Abstract
The green anole lizard (Anolis carolinensis) is a model organism for behavior and genomics that is native to the southeastern United States. It is currently thought that the ancestors of modern green anoles dispersed to peninsular Florida from Cuba. However, the climatic changes and geological features responsible for the early diversification of A. carolinensis in North America have remained largely unexplored. This is because previous studies (1) differ in their estimates of the divergence times of populations, (2) are based on a single genetic locus and (3) did not test specific hypotheses regarding the geologic and topographic history of Florida. Here we provide a multi-locus study of green anole genetic diversity and find that the Florida peninsula contains a larger number of genetically distinct populations that are more diverse than those on the continental mainland. As a test of the island refugia hypothesis in Pleistocene Florida, we use a coalescent approach to estimate the divergence times of modern green anole lineages. We find that all demographic events occurred during or after the Upper Pliocene and suggest that green anole diversification was driven by population divergence on interglacial island refugia in Florida during the Lower Pleistocene, while the region was often separated from continental North America. When Florida reconnected to the mainland, two separate dispersal events led to the expansion of green anole populations across the Atlantic Seaboard and Gulf Coastal Plain.
Keywords: Anolis carolinensis, Florida, green anole, island refugia, mitochondrial DNA, nuclear DNA, Pleistocene
INTRODUCTION
Geographic variation and the historical processes that underlie it are important factors in the study of any model organism. For example, recent adaptations (Williamson et al. 2007; Peng et al. 2011; Tishkoff et al. 2007) and certain disease susceptibilities (Tishkoff and Williams 2002) in humans only make sense in light of the geographic distribution and dispersal abilities of ancient populations. The first reptilian genomic model is the green anole lizard (Anolis carolinensis) (Squamata; Iguania, Polychrotidae) (Alfoldi et al. 2011), which has been studied for decades in neuroscience, reproductive biology and behavior (Wade 2012; Lovern et al. 2004) and more recently for developmental biology (Eckalbar et al. 2012). A. carolinensis occurs in the southeastern United States and ranges from Florida to Texas and northwards to North Carolina and Tennessee. Regional differences in life history traits within this species have been described (Michaud and Echternacht 1995; Bishop and Echternacht 2004; Goodman et al. 2013), yet it is not fully known to what degree any of these differences have been constrained by evolutionary history. Although they would inform any subsequent genetic studies of a genomic model, biogeographic hypotheses explaining the distribution patterns of green anoles have gone relatively untested.
The evolutionary history of green anoles in North America has recently undergone scrutiny (Tollis et al. 2012; Campbell-Staton et al. 2012). Analyzing patterns of intraspecific mitochondrial and nuclear DNA sequence variation, Tollis et al. (2012) described four populations: (1) one endemic to the northern Gulf coast of Florida (the Suwannee population hereafter), (2) an Everglades population, (3) a North Carolina population, and (4) a population ranging from the Atlantic Coast of South Carolina and Georgia and across the Gulf Coastal Plain to Texas (the Gulf-Atlantic population hereafter). Campbell-Staton et al. (2012) described a fifth population sampled along the Atlantic coast of central Florida (the Central Florida population hereafter). Florida harbors four out of these five major green anole populations while comprising a relatively small proportion of the species’ range. This complex genetic structure in Florida is in stark contrast to wider-ranging populations such as the Gulf-Atlantic which, despite ranging from South Carolina to Texas, shows a lack of isolation-by-distance and is depauperate in terms of genetic diversity (Tollis et al. 2012).
A. carolinensis is phylogenetically nested within a group of Cuban anoles known as A. porcatus, which suggests an ancient overwater dispersal to Florida from Cuba (Glor et al. 2005). It is established that A. carolinensis existed in Florida at least as long ago as the Pleistocene Era (Buth et al. 1980, Campbell-Staton et al. 2012 and Tollis et al. 2012), which began ~2.6 – 0.1 million years ago (Mya). Pleistocene glacial cycles began ~2 Mya, and the resultant climatic oscillations undoubtedly exposed green anole populations to significant landscape changes – including the repeated inundation of Florida and the emergence of a series of archipelagoes (Lane 1994; Petuch 2004). Patterns of genetic discontinuity in Florida observed in other taxa (Soltis et al. 2006) suggest that these vicariant events had profound effects on biodiversity, including increased endemism. It is possible that similar fragmentation occurred between A. carolinensis populations living on the Florida peninsula during the Pleistocene, and that island refugia drove the early diversification of the species. However, estimates of the time to most recent common ancestor (Tmrca) for modern green anole populations have ranged greatly, from as long ago as the Miocene between ~13 and 7 Mya (Campbell-Staton et al. 2012) to as recent as the Upper Pliocene or Lower Pleistocene between ~3 and 1.5 Mya (Tollis et al. 2012). In addition, the westward expansion of the Gulf-Atlantic population was estimated to have begun during the Pleistocene ~0.30 Mya (Tollis et al. 2012), but in another study its divergence from other populations was estimated to have begun during the Miocene or Pliocene ~6.1 – 2.5 Mya (Campbell-Staton et al. 2012). The older divergence times would suggest that A. carolinensis had already dispersed to mainland North America by the time of the first Pleistocene glaciation, while the younger estimates would suggest that most of the species demographic history occurred during or after it. It is therefore not yet established whether green anole intraspecific diversity was driven by Pleistocene climate change in Florida or by some more ancient phenomenon.
To date, estimates of the Tmrca for A. carolinensis have relied on clock-like phylogenetic methods using a single locus (Tollis et al. 2012; Campbell-Staton et al. 2012). While relaxed clock phylogenetics use fossil calibrations or informative substitution rate priors and have made the estimation of divergence times more accurate (Yang and Rannala 2006; Weir and Schluter 2008), the use of a single gene or the concatenation of multiple genes for divergence time estimation can lead to error because gene-tree heterogeneity is the rule rather than the exception (Knowles and Carstens 2007; Brito and Edwards 2009). In this study, we use a multi-locus coalescent approach that incorporates topological discord between sampled gene genealogies and provides a more accurate estimation of divergence histories in order to date important events in the history of green anoles. By cataloguing the genomic diversity of green anoles on the Florida peninsula, and incorporating what is known about the geologic record in Florida, we aim to elucidate key aspects of the early biogeographic history of a model organism.
MATERIALS AND METHODS
Sampling localities, genetic markers and sequence acquisition
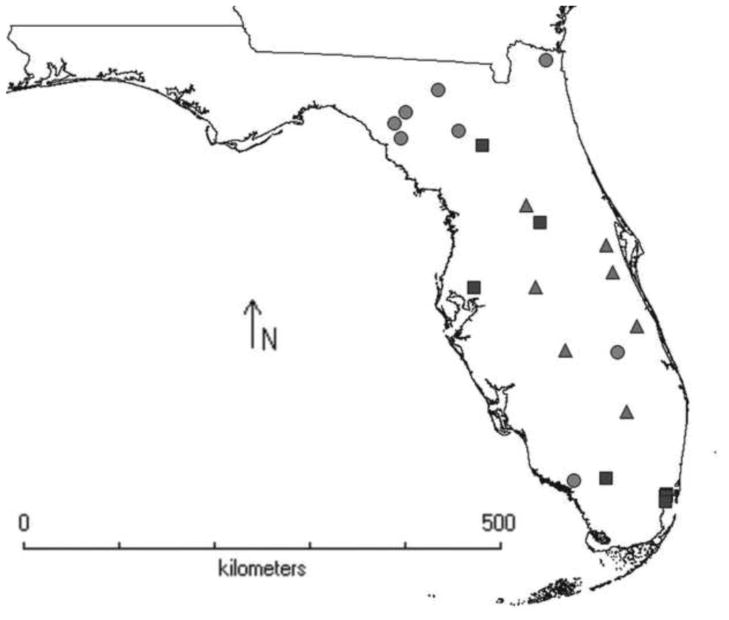
This study used green anole samples that were collected at 49 localities across nine U.S. states of Florida, Georgia, South Carolina, North Carolina, Tennessee, Alabama, Louisiana, Arkansas and Texas, reported previously in Tollis et al. (2012) (see Tollis et a. 2012, Figure 1 and Table S1 for locality information). In addition, we collected 35 additional anoles from six new localities along the Atlantic coast of Florida in September 2012. We were also given tail tissues of 15 anoles from 11 localities in Florida that were obtained from T. Hsieh of Temple University. A map of all Florida collecting localities used in this study is given in Figure 1, and the GPS coordinates of the samples new to this study are available in Supplementary Material Table 1. Genomic DNA was extracted from tissues using the Promega Genomic Wizard DNA Extraction kit. We amplified by PCR an 1172bp mtDNA fragment that includes the NADH-2 gene and two adjacent tRNAs with primers used in Tollis et al. (2012). We also amplified three nDNA loci using primers that were reported in Tollis et al. (2012): HMGCS (1288bp), RALGAPA (970bp), and TERT (1087bp). PCR protocols were repeated as in Tollis et al. (2012), and all products were sequenced in both forward and reverse directions at the High-Throughput Genomics Unit at the University of Washington in Seattle, WA. Chromatograms were imported into Geneious v5.5 (Drummond et al. 2010), where poor-quality regions were trimmed and heterozygous sites were called using the Find Heterozygotes plugin. Forward and reverse reads were assembled into contigs and consensus sequences were extracted and aligned using ClustalW (Larkin et al. 2007) in BioEdit (Hall 1999), where alignments were further edited by eye. Gametic phases of each nDNA haplotype were resolved computationally using PHASE 2.1 (Stephens et al. 2001) with a 90% probability cutoff.
Figure 1.
Map of Florida sampling localities. Circles indicate localities from Tollis et al. (2012). Boxes (collected by Boissinot lab in 2012) and triangles (obtained from the Hsieh lab in 2012) indicate field localities new to this study.
Phylogeographic Inference
Phylogenetics
Using MrBayes3.2 (Ronquist et al. 2012), we reconstructed a phylogeny of green anoles based on an mtDNA dataset comprised of the following: the NADH-2 sequences from Tollis et al. (2012), 25 additional sequences available from GenBank, (see Supplementary Material Table 2) and 45 sequences new to this study. The outgroups used were the same as in Tollis et al. (2012), including A. isoleps, A. altitudinalis, and three A. porcatus, plus an additional A. porcatus collected in Florida by T. Hsieh. The total sample size for the mtDNA phylogenetic analysis was n=299. Nucleotide substitution models were tested using Bayes Factors in MEGA 5.0 (Tamura et al. 2011) and most likely model was HKY + Gamma + Invariant sites. The phylogenetic analysis was run for 20,000,000 generations, and we sampled 10,000 trees, discarding 10% as burn-in. We also reconstructed phylogenies for the HMGCS (n=109), RALGAPA (n=104) and TERT (n=100) genes using Maximum Likelihood (ML) estimation as implemented with PhyML (Guindon et al. 2009). We used the TN93 model of sequence evolution, which was also determined in MEGA, and completed 500 bootstrap replicates to assess node support. For the nDNA trees, we used A. porcatus as an outgroup.
Clustering of nDNA haplotypes
In order to assess the number of populations in our sample without using a priori information about population structure (i.e. from the gene tree phylogenies), the entire nDNA dataset for 158 anoles was loaded into the Bayesian clustering program Structure 2.3.3 (Pritchard et al. 2000), which can estimate the likelihood of a user-set number of K clusters as well as assign individuals to each cluster. We ran the admixture model for 60,000 generations, with 10,000 discarded as burn-in. Analyses for a range of K values from 1 to 10 were repeated three times each, and the results files were submitted to Structure Harvester (Earl and Vonholdt 2011), which chooses the most likely number of clusters using the ΔK method (Evanno et al. 2005). Individual cluster assignments were then compared to their placement on the mtDNA phylogeny in order to delimit populations.
Genetic Variation and Historical Demography
As a comparison of genetic variation between Florida and the mainland, we computed the average pair wise Kimura 2-Parameter genetic distance both within Florida and within the continental mainland in MEGA 5.0. Taking into account where there was agreement between the phylogenetic and cluster assignments, we computed standard measurements of sequence diversity within all populations for each gene using DnaSP version 5 (Librado and Rozas 2009) including the number of haplotypes, hapoltype diversity (Hd), nucleotide diversity (π), Theta per site (Θ = 4 Neu), Tajima’s D (Tajima 1989) and Fu’s Fs (Fu 1997). We also calculated FST and Nm (Hudson et al. 1992) for each gene to estimate the extent of population subdivision and the extent of migration, respectively. We also calculated pair wise FST between all populations for each gene in DnaSP and for the entire nDNA dataset in Arlequin v3.5 (Excoffier and Lischer 2010). To examine evidence of past demographic expansions in a population, we examined the mismatch distribution of nDNA polymorphisms with a concatenated data set in Arlequin, comparing the observed data to what is expected under a model of population size expansion (Harpending 1994). We also constructed an Extended Bayesian Skyline plot (Heled and Drummond 2008), which uses the coalescent histories of multiple genomic loci to estimate the number and extent of population size changes in the past. As mismatch distribution analyses and skyline plots were reported for four populations in Tollis et al. (2012), we did not repeat these analyses here and only extended them to any newly described populations.
Species Tree Estimation
To account for mito-nuclear discordance as well as stochastic differences in the coalescent histories of the sampled gene genealogies while making inferences about the evolutionary history of A. carolinensis, we used *BEAST (Heled and Drummond 2010). *BEAST implements a probabilistic framework that uses sequence information from different loci and multiple individuals per taxon (or population). By incorporating prior probabilities on substitution rates, *BEAST will jointly estimate the species tree as well as the individual gene trees, taking deeper gene tree coalescence into account and providing better estimates of divergence times (McCormack et al. 2011). The goals of this analysis were twofold: (1) to obtain a population phylogeny representing the intraspecific evolutionary history, and (2) to estimate the divergence times of green anole populations. Partitions included the NADH-2, HMGCS, RALGAPA and TERT alignments. DNA substitution models used were HKY + Gamma + Invariant Sites for NADH-2, and TN93 for the nDNA partitions, as determined previously in MEGA. We used A. porcatus as an outgroup to root the species tree. We assigned each A. carolinensis individual a discrete trait representing one of the green anole populations as determined by two criteria: (1) its mtDNA clade and (2) >95% of its genome belonging to a specific nDNA cluster. Each partition contained eight individuals per population. For divergence time estimation, we used the NADH-2 mutation rate of 1.3% per million years (My) for lizards (Macey et al. 1999) and placed normally distributed prior probabilities around the mutation rate parameters for each nuclear gene with a starting mean of 1. This analysis used a Yule prior, which assumes an unknown yet constant birth rate of lineages.
The *BEAST results were obtained by combining the parameter files from two independent MCMC chains of length 500,000,000 each. To assess convergence between these runs we monitored the effective sample size (ESS) values and consistency of parameter estimates using Tracer v1.5, and once confirmed, the separate runs were combined using LogCombiner. The initial 10% of each run was ignored as burn-in. From the combined results, we sampled parameters and trees every 100,000 for a total of 10,000. Out of the 10,000 sampled genealogies, we obtained the maximum-clade credibility tree with divergence time estimates for the species tree using TreeAnnotator, discarding 10% as burn-in.
Coalescent-Based Demographic Parameter Estimation
Using a different coalescent framework, we estimated key population genetic parameters including population sizes (Θ), population divergence times (τ), and migration rates (m) with G-PHoCS (Gronau et al. 2011). G-PHoCS accepts multiple sequences from independently evolving neutral loci across the genome and uses a MCMC sampling algorithm for Bayesian parameter estimation. A particularly useful feature of this package is its ability to integrate over all possible phases of diploid genotype data, thus removing the need for a priori computational haplotype inference, which can be a potential source of error in phylogeographic analyses (Garrick et al. 2010). We used six to ten randomly sampled individuals from each population and the three nuclear loci. Total sample sizes per gene were n=37 for HMGCS, n=32 for RALGAPA and n=30 for TERT. The assumed population history was the species tree from the *BEAST analysis, and we estimated the two-way migration rates of geographically adjacent populations. We evaluated the effect of different priors of the gamma distribution G(α,β) for the Θ and τ parameters, which were ~G(2,10), ~G(2,1000) and ~G(2,2000). For each set of priors, we ran five replicate analyses with different seeds for 500,000 generations with a sampling interval of 50 generations, and we assessed the convergence of separate runs using Tracer v1.5. Demographic parameter estimates in G-PHoCS are given as relative values as they are scaled by the mutation rate. Therefore, the absolute values for population sizes and divergence times were obtained through a calibration as outlined by Gronau et al. (2011) which relies on an external estimate of ancestral divergence time, τdiv. For this purpose, we evaluated calibrations using divergence time estimates for A. porcatus and A. carolinensis from the *BEAST analysis in the present study as well as from the literature (Buth et al. 1980; Campbell-Staton et al. 2012). We assumed a one-year generation time based on knowledge of the species natural history, which is well conserved across the Anolis genus (Losos 2009).
RESULTS
Phylogeographic analysis
Phylogenetics
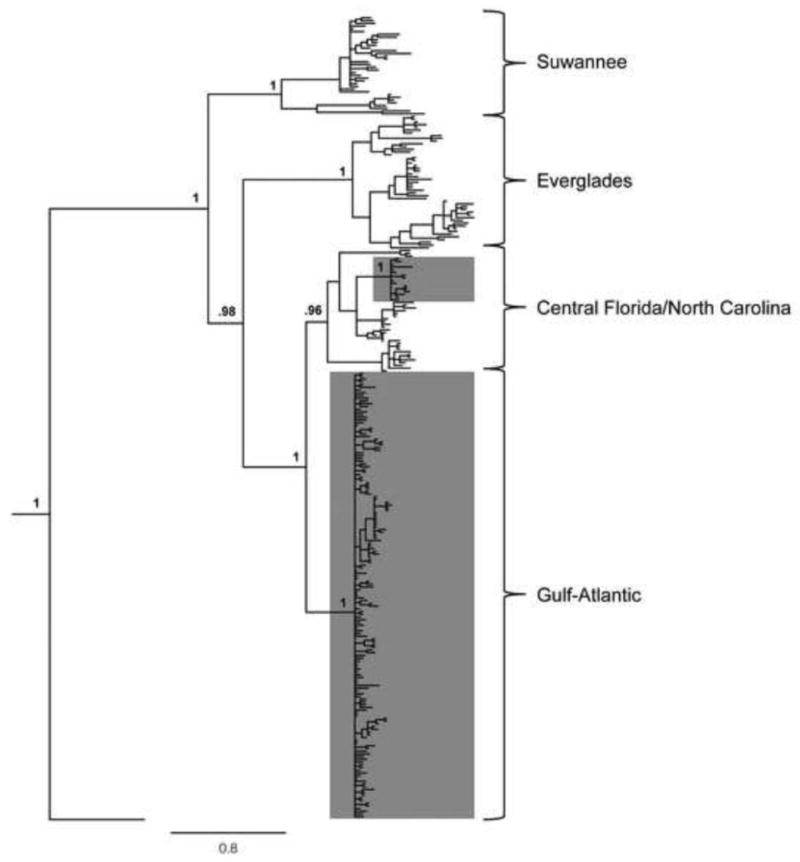
All new sequence data has been deposited in GenBank (accession numbers KF872494-KF872618). The mtDNA phylogeny recovered four major clades, and the topology and geographic distribution was highly congruent with the results published in Tollis et al. (2012) and Campbell-Staton et al. (2012) (Figure 2). The four clades include: (1) the Suwannee clade endemic to the central and northern Gulf Coast of Florida up to the eastern side of the Apalachicola River, (2) the Everglades clade centered around the Gulf and Atlantic coasts at the southern tip of the Florida peninsula and northwards up to Lake Okeechobee, (3) a clade featuring a paraphyletic Central Florida lineage from the central Atlantic Florida coast, with a North Carolina lineage nested within it, and (4) the Gulf-Atlantic clade which includes individuals from localities along the Atlantic coasts of South Carolina and Georgia, across the Gulf Coastal Plain and the Mississippi River into Texas. This wide-ranging clade includes some populations from northern Florida (see below). All of these nodes featured >95% posterior probability support.
Figure 2.
Majority consensus tree from a phylogenetic analysis conducted in MrBayes3.2 on the mitochondrial NADH-2 region (n=299). The four major mitochondrial lineages of Anolis carolinensis are indicated at right with labeled brackets. Monophyletic groups restricted to mainland North America (the Gulf-Atlantic and North Carolina lineages) are indicated by shaded boxes. The posterior probabilities of important nodes are given. A. porcatus is the only outgroup shown, although A. isolepis and A. altitudinalis were also used in the analysis.
Florida contains (1) the highest number of mitochondrial clades and (2) a considerable amount of within-clade diversity. For instance, the Suwannee clade consists of two well-supported minor clades (both with 100% posterior probability), the first of which ranges primarily along the Gulf of Mexico coastline from Tampa Bay to just east of the Apalachicola River, while the second one is centered along the Central Highlands that run inland down the northwestern part of the peninsula. There is also considerable structure within the Everglades clade: a clade from the Gulf Coast of very southern Florida, near Everglades City (96% posterior support), and a more geographically widespread clade (98% posterior support) that consists of individuals from the Atlantic Coast in and around Miami as well as a nested clade from South Bay just south of Lake Okeechobee (100% posterior support). The third major Floridian clade, Central Florida, was described in Campbell-Staton et al. (2012), and our phylogenetic results here are very similar. The Central Florida clade is perhaps the most complex because it consists of two deep mitochondrial lineages (both with 100% posterior support) across its relatively limited geographic range in Florida; one of which contains a nested clade (100% posterior support) that is endemic to North Carolina. Interestingly, anoles sampled near Jacksonville, Florida in Nassau County, fall within the Gulf-Atlantic clade. These anoles are more closely related to conspecifics greater than 1000km away in Texas than they are to conspecifics less than 50km to the south in neighboring Florida counties. There are also individual mtDNA haplotypes sampled from near Panama City, west of the Apalachicola River, which were assigned to the Gulf-Atlantic clade. Figure 3a shows the distribution of mtDNA clades on the Florida peninsula.
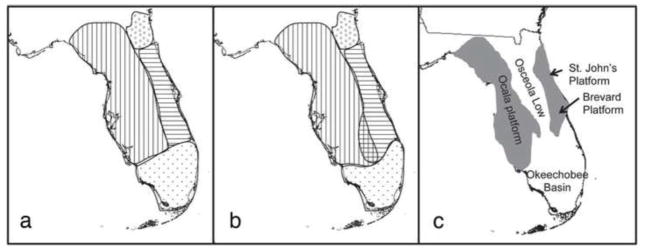
Figure 3.
(a) and (b): Geographic distribution of genetic populations of A. carolinensis in Florida. Collecting localities are indicated by black dots. Patterned shapes depict the geographic distribution of mitochondrial lineages (a) and schematic representation of the nDNA clusters (b). Open circles the – Gulf-Atlantic lineage is present in Nassau County near Jacksonville; vertical lines – Suwannee; horizontal lines – Central Florida; black dots – Everglades. (c): Major geologic structural elements of the Florida peninsula. Positive structures of the Ocala, St. John’s, and Brevard platforms are indicated in grey. Lower elements of the Osceola Low and the Okeechobee basin are indicated. Adapted from Lane (1994).
Due to random effects of coalescence and the slower mutation rates of nuclear genes, we did not expect a high degree of congruence between the nDNA phylogenies. There was significant discordance across the nDNA gene tree topologies and lower statistical support when compared to the mtDNA phylogeny (see Supplementary Material Figures 1 – 3). Nonetheless, all of the groups that were recovered by the mitochondrial analysis appeared on at least one of the nDNA gene trees, and there were several statistically supported clades that were dominated by individuals from a single geographic region. Across nDNA gene trees, reciprocally monophyletic groups included individuals from North Carolina, the Everglades, Central Florida, Suwannee, and the Gulf-Atlantic mitochondrial clades. Samples from the Gulf-Atlantic mitochondrial clade formed distinct polytomies in each nDNA tree, similar to the mtDNA phylogeny. Much of the gene tree discordance was due to differential placement of sequences from individuals across Florida, most notably from South Bay, which phylogenetically groups with the Everglades mtDNA clade but shows very little evidence of affinity to the Everglades based on the nDNA.
Statistical Clustering of the nDNA
The ΔK method from our Structure analysis indicated that the most likely number of clusters in our nDNA dataset was five (Figure 4). Although there is more variability in the nDNA, the distribution of genetic clusters in Florida closely mirrored that of the lineages recovered from the mtDNA phylogenetic analysis, and different clusters dominate different geographic regions (Figure 3b). This is especially the case for the Everglades, where only one haplotype cluster is found. Unique haplotype clusters are found in the regions of the Florida Gulf Coast and Central Florida, respectively, although both of these regions contain haplotypes from every cluster, adding to the evidence that Florida has a high degree of genetic variation for this species. Individuals collected in South Bay were assigned to both the Central Florida and Suwannee nDNA clusters, with little evidence of persistent Everglades haplotypes in this area. This differs from the mtDNA phylogeny, in which all South Bay mtDNA haplotypes group with the Everglades, but is similar to the nDNA phylogenies. Another point of mito-nuclear discordance was the detection of significant admixture between the Gulf-Atlantic and North Carolina nDNA clusters that extends into southeastern Georgia, which is notably absent in the mtDNA phylogeny. Individuals sampled near Nassau, Florida cluster with individuals across the Gulf Coastal Plain, rather than with other Florida clusters, as in the mtDNA phylogeny. The slight differences in geographic pattern between the mtDNA phylogeny and the nDNA Structure analysis do not discount the fact that mtDNA phylogenetic placement, patterns in the nDNA gene trees, and nDNA cluster assignment can be used in concert to assign individuals to five populations: Everglades, Suwannee, Central Florida, North Carolina, and Gulf-Atlantic.
Figure 4.
Visualization of the Bayesian clustering analysis, with the most likely number of nDNA genetic clusters K=5 in 158 A. carolinensis samples collected across nine U.S. states. Bottom bar: x-axis represents each individual (arranged by geographic region, labeled below) and shading along the y-axis represents the proportion each individual’s genome derived from one of the K clusters. Top bar: horizontal shading in boxes represent the proportion of each of the five nDNA clusters found in each geographic region. Barplots were produced using the program DISTRUCT (Rosenberg 2004)
Genetic Differentiation and Historical Demography in Florida
The average genetic distances between individuals within Florida are much greater than those of the continental mainland (0.0538 versus 0.0113 mitochondrial and 0.0029 versus 0.0014 nuclear). Table 1 shows the genetic diversity overview for each gene. In general, genetic diversity (measured by Hd, π or Θ) is greatest in the Central Florida population, with some exceptions where diversity estimates in the Everglades and Suwannee are greater. Non-significant negative values of Tajima’s D and high negative values of Fu’s Fs across all genes suggest a lack of departures from neutrality due to population bottlenecks or expansions in Florida. The fact that Central Florida harbors a relatively large amount of genetic diversity is consistent with the mismatch distributions (not shown), which reject a model of expansion, and the EBSP (see Supplementary Material Figure 4), which places most of posterior probability under a model of zero past population size changes. Across all five green anole populations, FST calculations are significant and Nm is consistently small (≪1) (Table 2), suggesting strong population subdivisions with relatively low amounts of gene flow and few migrants. The pair wise calculations FST for each gene show highly significant values for every population comparison (Table 3).
Table 1.
Polymorphism overview for each gene within five green anole populations.
| Gene | Population | # haps | Hd | π | Θ | D | Fs |
|---|---|---|---|---|---|---|---|
| HMGCS | Central Florida | 16 | 0.893 | 0.00499 | 0.00506 | −0.04405 | −1.275 |
| Everglades | 2 | 0.264 | 0.00021 | 0.00024 | −0.34144 | 0.186 | |
| Suwannee | 6 | 0.675 | 0.00262 | 0.00329 | −0.79339 | 0.728 | |
| North Carolina | 4 | 0.6 | 0.00114 | 0.00197 | −1.46261 | 0.907 | |
| Gulf-Atlantic | 13 | 0.753 | 0.00225 | 0.00244 | −0.22659 | −1.187 | |
| RALGAPA | Central Florida | 8 | 0.656 | 0.00086 | 0.00168 | −1.32791 | −4.071 |
| Everglades | 6 | 0.893 | 0.0014 | 0.00159 | −0.52474 | −3.746 | |
| Suwannee | 7 | 0.66 | 0.00107 | 0.00227 | −1.59816 | −2.404 | |
| North Carolina | 2 | 0.356 | 0.0011 | 0.00109 | 0.02107 | 2.338 | |
| Gulf-Atlantic | 3 | 0.076 | 0.00023 | 0.00084 | −1.49607 | −0.947 | |
| TERT | Central Florida | 16 | 0.905 | 0.00292 | 0.0048 | −1.35152 | −7.19 |
| Everglades | 4 | 0.867 | 0.00239 | 0.00242 | −0.06042 | −0.024 | |
| Suwannee | 10 | 0.863 | 0.00223 | 0.00363 | −1.41478 | −3.683 | |
| North Carolina | 2 | 0.44 | 0.0004 | 0.00029 | 0.84228 | 0.944 | |
| Gulf-Atlantic | 5 | 0.141 | 0.00013 | 0.00074 | −1.68341 | −5.44 | |
| NADH-2 | Central Florida | 21 | 0.992 | 0.03576 | 0.02862 | 1.00251 | −1.893 |
| Everglades | 12 | 0.987 | 0.01684 | 0.01896 | −0.50537 | −1.546 | |
| Suwannee | 25 | 0.997 | 0.02706 | 0.03702 | −1.06254 | −5.925 | |
| North Carolina | 12 | 0.958 | 0.00328 | 0.00547 | −1.61885 | −5.61 | |
| Gulf-Atlantic | 80 | 0.978 | 0.00531 | 0.02155 | −2.41167 | −112.233 |
#haps – number of haplotypes
Hd – haplotype diversity
π - nucleotide diversity
Θ - Theta per site (4Neu)
D – Tajima’s D. Statistically significant values in bold.
Fs – Fu’s Fs
Table 2.
Estimates of genetic differentiation and gene flow across Florida for four genes.
| Gene | FST | Nm |
|---|---|---|
| ND2 | 0.65285 | 0.13 |
| HMGCS | 0.52482 | 0.23 |
| RALGAPA | 0.39908 | 0.38 |
| TERT | 0.36801 | 0.43 |
Table 3.
Pair wise FST between each green anole population per gene calculated in DnaSP.
| Population 1 | Population 2 | HMGCS | RALGAPA | TERT | NADH-2 |
|---|---|---|---|---|---|
| Gulf-Atlantic | Everglades | 0.7686 | 0.55498 | 0.46658 | 0.80269 |
| Gulf-Atlantic | Suwannee | 0.0825 | 0.61162 | 0.62357 | 0.68086 |
| Gulf-Atlantic | North Carolina | 0.57286 | 0.04871 | 0.1854 | 0.86165 |
| Gulf-Atlantic | Central Florida | 0.09638 | 0.68025 | 0.4702 | 0.50528 |
| Everglades | Suwannee | 0.76766 | 0.10532 | 0.28301 | 0.64174 |
| Everglades | North Carolina | 0.86352 | 0.36216 | 0.31365 | 0.81747 |
| Everglades | Central Florida | 0.5628 | 0.17292 | 0.13067 | 0.47306 |
| Suwannee | North Carolina | 0.60442 | 0.41168 | 0.54353 | 0.73003 |
| Suwannee | Central Florida | 0.10604 | 0.27986 | 0.1323 | 0.48819 |
| North Carolina | Central Florida | 0.40845 | 0.42515 | 0.35287 | 0.46326 |
Species Tree Estimation
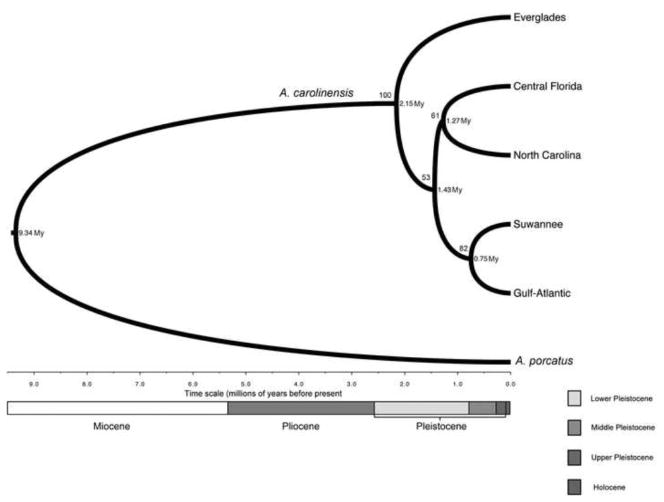
Parameter estimates between the separate *BEAST runs were highly concordant, suggesting convergence, and the ESS values were very high for all parameters (mostly over 4000), suggesting proper mixing of the MCMC chain. The estimated mutation rates for the nDNA loci (0.025% per My for HMGCS, 0.016% per My for RALGAPA, and 0.030% per My for TERT) were an order of magnitude slower than that of the mitochondrial rate and fall within expectations for nuclear genes. The species tree topology (Figure 5) differs significantly from that of the mtDNA phylogeny. Most notably, in the species tree there is the early divergence of the Everglades population (100% posterior support), followed by a split into two lineages (53% posterior support): one lineage includes the Central Florida and North Carolina populations (61% posterior support), and then includes the Suwannee and Gulf-Atlantic populations (82% posterior support). The divergence time estimates are more similar to the calibrated mtDNA-based results published in Tollis et al (2012) than to those of the relaxed clock analysis Campbell-Staton et al (2012), providing more support for the hypothesis that the major evolutionary events in this species’ history occurred during the Pleistocene. Our *BEAST analysis placed the Tmrca for A. carolinensis and the branching off of the Everglades population at ~2.2 Mya (1.3 – 3.1 95% highest posterior density or HPD). The next divergence was between populations that live along either the Atlantic Coast, or the Gulf Coast and more inland across the Gulf Coastal Plain, and occurred ~1.4 Mya (0.83 – 2.3 95% HPD). Along the Atlantic Coast, the split between the Central Florida and North Carolina populations occurred ~1.3 Mya (0.74 – 1.8 95% HPD). Meanwhile, the Gulf-Atlantic population split off from the Suwannee 0.75 Mya (0.45– 1.1 95% HPD).
Figure 5.
Maximum clade credibility tree from a *BEAST analysis using 1 mtDNA and 3 nDNA loci showing the inferred branching order and divergence times for five populations of green anoles and their outgroup A. porcatus. Within A. carolinensis, percent posterior probabilities and divergence time estimates (My = million years) are given to the left and right of each node, respectively.
Coalescent-Based Demographic Parameter Estimation
When we evaluated the three sets of G-PHoCS analyses, we found that a prior distribution of ~G(2,10) produced inconsistent results that failed to converge between runs, while prior distributions of ~G(2,1000) and ~G(2,2000) produced highly similar results and did converge. The parameter estimates and their 95% HPD, calibrated with the A. porcatus – A. carolinensis Tmrca estimate of 9.3 Mya, are shown in Table 4. The parameter τroot, or the divergence time of all A. carolinensis populations, was estimated to be ~1.8 Mya. The split between the Atlantic Coast (Central Florida and North Carolina) and Gulf Coast (Suwannee and Gulf-Atlantic) populations was ~1.4 Mya. The split between Central Florida and North Carolina was estimated to be ~0.79 Mya and the split between the Suwannee and Gulf-Atlantic was estimated to be ~0.73 Mya. These estimates for all green anole demographic events overlap closely with those from *BEAST, and are well within the Pleistocene. Using this calibration, the mean estimated mutation rate for the three nuclear genes was 0.021% per My, which was also very similar to *BEAST. The timing of the split between A. porcatus and A. carolinensis has differed across the literature, ranging from ~5 Mya (Buth et al. 1980) to ~6.8 – 17.8 Mya (Campbell-Staton et al. 2012). If we use the central value of the distribution estimated in Campbell-Staton et al. (2012) (12.3 Mya), then the mean estimate of τroot is ~2.4 Mya (~1.3 – 3.5 95% HPD). Thus, using an older A. porcatus – A. carolinensis Tmrca places the ancestor of all A. carolinensis as possibly pre-Pleistocene, but all within-species divergences still remain within the Pleistocene. The Θ estimates for ancestral populations suggest an ancient population size expansion in Florida, as Θroot is less than Θalpha. The Θ estimates also suggest that the extant Florida populations are much larger than those on the continental mainland. In fact, all of the Florida Θ estimates from G-PHoCS are at least double the mean Θ estimates for both the Gulf-Atlantic and North Carolina. Patterns of gene flow inferred from this analysis suggest that between adjacent populations migration is more likely to be occurring from geographically wide-ranging populations to more geographically restricted populations (i.e., from the Gulf-Atlantic to North Carolina).
Table 4.
Coalescent population genetic parameter estimates for A. carolinesnsis from G-PHoCS. Shown are the values for divergence time (τ) in years and population size (Θ) in individuals, calibrated with an A. porcatus – A. carolinensis Tmrca of 9.3 Mya, and the raw relative values for specific one-way migration bands (m) averaged across five separate analyses which used prior distributions of ~G(2,2000). τroot refers to the divergence time of all five green anole populations and τalpha refers to the time of the split between the Everglades and all other populations. Θroot refers to the size of the ancestral A. carolinensis population and Θalpha the size of the ancestral population that gave rise to the Suwannee, Gulf-Atlantic, Central Florida and North Carolina populations. Parameters labeled Suwannee/Gulf-Atlantic and Central Florida/North Carolina refer to the respective ancestral population of each pair. Arrows indicate directionality of migration between geographically adjacent populations.
| Parameter | Mean | 95% HPD Lower | 95% HPD Upper |
|---|---|---|---|
| τroot | 1,822,441 | 1,000,776 | 2,651,589 |
| τalpha | 1,408,803 | 633,669 | 2,102,097 |
| τSuwannee/Gulf-Atlantic | 730,800 | 189,399 | 1,529,223 |
| τCentral Florida/North Carolina | 786,919 | 229,150 | 1,384,251 |
| Θroot | 1,025,117 | 44,427 | 2,272,791 |
| Θalpha | 1,587,212 | 285,268 | 3,458,289 |
| ΘSuwannee/Gulf-Atlantic | 1,651,047 | 137,957 | 3,308,641 |
| ΘCentral Florida/North Carolina | 1,385,654 | 56,118 | 3,095,859 |
| ΘGulf-Atlantic | 651,066 | 72,486 | 1,323,456 |
| ΘNorth Carolina | 514,277 | 81,839 | 1,082,615 |
| ΘCentral Florida | 2,521,815 | 965,702 | 4,269,666 |
| ΘSuwannee | 2,859,460 | 1,096,645 | 4,889,306 |
| ΘEverglades | 1,612,933 | 484,020 | 2,922,827 |
| mGulf-Atlantic → North Carolina | 1497 | 0 | 3883 |
| mNorth Carolina → Gulf-Atlantic | 186 | 0 | 1016 |
| mGulf-Atlantic → Suwannee | 92 | 0 | 465 |
| mSuwannee → Gulf-Atlantic | 605 | 0 | 2357 |
| mCentral Florida → Suwannee | 24 | 0 | 134 |
| mSuwannee → Central Florida | 1533 | 0 | 3239 |
| mSuwannee → Everglades | 298 | 0 | 1137 |
| mEverglades → Suwannee | 16 | 0 | 86 |
| mCentral Florida → Everglades | 121 | 0 | 671 |
| mEverglades>Central Florida | 31 | 0 | 179 |
DISCUSSION
Here we use phylogenetic and clustering methods to delimit five distinct populations of green anoles. We also use multi-locus coalescent approaches in order to understand the historical biogeography of these populations. We provide evidence that the green anole populations of southern Florida split from the rest of the species very early in its history, which contrasts with previous reports suggesting the most phylogenetically ancient populations live today along the northern Gulf coast of Florida (Tollis et al. 2012; Campbell-Staton et al. 2012). We also demonstrate that most of the genetic variation within A. carolinensis, including a relatively high number of genetic populations, resides in peninsular Florida, and most of the divergence time estimates for the major lineages of green anoles fall within the Pleistocene. Taken as a whole, these observations are consistent with the hypothesis that climatic changes brought by rising sea levels during periods of Pleistocene warming created island refugia, and facilitated the early diversification of green anoles. Our findings provide a framework for future studies of intraspecific genomic variation in this genomic model species.
Due to the discordance across the mtDNA and nDNA gene tree topologies demonstrated here, elucidating the branching order of green anole populations in time has proven to be complicated and requires more than a simple interpretation of the mtDNA phylogeny. However, there are aspects of A. carolinensis evolutionary history that can be inferred from the gene trees. For instance, there are reciprocally monophyletic lineages across both mtDNA and nDNA gene trees, including the Everglades and North Carolina, which suggests the long-term geographic isolation of these populations. Also, in addition to the geographic patterns in the phylogenies, there is strong structure present across all genes if one examines their global and pair wise FST, suggesting that all of these data sets contain valuable signatures of population history (Tables 2 and 3). The early divergence of the Everglades population has 100% posterior support in our *BEAST results, and it is also supported by an analysis in Tollis et al. (2012), in which a neighbor-joining tree constructed from a pairwise FST matrix features a long branch length leading to the Everglades and suggests long-term isolation (Tollis et al. 2012, Figure 4). It is obvious that the nuclear genes are driving the topology differences with respect to the mtDNA phylogeny. This may be due to possible artifacts of mtDNA evolution such as selective sweeps or incomplete lineage sorting resulting from relatively rapid population fragmentation during the diversification process.
In addition to the order of population splits within A. carolinensis, our estimates of the timing of the demographic events have differed somewhat from previous studies. Tollis et al. (2012) used a calibrated phylogenetic approach that centered on previous divergence time estimates within the carolinensis subgroup in conjunction with the pair wise genetic distances of the major lineages within A. carolinensis and a strict molecular clock to estimate a Tmrca that straddles the Plio-Pleistocene boundary at ~1.3 – 2.9 Mya. Campbell-Staton et al. (2012) used a relaxed molecular clock model with the same mtDNA region and a fixed mutation rate of 1.3%/My to estimate a Tmrca of ~6.8 – 12.6 Mya, which is well before the Pliocene and sometime during the Miocene (which ended ~5.3 Mya). These widely different estimates require reconciliation if the Anolis genome community is to converge on a realistic model of the species historical biogeography. We suggest that previous results are less robust due to problems that stem from the use of a single mtDNA locus to reconstruct the timing of population divergences within a species. The risk of over-interpretation when using one gene lies in many factors. First, it is well established that during lineage diversification, gene histories will diverge before populations. Since gene trees are embedded within species trees, it is expected that dates estimated from gene trees will overestimate the true divergence times. Another confounding factor in single-gene phylogeographic inference is gene tree heterogeneity due to random effects of coalescence in finite populations (Brito and Edwards 2009). This is obviously the case for A. carolinensis and is apparent when one compares the topologies of the mtDNA and nDNA phylogenies. Other problems, related to using a relaxed clock model explicitly, include the relatively rapid substitution rates in mitochondrial genomes, which may cause over-saturation in more ancient lineages and severely bias divergence time estimates at larger time scales (Zheng et al. 2011). At the other temporal end, applying a relaxed clock within species may be confounded by ancestral sequence polymorphisms that have not fully sorted between lineages, leading to an apparent yet false inflation of more recent molecular clocks (Ho et al. 2005; Peterson and Masel 2009).
While the current study aims to resolve some of the timing of diversification events in the early history of A. carolinensis, there are a few caveats and potential sources of bias in our analyses that are worth exploring. For instance, our findings are consistent with a previous study in birds (jays of the genus Aphelocoma) (McCormack et al. 2011), which compared *BEAST to BEAST and concluded that the multi-locus coalescent analysis produced divergence time estimates that were more shifted towards the present. However, the Aphelocoma study investigated multiple species while the present study is an intra-specific analysis, and the *BEAST model assumes the sampled taxa represent separate biological species with little to no gene flow between them (Heled and Drummond 2010). Our estimates of gene flow based on FST and Nm were highly significant between the five green anole populations, suggesting long-term isolation. Therefore, it was reasonable to treat them as independently evolving lineages that were suitable for the model used in *BEAST, which has been used to investigate relationships at the population-species boundary (Salicini et al. 2011). Regardless, recent simulation studies have shown that divergence time estimates in species tree analyses will be biased towards the present if there is gene flow (Leaché et al. 2013), and for populations below the species level it will remain difficult to determine whether certain shared genetic variation is the result of either migration or ancestral polymorphisms. This is why we designed the *BEAST analysis to include individuals that were unlikely to be introgressed. It is also important to note that the model in G-PHoCS does allow for migration between populations and accounts for it in demographic parameter estimation, and produced even more recent divergence time estimates than *BEAST. It is therefore possible that migration plays less of a confounding role in this system than other factors such as the mito-nuclear discordances, although further work is needed to fully resolve this. With or without migration, we believe there is a strong argument to be made for a within-Pleistocene timing of most green anole demographic events, and that deep mitochondrial coalescence has probably contributed to an over-estimation of the A. carolinensis Tmrca. Indeed, the earliest instances of A. carolinensis in the North American fossil record date to the Upper Pleistocene (Holman 1995); our results here suggest that the reason for this may be that the species history on the continent does not reach much farther back than the Lower Pleistocene.
Florida currently comprises less than 15% of A. carolinensis total natural range, but it is the cradle of green anole diversity. The populations found along the Florida peninsula are the most ancient according to the phylogenetics, and by every measure genetic variation is greatest in all of these populations when compared to more wide-ranging conspecifics across the mainland. The evidence supports an ancient population size expansion in Florida, which suggests long periods with suitable habitat and refuge for genetic diversity. This is most apparent in the Central Florida population, which is comprised of two deeply divergent mitochondrial lineages, contains individuals with haplotypes from three of the nDNA clusters, and harbors elevated sequence diversity when compared to all other populations. The Everglades and Suwannee populations also contain multiple mitochondrial lineages and are genetically diverse when compared to the mainland. While most populations show at least some evidence of effective size changes at some point in the past, the Central Florida population in particular seems to have been demographically stable for most if not all of its history. Population size changes that have been inferred for the Suwannee and Everglades populations are much more ancient than the ones inferred for the mainland populations (Tollis et al. 2012). In contrast to this relative stability, continental mainland populations are either isolated, as in the North Carolina population, or have much more recently undergone dramatic range expansions, as in the Gulf-Atlantic population (Tollis et al. 2012; Campbell-Staton et al. 2012), and have reduced estimated effective sizes.
If the “Out of Cuba” hypothesis is correct, then Florida provided the first stepping-stone for the invasion of Anolis lizards into the mainland southeastern United States (Glor et al. 2005). Florida has a dynamic geologic history (Lane 1994; Petuch 2004) and according to our estimates A. carolinensis arrived at a time of dramatic physiographic upheaval with possible vicariant effects. While Florida maintained its form as a peninsula throughout the Pliocene (~5.3 – 2.6 Mya), the Pleistocene was a period of recurrent glacial epochs. The first of these began ~2 Mya and caused the receding of the surrounding ocean and the augmentation of the Florida peninsula, particularly along its western edge. During interglacials throughout the Quaternary, the sea would rise hundreds of feet above its present level, effectively reducing Florida to a series of islands. The central Florida region corresponds to these archipelagoes, primarily along the backbone provided by the Central Highlands, which run north-to-south through the peninsula. West of the highlands in the northwestern part of the peninsula are other areas of isolated elevation. These insights into the geology have been invoked to explain the numerous taxonomic discontinuities that have observed between Florida and the mainland as well as within the Florida peninsula itself, both in the historical literature (Neill 1957) and more recently in the field of molecular phylogeography (Soltis et al. 2006). The geographic distribution of green anole lineages in Florida tightly coincides with the physiographic history: one needs only to look at the similar distribution of the divergent lineages and positive and negative geologic elements on the peninsula (Figure 3c). Our estimates of the timing of demographic events for A. carolinensis land squarely in one of the most complicated parts of Florida’s past, supporting the hypothesis that Pleistocene climatic upheaval was a driver of green anole diversity.
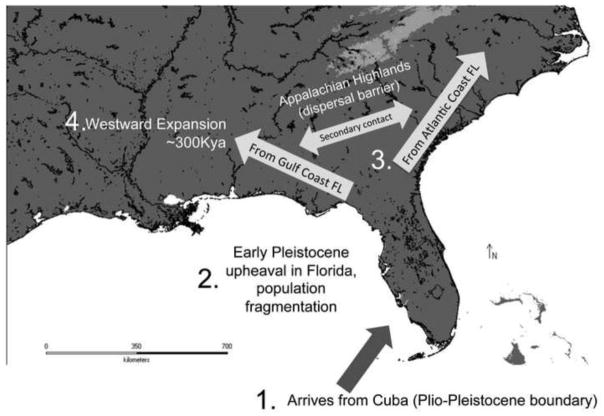
Given our divergence time estimates and the order of population splits within A. carolinensis, as well as the underlying geology of the region in which it lives, we may attempt to reconstruct the most likely historical biogeographic scenario (Figure 6). Green anoles dispersed to Florida from Cuba by the Upper Pliocene or Lower Pleistocene, when the peninsula was fully intact, allowing for dispersal across the peninsula. By ~2 Mya Florida was in physiographic upheaval due to interglacial seawater inundation. This process would have driven population fragmentation into island refugia, followed by gradual genetic divergence. These multiple green anole lineages persisted in Florida, and their diverse genetic signatures are observable today. Once the Florida peninsula reconnected to the mainland, populations living along the Atlantic Coast of Florida would have been able to disperse northward towards North Carolina; the barrier posed by the Appalachian Mountains would have prevented westward dispersal at this time. Populations on the Gulf coast of Florida near the modern-day panhandle would have dispersed along the river drainage systems of the Gulf Coastal Plain. The lack of mountainous barriers along this landscape resulted in the dramatic westward expansion of green anoles across the Mississippi River and into Texas, which began approximately 300,000 years ago (Tollis et al. 2012). More recently, secondary contact between Gulf-Coastal Plain and Atlantic Coast lineages produced admixed populations in the region just south of the Appalachian Mountains and along the coast of South Carolina and Georgia.
Figure 6.
Hypothesized biogeographic scenario depicting the major events in the history of A. carolinensis in the southeastern United States inferred in this chapter. (1) Green anoles disperse from Cuba to the southern tip of Florida, where the ancestor of modern populations lived near the Plio-Pleistocene boundary. (2) During the Early Pleistocene, physiographic upheaval driven by rising sea levels during glacial cycles cause population fragmentation and divergence. (3) During the late part of the Early Pleistocene, the peninsula connects to the mainland again, and green anole populations living along the Central Atlantic Coast of Florida are able to disperse northwards towards the Carolinas. High elevations along the Appalachian Highlands (lighter shading indicates >750 meters elevation) most likely provided a dispersal barrier westward as unsuitable habitat. (4) During the Middle Pleistocene, populations living along the Gulf Coast of Florida began to colonize watersheds along the Gulf Coastal Plain, resulting in a westward expansion of green anoles across the Mississippi River and into Texas.
CONCLUSION
A. carolinensis is widespread and abundant throughout the southeastern United States today, and our results suggest that the most recent common ancestor of modern populations did not live before the Miocene-Pliocene boundary as suggested by some studies. Upon establishment in Florida in the Upper Pliocene or Lower Pleistocene, the dynamic topographies and geological histories of the landscape strongly effected the modern geographic distribution of individuals. We have found evidence suggesting that island refugia in Florida drove the early diversification of green anole lineages. These Florida lineages show evidence of being the most ancient and the most stable in terms of population size over their demographic histories. Two different founding green anole populations most likely undertook separate migrations along the river drainage systems of the Atlantic Coast and the Gulf Coastal Plain, respectively. Using a multi-locus coalescent framework, we conclude that previous analyses based on single mtDNA gene trees and relaxed clock phylogenetic models may be biased towards older divergence time estimates. This study adds to the growing body of evidence that methods accounting for deeper gene tree coalescence are more desirable as they incorporate uncertainty and differences across all sampled loci.
Supplementary Material
Acknowledgments
We would like to thank Sela Sherr (Queens College) for his help collecting anoles in the field, and Tonia Hsieh (Temple University) for sharing tissue samples from Florida. This research was supported by PSC-CUNY grant 63799-00-41 and NIH grant R15GM096267-01 to S.B. Fieldwork was funded by a CUNY Doctoral Research Grant and an American Museum of Natural History Theodore Roosevelt Memorial Grant to M.T. The work was conducted in part with equipment from the Core Facility for Imaging, Cellular and Molecular Biology at Queens College and was supported, in part, under National Science Foundation Grants CNS-0958379 and CNS-0855217 and the City University of New York High Performance Computing Center.
References
- Alfoldi J, Di Palma F, Grabherr M, Williams C, Kong L, et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477 (7366):587–591. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DC, Echternacht AC. Emergence behavior and movements of winter-aggregated green anoles (Anolis carolinensis) and the thermal characteristics of their crevices in Tennessee. Herpetologica. 2004;60 (2):168–177. doi: 10.1655/02-34. [DOI] [Google Scholar]
- Brito PH, Edwards SV. Multilocus phylogeography and phylogenetics using sequence-based markers. Genetica. 2009;135 (3):439–455. doi: 10.1007/s10709-008-9293-3. [DOI] [PubMed] [Google Scholar]
- Buth DG, Gorman GC, Lieb CS. Genetic divergence between Anolis carolinensis and its Cuban progenitor, Anolis porcatus. Journal of Herpetology. 1980;14(3):279–284. [Google Scholar]
- Campbell-Staton SC, Goodman RM, Backstrom N, Edwards SV, Losos JB, et al. Out of Florida: mtDNA reveals patterns of migration and Pleistocene range expansion of the Green Anole lizard (Anolis carolinensis) Ecology and Evolution. 2012;2 (9):2274–2284. doi: 10.1002/ece3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, et al. Geneious v5.5. 2010 Available from http://www.geneious.com.
- Earl DA, Vonholdt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2011;4 (2):359–361. [Google Scholar]
- Eckalbar WL, Lasku E, Infante CR, Elsey RM, Markov GJ, et al. Somitogenesis in the anole lizard and alligator reveals evolutionary convergence and divergence in the amniote segmentation clock. Developmental Biology. 2012;363 (1):308–319. doi: 10.1016/j.ydbio.2011.11.021. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14 (8):2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resoures. 2010;10 (3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147 (2):915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick RC, Sunnucks P, Dyer RJ. Nuclear gene phylogeography using PHASE: dealing with unresolved genotypes, lost alleles, and systematic bias in parameter estimation. BMC Evolutionary Biology. 2010;10(1):118. doi: 10.1186/1471-2148-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glor RE, Losos JB, Larson A. Out of Cuba: overwater dispersal and speciation among lizards in the Anolis carolinensis subgroup. Molecular Ecology. 2005;14 (8):2419–2432. doi: 10.1111/j.1365-294X.2005.02550.x. [DOI] [PubMed] [Google Scholar]
- Goodman RM, Echternacht AC, Hall JC, Deng LD, Welch JN. Influence of geography and climate on patterns of cell size and body size in the lizard Anolis carolinensis. Integr Zool. 2013;8 (2):184–196. doi: 10.1111/1749-4877.12041. [DOI] [PubMed] [Google Scholar]
- Gronau I, Hubisz MJ, Gulko B, Danko CG, Siepel A. Bayesian inference of ancient human demography from individual genome sequences. Nature Genetics. 2011;43 (10):1031–1034. doi: 10.1038/ng.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods in Molecular Biology. 2009;537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Harpending HC. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Human Biology. 1994;66 (4):591–600. [PubMed] [Google Scholar]
- Heled J, Drummond AJ. Bayesian inference of population size history from multiple loci. BMC Evoutionary Biology. 2008;8:289. doi: 10.1186/1471-2148-8-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Molecular Biology and Evolution. 2010;27 (3):570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SY, Phillips MJ, Cooper A, Drummond AJ. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Molecular Biology and Evolution. 2005;22 (7):1561–1568. doi: 10.1093/molbev/msi145. [DOI] [PubMed] [Google Scholar]
- Holman JA. Pleistocene Amphibians and Reptiles in North America. Vol. 32. Oxford University Press; 1995. [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132 (2):583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles LL, Carstens BC. Estimating a geographically explicit model of population divergence. Evolution. 2007;61 (3):477–493. doi: 10.1111/j.1558-5646.2007.00043.x. [DOI] [PubMed] [Google Scholar]
- Lane E. Florida’s Geological History and Geological Resources. Special publication (Florida Geological Survey (1989)) Vol. 35. Published for the Florida Geological Survey; Tallahassee, FL: 1994. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23 (21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leaché AD, Harris RB, Rannala B, Yang Z. The Influence of Gene Flow on Species Tree Estimation: A Simulation Study. Syst Biol. 2013 doi: 10.1093/sysbio/syt049. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25 (11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Losos JB. Organisms and environments. Vol. 10. University of California Press; Berkeley: 2009. Lizards in an evolutionary tree : ecology and adaptive radiation of anoles. [Google Scholar]
- Lovern MB, Holmes MM, Wade J. The green anole (Anolis carolinensis): a reptilian model for laboratory studies of reproductive morphology and behavior. The Institute for Laboratory Animal Research Journal. 2004;45 (1):54–64. doi: 10.1093/ilar.45.1.54. [DOI] [PubMed] [Google Scholar]
- Macey JR, Schulte JA, 2nd, Larson A, Tuniyev BS, Orlov N, et al. Molecular phylogenetics, tRNA evolution, and historical biogeography in anguid lizards and related taxonomic families. Mol Phylogenet Evol. 1999;12 (3):250–272. doi: 10.1006/mpev.1999.0615. [DOI] [PubMed] [Google Scholar]
- McCormack JE, Heled J, Delaney KS, Peterson AT, Knowles LL. Calibrating divergence times on species trees versus gene trees: implications for speciation history of Aphelocoma jays. Evolution. 2011;65 (1):184–202. doi: 10.1111/j.1558-5646.2010.01097.x. [DOI] [PubMed] [Google Scholar]
- Michaud EJ, Echternacht AC. Geographic variation in the life history of the Lizard Anolis carolinensis and support for the pelvic constraint model. J Herpetol. 1995;29 (1):86–97. doi: 10.2307/1565090. [DOI] [Google Scholar]
- Neill W. Historical Biogeography of Present-Day Florida, vol 2. Bulletin of the Florida State Museum. Vol. 7. University of Florida; Gainesville, FL: 1957. [Google Scholar]
- Peng Y, Yang Z, Zhang H, Cui C, Qi X, et al. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Molecular Biology and Evolution. 2011;28 (2):1075–1081. doi: 10.1093/molbev/msq290. [DOI] [PubMed] [Google Scholar]
- Peterson GI, Masel J. Quantitative prediction of molecular clock and ka/ks at short timescales. Molecular Biology and Evolution. 2009;26 (11):2595–2603. doi: 10.1093/molbev/msp175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petuch EJ. Cenozoic Seas : the View from Eastern North America. CRC Press; Boca Raton: 2004. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155 (2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4(1):137–138. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61 (3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salicini I, Ibanez C, Juste J. Multilocus phylogeny and species delimitation within the Natterer’s bat species complex in the Western Palearctic. Mol Phylogenet Evol. 2011;61 (3):888–898. doi: 10.1016/j.ympev.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Morris AB, McLachlan JS, Manos PS, Soltis PS. Comparative phylogeography of unglaciated eastern North America. Molecular Ecology. 2006;15 (14):4261–4293. doi: 10.1111/j.1365-294X.2006.03061.x. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics. 2001;68 (4):978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123 (3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28 (10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nature Genetics. 2007;39 (1):31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nature Reviews Genetics. 2002;3 (8):611–621. doi: 10.1038/nrg865. [DOI] [PubMed] [Google Scholar]
- Tollis M, Ausubel G, Ghimire D, Boissinot S. Multi-locus phylogeographic and population genetic analysis of Anolis carolinensis: historical demography of a genomic model species. PLoS One. 2012;7 (6):e38474. doi: 10.1371/journal.pone.0038474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade J. Sculpting reproductive circuits: relationships among hormones, morphology and behavior in anole lizards. General and Comparative Endocrinology. 2012;176 (3):456–460. doi: 10.1016/j.ygcen.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Weir JT, Schluter D. Calibrating the avian molecular clock. Molecular Ecology. 2008;17 (10):2321–2328. doi: 10.1111/j.1365-294X.2008.03742.x. [DOI] [PubMed] [Google Scholar]
- Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, et al. Localizing recent adaptive evolution in the human genome. PLoS Genetics. 2007;3 (6):e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Molecular Biology and Evolution. 2006;23 (1):212–226. doi: 10.1093/molbev/msj024. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Peng R, Kuro-o M, Zeng X. Exploring patterns and extent of bias in estimating divergence time from mitochondrial DNA sequence data in a particular lineage: a case study of salamanders (order Caudata) Molecular Biology and Evolution. 2011;28 (9):2521–2535. doi: 10.1093/molbev/msr072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.