Abstract
TGFβ is a known driver of epithelial-mesenchymal transition (EMT) which is associated with tumor aggressiveness and metastasis. However, EMT has not been fully explored in clinical specimens of castration-resistant prostate cancer (CRPC) metastases. To assess EMT in CRPC, gene expression analysis was performed on 149 visceral and bone metastases from 62 CRPC patients and immunohistochemical analysis was performed on 185 CRPC bone and visceral metastases from 42 CRPC patients. In addition, to assess the potential of metastases to seed further metastases the mitochondrial genome was sequenced at different metastatic sites in one patient. TGFβ was increased in bone versus visceral metastases. While primarily cytoplasmic; nuclear and cytoplasmic Twist were significantly higher in bone than in visceral metastases. Slug and Zeb1 were unchanged, with the exception of nuclear Zeb1 being significantly higher in visceral metastases. Importantly, nuclear Twist, Slug, and Zeb1 were only present in a subset of epithelial cells that had an EMT-like phenotype. Underscoring the relevance of EMT-like cells, mitochondrial sequencing revealed that metastases could seed additional metastases in the same patient. In conclusion, while TGFβ expression and EMT-associated protein expression is present in a considerable number of CRPC visceral and bone metastases, nuclear Twist, Slug, and Zeb1 localization and an EMT-like phenotype (elongated nuclei and cytoplasmic compartment) was only present in a small subset of CRPC bone metastases. Mitochondrial sequencing from different metastases in a CRPC patient provided evidence for the seeding of metastases from previously established metastases, highlighting the biological relevance of EMT-like behavior in CRPC metastases.
Keywords: Prostate cancer, Epithelial mesenchymal transition, TGFβ, Bone metastasis
Introduction
Castration resistant prostate cancer (CRPC) has a propensity to metastasize to bone and often progresses rapidly in the skeletal system. Epithelial mesenchymal transition (EMT) is known to be responsible for the fibroblastoid and migratory properties of metastatic tumor cells and is a prerequisite for metastasis formation. Evidence of EMT has been observed in prostate cancer (PCa) cell lines [24, 42], and a number of factors including TGFβ, and transcription factors Twist, Zeb1/Zeb2, and Snail/Slug have been implicated in promoting this process in PCa [12, 15, 20, 40, 50]. The hallmarks of EMT have been shown to include the loss of epithelial markers, and the expression of mesenchymal markers. The majority of literature on EMT in PCa has been obtained from studies using PCa cell lines and in primary PCa, not in CRPC. We identified TGFB1 on a gene expression array derived from 62 rapid autopsy patients with metastatic CRPC and noted TGFB1 expression was higher in bone metastases relative to visceral metastases. We verified these findings and then assessed TGFβ, Twist, Slug, Zeb1, and vimentin expression on a tissue microarray of bone and visceral metastases from 42 CRPC patients. Each of these factors has been independently reported to be intimately involved in the process of EMT in the literature, but it is unclear if all of these factors play a role in EMT in CRPC [6, 15, 16, 34, 43, 47]. Furthermore, it has been suggested that expression profiles of these EMT-related proteins may be used to predict biochemical recurrence after radical prostatectomy for PCa [3]. We sought to (1) evaluate the presence and prevalence of cells undergoing EMT in CRPC metastases, (2) demonstrate the expression of these EMT-related proteins in CRPC metastases, (3) assess whether the site of metastasis could influence the prevalence of cells undergoing EMT in skeletal and visceral metastases and (4) use a mitochondrial DNA signature highlighting the biological significance of cells undergoing EMT in metastases that have the potential to seed additional metastases.
Materials and methods
Reagents
The antibodies used in this study and the working conditions are listed in Supplemental Table 1.
Tissue acquisition
Samples were obtained from patients who died of metastatic CRPC and who signed written informed consent for a rapid autopsy performed within 6 h of death, under the aegis of the Prostate Cancer Donor Program at the University of Washington [25].
Tissue microarray construction
Human tissue microarrays were constructed using the Beecher Instruments Tissue-Arrayer™ (Beecher Instruments); the first consisting of 27 benign, 25 Gleason pattern 3 prostate carcinoma, and 15 Gleason pattern 4 prostate carcinoma. The second consisted of two replicate 1 mm cores of 65 visceral metastases (35-lymph node; 18-liver; 6-lung; 2-adrenal; 1-brain; 1-periaortic mass) and 120 bone metastases from 42 patients with CRPC as described previously [49]. Thirty-one of the 42 patients had matched bone and visceral metastases, 4 had only visceral metastases represented, and 7 had only bone metastases represented.
RNA isolation
One hundred and forty-nine CRPC metastases from 63 patients were collected at rapid autopsy and stored at −80 °C in OCT (Tissue-Tek). Eight micron frozen sections were placed onto PEN Membrane Frame Slides (Life Technologies) and immediately fixed in 95 % ethanol, briefly stained with Mayer's hematoxylin solution (Sigma-Aldrich) then dehydrated in 100 % ethanol. Laser capture microdissection was performed with an Arcturus Veritas instrument, using both UV cutting and infrared capture lasers, with CapSure® Macro LCM Caps (Life Technologies) collecting 5000–10,000 tumor cells per sample. Digital photographs were taken of tissue sections before, during, and after LCM and assessed by a pathologist to confirm the tumor content. Following LCM, captured cells were lysed in Arcturus RNA Extraction Buffer. RNA was isolated using the Arcturus PicoPure RNA Isolation Kit and the samples were DNAse treated using the Qiagen RNase-Free DNase Set. Subsequently, RNA was amplified for two rounds using the Ambion MessageAmp aRNA Kit [22]. Bone metastases (n = 20) could not be laser captured, as the frozen tissue could not be sectioned. A 1 mm core was obtained from each of 20 bone metastases frozen in OCT that had been previously identified based upon review of H&E sections from corresponding paraffin embedded blocks, adjacent areas of tumor were cored out of the frozen tissue blocks using a 1 mm diameter tissue punch in a −20 °C cryostat. These cores were then stored at −80 °C. Following the manufacturer's instructions, RNA was isolated from the tissue cores using RNeasy Plus Micro Kit (Qiagen). Tissue cores were placed in Buffer RLT Plus with β-mercaptoethanol and homogenized with a disposable hard tissue homogenizer tip (Omni International). The purity and yield of the RNA were determined on a Nano-Drop by using the sample absorbance at 260 nm and 280 nm, both normalized to the background 340 nm. RNA integrity was assessed on an Agilent 2100 Bioanalyzer.
Microarray hybridization
Agilent 44K whole human genome expression oligonucleotide microarrays (Agilent Technologies Inc) were used. Probe labeling and hybridization was performed following the Agilent suggested protocols and fluorescent array images were collected using the Agilent DNA microarray scanner G2565BA.
Realtime PCR
TGFB1 transcript levels were measured using quantitative reverse transcription-PCR (qRT-PCR) in triplicate reactions using 5 ng first strand cDNA, 0.2 μmol/L of each primer, and Power SYBR Green PCR master mix (Applied Biosystems). We normalized mean cycle threshold (Ct) to a housekeeping gene, RPL13A, in the same sample using the ΔCT method. Primers used: TGFB1 Fwd 5′-AAC TCA TTC AGT CAC CAT AGC AAC-3′, TGFB1 Rev 5′-GGG CTT GTT TCC TCA CCT TTA AAA T-3′, RPL13A Fwd 5′-CCT GGA GGA GAA GAG GAA AGA GA-3′, RPL13A Rev 5′-TTG AGG ACC TCT GTG TAT TTG TCA A-3′.
Gene expression and pathway analyses
Data were loess normalized within arrays and quantile normalized between arrays by batch in R using the Limma Bioconductor package. Systematic batch effects were normalized by application of the ComBat function within the sva Bioconductor package to the log2 Cy3 signal intensities. Samples with poor microarray hybridization signals were removed. Data were filtered to remove probes of poor quality hybridization or with mean signal intensities below 200. Groups were compared using the limma package in R with an eBayes moderated t test including patient as a factor in the model. Microarray data are deposited in the Gene Expression Omnibus database under the accession number (GSE74685). Ingenuity pathway Analysis (IPA) was conducted on the 298 differentially expressed genes between bone and visceral tissues based on SAM score ≥3. Upstream regulator analysis was conducted in the IPA to identify the potential key upstream regulators of these 298 genes.
Immunohistochemistry (IHC)
All specimens were formalin fixed and embedded in paraffin respectively (bone metastases were decalcified in 10 % formic acid before paraffin embedding). Five-micron TMA sections were deparaffinized and rehydrated in sequential xylene and graded ethanol series. Antigen retrieval was performed in 10 mM citrate buffer (pH 6.0) in high pressure cooker (15 psi) for 30 min. Endogenous peroxidase and avidin/biotin were blocked respectively (Vector Laboratories Inc.). Sections were then incubated with 5 % normal goat-horse-chicken serum for 1 h at room temperature, followed by primary antibody (Supplemental Table 1), biotinylated secondary antibody (Vector Laboratories Inc.) and ABC reagent (Vector Laboratories Inc.) incubation. Stable DAB (Invitrogen Corp.) was used as chromogen. All sections were lightly counterstained with hematoxylin and mounted with Cytoseal XYL (Richard Allan Scientific). Mouse or rabbit IgG was used at the same concentration as the primary antibody as negative controls.
Immunohistochemical assessment
Immunostaining was assessed using a quasi-continuous scoring system, created by multiplying each optical density level (“0” for negative stain, “1” for faint/equivocal stain, and “2” for definitive stain) by the percentage of cells at each staining level. The sum of the 3 multiplicands provided a final score for each sample (score range was 0–200). The score for each sample was the average of the scores of each duplicate. Cytoplasm and nuclei were evaluated separately. The scores were categorized as “none” (score range: 0), “weak” (score range: 1–100), “moderate” (score range: 101–150) and “intense” (score range: 151–200). Samples with missing or damaged cores were excluded from analysis.
Mitochondrial genome DNA sequencing
The entire mitochondrial genome was sequenced in normal muscle, normal kidney, the primary prostate tumor, and 15 different metastases from one patient by first PCR amplifying the mtDNA with 28 pairs of primers, as previously described [37]. Clonally expanded mutations were scored only when the sequence of the primary or metastatic mtDNA differed from that of the normal tissue. All regions with detected mutations were reamplified and sequenced to rule out the possibility of the mutations being produced by polymerase errors during the PCR or sequencing processes. In addition, to guard against the sample mix-up and contamination that has confounded many mtDNA mutation studies [33], we compared sequences of patient-matched tissues to the revised Cambridge Reference Sequence (rCRS) to confirm they shared common polymorphisms.
Statistical analysis
The statistical analysis of microarray (SAM) program was used to analyze expression differences between groups using unpaired, two-sample t tests and controlled for multiple testing by estimation of q-values using the false discovery rate (FDR) method [38]. Significance of differences for the IHC analyses were calculated using a students t test, with p values ≤0.05 indicating statistical significance.
Results
TGFβ gene and protein is expressed at higher levels in bone metastases relative to visceral metastases in CRPC
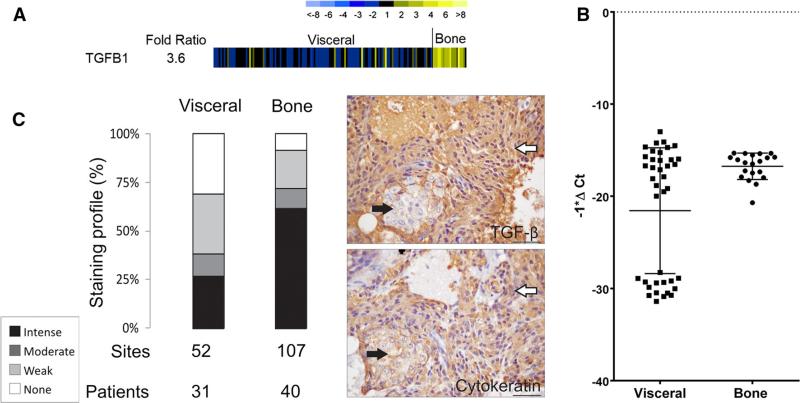
One hundred and forty-nine CRPC metastases divided into visceral metastases (n = 129) and bone metastases (n = 20) were available for gene expression analysis. The gene expression analysis demonstrated that TGFB1 was more highly expressed in bone versus visceral metastases (3.6 fold) (Fig. 1a). Realtime PCR of a subset of the CRPC metastases divided into visceral metastases (n = 39) and bone metastases (n = 20) verified the gene expression analysis data (p = 0.003) (Fig. 1b). Comparing the bone versus visceral metastases the TGFB1 gene was significantly higher in bone (ranked 35). Upstream regulator analysis showed TGFB1 was a top upstream regulator (z-score = 3.45) in bone metastases and predicted that TGFβ was activated in bone versus visceral tissues (p = 0.013, data not shown). There is broad experimental support for an important role of TGFβ in EMT associated with cancer progression [10, 13, 17] and several key transcription factors in the EMT pathway are downstream of TGFβ including TWIST, SLUG, and ZEB1 [8, 29, 45]. Using a TMA of 185 metastases (159 were available for analysis) from 42 CRPC patients we showed that TGFβ was significantly higher by IHC in bone metastases relative to visceral metastases (p < 0.001) (Fig. 1c). In addition, the expression of TGFβ was present throughout the epithelial component of a bone metastatic site, or was more highly expressed in regions containing cytokeratin positive cells that had a mesenchymal like phenotype (elongated nuclei and cytoplasm). This differential expression is highlighted in Fig. 1c and Supplemental Fig. 1. These data led us to conclude that TGFβ was more highly expressed in CRPC bone metastases, and suggested that other EMT-associated proteins could also be highly expressed in bone metastases relative to visceral metastases. However, it is important to note that there was a subset of visceral metastases (that were not tissue specific) that displayed little to no expression of TGFB1/TGFβ in the realtime-PCR and IHC analysis that may account for this difference (Fig. 1b, c).
Fig. 1.
a Agilent™ oligo array expression analysis of TGFB1 in CRPC metastases. There was a 3.6 fold difference in mean-centered ratios between visceral metastases versus bone metastases. Yellow indicates higher expression and blue lower expression. b Realtime PCR for TGFB1 between visceral metastases versus bone metastases. TGFB1 was higher in bone metastases (p = 0.003). c Cytoplasmic TGFβ expression is significantly higher in bone metastases than visceral metastases (p < 0.001). Staining intensities (intense, moderate, weak, or no staining) are described in the inset. Patients—represents the number of patients represented, sites—represents the number of metastases. To highlight the epithelial phenotype, images of serial sections of a bone metastasis stained for TGFβ and Pan-cytokeratin are shown to the right. White arrows highlight epithelial cells with elongated nuclei and the black arrows highlight cells with round nuclei. Bar = 40 microns. (Color figure online)
Twist is associated with a fibroblast-like epithelial phenotype in CRPC bone metastases
Increased expression of both TGFβ and Twist are associated with the cadherin switch in EMT and play an important role in the development of PCa [23]. The role of Twist in the down-modulation of E-cadherin is well-established and Twist expression has been shown to predict the metastatic potential of primary PCa [46]. Inactivation of Twist suppresses the migratory and invasive abilities of PCa cells and this is correlated with induction of E-cadherin expression as well as morphologic and molecular changes associated with mesenchymal to epithelial transition (MET) [21]. Nuclear Twist and Twist at the invasive margin of a primary tumor is associated with increasing Gleason grade and recurrence in primary PCa [21, 30]. To confirm nuclear and cytoplasmic Twist expression in primary PCa, benign (n = 27), Gleason pattern 3 prostate carcinoma (n = 25), and Gleason pattern 4 prostate carcinoma (n = 15) tissues on a TMA were stained for Twist. In primary PCa significantly more nuclear Twist was present in Gleason pattern 3 tumors when compared to benign prostate (p < 0.001) and in Gleason pattern 4 tumors when compared to Gleason pattern 3 tumors (p = 0.038) (Fig. 2a). Cytoplasmic Twist was significantly higher in Gleason pattern 3 tumors when compared to benign prostate (p = 0.007), with only a trend towards higher expression observed in Gleason pattern 4 tumors relative to benign tissue (p = 0.068) (Fig. 2a). Additionally, using TMAs of metastatic CRPC bone (n = 117) and visceral metastases (n = 65) we determined both nuclear and cytoplasmic Twist were significantly higher in bone when compared to visceral metastases (p < 0.05). Further, we observed that only a small proportion of samples had nuclear Twist (Fig. 2b). Importantly, the tumor cells in the bone metastases that expressed nuclear Twist had a more mesenchymal phenotype while retaining cytokeratin expression (Fig. 2b). We wanted to determine why only a small number of metastases on the TMA expressed nuclear Twist. Was it because (1) a limited number of CRPC metastases have nuclear Twist, or (2) nuclear Twist is present in a subset of tumor cells? To address this question, 1.1 cm (diameter) CRPC metastatic bone biopsies were stained for Twist. Regional expression of nuclear Twist could be observed in a subset of fibroblast-like cells. To ensure the subset of fibroblast-like cells expressing nuclear Twist were epithelial we cut and stained serial sections. The elongated fibroblast-like cells with elongated nuclei were PAN-cytokeratin positive and positive for nuclear Twist. The cells were generally associated with adjoining regions of epithelial cells with a more epithelial phenotype, rounded nuclei and in many cases displaying cytoplasmic rather than nuclear Twist expression (Supplemental Fig. 2).
Fig. 2.
Twist expression in human primary PCa and CRPC metastases. a Cytoplasmic and nuclear staining profile of Twist in benign/normal prostate (NP), Gleason (Gl) pattern 3 and 4 prostate carcinoma. Both nuclear and cytoplasmic Twist expression is higher with increasing Gleason grades. b Nuclear Twist is expressed in a subset of cells displaying EMT-like behavior in CRPC bone and visceral metastases. Cytoplasmic and nuclear Twist expression is higher bone metastases (p < 0.001). Staining intensities (intense, moderate, weak, or no staining) are described in the inset in Fig. 1. Patients—represents the number of patients represented, sites—represents the number of metastases. Examples of nuclear Twist and Pan-cytokeratin staining in a CRPC bone metastasis are shown to the lower right. White arrows highlight epithelial cells with elongated nuclei and the black arrows highlight cells with round nuclei. Bar = 40 microns
Slug and Zeb1 are also associated with a fibroblast-like epithelial phenotype in CRPC bone metastases
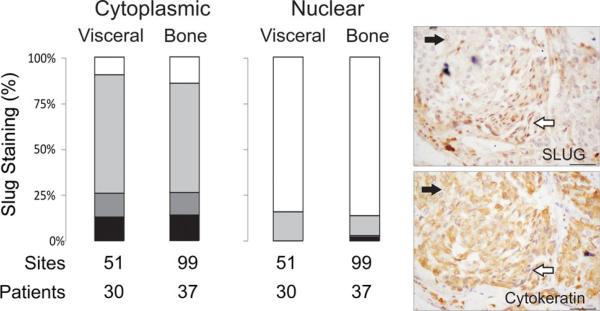
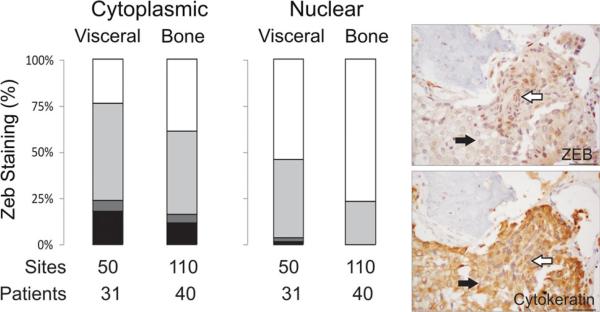
Snail/Slug and Zeb1/Zeb2 are transcription factors that repress E-cadherin [2, 6, 14, 39]. Slug and Zeb1 are well-known transcription factors downstream of TGFβ and Twist and there is a plethora of literature relating the role of Slug in EMT, cell proliferation and invasiveness of metastatic PCa cell lines [15, 41, 48]. Zeb1 expression is seen in highly aggressive PCa cells and correlates directly with Gleason grade in human prostate tumors [1]. Additionally, the reversal of the EMT phenotype in PC3 cells has been associated with the down-regulation of ZEB1, ZEB2, and SLUG [11, 12]. Similar to Twist, the majority of literature on Slug and Zeb1 activity is based on the analysis of primary PCa, cell culture, and xenograft studies. To our knowledge, it has not yet been assessed in a large cohort of CRPC bone and visceral metastases. Similar to Twist, the majority of bone and visceral metastases on the tissue microarrays expressed cytoplasmic Slug and Zeb1, with a minority displaying nuclear localization of the proteins. Unlike Twist we did not observe differential expression of Slug or Zeb1 in bone versus visceral metastases; with the exception of nuclear Zeb1 being significantly higher in visceral metastases (p < 0.05) (Figs. 3, 4). We further examined whole sections of CRPC bone metastases to determine if, similar to Twist, a population of epithelial cells with an EMT-like morphology displayed Slug and Zeb1 nuclear localization in CRPC bone metastases. Nuclear localization of Slug and Zeb1 was observed in a subset of fibroblast-like cells that were positive for cytokeratin, typically observed on the periphery of a tumor mass close to the stromal compartment (Supplemental Figs. 3, 4).
Fig. 3.
Slug, a known transcription factor for EMT-related proteins, is not differentially expressed in bone versus visceral metastases, but is seen in a subset of cells displaying EMT-like behavior. Expression of Cytoplasmic and nuclear Slug expression is not significantly different in bone metastases vs visceral metastases. Staining intensities (intense, moderate, weak, or no staining) are described in the inset in Fig. 1. Patients—represents the number of patients represented, sites—represents the number of metastases. To highlight the epithelial phenotype, images of serial sections of a bone metastasis stained for Slug and Pan-cytokeratin are shown to the right. White arrows highlight epithelial cells with elongated nuclei and the black arrows highlight cells with round nuclei. Bar = 40 microns
Fig. 4.
The transcription factor Zeb1, another important driver of EMT is also seen in the nuclei of a subset of cells displaying EMT-like behavior but was not differentially expressed in bone versus visceral metastases. Cytoplasmic and nuclear Zeb1 expression is not significantly different in bone metastases vs visceral metastases. Staining intensities (intense, moderate, weak, or no staining) are described in the inset in Fig. 1. Patients—represents the number of patients represented, sites—represents the number of metastases. To highlight the epithelial phenotype, images of serial sections of a bone metastasis stained for Zeb1 and Pan-cytokeratin are shown to the right. White arrows highlight epithelial cells with elongated nuclei and the black arrows highlight cells with round nuclei. Bar = 40 microns
Absence of vimentin in the epithelial compartment of CRPC metastases
In addition to demonstrating the absence of epithelial markers in EMT, the presence of certain mesenchymal cell markers is commonly used to identify cells that have completed the transition. Vimentin is one such marker that correlates with the down-modulation of epithelial cytokeratins and is said to be a marker of the mesenchymal state of ‘completely transitioned’ cells [4, 16]. However, while vimentin was present in the stromal compartment of the CRPC metastases, no vimentin was observed in the epithelial compartment (Supplemental Fig. 5).
Mitochondrial DNA sequencing underscores the biological relevance of EMT-like tumor cells in CRPC metastases
It could be argued that the EMT-like cells in the CRPC metastases are not highly relevant as all of the metastases in a given patient have already seeded from the original primary tumor. However, if metastases can seed new metastases then the presence of EMT-like cells in established metastases could be a potential target for therapy. Since mitochondrial DNA can accumulate mutations over time, we focused on using mitochondrial DNA to map the spread of tumor cells in a patient. We reasoned that any mitochondrial mutations present in the primary tumor would be present in all metastasis in a given patient. However, any additional mitochondrial mutations that accumulated in a given metastasis would only be present in other metastases if they were derived from that metastasis rather than the primary tumor. Therefore, if additional mitochondrial mutation(s) were present in multiple metastases this would provide evidence that metastases could seed new metastases. To this end, we used mitochondrial DNA mutational spectra to map the spread of tumor cells in a patient to determine whether metastases can seed further metastases in a given patient. To identify the mitochondrial DNA mutational spectra in CRPC metastases, mitochondrial DNA was sequenced in one patient from the primary tumor, 15 different metastases, and normal muscle and kidney to obtain the germ line consensus sequence in an individual patient (Table 1; Supplemental Fig. 6). No mutations were observed in the germ line consensus sequence from normal tissues. Importantly, a somatic mutation in the ND1 gene was identified in the primary prostate cancer in the patient. Five metastases displayed the same mutation in the ND1 gene. Eight further metastases possessed the same mutation in the ND1 gene and acquired an additional mutation in the ND5 gene. Two further independent metastases had acquired the ND1 and ND5 mutation and gained two independent mutations in a tRNA gene and the ND2 gene. These data point to the acquisition of novel mitochondrial mutations as the tumor cells metastasized from one site to another, underscoring the ability of an individual metastasis to seed further metastases in this CRPC patient.
Table 1.
Somatic mtDNA mutations in metastases from a CRPC patient
| Tissue | Somatic mtDNA mutation (position type) |
|||
|---|---|---|---|---|
| 4048 G>A | 12390 ins(C) | 4430 ins(G) | 4480 T>G | |
| Kidney (normal) | – | – | – | – |
| Muscle (normal) | – | – | – | – |
| Prostate (primary tumor) | + | – | – | – |
| Adrenal | + | – | – | – |
| Lung | + | – | – | – |
| Retroperitoneal lymph | + | – | – | – |
| Sternum | + | – | – | – |
| Vertebrae, T10 | + | – | – | – |
| Liver | + | + | – | – |
| Spleen | + | + | – | – |
| Vertebrae, L1 | + | + | – | – |
| Vertebrae, L2 | + | + | – | – |
| Vertebrae, L4 | + | + | – | – |
| Vertebrae, L5 | + | + | – | – |
| Vertebrae, T8 | + | + | – | – |
| Vertebrae, T9 | + | + | – | – |
| Pelvic lymph node | + | + | + | – |
| Tracheal lymph node | + | + | – | + |
The mitochondrial genome was sequenced from two normal tissues, the primary tumor, and fifteen separate metastases in a castration-resistant prostate cancer patient. Somatic mtDNA mutations were discovered in four genes among the metastases: ND1 (4048 G>A), ND5 (12390 insert C), tRNA M (4430 insert G), ND2 (4480 T>G). mtDNA sites are labeled as wild-type (“–”) or mutant (“+”) for each tissue
Discussion
EMT provides a cytoarchitecture that is permissive for greater epithelial cell motility. In PCa this process has been associated with increased migratory and invasive potential and has been interrogated in a considerable number of model systems. While there is ample evidence that EMT can occur in vitro with cancer cell lines, there is some debate on the occurrence of EMT in neoplasia and little evidence of EMT occurring in PCa patients [19, 26, 36]. Our data suggests that a subset of PCa cells in a CRPC metastasis, especially bone metastases, can undergo partial-EMT retaining aspects of their epithelial phenotype while morphologically resembling a more fibroblast-like cell. These EMT-like cells were associated with an epithelial component within the tumor mass that did not share the fibroblast-like phenotype. The epithelial component present in the metastases might have been derived from tumor cells that underwent an EMT-like transition, disseminated from the primary tumor to the site of metastasis and have reverted back to a more epithelial phenotype undergoing mesenchymal-epithelial transition (MET). There is evidence for MET in breast cancer [7]. However this is a topic for discussion in PCa, with conflicting publications suggesting higher and lower expression of E-cadherin in PCa bone metastases when compared to primary tumors [5, 9, 32] [27]. Putzke et al., in a more comprehensive set of CRPC bone metastases has shown significantly higher E-cadherin expression in bone versus lymph node or visceral metastases, with considerable variation in the expression of E-cadherin in bone metastases [28]. We propose that partial-EMT is occurring in a subset of epithelial cells that have disseminated to the bone, typically located on the periphery of the tumor mass.
TGFβ initiates EMT in mature epithelial cells through autocrine and paracrine activation of intracellular signaling molecules that trigger transcriptional reprogramming of the cell [47]. Maintenance of autocrine activity is required for TGFβ signaling which, along with other oncogenic pathways, has been suggested to sustain the mesenchymal phenotype of metastatic tumor cells [47]. A number of studies have shown that inhibition of TGFβ can reduce the metastatic and/or invasive properties of cancer presumably by preventing activation of EMT pathways [13, 35, 44]. As stated previously, TGFβ induces EMT to stimulate cancer progression. We observed significantly more TGFβ in CRPC bone metastases than in visceral metastases and TGFβ was associated with a more fibroblast-like epithelial phenotype. Twist is a transcription factor that down-regulates E-cadherin expression. Our IHC analysis of Twist in primary PCa is consistent with a previous report showing that nuclear Twist is associated with increasing aggressiveness in the epithelium of primary PCa [46]. Further, we have shown that the majority of CRPC bone metastases were Twist positive with a subset of nuclear Twist positive epithelial cells displaying a fibroblast-like phenotype. It is important to note that while Twist was expressed in the majority of CRPC bone metastases, as a nuclear transcription factor it might only be transcriptionally active in a subset of epithelial cells where Twist is localized to the nucleus. We hypothesize here that the fibroblast-like nuclear Twist positive epithelial cells in the bone metastases have the ability to seed secondary metastases in PCa.
In addition a similar subset of fibroblast-like epithelial cells also express EMT regulating transcription factors, Slug and Zeb1. Twist, Slug, and Zeb1 are all transcription factors that regulate E-cadherin expression during development [18]. It is noteworthy that Twist, Slug and Zeb1 were localized to the nucleus of fibroblast-like epithelial cells, highlighting their potential for repressing E-cadherin, loosening cancer cell adhesion therefore facilitating metastases during PCa progression.
Our data demonstrate that TGFβ and the EMT-related transcription factor Twist, but not EMT regulators Slug and Zeb1 are more highly expressed in CRPC bone metastases than visceral metastases, potentially explaining the high frequency and rapid progression of skeletal metastatic disease. More importantly our data show that while a considerable number of metastases express EMT-associated factors, that these factors may only be active in a subset of cells at any given time. Therefore when assessing the presence or absence of an EMT-like phenotype in patient samples it is important to further investigate the localization of the protein and the morphology of the tumor cells relative to their counterparts in the tumor mass.
We recognize that not all of the CRPC metastases in this study expressed high levels of Twist, Slug, and Zeb1. This heterogeneous phenotype in CRPC metastases is consistent with a previous report [31]. It may be that in some cases Twist may be expressed where Slug and/or Zeb1 are not, some metastases could be highly differentiated leading to decreased or limited expression of TGFβ, Twist, Slug or Zeb1. Whether the expression of these factors impacts the rate of metastatic spread remains to be seen.
It has been suggested that if EMT is an early event, one may not have an opportunity to intervene [26]. However, the staining of the CRPC bone metastases has provided evidence that in late stage disease, a subset of epithelial cells can retain the partial-EMT phenotype. Additionally, we detail using the mitochondrial genome in one CRPC patient with multiple metastases that the addition of new mutations to the mitochondrial genome maps out the stepwise progression in the evolution and development of new metastases from previously established metastases. The potential for a metastasis to seed further metastases highlights a potential role for the EMT-like cells in the seeding of new metastases in late stage disease. Our data suggest that while partial-EMT may occur during dissemination from the primary site, that there is a subset of epithelial cells in CRPC bone metastases that still retain the characteristics of motile epithelial cells that are undergoing partial-EMT with the potential to seed new metastases. This highlights the unique potential opportunity to target tumor cell dissemination and prevent the emergence of additional metastases in patients with disseminated disease.
Supplementary Material
Acknowledgments
We would like to thank the patients and their families who were willing to participate in the Prostate Cancer Donor Program, for without them research of this nature would not be possible. Additionally, we would also like to thank Khanhthy Doan, Funda Vakar-Lopez, Maria Tretiakova, Evan Yu, Elahe Mostaghel and the rapid autopsy teams in the Urology Department at the University of Washington. This material is the result of work supported by resources from the VA Puget Sound Health Care System, Seattle, Washington (RLV is a VA Biomedical Laboratory R&D Senior Research Career Scientist, PHL is a Staff Physician), the Pacific Northwest Prostate Cancer SPORE (P50CA97186), the PO1 NIH grant (PO1CA085859), the LUCAS Foundation, W81XWH-10-1-0563 from the CDMRP/U.S. Department of Defense, and an Outstanding New Environmental Scientist Award (ONES) (R01) from the National Institute of Environmental Health Sciences (R01 ES019319). CM is a recipient of a Career Development Award from Jim and Cathrine Allchin.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10585-015-9773-7) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest None.
References
- 1.Anose BM, LaGoo L, Schwendinger J. Characterization of androgen regulation of ZEB-1 and PSA in 22RV1 prostate cancer cells. Adv Exp Med Biol. 2008;617:541–546. doi: 10.1007/978-0-387-69080-3_55. [DOI] [PubMed] [Google Scholar]
- 2.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, De Garcia HA. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 3.Behnsawy HM, Miyake H, Harada K, Fujisawa M. Expression patterns of epithelial-mesenchymal transition markers in localized prostate cancer: significance in clinicopathological outcomes following radical prostatectomy. BJU Int. 2013;111:30–37. doi: 10.1111/j.1464-410X.2012.11551.x. [DOI] [PubMed] [Google Scholar]
- 4.Boyer B, Tucker GC, Valles AM, Franke WW, Thiery JP. Rearrangements of desmosomal and cytoskeletal proteins during the transition from epithelial to fibroblastoid organization in cultured rat bladder carcinoma cells. J Cell Biol. 1989;109:1495–1509. doi: 10.1083/jcb.109.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryden AA, Hoyland JA, Freemont AJ, Clarke NW, Schembri WD, George NJ. E-cadherin and beta-catenin are down-regulated in prostatic bone metastases. BJU Int. 2002;89:400–403. doi: 10.1046/j.1464-4096.2001.01712.x. [DOI] [PubMed] [Google Scholar]
- 6.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 7.Chao YL, Shepard CR, Wells A. Breast carcinoma cells re-express E-cadherin during mesenchymal to epithelial reverting transition. Mol Cancer. 2010;9:179. doi: 10.1186/1476-4598-9-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das S, Becker BN, Hoffmann FM, Mertz JE. Reversal of transforming growth factor-beta induced epithelial-to-mesenchymal transition and the ZEB proteins. Fibrogenesis Tissue Repair. 2012;5:S28. doi: 10.1186/1755-1536-5-S1-S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Marzo AM, Knudsen B, Chan-Tack K, Epstein JI. E-cadherin expression as a marker of tumor aggressiveness in routinely processed radical prostatectomy specimens. Urology. 1999;53:707–713. doi: 10.1016/s0090-4295(98)00577-9. [DOI] [PubMed] [Google Scholar]
- 10.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 11.Drake JM, Barnes JM, Madsen JM, Domann FE, Stipp CS, Henry MD. ZEB1 coordinately regulates laminin-332 and {beta}4 integrin expression altering the invasive phenotype of prostate cancer cells. J Biol Chem. 2010;285:33940–33948. doi: 10.1074/jbc.M110.136044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drake JM, Strohbehn G, Bair TB, Moreland JG, Henry MD. ZEB1 enhances transendothelial migration and represses the epithelial phenotype of prostate cancer cells. Mol Biol Cell. 2009;20:2207–2217. doi: 10.1091/mbc.E08-10-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumont N, Arteaga CL. Targeting the TGF beta signaling network in human neoplasia. Cancer Cell. 2003;3:531–536. doi: 10.1016/s1535-6108(03)00135-1. [DOI] [PubMed] [Google Scholar]
- 14.Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 15.Emadi BM, Soheili ZS, Essmann F, Deezagi A, Engers R, Goering W, Schulz WA. Slug/SNAI2 regulates cell proliferation and invasiveness of metastatic prostate cancer cell lines. Tumour Biol. 2010;31:297–307. doi: 10.1007/s13277-010-0037-5. [DOI] [PubMed] [Google Scholar]
- 16.Franke WW, Grund C, Kuhn C, Jackson BW, Illmensee K. Formation of cytoskeletal elements during mouse embryogenesis. III. Primary mesenchymal cells and the first appearance of vimentin filaments. Differentiation. 1982;23:43–59. doi: 10.1111/j.1432-0436.1982.tb01266.x. [DOI] [PubMed] [Google Scholar]
- 17.Gotzmann J, Mikula M, Eger A, Schulte-Hermann R, Foisner R, Beug H, Mikulits W. Molecular aspects of epithelial cell plasticity: implications for local tumor invasion and metastasis. Mutat Res. 2004;566:9–20. doi: 10.1016/s1383-5742(03)00033-4. [DOI] [PubMed] [Google Scholar]
- 18.Hugo HJ, Kokkinos MI, Blick T, Ackland ML, Thompson EW, Newgreen DF. Defining the E-cadherin repressor interactome in epithelial-mesenchymal transition: the PMC42 model as a case study. Cells Tissues Organs. 2011;193:23–40. doi: 10.1159/000320174. [DOI] [PubMed] [Google Scholar]
- 19.Keshamouni VG, Schiemann WP. Epithelial-mesenchymal transition in tumor metastasis: a method to the madness. Future Oncol. 2009;5:1109–1111. doi: 10.2217/fon.09.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, Kim HR, Sarkar FH. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27:1712–1721. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C, Zhang X, Chua CW, Chan KW, Chan FL, Glackin C, Wong YC, Wang X. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153–5162. doi: 10.1158/0008-5472.CAN-04-3785. [DOI] [PubMed] [Google Scholar]
- 22.Larson SR, Zhang X, Dumpit R, Coleman I, Lakely B, Roudier M, Higano CS, True LD, Lange PH, Montgomery B, Corey E, Nelson PS, Vessella RL, Morrissey C. Characterization of osteoblastic and osteolytic proteins in prostate cancer bone metastases. Prostate. 2013;73:932–940. doi: 10.1002/pros.22639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu GL, Yang HJ, Liu T, Lin YZ. Expression and significance of E-cadherin, N-cadherin, transforming growth factor-beta1 and Twist in prostate cancer. Asian Pac J Trop Med. 2014;7:76–82. doi: 10.1016/S1995-7645(13)60196-0. [DOI] [PubMed] [Google Scholar]
- 24.Luo Y, He DL, Ning L. Expression of “epithelial-mesenchymal transition” associated proteins in prostate cancer cell lines with different metastatic potentials and its significance. Zhonghua Nan Ke Xue. 2006;12:696–700. [PubMed] [Google Scholar]
- 25.Morrissey C, Roudier MP, Dowell A, True LD, Ketchanji M, Welty C, Corey E, Lange PH, Higano CS, Vessella RL. Effects of androgen deprivation therapy and bisphosphonate treatment on bone in patients with metastatic castration-resistant prostate cancer: results from the University of Washington Rapid Autopsy Series. J Bone Miner Res. 2013;28:333–340. doi: 10.1002/jbmr.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nauseef JT, Henry MD. Epithelial-to-mesenchymal transition in prostate cancer: paradigm or puzzle? Nat Rev Urol. 2011;8:428–439. doi: 10.1038/nrurol.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pontes J, Jr, Srougi M, Borra PM, Dall’ Oglio MF, Ribeiro-Filho LA, Leite KR. E-cadherin and beta-catenin loss of expression related to bone metastasis in prostate cancer. Appl Immunohistochem Mol Morphol. 2010;18:179–184. doi: 10.1097/PAI.0b013e3181640bca. [DOI] [PubMed] [Google Scholar]
- 28.Putzke AP, Ventura AP, Bailey AM, Akture C, Opoku-Ansah J, Celiktas M, Hwang MS, Darling DS, Coleman IM, Nelson PS, Nguyen HM, Corey E, Tewari M, Morrissey C, Vessella RL, Knudsen BS. Metastatic progression of prostate cancer and e-cadherin regulation by zeb1 and SRC family kinases. Am J Pathol. 2011;179:400–410. doi: 10.1016/j.ajpath.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao B, Johnson NW, Gao J. Epithelial-mesenchymal transition in oral squamous cell carcinoma triggered by transforming growth factor-beta1 is Snail family-dependent and correlates with matrix metalloproteinase-2 and -9 expressions. Int J Oncol. 2010;37:663–668. doi: 10.3892/ijo_00000715. [DOI] [PubMed] [Google Scholar]
- 30.Raatikainen S, Aaltomaa S, Palvimo JJ, Karja V, Soini Y. TWIST overexpression predicts biochemical recurrence-free survival in prostate cancer patients treated with radical prostatectomy. Scand J Urol. 2014;49:51. doi: 10.3109/21681805.2014.909529. [DOI] [PubMed] [Google Scholar]
- 31.Roudier MP, True LD, Higano CS, Vesselle H, Ellis W, Lange P, Vessella RL. Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum Pathol. 2003;34:646–653. doi: 10.1016/s0046-8177(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 32.Rubin MA, Mucci NR, Figurski J, Fecko A, Pienta KJ, Day ML. E-cadherin expression in prostate cancer: a broad survey using high-density tissue microarray technology. Hum Pathol. 2001;32:690–697. doi: 10.1053/hupa.2001.25902. [DOI] [PubMed] [Google Scholar]
- 33.Salas A, Yao YG, Macaulay V, Vega A, Carracedo A, Bandelt HJ. A critical reassessment of the role of mitochondria in tumorigenesis. PLoS Med. 2005;2:e296. doi: 10.1371/journal.pmed.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slabakova E, Pernicova Z, Slavickova E, Starsichova A, Kozubik A, Soucek K. TGF-beta1-induced EMT of non-transformed prostate hyperplasia cells is characterized by early induction of SNAI2/Slug. Prostate. 2011;71:1332–1343. doi: 10.1002/pros.21350. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian G, Schwarz RE, Higgins L, McEnroe G, Chakravarty S, Dugar S, Reiss M. Targeting endogenous transforming growth factor beta receptor signaling in SMAD4-deficient human pancreatic carcinoma cells inhibits their invasive phenotype1. Cancer Res. 2004;64:5200–5211. doi: 10.1158/0008-5472.CAN-04-0018. [DOI] [PubMed] [Google Scholar]
- 36.Tarin D, Thompson EW, Newgreen DF. The fallacy of epithelial mesenchymal transition in neoplasia. Cancer Res. 2005;65:5996–6000. doi: 10.1158/0008-5472.CAN-05-0699. [DOI] [PubMed] [Google Scholar]
- 37.Taylor RW, Taylor GA, Durham SE, Turnbull DM. The determination of complete human mitochondrial DNA sequences in single cells: implications for the study of somatic mitochondrial DNA point mutations. Nucleic Acids Res. 2001;29:E74. doi: 10.1093/nar/29.15.e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandewalle C, Comijn J, De CB, Vermassen P, Bruyneel E, Andersen H, Tulchinsky E, Van RF, Berx G. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33:6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallerand H, Robert G, Pasticier G, Ravaud A, Ballanger P, Reiter RE, Ferriere JM. The epithelial-mesenchymal transition-inducing factor TWIST is an attractive target in advanced and/or metastatic bladder and prostate cancers. Urol Oncol. 2010;28:473–479. doi: 10.1016/j.urolonc.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 41.Wu KJ, Zeng J, Zhu GD, Zhang D, Xue Y, Chen YL, Wang XY, He DL. Comparison of transcription factors repressing epithelial phenotype in two different prostate cancer EMT models and its significance. Zhonghua Nan Ke Xue. 2010;16:137–141. [PubMed] [Google Scholar]
- 42.Xu J, Wang R, Xie ZH, Odero-Marah V, Pathak S, Multani A, Chung LW, Zhau HE. Prostate cancer metastasis: role of the host microenvironment in promoting epithelial to mesenchymal transition and increased bone and adrenal gland metastasis. Prostate. 2006;66:1664–1673. doi: 10.1002/pros.20488. [DOI] [PubMed] [Google Scholar]
- 43.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Yingling JM, Blanchard KL, Sawyer JS. Development of TGF-beta signalling inhibitors for cancer therapy. Nat Rev Drug Discov. 2004;3:1011–1022. doi: 10.1038/nrd1580. [DOI] [PubMed] [Google Scholar]
- 45.Yu J, Xie F, Bao X, Chen W, Xu Q. miR-300 inhibits epithelial to mesenchymal transition and metastasis by targeting Twist in human epithelial cancer. Mol Cancer. 2014;13:121. doi: 10.1186/1476-4598-13-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuen HF, Chua CW, Chan YP, Wong YC, Wang X, Chan KW. Significance of TWIST and E-cadherin expression in the metastatic progression of prostatic cancer. Histopathology. 2007;50:648–658. doi: 10.1111/j.1365-2559.2007.02665.x. [DOI] [PubMed] [Google Scholar]
- 47.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 48.Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP. Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 2004;23:1155–1165. doi: 10.1038/sj.emboj.7600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Morrissey C, Sun S, Ketchandji M, Nelson PS, True LD, Vakar-Lopez F, Vessella RL, Plymate SR. Androgen receptor variants occur frequently in castration resistant prostate cancer metastases. PLoS ONE. 2011;6:e27970. doi: 10.1371/journal.pone.0027970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu ML, Kyprianou N. Role of androgens and the androgen receptor in epithelial-mesenchymal transition and invasion of prostate cancer cells. FASEB J. 2010;24:769–777. doi: 10.1096/fj.09-136994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.