Abstract
Degenerative retinal diseases, such as glaucoma, age-related macular degeneration, and diabetic retinopathy, have complex etiologies with environmental, genetic, and epigenetic contributions to disease pathology. Much effort has gone into elucidating both the genetic and the environmental risk factors for these retinal diseases. However, little is known about how these genetic and environmental risk factors bring about molecular changes that lead to pathology. Epigenetic mechanisms have received extensive attention of late for their promise of bridging the gap between environmental exposures and disease development via their influence on gene expression. Recent studies have identified epigenetic changes that associate with the incidence and/or progression of each of these retinal diseases. Therefore, these epigenetic modifications may be involved in the underlying pathological mechanisms leading to blindness. Further genome-wide epigenetic studies that incorporate well-characterized tissue samples, consider challenges similar to those relevant to gene expression studies, and combine the genome-wide epigenetic data with genome-wide genetic and expression data to identify additional potentially causative agents of disease are needed. Such studies will allow researchers to create much-needed therapeutics to prevent and/or intervene in disease progression. Improved therapeutics will greatly enhance the quality of life and reduce the burden of disease management for millions of patients living with these potentially blinding conditions.
Keywords: epigenetics, aging, neurodegeneration, glaucoma, age-related macular degeneration, diabetic retinopathy, genome-wide, chromatin, DNA methylation, histone modification
Background
Proper control of gene expression is fundamental to an organism’s viability, affecting every function of each cell, from maintaining homeostasis to responding to environmental changes. One level of control used by cells to regulate gene expression is to alter the structure of the DNA–protein–RNA complex known as chromatin (Table 1). Gene expression changes in response to environmental stimuli do not necessarily require remodeling of chromatin. However, mechanisms that underlie such gene expression changes by modifying the chromatin structure without changing the nucleotide sequence are called epigenetic mechanisms. To better understand the role of epigenetic mechanisms in mammalian gene regulation, one must first understand the role of chromatin structure in the eukaryotic cell.
Table 1.
Definitions of key terminology.
| TERM | DEFINITION |
|---|---|
| Acetylation | The addition of an acetyl group to lysine amino acid residues |
| Chromatin | The DNA molecule in combination with the proteins and RNA bound to it |
| Epigenetic modifications | Stable changes to chromatin structure that do not change the nucleic acid sequence, such as DNA methylation and histone modifications |
| Epigenetics | The study of epigenetic modifications |
| Fundus photograph | Picture of the back of the eye, including the retina, macula, fovea, and optic disk |
| H3K9me3 | Histone modification associated with transcriptionally inactive chromatin: three methyl groups attached to the lysine at amino acid position 9 of histone H3 |
| Histone | One of the five proteins that are the primary protein components of chromatin |
| Heterochromatin | Structurally condensed, transcriptionally inactive regions of chromatin |
| HP1 | Heterochromatin protein 1, protein associated with heterochromatin which maintains the heterochromatic state |
| Methylation | The addition of a methyl group to cytosine DNA residues; or the addition of one or more methyl groups to histone lysine or arginine amino acid residues |
| Optical coherence tomography | A method for capturing a cross-sectional image of tissue |
| Phosphorylation | The addition of a phosphoryl group to lysine, threonine, or tyrosine amino acid residues |
| Pioneering transcription factors | Transcription factors which bind to repressed, heterochromatic DNA to facilitate activation of a gene |
| Transcription factor | 1) any protein involved in transcription; 2) sequence-specific DNA binding proteins which facilitate the transcriptional activation or repression of target genes |
| Ubiquitination | The addition of one or more ubiquitin groups to lysine amino acid residues |
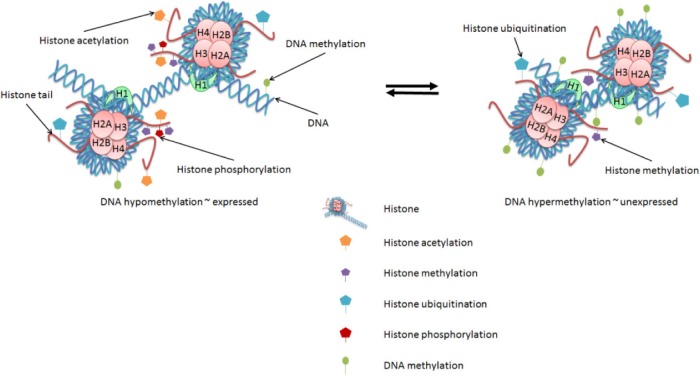
In eukaryotic cells, various proteins play important roles in establishing and maintaining the structure of DNA within a cell. Histone proteins are the primary protein components of chromatin and are responsible for condensing the DNA into nucleosomes that form a beads-on-a-string conformation (Fig. 1). Nucleosomes are formed by the wrapping of ~150 bp of the DNA double helix around histone octamers comprising two each of the core histone proteins H2A, H2B, H3, and H4 (Fig. 1). An additional histone, H1, is known as the linker histone because it binds to the DNA between the histone octamer beads and contributes to further condensation of the DNA. The positions of nucleosomes along the DNA helix affect the ability of other DNA-binding proteins, such as transcription factors (TFs), to access specific sequences of DNA, thereby influencing the expression of genes. Nucleosomes are highly repressive to transcription by their restriction of DNA accessibility.1
Figure 1.
Chromatin modifications affect gene expression. Changes in epigenetic marks, such as DNA methylation and histone modifications, each contribute to the regulation of gene expression. DNA methylation in the promoter regions of genes is generally associated with decreased gene expression. Histone modifications can be either activating or repressive. Histone acetylation and phosphorylation are generally associated with active genes; histone methylation and ubiquitination arrangements are associated with either active or repressed genes.
Additionally, the N-terminal ends, or tails, of the core histones are less structured and have regulatory roles (Fig. 1). Posttranslational modifications (PTMs) primarily to the N-terminal tails of histone proteins—including methylation, phosphorylation, acetylation, and ubiquitination—affect the conformation of the histone-DNA complex and, therefore, the accessibility of the DNA. For example, acetylation of histone tails is associated with actively expressed genes and generally results in the histones being more negatively charged and, therefore, binding to the DNA and each other less tightly.2 This allows more TF–DNA binding and local expression of relevant genes.
Histone modifications also serve as markers recognized by specific TFs involved in activation or repression of eukaryotic gene expression. Much effort of late has gone into deciphering the so-called histone code, and thereby understanding the roles of each of these marks at each of their many respective positions on histone tails within the context of the genome.3 Acetylation marks are associated with active promoters and are among the most well-studied histone modifications. Histone phosphorylation is also associated with active transcription.4 Methylation or ubiquitination of histone tails is often associated with either activation or repression of gene expression, depending on the specific amino acid residue being modified, the extent to which that given residue is modified, and the position of that residue within a gene or extragenic region.4,5
A number of writers, readers, and erasers are involved in adding, interpreting, and/or removing epigenetic modifications on chromatin. For example, histone acetyl-transferases add (write) and histone deacetylases (HDACs) remove (erase) acetyl groups to/from histone lysine residues. Bromodomain-containing DNA-binding proteins, such as the TFIID subunit TAF1, specifically recognize acetylated lysine residues on histone tails (read) to facilitate promoter recognition and transcriptional activation.6,7 Aberrant histone acetylation has been implicated in various pathologies, and HDAC inhibitors are in clinical trial to treat cancer and have been suggested for the treatment of retinal degenerative diseases.8 One class of HDACs known as sirtuins have been implicated in the aging process, including evidence that suggests that they facilitate the lifespan-extending effects of calorie restriction in model organisms and that their activation may be beneficial for age-related diseases, including neurode-generative diseases.9
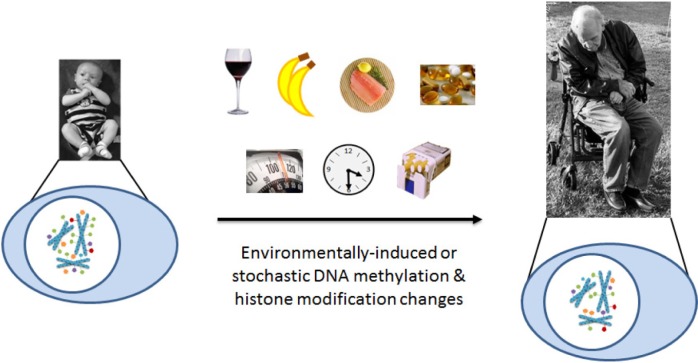
In addition to histone modifications, modifications to the DNA itself have profound effects on gene expression. DNA methylation—the addition of a methyl group to the C5 position of cytosine bases that are followed by guanosine bases within the DNA sequence (5′-CpG-3′)—is strongly associated with the repression of gene expression. CpG-rich regions of the genome, referred to as CpG islands, are often found in the 5′ regulatory regions of genes.10 Actively expressed genes generally have unmethylated CpG islands near their transcription start sites, whereas unexpressed genes generally have methylated CpG islands near their transcription start sites. Methylation patterns can be heritable across both meiotic and mitotic cell divisions. In genomic imprinting, for example, methylation is used to ensure that single copies of particular genes are repressed during egg or sperm cell generation in a maternal- or paternal-specific pattern, which persists into the adult.11,12 DNA methylation is also used during development to program cell differentiation by specifying the particular subset of genes to be expressed by each cell type.13 Of particular relevance to the development of age-related disease is that environmental factors induce DNA methylation changes throughout an organism’s lifespan.14 Figure 2 illustrates a number of environmental factors associated with retinal degenerations such as age-related macular degeneration (AMD) that have also been shown to associate with changes in DNA methylation, including tobacco smoke, alcohol, diet (particularly omega-3 fatty acids and antioxidants), and obesity.15–27 The cumulative effects of each of these factors on the epigenetic landscape of an individual’s genome may gradually perturb the expression profile of a tissue sufficiently to transform it from an otherwise healthy state to a diseased state.
Figure 2.
Epigenetics in aging and age-related disease. Stable epigenetic marks may be the link between genetic and environmental processes involved in the development of age-related diseases, such as AMD. Environmental influences contribute to epigenetic changes that accumulate with age. Risk factors, such as diet, obesity, smoking, sun exposure, and age, may elicit epigenetic changes that accumulate over a lifetime, eventually resulting in altered expression of genes involved in the disease process. These environmental influences contribute to epigenetic modifications, such as DNA methylation (green ovals), histone methylation (purple pentagons), histone acetylation (orange pentagons), histone ubiquitination (blue pentagons), and histone phosphorylation (red pentagons). The epigenetic changes that accumulate throughout the genome may associate with transcriptional changes at the affected genomic loci. Such expression changes at disease-relevant loci may promote either protection against or progression of age-related disease. The sum of these effects over time may then perturb the normal, healthy homeostasis enough to result in the development and/or progression of diseases such as AMD.
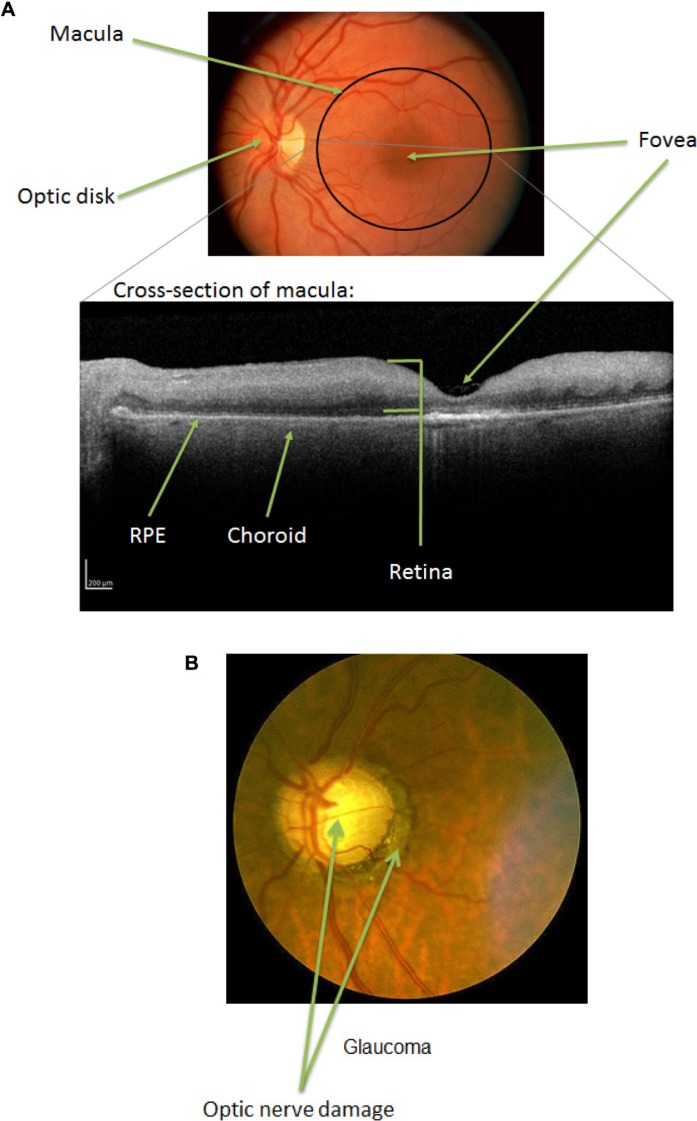
Such gradual epigenetic modifications may play significant roles in the development of age-related retinal diseases. The retina is a region at the back of the eye onto which the lens focuses an image and which contains the photoreceptor cells that transmit the image as a signal to the brain. The macula is the region of the retina responsible for central vision, with the fovea at its center. Figure 3A shows a fundus image (photograph of the back of the eye) and an optical coherence tomography (OCT) image (which shows a cross-section of the macula) of a healthy retina. In the OCT image, the retina is the upper cellular layer, with the retinal pigment epithelium (RPE) and choroid below (posterior to) the retina. The choroid comprises blood vessels that support the retina, with the RPE serving as the inner blood–retinal barrier through which nutrients and waste must pass to transfer between the retina and the choroidal blood supply.28
Figure 3.
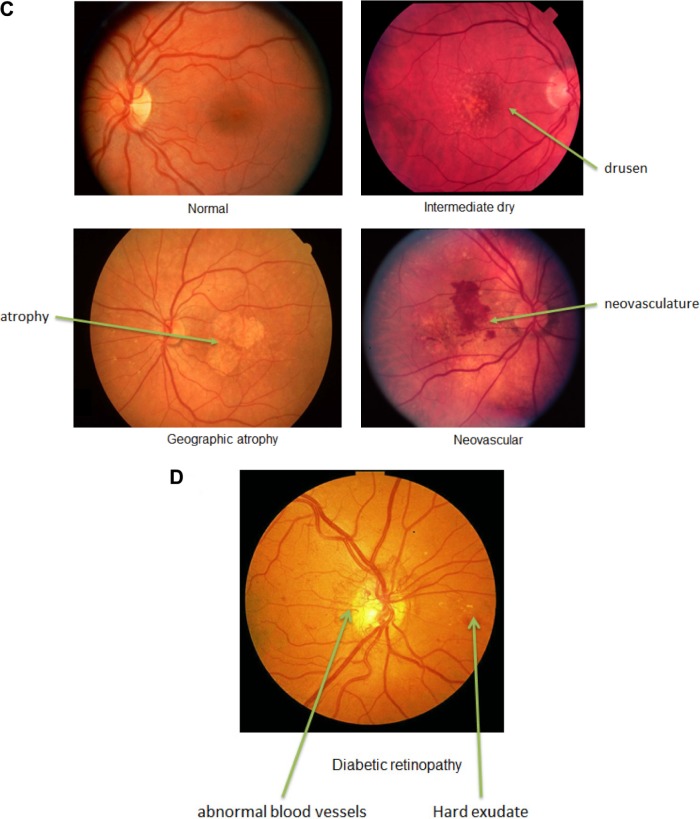
Progression of retinal disease. (A) Fundus and OCT images of normal eye with anatomy labels. (B) Fundus image of an eye with glaucoma. Note the optic nerve damage. (C) Fundus images of normal, intermediate, and advanced (GA and neovascular) AMD. Note the drusen deposits, atrophy, and neovascularization. (D) Fundus image of an eye with DR. Note the abnormal blood vessels and hard exudates. Photographs taken from DeAngelis laboratory patient cohorts. The study protocol was reviewed and approved by the Institutional Review Board at the University of Utah and conforms to the tenets of the Declaration of Helsinki.
Definition of Epigenetics
As stated earlier, changes to DNA structure that do not involve changes to the DNA sequence are considered epigenetic. The definition of the term epigenetic has been met with some controversy as the study of chromatin modifications has advanced. The more conservative definitions limit the term to refer specifically to inheritable changes in chromatin structure, and the more liberal definitions use the term to encompass any nonsequence changes to chromatin structure. Two distinct definitions are worth noting. A consensus definition of epigenetics from a 2008 meeting at Cold Spring Harbor Laboratories resulted in the following: “An epigenetic trait is a stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence.”29 Note the requirement in this definition for the changes to be stably heritable. In contrast, the NIH Roadmap Epigenomics Mapping Consortium defines epigenetics for their purposes as, “both heritable changes in gene activity and expression (in the progeny of cells or of individuals) and also stable, long-term alterations in the transcriptional potential of a cell that are not necessarily heritable.”30 The use of the term epigenetics to refer to non-heritable chromatin changes in postmitotic cells, as discussed herein, violates the conservative requirement for heritability and therefore requires a more liberal application of the term in line with the NIH Roadmap Consortium definition.
The epigenetic landscape of a particular gene or genome can be affected by various factors, including genetic and environmental influences, as noted earlier. The activation and repression of gene expression is a complex interplay of numerous factors that must be tightly regulated to ensure proper response to environmental signals. Upon detection of an environmental signal, sequence-specific DNA-binding transcription activators or repressors (TFs) are activated alternatively by various methods, such as: (1) Directly binding to the signal, e.g., the nuclear receptor family of TFs in metazoans, including the estrogen receptor alpha and the retinoic acid receptor.31,32 (2) Sensing a disruption to the cellular homeostasis caused by the environmental signal, e.g., protein kinase R-like endoplasmic reticulum kinase (PERK) sensing misfolded proteins in the endoplasmic reticulum of eukaryotes, leading to activation of TFs involved in apoptosis.33 (3) Through a signaling cascade propagated from the extracellular signal-binding receptor, e.g., vascular endothelial growth factor (VEGF) binding to VEGF receptors to induce downstream activation of several TFs in metazoans.34 Once activated, the TFs bind to target sequences within regulatory regions of the genome to modulate the transcription of target genes. Transcriptional activators recruit RNA polymerase II (RNA pol II) and other transcriptional machinery, including chromatin remodelers, to the gene to initiate transcription. The so-called pioneering TFs bind to their target sequence even when the chromatin is in a repressed heterochromatic state, in which the chromatin is structurally condensed and transcriptionally inactive, and can initiate events that lead to the opening of the chromatin and activation of the affiliated gene(s).35
Alternatively, transcriptional repressors can bind to their target sequences and recruit chromatin remodeling proteins to convert the local chromatin to, or maintain it in, a heterochromatic state. For example, the Krüppel-associated box (KRAB) zinc finger proteins are sequence-specific DNA-binding transcriptional repressors that induce the formation of heterochromatin through the recruitment of KRAB-associated protein 1 (KAP1), which then recruits histone methyltransferases (ie, SETDB1) and deacetylases and promotes the binding of heterochromatin protein 1 (HP1), which maintains the heterochromatic state.36–38
In addition to the function of gene-specific TFs, the process of transcription itself is generally associated with the exchange of histones in chromatin. This histone exchange is mediated by adenosine triphosphate (ATP)-dependent chromatin remodelers, such as SWI/SNF, FACT, and Spt6, and facilitates the progression of RNA pol II through the chromatin.39 In addition to allowing the progression of RNA pol II along the DNA template, the dynamic deposition and removal of histones from the DNA allows for dynamic control over nucleosome composition. The replication-independent, transcription-dependent deposition of noncanonical histones, such as H3.3 and H2A.Z, provides one example of changing histone composition.40,41 Additionally, the continued removal and deposition of nucleosomes requires the continual proximity of protein complexes involved in that process, therefore providing a mechanism for rapid changes in response to environmental changes, either continued induction through continual recruitment of nucleosome remodelers or rapid shutoff upon loss of chromatin remodeler recruitment. The turnover of the nucleosomes also provides a potential mechanism for conservation of a histone modification pattern by copying a pattern from a redeposited H3/H4 heterotetramer or H2A/H2B dimer to a newly deposited counterpart. The turnover also allows for the dilution of histone marks in the absence of the continued activity of histone-modifying enzymes by incorporation of unmodified histones.
The expression of canonical histones (H3, H4, H2A, and H2B) is limited to the S-phase of the cell cycle, when the histone proteins are needed for incorporation into the daughter chromosomes.42,43 The noncanonical histones, such as H3.3 and H2A.Z, are expressed throughout the cell cycle and are deposited in the wake of RNA pol II during transcription-dependent histone exchange.44 This likely contributes to the observation that, in postmitotic cells, the noncanonical histones accumulate, while the canonical histones are gradually lost from the cell with age.39
The continued development of robust methods for the genome-wide assessment of DNA methylation and other epigenetic marks is providing a means for extensive analysis of differential epigenetic patterns in various tissues at various ages and disease states.45,46 Table 2 presents a list of genome-wide epigenetic studies in human complex disease to date that may have overlapping pathophysiology with diseases of the posterior eye, including glaucoma, AMD, and/or diabetic retinopathy (DR). Also, several resources are available online to access and analyze genome-wide epigenetic data (Table 3). However, this field remains dynamic with more work to be done, particularly with respect to the role postmortem time plays in tissue integrity for whole-genome epigenetic studies. The quality of tissue, for example, would influence downstream analysis of epigenomic data. The fresher the donor tissue is, the more meaningful the downstream interpretation of the statistical analysis will be.47–50 This is similar to studies of human gene expression from donor eye tissue.51
Table 2.
Genome-wide epigenetic studies of retinal disease, risk factors, and related pathologies.
| STUDY | SYSTEM | DISEASE | METHODOLOGY | RNA EXPRESSION | SAMPLE SIZE | TISSUE SAMPLE(S) | DEATH/COLLECTION TO PRESERVATION TIME | NUMBER OF DIFFERENTIALLY METHYLATED PROBES OR REGIONS (DMPs OR DMRs)/GENES | PATHWAYS IDENTIFIED IN DMR LOCI | VALIDATED LOCI WITH DIFFERENTIAL METHYLATION |
|---|---|---|---|---|---|---|---|---|---|---|
| Aavik et al, 2015173 | Cardiovascular | Atherosclerosis | MeCP2-seq | Affymetrix HGU133 Plus2 microarray; qPCR; immunohisto-chemistry; western blots | Femoral artery ather-ectomy samples (22); mammary arteries (9) | Femoral artery atherectomy samples; mammary arteries | Not provided | 4779 promoter DMPs/3740 genes, 8604 exonic DMPs/4939 genes, 17782 intronic DMPs/7972 genes | Intermediate filament-related genes, ATP-binding proteins, cytoskeleton, chromatin regulators, cell adhesion, morphogenesis | RTL1 |
| Allione et al, 2015174 | Multiple | Smoking | Illumina Infinium Human-Methylation450K BeadChip | None | 20 MZ twin pairs | Whole blood | Not provided | 22 DMPs | GTPase regulatory activity, phospholipid binding, vitamin D response element binding, transcriptional repressor complex | None |
| Bell et al, 2010175 | Endocrine | Type 2 diabetes | MeDIP-chip Nimblegen; H3K4me1/2/3, CTCF, and H3K9me1 ChIP-chip | RNAseq | Discovery: 30 cases, 30 controls; validation: original 60 + 20 plus 14 donor brain tissue | Whole blood; donor brain tissue | Not provided | 0 DMR between T2D and controls | None | None |
| Binder et al, 2015176 | Endocrine | Gestational diabetes mellitus (GBM) | Discovery: Illumina Infinium HumanMethylation450K BeadChip; targeted replication with bisulfite-pyrosequencing | Affymetrix Human Transcriptome Array 2.0 | Discovery: 41 cases, 41 controls; validation: not provided | Placental tissues (maternal side) | Not provided | Not provided | Immune response, cellular metabolism, response to external stimuli | CCDC181, HLA-H/HLA-J, HLA-DOA, SNRPN/SNURF |
| Boks et al, 200963 | Multiple | Age, gender, genotype | Illumina GoldenGate DNA Methylation Cancer Panel 1 assay | None | 23 MZ twin pairs, 23 DZ twin pairs, 96 controls | Whole blood | Not provided | Age: 58 DMPs; gender: 56 auto-somal DMPs; heritability: 96 DMPs; genotype: 11 DMPs | None | None |
| Dayeh et al, 2014155 | Endocrine | Type 2 diabetes | Illumina Infinium Human-Methylation450K BeadChip | Affymetrix GeneChip Human Gene 1.0 ST arrays | 15 cases, 34 controls | Donor pancreatic islets | Cultured 2.7 days prior to DNA/RNA extraction | 1649 DMPs/853 genes | Cancer, axon guidance, MAPK signaling, focal adhesion, ECM-receptor interactions, regulation of actin cytoskeleton | None |
| Guida et al, 2015178 | Multiple | Smoking | Illumina Infinium Human-Methylation450K BeadChip | Illumina HumanWG-6 chip v3; Illumina HT-12 chip | 745 women | Whole blood | Not provided | 461 DMPs between current smokers and never smokers; 3 DMPs between former smokers and never smokers; 751 DMPs associated with time since cessation of smoking | None | None |
| Haas et al, 2013179 | Cardiovascular | Dilated cardiomyopathy | Discovery: Illumina Infinium HumanMehtylation27 Bead-Chip; replication: MassArray and bisulfite sequencing | RT-qPCR | Discovery: 9 cases, 8 controls; replication: 30 cases, 128 controls | Left ventricular tissue | Immediately washed and flash frozen | 90 genes | Cardiovascular disease, nutritional & metabolic diseases, pathological conditions, signs and symptoms | LY75, ERBB3, HOXB13, ADORA2A |
| Hannum et al, 201365 | Multiple | Aging | Illumina Infinium Human-Methylation450K BeadChip | Used data from Emilsson et al, 2008177 | Discovery: 482; validation: 174 additional individuals; verification: Heyn et al, 201264 data | Whole blood | Not provided | 70,387 | AD, cancer, tissue degradation, DNA damage, oxidative stress | None-used validation sets to validate their model of aging from initial set |
| Heyn et al, 201264 | Multiple | Aging | Discovery: whole genome bisulfite sequencing (WGBS); replication 1: WGBS and Illumina Infinium HumanMethylation450K BeadChip; replication 2: Illumina Infinium Human-Methylation450K BeadChip: validation of discovery + replication 1 combined and replication 2: targeted bisulfite sequencing validation | RT-qPCR | Discovery: 1 new-born, 1 centenarian; replication 1: 1 middle aged person; replication 2: 19 newborns, 19 nonagenarians | Discovery & replication 1: CD4+ T cells; replication 2: cord blood for newborns and peripheral blood mononuclear cells for nonagenarians | Not provided | Discovery & replication 1: 1149 DMPs/615 genes; replication 2: confirmed 214 DMPs plus additional 5774 DMPs | Not provided | AIM2, TNFRSF9, IGSF9B, PTPRE, ZRP-1, FHL2, miR-21 |
| Hidalgo et al, 2014180 | Renal | Diabetes | Illumina Infinium Human-Methylation450K BeadChip | None | Discovery: 544; replication: 293 | CD4+ T cells | From frozen | Presented top 5 DMPs | Not provided | ABCG1 |
| Irvin et al, 2014182 | Multiple | Fasting VLDL and TGs | Illumina Infinium Human-Methylation450K BeadChip | Discovery: RT-qPCR of single hit; replication: Affymetrix HumanExon 1.0 ST GeneChip | Discovery: 991; replication 1261 | CD4+ T cells | Not provided | 4 DMPs/1 gene | Lipid metabolism | CPT1A |
| Ko et al, 2013183 | Renal | Kidney fibrosis | Discovery: HELP-Nimblegen whole genome covering microarray; validation: bisulfite-Sequenom MassArray EpiTYPER; replication: Illumina Infinium HumanMethylation450K BeadChip | None | Discovery/validation: 12 hypertensive or diabetic chronic kidney disease, 14 controls; replication: 21 diabetic chronic kidney disease, 66 controls | Human kidney tubule epithelial cell tissue samples from healthy living transplants or surgical nephrectomies | Not provided | Discovery: 4751 DMRs/1535 genes; replication: 1061 genes | Cell adhesion, development | COLIVA1, DSCAM, DPT |
| Markunas et al, 2014184 | Multiple | Maternal smoking | Discovery: Illumina Infinium HumanMethylation450K BeadChip; replication using Joubert et al, 2012181 published data | None | 287 infants whose mothers smoked during 1st trimester, 602 infants whose mothers did not smoke during pregnancy | Whole blood | Not provided | 185 DMPs/110 genes | Nicotine dependence, smoking cessation, placental & embryonic development | FRMD4A, ATP9A, GALNT2, MEG3, GNG12, Loc284998, VGLL4, CUX2, FTO, KIF26B |
| McClay et al, 201467 | Multiple | Aging | Discovery: MBD-seq; replication: pyrosequencing | None | Discovery: 718 aged 25–92 years; replication: 558 | Whole blood | Not provided | 11 DMRs | Protocadherins, homeo-box genes, MAPKs, ryanodine receptors | LSAMP, ZEB2, MEIS7, GRIA2 |
| Menni et al, 201366 | Multiple | Aging | Discovery: Illumina Infinium HumanMehtylation27 BeadChip; replication: Illumina Infinium Human-Methylation450K BeadChip | None | Discovery: 172 female twins; replication: 350 twins | Fasting whole blood | Not provided | 3 DMPs associated with C-glyTrp levels (an age-associated metabolite) | Protein synthesis, cell cycle, embryonic development, renal inflammation & hypertension, age-related retinal degeneration | WDR85 |
| Miao et al, 2008185 | Endocrine | Type 1 diabetes | H3K9me2 ChIP-chip, including human 12 k cDNA array, human 12 k CpG island array, and Nimblegen human promoter tiling array | None | 9 T1D, 7 controls | Blood lymphocytes and monocytes | Not provided | Lymphoblasts: 193 DMPs 12 k cDNA array, 213 DMPx 12 k CpG island array, not specified for the tiling array; monocytes: none | Autoimmunity, inflammation, TGFbeta, NFkappa-B, p38 MAPK, toll-like receptor, IL-6 pathways, T1D genes | HOX13B, NKXB, CPLX4, CXCR4, FAM19A5, AR, PLAZG4B, CD55, IGSF4B, CTLA4 |
| Milenkovic et al, 2014186 | Cardiovascular | Flavanols & cardiovascular disease | Discovery: Illumina Infinium HumanMethylation450K BeadChip; validation with pyrosequencing | Agilent G4845A Human GE 4×44K v2 microarray | 13 male smokers before and after flavanol treatment | Leukocytes | Freshly stored in RNALater | None | None | CXCL12, SCRIB, PDGFRL, FERMT3, ICAM1, VCAM1 (not significantly DM at 10% change) |
| Monick et al, 2012187 | Multiple | Smoking | Illumina Infinium Human-Methylation450K BeadChip | RT-qPCR (transformed lymphoblasts) | 10 smokers, 9 controls | Lung alveolar macrophage | Not provided | 1381 DMPs | Wound healing, inflammation, G-protein/Ras signaling | AHRR |
| Movassagh et al, 2010188 | Cardiovascular | Cardiomyopathy | Discovery: MeDIP-chip with the Nimblegen CpG island and promoter microarray; validation: bisulfite PCR and sequencing | RT-qPCR | Discovery: 3 (ischemic & idiopathic) cardiomyopathic tissue from transplant surgeries; 1 normal tissue from donors; validation: 4 (ischemic & idiopathic) cardiomyopathic tissue from transplant surgeries; 4 normal tissue from donors | Left ventricular tissue | Not provided | 3 genes | Angiogenesis | AMOTL2, ARH-GAP24, PECAM1 |
| Movassagh et al, 2011189 | Cardiovascular | Cardiomyopathy | MeDIP-seq; H3K36me3 ChIP-seq | RT-qPCR | 4 (ischemic & idiopathic) cardiomyopathic tissue from transplant surgeries; 4 normal tissue from donors | Left ventricular tissue | Not provided | Not provided | Not provided | DUX4 |
| Nazarenko et al, 2015190 | Cardiovascular | Coronary Heart Disease | Illumina Infinium HumanMehtylation27 BeadChip; pyrosequencing validation | No | 3 tissues from 6 patients for microarray; 3 tissues from 21 patients for validation | Human right coronary artery in the area of advanced atherosclerotic plaques (CAP); internal mammary arteries (IMA); great saphenous veins (GSV) | Processed and snap frozen immediately after surgery | 22 (CAP-IMA); 27 (GSV-IMA) | Inflammation and immune response; development | HOXD4 |
| Nilsson et al, 2014191 | Endocrine | Type 2 diabetes | Illumina Infinium Human-Methylation450K BeadChip | RT-qPCR; Affymetrix GeneChip Human Gene 1.0 ST arrays | 14 discordant MZ twin pairs; cohort 1: 120 case-controls; cohort 2: 28 cases, 28 controls | Subcutaneous adipose tissue biopsies | Frozen immediately in liquid nitrogen | 15,627 DMPs/7,046 genes in cohort 2, 1410 of which DMPs also in MZ twins | Oxidative phosphorylation; mitogen-activated protein kinase signaling; carbohydrate, amino acid, and lipid metabolism; inflammation; Wnt signaling; glycan degradation; cancer | None |
| Nitert et al, 2012192 | Endocrine & musculoskel et al | Diabetes & exercise | Discovery: MeDIP-chip; replication: Illumina Infinium HumanMethylation450K BeadChip; validation: bisulfite-Sequenom Epi-TYPER, luciferase assays with or without methylation | Affymetrix Custom-Array NuGO-Hs1a520180 GeneChip | Discovery: 15 men with 1st degree relative with diabetes (FH+)/13 men w/o relative with diabetes (FH-); replication: 9 MZ twin pairs discordant for T2D | Skeletal muscle biopsies from vastus lateralis after fasting and without exercise for 48 hrs prior to biopsy | Not provided | 65 genes DM FH+/FH-before exercise; 38 genes DM FH+/FH-after exercise; 134 genes DM before/after exercise combined FH+/− | FH+/−: MAP signaling, insulin signaling, calcium signaling, Wnt signaling, starch and sucrose metabolism, sphingolipid metabolism; before/after exercise: purine metabolism, retinol metabolism, calcium signaling, muscle, T2D, gly/ser/thr metabolism, glycolysis, gluconeogenesis, starch and sucrose metabolism, insulin signaling | MSI2, THADA, MEF2A, RUNX1, NDUFC2 |
| Olsson et al, 2014193 | Endocrine | Insulin secretion | Discovery: Illumina Infinium HumanMethylation450K BeadChip; validation: pyrosequencing | Affymetrix GeneChip Human Gene 1.0 ST | 89 pancreatic islet donors | Human pancreatic islets | Cultured in vitro 4 days | 14 DMPs potentially mediating relationship between SNPs and mRNA level | Pancreatic islet function, type 1 diabetes, cell adhesion, ECM interaction, folate biosynthesis | GPX7, GST-1, SNX19 |
| Rakyan et al, 2011194 | Endocrine | Type 1 diabetes | Illumina Infinium HumanMehtylation27 BeadChip | None | 19 discordant MZ twin pairs; 7 singletons before and after diagnosis | CD14+ monocytes | Not provided | 132 DMPs | Type 2 diabetes, immune responses | HLA-DQB1, RFXAP, NFKB1A, TNF, GAD2 |
| Ronn et al, 2015195 | Endocrine, etc. | Aging, BMI, HbA1c | Discovery: Illumina Infinium HumanMethylation450K BeadChip; validation: pyrosequencing | Affymetrix GeneChip Human Gene 1.0 ST | Discovery: 96 males, 94 females; validation: 37 males, 67 females | Discovery: human adipose tissue biopsies (fasted state); validation: human adipose tissue biopsies and blood (fasted state) | Not provided | Age: 1050 genes methylation and expression; BMI: 2825 genes methylation and expression; HbA1c: 711 genes methylation | Cancer, type 2 diabetes, cardiovascular disease; signal transduction | None |
| Sapienza et al, 2011196 | Endocrine | Diabetes | Illumina Infinium HumanMehtylation27 BeadChip | None | 23 diabetics with end stage renal disease; 23 diabetics without nephropathy | Saliva | Not provided | 389 DMPs | Inflammation, oxidative stress, ubiquitination, fibrosis, drug metabolism, cancer, cardiotoxicity, hepatotoxicity, nephrotoxicity | None |
| Sharma et al, 2014197 | Cardiovascular | Coronary artery disease | Discovery: 12 k Human CpG Island Microarray; replication: bisulfite deep sequencing | None | Discovery: 18 cases, 18 controls; replication: 48 cases, 48 controls | Peripheral blood | Within 1 hour | 72 DMRs | Signal transduction, transcription regulation, organelle development, transport, cell regulation, ion channel activity, E-box binding, mitochondria, plasma membrane, nuclear chromatin | STRADA, C1QL4, HSP90B3P |
| Steenaard et al, 2015198 | Cardiovascular | Tobacco smoking (coronary artery disease) | Illumina Infinium HumanMethylation450K BeadChip | Illumina HumanHT12v4 Expression BeadChip | 724 subjects: 195 current smokers, 201 never smokers, 328 prior smokers | Whole blood | Not provided | 15 DMPs of 3669 CpGs/1169 CAD genes analyzed | Only looked at CAD related genes | None |
| Stringhini et al, 2015199 | Immune System | Inflammation | Illumina Infinium Human-Methylation450K BeadChip | None | 857 | Whole blood | Not provided | 41 DMPs/10 genes associated with household occupational position; 12 DMPs/6 genes associated with socioeconomic trajectories | Only looked at candidate genes involved in socio-economic status-related inflammation | None |
| Sun et al, 201468 | Multiple | Aging | Illumina Infinium HumanMethylation450K BeadChip | None | 100 male, 100 female nonagenarians/centenarians | Peripheral blood mononuclear cells | Not provided | 850 DMPs/564 genes | Hormone regulation, neuron projection, disease-related pathways, cellular component organization, cell morphogenesis, cell-cell junctions, CNS development, ECM-receptor interactions, axon guidance, cell adhesion molecules | None |
| Suter et al, 2011200 | Multiple | Maternal tobacco exposure | Discovery: Illumina Infinium HumanMehtylation27 BeadChip; validation: bisulfite PCR sequencing | Illumina HG-12, RT-qPCR | Discovery: 18 smokers, 18 non-smokers; validation: 9 smokers, 9 non-smokers | Placental tissues | Collected immediately after delivery and flash frozen | 1024 DMPs | Oxidative stress pathways, oxidative phosphorylation, mitochondrial dysfunction, HIF1alpha signaling, cell death, cell morphology, cell-to-cell signaling | PURA, GTF2H2, GCA, GPR135, HKR1 |
| Tsaprouni et al, 2014201 | Multiple | Smoking | Illumina Infinium HumanMethylation450K BeadChip | RNAseq | Discovery: 22 current smokers, 263 former smokers, 179 never smokers; replication: 41 current, 104 former, 211 never | Peripheral blood | Not provided | 30 DMPs/15 loci | Cardiovascular disease; cancer; connective tissue and developmental disorders; cell death, cell survival, and cell-cell interactions; heme biosynthesis; aryl hydrocarbon receptor signaling | None |
| Wan et al, 2012202 | Multiple | Smoking | Discovery: Illumina Infinium HumanMethylation27 BeadChip: validation: bisulfite pyrosequencing | None | Discovery: 1085; replication: 369 | Peripheral blood leukocytes | Not provided | 15 DMPs current vs. former smokers; 2 DMPs cumulative exposure; 3 DMPs time since quitting | Not provided | F2RL3, GPR15 |
| Wang et al, 2013203 | Cardiovascular | Hypertension | Discovery: Illumina Infinium HumanMehtylation27 BeadChip; replication: bisulfite sequencing | None | Discovery: 8 cases, 8 controls 14–23 years old; replication 1: 36 cases, 60 controls 14–30 years old; replication 2: 36 cases, 34 controls 15.8–40 years old | Blood leukocytes | Not provided | 1 gene | Inflammation | SULF1 |
| Xiao et al, 201569 | Multiple | Aging | Discovery: MeDIP-seq; replication: whole genome bisulfite sequencing | None | Discovery: 4 centenarians, 4 middle aged; replication: additional 1 centenarian, 1 middle aged | Peripheral blood | Not provided | Discovery: 626 DMRs/251 genes; replication: 156 genes | Age-related diseases: T2D, cardiovascular disease, stroke, coronary artery disease, AD; developmental processes; cell adhesion; signal transduction; cell communication; cell adhesion; cadherin signaling; Wnt signaling; AD-presenilin | None |
| Xu et al, 2013204 | Multiple | Obesity | Illumina Infinium Human-Methylation450K BeadChip | None | 48 obese cases, 28 lean controls, all 14–20 years old | Fasting peripheral blood leukocytes | Not provided | 23305 DMPs | DNA binding, development, regulation of neurogenesis, cell differentiation, transcription regulation, obesity-related diseases | None |
| Yang et al, 201570 | Neurological | Aging | Illumina Infinium Human-Methylation450K BeadChip | None | 740 aged 66 to 108 years old | Postmortem dorsolateral prefrontal cortex | Not provided | 4263 DMPs associate with age | Not provided | None |
| Yuan et al, 2014205 | Endocrine | Type 2 diabetes | Discovery: MeDIP-seq; replication: Illumina Infinium HumanMethylation450K BeadChip | Illumina HumanHT12v3 Expression BeadChip | Discovery: 27 MZ twin pairs (17 T2D discordant, 2 T2D concordant, 7 control concordant pairs); replication: 42 unrelated T2D cases, 221 controls | Whole blood | Blood drawn into EDTA tubes | 1355 DMRs | T2D GWAS genes, imprinted genes | GPR61, PRKCB, MALT1 |
| Zaina et al, 2014206 | Cardiovascular | Atherosclerosis | Discovery: whole-genome shotgun bisulfite sequencing; replication: Illumina Infinium HumanMethylation450K BeadChip; validation: pyrosequencing and conventional bisulfite sequencing | RT-qPCR | Discovery: 1 matched pair of atherosclerotic and normal tissue from the same donor; replication: 15 matched pairs of atherosclerotic and normal tissues each from the same donor; validation: (multiple) 5 aortas/24 donor pairs/15 donor pairs from original cohort/1 donor pair | Postmortem aortas and carotid tissues | 3–26 hours | Discovery: 54625 DMRs; replication: 1895 DMPs | Genes related to processes related to endothelial cells and vascular smooth muscle cells; vascular smooth muscle cell contraction; extracellular matrix formation | HOX family members, PDGFA, PLAT, PRRX1, PXDN, MIR23b |
| Zawada et al, 2012207 | Cardiovascular | Chronic kidney disease-associated cardiovascular disease | Discovery: SuperTAG methylation-specific digital karyotyping; validation: bisulfite pyrosequencing | RT-qPCR | 10 cases, 10 controls | Peripheral blood | Not provided | 4288 DMRs | Lipid metabolism & transport, cell proliferation, cell-cycle regulation, angiogenesis, inflammation | METTL2B |
Table 3.
Epigenetic resource databases.
| DATABASE | WEBSITE | TISSUE DATA SET | SOURCE |
|---|---|---|---|
| Epigenome Browser | http://www.epigenomebrowser.org/ | Varied | ENCODE |
| Roadmap Epigenomics Visualization Hub (VizHub) | http://vizhub.wustl.edu/ | Stem cells & healthy primary tissues | NIH Roadmap Epigenomics Mapping Consortium |
| WashU EpiGenome Browser | http://epigenomegateway.wustl.edu/browser/ | Varied | Roadmap Epigenomics; ENCODE |
| NCBI | http://www.ncbi.nlm.nih.gov/epigenomics | Varied | Varied |
| Emsembl | http://useast.ensembl.org/info/website/tutorials/encode.html | Varied cell lines | Varied |
| MethylomeDB | http://www.neuroepigenomics.org/methylomedb/ | Human and mouse brain | Haghighi Lab |
| MethBase | http://smithlabresearch.org/software/methbase/ | Varied | Smith lab |
| NGSMethDB | http://bioinfo2.ugr.es:8080/NGSmethDB/ | Varied | Repository |
| DiseaseMeth | http://202.97.205.78/diseasemeth/ | Varied human disease | Varied |
| Gene Expression Omnibus (GEO) | http://www.ncbi.nlm.nih.gov/geo/roadmap/epigenomics/ | Varied tissues and cell lines | NIH Roadmap Epigenomics data |
| The Epigenome Atlas | http://www.genboree.org/epigenomeatlas/index.rhtml | Human reference epigenomes | NIH Roadmap Epigenomics Mapping Consortium |
| Canadian Epigenomics, Environment and Health Research Consortium Platform (CEEHRC) | http://epigenomes.ca/ | Varied | CEEHRC |
| Epigenie | http://epigenie.com/epigenetic-tools-and-databases/ | List of epigenetic resources | Varied |
Aging
Throughout development, cells acquire chromatin changes that regulate the transcriptional output of each cell. In gametogenesis and embryogenesis, DNA methylation is erased and reset at specific reprograming stages to ensure appropriate gene expression and totipotency of embryonic cells.52 This reprograming is used by the cell for processes such as X-chromosome inactivation for dosage compensation, repression of invasive DNA such as retrotransposons, imprinting to ensure parental allele-specific expression, and to otherwise control the expression of genes relevant to the developmental process. Additionally, the process of cellular differentiation, in which individual cells acquire specialized gene expression profiles specific to their individual tissue types and functions, requires further changes to the chromatin landscape (reviewed for various tissues such as skeletal muscle,53 myeloid cells,54 and the lens and retina55). Much effort of late has focused on defining the tissue-specific differential methylation patterns in differentiated tissues in relation to differential gene expression patterns in an attempt to understand the underlying mechanisms of tissue-specific processes.56–60 Even after a cell has reached its terminally differentiated state, further changes that associate with the aging process can continue to accrue throughout the lifespan of the cell. Methylation levels generally decrease with aging, although individual genes may have increased methylation levels with age.61–70 Replication-independent histone exchange results in the accumulation of histone variants, such as H3.3 and H2A.Z, in older cells.39 Cells from aged human beings have been shown to have reduced levels of both the heterochromatin maintenance protein HP1 and the H3K9me3 histone modification known to recruit HP1 to chromatin.71 Experiments in model organisms showing that the reversal of age-dependent chromatin changes results in extension of lifespan suggest that at least some of the age-related chromatin changes are indeed causative of aging, rather than just a result of aging.72–74 Much work is still needed to fully understand the role of acquired chromatin changes in the aging process.
Sirtuins are a class of HDACs implicated in lifespan regulation and the promotion of healthy aging through various epigenetic and nonepigenetic cellular roles, including telomere maintenance, DNA repair, metabolism, stress tolerance, cellular differentiation, apoptosis, and inflammation.75,76 Sirtuins have also been implicated in age-related diseases, such as diabetes, cardiovascular disease (CVD), neurodegenerative diseases, and some cancers.9 Roles for sirtuins have been observed in ocular aging. The involvement of sirtuin 1 (SIRT1) in ocular aging, including retinal degeneration, has been the most well characterized of the seven mammalian sirtuins. SIRT1 has been associated with retinal stem cell self-renewal, and decreased expression of SIRT1 with age has been suggested to contribute to the aging process.77 SIRT1 also exhibits neuroprotective effects in the retina at least partially through its antioxidant, energy balancing, and antiapoptotic functions.78 Abnormal SIRT1 localization is also thought to promote the apoptosis of photoreceptor cells and precocious aging in the rd10 mouse model of retinal degeneration.79 SIRT6, which is regulated by SIRT1, has also been implicated in retinal aging, and SIRT6 deficiency in mice results in increased levels of retinal cell apoptosis.80,81 Altered TF functions have also been observed in the aging mammalian retina. For example, the function of the oxidative stress response master regulator nuclear factor (erythroid-derived 2)-like 2 (Nrf2) was shown to be altered in aged C57Bl6/J mice compared with young C57Bl6/J mice.82 The aged mice in this study exhibited higher uninduced (basal) expression levels of Nrf2 antioxidant targets compared with the young mice. Upon induction with an oxidative stressor, the Nrf2-dependent antioxidant genes were robustly induced in young mice, but not in aged mice. Nrf2 was also induced by the oxidative stressor in young mice, but not in aged mice. Expression of Nrf2 is regulated by DNA methylation and modulated by dietary factors such as sulforaphane; differential methylation of the Nrf2 promoter is also associated with various diseases.83
The question of whether complex age-related diseases are the inevitable eventual outcome of the aging process or whether they are, in fact, the result of aberrant aging is yet to be elucidated and is of great interest in the field today. Along with the extension of lifespan achieved to date by modern medical advances has come an increase in the prevalence of age-related conditions that severely impact the quality of life of older adults. Degenerative diseases of the retina, such as glaucoma, AMD, and DR, affect >15 million people >40 years old in the US and are the leading causes of blindness in older adults.84–86 As of 2013, 11 million individuals were affected with AMD, per year, in the United States alone. Because aging is the greatest risk factor, this number is anticipated to increase to 22 million by 2050 as a result of the population aging (http://www.brightfocus.org/).
We present here what is known of epigenetic effects in major retinal diseases associated with aging: glaucoma, AMD, and DR. Further work in this field has promise to revolutionize the way we approach therapeutic interventions by revealing points of intervention with potential long-term influences on the progression of disease.
Glaucoma
Glaucoma is the leading cause of irreversible blindness in the US and worldwide, according to the World Health Organization (2015); it ranks second overall after cataract, which is reversible.87–89 Primary open-angle glaucoma is considered the silent thief of vision, as it is initially asymptomatic, progresses slowly, and eventually, causes irreversible vision loss due to optic nerve damage and loss of retinal ganglion cells (Fig. 3B). Early diagnosis and treatment can control glaucoma before vision loss occurs. At present, lowering intraocular pressure (IOP) is the only proven method of slowing glaucoma-associated visual field deficit progression. Glaucoma is a progressive neurodegenerative disease and is thought to progress as follows: fibrosis in the trabecular meshwork (TM) and in the lamina cribrosa at the optical nerve head reduces outflow of the aqueous humor (AH), which increases the IOP and induces hypoxia. Subsequent damage to the optic nerve head is associated with retinal ganglion cell loss and vision loss.90,91 The molecular details of disease etiology remain unclear to date. Risk factors for glaucoma include age, IOP, and central corneal thickness.92 Oxidative stress, systemic hypertension, and family history have also been associated with the development of glaucoma.93,94 At least 29 loci have been associated with genetic risk of glaucoma, of which the best characterized are myocilin (MYOC), optineurin (OPTN), and WD repeat domain 36 (WDR36).95 MYOC acts in cyto-skeletal organization, extracellular matrix remodeling, and mitochondrial function.96,97 Accumulation of MYOC aggregates within the TM is thought to cause TM cell apoptosis and ECM changes, which blocks the AH outflow and leads to increased IOP.98 OPTN is protective against oxidative stress and stabilizes MYOC mRNA.99,100 WDR36 variants in mouse alter retinal ganglion cell axon growth that leads to retinal degeneration.101 However, the role of this gene in the development of human disease remains controversial.95
Studies of gene expression in donor eye samples affected with glaucoma have shown upregulation of profibrotic factors in the TM and AH, including transforming growth factor beta (TGF-β), thrombospondin-1 (TSP1), and connective tissue growth factor, compared with unaffected controls.102–104 Extensive investigations of epigenetic mechanisms have yet to be performed in human tissues affected by glaucoma. However, epigenetic processes involved in fibrosis in other cell types/disease processes give insight into likely processes involved in the glaucoma-associated fibrosis. Hypermethylation of TSP1 in colorectal cancer suppresses TSP1-mediated activation of TGF-β.105 TGF-β-mediated regulation, in turn, is sensitive to the chromatin modifications, including DNA methylation and histone acetylation, of its downstream target genes.106–108
Hypoxia is thought to contribute to the degeneration of retinal ganglion cells in glaucoma.109 The expression of hypoxia-induced factor 1 alpha (HIF1α), which induces the adaptive transcriptional response to hypoxia,110,111 is greater in the retina and optic nerve head of glaucomatous eyes compared with controls.110 HIF1α is stabilized under hypoxic conditions by the loss of a specific oxygen-dependent degradation mechanism involving proline hydroxylation of HIF1α, 112,113 which allows HIF1α to dimerize with HIF1β to form an active HIF1 heterodimer.114 Hypoxia induces the translocation of HIF1α from the cytoplasm to the nucleus, where it recruits the histone acetyltransferase CBP/p300 coactivator and regulates transcription of target genes.115 HIF1α activity is regulated epigenetically in a few ways. The HIF1α promoter contains a hypoxia response element with a CpG that is methylated under hypoxic conditions; a DNA methyltransferase (DNMT) inhibitor increased hypoxia-induced gene expression; methylation of the hypoxia response element influences HIFα binding; and various histone modifications at hypoxia responsive genes are specific to hypoxic conditions.116
Further evidence for epigenetic effects in hypoxia has been provided by Shahrzad et al who showed an inverse relationship between DNA methylation and hypoxia in primary cancer cells and in normal human dermal fibroblasts.117 Also, under chronic hypoxia in prostate epithelial cells, the induced HIF1α stability is lost, and both H3K9 hyperacetylation and DNA hypermethylation accumulate, which are proposed to maintain the hypoxia-conditioned state in the absence of HIF1α.111
Genome-wide epigenetic studies utilizing well-characterized/phenotyped glaucoma-affected tissues and matched normal controls will be important for elucidating the epigenetic changes that occur within the eye throughout the progression of the disease, from normal to the onset of increased IOP, to the damaging effects of that pressure on the optic nerve. Additionally, the use of monozygotic twin pairs discordant for glaucoma for genome-wide epigenetic analysis will further highlight epigenetic processes associated with the development of glaucoma. The information gained by combining rigorous genome-wide epigenetic, genetic, and expression data will lead to improved understanding of the disease and, therefore, better means to treat it.
Age-related Macular Degeneration
Similar to glaucoma, AMD is also a multifactorial, progressive, neurodegenerative disease. AMD is characterized by the sub-RPE accumulation of yellowish lipid-rich, protein-containing drusen deposits and the progressive degeneration of the RPE and the neural retina. Although the whole retina can be affected, the disease preferentially affects the macular region with or without foveal involvement (Fig. 3C). The presence, size, and number of drusen, along with the presence or absence of hypo- or hyperpigmentary changes in the RPE, determine the severity of early or intermediate stages of the disease and the risk for the development of either form of advanced AMD: geographic atrophy (GA; dry AMD) or neovascular (wet) AMD.118–121 GA involves the regional loss of RPE and choroid accompanied by a gradual loss of photoreceptors and central vision.122,123 Neovascular AMD involves the growth of new abnormal blood vessels from the choroid into the normally avascular sub-RPE and subretinal regions. These new vessels are prone to hemorrhage and exudation, which can lead to rapid loss of vision and accounts for the majority of blindness associated with AMD.124 One model for the development of neovascularization in AMD involves the accumulation of drusen, which disrupts the connection between the RPE and the choroidal blood supply, causing hypoxia. The hypoxia in turn induces the expression of VEGF and the formation of new vessels.125 Current therapies for AMD target the advanced neovascular stage, have modest results, and require frequent in-office intraocular injections. Much effort is ongoing in the development of new therapeutics that may ultimately improve outcomes as well as reduce the burden of treatment on affected individuals. Advances in genetic and epigenetic studies have the potential to reveal additional targets for intervention. Additionally, the discovery of molecular mechanisms for the progression from early to intermediate to advanced forms of the disease will provide points for earlier interventions to prevent advancement to late-stage disease and the accompanying vision loss.
Genome-wide association studies (GWAS) for AMD risk alleles have identified 19 genetic variants that associate with AMD at genome-wide significance levels, including genes involved in the complement pathway, lipid metabolism, angiogenesis, and atherosclerosis, among other cellular processes.126 A recent exome chip study identified 34 loci representing both common and rare variants independently associated with AMD.127 This study found evidence for causal roles for three very rare coding variants in three genes (complement factor H [CFH], CHI, and TIMP3) and one splice variant (SC16A8). The two loci with the strongest association with AMD risk are the CFH and ARMS2/HTRA1 loci. The role of CFH in the complement pathway is well characterized.128 However, the role of the ARMS2/HTRA1 locus remains elusive, with ongoing debates regarding which of the two genes at that locus is responsible for the associated genetic risk for AMD.129
Twin studies estimate the genetic component of AMD risk at ~40%–70% of the total risk.130 Known environmental risks include smoking, obesity, and diet.25 Environmental contributions to disease potentially involve epigenetic changes that affect the expression of genes involved in the generation of disease and may also convert environmental exposures into heritable phenotypes.131 This relationship between epigenetics, environment, and heritability may also lead to increased measures of heritability in monozygotic twin studies—due to the shared zygotic epigenetic history of monozygotic twins— than would arise from genetic heritability alone.131 Therefore, epigenetic factors may contribute to both the heritable and nonheritable components of risk. This is consistent with observations that some chromatin changes are conserved through gametogenesis, whereas others are erased in the process.52 The idea that environmental forces introduce small epigenetic effects that accumulate with time (illustrated in Fig. 2) is also consistent with both the observations that age is the primary risk factor for AMD and that siblings with identical AMD risk profiles from major AMD genetic loci, smoking, body mass index (BMI), and CVD can still be extremely discordant for disease development (Fig. 4).
Figure 4.
Siblings extremely discordant for AMD. These fundus images illustrate the retinas of discordant siblings with identical risk profiles for major AMD genetic loci, smoking, BMI, and CVD. Note the severe neovascular AMD in one sibling and normal macula in the other at a comparable age. Environmentally induced epigenetic changes may explain the occurrence of such discordance for AMD or other age-related disease. OD, oculus dexter (right eye); OS, oculus sinister (left eye). Photographs taken from DeAngelis laboratory patient cohorts. The study protocol was reviewed and approved by the Institutional Review Board at the University of Utah and conforms to the tenets of the Declaration of Helsinki.
Evidence for the involvement of epigenetic processes associated with macular degeneration has been accumulating in the field. Early work looking at proteins involved in the modification of chromatin showed requirements for these functions in relation to AMD. Suuronen et al demonstrated that HDAC inhibitors increased the expression and secretion of clusterin, the major protein component of drusen, in a human RPE cell line,132 suggesting that histone deacetylation activity is important to limit the amount of clusterin produced in RPE cells to prevent its accumulation in drusen. Chen and Cepko provided further evidence for the role of histone deacetylation in the maintenance of a healthy retina when they showed that HDAC4 promotes the survival of retinal neurons in mouse.133(p4) Given that neovascularization is thought to be a response to hypoxia caused by drusen deposits interrupting the flow of oxygen from the choroid through the RPE and into the retinal cells, it is also relevant that knockout of the noncanonical H2A.X histone in mice reduced the neovascular response to hypoxia.134
In addition to functional studies looking at the effects of histone-modifying activities, studies have looked at the association of chromatin modifications at genes relevant to the AMD disease process in human tissue samples. Using a DNA methylation microarray and bisulfite pyrosequencing of frozen human donor RPE/choroid samples with a postmortem interval of up to nine hours, Hunter et al identified the glutathione S-transferase isoforms mu1 and mu5 (GSTM1 and GSTM5) as hypermethylated in AMD samples compared with age-matched controls.135 The function of these two proteins in reducing oxidative stress is likely relevant to the development of AMD, as oxidative stress is hypothesized to contribute to the underlying pathophysiology of AMD. In addition to differential methylation of GSTM1 and GSTM5, the study also looked at exon expression microarray data from the same RPE/choroid samples plus the corresponding neural retinas and observed reduced mRNA levels of both genes in AMD samples compared with controls. DNA hypermethylation at the GSTM1 promoter in RPE/choroid samples correlated significantly with the reduced expression of both the GSTM1 gene and the adjoining, downstream GSTM5 gene.
Another early example of differential methylation in AMD correlating inversely with gene expression came from Wei et al’s report of hypomethylation in the promoter of IL17RC from peripheral blood mononuclear cells of patients with AMD compared with their unaffected twins, which corresponded to increased expression of IL17RC in the retina and RPA/choroid from formalin-fixed, paraffin-embedded ocular sections of donor eyes.136 However, Oliver et al were unable to replicate this finding in peripheral blood of patients with AMD compared with controls.137 This question will need to be settled by utilizing tissue samples from the RPE/choroid, neural retina, and blood from the same donors to accurately assess the relationship between differential methylation and AMD disease status, as well as through further molecular characterization of the role of IL17RC in AMD.
As a step in that direction, Oliver et al investigated DNA methylation levels in peripheral blood samples and frozen sucrose gradient-treated peripheral retinas (postmortem interval of donor eye tissue was not reported) of AMD patients with either GA or neovascularization compared with unaffected control patients.138 In the only genome-wide epigenetic study of AMD to date, they observed hypomethylation at the ARMS2/HTRA1 locus and hypermethylation at the PRSS50 locus in AMD patients compared with controls. As described above, the ARMS2/HTRA1 locus is one of the top two loci genetically associated with AMD. The finding that hypomethylation at the ARMS2/HTRA1 locus associates with AMD supports a role for either or both of those genes in the development of disease. The PRSS50 locus had not previously been associated with AMD risk.
The genome-wide methylation study by Oliver et al in AMD-affected RPE/choroid and retina might be a step toward understanding the epigenetic mechanisms underlying gene expression of AMD. Building on their study, researchers will need to go on to evaluate the epigenetic changes that occur throughout the progression from normal to severe disease. This will require well-characterized tissue samples with short postmortem intervals to tease out epigenetic changes at each stage of the disease: preclinical through advanced stages. This will allow for the identification of early changes that may influence the progression of AMD and, therefore, present early therapeutic interventions to prevent the progression to severe forms of the disease.
Further methodological improvements will also need to be incorporated to accomplish this goal. Increased quality of well-characterized donor eyes for each stage of the disease, along with well-characterized phenotypically normal controls, will increase the power of the study to identify changes associated with each step of the disease process. Also, the postmortem interval between death and preservation of the tissue must be carefully selected to preserve the integrity of the sample and minimize the noise in the genome-wide data.48 It is also important that these studies be carried out in a fresh tissue that is directly involved and relevant for the specific disease process being studied. Similar to gene expression profiles, DNA methylation profiles differ among tissues.13,14 Therefore, it cannot be taken for granted that an easily accessible tissue, such as peripheral blood, will adequately substitute or replicate for the relevant, disease-involved tissue.
Diabetic Retinopathy
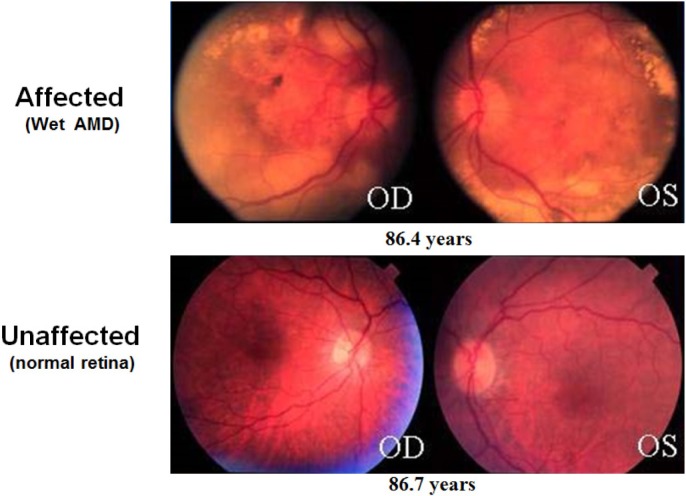
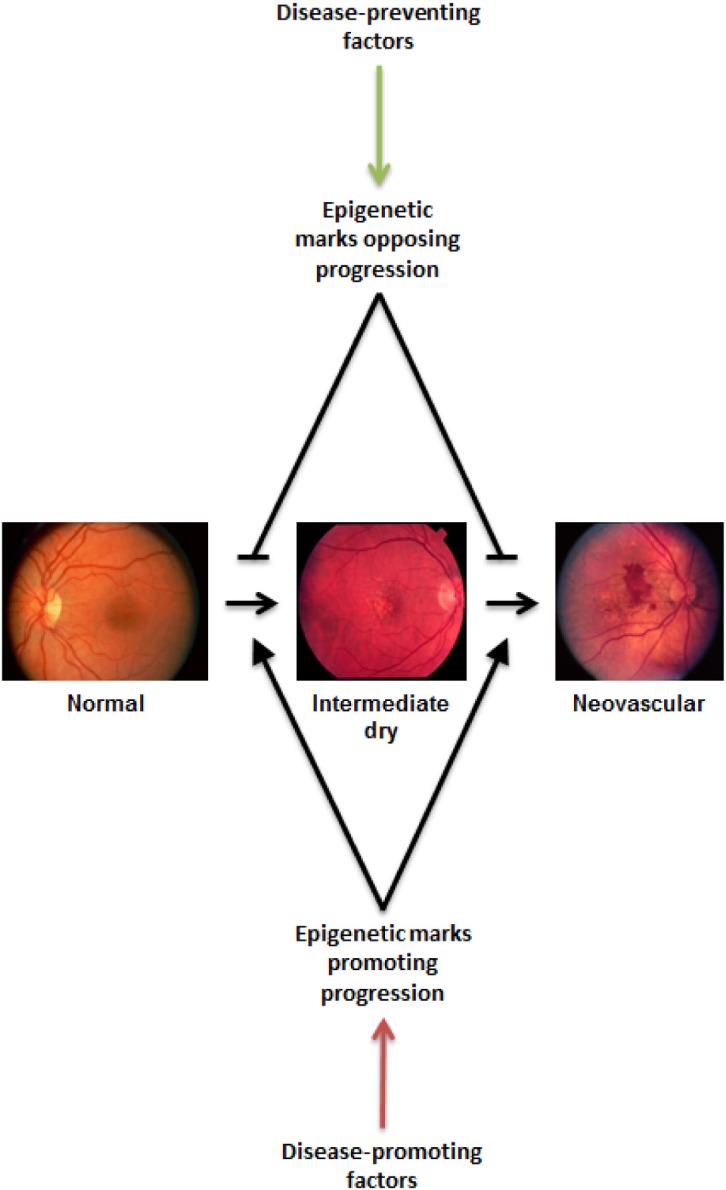
In 2012, ~10% of Americans had diabetes, according to the American Diabetes Association.139 Every diabetic patient is at risk of developing DR, which is the most common cause of blindness among working-age adults in developed countries.140 The risk for DR increases with increasing time since diagnosis of diabetes.141,142 Similar to the age-related risk for AMD and glaucoma, the time component of risk for DR is also consistent with the idea that environmental influences introduce small epigenetic effects that accumulate with time (Fig. 5).
Figure 5.
Why do some patients progress beyond and others stop at an intermediate stage of disease? The complex functional networks at play within a cell or tissue are likely susceptible to small perturbations exerted by environmental influences over long periods of time. The gradual accumulation of altered expression or function of key factors within a system may then lead to dysfunction and disease. Disease-promoting factors (such as smoking, obesity, and poor diet) work to alter the networks in ways that lead to pathological states, whereas disease-preventing factors (such as antioxidant consumption and healthy BMI) oppose the progression of disease by keeping the system within the bounds of healthy function. The ratio of disease-promoting and -preventing influences may balance at different equilibrium points, leading to individual cases that progress only so far along the pathological pathway before reaching that balance point. By discovering the various promoting and preventing factors, along with their respective significances at each point along the disease progression timeline, researchers (and eventually clinicians and patients) will be able to balance the networks to prevent or reverse the progression of disease. Photographs taken from the DeAngelis laboratory cohorts. The study protocol was reviewed and approved by the Institutional Review Board at the University of Utah and conforms to the tenets of the Declaration of Helsinki.
Type 2 diabetes is characterized by hyperglycemia due to impaired insulin secretion by pancreatic β-cells and/or to insulin resistance in peripheral tissues, including skeletal muscle, adipose, and liver. Aging is a major risk factor for type 2 diabetes, along with obesity, low physical activity levels, poor diet, and genetics. DNA methylation at the type 2 diabetes risk genes, NDUFB6 and COX7A1, which encode components of the mitochondrial electron transport chain complexes one and four, respectively, were found to increase with age in skeletal muscle of unaffected individuals.143,144 This increased DNA methylation was also associated with decreased gene expression (RNA and protein for NDUFB6, RNA for COX7A1) in nondiabetic elderly patients, consistent with the DNA methylation having a functional effect on gene expression. Both NDUFB6 and COX7A1 were also shown to have decreased expression in skeletal muscle of type 2 diabetes patients compared with controls.145 Ling et al found that a single-nucleotide polymorphism (SNP, rs629566) that introduces an additional CpG methylation site in the NDUFB6 promoter accounted for the increased methylation in non-diabetic elderly subjects.143 Although decreased expression of NDUFB6 is associated with insulin insensitivity in skeletal muscle, the NDUFB6 expression is decreased in type 2 diabetic patients,145 the authors were unable to find an association between the rs629566 SNP and the risk of developing type 2 diabetes. However, their data did suggest that rs629566 may be associated with insulin-stimulated glucose uptake in elderly subjects. Additionally, a small association with disease was found for another NDUFB6 locus SNP (rs540467) in their data and for two other NDUFB6 locus SNPs in GWAS data mined from other groups. Further work is still needed to elucidate the mechanistic relationships between DNA methylation and genetic variants of NDUFB6 and the development of type 2 diabetes. This gene exemplifies the complex relationships among genetic, epigenetic, and nongenetic contributions to gene expression in aging and the development of chronic pathology.
Many studies have shown links between epigenetics and the development of type 2 diabetes. An early study linked nutrition to heritable, potentially epigenetic changes that increased or decreased the risk for type 2 diabetes.146 Exposure to famine during the paternal grandfather’s childhood is protective against, whereas access to abundant food supplies associates with increased risk for diabetes as a cause of death for his grandchild. Also, studies have shown that diabetes during pregnancy increases the risk for that child to be diagnosed with diabetes as an adult.147
In addition to the multigenerational heritability of epigenetic changes involved in the development of type 2 diabetes, many studies of late have shown evidence for long-term epigenetic changes within an individual with hyperglycemia. The observation of a metabolic memory in which diabetic patients continue to develop complications even after the onset of glycemic control following prior hyperglycemia is consistent with stable epigenetic changes in response to hyperglycemia.148,149 A prospective study comparing patients with an intensive therapy for glycemic control compared to a traditional therapy showed cardiovascular benefits of intensive therapy. These benefits continued in the intensive therapy group 10 years after the trial even though mean glycated hemoglobin levels equilibrated between the two groups within one year following the trial.150 Thus, the early hypoglycemia initiated prephenotypic events that would lead to later diabetic complications independent of later glycemic control.
A genome-wide methylation study looking at differential DNA methylation in skeletal muscle biopsies from type 2 diabetic patients and age-matched controls identified 838 promoter regions that were differentially methylated between the two groups, including the promoter for PGC-1α,151 which functions as a coactivator for the transcription of genes involved in mitochondrial function and biogenesis.152 Further characterization of the PGC-1α promoter by Barrès et al showed hypermethylation in type 2 diabetic patients compared with controls, which negatively correlated with expression of PGC-1α and mitochondrial density.151 Interestingly, the hypermethylation at this locus was found mostly at non-CpG cytosines, which is less common and less well characterized than CpG methylation.151,153 Furthermore, the hypermethylation and associated expression changes were responsive to environmental influences, such as free fatty acid exposure and exercise.151,154
A more recent genome-wide methylation study identified 1,649 genes with differential DNA methylation in pancreatic islets between subjects with type 2 diabetes and normal controls, most of which involved modest changes in percent methylation in the range of 5%–10%.155 Of these 1,649 differentially methylated genes, 102 also showed differential gene expression in type 2 diabetics compared with controls. Pathway clustering of these genes identified pathways enriched within these gene sets, providing insight into the possible pathological mechanisms. Genome-wide studies such as this provide targets for further functional analysis that will lead both to a better understanding of the disease process and to novel therapeutic targets for improved management of the disease.
Numerous PTMs to histone proteins have also been associated with the development of type 2 diabetes and accompanying complications.148,149 One example is that the TNF-α and COX2 promoters have increased H3K9 acetylation in blood monocytes from patients with either type 1 or type 2 diabetes compared with healthy controls.156 H3K9 acetylation is associated with open chromatin structures and increased gene expression. These in vivo data, along with cell culture data, show that diabetic conditions are capable of inducing chromatin modifications to induce the expression of inflammatory genes involved in the diabetic disease process. The complicated interplay of causes and effects in metabolic and epigenetic changes involved in diabetes is likely to be an ongoing research focus for some time that will ultimately provide additional therapeutic targets not only for the stabilization of blood glucose levels but also possibly for the prevention of disease progression and the corresponding complications.
DR is a common complication of diabetes, affecting greater than half of patients having type 2 diabetes for ≥10 years and is the leading cause of blindness among working-age adults.142 Although glaucoma and AMD each have associations with processes, including immune function, which suggest that they may have systemic contributions to the disease processes, DR is clearly the result of systemic disease. DR develops via hyperglycemic induction of vasoactive and inflammatory factors that leads to vascular damage and neuronal injury, further leading to ischemia and the subsequent formation of new vessels, which are prone to bleeding and lead to retinal detachment and ultimately vision loss (Fig. 3D).157,158 Specific epigenetic modifications associated with the development of DR include decreased H3K4me2 in the promoter and enhancer regions of the manganese superoxide dismutase gene, Sod2, in retinas from human donors with DR compared with age-matched nondiabetic controls.159 H3K4me2 is associated with transcriptional activation, and Sod2 expression is also decreased in retinas from human donors with DR compared with age-matched nondiabetic controls. Additionally, the expression of lysine-specific histone demethylase-1, which was shown to demethylate H3K4 at the Sod2 promoter in bovine retinal endothelial cells, was increased in the retina of diabetic donor eyes. Zhong and Kowluru also observed increased H3K9 acetylation and the accompanying NFκB subunit p65 binding at the matrix metalloproteinase-9 (MMP9) promoter in retinas of human donors with DR compared with age-matched controls, consistent with MMP9 expression being increased in DR retinas compared with controls.160 Another study from the Kowluru laboratory used donor retinas with a postmortem time of six to eight hours from patients with established DR and age-matched nondiabetic controls and observed increased expression of Kelch-like erythroid cell-derived protein with Cap’n’colar (CNC) homology (ECH)-associated protein 1 (Keap1), which is an inhibitor of the antioxidative stress agent Nrf2 in the DR retinas compared with controls.161 The increased expression of Keap1 was accompanied by increased Sp1 binding at the Keap1 promoter, which the authors showed to be dependent on the methyltransferase SetD7 in bovine retinal endo-thelial cells, suggesting that monomethylation of H3K4 at the Keap1 promoter is necessary for the increased Keap1 expression in DR.
These studies have provided insights into the epigenetic mechanisms that may underlie gene expression in type 2 diabetes and DR. Genome-wide epigenetic studies are needed to further elucidate the epigenetic processes involved in the progression of DR. In particular, investigations comparing the epigenetic profiles of monozygotic twins discordant for DR will potentially highlight differentially modified loci with significant influences on disease development. Such studies will improve the understanding of disease progression and identify potential opportunities for therapeutic intervention.
Future Directions: Epigenetics as Therapeutics
Studies seeking to understand the various epigenetic changes—and their relationship to gene expression—involved in retinal diseases will inform researchers regarding the intricate molecular mechanisms and processes that lead to the development and progression of complex blinding diseases. With an understanding of the disease mechanisms, researchers will develop treatments to effectively manage the full spectrum from preclinical to advanced stages of disease, including both interventions that prevent clinical manifestations in asymptomatic patients and those that prevent the progression to blinding advanced stages and/or promote the regression of existing disease phenotypes.
Data indicating which genes undergo epigenetic modifications associated with the development of retinal disease will provide potential therapeutic targets for interventions that alternatively fix the aberrant chromatin modification(s), target the particular gene/gene product whose expression is pathologically altered as a result of the aberrant chromatin modification(s), or target proteins or RNAs that interact with or facilitate the effects of the affected gene/gene product. Therapeutics that target chromatin-modifying enzymes, such as HDAC inhibitors and DNMT inhibitors, were initially developed as treatments for cancers, and some are currently in clinical use or clinical trials (see the NIH website for current clinical trial information: ClinicalTrials.gov).162–164 More recently, epigenetic therapeutic agents are being considered as potential treatments for noncancerous complex diseases, such as CVD and diabetes, which share common risk factors with age-related retinal diseases.165–168 Additionally, preclinical studies are investigating the potential use of epigenetic therapeutics for the treatment of AMD and DR.169 However, at the time of writing this report, clinical studies of therapeutic agents that directly modulate epigenetic processes have yet to be executed.
A potential epigenetic intervention was observed in a recent clinical trial looking at specific B-vitamin supplementation. The study looked at the association of folic acid, vitamin B6, and vitamin B12 supplementation on the development of AMD in women with either preexisting CVD or at least three risk factors for CVD, which are also risk factors for AMD.170 The authors found an association between folic acid/vitamin B6/vitamin B12 supplementation and protection against AMD development. An ongoing clinical study is also investigating the effects of folic acid, vitamin B6, and vitamin B12 supplementation on the progression of DR.171 Folic acid is an upstream methyl donor for DNA methylation; vitamins B6 and B12 are involved as cofactors in the process of transferring the methyl group indirectly from folic acid to the DNA. Recent data suggest an association between dietary folic acid intake and global methylation levels.172 Therefore, the association of folic acid and vitamins B6 and B12 supplementation with decreased incidence of AMD may indicate an epigenetic influence on disease progression and, therefore, an epigenetic modulation of disease risk via supplementation. However, the effect of folic acid and vitamins B6 and B12 supplementation on disease risk may result from alternative molecular mechanisms, such as direct antioxidant effects.170 Therefore, additional studies are needed to elucidate the molecular mechanism of folic acid and vitamins B6 and B12 supplementation in the progress of AMD and DR.
Further studies into the epigenetics of blinding retinal diseases will provide therapeutic targets for improved treatment strategies. Current treatments for neovascular AMD have limited results and require frequent intraocular injections, which are not ideal for the patient. The development of therapies that reduce or eliminate the need for injections will greatly reduce the burden of care on patients and the health-care system at large. For example, administration of pharmacological agents orally or via eye drops will not only reduce the pain and trouble associated with current therapies but could also improve vision over the long term.
Perspectives
The progression of complex disease is a multifactorial process influenced by various inputs, including genetic, epigenetic, and environmental elements. Extensive genome-wide analyses are insufficient to explain the development and progression of the complex disease from phenotypically normal through advanced disease. Studies utilizing well-characterized collections of tissue, which take into account tissue integrity and represent each stage of the disease, may help us to understand the molecular changes taking place as the disease progresses. Genome-wide epigenetic and expression data for each stage of the disease will provide candidates for involvement in the progression of disease. However, whether these factors are causes or effects of disease progression will have to be teased out with additional studies designed to determine the mechanistic role of each of these factors.
An ideal experimental setup for genome-wide epigenetic experiments will need to incorporate various features to ensure sufficient power and noise reduction in the resulting data (Fig. 6). Each of the nine characteristics illustrated in Figure 6 contributes to the production of meaningful epigenetic data that can be used to identify genes that are differentially methylated/modified in specific disease states. The generation of high-quality genome-wide epigenetic data pertinent to the development or progression of disease requires that relevant tissue(s) be sampled from both thoroughly phenotyped diseased subjects and well-characterized control subjects who are appropriately age and gender matched. Sampling from both affected and unaffected tissues for both the diseased and the control subjects will also allow for determination of changes that are isolated to the relevant tissue(s). Furthermore, the postmortem interval (time from death to preservation of the tissue sample) is critical for the extraction of accurate information. Both the number and quality of the samples influence the power to identify statistically significant differences associated with disease.
Figure 6.
Ideal experimental setup for genome-wide epigenetic experiments. Each of these nine characteristics contributes to the production of meaningful epigenetic data that can be used to identify genes that are differentially methylated/modified in specific disease states. The combination of robust epigenetic data with RNAseq expression data provides additional information pertaining to the mechanism of disease by correlating expression changes with underlying chromatin modifying processes and providing initial insights into the mechanisms of disease. These studies identify genes and/or regulatory regions that potentially have causative roles in the development or progression of disease. Further mechanistic studies focusing on the roles of each correlated gene/regulatory region will then provide additional understanding of the disease and targets for therapeutic interventions. Incorporation of genetic and lifestyle data will also improve analysis by integrating the full spectrum of risk into studies of disease mechanisms.
The combination of robust high-resolution epigenetic data with RNAseq expression data provides additional information pertaining to the mechanism of disease by correlating expression changes with underlying chromatin modifying processes and providing initial insights into the mechanisms of disease. These studies identify genes and/or regulatory regions that potentially have causative roles in the development or progression of disease. Further mechanistic studies focusing on the roles of each correlated gene/regulatory region will then provide additional understanding of the disease pathways, networks, and points for therapeutic interventions. Incorporation of genetic, lifestyle, and other environmental data will also improve analysis by integrating the full spectrum of risk into studies of disease mechanisms.
Why do some patients’ diseases progress and others’ stop at an intermediate stage? The complex functional networks at play within a cell or tissue are likely susceptible to small perturbations exerted by environmental influences over long periods of time (Fig. 5). The gradual accumulation of altered expression or function of key factors within a system may then lead to dysfunction and disease. Disease-promoting factors (such as smoking, obesity, and poor nutrition) work to alter the networks/pathways in ways that lead to pathological states, whereas disease-preventing factors (such as antioxidant consumption and healthy BMI) oppose the progression of disease by keeping the system within the bounds of healthy function. The ratio of disease-promoting and -preventing influences may balance at different equilibrium points, leading to individual cases that progress only so far along the pathological pathway before reaching that equilibrium point. By discovering the various promoting and preventing factors, along with their respective significances at each point along the disease progression timeline, researchers (and eventually clinicians and patients) will be able to balance the networks and pathways to prevent or reverse the progression of disease. Such interventions are desperately needed to address the growing costs of complex diseases on the quality and duration of life of millions of people. Additionally, therapeutics capable of preventing progression to advanced, debilitating stages of disease will also decrease the overall financial cost of managing these patients’ diseases.
Summary
Scientists in the field need to tease out the various relationships of cause and effect among genetic, environmental, and epigenetic contributions on both gene expression changes and protein interactions which lead to or result from pathological processes, including potentially circular effects of self-propagating cycles of decline.
With an understanding of the complex relationships between genetic, epigenetic, and environmental factors involved in the development of complex diseases, such as degenerative retinal diseases, preventative and therapeutic interventions will follow that will both decrease incidence of debilitating diseases and also reduce the severity of and complications arising from such diseases, thereby improving the quality of life for older patients.
Footnotes
ACADEMIC EDITOR: Lora Talley Watts, Editor in Chief
PEER REVIEW: Three peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1541 words, excluding any confidential comments to the academic editor.
FUNDING: This work was supported by the National Institutes of Health, National Eye Institute (EY014800), an unrestricted grant from Research to Prevent Blindness, Inc., New York, NY, to the Department of Ophthalmology & Visual Sciences, University of Utah, the ARVO Foundation for Eye Research, The Skaggs Foundation for Research, The Carl Marshall Reeves & Mildred Almen Reeves Foundation, Inc., and the Macular Degeneration Foundation, Inc. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Analyzed the data: KLP and MMD. Wrote the first draft of the manuscript: KLP. Contributed to the writing of the manuscript: KLP and MMD. Agree with manuscript results and conclusions: KLP and MMD. Jointly developed the structure and arguments for the paper: KLP and MMD. Made critical revisions and approved final version: KLP and MMD. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Piña B, Brüggemeier U, M Beato. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990;60(5):719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- 2.Gregory PD, Hörz W. Chromatin and transcription—how transcription factors battle with a repressive chromatin environment. Eur J Biochem. 1998;251:1–2. 9–18. doi: 10.1046/j.1432-1327.1998.2510009.x. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications— writers that read. EMBO Rep. 2015;16(11):1467–1481. doi: 10.15252/embr.201540945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wozniak GG, Strahl BD. Hitting the “mark”: interpreting lysine methylation in the context of active transcription. Biochim Biophys Acta. 2014;1839(12):1353–1361. doi: 10.1016/j.bbagrm.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Morinière J, Rousseaux S, Steuerwald U, et al. Cooperative binding of two acety-lation marks on a histone tail by a single bromodomain. Nature. 2009;461(7264):664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- 7.Yang XJ. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays. 2004;26(10):1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Dai X, Qi Y, He Y, Du W, Pang JJ. Histone deacetylases inhibitors in the treatment of retinal degenerative diseases: overview and perspectives. J Ophthalmol. 2015;2015:250812. doi: 10.1155/2015/250812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wątroba M, Szukiewicz D. The role of sirtuins in aging and age-related diseases. Adv Med Sci. 2015;61(1):52–62. doi: 10.1016/j.advms.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 11.Kalish JM, Jiang C, Bartolomei MS. Epigenetics and imprinting in human disease. Int J Dev Biol. 2014;58:2–4. 291–298. doi: 10.1387/ijdb.140077mb. [DOI] [PubMed] [Google Scholar]
- 12.Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet. 2014;15(8):517–530. doi: 10.1038/nrg3766. [DOI] [PubMed] [Google Scholar]
- 13.Senner CE. The role of DNA methylation in mammalian development. Reprod Biomed Online. 2011;22(6):529–535. doi: 10.1016/j.rbmo.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 14.Kim M, Bae M, Na H, Yang M. Environmental toxicants—induced epigenetic alterations and their reversers. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2012;30(4):323–367. doi: 10.1080/10590501.2012.731959. [DOI] [PubMed] [Google Scholar]
- 15.Gao X, Jia M, Zhang Y, Breitling LP, Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin Epigenetics. 2015;7:113. doi: 10.1186/s13148-015-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lardenoije R, Iatrou A, Kenis G, et al. The epigenetics of aging and neurodegeneration. Prog Neurobiol. 2015;131:21–64. doi: 10.1016/j.pneurobio.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zakhari S. Alcohol metabolism and epigenetics changes. Alcohol Res. 2013;35(1):6–16. [PMC free article] [PubMed] [Google Scholar]
- 18.Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S. Emerging role of epigenetics in the actions of alcohol. Alcohol Clin Exp Res. 2008;32(9):1525–1534. doi: 10.1111/j.1530-0277.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- 19.Mazzio EA, Soliman KFA. Epigenetics and nutritional environmental signals. Integr Comp Biol. 2014;54(1):21–30. doi: 10.1093/icb/icu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drong AW, Lindgren CM, McCarthy MI. The genetic and epigenetic basis of type 2 diabetes and obesity. Clin Pharmacol Ther. 2012;92(6):707–715. doi: 10.1038/clpt.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khaire AA, Kale AA, Joshi SR. Maternal omega-3 fatty acids and micronutrients modulate fetal lipid metabolism: a review. Prostaglandins Leukot Essent Fatty Acids. 2015;98:49–55. doi: 10.1016/j.plefa.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Malireddy S, Kotha SR, Secor JD, et al. Phytochemical antioxidants modulate mammalian cellular epigenome: implications in health and disease. Antioxid Redox Signal. 2012;17(2):327–339. doi: 10.1089/ars.2012.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodell A, Rohrer B. A mechanistic review of cigarette smoke and age-related macular degeneration. Adv Exp Med Biol. 2014;801:301–307. doi: 10.1007/978-1-4614-3209-8_38. [DOI] [PubMed] [Google Scholar]
- 24.Chong EW-T, Kreis AJ, Wong TY, Simpson JA, Guymer RH. Alcohol consumption and the risk of age-related macular degeneration: a systematic review and meta-analysis. Am J Ophthalmol. 2008;145(4):707–715. doi: 10.1016/j.ajo.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Sobrin L, Seddon JM. Nature and nurture-genes and environment-predict onset and progression of macular degeneration. Prog Retin Eye Res. 2014;40:1–15. doi: 10.1016/j.preteyeres.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koo E, Neuringer M, SanGiovanni JP. Macular xanthophylls, lipoprotein-related genes, and age-related macular degeneration. Am J Clin Nutr. 2014;100(suppl 1):336S–346S. doi: 10.3945/ajcn.113.071563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Querques G, Souied EH. The role of omega-3 and micronutrients in age-related macular degeneration. Surv Ophthalmol. 2014;59(5):532–539. doi: 10.1016/j.survophthal.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Kay P, Yang YC, Paraoan L. Directional protein secretion by the retinal pigment epithelium: roles in retinal health and the development of age-related macular degeneration. J Cell Mol Med. 2013;17(7):833–843. doi: 10.1111/jcmm.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23(7):781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roadmap Epigenomics Project—Overview. [Accessed August 11, 2015]. Available at: http://www.roadmapepigenomics.org/overview.
- 31.Cvekl A, Wang WL. Retinoic acid signaling in mammalian eye development. Exp Eye Res. 2009;89(3):280–291. doi: 10.1016/j.exer.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vrtačnik P, Ostanek B, Mencej-Bedrač S, Marc J. The many faces of estrogen signaling. Biochem Med. 2014;24(3):329–342. doi: 10.11613/BM.2014.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z, Lv Y, Zhao N, Guan G, Wang J. Protein kinase R-like ER kinase and its role in endoplasmic reticulum stress-decided cell fate. Cell Death Dis. 2015;6:e1822. doi: 10.1038/cddis.2015.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossant J, Hirashima M. Vascular development and patterning: making the right choices. Curr Opin Genet Dev. 2003;13(4):408–412. doi: 10.1016/s0959-437x(03)00080-7. [DOI] [PubMed] [Google Scholar]
- 35.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25(21):2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., III SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16(8):919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultz DC, Friedman JR, Rauscher FJ. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev. 2001;15(4):428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol. 2006;26(22):8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das C, Tyler JK. Histone exchange and histone modifications during transcription and aging. Biochim Biophys Acta. 2013;1819(0):332–342. doi: 10.1016/j.bbagrm.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janicki SM, Tsukamoto T, Salghetti SE, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116(5):683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9(6):1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 42.Prescott DM. The syntheses of total macronuclear protein, histone, and DNA during the cell cycle in Euplotes eurystomus. J Cell Biol. 1966;31(1):1–9. doi: 10.1083/jcb.31.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stein GS, Park WD, Thrall CL, Mans RJ, Stein JL. Cell cycle stage-specific transcription of histone genes. Biochem Biophys Res Commun. 1975;63(4):945–949. doi: 10.1016/0006-291x(75)90660-9. [DOI] [PubMed] [Google Scholar]
- 44.Biterge B, Schneider R. Histone variants: key players of chromatin. Cell Tissue Res. 2014;356(3):457–466. doi: 10.1007/s00441-014-1862-4. [DOI] [PubMed] [Google Scholar]
- 45.Zentner GE, Henikoff S. High-resolution digital profiling of the epigenome. Nat Rev Genet. 2014;15(12):814–827. doi: 10.1038/nrg3798. [DOI] [PubMed] [Google Scholar]
- 46.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155(1):39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrachina M, Ferrer I. DNA methylation of Alzheimer disease and tauopathy-related genes in postmortem brain. J Neuropathol Exp Neurol. 2009;68(8):880–891. doi: 10.1097/NEN.0b013e3181af2e46. [DOI] [PubMed] [Google Scholar]
- 48.Rhein M, Hagemeier L, Klintschar M, Muschler M, Bleich S, Frieling H. DNA methylation results depend on DNA integrity—role of post mortem interval. Front Genet. 2015;6:182. doi: 10.3389/fgene.2015.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coppieters N, Dieriks BV, Lill C, Faull RL, Curtis MA, Dragunow M. Global changes in DNA methylation and hydroxymethylation in Alzheimer’s disease human brain. Neurobiol Aging. 2014;35(6):1334–1344. doi: 10.1016/j.neurobiolaging.2013.11.031. [DOI] [PubMed] [Google Scholar]
- 50.Hodgetts SI, Stagg K, Sturm M, Edel M, Blancafort P. Long live the stem cell: the use of stem cells isolated from post mortem tissues for translational strategies. Int J Biochem Cell Biol. 2014;56:74–81. doi: 10.1016/j.biocel.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 51.Morgan DJ, DeAngelis MM. Differential gene expression in age-related macular degeneration. Cold Spring Harb Perspect Med. 2014;5:a017210. doi: 10.1101/cshperspect.a017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monk D. Germline-derived DNA methylation and early embryo epigenetic reprogramming: the selected survival of imprints. Int J Biochem Cell Biol. 2015;67:128–138. doi: 10.1016/j.biocel.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 53.Moresi V, Marroncelli N, Coletti D, Adamo S. Regulation of skeletal muscle development and homeostasis by gene imprinting, histone acetylation and microRNA. Biochim Biophys Acta. 2015;1849(3):309–316. doi: 10.1016/j.bbagrm.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 54.Álvarez-Errico D, Vento-Tormo R, Sieweke M, Ballestar E. Epigenetic control of myeloid cell differentiation, identity and function. Nat Rev Immunol. 2015;15(1):7–17. doi: 10.1038/nri3777. [DOI] [PubMed] [Google Scholar]
- 55.Cvekl A, Mitton KP. Epigenetic regulatory mechanisms in vertebrate eye development and disease. Heredity. 2010;105(1):135–151. doi: 10.1038/hdy.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ollikainen M, Smith KR, Joo EJ-H, et al. DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum Mol Genet. 2010;19(21):4176–4188. doi: 10.1093/hmg/ddq336. [DOI] [PubMed] [Google Scholar]
- 57.Wan J, Oliver VF, Zhu H, Zack DJ, Qian J, Merbs SL. Integrative analysis of tissue-specific methylation and alternative splicing identifies conserved transcription factor binding motifs. Nucleic Acids Res. 2013;41(18):8503–8514. doi: 10.1093/nar/gkt652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taghdouini AE, Sørensen AL, Reiner AH, et al. Genome-wide analysis of DNA methylation and gene expression patterns in purified, uncultured human liver cells and activated hepatic stellate cells. Oncotarget. 2015;6(29):26729–26745. doi: 10.18632/oncotarget.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schultz MD, He Y, Whitaker JW, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523(7559):212–216. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suderman M, Pappas JP, Borghol N, et al. Lymphoblastoid cell lines reveal associations of adult DNA methylation with childhood and current adversity that are distinct from whole blood associations. Int J Epidemiol. 2015;44(4):1331–1340. doi: 10.1093/ije/dyv168. [DOI] [PubMed] [Google Scholar]
- 61.Bollati V, Schwartz J, Wright R, et al. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130(4):234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brunet A, Berger SL. Epigenetics of aging and aging-related disease. J Gerontol A Biol Sci Med Sci. 2014;69(suppl 1):S17–S20. doi: 10.1093/gerona/glu042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boks MP, Derks EM, Weisenberger DJ, et al. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS One. 2009;4(8):e6767. doi: 10.1371/journal.pone.0006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heyn H, Li N, Ferreira HJ, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A. 2012;109(26):10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Menni C, Kastenmüller G, Petersen AK, et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int J Epidemiol. 2013;42(4):1111–1119. doi: 10.1093/ije/dyt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McClay JL, Aberg KA, Clark SL, et al. A methylome-wide study of aging using massively parallel sequencing of the methyl-CpG-enriched genomic fraction from blood in over 700 subjects. Hum Mol Genet. 2014;23(5):1175–1185. doi: 10.1093/hmg/ddt511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun L, Lin J, Du H, et al. Gender-specific DNA methylome analysis of a Han Chinese longevity population. Biomed Res Int. 2014;2014:396727. doi: 10.1155/2014/396727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao F-H, He Y-H, Li Q-G, Wu H, Luo LH, Kong QP. A genome-wide scan reveals important roles of DNA methylation in human longevity by regulating age-related disease genes. PLoS One. 2015;10(3):e0120388. doi: 10.1371/journal.pone.0120388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang J, Yu L, Gaiteri C, et al. Association of DNA methylation in the brain with age in older persons is confounded by common neuropathologies. Int J Biochem Cell Biol. 2015;67:58–64. doi: 10.1016/j.biocel.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312(5776):1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feser J, Truong D, Das C, et al. Elevated histone expression promotes lifespan extension. Mol Cell. 2010;39(5):724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Greer EL, Maures TJ, Hauswirth AG, et al. Members of the histone H3 lysine 4 trimethylation complex regulate lifespan in a germline-dependent manner in C elegans. Nature. 2010;466(7304):383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dang W, Steffen KK, Perry R, et al. Histone H4 lysine-16 acetylation regulates cellular lifespan. Nature. 2009;459(7248):802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kugel S, Mostoslavsky R. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem Sci. 2014;39(2):72–81. doi: 10.1016/j.tibs.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finkel T, Deng C-X, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460(7255):587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peng C-H, Chang Y-L, Kao C-L, et al. SirT1—a sensor for monitoring self-renewal and aging process in retinal stem cells. Sensors. 2010;10(6):6172–6194. doi: 10.3390/s100606172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mimura T, Kaji Y, Noma H, Funatsu H, Okamoto S. The role of SIRT1 in ocular aging. Exp Eye Res. 2013;116:17–26. doi: 10.1016/j.exer.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 79.Jaliffa C, Ameqrane I, Dansault A, et al. Sirt1 involvement in rd10 mouse retinal degeneration. Invest Ophthalmol Vis Sci. 2009;50(8):3562–3572. doi: 10.1167/iovs.08-2817. [DOI] [PubMed] [Google Scholar]
- 80.Silberman DM, Ross K, Sande PH, et al. SIRT6 is required for normal retinal function. PLoS One. 2014;9(6):e98831. doi: 10.1371/journal.pone.0098831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim H-S, Xiao C, Wang R-H, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12(3):224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sachdeva MM, Cano M, Handa JT. Nrf2 signaling is impaired in the aging RPE given an oxidative insult. Exp Eye Res. 2014;119:111–114. doi: 10.1016/j.exer.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guo Y, Yu S, Zhang C, Kong AN. Epigenetic regulation of Keap1-Nrf2 signaling. Free Radic Biol Med. 2015;88(pt B):337–349. doi: 10.1016/j.freeradbiomed.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.The Eye Diseases Prevalence Research Group Prevalence of open-angle glaucoma among adults in the United States. Arch Ophthalmol. 2004;122(4):532–538. doi: 10.1001/archopht.122.4.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.The Eye Diseases Prevalence Research Group The prevalence of diabetic retinopathy among adults in the United States. Arch Ophthalmol. 2004;122(4):552–563. doi: 10.1001/archopht.122.4.552. [DOI] [PubMed] [Google Scholar]
- 86.The Eye Diseases Prevalence Research Group Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122(4):564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 87.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90(3):262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Resnikoff S, Pascolini D, Etya’ale D, et al. Global data on visual impairment in the year 2002. Bull World Health Organ. 2004;82(11):844–851. [PMC free article] [PubMed] [Google Scholar]
- 89.Kingman S. Glaucoma is second leading cause of blindness globally. Bull World Health Organ. 2004;82(11):887–888. [PMC free article] [PubMed] [Google Scholar]
- 90.Doucette LP, Rasnitsyn A, Seifi M, Walter MA. The interactions of genes, age, and environment in glaucoma pathogenesis. Surv Ophthalmol. 2015;60(4):310–326. doi: 10.1016/j.survophthal.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 91.McDonnell F, O’Brien C, Wallace D. The role of epigenetics in the fibrotic processes associated with glaucoma. J Ophthalmol. 2014;2014:750459. doi: 10.1155/2014/750459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang L, Xu L, Yang H. Risk factors and the progress of primary open-angle glaucoma. China J Ophthalmol. 2009;45(4):380–384. [PubMed] [Google Scholar]
- 93.Sommer A. Glaucoma risk factors observed in the Baltimore Eye Survey. Curr Opin Ophthalmol. 1996;7(2):93–98. doi: 10.1097/00055735-199604000-00016. [DOI] [PubMed] [Google Scholar]
- 94.Kumar DM, Agarwal N. Oxidative stress in glaucoma: a burden of evidence. J Glaucoma. 2007;16(3):334–343. doi: 10.1097/01.ijg.0000243480.67532.1b. [DOI] [PubMed] [Google Scholar]
- 95.Kumar S, Malik MA, Goswami S, Sihota R, Kaur J. Candidate genes involved in the susceptibility of primary open angle glaucoma. Gene. 2015;577(2):119–131. doi: 10.1016/j.gene.2015.11.032. [DOI] [PubMed] [Google Scholar]
- 96.Fautsch MP, Vrabel AM, Johnson DH. The identification of myocilin-associated proteins in the human trabecular meshwork. Exp Eye Res. 2006;82(6 SPEC ISS):1046–1052. doi: 10.1016/j.exer.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 97.He Y, Leung KW, Zhuo Y-H, Ge J. Pro370Leu mutant myocilin impairs mitochondrial functions in human trabecular meshwork cells. Mol Vis. 2009;15:815–825. [PMC free article] [PubMed] [Google Scholar]
- 98.Yam GH-F, Gaplovska-Kysela K, Zuber C, Roth J. Aggregated myocilin induces russell bodies and causes apoptosis: implications for the pathogenesis of myocilin-caused primary open-angle glaucoma. Am J Pathol. 2007;170(1):100–109. doi: 10.2353/ajpath.2007.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Park B-C, Tibudan M, Samaraweera M, Shen X, Yue BY. Interaction between two glaucoma genes, optineurin and myocilin. Genes Cells. 2007;12(8):969–979. doi: 10.1111/j.1365-2443.2007.01102.x. [DOI] [PubMed] [Google Scholar]
- 100.Marco ND, Buono M, Troise F, Diez-Roux G. Optineurin increases cell survival and translocates to the nucleus in a Rab8-dependent manner upon an apoptotic stimulus. J Biol Chem. 2006;281(23):16147–16156. doi: 10.1074/jbc.M601467200. [DOI] [PubMed] [Google Scholar]
- 101.Chi Z-L, Yasumoto F, Sergeev Y, et al. Mutant WDR36 directly affects axon growth of retinal ganglion cells leading to progressive retinal degeneration in mice. Hum Mol Genet. 2010;19(19):3806–3815. doi: 10.1093/hmg/ddq299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Agarwal P, Daher AM, Agarwal R. Aqueous humor TGF-β2 levels in patients with open-angle glaucoma: a meta-analysis. Mol Vis. 2015;21:612–620. [PMC free article] [PubMed] [Google Scholar]
- 103.Browne JG, Ho SL, Kane R, et al. Connective tissue growth factor is increased in pseudoexfoliation glaucoma. Invest Ophthalmol Vis Sci. 2011;52(6):3660–3666. doi: 10.1167/iovs.10-5209. [DOI] [PubMed] [Google Scholar]
- 104.Flügel-Koch C, Ohlmann A, Fuchshofer R, Welge-Lüssen U, Tamm ER. Thrombospondin-1 in the trabecular meshwork: localization in normal and glaucomatous eyes, and induction by TGF-beta1 and dexamethasone in vitro. Exp Eye Res. 2004;79(5):649–663. doi: 10.1016/j.exer.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 105.Rojas A, Meherem S, Kim Y-H, et al. The aberrant methylation of TSP1 suppresses TGF-beta1 activation in colorectal cancer. Int J Cancer. 2008;123(1):14–21. doi: 10.1002/ijc.23608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bruna A, Darken RS, Rojo F, et al. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11(2):147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 107.Yang L, Qu M, Wang Y, et al. Trichostatin A inhibits transforming growth factor-β-induced reactive oxygen species accumulation and myofibroblast differentiation via enhanced NF-E2-related factor 2-antioxidant response element signaling. Mol Pharmacol. 2013;83(3):671–680. doi: 10.1124/mol.112.081059. [DOI] [PubMed] [Google Scholar]
- 108.Zhou Q, Yang L, Wang Y, et al. TGFbeta mediated transition of corneal fibroblasts from a proinflammatory state to a profibrotic state through modulation of histone acetylation. J Cell Physiol. 2010;224(1):135–143. doi: 10.1002/jcp.22110. [DOI] [PubMed] [Google Scholar]
- 109.Kitano S, Morgan J, Caprioli J. Hypoxic and excitotoxic damage to cultured rat retinal ganglion cells. Exp Eye Res. 1996;63(1):105–112. doi: 10.1006/exer.1996.0096. [DOI] [PubMed] [Google Scholar]
- 110.Tezel G, Wax MB. Hypoxia-inducible factor 1alpha in the glaucomatous retina and optic nerve head. Arch Ophthalmol. 2004;122(9):1348–1356. doi: 10.1001/archopht.122.9.1348. [DOI] [PubMed] [Google Scholar]
- 111.Watson JA, Watson CJ, McCrohan A-M, et al. Generation of an epigenetic signature by chronic hypoxia in prostate cells. Hum Mol Genet. 2009;18(19):3594–3604. doi: 10.1093/hmg/ddp307. [DOI] [PubMed] [Google Scholar]
- 112.Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 113.Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 114.Sharp FR, Bernaudin M. HIF1 and oxygen sensing in the brain. Nat Rev Neurosci. 2004;5(6):437–448. doi: 10.1038/nrn1408. [DOI] [PubMed] [Google Scholar]
- 115.Kallio PJ, Okamoto K, O’Brien S, et al. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. EMBO J. 1998;17(22):6573–6586. doi: 10.1093/emboj/17.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhan L, Huang C, Meng X-M, et al. Hypoxia-inducible factor-1alpha in hepatic fibrosis: a promising therapeutic target. Biochimie. 2015;108:1–7. doi: 10.1016/j.biochi.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 117.Shahrzad S, Bertrand K, Minhas K, Coomber BL. Induction of DNA hypomethylation by tumor hypoxia. Epigenetics. 2007;2(2):119–125. doi: 10.4161/epi.2.2.4613. [DOI] [PubMed] [Google Scholar]
- 118.Age-Related Eye Disease Study Research Group A simplified severity scale for age-related macular degeneration: AREDS report no. 18. Arch Ophthalmol. 2005;123(11):1570–1574. doi: 10.1001/archopht.123.11.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yonekawa Y, Kim IK. Clinical characteristics and current treatment of age-related macular degeneration. Cold Spring Harb Perspect Med. 2015;5(1):a017178. doi: 10.1101/cshperspect.a017178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Deangelis MM, Silveira AC, Carr EA, Kim IK. Genetics of age-related macular degeneration: current concepts, future directions. Semin Ophthalmol. 2011;26(3):77–93. doi: 10.3109/08820538.2011.577129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jager RD, Mieler WF, Miller JW. Age-related macular degeneration. N Engl J Med. 2008;358(24):2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- 122.Curcio CA, Medeiros NE, Millican CL. Photoreceptor loss in age-related macular degeneration. Invest Ophthalmol Vis Sci. 1996;37(7):1236–1249. [PubMed] [Google Scholar]
- 123.Klein R, Klein BEK, Jensen SC, Meuer SM. The five-year incidence and progression of age-related maculopathy. Ophthalmology. 1997;104(1):7–21. doi: 10.1016/s0161-6420(97)30368-6. [DOI] [PubMed] [Google Scholar]
- 124.Shah AR, Del Priore LV. Progressive visual loss in subfoveal exudation in age-related macular degeneration: a meta-analysis using Lineweaver-Burke plots. Am J Ophthalmol. 2007;143(1):83.e–89.e. doi: 10.1016/j.ajo.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 125.Stefánsson E, Geirsdóttir A, Sigurdsson H. Metabolic physiology in age related macular degeneration. Prog Retin Eye Res. 2011;30(1):72–80. doi: 10.1016/j.preteyeres.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 126.Consortium TAG. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45(4):433–439. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fritsche LG, Igl W, Bailey JN, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48(2):134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pouw RB, Vredevoogd DW, Kuijpers TW, Wouters D. Of mice and men: the factor H protein family and complement regulation. Mol Immunol. 2015;67(1):12–20. doi: 10.1016/j.molimm.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 129.Kortvely E, Ueffing M. Gene structure of the 10q26 locus: a clue to cracking the ARMS2/HTRA1 riddle? Adv Exp Med Biol. 2016;854:23–29. doi: 10.1007/978-3-319-17121-0_4. [DOI] [PubMed] [Google Scholar]
- 130.Seddon JM, Cote J, Page WF, Aggen SH, Neale MC. The US twin study of age-related macular degeneration: relative roles of genetic and environmental influences. Arch Ophthalmol. 2005;123(3):321–327. doi: 10.1001/archopht.123.3.321. [DOI] [PubMed] [Google Scholar]
- 131.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465(7299):721–727. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- 132.Suuronen T, Nuutinen T, Ryhänen T, Kaarniranta K, Salminen A. Epigenetic regulation of clusterin/apolipoprotein J expression in retinal pigment epithelial cells. Biochem Biophys Res Commun. 2007;357(2):397–401. doi: 10.1016/j.bbrc.2007.03.135. [DOI] [PubMed] [Google Scholar]
- 133.Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009;323(5911):256–259. doi: 10.1126/science.1166226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Economopoulou M, Langer HF, Celeste A, et al. Histone H2AX is integral to hypoxia-driven neovascularization. Nat Med. 2009;15(5):553–558. doi: 10.1038/nm.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hunter A, Spechler PA, Cwanger A, et al. DNA methylation is associated with altered gene expression in AMD. Invest Ophthalmol Vis Sci. 2012;53(4):2089–2105. doi: 10.1167/iovs.11-8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wei L, Liu B, Tuo J, et al. Hypomethylation of the IL17RC promoter associates with age-related macular degeneration. Cell Rep. 2012;2(5):1151–1158. doi: 10.1016/j.celrep.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Oliver VF, Franchina M, Jaffe AE, et al. Hypomethylation of the IL17RC promoter in peripheral blood leukocytes is not a hallmark of age-related macular degeneration. Cell Rep. 2013;5(6):1527–1535. doi: 10.1016/j.celrep.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Oliver V, Jaffe AE, Song J, et al. Differential DNA methylation identified in the blood and retina of AMD patients. Epigenetics. 2015;10(8):698–707. doi: 10.1080/15592294.2015.1060388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.American Diabetes Association® Statistics About Diabetes. [Accessed October 23, 2015]. Available at: http://www.diabetes.org/diabetes-basics/statistics/
- 140.National Eye Institute Facts About Diabetic Eye Disease. [Accessed October 23, 2015]. Available at: https://nei.nih.gov/health/diabetic/retinopathy.
- 141.Stefánsson E, Bek T, Porta M, Larsen N, Kristinsson JK, Agardh E. Screening and prevention of diabetic blindness. Acta Ophthalmol Scand. 2000;78(4):374–385. doi: 10.1034/j.1600-0420.2000.078004374.x. [DOI] [PubMed] [Google Scholar]
- 142.Yau JWY, Rogers SL, Kawasaki R, et al. Meta-Analysis for Eye Disease (META-EYE) Study Group Global prevalence and major risk factors of diabetic retinopathy. Diabetes Care. 2012;35(3):556–564. doi: 10.2337/dc11-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ling C, Poulsen P, Simonsson S, et al. Genetic and epigenetic factors are associated with expression of respiratory chain component NDUFB6 in human skeletal muscle. J Clin Invest. 2007;117(11):3427–3435. doi: 10.1172/JCI30938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rönn T, Poulsen P, Hansson O, et al. Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia. 2008;51(7):1159–1168. doi: 10.1007/s00125-008-1018-8. [DOI] [PubMed] [Google Scholar]
- 145.Mootha VK, Lindgren CM, Eriksson K-F, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 146.Kaati G, Bygren LO, Edvinsson S. Cardiovascular and diabetes mortality determined by nutrition during parents’ and grandparents’ slow growth period. Eur J Hum Genet. 2002;10(11):682–688. doi: 10.1038/sj.ejhg.5200859. [DOI] [PubMed] [Google Scholar]
- 147.Nielsen JH, Haase TN, Jaksch C, et al. Impact of fetal and neonatal environment on beta cell function and development of diabetes. Acta Obstet Gynecol Scand. 2014;93(11):1109–1122. doi: 10.1111/aogs.12504. [DOI] [PubMed] [Google Scholar]
- 148.Schones DE, Leung A, Natarajan R. Chromatin modifications associated with diabetes and obesity. Arterioscler Thromb Vasc Biol. 2015;35(7):1557–1561. doi: 10.1161/ATVBAHA.115.305041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Villeneuve LM, Natarajan R. The role of epigenetics in the pathology of diabetic complications. Am J Physiol Renal Physiol. 2010;299(1):F14–F25. doi: 10.1152/ajprenal.00200.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Holman RR, Paul SK, Bethel MA, Matthews DR, Neil HA. 10-year followup of intensive glucose control in type 2 diabetes. N Engl J Med. 2008;359(15):1577–1589. doi: 10.1056/NEJMoa0806470. [DOI] [PubMed] [Google Scholar]
- 151.Barrès R, Osler ME, Yan J, et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10(3):189–198. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 152.Wu Z, Puigserver P, Andersson U, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 153.Patil V, Ward RL, Hesson LB. The evidence for functional non-CpG methylation in mammalian cells. Epigenetics. 2014;9(6):823–828. doi: 10.4161/epi.28741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Barrès R, Yan J, Egan B, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012;15(3):405–411. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 155.Dayeh T, Volkov P, Salö S, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014;10(3):e1004160. doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Miao F, Gonzalo IG, Lanting L, Natarajan R. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem. 2004;279(17):18091–18097. doi: 10.1074/jbc.M311786200. [DOI] [PubMed] [Google Scholar]
- 157.Chistiakov DA. Diabetic retinopathy: pathogenic mechanisms and current treatments. Diabetes Metab Syndr. 2011;5(3):165–172. doi: 10.1016/j.dsx.2012.02.025. [DOI] [PubMed] [Google Scholar]
- 158.Singh LP, Perrone L. Thioredoxin interacting protein (TXNIP) and pathogenesis of diabetic retinopathy. J Clin Exp Ophthalmol. 2013;4:287. doi: 10.4172/2155-9570.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhong Q, Kowluru RA. Epigenetic modification of Sod2 in the development of diabetic retinopathy and in the metabolic memory: role of histone methylation. Invest Ophthalmol Vis Sci. 2013;54(1):244. doi: 10.1167/iovs.12-10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhong Q, Kowluru RA. Regulation of matrix metalloproteinase-9 by epigenetic modifications and the development of diabetic retinopathy. Diabetes. 2013;62(7):2559–2568. doi: 10.2337/db12-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mishra M, Zhong Q, Kowluru RA. Epigenetic modifications of Keap1 regulate its interaction with the protective factor Nrf2 in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2014;55(11):7256–7265. doi: 10.1167/iovs.14-15193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ramachandran S, Ient J, Göttgens E-L, Krieg AJ, Hammond EM. Epigenetic therapy for solid tumors: highlighting the impact of tumor hypoxia. Genes. 2015;6(4):935–956. doi: 10.3390/genes6040935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. ClinicalTrials.gov Search of: DNMT Inhibitor—List Results. [Accessed October 26, 2015]. Available at: https://clinicaltrials.gov/ct2/results?term=DNMT+inhibitor&Search=Search.
- 164. ClinicalTrials.gov Search of: HDAC Inhibitor—List Results. [Accessed October 26, 2015]. Available at: https://clinicaltrials.gov/ct2/results?term=HDAC+inhibitor&Search=Search.
- 165.D’Onofrio N, Vitiello M, Casale R, Servillo L, Giovane A, Balestrieri ML. Sirtuins in vascular diseases: emerging roles and therapeutic potential. Biochim Biophys Acta. 2015;1852(7):1311–1322. doi: 10.1016/j.bbadis.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 166.Byrne MM, Murphy RT, Ryan AW. Epigenetic modulation in the treatment of atherosclerotic disease. Front Genet. 2014;5:364. doi: 10.3389/fgene.2014.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Christensen DP, Dahllöf M, Lundh M, et al. Histone deacetylase (HDAC) inhibition as a novel treatment for diabetes mellitus. Mol Med. 2011;17:5–6. 378–390. doi: 10.2119/molmed.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rönn T, Ling C. DNA methylation as a diagnostic and therapeutic target in the battle against type 2 diabetes. Epigenomics. 2015;7(3):451–460. doi: 10.2217/epi.15.7. [DOI] [PubMed] [Google Scholar]
- 169.Kwa FAA, Thrimawithana TR. Epigenetic modifications as potential therapeutic targets in age-related macular degeneration and diabetic retinopathy. Drug Discov Today. 2014;19(9):1387–1393. doi: 10.1016/j.drudis.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 170.Christen WG, Glynn RJ, Chew EY, Albert CM, Manson JE. Folic acid, vitamin B6, and vitamin B12 in combination and age-related macular degeneration in a ran-domized trial of women. Arch Intern Med. 2009;169(4):335–341. doi: 10.1001/archinternmed.2008.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Malaguarnera G, Gagliano C, Salomone S, et al. Folate status in type 2 diabetic patients with and without retinopathy. Clin Ophthalmol. 2015;9:1437–1442. doi: 10.2147/OPTH.S77538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Crider KS, Yang TP, Berry RJ, Bailey LB. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role 12. Adv Nutr. 2012;3(1):21–38. doi: 10.3945/an.111.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Aavik E, Lumivuori H, Leppänen O, et al. Global DNA methylation analysis of human atherosclerotic plaques reveals extensive genomic hypomethylation and reactivation at imprinted locus 14q32 involving induction of a miRNA cluster. Eur Heart J. 2015;36(16):993–1000. doi: 10.1093/eurheartj/ehu437. [DOI] [PubMed] [Google Scholar]
- 174.Allione A, Marcon F, Fiorito G, et al. Novel epigenetic changes unveiled by mono-zygotic twins discordant for smoking habits. PLoS One. 2015;10(6):e0128265. doi: 10.1371/journal.pone.0128265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bell CG, Finer S, Lindgren CM, et al. Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus. PLoS One. 2010;5(11):e14040. doi: 10.1371/journal.pone.0014040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Binder AM, LaRocca J, Lesseur C, Marsit CJ, Michels KB. Epigenome-wide and transcriptome-wide analyses reveal gestational diabetes is associated with alterations in the human leukocyte antigen complex. Clin Epigenetics. 2015;7(1):79. doi: 10.1186/s13148-015-0116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Emilsson V, Thorleifsson G, Zhang B, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452(7186):423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 178.Guida F, Sandanger TM, Castagné R, et al. Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Hum Mol Genet. 2015;24(8):2349–2359. doi: 10.1093/hmg/ddu751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Haas J, Frese KS, Park YJ, et al. Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO Mol Med. 2013;5(3):413–429. doi: 10.1002/emmm.201201553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hidalgo B, Irvin MR, Sha J, et al. Epigenome-wide association study of fasting measures of glucose, insulin, and HOMA-IR in the genetics of lipid lowering drugs and diet network study. Diabetes. 2014;63(2):801–807. doi: 10.2337/db13-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Joubert BR, Håberg SE, Nilsen RM, et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120(10):1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Irvin MR, Zhi D, Joehanes R, et al. Epigenome-wide association study of fasting blood lipids in the genetics of lipid-lowering drugs and diet network study. Circulation. 2014;130(7):565–572. doi: 10.1161/CIRCULATIONAHA.114.009158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ko Y-A, Mohtat D, Suzuki M, et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 2013;14(10):R108. doi: 10.1186/gb-2013-14-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Markunas CA, Xu Z, Harlid S, et al. Identification of DNA methylation changes in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2014;122(10):1147–1153. doi: 10.1289/ehp.1307892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Miao F, Smith DD, Zhang L, Min A, Feng W, Natarajan R. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetes. Diabetes. 2008;57(12):3189–3198. doi: 10.2337/db08-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Milenkovic D, Vanden Berghe W, Boby C, et al. Dietary flavanols modulate the transcription of genes associated with cardiovascular pathology without changes in their DNA methylation state. PLoS One. 2014;9(4):e95527. doi: 10.1371/journal.pone.0095527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Monick MM, Beach SRH, Plume J, et al. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(2):141–151. doi: 10.1002/ajmg.b.32021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Movassagh M, Choy M-K, Goddard M, Bennett MR, Down TA, Foo RS. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PLoS One. 2010;5(1):e8564. doi: 10.1371/journal.pone.0008564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Movassagh M, Choy M-K, Knowles DA, et al. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124(22):2411–2422. doi: 10.1161/CIRCULATIONAHA.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nazarenko MS, Markov AV, Lebedev IN, et al. A comparison of genome-wide DNA methylation patterns between different vascular tissues from patients with coronary heart disease. PLoS One. 2015;10(4):e0122601. doi: 10.1371/journal.pone.0122601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nilsson E, Jansson PA, Perfilyev A, et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes. 2014;63(9):2962–2976. doi: 10.2337/db13-1459. [DOI] [PubMed] [Google Scholar]
- 192.Nitert MD, Dayeh T, Volkov P, et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes. 2012;61(12):3322–3332. doi: 10.2337/db11-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Olsson AH, Volkov P, Bacos K, et al. Genome-wide associations between genetic and epigenetic variation influence mRNA expression and insulin secretion in human pancreatic islets. PLoS Genet. 2014;10(11):e1004735. doi: 10.1371/journal.pgen.1004735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rakyan VK, Beyan H, Down TA, et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. 2011;7(9):e1002300. doi: 10.1371/journal.pgen.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rönn T, Volkov P, Gillberg L, et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet. 2015;24(13):3792–3813. doi: 10.1093/hmg/ddv124. [DOI] [PubMed] [Google Scholar]
- 196.Sapienza C, Lee J, Powell J, et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics. 2011;6(1):20–28. doi: 10.4161/epi.6.1.13362. [DOI] [PubMed] [Google Scholar]
- 197.Sharma P, Garg G, Kumar A, et al. Genome wide DNA methylation profiling for epigenetic alteration in coronary artery disease patients. Gene. 2014;541(1):31–40. doi: 10.1016/j.gene.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 198.Steenaard RV, Ligthart S, Stolk L, et al. Tobacco smoking is associated with methylation of genes related to coronary artery disease. Clin Epigenetics. 2015;7(1):54. doi: 10.1186/s13148-015-0088-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Stringhini S, Polidoro S, Sacerdote C, et al. Life-course socioeconomic status and DNA methylation of genes regulating inflammation. Int J Epidemiol. 2015;44(4):1320–1330. doi: 10.1093/ije/dyv060. [DOI] [PubMed] [Google Scholar]
- 200.Suter M, Ma J, Harris A, et al. Maternal tobacco use modestly alters correlated epigenome-wide placental DNA methylation and gene expression. Epigenetics. 2011;6(11):1284–1294. doi: 10.4161/epi.6.11.17819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tsaprouni LG, Yang T-P, Bell J, et al. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics. 2014;9(10):1382–1396. doi: 10.4161/15592294.2014.969637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wan ES, Qiu W, Baccarelli A, et al. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum Mol Genet. 2012;21(13):3073–3082. doi: 10.1093/hmg/dds135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wang X, Falkner B, Zhu H, et al. A genome-wide methylation study on essential hypertension in young African American males. PLoS One. 2013;8(1):e53938. doi: 10.1371/journal.pone.0053938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Xu X, Su S, Barnes VA, et al. A genome-wide methylation study on obesity: differential variability and differential methylation. Epigenetics. 2013;8(5):522–533. doi: 10.4161/epi.24506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yuan W, Xia Y, Bell CG, et al. An integrated epigenomic analysis for type 2 diabetes susceptibility loci in monozygotic twins. Nat Commun. 2014;5:5719. doi: 10.1038/ncomms6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zaina S, Heyn H, Carmona FJ, et al. DNA methylation map of human atherosclerosis. Circ Cardiovasc Genet. 2014;7(5):692–700. doi: 10.1161/CIRCGENETICS.113.000441. [DOI] [PubMed] [Google Scholar]
- 207.Zawada AM, Rogacev KS, Hummel B, et al. SuperTAG methylation-specific digital karyotyping reveals uremia-induced epigenetic dysregulation of atherosclerosis-related genes. Circ Cardiovasc Genet. 2012;5(6):611–620. doi: 10.1161/CIRCGENETICS.112.963207. [DOI] [PubMed] [Google Scholar]