | L296F | MAPK, p38 |
Presented is a list of isolates identified in a screen for mutants that are able to develop faster in the presence of the immunostimulatory anti-infective xenobiotic R24 compared to controls.
To characterize these newly-isolated p38 MAPK pathway mutants, we conducted pathogenesis assays with P. aeruginosa and found that, as with the classic loss-of-function mutants in p38 MAPK pathway components, four of the five mutants [tir-1(ums2), tir-1(ums3), pmk-1(ums4) and sek-1(ums5)] had an enhanced susceptibility to pathogens (Esp) phenotype (P < 0.001, Figure 1C). nsy-1(ums1), the weakest toxicity suppressor of these five mutants, did not have an obvious Esp phenotype. In addition to conferring an Esp phenotype, loss- and reduction-of-function mutations in upstream components of the p38 MAPK PMK-1 pathway cause a reduction in the amount of activated PMK-1, which can be detected in an immunoblot experiment using an antibody that specifically recognizes the doubly phosphorylated TGY motif of PMK-1 (Kim et al. 2002; Liberati et al. 2004). We found that nsy-1(ums1), tir-1(ums2), tir-1(ums3), pmk-1(ums4) and sek-1(ums5) each had reduced levels of the active form of PMK-1 compared to wild-type controls (Figure 1D). These data indicate that the forward genetic screen for mutations that confer resistance to the toxic effects of the immunostimulatory xenobiotic R24 identified hypomorphic alleles in the p38 MAPK pathway. Interestingly, a previous forward genetic screen for loss-of-function mutations in the p38 MAPK pathway components identified two alleles, pmk-1(qd9) and sek-1(qd37), which had the identical missense mutation as pmk-1(ums4) and at the same amino acid as sek-1(ums5), respectively (Shivers et al. 2010).
Together, these data demonstrate that the toxicity of R24 can be suppressed by hypomorphic mutations in the p38 MAPK pathway, although no single mutation was identified that resulted in growth progression identical to worms in the absence of the compound. To confirm this observation, we studied the previously characterized null alleles tir-1(qd4), nsy-1(ag3) and pmk-1(km25) (Kim et al. 2002; Shivers et al. 2009) (Figure 1, A and B) and found that these mutations also suppressed the R24-induced developmental delay to a degree comparable to the pmk-1(ums4) mutant, and the tir-1(ums2) and tir-1(ums3) alleles, respectively (Figure 1, A and B).
In summary, these data indicate that hyperactivation of p38 MAPK immune defenses is toxic to developing nematodes.
Forward genetic screen uncovers a gain-of-function allele in the MAPKKK nsy-1
To explore further the physiological consequences of innate immune hyperactivation in C. elegans, we designed a forward genetic screen for endogenous activators of p38 MAP kinase PMK-1 signaling. The innate immune transcriptional reporter F08G5.6::GFP is induced during P. aeruginosa infection, and robustly by the anti-infective xenobiotic R24, in a manner dependent on the p38 MAPK PMK-1 pathway (Pukkila-Worley et al. 2012, 2014). We therefore reasoned that a screen for dominant activators of F08G5.6::GFP would identify mutations that cause constitutive activation of the p38 MAPK pathway. We screened the F1 progeny of mutagenized F08G5.6::GFP animals, and identified a single mutant allele, ums8, from approximately 170,000 mutagenized haploid genomes. Following both 1X and 2X outcrosses to wild-type N2 animals, we pooled F2 recombinants that were homozygous for the mutant phenotype, and sequenced the genomes of these samples using next-generation sequencing technology (Sarin et al. 2008; Minevich et al. 2012). This study revealed that the ums8 mutant contained a G→A missense mutation in the coding region of nsy-1 that resulted in the substitution of a strongly conserved Arg246 to Gln246 (Figure 2A). NSY-1/ASK1 is a conserved MAPKKK that acts upstream of the p38 MAPK PMK-1 to regulate immune and stress responses in C. elegans (Kim et al. 2002). We outcrossed the ums8 strain a total of six times to wild-type N2, and confirmed that the R246Q mutation was present and F08G5.6::GFP expression was activated in the outcrossed strain.
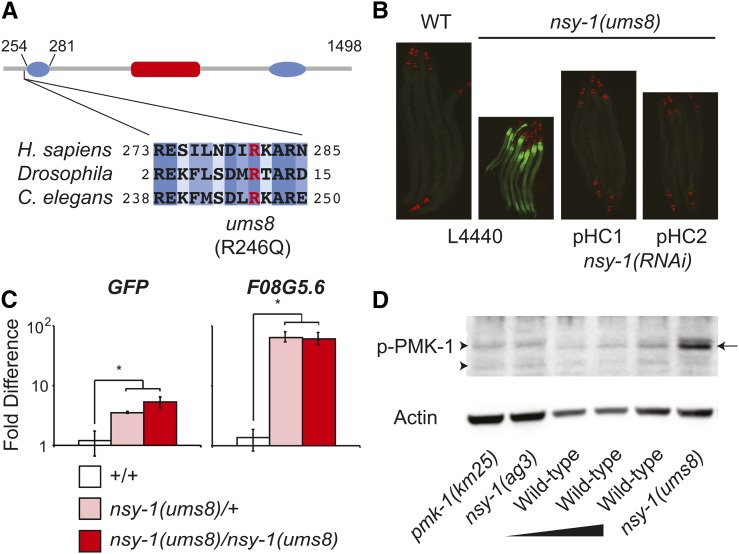
Figure 2.
The nsy-1(ums8) allele encodes a gain-of-function mutation in nsy-1. (A) The putative domain architecture of NSY-1, based on homology to mammalian ASK1, is presented. This schematic was adapted from Bunkoczi et al. (2007). The putative location of the central serine-threonine kinase domain, and two coiled-coil domains in the N and C termini are shown in red and blue, respectively. The boundary of the N-terminal negative regulatory domain relative to the NSY-1 protein is presented above the diagram. The Arg that was mutated to Gln in the ums8 strain (R246Q) is highlighted in red. Amino acid sequence alignment of human ASK1, Drosophila Pk92B (the ASK1 homolog) and C. elegans NSY-1 demonstrates that the ums8 mutation is located in a strongly conserved region. Dark blue shading indicates identical amino acids, with progressive lighter blue shading indicating the level of similarity in amino acid class as determined by the software ClustalW2. (B) Wild-type (WT) and nsy-1(ums8) animals were exposed to the feeding RNAi bacteria strain transformed with the control vector (L4440), or with two separate RNAi constructs (pHC1 and pHC2) that target different areas of coding region in the nsy-1 gene and photographed. Green is F08G5.6::GFP induction and red is myo-2::mCherry, which was used as the co-injection marker. (C) The expression of the indicated genes was determined using qRT-PCR in wild-type (+/+), nsy-1(ums8)/+ heterozygotes, and nsy-1(ums8)/nsy-1(ums8) homozygotes. F08G5.6::GFP was used as the wild-type strain. Data are the average of three replicates, each normalized to a control gene with error bars representing SEM, and are presented as the value relative to the average expression of the indicated gene in wild-type animals. * equals P < 0.05. There was no statistical difference in the levels of induction between the heterozygous and homozygous samples for either gene. For additional genes tested, see Figure S1A. (D) Immunoblot analysis of lysates from L4 larval stage animals of the indicated genotype using antibodies that recognize the doubly phosphorylated TGY motif of PMK-1 (p-PMK-1) and actin. Thirty μg of nsy-1(ums8), pmk-1(km25) and nsy-1(ag3) total protein were loaded on the gel alongside a dilution series of wild-type template [15 μg, 20 μg, and 30 μg of total protein (left to right)] to control for the ability of the p-PMK-1 antibody to detect different concentrations of substrate. The arrow on the right highlights the PMK-1 band, which is absent in the pmk-1(km25) and nsy-1(ag3) mutants. The arrowheads on the left point to nonspecific bands. Data are representative of two biological replicates.
To determine if the constitutive expression of the immune reporter F08G5.6::GFP in the ums8 mutant is caused by the G→A mutation in the coding region of nsy-1, we used RNAi to knockdown the expression of nsy-1 in the ums8 mutant and found that this treatment suppressed F08G5.6::GFP induction (Figure 2B). Moreover, RNAi-mediated knockdown of nsy-1 in wild-type animals did not cause F08G5.6 induction (see qRT PCR data discussed in the next section). In addition, F08G5.6::GFP induction in the nsy-1(ums8) strain was suppressed by RNAi-mediated knockdown of pmk-1, the p38 MAPK that is downstream of nsy-1 (Figure S2A). We also used qRT-PCR to confirm that nsy-1(ums8) is a dominant, gain-of-function allele that drives both the constitutive activation of the F08G5.6::GFP and the F08G5.6 gene itself (Figure 2C). F08G5.6 and GFP were induced to a similar degree in nsy-1(ums8)/+ heterozygotes and in nsy-1(ums8)/nsy-1(ums8) homozygotes compared to wild-type controls that carried the transgene F08G5.6::GFP (+/+).
Consistent with our studies suggesting that nsy-1(ums8) is a gain-of-function allele of nsy-1, we performed an immunoblot analysis of protein lysates from nsy-1(ums8) animals and found that the nsy-1(ums8) animals had greater amounts of activated p38 MAPK PMK-1 than control animals (Figure 2D).
In summary, these data characterize a gain-of-function allele of the MAPKKK nsy-1 that causes hyperactivation of the p38 MAPK pathway.
The nsy-1(ums8) gain-of-function allele causes the constitutive activation of p38 MAPK PMK-1-dependent innate immune effectors
To determine if the basal expression of other genes was hyperactivated in the nsy-1(ums8) allele, we used the nanoString nCounter system to compare the expression profile of 118 innate immune and stress response genes in nsy-1(ums8) gain-of-function, nsy-1(ag3) loss-of-function and wild-type animals when the strains were feeding on the normal laboratory food source E. coli OP50 (Figure 3A). Of the 118 genes in this codeset, 24 were transcriptionally upregulated at least fivefold in nsy-1(ums8) mutants compared to wild-type animals (Table S1). Seven of the 24 genes most strongly upregulated in the nsy-1(ums8) gain-of-function allele were among the 12 most strongly reduced in the nsy-1(ag3) loss-of-function allele (F08G5.6, F35E12.5, C09H5.2, T24B8.5, clec-67, C32H11.12, and oac-6, Figure 3A and Table S1). Indeed, this group of seven genes includes F08G5.6 and five other putative immune effectors (F35E12.5, C09H5.2, T24B8.5, C32H11.12 and clec-67) that are previously characterized targets of the p38 MAPK PMK-1 (Table S1) (Troemel et al. 2006). Of note, the codeset included four additional genes whose basal or pathogen-induced expression requires the p38 MAPK PMK-1 (F49F1.6, C17H12.8, C32H11.1, K08D8.5, lys-2), and each of these genes was induced at least twofold in the nsy-1(ums8) mutants, and repressed at least fourfold in the nsy-1(ag3) mutants (Table S1). In addition, we previously used this codeset to identify 13 genes whose induction by the immunostimulatory anti-infective xenobiotic R24 was dependent on the p38 MAPK PMK-1 (Pukkila-Worley et al. 2014). Nine of these 13 genes were upregulated more than fivefold, and all of these genes were induced at least twofold in the nsy-1(ums8) gain-of-function mutant (Table S1). Thus, the nsy-1(ums8) gain-of-function allele drives the constitutive activation of genes whose basal, pathogen-induced or R24-induced expression is dependent the p38 MAPK pathway.
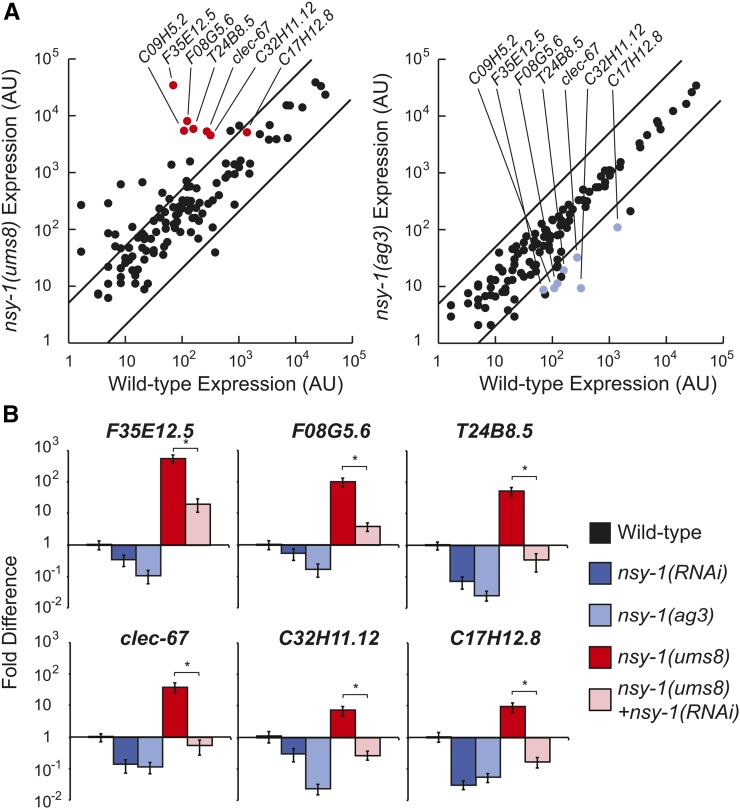
Figure 3.
p38 MAPK-dependent putative immune effectors are constitutively activated in the nsy-1(ums8) mutant. (A) A scatter plot compares the expression of 118 C. elegans genes, which were analyzed using nanoString nCounter gene expression system in wild-type, nsy-1(ums8) and nsy-1(ag3) animals. Data are the average of two replicates for nsy-1(ums8), and nsy-1(ag3), and are from one sample for wild-type. The expression of each gene was normalized to the geometric mean of the expression of three control genes. Genes that are outside the two parallel lines on each graph are differentially regulated more than fivefold from the expression in wild-type animals. Genes that are previously characterized targets of the p38 MAPK PMK-1, and were strongly differentially regulated in this experiment, are highlighted. See Table S1 for the expression levels of all the genes. (B) qRT-PCR was used to study the expression of six putative immune effectors in RNAi-treated, mixed-stage animals of the indicated genotypes. All animals were grown on the RNAi bacteria feeder strain HT115 expressing the empty vector L4440, except for the two indicated samples that were exposed to bacteria expressing the nsy-1(RNAi) construct. The location on the scatter plots of these genes is indicated in (A) with red (left) and blue (right) dots. Data are the average of three replicates, each normalized to a control gene with error bars representing SEM, and are presented as the value relative to the average expression of the indicated gene in wild-type animals. * equals P < 0.05.
We confirmed these data in three biological replicate samples using qRT-PCR to study six putative immune effectors that were differentially regulated in the nanoString analysis and are known targets of the p38 MAPK pathway. All six genes were transcriptionally repressed in nsy-1(RNAi) and nsy-1(ag3) loss-of-function mutants, and were upregulated in the nsy-1(ums8) allele (Figure 3B). RNAi-mediated knockdown of nsy-1 partially suppressed the constitutive activation of these six putative immune effectors in the nsy-1(ums8) allele (Figure 3B). In addition, we found that nsy-1(ums8)/+ heterozygotes caused the induction of three p38 MAPK-dependent immune effectors to levels equivalent to that in nsy-1(ums8) homozygous animals (Figure S1A), as we observed for F08G5.6 (Figure 2C). Of note, we also exposed the nsy-1(ums8) mutant to R24, and found that F08G5.6::GFP expression was markedly increased compared to untreated nsy-1(ums8) animals, suggesting that the effects of R24 and the nsy-1 gain-of-function allele on gene expression are additive (Figure S1B). Consistent with this observation, we previously found that R24 also further enhanced the expression of P. aeruginosa immune effectors that are induced during bacterial infection (Pukkila-Worley et al. 2012).
Previous whole genome, transcriptome profiling analyses have identified a group of putative immune effectors whose basal expression is strongly repressed in pmk-1(km25) mutants (Troemel et al. 2006). The term basal regulation has been used in this context to describe the expression of genes in animals that are growing on the laboratory food source (E. coli OP50) and is used to distinguish from the pathogen-induced expression of putative immune effectors that is observed during microbial infection. In addition to controlling the basal expression of putative immune effectors, a number of studies have demonstrated that the p38 MAPK PMK-1 is also required for the pathogen-induced expression of putative immune effectors (Troemel et al. 2006; Bolz et al. 2010; Pukkila-Worley et al. 2012). It also is clear, however, that there is a group of genes, the identities of which have not been comprehensively defined, that require the p38 MAPK PMK-1 pathway for their induction, but not for their basal expression. The nanoString data of nsy-1(ums8) mutant indicates that this gain-of-function allele can be used to identify this group of innate immune effectors. Of the 24 genes in our codeset that were most strongly induced in the nsy-1(ums8) mutant, 17 were not dependent on NSY-1 for their basal expression—a group which includes both pathogen and stress-response genes (Table S1).
It is also interesting to note that genes involved in the detoxification of small molecule toxins, such as cytochrome P450s (CYPs), glutathione-s-transferases (GSTs), and UDP-glucuronosyltransferases (UDPs), were not among those transcriptionally upregulated in the nsy-1(ums8) gain-of-function allele (Table S1). We previously observed that the anti-infective xenobiotic R24 caused the robust induction of these gene classes, in addition to genes involved in the response to pathogens (Pukkila-Worley et al. 2012). Unlike the induction of antimicrobial immune effectors, however, the upregulation of detoxification genes by R24 was not dependent on the p38 MAPK PMK-1 pathway (Pukkila-Worley et al. 2014). Together with the findings in this study, these data argue that p38 MAPK innate immune hyperstimulation itself does not lead to the induction small molecule detoxification responses, and that other cellular mechanisms are engaged to recognize and respond to xenobiotic toxins in C. elegans.
Endogenous hyperactivation of the p38 MAPK PMK-1 pathway protects nematodes from bacterial infection, but is toxic to C. elegans
Stimulation of the p38 MAPK pathway by the small molecule xenobiotic R24 protects C. elegans from bacterial infection by stimulating immune effector expression through the p38 MAPK pathway (Pukkila-Worley et al. 2012, 2014). Immune hyperactivation in this context has negative physiological consequences to nematodes developing under normal laboratory conditions (Figure 1). We therefore hypothesized that endogenous hyperactivation of the p38 MAPK pathway, by means of a gain-of-function mutation in the MAPKKK NSY-1, would display similar phenotypes as worms treated with R24.
We tested the susceptibility of nsy-1(ums8) gain-of-function mutant animals to P. aeruginosa infection compared to wild-type animals, and found that nsy-1(ums8) animals were resistant to killing by P. aeruginosa (Figure 4A and Figure S3). RNAi-mediated knockdown of nsy-1 suppressed the resistance phenotype of nsy-1(ums8) animals (Figure 4A). Interestingly, the development of nsy-1(ums8) larvae was markedly delayed compared to wild-type controls, which was quantified by determining the number of mutant and wild-type animals that reached the L4 stage from synchronized eggs after 3 d of incubation at 20° (Figure 4B). Knockdown of nsy-1 in nsy-1(ums8) animals partially suppressed this phenotype (Figure 2B and Figure 4B). We also observed that nsy-1(ums8) animals had markedly smaller brood sizes than wild-type controls (Figure S2B), data that are reminiscent of our observations with the immunostimulatory xenobiotic R24 (Pukkila-Worley et al. 2012). In addition, RNAi-mediated knockdown of pmk-1 also suppressed the delayed development of nsy-1(ums8) animals (Figure S2C).
In this study, we provide direct evidence from two forward genetic screens that aberrant stimulation of p38 MAPK PMK-1-mediated innate immune responses via a gain-of-function mutation in nsy-1 or the exogenous addition of the anti-infective R24 drives immune responses that are protective during infection, but are toxic to nematodes under normal growth conditions. It is interesting to note that the alleles with the most penetrant Xts phenotypes all had hypomorphic mutations in the known components of the p38 MAPK pathway. Of note, a forward genetic screen for p38 MAPK pathway components also did not identify mutations in any gene that functions upstream of tir-1 (Shivers et al. 2010). Together, these data suggest that the p38 MAPK signaling cassette receives multiple inputs to regulate protective host responses in C. elegans.
In addition to promoting clearance of invading pathogens, an emerging body of literature has established that host systems also function to promote tolerance to infection, thereby reducing the negative impact that infection can have on organismal fitness (Ayres and Schneider 2008; 2009). Conceptually, host tolerance mechanisms function to mitigate the damage caused by the pathogen and also limit collateral injury associated with immune activation (Ayres and Schneider 2012; Medzhitov et al. 2012). Our studies of the immunostimulatory xenobiotic R24 and the nsy-1(ums8) gain-of-function allele suggest that host tolerance mechanisms may function in nematodes to limit the immunopathology associated with aberrant p38 MAPK activation and the ensuing hyperproduction of immune effectors. In C. elegans, the unfolded protein response (UPR) is one such mechanism that has been shown to promote tolerance during immune activation (Richardson et al. 2010; Sun et al. 2011). The IRE1-XBP1/Hac1 branch of the UPR is required to handle the physiological stress associated with an increase in p38 MAPK PMK-1 activity, which was conferred experimentally by exposure to P. aeruginosa or through RNAi-mediated knockdown of the MAPK phosphatase vhp-1, a negative regulator of PMK-1 (Richardson et al. 2010; Mizuno et al. 2004; Kim et al. 2004). Consistent with these data, we found that xbp-1(zc12) mutants are dramatically susceptible to the toxic effects of the immunostimulatory xenobiotic R24 in a developmental assay compared to control animals (Figure S4). Thus, there are likely at least two mechanisms in C. elegans to ensure immune homeostasis and promote tolerance during infection. The UPR functions to protect the host from the ER stress, which occurs as a consequence of p38 MAPK activation (Richardson et al. 2010). In addition, the isolation of a gain-of-function allele in nsy-1 argues that p38 MAPK PMK-1-mediated transcriptional responses are controlled via upstream mechanisms that negatively regulate immune activation at the level of NSY-1.
The human homolog of NSY-1, ASK1, regulates p38 activity through at least three conserved protein domains: a central serine-threonine kinase domain and two coiled-coil domains in the N and C termini, respectively (Figure 2A) (Bunkoczi et al. 2007). The N-terminal domain binds thioredoxin (TRX), which inhibits the function of NSY-1. Interestingly, the gain-of-function ums8 mutation, in which the negatively charged Arg is substituted for Gln, an uncharged amino acid with a large polar side chain, is located in a strongly conserved region, eight amino acids upstream of the region in NSY-1 predicted by homology to encode this N-terminal regulatory domain (Figure 2A). We hypothesize that the activity of C. elegans NSY-1 is normally controlled by a negative regulatory factor, which binds to the N-terminal coiled-coil domain. Thus, disruption of this region by the ums8 mutation could account for the constitutive activation of p38 MAPK immune effectors resulting in a pathogen resistance phenotype and toxicity to developing nematodes. There are five thioredoxin (TRX) homologs in C. elegans and it will be interesting to determine if any of these genes function as negative regulators of NSY-1.
Supplementary Material
Acknowledgments
We thank Neal Silverman and Amy Walker for helpful discussions and Richa Chhaya for technical assistance. This study was supported by National Institute of Allergy and Infectious Diseases Award K08 AI081747 (to R.P.W.). R.L.F. and A.L.C. were supported by National Institute of Allergy and Infectious Diseases Awards R01 AI085581 and P01 AI083214.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.025650/-/DC1
Communicating editor: D. Schneider
Literature Cited
- Aggarwal K., Silverman N., 2008. Positive and negative regulation of the Drosophila immune response. BMB Rep. 41: 267–277. [DOI] [PubMed] [Google Scholar]
- Ayres J. S., Schneider D. S., 2008. A signaling protease required for melanization in Drosophila affects resistance and tolerance of infections. PLoS Biol. 6: e305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres J. S., Schneider D. S., 2009. The role of anorexia in resistance and tolerance to infections in Drosophila. PLoS Biol. 7: e1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres J. S., Schneider D. S., 2012. Tolerance of infections. Annu. Rev. Immunol. 30: 271–294. [DOI] [PubMed] [Google Scholar]
- Bolz D. D., Tenor J. L., Aballay A., 2010. A conserved PMK-1/p38 MAPK is required in Caenorhabditis elegans tissue-specific immune response to Yersinia pestis infection. J. Biol. Chem. 285: 10832–10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunkoczi G., Salah E., Filippakopoulos P., Fedorov O., Müller S., et al. , 2007. Structural and functional characterization of the human protein kinase ASK1. Structure 15: 1215–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., et al. , 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415: 92–96. [DOI] [PubMed] [Google Scholar]
- Cohen L. B., Troemel E. R., 2015. Microbial pathogenesis and host defense in the nematode C. elegans. Curr. Opin. Microbiol. 23: 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W., Smulan L. J., Hou N. S., Taubert S., Watts J. L., et al. , 2015. s-Adenosylmethionine levels govern innate immunity through distinct methylation-dependent pathways. Cell Metab. 22: 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui J. E., Urbach J. M., Ausubel F. M., 2010. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 10: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen E. M., Mango S. E., 2002. The art and design of genetic screens: Caenorhabditis elegans. Nat. Rev. Genet. 3: 356–369. [DOI] [PubMed] [Google Scholar]
- Kim D. H., Feinbaum R., Alloing G., Emerson F. E., Garsin D. A., et al. , 2002. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297: 623–626. [DOI] [PubMed] [Google Scholar]
- Kim D. H., Liberati N. T., Mizuno T., Inoue H., Hisamoto N., et al. , 2004. Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proc. Natl. Acad. Sci. USA 101: 10990–10994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakis J. M., Avruch J., 2012. Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol. Rev. 92: 689–737. [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., et al. , 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Liberati N. T., Fitzgerald K. A., Kim D. H., Feinbaum R., Golenbock D. T., et al. , 2004. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc. Natl. Acad. Sci. USA 101: 6593–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Schneider D. S., Soares M. P., 2012. Disease tolerance as a defense strategy. Science 335: 936–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minevich G., Park D. S., Blankenberg D., Poole R. J., Hobert O., 2012. CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics 192: 1249–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Hisamoto N., Terada T., Kondo T., Adachi M., et al. , 2004. The Caenorhabditis elegans MAPK phosphatase VHP-1 mediates a novel JNK-like signaling pathway in stress response. EMBO J. 23: 2226–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson L. W., Artis D., 2014. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14: 141–153. [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W., 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R., Ausubel F. M., 2012. Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Curr. Opin. Immunol. 24: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R., Ausubel F. M., Mylonakis E., 2011. Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog. 7: e1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R., Feinbaum R., Kirienko N. V., Larkins-Ford J., Conery A. L., et al. , 2012. Stimulation of host immune defenses by a small molecule protects C. elegans from bacterial infection. PLoS Genet. 8: e1002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R., Feinbaum R. L., McEwan D. L., Conery A. L., Ausubel F. M., 2014. The evolutionarily conserved mediator subunit MDT-15/MED15 links protective innate immune responses and xenobiotic detoxification. PLoS Pathog. 10: e1004143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C. E., Kooistra T., Kim D. H., 2010. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature 463: 1092–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S., Prabhu S., O’Meara M. M., Pe’er I., Hobert O., 2008. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat. Methods 5: 865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivers R. P., Kooistra T., Chu S. W., Pagano D. J., Kim D. H., 2009. Tissue-specific activities of an immune signaling module regulate physiological responses to pathogenic and nutritional bacteria in C. elegans. Cell Host Microbe 6: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivers R. P., Pagano D. J., Kooistra T., Richardson C. E., Reddy K. C., et al. , 2010. Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet. 6: e1000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Singh V., Kajino-Sakamoto R., Aballay A., 2011. Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science 332: 729–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M. W., Mahajan-Miklos S., Ausubel F. M., 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96: 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A., 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112. [DOI] [PubMed] [Google Scholar]
- Troemel E. R., Chu S. W., Reinke V., Lee S. S., Ausubel F. M., et al. , 2006. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2: e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvikis O., Ihuegbu N., Labed S. A., Luhachack L. G., Alves A.-M. F., et al. , 2014. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity 40: 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waetzig G. H., Seegert D., Rosenstiel P., Nikolaus S., Schreiber S., 2002. p38 Mitogen-activated protein kinase is activated and linked to TNF-α signaling in inflammatory bowel disease. J. Immunol. 168: 5342–5351. [DOI] [PubMed] [Google Scholar]
- Xavier R. J., Podolsky D. K., 2007. Unravelling the pathogenesis of inflammatory bowel disease. Nature 448: 427–434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available upon request. Accession numbers for genes and gene products are given for the publically available database Wormbase (http://www.wormbase.org). The accession numbers for the principal genes mentioned in this paper are: C17H12.8, C32H11.12, F08G5.6, F35E12.5, F56D6.2, nsy-1 (F59A6.1), pmk-1 (B0218.3), sek-1 (R03G5.2), tir-1(F13B10.1), T24B8.5. Other accession numbers are given in Table S1.