Abstract
Winter damage, especially in northern climates, is a major limitation of the utilization of perennial forages such as alfalfa. Therefore, improving freezing tolerance is imperative in alfalfa genetic breeding. However, freezing tolerance is a complex trait that is determined by many genes. To understand the complex regulation mechanisms of freezing tolerance in alfalfa, we performed small RNA sequencing analysis under cold (4°) and freezing (−8°) stress. The sequencing results revealed that 173 known, and 24 novel miRNAs were expressed, and that the expression of 35 miRNAs was affected by cold and/or freezing stress. Meanwhile, 105 target genes cleaved by these miRNAs were characterized by degradome sequencing. These targets were associated with biological regulation, cellular processes, metabolic processes, and response to stress. Interestingly, most of them were characterized as transcription factors (TFs), including auxin response factors, SBP, NAC, AP2/ERF, and GRF, which play important roles in plant abiotic responses. In addition, important miRNAs and mRNAs involved in nodulation were also identified, for example, the relationship between miR169 and the TF CCAAT (also named as NF-YA/HAP2), which suggested that nodulation has an important function in freezing tolerance in alfalfa. Our results provide valuable information to help determine the molecular mechanisms of freezing tolerance in alfalfa, which will aid the application of these miRNAs and their targets in the improvement of freezing tolerance in alfalfa and related plants.
Keywords: Medicago sativa, cold acclimation, freezing stress, microRNA, degradome sequencing
Lack of tolerance to freezing is a major environmental limitation of the survival, productivity, and ecological distribution of plants. However, freezing tolerance is a complex trait that is determined by numerous factors from plants and the environment. Among these factors, cold acclimation, the exposure of plants to low, subfreezing temperatures, plays an important role in conferring freezing tolerance (Thomashow 2010). During the cold acclimation process, several biochemical and physiological modifications occur, including accumulation of soluble sugars, free amino acids, and the expression of cold-regulated (COR) genes, which potentially improving freezing tolerance in plants. To date, the identification and characterization of C-repeat (CRT)-binding factors (CBFs) has shown that they play critical roles in the cold acclimation process (Gilmour et al. 1998; Thomashow 2010), by regulating downstream functional genes, such as COR genes (Hajela et al. 1990; Thomashow 2010). Regulation of COR genes by CBFs constitutes the central component of cold signaling pathways that confer freezing tolerance on plants. In addition, the CBF signaling pathway is also regulated by other factors, for example, ICE1 (Inducer of CBF Expression 1) (Chinnusamy et al. 2003; Lee et al. 2005; Miura and Hasegawa 2008), and HOS1 (High Expression of Osmotically Responsive Gene 1) (Jung and Park 2013; Lee et al. 2001), which are important components in the plant cold acclimation process.
MicroRNAs are a class of noncoding small RNAs of approximately 21–24 nt, which bind to complementary sequences in the mRNAs of target genes (Mallory and Vaucheret 2006). This binding results in the regulation of gene expression at the posttranscriptional level by cleavage-induced degradation of the mRNA, or suppression of its translation (Mallory and Vaucheret 2006). In recent years, many miRNAs have been demonstrated to have important regulatory functions in plant growth, development, and stress responses (Babar et al. 2008; Khraiwesh et al. 2012; Sunkar et al. 2012). A number of miRNAs are involved in the cold response process in Arabidopsis (Liu et al. 2008; Sunkar and Zhu 2004; X. Zhou et al. 2008), poplar (Chen et al. 2012; Lu et al. 2008), Brachypodium distachyon (Zhang et al. 2009), rice (Lv et al. 2010), and wheat (Tang et al. 2011, 2012), including miR156/157, miR169, miR393, miR396, miR394, and miR398 (Rajwanshi et al. 2014). With the development of high-throughput sequencing technology, numerous miRNAs have been identified and characterized in plants. MiRNAs biological functions have been deduced by the identification and characterization of their target genes. Degradome sequencing, also termed parallel analysis of RNA ends (PARE), was developed based on high-throughput sequencing technology for the genome-wide identification of miRNA target genes (Folkes et al. 2012). Using high throughput sequencing methods, many new miRNA–mRNA target pairs have been identified successfully in Arabidopsis (Addo-Quaye et al. 2008), rice (Sun et al. 2015), wheat (Chen et al. 2015a), soybean (Shamimuzzaman and Vodkin 2012), maize (Zhao et al. 2013), and grapevine (Pantaleo et al. 2010), which has helped lucidate the regulatory relationships between miRNAs and their target genes.
Alfalfa (Medicago sativa L.), which is grown worldwide, is a highly productive perennial forage species with the capacity for biological fixation of atmospheric nitrogen. However, because of insufficient freezing tolerance, hard winters (with extremely low temperatures) are a major limitation to alfalfa production (Castonguay et al. 2013; Pennycooke et al. 2008). Thus, improvement of freezing tolerance is an important breeding aim for high yield and longer production periods in alfalfa, especially in northern climates, for example, in the United States, Canada, and China (Chen et al. 2015b). M. sativa L. cv. Zhaodong was domesticated and bred from wild M. sativa by the Institute of Animal Husbandry of Heilongjiang Province (IAH-HLJ, China), and has high tolerance to freezing stress, enabling it to survive during the winter season in the fields of Heilongjiang Province, China (average temperature –35°). The high freezing tolerance of M. sativa L. cv. Zhaodong is determined by its specific gene regulation network (Luo et al. 2004). Determining the specific expression patterns of miRNAs and mRNAs would be helpful to understand the complex molecular mechanism of freezing tolerance in this species.
In the present study, miRNAs and their targets that are involved in cold and/or freezing stress were investigated using high-throughput sequencing. Freezing-stress-responsive miRNAs were selected and validated by quantitative real-time reverse transcription (qRT-PCR) experiments. Meanwhile, the potential miRNA targets were predicted and confirmed by degradome sequencing.
Materials and Methods
Plant growth and treatment
Seeds of M. sativa (cv. Zhaodong), kindly provided by Prof. Hong Li (IAH-HLJ, China), were germinated and transferred onto a mix of perlite and sand (3:1, v/v). All seedlings were grown in a growth chamber (Conviron E15, Canada), at a temperature between 18° (night) and 24° (day), with humidity ranging from 60% to 80%, and a light period of 14 hr/10 hr (daytime, 06:00–20:00). The seedlings were irrigated with half-strength Hoagland solution once every other day, and after 8 wk they were randomly divided into three groups for stress treatments. For the control group (untreated, A group), the seedlings continued to grow at 18° (night) to 24° (day). For cold stress (B group) and freezing stress (C group), the seedlings were transferred into another chamber with the temperature set at 4° or −8°, respectively. According to our previous research (Shu et al. 2015), all seedlings were harvested at 3 hr after stress treatments; five whole seedlings from each group were bulked separately. All samples were frozen in liquid nitrogen, and stored at −80° until use.
Small RNA library construction and sequencing
Small RNAs were extracted from samples from the three treatments (control, cold, and freezing) using the TRIzol method (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions. Small RNAs were ligated sequentially to 5′- and 3′- RNA/DNA chimeric oligonucleotide adaptors; the resulting ligation products were gel-purified by 15% denaturing PAGE, and reverse-transcribed to produce cDNAs. The cDNAs were sequenced using a Genome Analyzer IIx System, according to the manufacturer’s instructions (BGI-Shenzhen Co. Ltd., Shenzhen, China).
Identification of conserved and novel miRNAs
Raw data were first processed by filtering out low-quality reads, trimming the adaptors, and removing other noise reads, to obtain clean reads. The clean reads were then aligned to the Rfam database to remove other noncoding RNAs, including rRNA, tRNA, and snRNA. The remaining reads were mapped to assembled transcriptome sequences from M. sativa cv. Zhaodong (as described in the section below on transcriptome sequencing, RNA-seq library construction, sequencing and analysis). Mapping reads were retrieved and aligned to plant miRNA sequences from miRBase V21 (Kozomara and Griffiths-Jones 2014) using Bowtie (Langmead and Salzberg 2012), which identified and annotated conserved miRNA genes. To identify novel miRNA genes, first, miRDeep-P retrieved the flanking assembled transcriptome sequences of mapping reads, which were identified as candidate precursors of miRNA (Yang and Li 2011). Second, based on the precursors, the novel miRNAs were identified using MIREAP, and their secondary structures were verified by the software RNAfold, as previously reported (Denman 1993). In addition, plant miRNA criteria mentioned for Arabidopsis (Meyers et al. 2008), and rice (Jeong et al. 2011), were also considered for alfalfa novel miRNA identification.
All miRNA abundances were evaluated and normalized using the tags per million reads (TPM) method, based on BLAST mapping results. The TPM values were calculated as follows: TPM = number of mapped miRNA reads × 106 / number of clean sample reads. The normalized expression was adjusted to 0.01 when miRNA expression (TPM) was zero to avoid negating the subsequent calculation of fold change. The miRNA expression fold changes between stress and control groups (Cold/Control, Freezing/Control) were computed, and chi-squared tests were performed to determine the significance of miRNA expression for each comparison using the R software. The miRNAs with fold change (TPM ratios) ≥ 2 or ≤ 0.5, and p-value ≤ 0.05 were deemed differentially expressed in response to cold and/or freezing stresses, as described by Xie et al. (2014).
Quantitative real-time reverse transcription PCR (qRT-PCR) analysis of miRNA expression
The expression of 12 selected miRNAs from the three conditions were assayed using stem-loop quantitative reverse transcription PCR (qRT-PCR). Primers for all miRNAs, and the reference gene (U6), were designed as shown in Supporting Information, Table S1. Total RNA was extracted from alfalfa grown under the three conditions (control, cold, and freezing) with TRIZOL reagent, according to the manufacturer’s instructions (Invitrogen). These RNAs were reverse transcribed to cDNA, according to the manufacturer’s protocol. qRT-PCR was performed using the LightCycler 480II Detection System (Roche) with a total reaction volume of 20 μl, containing 1 μl of cDNA templates, 8 μM of primer mix, 10.0 μl of 2 × SYBR Green Mix, and 8.7 μl ddH2O. The PCR conditions were set as follows: 95° for 2 min; 40 cycles of 95° for 10 sec, 60° for 30 sec, and 60° for 45 sec. The miRNA expression abundances were determined based on the 2–ΔΔCT method, and relative changes in miRNA expression from the qRT-PCR experiments were calculated. Three biological replicates for each group were run, and each reaction was performed with three technical replicates.
Degradome library construction and target identification
To investigate the potential target mRNAs, two degradome libraries from cold stress and freezing stress samples were constructed, as previously described (Folkes et al. 2012). In brief, poly(A)-enriched RNAs were isolated, and ligated to an RNA oligo-nucleotide adaptor containing a 3′-MmeI recognition site; the ligated products were used to synthesize first-strand cDNA. A short PCR (five cycles) reaction was used to amplify the cDNA, and the product was ligated to a double-stranded DNA adaptor, before being subjected to gel purification again for PCR amplification. The final cDNA library was purified and sequenced on an Illumina GAII by BGI-Shenzhen Co. Ltd (Shenzhen, China).
Adaptor sequences and low quality sequencing reads were removed from the raw reads, and the clean reads were used to identify potentially cleaved targets based on M. sativa assembled transcriptome sequences (described below) by the CleaveLand4 pipeline (Addo-Quaye et al. 2009). Meanwhile, the psRNATarget tool was also used to predict miRNA targets using a set of default parameters (Dai and Zhao 2011). MiRNA target genes were also predicted from M. sativa assembled transcriptome sequences. Prediction targets were used to cross-check the degradome sequencing results.
RNA-seq library construction, sequencing, and analysis
Total RNAs were extracted from three samples using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA), and transcriptome sequencing libraries were constructed according to the manufacturer’s instructions. In brief, short fragments were purified using a MinElute PCR Purification Kit (Qiagen), and eluted in 10 μl of EB buffer (Qiagen). The short fragments were ligated with sequencing adapters, and the desired fragments (around 250 bp) were separated by agarose gel electrophoresis, and purified using a gel extraction kit. Finally, the sequencing library was constructed by linear PCR amplification (15 cycles), and sequenced using the Illumina GAII platform by BGI-Shenzhen Co. Ltd (Shenzhen, China), generating 100-bp pair-end reads. Processing of raw data, removal of adapter sequences, base-calling, and quality value calculations, were performed to produce clean data. Clean reads from three libraries were assembled de novo into contigs using Trinity software with the following parameter: “min_kmer_cov 2” (Haas et al. 2013). To remove redundancy among the Trinity-generated contigs, they were further assembled de novo using iAssembler, with the minimum percent identify (−p) set to 97 (Zheng et al. 2011). The resulting unique transcripts were identified as M. sativa transcriptome unique assembled mRNA sequences. These assembled transcripts were BLAST-searched against combined databases of Arabidopsis, rice, soybean, and Medicago truncatula protein sequences for functional annotation (the e-value was set at 1E-5). Gene ontology (GO) annotations were assigned to the targets based on the GO terms annotated to their corresponding homologs in the combined database, and the GO enrichment analysis of miRNA targets was performed using package topGO on the R platform. Meanwhile, plant transcription factors (TFs) from M. sativa were identified and classified into different families using the iTAK pipeline (http://bioinfo.bti.cornell.edu/tool/itak) (Jin et al. 2014).
Clean reads from the three samples were mapped to the M. sativa assembled transcripts generated by RNA-seq, as previously described, using the TopHat software (Trapnell et al. 2009), and mRNA target gene expressions [as estimated by the fragments per kilobase of exon per million fragments mapped (FPKM) method] across the control, cold, and freezing samples were evaluated using the Cufflinks software (Trapnell et al. 2012). Differential expression analysis was performed using the edgeR package (Robinson et al. 2010) on the R platform, and target genes with fold changes ≥ 2 or ≤ 0.5, with an adjusted p-value ≤ 0.01, were identified as differentially expressed in response to cold, and/or freezing stress.
Data availability
The Medicago sativa (cv. Zhaodong) small RNA sequences, degradome sequences, and transcriptome sequences are all deposited into NCBI SRA with accession number: SRP064230.
Results
High-throughput sequencing of small RNA libraries
Raw reads of three libraries were obtained by high-through sequencing, and were deposited with NCBI SRA under accession number SRP064230. After removing low-quality reads, poly(A) reads, oversized insertions, reads shorter than 18 nt, and adaptor-contaminated reads, 10,823,011, 10,833,023, and 10,781,132 clean reads were generated from the control, cold, and freezing libraries, respectively. These reads included unique reads, and the length distribution of small RNA reads from the three libraries ranged from 18 nt to 28 nt (see Figure S1). The majority of small RNA reads were 20–24 nt sequences, comprising over 80% of the reads. The 21 nt and 24 nt small RNAs comprised the two major classes, which was consistent with previous publications (Fan et al. 2014; Tang et al. 2012; Zhang et al. 2009). Other noncoding RNAs, including rRNA, tRNA, and snRNA, were removed by mapping reads to the Rfam database. The remaining reads were mapped to the alfalfa transcriptome assembled sequences; about 41% of the reads were mappable (Table 1). To identify conserved miRNAs in alfalfa, the mappable reads were aligned to known plant miRNAs in the miRBase database, using Bowtie with no more than one mismatch. In total, 1,597,215, 1,287,494, and 987,515 reads were identified as homologous to known miRNAs from the control, cold, and freezing libraries, respectively. They were identified as 173 conserved miRNA genes from 112 miRNA families (see Table S2). The flanking sequences of the remaining mappable reads were retrieved and analyzed by MIREAP using plant default parameters. In total, 24 novel miRNAs among the predicted RNA hairpins were identified in alfalfa, most of them were 21 nt and 24 nt in length (Table S3).
Table 1. Overview of small RNA sequences in three alfalfa libraries.
| Data Type | Control | Cold | Freezing | |||
|---|---|---|---|---|---|---|
| Total Reads | Unique Reads | Total Reads | Unique Reads | Total Reads | Unique Reads | |
| Clean reads | 10,823,011 | 4,415,425 | 10,833,023 | 4,746,716 | 10,781,132 | 4,296,444 |
| Rfam | 229,925 | 18,696 | 400,604 | 28,495 | 1,947,224 | 38,460 |
| Transcriptome | 4,933,169 | 1,201,107 | 4,509,305 | 1,232,313 | 3,892,472 | 1,219,439 |
| Known miRNA | 1,597,215 | 24,334 | 1,287,494 | 27,292 | 987,515 | 24,278 |
| Novel miRNA | 172,273 | 1355 | 82,573 | 1121 | 62,290 | 944 |
To identify miRNAs involved in alfalfa response to cold and freezing stress, the miRNA expressions of three groups were evaluated and normalized (see Table S4). Compared with the control group, miRNAs with a log2 fold change higher than 1, or less than –1, combined with a p-value less than 0.05, were identified as differentially expressed miRNAs. There were 35 differentially expressed miRNAs in response to cold, and/or freezing stress (Table 2). Among these miRNAs, 12 were regulated by cold stress, while 30 responded to freezing stress. Nine miRNAs responded to cold and freezing stress, three miRNAs were specifically regulated by cold stress, and 23 miRNAs specifically responded to freezing stress. In addition, most (29 miRNAs) were downregulated by cold and/or freezing stress; only six miRNAs were induced by cold and/or freezing stress.
Table 2. The differential expression of miRNA genes in alfalfa in response to cold and/or freezing stresses.
| miRNA | Fold Change | Statistical Significance | ||
|---|---|---|---|---|
| Cold | Freezing | Cold | Freezing | |
| miR156c-3p | −0.62 | −1.02 | ** | ** |
| miR156i-5p | −0.78 | −1.43 | ** | ** |
| miR159a | −1.85 | −1.15 | ** | ** |
| miR159b | 0.00 | 9.80 | ** | |
| miR160e | 0.11 | 1.50 | ** | |
| miR166e-5p | −0.58 | −1.64 | ** | |
| miR166f | 1.52 | 2.81 | ** | ** |
| miR166g-5p | −0.75 | −1.97 | ** | ** |
| miR167a | −1.11 | −1.53 | ** | ** |
| miR167b-5p | −0.94 | −1.99 | ** | ** |
| miR168c-3p | −0.19 | −1.04 | ** | |
| miR171a | −0.99 | 1.43 | ** | |
| miR172a | −1.77 | −2.30 | ** | ** |
| miR172c-3p | −1.21 | −1.82 | ** | ** |
| miR172d-3p | −1.31 | −0.69 | ** | ** |
| miR2119 | −0.90 | 2.08 | ** | |
| miR396a-5p | −1.74 | −2.31 | ** | ** |
| miR396b-3p | −0.71 | −1.16 | * | ** |
| miR398a-5p | −2.28 | 0.17 | ** | |
| miR5037c | 2.01 | 1.63 | ** | * |
| miR5231 | −0.93 | −1.94 | ** | ** |
| miR5232 | 0.10 | −1.01 | * | |
| miR5234 | −0.63 | −1.37 | * | ** |
| miR5239 | −0.85 | −1.58 | ** | ** |
| miR5287b | −0.30 | −1.83 | ** | |
| NmiR0018 | −1.01 | −0.28 | ** | * |
| NmiR0019 | −0.62 | −3.60 | ** | ** |
| NmiR0026 | −0.69 | −1.37 | ** | ** |
| NmiR0028 | −0.84 | −1.07 | ** | ** |
| NmiR0029 | −0.98 | −1.85 | ** | ** |
| NmiR0043 | −0.51 | −1.92 | ** | |
| NmiR0047 | −0.45 | −2.18 | * | ** |
| NmiR0049 | −1.79 | −2.58 | ** | ** |
| NmiR0051 | −2.04 | −2.21 | ** | ** |
| NmiR0053 | −0.52 | −2.20 | * | |
The miRNA abundance was evaluated and normalized using the TPM method, and the expression values of miRNAs whose expression was zero were adjusted to 0.01. The fold changes were computed from cold [log2(Cold/Control)] and freezing [log2(Freezing/Control)] based expression. Meanwhile, chi-squared tests were also performed to test the significance of each comparison, using the R software. A p-value no more than 0.01 was set as the extremely significant level (**), while a p-value greater than 0.01 and no more than 0.05 was defined as significant (*).
qRT-PCR validation of miRNA expression
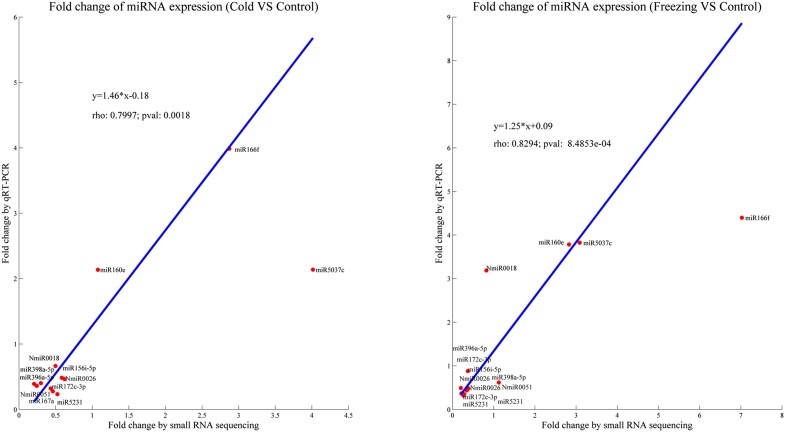
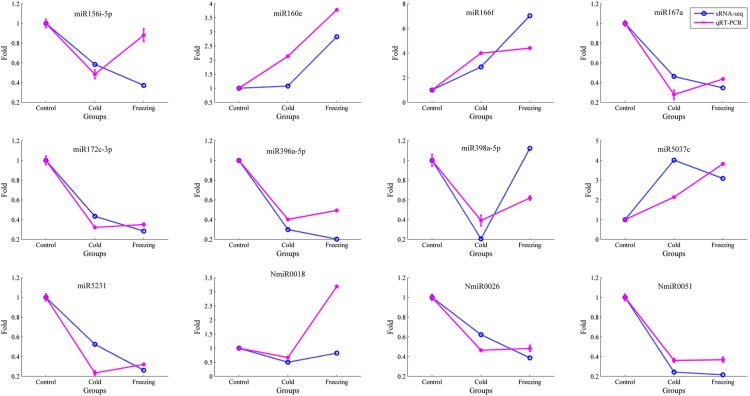
To validate an miRNAs’ function in alfalfa in response to cold and freezing stress, 12 differentially expressed miRNAs were selected for qRT-PCR detection. The means of the correlation coefficients of the qRT-PCR validations, and the high-throughput sequencing results for the miRNAs were as high as 0.80 and 0.83 under cold and freezing stress, respectively, which implied that our miRNA sequencing results were highly reliable (Figure 1). For example, miR167a, miR172c-3p, miR396a-5p, and miR5231 were identified as downregulated by both cold and freezing stress by Illumina sequencing, and qRT-PCR showed the same expression profile in response to cold and freezing stress (Figure 2). Similarly, the qRT-PCR results also validated the expression patterns of miR160e and miR166f, which were upregulated by cold/freezing stress. Notably, the expression of three novel miRNAs (NmiR0018, NmiR0026, and NmiR0051) were characterized and confirmed by qRT-PCR detection. However, there were some miRNAs that showed qRT-PCR results that were inconsistent with the sequencing results, i.e., miR156i-5p, miR398a-5p, and miR5037c. This possibly reflected the different sensitivities of high-throughput sequencing and the qRT-PCR detection method for specific miRNAs.
Figure 1.
Comparison of the expression of 12 miRNAs between small RNA sequencing and qRT-PCR in response to cold and freezing stress. Red dots are plot-based fold changes of each miRNA gene between the abundance from RNA sequencing and qRT-PCR detection. The line correction relationship was computed based on the expression of 12 miRNA genes (blue line).
Figure 2.
Validation of the expression of 12 miRNAs in alfalfa using qRT-PCR. Expressional abundance of each miRNA gene in the control sample was set as 1, and fold changes of each miRNA gene relative to the control sample were calculated. Fold change values greater than 2 or less than 0.5 indicate upregulated or downregulated miRNAs. The blue plots are small RNA sequencing results, and pink represents the qRT-PCR results.
Analysis of alfalfa transcriptome sequences
Three transcriptome libraries were sequenced, and 19,808,866, 23,870,694, 22,350,578 reads were collected from the control, cold, and freezing samples, respectively. After discarding the low-quality raw reads, the remaining reads were assembled de novo using the Trinity software. We obtained 124,821 assembled transcripts, with an N50 of 1392 bp, and an average length of 828 bp; detailed information is provided in Table 3. The assembled alfalfa transcripts were annotated by BLASTX analysis against Arabidopsis, rice, soybean, and M. truncatula proteins, revealing 73,993 (59.3%) transcripts with significant hits. The results showed that the percentage of genes that could be annotated was positively correlated with the length of the genes, as shown in Figure S2. In addition, a BLASTN search was performed against the Mt4.0v1 mRNAs (http://jcvi.org/medicago/) to estimate possible differences in transcript sequences between Medicago sativa and the model plant Medicago truncatula. The results showed that 54.1% (67,492/124,821) of assembled alfalfa transcripts had significant matches with M. truncatula transcripts, most of them having high identity percentages, as shown in Figure S3, which indicated high genetic similarity between the two species.
Table 3. Summary of the de novo assembled alfalfa transcriptome.
| Data Type | Number |
|---|---|
| Total sequence | 124,821 |
| Number of sequences in 200–500 bp | 64,650 |
| Number of sequences in 500–1000 bp | 26,072 |
| Number of sequences more than 1000 bp | 34,099 |
| Minimal length (bp) | 201 |
| Maximal length (bp) | 14,177 |
| N50 (bp) | 1392 |
| Average length (bp) | 828 |
Identification of miRNA targets in alfalfa
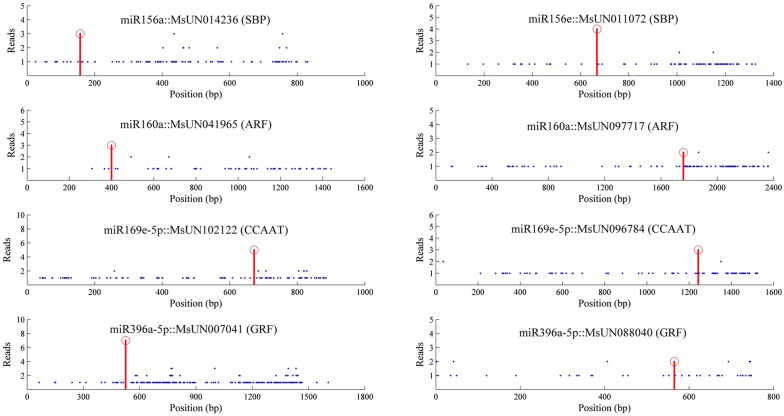
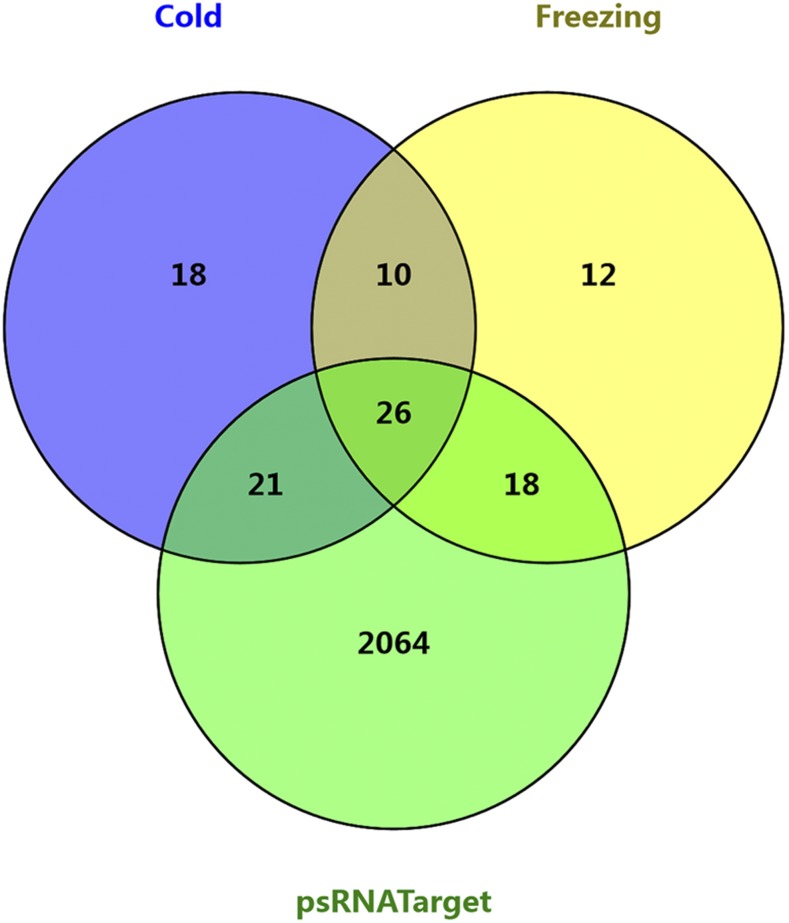
To determine the function of miRNAs in alfalfa, degradome sequencing was used to identify the miRNA targets. After removing reads without adaptor sequences, 10,878,175 and 10,849,379 clean reads were obtained from the two degradome libraries, respectively (cold and freezing libraries); most of them were 20 nt or 21 nt in length (Figure S4). The reads were aligned to the alfalfa transcriptome sequences: 3,008,989 and 2,441,179 reads mapped to the assembled transcriptome sequences, respectively. The CleaveLand pipeline was used for further analysis, and 105 target mRNAs that were potentially cleaved by known miRNAs, and novel miRNAs, were identified from the two degradome libraries (Figure 3, File S1, File S2, and Table S5). Among these genes, 75 targets were identified in the cold stress library, 66 targets were present in freezing stress, and 36 targets were cleaved during both cold and freezing stress (Figure 4). According to the relative abundance of reads at the target sites, they were classified into five categories, 0–4, (see Figure S5). Category 0 has more than one raw read at the position, and the abundance at the position is equal to the maximum on the transcript, only one maximum. Category 1 has a similar definition to Category 0, but there is more than one maximum on the transcript. Category 2 has more than one raw read at the position. The abundance at the position is less than the maximum, but higher than the median for the transcript. Category 3 has more than one raw read at the position. The abundance at the position is equal to, or less than, the median for the transcript. Category 4 has only one raw read at the position. In this study, Category 0 had the most members under both cold and freezing stress, providing high confidence in the degradome sequencing results. To validate target genes from degradome sequencing, miRNA and transcripts were submitted to psRNATarget to identify cleavage using default parameters. There were 65 (65/105, 61.9%) targets from the degradome that were present in the psRNATarget results list (Figure 4). The results indicated that degradome sequencing was an effective method to identify cleaved targets of miRNAs.
Figure 3.
Target plots (t-plots) of miRNA targets identified by degradome sequencing in alfalfa. The values of the Reads axis indicate signature abundances of cleavage sites. Red circles on the Position axis indicate predicted cleavage sites, and red lines indicate signatures produced by miRNA-directed cleavage-based analysis by the CleaveLand4 software.
Figure 4.
Distribution of targets genes identified by two degradome sequencing libraries, and psRNATarget. Target genes of miRNAs from cold and freezing libraries were identified by degradome sequencing, while target genes of psRNATarget were mined by scanning assembled alfalfa transcripts using psRNATarget software with default parameters.
GO analysis and expression analysis of target genes
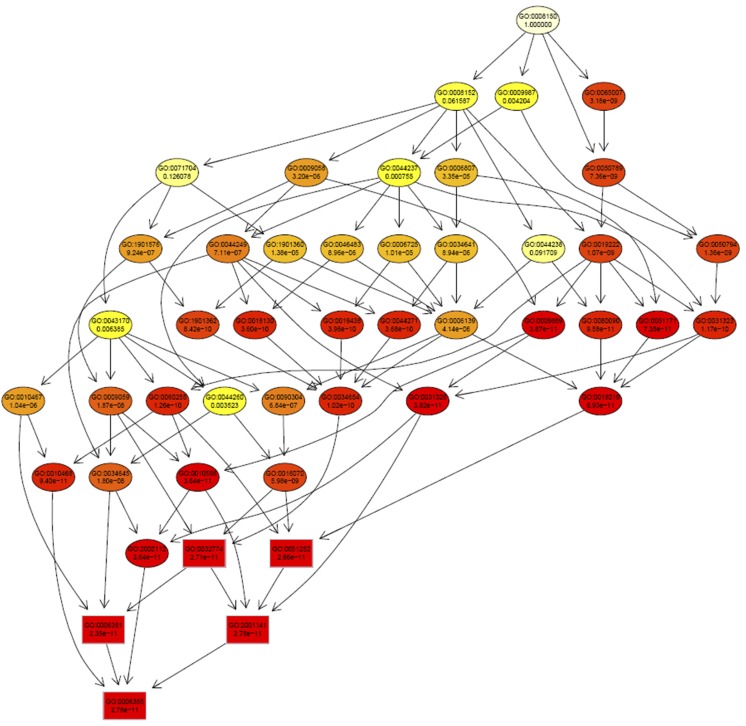
To further understand the functions of miRNAs, the target genes were BLAST searched against a combined database of Arabidopsis, rice, soybean, and M. truncatula. In total, 94 target genes were identified as homologous to functional genes from model plants, and enrichment analysis of GO annotation was performed using the topGO package (Figure 5 and Table S6). As expected, GO terms in the biological process category were highly enriched, including GO:0006355 (regulation of transcription), GO:0031323 (regulation of cellular metabolic process), GO:0050896 (response to stimulus), and GO:0009725 (response to hormone). The most enriched molecular function was GO:0003677 (DNA binding), which is consistent with the results in biological process. Similarly, the most enriched cellular components were GO:0005634 (nucleus), GO:0044424 (intracellular part), and GO:0044464 (cell part). These results implied the possible function of miRNAs in the regulation of transcription during cold and freezing stresses. To investigate transcriptional regulation in detail, we used the iTAK pipeline to scan for TFs: we identified 28 TFs as miRNA target genes (Table 4) that were highly enriched (hypergeometric test, p-value is 6.7E-20). The results were consistent with, and confirmed the results of, the GO enrichment analysis.
Figure 5.
Enrichment analysis results of biological processes involving miRNA target genes in alfalfa. Biological processes were scanned by Fisher’s test using the topGO package, and each GO term was shown in yellow or red according to the p-value (yellow for a high p-value; red for a low p-value). The rectangular boxes represent the top five significant GO terms.
Table 4. TF targets of miRNAs identified by degradome sequencing.
| miRNA Family | Target Gene | TF Family |
|---|---|---|
| miR156 | MsUN014236 | SBP |
| MsUN046666 | SBP | |
| MsUN050762 | SBP | |
| MsUN011072 | SBP | |
| MsUN049469 | SBP | |
| MsUN086910 | SBP | |
| miR160 | MsUN041965 | ARF |
| MsUN097717 | ARF | |
| miR164 | MsUN043080 | NAC |
| MsUN045895 | NAC | |
| miR167 | MsUN007721 | AUX/IAA |
| miR169 | MsUN004554 | CCAAT |
| MsUN014711 | CCAAT | |
| MsUN030848 | CCAAT | |
| MsUN044017 | CCAAT | |
| MsUN096784 | CCAAT | |
| MsUN102122 | CCAAT | |
| miR396 | MsUN007041 | GRF |
| MsUN088040 | GRF | |
| MsUN104762 | C3HC4 | |
| MsUN104763 | C3HC4 | |
| MsUN101411 | MADS-box | |
| miR2645 | MsUN102295 | bZIP |
| miR5205 | MsUN046011 | AP2-EREBP |
| miR5249 | MsUN104998 | bHLH |
| miR530 | MsUN047511 | NAC |
| NmiR0007 | MsUN030863 | GRAS |
| NmiR0028 | MsUN045647 | G2-like |
The assembled alfalfa transcript sequences were submitted to the iTAK software for TF identification using default parameters; 28 TFs from 13 families were identified, and the results of hypergeometric test showed that TF genes were highly enriched among these targets.
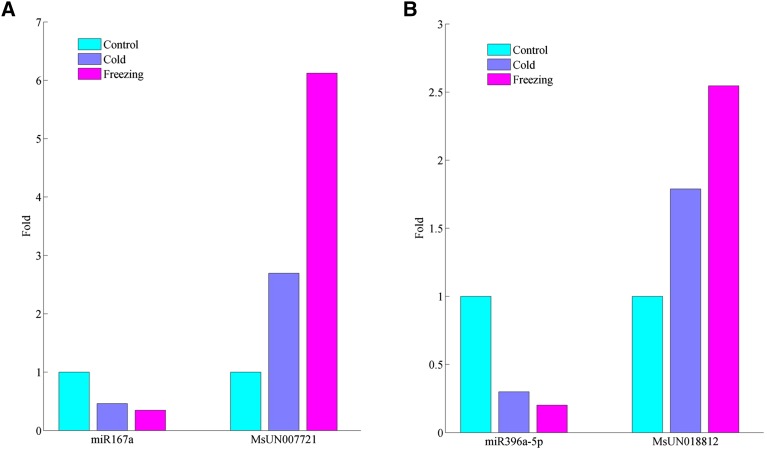
Based on transcriptome sequencing, we evaluated the expression of the target genes under cold and freezing stresses. Among them, six target genes are regulated by four differentially expressed miRNAs (Table S7). Compared with control conditions, the expression of these four miRNAs was depressed under cold and freezing stress, and their targets (except MsUN037724, cleaved by miR396a-5p) were markedly upregulated under cold and freezing stresses, i.e., MsUN007721, and MsUN018812 (Figure 6). In addition, correlation coefficients between miRNAs and their target genes were computed. Negative results (mean value close to –0.41) implied repressive regulation between an miRNA and its target gene. In particular, expression of the transcript MsUN007721 (an AUX/IAA TF), targeted by miRNA167a, was induced significantly under cold and freezing stress, which is consist with the observed repression of miRNA167a. These results suggested that miRNA167 plays an important role by regulating the functions of genes of the auxin signaling pathway (i.e., MsUN007721) in alfalfa in response to cold and freezing stresses. A similar regulation function between miR167 and ARF TF has been confirmed as essential for soybean nodulation (Wang et al. 2015).
Figure 6.
Differential expression of miRNAs and their target genes in response to cold and/or freezing stresses. (A) miRNA167a vs. MsUN007721; (B) miRNA396a-5p vs. MsUN018812. The miRNA expression abundances were evaluated by small RNA sequencing using the TPM method, while target genes were estimated by RNA-seq data using the FPKM method. Their expression levels were adjusted by comparing them with control samples, and relative fold change (FC) values were explored. FCs greater than 2 or less than 0.5 were identified as upregulated or downregulated, respectively.
Discussion
Plant growth and development are threatened by extreme environmental conditions worldwide. In response to stress, plants employ complex systems to adapt to environmental stress, undergoing physiological and biochemical changes in response to unfavorable conditions. MiRNAs are important regulators involved in various stress responses that have received increasing attention, especially with the development of high-throughput sequencing technology. Freezing stress is a common environmental stress of plants that affects plant growth, development, and survival, and particular limits perennial plants. Recently, miRNAs involved in freezing stress have been investigated in many plants, mainly in model plants such as Arabidopsis (Mallory and Vaucheret 2006; Sunkar and Zhu 2004), rice (Lv et al. 2010), wheat (Tang et al. 2012; Tang et al. 2011), and M. truncatula (Formey et al. 2014; Lelandais-Brière et al. 2009; Reynoso et al. 2013; Z. Zhou et al. 2008); however, few reports have been published about alfalfa, a perennial legume forage (Fan et al. 2014; Long et al. 2015). Here, we presented a comprehensive analysis of miRNAs, and their targets, in the freezing tolerant alfalfa cultivar Zhaodong, under cold and freezing stress, using high-throughput sequencing. The results revealed that miRNAs are indeed affected by freezing stress, implying their role in this process. In addition, targets of miRNAs, including TFs, enzymes, and nodulation genes, were identified by degradome sequencing. Taken together, these results provide novel insights into the regulatory mechanism of cold acclimation and freezing tolerance in alfalfa, mediated by miRNAs.
Different expression patterns of miRNAs involving in freezing stress in alfalfa
Based on our sequencing results, 35 miRNAs were differentially regulated by cold and/or freezing stress in alfalfa. Notably, most miRNAs (29/35, 83%) were downregulated by cold and/or freezing stress, which is consist with previous reports in other plants (Chen et al. 2012; Long et al. 2015; Lv et al. 2010; Zhang et al. 2009; X. Zhou et al. 2008). Among these miRNAs, miRNAs responsive to cold stress were more conserved across plants, for instance, miR156, miR159, miR167, miR172, miR396, and miR398, implying their consistent function in cold stress. For example, the Cu/Zn superoxide dismutase-encoding gene, which is cleaved by miR398 in plants, acts as a scavenger of reactive oxygen species (ROS), playing an important role in plant abiotic stress (Dugas and Bartel 2008; Sunkar et al. 2006). Other miRNAs that were specifically regulated by freezing stress, and which were mainly Medicago-specific, included miR5231, miR5232, miR5234, miR5239, miR5287, and novel miRNAs (NmiR0019, NmiR0026, NmiR0028, NmiR0029, NmiR0043, NmiR0047, NmiR0053). The results suggested that they play important roles in the freezing tolerance of alfalfa. However, their functions have been reported rarely in Medicago or other plants, some were even identified in the present study for the first time. Thus, their regulatory functions need to be determined in depth, which would be valuable for breeding alfalfa with improved freezing tolerance. In addition, there were also some miRNAs that were upregulated by cold and/or freezing stress, such as miR160, miR166, miR171, miR2119, and miR5037, which might regulate genes negatively involved in the freezing tolerance of alfalfa.
TF targeted by alfalfa miRNAs involved in freezing tolerance
With development of high-throughput sequencing technology, degradome sequencing has been used widely to identify targets of miRNAs in plants. In the present study, we identified 105 targets from cold stress, and freezing stress libraries. Based on the targets’ functional annotations, we found that conserved miRNAs were more likely to target TFs involved in regulating plant growth and stress response, which is consistent with previous reports in other plants (Aung et al. 2015; Fang et al. 2014; Tang et al. 2012; Wang et al. 2015) (see Table 4). Degradome sequencing identified TF members from families characterized as targets of miRNAs, including SBP, ARF, NAC and GRF. In Arabidopsis and rice, miR156 participates in plant growth and development by cleaving SBP TFs (Ling and Zhang 2012; Xing et al. 2010). Similarly, miR160 and miR167 target auxin response factors (ARF and AUX/IAA), involved in plant development process (Rubio-Somoza et al. 2009; Wang et al. 2015; Yang et al. 2006). In this study, eight SBP, ARF and AUX/IAA TFs were identified by high-throughput sequencing, which indicated that the conserved miRNAs might regulate alfalfa cold and freezing responses by controlling alfalfa developmental process, which would correlate with their functions in other model plants. Similarly, NAC TFs are plant-specific, with important roles in plant development and stress response processes. Two NAC TFs were identified as targets of miR164, which was slightly suppressed by cold stress, implying that NAC TFs were positively regulated during the cold response (Fang et al. 2014). In addition to targeting TFs, some genes involved in stress responses and metabolic processes were also cleaved by miRNAs in alfalfa. For instance, the pentatricopeptide repeat (PPR) protein is a negative regulator of abscisic acid (ABA) signaling, which has a positive on plant responses to abiotic stresses (Jiang et al. 2014; Sechet et al. 2015). In total, eight PPR genes were identified as targets of NmiR0018 and NmiR0041 (see Table S8), implying that these miRNAs function in cold and freezing stress by controlling the expression of PPR genes.
Freezing response miRNAs and their targets involved in the nodulation process
As a legume crop, alfalfa is able to establish a symbiotic association with nitrogen-fixing bacteria, resulting in development of a novel plant organ, termed the root nodule (Soyano and Hayashi 2014). Within nodules, the symbiotic nitrogen fixation relationship between the plant and bacteria is sensitive to environmental stress, such as salt, drought, and cold stress (Kaur et al. 2014; Zhang et al. 2014). In particular, nodules of alfalfa are perennial, and are capable of continuous growth, helping alfalfa to undergo dormancy, and to survive hard winters. However, the molecular mechanisms of symbiotic nitrogen fixation (SNF) in response to cold stress have received relatively little attention. In Medicago, some TFs were characterized as being involved in nodulation, including GRAS (Cerri et al. 2012; Heckmann et al. 2006; Hirsch et al. 2009; Liu et al. 2011), AP2/ERF (Middleton et al. 2007), and CCAAT TFs (HAP2-1/NF-YA) (Battaglia et al. 2014; Laporte et al. 2014), etc. Recently, miRNAs were also demonstrated to participate in the nodulation process, such as miR169 and miR172, which target the NF-YA/HAP2 and AP2 TFs (Combier et al. 2006; Reynoso et al. 2013; Wang et al. 2014). In this study, we confirmed the relationship between miRNAs and mRNAs; for instance, GRAS, AP2/ERF, and CCAAT TFs were regulated by NmiR0007, miR5205, and miR169. Significantly, six CCAAT TFs were cleaved by miR169, as identified by degradome sequencing (see Table 2). The results confirmed the function of miR169 as regulating the expression of CCAAT TFs (NF-YA/HAP2), which implied that miR169 responded to cold stress via the SNF process (Lelandais-Brière et al. 2009; Zhang et al. 2014).
Conclusions
In summary, we investigated miRNAs and their targets by high-throughput sequencing technology during the cold and freezing response of alfalfa. We identified the expression profiles of 197 miRNAs (173 known miRNAs, and 24 novel miRNAs); 35 miRNAs were identified as cold- and/or freezing-responsive miRNAs. Using degradome sequencing, 105 functional genes were found to be cleaved by these miRNAs; most of them were transcription factors involved in plant development and abiotic response processes, which implied important roles in alfalfa freezing tolerance. Some differentially expressed miRNAs and their targets were identified as participating in SNF, which indicated that SNF might aid freezing tolerance in alfalfa. These findings provided valuable information for exploring the molecular mechanisms of the cold and freezing response, and also represent a foundation for future application of miRNAs to improve freezing tolerance in alfalfa or related plants.
Supplementary Material
Acknowledgments
This work was supported by grants from the Natural and Science Foundation of China (nos. 31302019 and 31470571), the China Postdoctoral Science Foundation (2015M571430), the MOST 863 project (2013AA102607-5), and the Heilongjiang Province Postdoctoral Science Foundation (no. LBH-Z14126).
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.025981/-/DC1
Communicating editor: M. Warburton
Literature Cited
- Addo-Quaye C., Eshoo T. W., Bartel D. P., Axtell M. J., 2008. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18: 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C., Miller W., Axtell M. J., 2009. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25: 130–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung B., Gruber M. Y., Amyot L., Omari K., Bertrand A., et al. , 2015. MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnol. J. 13: 779–790. [DOI] [PubMed] [Google Scholar]
- Babar I. A., Slack F. J., Weidhaas J. B., 2008. miRNA modulation of the cellular stress response. Future Oncol. 4: 289–298. [DOI] [PubMed] [Google Scholar]
- Battaglia M., Rípodas C., Clúa J., Baudin M., Aguilar O. M., et al. , 2014. A nuclear factor Y interacting protein of the GRAS family is required for nodule organogenesis, infection thread progression, and lateral root growth. Plant Physiol. 164: 1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castonguay Y., Dube M. P., Cloutier J., Bertrand A., Michaud R., et al. , 2013. Molecular physiology and breeding at the crossroads of cold hardiness improvement. Physiol. Plant. 147: 64–74. [DOI] [PubMed] [Google Scholar]
- Cerri M. R., Frances L., Laloum T., Auriac M. C., Niebel A., et al. , 2012. Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol. 160: 2155–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Zhang X., Zhang N., Wang S., Yin G., et al. , 2015a Combined small RNA and degradome sequencing reveals novel miRNAs and their targets in the high-yield mutant wheat strain Yunong 3114. PLoS One 10: e0137773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Han G., Shang C., Li J., Zhang H., et al. , 2015b Proteomic analyses reveal differences in cold acclimation mechanisms in freezing-tolerant and freezing-sensitive cultivars of alfalfa. Front. Plant Sci. 6: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhang Y., Ren Y., Xu J., Zhang Z., et al. , 2012. Genome-wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem. Biophys. Res. Commun. 417: 892–896. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B. H., Hong X., et al. , 2003. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17: 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier J.-P., Frugier F., de Billy F., Boualem A., El-Yahyaoui F., et al. , 2006. MtHAP2–1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 20: 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Zhao P. X., 2011. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39: W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denman R. B., 1993. Using RNAFOLD to predict the activity of small catalytic RNAs. Biotechniques 15: 1090–1095. [PubMed] [Google Scholar]
- Dugas D. V., Bartel B., 2008. Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol. Biol. 67: 403–417. [DOI] [PubMed] [Google Scholar]
- Fan W., Zhang S., Du H., Sun X., Shi Y., et al. , 2014. Genome-wide identification of different dormant Medicago sativa L. microRNAs in response to fall dormancy. PLoS One 9: e114612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Xie K., Xiong L., 2014. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J. Exp. Bot. 65: 2119–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkes L., Moxon S., Woolfenden H. C., Stocks M. B., Szittya G., et al. , 2012. PAREsnip: a tool for rapid genome-wide discovery of small RNA/target interactions evidenced through degradome sequencing. Nucleic Acids Res. 40: e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formey D., Sallet E., Lelandais-Briere C., Ben C., Bustos-Sanmamed P., et al. , 2014. The small RNA diversity from Medicago truncatula roots under biotic interactions evidences the environmental plasticity of the miRNAome. Genome Biol. 15: 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S. J., Zarka D. G., Stockinger E. J., Salazar M. P., Houghton J. M., et al. , 1998. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16: 433–442. [DOI] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., et al. , 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8: 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajela R. K., Horvath D. P., Gilmour S. J., Thomashow M. F., 1990. Molecular cloning and expression of cor (Cold-Regulated) genes in Arabidopsis thaliana. Plant Physiol. 93: 1246–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann A. B., Lombardo F., Miwa H., Perry J. A., Bunnewell S., et al. , 2006. Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142: 1739–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Muñoz A., Heckmann A. B., Downie J. A., et al. , 2009. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D. H., Park S., Zhai J., Gurazada S. G., De Paoli E., et al. , 2011. Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23: 4185–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S. C., Mei C., Wang X. F., Zhang D. P., 2014. A hub for ABA signaling to the nucleus: significance of a cytosolic and nuclear dual-localized PPR protein SOAR1 acting downstream of Mg-chelatase H subunit. Plant Signal. Behav. 9: e972899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Zhang H., Kong L., Gao G., Luo J., 2014. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42: D1182–D1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J. H., Park C. M., 2013. HOS1-mediated activation of FLC via chromatin remodeling under cold stress. Plant Signal. Behav. 8: e27342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S., Sharma B., Gupta A., Kaur S., Kaur J., 2014. Nodule metabolism in cold stress tolerant and susceptible chickpea cultivars. Symbiosis 64: 33–42. [Google Scholar]
- Khraiwesh B., Zhu J. K., Zhu J., 2012. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S., 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte P., Lepage A., Fournier J., Catrice O., Moreau S., et al. , 2014. The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J. Exp. Bot. 65: 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. H., Henderson D. A., Zhu J. K., 2005. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17: 3155–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Xiong L., Gong Z., Ishitani M., Stevenson B., et al. , 2001. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo–cytoplasmic partitioning. Genes Dev. 15: 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelandais-Brière C., Naya L., Sallet E., Calenge F., Frugier F., et al. , 2009. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell 21: 2780–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling L. Z., Zhang S. D., 2012. Exploring the evolutionary differences of SBP-box genes targeted by miR156 and miR529 in plants. Genetica 140: 317–324. [DOI] [PubMed] [Google Scholar]
- Liu H. H., Tian X., Li Y. J., Wu C. A., Zheng C. C., 2008. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14: 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Kohlen W., Lillo A., Op den Camp R., Ivanov S., et al. , 2011. Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23: 3853–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R. C., Li M. N., Kang J. M., Zhang T. J., Sun Y., et al. , 2015. Small RNA deep sequencing identifies novel and salt-stress-regulated microRNAs from roots of Medicago sativa and Medicago truncatula. Physiol. Plant. 154: 13–27. [DOI] [PubMed] [Google Scholar]
- Lu S., Sun Y. H., Chiang V. L., 2008. Stress-responsive microRNAs in Populus. Plant J. 55: 131–151. [DOI] [PubMed] [Google Scholar]
- Luo X., Feng C., Li H., Sha W., 2004. Study on changes of SOD and proline activity during low temperature stress on Medicago sativa L. cv. Zhaodong. Grassland China 26: 79–81. [Google Scholar]
- Lv D. K., Bai X., Li Y., Ding X. D., Ge Y., et al. , 2010. Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 459: 39–47. [DOI] [PubMed] [Google Scholar]
- Mallory A. C., Vaucheret H., 2006. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 38(Suppl): S31–S36. [DOI] [PubMed] [Google Scholar]
- Meyers B. C., Axtell M. J., Bartel B., Bartel D. P., Baulcombe D., et al. , 2008. Criteria for annotation of plant MicroRNAs. Plant Cell 20: 3186–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton P. H., Jakab J., Penmetsa R. V., Starker C. G., Doll J., et al. , 2007. An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Hasegawa P. M., 2008. Regulation of cold signaling by sumoylation of ICE1. Plant Signal. Behav. 3: 52–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo V., Szittya G., Moxon S., Miozzi L., Moulton V., et al. , 2010. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. 62: 960–976. [DOI] [PubMed] [Google Scholar]
- Pennycooke J. C., Cheng H., Stockinger E. J., 2008. Comparative genomic sequence and expression analyses of Medicago truncatula and alfalfa subspecies falcata COLD-ACCLIMATION-SPECIFIC genes. Plant Physiol. 146: 1242–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajwanshi R., Chakraborty S., Jayanandi K., Deb B., Lightfoot D. A., 2014. Orthologous plant microRNAs: microregulators with great potential for improving stress tolerance in plants. Theor. Appl. Genet. 127: 2525–2543. [DOI] [PubMed] [Google Scholar]
- Reynoso M. A., Blanco F. A., Bailey-Serres J., Crespi M., Zanetti M. E., 2013. Selective recruitment of mRNAs and miRNAs to polyribosomes in response to rhizobia infection in Medicago truncatula. Plant J. 73: 289–301. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I., Cuperus J. T., Weigel D., Carrington J. C., 2009. Regulation and functional specialization of small RNA–target nodes during plant development. Curr. Opin. Plant Biol. 12: 622–627. [DOI] [PubMed] [Google Scholar]
- Sechet J., Roux C., Plessis A., Effroy D., Frey A., et al. , 2015. The ABA-deficiency suppressor locus HAS2 encodes the PPR protein LOI1/MEF11 involved in mitochondrial RNA editing. Mol. Plant 8: 644–656. [DOI] [PubMed] [Google Scholar]
- Shamimuzzaman M., Vodkin L., 2012. Identification of soybean seed developmental stage-specific and tissue-specific miRNA targets by degradome sequencing. BMC Genomics 13: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., Liu Y., Zhang J., Song L., Guo C., 2015. Genome-wide analysis of the AP2/ERF superfamily genes and their responses to abiotic stress in Medicago truncatula. Front. Plant Sci. 6: 1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T., Hayashi M., 2014. Transcriptional networks leading to symbiotic nodule organogenesis. Curr. Opin. Plant Biol. 20: 146–154. [DOI] [PubMed] [Google Scholar]
- Sun Z., He Y., Li J., Wang X., Chen J., 2015. Genome-wide characterization of rice black streaked dwarf virus-responsive microRNAs in rice leaves and roots by small RNA and degradome sequencing. Plant Cell Physiol. 56: 688–699. [DOI] [PubMed] [Google Scholar]
- Sunkar R., Zhu J. K., 2004. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Kapoor A., Zhu J. K., 2006. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18: 2051–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Li Y. F., Jagadeeswaran G., 2012. Functions of microRNAs in plant stress responses. Trends Plant Sci. 17: 196–203. [DOI] [PubMed] [Google Scholar]
- Tang Z., Zhang L., Yang D., Zhao C., Zheng Y., 2011. Cold stress contributes to aberrant cytokinesis during male meiosis I in a wheat thermosensitive genic male sterile line. Plant Cell Environ. 34: 389–405. [DOI] [PubMed] [Google Scholar]
- Tang Z., Zhang L., Xu C., Yuan S., Zhang F., et al. , 2012. Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol. 159: 721–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M. F., 2010. Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 154: 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang L., Zou Y., Chen L., Cai Z., et al. , 2014. Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell 26: 4782–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li K., Chen L., Zou Y., Liu H., et al. , 2015. MicroRNA167-directed regulation of the auxin response factors GmARF8a and GmARF8b is required for soybean nodulation and lateral root development. Plant Physiol. 168: 984–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Wang Q., Sun R., Zhang B., 2014. Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J. Exp. Bot. 66: 789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S., Salinas M., Hohmann S., Berndtgen R., Huijser P., 2010. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22: 3935–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. H., Han S. J., Yoon E. K., Lee W. S., 2006. Evidence of an auxin signal pathway, microRNA167–ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 34: 1892–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Li L., 2011. miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics 27: 2614–2615. [DOI] [PubMed] [Google Scholar]
- Zhang J., Xu Y., Huan Q., Chong K., 2009. Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genomics 10: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Wang Y., Li K., Zou Y., Chen L., et al. , 2014. Identification of cold-responsive miRNAs and their target genes in nitrogen-fixing nodules of soybean. Int. J. Mol. Sci. 15: 13596–13614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Xu Z., Mo Q., Zou C., Li W., et al. , 2013. Combined small RNA and degradome sequencing reveals novel miRNAs and their targets in response to low nitrate availability in maize. Ann. Bot. (Lond.) 112: 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Zhao L., Gao J., Fei Z., 2011. iAssembler: a package for de novo assembly of Roche-454/Sanger transcriptome sequences. BMC Bioinformatics 12: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Wang G., Sutoh K., Zhu J. K., Zhang W., 2008. Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim. Biophys. Acta 1779: 780–788. [DOI] [PubMed] [Google Scholar]
- Zhou Z. S., Huang S. Q., Yang Z. M., 2008. Bioinformatic identification and expression analysis of new microRNAs from Medicago truncatula. Biochem. Biophys. Res. Commun. 374: 538–542. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Medicago sativa (cv. Zhaodong) small RNA sequences, degradome sequences, and transcriptome sequences are all deposited into NCBI SRA with accession number: SRP064230.