Abstract
Nicotinic acetylcholine (ACh) receptor (nAChR) agonists are potential therapeutic agents for neurological dysfunction. In the present study, the homopentameric mollusk ACh binding protein (AChBP), used as a surrogate for the extracellular ligand-binding domain of the nAChR, was specifically derivatized by the highly potent agonist azidoepibatidine (AzEPI) prepared as a photoaffinity probe and radioligand. One EPI-nitrene photoactivated molecule was incorporated in each subunit interface binding site based on analysis of the intact derivatized protein. Tryptic fragments of the modified AChBP were analyzed by collision-induced dissociation and Edman sequencing of radiolabeled peptides. Each specific EPI-nitrene-modified site involved either Tyr195 of loop C on the principal or (+)-face or Met116 of loop E on the complementary or (−)-face. The two derivatization sites were observed in similar frequency, providing evidence of the reactivity of the azido/nitrene probe substituent and close proximity to both residues. [3H]AzEPI binds to the α4β2 nAChR at a single high-affinity site and photoaffinity-labels only the α4 subunit, presumably modifying Tyr225 spatially corresponding to Tyr195 of AChBP. Phe137 of the β2 nAChR subunit, equivalent to Met116 of AChBP, conceivably lacks sufficient reactivity with the nitrene generated from the probe. The present photoaffinity labeling in a physiologically relevant condition combined with the crystal structure of AChBP allows development of precise structural models for the AzEPI interactions with AChBP and α4β2 nAChR. These findings enabled us to use AChBP as a structural surrogate to define the nAChR agonist site.
Neurotransmitter receptors and their intrinsic or associated channels or coupling proteins are the targets for many drugs of therapy and abuse in the central nervous system. Nicotinic acetylcholine (ACh) receptors (nAChRs) are prototypical agonist-gated ion channels responsible for rapid excitatory neurotransmission. Vertebrate nAChRs consist of diverse subtypes assembled from different sets of subunits expressed in skeletal muscle [α1, β1, γ (or ε), and δ], neurons (α2–α10 and β2–β4), and sensory epithelia (α9 and α10) (1,2). Among them the α4β2 nAChR, consisting of two α4 and three β2 subunits, is the most abundant and widespread in brain (3–6). The α4β2 nicotinic receptor is a major target for potential therapeutic agents for neuropathic diseases, cognitive enhancement, analgesia, and smoking cessation (2, 7) and is also important in considering the selective toxicity of insecticides (8).
Electron microscopy, X-ray crystallography, and fluorescence and NMR spectroscopy approaches give important but limited understanding of the recognition properties of the binding sites and their capacity to elicit a signal. Affinity-labeling and site-directed mutation studies on Torpedo or skeletal muscle nAChR have identified several amino acid residues delineating the ligand-binding pocket (loops A–F) that are localized at the interfacial region between subunits (1, 9–12). Specific subunit combinations confer differences in sensitivity to ACh and pharmacological profiles among the nAChR subtypes. The ligand-binding site in all subtypes contains a conserved core of aromatic amino acid residues (1, 13–16). Neighboring variable residues are considered to confer individual pharmacological properties to each nAChR subtype (1, 10).
The knowledge of agonist-nAChR interactions was greatly advanced by the discovery of epibatidine (EPI) isolated from the skin of an Ecuadoran frog (17) which has exceptionally high affinity to the nAChRs, including α4β2, and profound analgesic activity (18). Another major step forward was the identification and crystallization of soluble ACh binding proteins (AChBPs) from the snail Lymnaea stagnalis (19, 20) and apo and EPI-bound forms from a saltwater mollusk, Aplysia californica (21, 22), as surrogates for the extracellular ligand-binding domain of the nAChR. EPI and AChBP offer a signature high-affinity complex that prompts this study using the photoaffinity-labeling approach combined with mass spectrometry (MS), which is a physiologically compatible chemical method for three-dimensional structural investigations. The first objective of this research is to develop 5-azidoepibatidine (AzEPI) as a radioligand and photoaffinity probe. The second is to identify the AzEPI-labeled site by specific derivatization of AChBP and collision-induced dissociation (CID) analysis. The third employs [3H]AzEPI to photoaffinity label the α4β2 nAChR and predict potential modification sites. Finally, structural models are presented for the agonist binding domains of AChBP and α4β2 nAChR.
EXPERIMENTAL PROCEDURES
Chemicals
Sources were as follows: (±)-EPI HCl from TOCRIS (Ellisville, MO); carbachol (Carb) chloride, (−)-nicotine (NIC) hydrogen tartrate, and α-bungarotoxin (R-BGT) from Sigma (St. Louis, MO); unlabeled (±)-5-azidoepibatidine (AzEPI) from our previous study (23) using a different preparation methodology from the radiosynthesis of [3H]AzEPI described below. [3H]EPI was purchased from Amersham Biosciences (Piscataway, NJ).
Synthesis of AzEPI Intermediate 3 (Scheme 1)
Scheme 1. Radiosynthesis of [3H]AzEPIa.
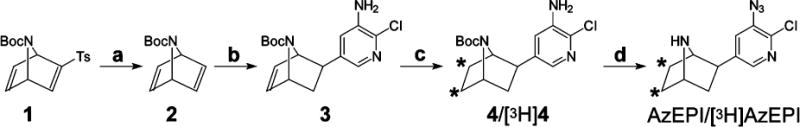
aConditions: (a) 5% Na/Hg, MeOH; (b) 3-amino-2-chloro-5-iodopyridine, Pd(OAc)2, PPh3, DMF, piperidine, HCOOH, room temperature, 4 days; (c) T2 or H2, Pd/C (10%), EtOAc, room temperature, 4 h; (d) (i) 37% HCl; (ii) NaNO2, 0 °C to room temperature, 30 min; (iii) NH2OH·HCl, 0 °C; (iv) 2 N NaOH, 0 °C, 4 h. Asterisks indicate positions of tritium.
N-Boc-2-tosyl-7-azahepta-2,5-diene (1) was synthesized by the Diels-Alder reaction of N-Boc-pyrrole and tosylacetylene (24). Into a solution of 1 (700 mg, 2.0 mmol) in THF and methanol (1:1, 40 mL) was added Na2HPO4 (4.25 g, 30 mmol). The solution was cooled to −15 °C, and 5% sodium amalgam (2 × 2.0 g) was added with further stirring for 1 h. The reaction was quenched with saturated NH4Cl solution and then extracted with EtOAc. Removal of the solvent provided 2 (125 mg, 30%). Into a vial with 2 (125 mg, 0.65 mmol) were added sequentially 3-amino-2-chloro-5-iodopyridine (23) (149 mg, 0.58 mmol), Pd(OAc)2 (14 mg, 0.062 mmol), PPh3 (34 mg, 0.13 mmol), DMF (1.0 mL), piperidine (166 mg, 193 μL, 1.95 mmol), and formic acid (75 mg, 61 μL, 1.63 mmol). The solution was then stirred at room temperature for 4 days. Workup as usual (23) followed by column chromatography on silica gel with ether and hexane (2:3) provided 3 (30 mg, 19%). 1H NMR: δ 7.70 (s, 1H), 7.13 (s, 1H), 6.45 (br s, 2H), 4.84 (br s, 1H), 4.54 (br s, 1H), 4.05 (s, 2H), 2.59 (dd, J = 4.1, 8.2 Hz), 1.86–1.74 (m, 2H), 1.41 (s, 9H).
Conversion of 3 to AzEPI and [3H]AzEPI (Scheme 1)
Into a solution of 3 (12 mg, 0.037 mmol) in EtOAc (1.5 mL) was added 10% Pd/C (2.5 mg). The atmosphere was replaced with hydrogen, and the solution was stirred at room temperature for 4 h. Hydrogen was then evacuated, and the solution was passed through a syringe membrane filter into a vial. The solvent was removed by flushing with nitrogen, providing a residue of 4 which was treated with 37% HCl (0.16 mL, 1.92 mmol) and stirred at room temperature for 30 min followed by addition of H2O (0.4 mL). The mixture was then cooled to 0 °C, and NaNO2 (125 μL of 100 mg/mL solution precooled to 0 °C, 0.18 mmol) was added with continuous stirring for 15 min at 0 °C and an additional 15 min at room temperature. Solid NH2OH·HCl salt (15 mg, 0.22 mmol) was added, and the mixture was cooled to 0 °C again. NaOH (2 N, 1.0 mL, 2.0 mmol) was added dropwise, and the solution was stirred at 0 °C for 4 h. HPLC indicated the desired product AzEPI (23). Tritiation was performed by ViTrax (Placentia, CA). Following the above hydrogenation procedure, replacing hydrogen with tritium gas, and properly modifying the reactor to handle the radioactive atmosphere provided [3H]AzEPI, which was purified by HPLC with an ODS1 150 × 4.6 mm C18 reverse-phase column (mobile phases, 0.01% trifluoroacetic acid in water and acetonitrile; gradient, 10–90% acetonitrile over 10 min at 5 mL/min). The specific activity and radiochemical purity were determined by HPLC cochromatography and radioflow chromatography.
Preparation of AChBP
Aplysia AChBP (flanked with an N-terminal FLAG epitope) was expressed as a soluble exported protein from stably transfected HEK293S cells lacking the N-acetylglucosaminyltransferase I gene and selected for geneticin resistance (21, 22). Culture medium containing the secreted AChBP was harvested from multi-layered flasks to obtain several liters of media over a 1–2 week period. Media were stored at 4 °C with 0.02% sodium azide. AChBP was purified on immobilized anti-FLAG antibody and dialyzed against 50 mM Tris-HCl buffer (pH 7.4) containing 150 mM NaCl and 0.02% sodium azide. The dialysate was concentrated by ultrafiltration using the YM-50 Centricon unit (Millipore, Bedford, MA), and the Tris buffer was finally exchanged by phosphate-buffered saline (PBS) [50 mM sodium phosphate buffer (pH 7.5) containing 50 mM NaCl] using the Centricon to give 2–3 mg of protein/mL.
Preparation of the α4β2 Receptor
Chick α4β2 nAChR was prepared from the M10 cell line of mouse fibroblasts stably transfected with chick α4 and β2 subunits under the control of a dexamethasone-sensitive promoter (25). The 40000g pellet (30 min, 4 °C) from a homogenate of the harvested cells was resuspended in PBS and immediately used for radioligand binding. Alternatively, the pellet for photoaffinity labeling was solubilized with 2% Triton X-100 in PBS containing 2 mM phenylmethanesulfonyl fluoride (solubilization medium) for 20 min at 4 °C on a rocking platform. Unsolubilized material was removed by centrifugation as above.
Radioligand Binding
Different types of procedures were used for assays with AChBP and α4β2 nAChR. AChBP determination of AzEPI potency involved an adaptation of a scintillation proximity assay with [3H]EPI (26). The α4β2 receptor preparation (200–300 μg of membrane protein) in PBS (500 μL final volume) was incubated for 60 min at 25 °C with appropriate concentrations of [3H]AzEPI alone or with an unlabeled displacer. The assay was terminated by rapid filtration and three rinses with ice-cold saline and then scintillation counting. Nonspecific binding was determined with 2 μM EPI. Binding parameters [dissociation constant (KD), maximum binding capacity (Bmax), and Hill coefficient (nH)] were obtained from saturation experiments. IC50 values, molar concentrations for 50% displacement, were converted to inhibition constant (Ki) values using the equation of Cheng and Prusoff (27).
Photoaffinity Labeling
AChBP has five interfacial binding sites in the homopentamer. A solution of AChBP (38 or 76 pmol sites) in PBS [50 or 100 μL final volume, respectively (760 nM sites final concentration)] was incubated with 1.2 equiv of [3H]AzEPI for 60 min at 25 °C in the dark without or with 10000 pmol (10 μM final concentration) of EPI (for nonspecific labeling). After irradiation with 300 nm lamps for 20 s (AzEPI completely photoreacted in this condition), the radiolabeled AChBP recovered by ultrafiltration with the YM-30 Centricon unit was subjected to SDS–PAGE with detection of proteins by Coomassie dye and labeled bands with the Amplify fluorographic reagent (Amersham Biosciences) followed by gel slicing and scintillation counting. For MS analysis AChBP (200 pmol sites, 2 μM sites final concentration) in PBS (100 μL) was treated with 1.5 equiv of nonradioactive AzEPI as above. Derivatized AChBP was purified by the YM-30 Centricon unit and PBS exchanged to 25 mM ammonium bicarbonate buffer (pH 7.8).
For α4β2 nAChR, the solubilized receptor preparation (10 pmol of [3H]AzEPI binding sites, about 10 mg of total protein) in solublization medium (1 mL) was incubated at 25 °C for 60 min in the dark with 2 equiv of [3H]AzEPI (20 nM final concentration) without or with 10000 pmol (10 μM final concentration) of EPI (for nonspecific labeling). The reaction mixture was irradiated as above, incubated overnight at 4 °C with monoclonal antibody mAb270 against the β2 subunit (Sigma) (dilution ratio 1:100), and finally reacted with protein G PLUS-agarose (Santa Cruz Biotechnology Inc., Santa Cruz, CA) for 120 min at 4 °C. After recovery of the agarose beads and five washes with solubilization medium, the proteins were subjected to SDS–PAGE followed by fluorographic detection and radioactivity quantification.
Edman Sequencing of Radiolabeled Peptides
[3H]AzEPI-labeled AChBP was denatured with urea, reduced with dithiothreitol, and alkylated with iodoacetamide. The sample was desalted by the YM-3 Centricon column and washed twice with 25 mM ammonium bicarbonate buffer (pH 7.8). The radiolabeled protein was deglycosylated with peptide N-glycosidase F (Sigma) and then digested with trypsin from Promega (Madison, WI). The digested fragments were separated by HPLC with a Vydac 150 × 1 mm C18 reverse-phase column (Sorbent Technologies, Atlanta, GA). The radioactive peptide fractions defined by liquid scintillation counting were analyzed with the ABI 494-HT Procise Edman sequencer (Applied Biosystems, Foster City, CA).
MS Analysis
Molecular weights of the intact AChBP subunit were measured on an Applied Biosystems MDS Sciex QSTAR hybrid quadrupole time-of-flight mass spectrometer. Samples in 10 mM ammonium bicarbonate were injected onto a Phenomenex 150 × 0.1 mm Onyx monolithic column (Torrance, CA) flowing at 500 nL/min using 50% acetonitrile in water containing 0.1% formic acid as the mobile phase. Data were acquired and the charge state distributions converted to a normalized zero charge spectrum using the Analyst 1.1 software. Protein standards of about the same size showed a mass accuracy of approximately 1–2 Da.
For CID analysis, intact proteins in buffer with an equal volume of acetonitrile were reduced with dithiothreitol, alkylated with iodoacetamide, and digested with trypsin (Promega) for 4 h at pH 7.8 and 37 °C. After removal of the acetonitrile, digested samples were analyzed by nanoflow LC/MS/MS on the QSTAR instrument as above with a self-packed Ultro 120 (5 μm particle size) (Peeke Scientific, Redwood City, CA) 150 × 0.1 mm capillary column. The flow rate was 300–330 nL/min with a gradient from 5% to 50% acetonitrile in 60 min (all solvents contained 0.1% formic acid). The Information-Dependent Acquisition mode was used with a 1.0 s survey scan from m/z 310 to m/z 1400, followed by one 3 s CID scan on the most abundant multiply charged ion in the survey scan. Data were analyzed with Protein Prospector v4.23.4, an in-house developed program, to identify peptides that shifted in mass reflecting the photolabel incorporation.
Accurate masses of the labeled peptides were obtained from LC/MS/MS analysis on a Thermo Electron LTQ FT hybrid linear ion trap FTICR mass spectrometer (Waltham, MA). Chromatography was as described above for LC/MS/MS on the QSTAR. The data-dependent acquisition method consisted of one full mass survey scan in the FTICR at 25000 resolution. The three most intense multiply charged target ions were isolated for accurate mass measurement in the FTICR in SIM mode, using a 10 Da mass window at 50000 resolution. The accuracy of the mass measurements is typically within 2 ppm under such conditions. These three ions were then fragmented in the linear ion trap using CID. Target values for the three different scan modes were 3000000, 50000, and 30000, respectively.
Modeling and Calculations
The X-ray structure for Aplysia AChBP (22) as the EPI-bound (PDB code 2BYQ) form was used as template for building the α4β2 nAChR homology model. The fasta protein sequences of the α4 and β2 subunits for Gallus gallus (5, 28, 29) were downloaded from UniProt (30) (accession numbers P09482 for α4 and P09484 for β2) and then aligned with 2BYQ using the CLUSTAL W (31) web server at the European Bioinformatics Institute (32, 33). Chain A of the PDB structure was chosen for sequence alignment with the ligand-binding domain (residues α4 1–240 versus 2BYQ:A, β2 1–240 versus 2BYQ:A). These alignments were input to the SWISS-MODEL protein homology server (34). The resulting model subunit structures were imported into Maestro 6.5 (Schrodinger, L.L.C., Portland, OR). The α4 and β2 model structures were aligned via comparison of backbone atoms onto the corresponding subunits in 2BYQ, yielding a complete pentameric α4β2 model structure. Further optimization was limited to a single α4/β2 subunit pair and, hence, a single binding interface. This α4β2 model was subjected to cycles of energy minimization using the OPLS_2005 force field implemented in Macromodel (35, 36). Up to 5000 steps per minimization were run to achieve a gradient of 0.5 with respect to energy. In the initial minimization cycles, the backbone was held constant. In subsequent minimization cycles, the region within 15 Å of the binding pocket was free to move with progressive constraints on the remainder of the structure. Docking calculations were carried out using the GALS algorithm in AutoDock 3 (37, 38). Docked molecules were constrained to a 40 point grid in the region of the binding pocket.
RESULTS
Radiosynthesis
The reaction sequence to prepare [3H]-AzEPI via 1–4 was designed to introduce the tritium label at the penultimate step and finally convert aminopyridine [3H]-4 to azidopyridine [3H]AzEPI. Detosylation of 1 with sodium–mercury amalgam (a) provided diene 2, which was treated with 3-amino-2-chloro-5-iodopyridine under reductive Heck coupling conditions (b) to provide the desired exo isomer 3. Compound 3 was hydrogenated or tritiated with tritium gas using Pd/C as the catalyst (c). The amino group was converted to azido by sequential treatment with concentrated HCl and NH2OH (d), which also removed the Boc group to provide [3H]AzEPI after HPLC with a specific activity of 36 Ci/mmol and radiochemical purity of >99% by radioflow chromatography. Little or no radiochemical decomposition was observed for [3H]AzEPI stored in ethanol at −20 °C for at least 3 years.
Ligand-Binding Measurements (Figure 1)
FIGURE 1.
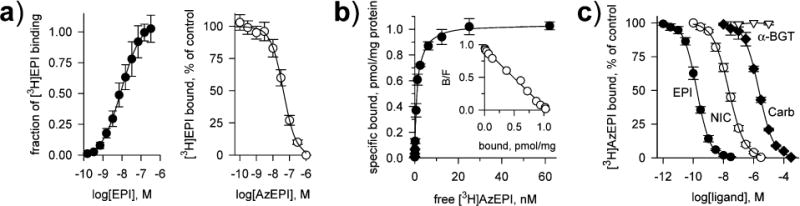
Ligand binding with AChBP (a) and α4β2 nAChR (b and c). (a) Concentration-dependent response curves for direct titration of [3H]EPI and for AzEPI competing with [3H]EPI binding to AChBP. KD for [3H]EPI is 8.6 ± 2.0 nM (nH 0.90 ± 0.05), and Ki for AzEPI is 10 ± 1 nM (nH 1.13 ± 0.08) (±SD, n = 3–4). (b) Saturation isotherm and Scatchard plot (insert) for specific [3H]AzEPI binding to α4β2 nAChR. The binding parameters (±SD, n = 3) are KD 1.0 ± 0.1 nM, Bmax 1.0 ± 0.02 pmol/mg of membrane protein, and nH 0.99 ± 0.03. (c) Displacement curves of specific [3H]AzEPI binding to α4β2 nAChR by three nicotinic agonists and the antagonist α-BGT. Ki values (and nH) are as follows (±SD, n = 3): 0.17 ± 0.02 nM (1.02 ± 0.02) for EPI, 14 ± 2 nM (0.96 ± 0.01) for NIC, and 820 ± 100 nM (0.98 ± 0.01) for Carb.
For Aplysia AChBP, affinities for EPI and AzEPI were 8.6 and 10 nM, respectively. For α4β2 nAChR expressed in M10 cells, specific [3H]AzEPI binding was saturable and exhibited a single high-affinity interaction with KD 1.0 nM, Bmax 1.0 pmol/mg of protein, and nH 0.99. In competition experiments, EPI, NIC, and Carb displaced [3H]AzEPI binding with Ki values of 0.2, 14, and 820 nM, respectively, while α-BGT failed to show any inhibition even at 10000 nM.
AChBP Binding Site. Photoaffinity Labeling at Two Distinct Sites in Similar Frequency
[3H]AzEPI photoreacted with 19.5 ± 1.2 pmol sites with nonspecific labeling of 0.43 ± 0.002 pmol (specific labeling 98%), occupying 24% of the total AChBP (76 pmol sites) (Figure 2). EPI protected against specific incorporation of [3H]AzEPI into AChBP by 50% at approximately 500 pmol. Tryptic fragments of [3H]-AzEPI-derivatized AChBP were subjected to HPLC separation, revealing only two labeled peptides with nearly equivalent radioactivity (Figure 3). Edman sequencing then established that the two radioactive peptides were Ser189−Lys203 (a chymotryptic fragment from trace chymotrypsin impurity) and Thr80−Arg122 (a tryptic fragment) (data not shown), establishing that there were two sites of labeling in similar frequency.
FIGURE 2.
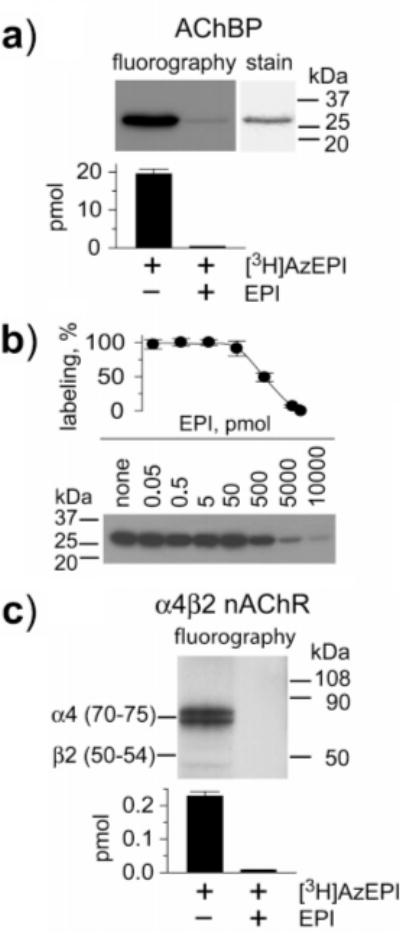
[3H]AzEPI photoaffinity labeling of AChBP and α4β2 nAChR. (a) Fluorography of [3H]AzEPI-labeled AChBP compared with Coomassie blue-stained protein and quantification by scintillation counting (bar graph with SD of three experiments). AChBP (76 pmol sites) was photoaffinity labeled by [3H]AzEPI (92 pmol) alone (−) and protected with EPI (10000 pmol) (+). (b) Quantification for protection of photoaffinity labeling of AChBP by EPI. AChBP (38 pmol sites) was reacted with [3H]AzEPI (46 pmol). The titration curve is plotted by radioactivity determination for the protein band as above in separate experiments from the fluorography image. (c) The α4β2 receptor (10 pmol sites) was subjected to photoaffinity labeling by [3H]AzEPI (20 pmol) in the absence (−) and the presence (+) of EPI (10000 pmol). Fluorography shows exclusive labeling of the α4 subunit. Photoincorporation is defined by radioactivity in the corresponding region on the SDS–PAGE, and the labeled amount is the summation of the doublet which is due to different states of glycosylation (25).
FIGURE 3.
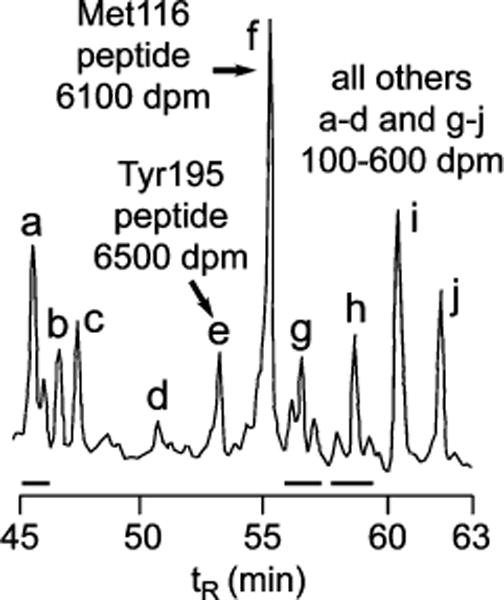
Liquid chromatogram of tryptic fragments of deglycosylated AChBP derivatized by [3H]AzEPI. The entire chromatogram (except for an area around the column void volume) contained two significant radioactive peaks in the positions illustrated. Peak areas are based on UV (214 nm) and radioactivity measurements (dpm) on equal sized aliquots. All other regions of the chromatogram have the 100–600 dpm basal level of radioactivity, i.e., the same in the absence (specific labeling) and the presence of 10000 pmol of EPI (nonspecific labeling).
Labeling at the Subunit Level
Mass measurement of AChBP treated with AzEPI yielded 25352 Da for the unmodified protein and 25574 Da for the derivative (Figure 4). The calculated MW for the FLAG-tagged [nine amino acids numbered as (−8)DYKDDDDKL(0)] protein is 25944 Da. MS-Nonspecific Search in the ProteinProspector software package revealed that 10 different sequences may correspond to this mass within 0.35%, where the N-terminus could be any amino acid from 1 to 6 [Asp(−8) to Asp(−3)] and the C-termini vary from 223 to full length, i.e., 228. At the same time LC/MS/MS analysis of the tryptic digest verified the presence of the N-terminal peptide (data not shown), and Arg215 was the last residue detected for the C-terminus. Considering the mass accuracy achieved for standard proteins, it was concluded that most likely a mixture with ragged termini was present. Tyr(−7) to Arg215 features the closest mass to the measured value: 25324 Da (−0.1% deviation). There were other components observed at 25368 and 25385 Da, i.e., with 16 and 33 Da higher mass, respectively. Since the sequence contains five Met, four Trp, and four Cys residues, it is reasonable to assume that these masses indicate partial oxidation most likely to methionine mono- and bissulfoxides. The second molecular mass cluster (with 25574 and 25590 Da) clearly indicates the incorporation of a single EPI-nitrene. The added mass relative to the unmodified protein was 222 Da, while the expected mass increment was 221 Da; this difference was verified at the peptide level (see below) where it could be more accurately measured. Very weak ions yielding an intact MW of 25795 Da indicated the possibility of trace double labeling.
FIGURE 4.
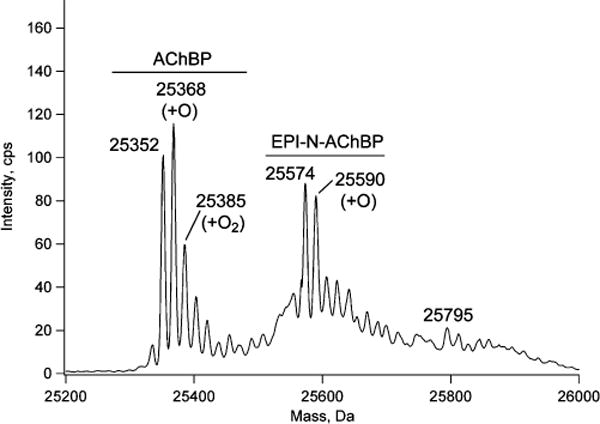
Molecular mass measurement of an AChBP subunit after labeling with AzEPI. The 25352 cluster represents the unmodified protein (AChBP) within which the mass increases of 16 and 33 Da may indicate the presence of methionine mono- and bissulfoxides. The second cluster corresponds to the singly EPI-nitrene-modified protein (EPI-N-AChBP) and a monooxidized derivative. The third very weak cluster indicates trace double labeling.
Labeled Positions Are Tyr195 and Met116
Once the presence of the label was ascertained at the protein level, the mixture was reduced, alkylated, and digested with trypsin. The resulting digest was subjected to LC/MS/MS analysis in a data-dependent fashion, where the computer selected the multiply charged ions for CID analysis. All MS/MS data were subjected to a two-step database search using the in-house ProteinProspector v4.23.4. First, with very strict search parameters the identity of the protein was verified, as well as the lack of contaminating proteins established. Then a second search was conducted that permitted a +300 Da modification on any amino acid in the sequence. The +221 Da shift identified three EPI-nitrene-modified peptides: (1) Gln184−Lys203 and two overlapping peptides, (2) Pro98−Arg122, and (3) Thr80−Arg122. Accurate mass measurements were obtained for the peptide precursor ions (m/z 668.1 and 970.3) for the two peptides, Gln184−Lys203 and Pro98−Arg122 (Table 1). The CID data identified the modified residues as Tyr195 (Figure 5) and Met116 (Figure 6).
Table 1.
Accurate Masses of the Labeled Peptides
| peptide | modified residue | precursor m/z (z) | MW (Da)
|
∆M (ppm) | |
|---|---|---|---|---|---|
| measured | calculated | ||||
| Gln184−Lys203 | Tyr195 | 668.0656 (4) | 2668.2311 | 2668.2336 | 0.9 |
| Pro98−Arg122a | Met116 | 970.2933 (5)a | 4846.4274 | 4846.4303 | 0.5 |
m/z 728.4 (+4 ion) for the peptide Pro98−Arg122 (see legend for Figure 6) was not observed in the LTQFT LCMSMS run. However, m/z 970.2933 (+5 ion) for the peptide Thr80-Arg122 was seen. The CID obtained on the LTQFT for 970.2933 confirmed its identity.
FIGURE 5.
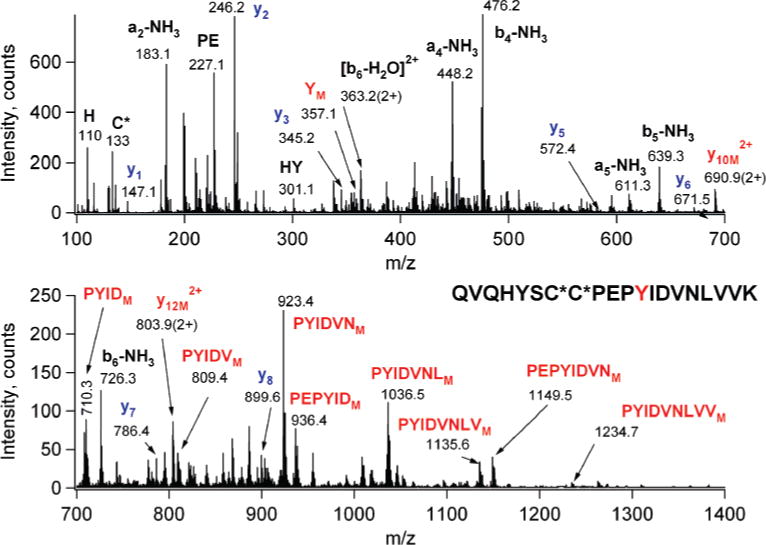
Low-energy CID spectrum of the EPI-nitrene-modified Gln184−Lys203 tryptic peptide, precursor ion at m/z 668.1 (4+). Assignments were made according to the nomenclature of Biemann (39). The modification occurred at Tyr195 as indicated by the unmodified N-terminal b ions (up to b6 in black) and C-terminal y fragments (up to y8 in blue) and the modified immonium y ion and internal fragments (in red). For simplicity labeling involves mostly sequence ions and internal fragments containing the modified residue. Carbamidomethylcysteines are designated as C*.
FIGURE 6.
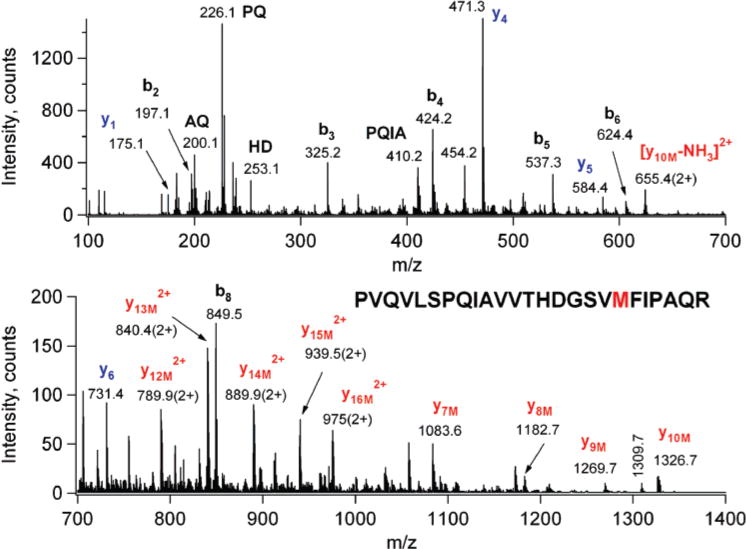
Low-energy CID spectrum of the EPI-nitrene-modified Pro98−Arg122 tryptic peptide, precursor ion at m/z 728.4 (4+). Met116 is modified. The C-terminal y ions were detected unmodified up to y6 (fragments in blue), while all y fragments from y7 (in red) show the 221 Da mass shift. Labeling involves the sequence ions and only a few other fragments.
α4β2 nAChR Binding Site. Photoaffinity Labeling Exclusively at the α4 Subunit
The α4β2 receptor was selectively labeled by [3H]AzEPI at the α4 subunit (as a doublet), but no labeling was evident on the β2 subunit (Figure 2). The labeling amount on the α4 subunit was 0.23 ± 0.01 and 0.008 ± 0.0002 pmol in the absence and the presence of unlabeled EPI, respectively, establishing 97% specific incorporation. The occupancy rate was 2.2% of the total sites (10 pmol). The existence of the α4 subunit in the corresponding region on the SDS−PAGE gel was confirmed by LC/MS/MS analysis of an unlabeled sample after immunoprecipitation (data not shown).
Binding Site Models
The structure of AzEPI was first optimized in Gaussian 03 at B3LYP/6-31G. The N=N=N angles for phenyl azides in the Cambridge Structural Database average around 170°, presumably due to resonance forms wherein the central nitrogen is not entirely sp. The electronic and steric environment also affects the angle, and AzEPI has a calculated N=N=N angle of 165°. The azido moiety can rotate about the C–N bond, resulting in two optimized conformers with similar energy levels, suggesting that both exist at room temperature. This bond was allowed to rotate in the docking calculations.
AzEPI is calculated to dock into the AChBP (2BYQ structure without EPI itself) and the present α4β2 nAChR model with energies of −9.69 and −7.87 kcal/mol, respectively (Figure 7). The AzEPI-bound AChBP structure is superimposable to the EPI-bound crystal (2BYQ). Two critical interactions are predicted for the AzEPI ammonium with Trp147 of AChBP and Trp177 of α4, i.e., hydrogen bonding with the backbone carbonyl oxygen (1.8 and 2.0 Å, respectively) and cation–π contact to the indole side chain (3.2 and 3.6 Å, respectively). The 6-chloro substituent of AzEPI makes van der Waals contacts to the backbone oxygen atoms of Ile106 (3.6 Å) and Met116 (3.6 Å) in AChBP and to Asn127 (3.7 Å) and Phe137 (3.3 Å) in α4β2. The AzEPI pyridine nitrogen atom probably undergoes hydrogen bonding with Ile118 (4.1 Å) and Trp147 (3.7 Å) via a solvent bridge in AChBP, and the equivalent targets would be Leu139 (4.6 Å) and Trp177 (3.5 Å) in α4β2. The AzEPI azido group is positioned between Tyr195/Met116 of AChBP and Tyr225 of α4 and Phe137 of β2, allowing the derivatization by the EPI-nitrene (e.g., the distance between the EPI-nitrene nitrogen to the hydroxyl oxygen of Tyr195 or sulfur of Met116 is presumably about 3–4 Å), although the β2 subunit was not modified. The azido conformer pointing toward the chlorine atom is closer to Met116, and the one pointing away is closer to Tyr195 (Figure 7). Further, Tyr195 and Met116 in the AzEPI-bound holo structure are 3–4 Å more closely positioned relative to the agonist molecule than those in the apo form (Figure 8).
FIGURE 7.
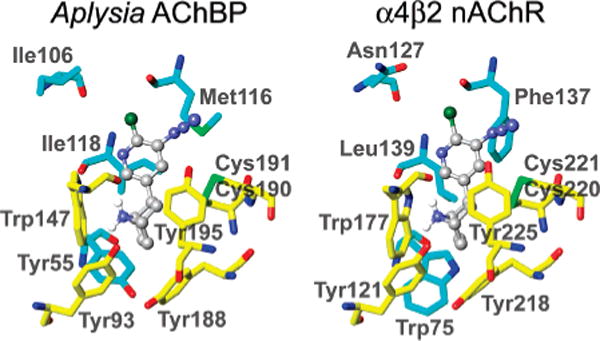
AzEPI-bound structures of Aplysia AChBP and chick α4β2 nAChR. Relevant residues in yellow are from the (+)-face of AChBP or the α4 subunit while relevant residues in aquamarine are from the (−)-face of AChBP or the β2 subunit. The AzEPI-docked AChBP model was calculated on the basis of the corresponding crystal structure (2BYQ) (22). The α4β2 interface was constructed using 2BYQ as the scaffold.
FIGURE 8.
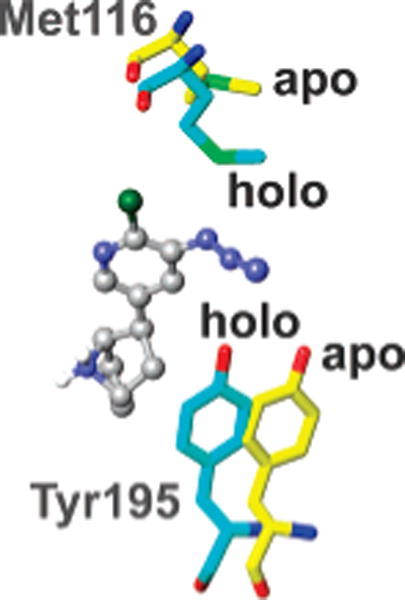
Comparisons of geometries for Tyr195 and Met116 between the AzEPI−AChBP complex and the apo form. Tyr195 and Met116 (in aquamarine) in the AzEPI-bound conformation (holo) are overlaid with those in the apo structure (in yellow) (2BYN) (22).
DISCUSSION
[3H]AzEPI, an Optimized Photoaffinity Radioligand
The photoreactive 5-azido substituent was introduced at a structural position selected to minimize any change in potency (23, 40, 41). AzEPI and [3H]AzEPI show similar affinity to EPI itself in the 1–10 nM range, and fortunately [3H]AzEPI has excellent radiochemical stability.
AChBP Agonist Binding Site
Structural information regarding the binding domains and subunit interfaces was expanded by the discovery and crystallization of AChBPs liganded with several agonists and antagonists (19, 20, 22, 42–44). The refined 4 Å resolution electron microscopy structure of the heteropentameric Torpedo nAChR established considerable structural similarity of AChBP with this nAChR (45). Aplysia AChBP is shown here to bind specifically and photoreact with AzEPI, giving the incorporation ratio of one probe molecule (EPI-nitrene) per AChBP subunit (binding site), and both Tyr195 on loop C and Met116 on loop E (in similar frequency) are revealed as the sites for derivatization. The nearly equivalent labeling on the primary loop C face and the complementary loop E on the neighboring subunit shows the close proximity of these residues on the two subunits. These results precisely define the orientation of EPI in the binding pocket under a physiologically compatible condition.
Comparing Aplysia AChBP–ligand complexes with an apo structure provides direct evidence of differential conformational rearrangements (particularly of loop C) induced by antagonist and agonist occupation (22). The agonist-elicited conformational changes are also supported by fluorescence spectroscopy, deuterium-hydrogen exchange MS, and NMR spectroscopy analyses of AChBP (46–48). The present photoaffinity labeling clearly indicates that loop C Tyr195 is oriented toward loop E Met116, and upon agonist binding loop C draws inward to cap the binding pocket and envelop the agonist molecule (Figure 8). A similar proposal has been made from photolabeling studies for tightening the interfacial binding domain (involving primary and complementary subunits) after ligand interaction in Torpedo nAChR (12, 49, 50). This conformational change serves as an initial event for the ligand-induced channel opening mechanism of the nAChR, but it remains unclear how agonist occupation is coupled to the distant transmembrane-localized ion gating site.
α4β2 nAChR Binding Sites
A major quest in developing nicotinic agents is the discovery of highly subtype-selective agonists and antagonists particularly for the α4β2 nAChR. The present study for the first time succeeds in photoaffinity labeling the specific binding site of the α4β2 receptor, and [3H]AzEPI exclusively labels the α4 subunit. Tyr225 of the α4 subunit is equivalent to Tyr195 of AChBP loop C. Phe137 of the β2 subunit corresponds to Met116 of AChBP loop E. Therefore, even though they are similarly positioned, the difference between Met116 of AChBP and Phe137 of the β2 subunit appears to determine whether loop E is labeled or not, presumably depending on reactivity of the side chain. For instance, the Met residue is a preferred site for photoderivatization by the benzophenone radical (51, 52). The first α4β2 nAChR binding site model was constructed on the basis of the crystal structure of HEPES-bound Lymnaea AChBP (53, 54). An improved α4β2 model docked with dechloro-EPI was generated using the NIC-bound Lymnaea AChBP template (55). The present model combines our photoaffinity labeling result and the X-ray structure of EPI-bound Aplysia AChBP. The cationic moiety of AzEPI interacts at a subsite surrounded by aromatic amino acid residues including the critical Trp177 on the α4β2 loop B segment. The backbone carbonyl oxygen and aromatic residue of Trp make hydrogen-bonding and cation–π contacts, respectively, on analogy with the findings on the R1-containing nAChR subtype and the Lymnaea and Aplysia AChBPs (14, 16, 20, 22). Substitution of Tyr55 (loop D) in Aplysia AChBP by Trp, as found in the β2 subunit (Figure 7), improves the affinity of EPI, suggesting that the nature or size of the aromatic side chain at this position is an important factor for EPI binding (22).
Concluding Remarks
Photoaffinity labeling in a physiologically relevant condition combined with MS analysis and the crystal structure of AChBP offers a precise molecular picture of the drug–receptor interaction and allows development of a structural model for the agonist binding site on the α4β2 subunit interface. These findings further define the surfaces with which agonists associate and should help to facilitate rational design of therapeutic agents for neurological disorders and analgesia.
Supplementary Material
Acknowledgments
The authors received valuable advice and assistance from Kwok-Yiu Ho (University of California, San Diego), Xin Zhang (University of California, San Francisco), Roselle Visaya (University of California, Davis), and Kazutoshi Fujioka (University of California, Berkeley).
Footnotes
Financial support: J.E.C. by the William Muriece Hoskins Chair in Chemical and Molecular Entomology; M.T., N.Z., and J.E.C. by NIH NIEHS Grant R01 ES08424; D.M., K.F.M., and A.L.B. by NIH NCRR Grants RR015084, RR001614, and RR019934; K.A.D. by NSF Grant CHE-0233882; T.T.T. and P.T. by USPHS Grants R37-GM18360 and UO-1 NS05846.
SUPPORTING INFORMATION AVAILABLE
Protein sequence alignment between Aplysia AChBP and chick α4 and β2 nAChR subunits. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Changeux JP, Edelstein SJ. Nicotinic Acetylcholine Receptors: From Molecular Biology to Cognition. Odile Jacob; New York: 2005. p. 284. [Google Scholar]
- 2.Cassels BK, Bermúdez I, Dajas F, Abin-Carriquiry JA, Wonnacott S. From ligand design to therapeutic efficacy: The challenge for nicotinic receptor research. Drug Discov Today. 2005;10:1657–1665. doi: 10.1016/S1359-6446(05)03665-2. [DOI] [PubMed] [Google Scholar]
- 3.Whiting PJ, Lindstrom JM. Characterization of bovine and human neuronal nicotinic acetylcholine receptors using monoclonal antibodies. J Neurosci. 1988;8:3395–3404. doi: 10.1523/JNEUROSCI.08-09-03395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anand R, Conroy WG, Schoepfer R, Whiting P, Lindstrom J. Neuronal nicotinic acetylcholine receptors expressed in Xenopus oocytes have a pentameric quaternary structure. J Biol Chem. 1991;266:11192–11198. [PubMed] [Google Scholar]
- 5.Cooper E, Couturier S, Ballivet M. Pentameric structure and subunit stoichiometry of a neuronal nicotinic acetylcholine receptor. Nature. 1991;350:235–238. doi: 10.1038/350235a0. [DOI] [PubMed] [Google Scholar]
- 6.Flores CM, Rogers SW, Pabreza LA, Wolfe BB, Kellar KJ. A subtype of nicotinic cholinergic receptor in rat brain is composed of α4 and β2 subunits and is up-regulated by chronic nicotine treatment. Mol Pharmacol. 1992;41:31–37. [PubMed] [Google Scholar]
- 7.Bunnelle WH, Dart MJ, Schrimpf MR. Design of ligands for the nicotinic acetylcholine receptors: The quest for selectivity. Curr Top Med Chem. 2004;4:299–334. doi: 10.2174/1568026043451438. [DOI] [PubMed] [Google Scholar]
- 8.Tomizawa M, Casida JE. Neonicotinoid insecticide toxicology: Mechanism of selective action. Annu Rev Pharmacol Toxicol. 2005;45:247–268. doi: 10.1146/annurev.pharmtox.45.120403.095930. [DOI] [PubMed] [Google Scholar]
- 9.Arias HR. Localization of agonist and competitive antagonist binding sites on nicotinic acetylcholine receptors. Neurochem Int. 2000;36:595–645. doi: 10.1016/s0197-0186(99)00154-0. [DOI] [PubMed] [Google Scholar]
- 10.Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nat ReV Neurosci. 2002;3:102–114. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- 11.Grutter T, Ehert-Sabatier L, Kotzyba-Hibert F, Goeldner M. Photoaffinity labeling of Torpedo nicotinic receptor with the agonist [3H]DCTA: Identification of amino acid residues which contribute to the binding of the ester moiety of acetylcholine. Biochemistry. 2000;39:3034–3043. doi: 10.1021/bi992393o. [DOI] [PubMed] [Google Scholar]
- 12.Nirthanan S, Ziebell MR, Chiara DC, Hong F, Cohen JB. Photolabeling the Torpedo nicotinic acetylcholine receptor with 4-azido-2,3,5,6-tetrafluorobenzoylcholine, a partial agonist. Biochemistry. 2005;44:13447–13456. doi: 10.1021/bi051209y. [DOI] [PubMed] [Google Scholar]
- 13.Dougherty DA. Cation-π interactions in chemistry and biology: A new view of benzene, Phe, Tyr, and Trp. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 14.Zhong W, Gallivan JP, Zhang Y, Li L, Lester HA, Dougherty DA. From ab initio quantum mechanics to molecular biology: A cation-π binding site in the nicotinic receptor. Proc Natl Acad Sci USA. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dougherty DA, Lester HA. Snail, synapses and smokers. Nature. 2001;411:252–255. doi: 10.1038/35077192. [DOI] [PubMed] [Google Scholar]
- 16.Cashin AL, Petersson EJ, Lester HA, Dougherty DA. Using physical chemistry to differentiate nicotinic from cholinergic agonists at the nicotinic acetylcholine receptor. J Am Chem Soc. 2005;127:350–356. doi: 10.1021/ja0461771. [DOI] [PubMed] [Google Scholar]
- 17.Spande TF, Garraffo HM, Edwards MW, Yeh HJC, Pannell L, Daly JW. Epibatidine: A novel (chloropyridyl)azabicycloheptane with potent analgesic activity from an Ecuadoran poison frog. J Am Chem Soc. 1992;114:3475–3478. [Google Scholar]
- 18.Badio B, Daly JW. Epibatidine, a potent analgetic and nicotinic agonist. Mol Pharmacol. 1994;45:563–569. [PubMed] [Google Scholar]
- 19.Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptor. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 20.Celie PHN, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 21.Hansen SB, Talley TT, Radić Z, Taylor P. Structural and ligand recognition characteristics of an acetylcholine-binding protein from Aplysia californica. J Biol Chem. 2004;279:24197–24202. doi: 10.1074/jbc.M402452200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen SB, Sulzenbacher G, Huxford T, Marchot P, Taylor P, Bourne Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 2005;24:3635–3646. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang N, Tomizawa M, Casida JE. 5-Azidoepibatidine: An exceptionally potent photoaffinity ligand for neuronal α4β2 and α7 nicotinic acetylcholine receptors. Bioorg Med Chem Lett. 2003;13:525–527. doi: 10.1016/s0960-894x(02)00936-8. [DOI] [PubMed] [Google Scholar]
- 24.Carroll FI, Liang F, Navarro HA, Brieaddy LE, Abraham P, Damaj MI, Martin BR. Synthesis, nicotinic acetylcholine receptor binding, and antinociceptive properties of 2-exo-2-(2′-substituted 5′-pyridinyl)-7-azabicyclo[2.2.1]heptanes. Epibatidine analogues. J Med Chem. 2001;44:2229–2237. doi: 10.1021/jm0100178. [DOI] [PubMed] [Google Scholar]
- 25.Whiting P, Schoepfer R, Lindstrom J, Priestley T. Structural and pharmacological characterization of the major brain nicotinic acetylcholine receptor subtype stably expressed in mouse fibroblasts. Mol Pharmacol. 1991;40:463–472. [PubMed] [Google Scholar]
- 26.Talley TT, Yalda S, Ho KY, Tor Y, Soti FS, Kem WR, Taylor P. Spectroscopic analysis of benzylidene anabaseine complexes with acetylcholine binding proteins as models for ligand-nicotinic receptor interactions. Biochemistry. 2006;45:8894–8902. doi: 10.1021/bi060534y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng YC, Prusoff WH. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 28.Nef P, Oneyser C, Alliod C, Couturier S, Ballivet M. Genes expressed in the brain define 3 distinct neuronal nicotinic acetylcholine-receptors. EMBO J. 1988;7:595–601. doi: 10.1002/j.1460-2075.1988.tb02852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoepfer R, Whiting P, Esch F, Blacher R, Shimasaki S, Lindstrom J. cDNA clones coding for the structural subunit of a chicken brain nicotinic acetylcholine receptor. Neuron. 1988;1:241–248. doi: 10.1016/0896-6273(88)90145-6. [DOI] [PubMed] [Google Scholar]
- 30.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O’Donovan C, Redaschi N, Yeh LSL. The universal protein resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lassmann T, Sonnhammer ELL. Kalignsan accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lassmann T, Sonnhammer ELL. Kalign, Kalignvu and Mumsa: Web servers for multiple sequence alignment. Nucleic Acids Res. 2006;34:W596–W599. doi: 10.1093/nar/gkl191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohamadi F, Richard NGJ, Guida WC, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still WC. Macromodelssan integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J Comput Chem. 1990;11:440–467. [Google Scholar]
- 36.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 37.Morris GM, Goodsell DS, Huey R, Olson AJ. Distributed automated docking of flexible ligands to proteins: Parallel applications of AutoDock 2.4. J Comput-Aided Mol Des. 1996;10:293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 38.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 39.Biemann K. Nomenclature for peptide fragment ions (positive ions) Methods Enzymol. 1990;193:886–887. doi: 10.1016/0076-6879(90)93460-3. [DOI] [PubMed] [Google Scholar]
- 40.Kagabu S, Maienfisch P, Zhang A, Granda-Minones J, Haettenschwiler J, Kayser H, Maetzke T, Casida JE. 5-Azidoimidacloprid and an acyclic analogue as candidate photoaffinity probes for mammalian and insect nicotinic acetylcholine receptors. J Med Chem. 2000;43:5003–5009. doi: 10.1021/jm000240p. [DOI] [PubMed] [Google Scholar]
- 41.Zhang N, Tomizawa M, Casida JE. Structural features of azidopyridinyl neonicotinoid probes conferring high affinity and selectivity for mammalian α4β2 and Drosophila nicotinic receptors. J Med Chem. 2002;45:2832–2840. doi: 10.1021/jm010508s. [DOI] [PubMed] [Google Scholar]
- 42.Bouzat C, Gumilar F, Spitzmaul G, Wang HL, Rayes D, Hansen SB, Taylor P, Sine SM. Coupling of agonist binding to channel gating in an ACh-binding protein linked to an ion channel. Nature. 2004;430:896–900. doi: 10.1038/nature02753. [DOI] [PubMed] [Google Scholar]
- 43.Celie PHN, Klaassen RV, van Rossum-Fikkert SE, van Elk R, van Nierop P, Smit AB, Sixma TK. Crystal structure of acetylcholine-binding protein from Bulinus truncates reveals the conserved structural scaffold and sites of variation in nicotinic acetylcholine receptors. J Biol Chem. 2005;280:26457–26466. doi: 10.1074/jbc.M414476200. [DOI] [PubMed] [Google Scholar]
- 44.Bourne Y, Talley TT, Hansen SB, Taylor P, Marchot P. Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake α-neurotoxins and nicotinic receptors. EMBO J. 2005;24:1512–1522. doi: 10.1038/sj.emboj.7600620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 46.Gao F, Bren N, Burghardt TP, Hansen S, Henchman RH, Taylor P, McCammon JA, Sine SM. Agonist-mediated conformational changes in acetylcholine-binding protein revealed by simulation and intrinsic tryptophan fluorescence. J Biol Chem. 2005;280:8443–8451. doi: 10.1074/jbc.M412389200. [DOI] [PubMed] [Google Scholar]
- 47.Gao F, Mer G, Tonelli M, Hansen SB, Burghardt TP, Taylor P, Sine SM. Solution NMR of acetylcholine binding protein reveals agonist-mediated conformational change of the C-loop. Mol Pharmacol. 2006;70:1230–1235. doi: 10.1124/mol.106.027185. [DOI] [PubMed] [Google Scholar]
- 48.Shi J, Koeppe JR, Komives EA, Taylor P. Ligand-induced conformational changes in the acetylcholine-binding protein analyzed by hydrogen-deuterium exchange mass spectrometry. J Biol Chem. 2006;281:12170–12177. doi: 10.1074/jbc.M600154200. [DOI] [PubMed] [Google Scholar]
- 49.Grutter T, Bertrand S, Kotzyba-Hibert F, Bertrand D, Goeldner M. Structural reorganization of the acetylcholine binding site of the Torpedo nicotinic receptor as revealed by dynamic photoaffinity labeling. ChemBioChem. 2002;3:652–658. doi: 10.1002/1439-7633(20020703)3:7<652::AID-CBIC652>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 50.Mourot A, Rodrigo J, Kotzyba-Hibert F, Bertrand S, Bertrand D, Goeldner M. Probing the reorganization of the nicotinic acetylcholine receptor during desensitization by time-resolved covalent labeling using [3H]AC5, a photoactivatable agonist. Mol Pharmacol. 2006;69:452–461. doi: 10.1124/mol.105.017566. [DOI] [PubMed] [Google Scholar]
- 51.Rihakova L, Deraët M, Auger-Messier M, Pérodin J, Boucard AA, Guillemette G, Leduc R, Lavigne P, Escher E. Methionine proximity assay, a novel method for exploring peptide ligand-receptor interaction. J Recept Signal Transduction. 2002;22:297–313. doi: 10.1081/rrs-120014603. [DOI] [PubMed] [Google Scholar]
- 52.Wittelsberger A, Thomas BE, Mierke DF, Rosenblatt M. Methionine acts as a “magnet” in photoaffinity crosslinking experiments. FEBS Lett. 2006;580:1872–1876. doi: 10.1016/j.febslet.2006.02.050. [DOI] [PubMed] [Google Scholar]
- 53.Le Novère N, Grutter T, Changeux JP. Models of the extracellular domain of the nicotinic receptors and of agonist- and Ca2+-binding sites. Proc Natl Acad Sci USA. 2002;99:3210–3215. doi: 10.1073/pnas.042699699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schapira M, Abagyan R, Totrov M. Structural model of nicotinic acetylcholine receptor isotypes bound to acetylcholine and nicotine. BMC Struct Biol. 2002;2:1. doi: 10.1186/1472-6807-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang X, Zheng F, Crooks PA, Dwoskin LP, Zhan CG. Modeling multiple species of nicotine and deschloroepibatidine interacting with α4β2 nicotinic acetylcholine receptor: From microscopic binding to phenomenological binding affinity. J Am Chem Soc. 2005;127:14401–14414. doi: 10.1021/ja052681+. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


