Abstract
The genome of the budding yeast Saccharomyces cerevisiae is sequenced and the location and dynamic of activation of DNA replication origins are known. G1 synchronized yeast cells can be released into S-phase in the presence of hydroxyurea (HU) (1), which slows down DNA replication and retains replication forks in proximity of DNA replication origins. In this condition, the Chromatin Immuno-Precipitation on chip (ChIP on chip) (2–4) of replisome components allows the precise localization of all active DNA replication forks. This analysis can be coupled with the ssDNA-BromodeoxyUridine (ssDNA-BrdU) Immuno-Precipitation on chip (ssDNA-BrdU IP on chip) technique (5–7), which detects the location of newly synthesized DNA. Comparison of binding and BrdU incorporation profiles allows to locate a factor of interest at DNA replication forks genome wide. We present datasets deposited in the gene expression omnibus (GEO) database under accession number GSE68214, which show how the DNA helicases Rrm3 and Pif1 (8) associate to active and inactive DNA replication forks.
Keywords: Rrm3 and Pif1, DNA replication fork, Rad53 and hydroxyurea, DNA replication stress, ChIP on chip and ssDNA-BrdU IP on chip
| Specifications | |
|---|---|
| Organism/cell line/tissue | S. cerevisiae |
| Connected publication | Rossi et al. Cell Reports 2015 Oct 6;13(1):80–92 |
| Sequencer or array type | [Sc03b_MR] Affymetrix GeneChip S. cerevisiae Tiling 1.0R Array |
| Data format | CEL, bar |
| Experimental factors | DNA polymerase α, Rrm3 and Pif1.BrdU Incorporation. |
| Experimental features | ChIP on chip and ssDNA-BrdU IP on chip experiments. |
| Consent | Reuse and publication of datasets under GSE68214 accession number is under authorization of FIRC institute of molecular Oncology foundation (IFOM), Milano, Italy. |
| Sample source location | FIRC institute of molecular Oncology foundation (IFOM), Milano, Italy. |
1. Direct link to deposited data
2. Experimental design, materials and methods
Indicated yeast strains (see GSE68214 and [9]), were synchronized in G1 with 4 μg/ml of α-factor at 28 °C for 2 h in YP + 2% glucose and released in S-phase in the presence of HU (150 mM [1]) and, when required for the analysis of newly synthesized DNA, BrdU (200–500 μg/ml) [5], [6], [7]. ChIP on chip experiments in the GSE68214 series were conducted as described [2], [3], [4] with the following modifications: zirconium beads were used for cell breakage and amplified DNA with the whole genome amplification kit (WGA-SIGMA) has been purified using the Qiagen QIAquick PCR purification kit instead of the YM130 cartridges [3]. The amplification steps in the ChIP on chip protocol were conducted in non-saturating conditions and the amount of DNA used to hybridize the affymetrix chips was normalized to 4 μg within the different samples to preserve quantitative ratios [3]. Briefly, CEL files obtained by scanning of the hybridized affymetrix chips were analyzed using a modified version of the Tiling Array Suite software (TAS) from affymetrix. The software does a linear scale normalization of input CEL files (IP and Sup) intensity so that the median value is equal to a selected target intensity of 500. Signals and the p-value changes obtained from TAS per each probe position were subsequently used by the software to detect clusters of enriched signals as ranges within the chromosomes. Conditions for clusters detection in whole range (at least 600 bps), except for segments within the range shorter than 600 bps, were: log2 signal (IP/SUP binding ratio) positive and change in p-value (evaluated using Wilcoxon signed rank test) < 0.2 [2]. Specifically engineered yeast strains capable of incorporating BrdU in to the DNA [10], [11] have been used to conduct ssDNA-BrdU IP on chip experiments [5], [6], [7]. BrdU incorporation profiles have been generated as described for protein binding profiles. Genome wide binding and BrdU incorporation profiles can be superimposed and the statistical significance of binding and BrdU cluster overlappings can be calculated using a confrontation against a null hypothesis model generated with a Montecarlo-like simulation [2]. Average profiling of DNA binding and BrdU incorporation signals within specific genomic loci can be obtained using sitepro script of CEAS package (Cis-Regulatory Element Annotation System) [12]. Briefly, log2 signal (IP/SUP binding ratio) bed files obtained from protein binding and BrdU incorporation analysis were wig converted and used to draw average signals around 141 active DNA replication origins (Autonomously Replicating Sequences, ARSs), setting 50 bps as the profiling resolution and 20 kbps as the size of flanking regions from the center of each ARS. For the calculation of average binding or BrdU incorporation signals, negative values were set to zero. Total average binding or BrdU incorporation signals around 141 ARSs have been derived as average of the average of signals from the 50 bp bins created by the sitepro CEAS script where negative values were set to zero.
3. Results
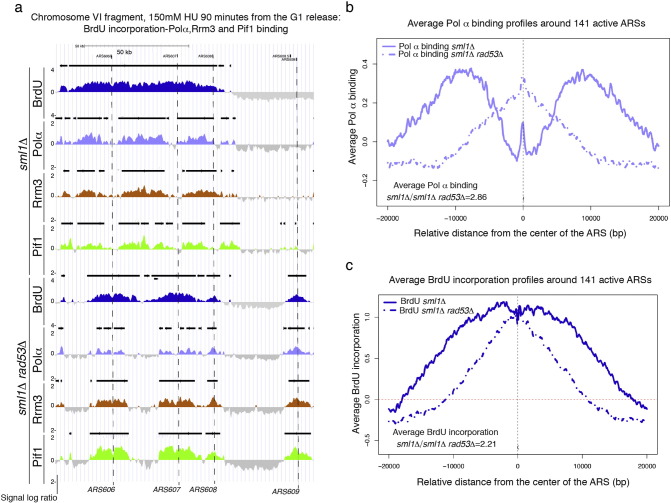
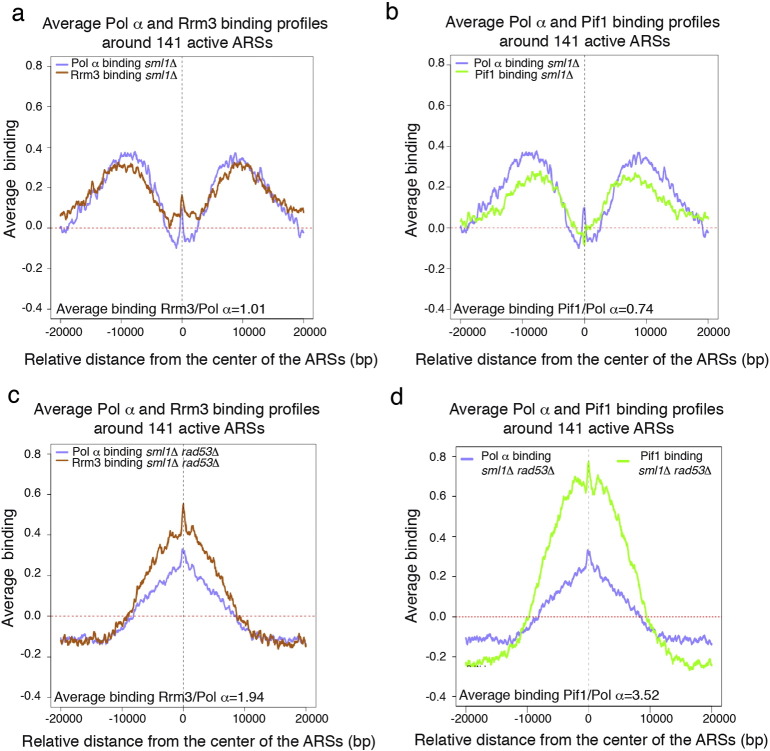
In Fig. 1a, DNA polymerase α (Pol α), Rrm3, Pif1 [8] binding and BrdU incorporation clusters were determined in sml1Δ (control strain) and sml1Δ rad53Δ cells released from G1 into S-phase in the presence of 150 mM of HU for 90 min. Pol α binding clusters overlapped with Rrm3 and Pif1 clusters in the two strains in a statistically significative way, suggesting either that Rrm3 and Pif1 are replisome-replication fork components in this experimental condition or that the absence of RAD53 does not influence the distribution of their binding sites to the forks (Fig. 1a). Interestingly, binding clusters of Pol α, Rrm3 and Pif1 localized to the borders of the BrdU clusters surrounding active DNA replication origins in sml1Δ cells as expected for proteins, which move with the DNA replication forks (Fig. 1a–c and 2a, b). rad53 cells fire late and dormant DNA replication origins in HU [13], [14]. According to their localization at replication forks, Pif1 and Rrm3 binding clusters co-localized with Pol α and BrdU clusters at the dormant origin ARS609, which is specifically fired in the absence of RAD53 in HU (Fig. 1a). RAD53 deletion inactivates DNA replication forks in the presence of HU leading either to dissociation of DNA polymerase α from the DNA template or to the accumulation of aberrant cruciform DNA structures at fork branching points [9], [15], [16], [17]. These fork-associated transitions strongly impair DNA replication fork progression in rad53 cells treated with HU. Consistently, BrdU, Pol α, Rrm3 and Pif1 clusters did not extend from the DNA replication origins in rad53 cells treated with HU while they were more extended in the sml1Δ control cells (Fig. 1a–c and 2a–d). Accordingly, the average binding signal of DNA polymerase α to a 40 kbps window centered on 141 active ARSs is 2.86 folds less in the absence of RAD53 (Fig. 1b) and BrdU incorporation around the same ARSs is reduced of 2.21 folds without the checkpoint kinase (Fig. 1c). Moreover while Pol α, Rrm3 and Pif1 binding clusters show overlapping bimodal distributions around 141 active ARSs in sml1Δ cells (Fig. 2a, b), consistent with forks moving away from the DNA replication origins, they remain close to the origin points in the absence of RAD53 (Fig. 2c, d). These evidences strongly support the previously proposed idea that DNA replication forks do not proceed and progressively collapse in rad53 cells treated with HU (Figs. 1a–c and 2a–d) [9], [15], [16], [17]. Intriguingly, while Rrm3 and Pif1 bind with the same magnitude of Pol α to 141 early ARSs in sml1Δ cells (Fig. 2a, b), the magnitude of their binding to the same ARSs is higher than Pol α binding in rad53 cells (Fig. 2c, d). These evidences suggest that additional substrates for Pif1 and Rrm3 may be created at collapsing forks of rad53 cells treated with HU leading to an increased recruitment of Rrm3 and Pif1 at inactivated forks.
Fig. 1.
) a) BrdU incorporation (dark blue) and Polα-Flag (light blue), Rrm3-13Myc (brown) and Pif1-Flag (green) binding profiles were determined, respectively, by ssDNA-BrdU IP on chip and ChIP on chip in the strains CY12488, CY13284, CY12470, CY13074, CY12493, CY13282, CY12422 and CY13073 released from G1 into 150 mM of HU for 90 min [9]. The y-axis shows the enrichment signals expressed as ratio log2 IP/SUP of loci significantly enriched in the IP fractions. The horizontal black bars above the picks indicate statistically significative BrdU or binding clusters. X-axis represents chromosomal coordinates. Early DNA replication origins (ARS606, ARS607) and the dormant origins (ARS608 and ARS609) are marked by dashed black lines. A black scale bar on the chromosome map indicates 50 kbps. b) Average binding profiles of DNA polymerase α in sml1Δ and sml1Δ rad53Δ cells (from the experiment shown in panel a), in a window of 40 kbps centered on each of the 141 active ARSs are shown. The ratio of average Pol α binding signals in sml1Δ versus sml1Δ rad53Δ cells is 2.86. c) Average BrdU incorporation profiles in sml1Δ and sml1Δ rad53Δ cells in a window of 40 kbps centered on 141 ARSs have been determined in the experiment shown in panel a. The ratio of BrdU incorporation signals in sml1Δ versus sml1Δ rad53Δ cells around 141 ARSs is 2.21.
Fig. 2.
) (a–d) Average binding profiles of the indicated proteins in sml1Δ (a, b) and sml1Δ rad53Δ cells (c, d) from the experiment shown in figure 1 panel a, in a window of 40 kbps centered on each of the 141 active ARSs are shown. Ratio of indicated total average binding signals is reported in each graph.
4. Conclusions
DNA binding profiles of replisome-DNA replication fork components by ChIP on chip and BrdU incorporation profiles by ssDNA-BrdU IP on chip allow the precise localization of all active DNA replication forks in the genome of Saccharomyces cerevisiae. Superimposition analysis of binding and BrdU incorporation profiles (or profiles of other genome features) can be used to locate a factor of interest at active DNA replication forks or to study the relationships between DNA replication and other genome wide regulated processes. Calculation of average binding or BrdU incorporation signals at specific genome loci in different genetic backgrounds allows uncovering roles in the regulation of specific chromosome processes. Ratio of total average binding signals provides a general indication of the relative strength of the binding of the considered factor to specific chromosome locations in different genetic backgrounds.
Acknowledgments
We thank Simone Minardi and the Genomics facility (Cogentech) at IFOM for hybridization of affymetrix chips and production of raw data files.
References
- 1.Krakoff I.H., Brown N.C., Reichard P. Inhibition of ribonucleoside diphosphate reductase by hydroxyurea. Cancer Res. 1968;28:1559–1565. [PubMed] [Google Scholar]
- 2.Bermejo R., Capra T., Gonzalez-Huici V., Fachinetti D., Cocito A., Natoli G., Katou Y., Mori H., Kurokawa K., Shirahige K. Genome-organizing factors Top2 and Hmo1 prevent chromosome fragility at sites of S phase transcription. Cell. 2009;138:870–884. doi: 10.1016/j.cell.2009.06.022. [DOI] [PubMed] [Google Scholar]
- 3.Bermejo R., Katou Y.M., Shirahige K., Foiani M. ChIP-on-chip analysis of DNA topoisomerases. Methods Mol. Biol. 2009;582:103–118. doi: 10.1007/978-1-60761-340-4_9. [DOI] [PubMed] [Google Scholar]
- 4.Katou Y., Kaneshiro K., Aburatani H., Shirahige K. Genomic approach for the understanding of dynamic aspect of chromosome behavior. Methods Enzymol. 2006;409:389–410. doi: 10.1016/S0076-6879(05)09023-3. [DOI] [PubMed] [Google Scholar]
- 5.Fachinetti D., Bermejo R., Cocito A., Minardi S., Katou Y., Kanoh Y., Shirahige K., Azvolinsky A., Zakian V.A., Foiani M. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol. Cell. 2010;39:595–605. doi: 10.1016/j.molcel.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katou Y., Kanoh Y., Bando M., Noguchi H., Tanaka H., Ashikari T., Sugimoto K., Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 7.Viggiani, C.J., Knott, S.R. and Aparicio, O.M. (2010) Genome-wide analysis of DNA synthesis by BrdU immunoprecipitation on tiling microarrays (BrdU-IP-chip) in Saccharomyces cerevisiae. Cold Spring Harb. Protoc., 2010, pdb prot5385. [DOI] [PubMed]
- 8.Bochman M.L., Sabouri N., Zakian V.A. Unwinding the functions of the Pif1 family helicases. DNA Repair. 2010;9:237–249. doi: 10.1016/j.dnarep.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossi S.E., Ajazi A., Carotenuto W., Foiani M., Giannattasio M. Rad53-mediated regulation of Rrm3 and Pif1 DNA helicases contributes to prevention of aberrant fork transitions under replication stress. Cell Rep. 2015;13:80–92. doi: 10.1016/j.celrep.2015.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viggiani C.J., Aparicio O.M. New vectors for simplified construction of BrdU-incorporating strains of Saccharomyces cerevisiae. Yeast. 2006;23:1045–1051. doi: 10.1002/yea.1406. [DOI] [PubMed] [Google Scholar]
- 11.Lengronne A., Pasero P., Bensimon A., Schwob E. Monitoring S phase progression globally and locally using BrdU incorporation in TK(+) yeast strains. Nucleic Acids Res. 2001;29:1433–1442. doi: 10.1093/nar/29.7.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin H., Liu T., Manrai A.K., Liu X.S. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 13.Santocanale C., Diffley J.F. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 14.Shirahige K., Hori Y., Shiraishi K., Yamashita M., Takahashi K., Obuse C., Tsurimoto T., Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 15.Lucca C., Vanoli F., Cotta-Ramusino C., Pellicioli A., Liberi G., Haber J., Foiani M. Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene. 2004;23:1206–1213. doi: 10.1038/sj.onc.1207199. [DOI] [PubMed] [Google Scholar]
- 16.Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C.S., Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 17.Sogo J.M., Lopes M., Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]