SUMMARY
Microorganisms can facilitate their survival in stressful environments by entering a state of metabolic inactivity or dormancy [1]. However, this state impairs the function of the very sensory systems necessary to detect favorable growth conditions. Thus, how can a metabolically quiescent cell accurately monitor environmental conditions in order to best decide when to exit dormancy? One strategy employed by microbes to deal with changing environments is the generation of phenotypes that may be less well adapted to a current condition but might confer an advantage in the future [2, 3]. This bet-hedging depends on phenotypic diversity in the population [4], which itself can derive from naturally occurring stochastic differences in gene expression [5, 6]. In the case of metabolic dormancy, a bet-hedging strategy that has been proposed is the “scout model” where cells comprising a fraction of the dormant population reinitiate growth stochastically, independent of environmental cues [7, 8]. Here, we provide experimental evidence that such a mechanism exists in dormant spores produced by the ubiquitous soil bacterium Bacillus subtilis. We observe that these spores reinitiate growth at a low but measureable frequency even in the absence of an inducing signal. This phenomenon is the result of phenotypic variation in the propensity of individual spores to reinitiate growth spontaneously. Since this bet-hedging mechanism produces individuals that will either grow under favorable conditions or die under unfavorable conditions, a population can properly respond to environmental changes despite the impaired sensory ability of individual cells.
RESULTS
Spontaneous Germination of B. subtilis
Many bacteria of the classes Bacilli and Clostridia produce metabolically quiescent, dormant spores that exhibit greatly reduced sensitivity to growth-inhibitory molecules like antibiotics or environmental stressors like heat [9]. Dormant B. subtilis spores respond to high (mM) levels of specific nutrients by efficiently reinitiating metabolic activity in a process termed induced germination [10] (Figure 1A, “induced”). This deterministic strategy is likely to be useful in nutrient-rich and predictable environments but may not be appropriate in fluctuating environments that are non-optimal but still permissive for growth. In contrast, germination that occurs in the absence of any inducing signal, “spontaneous germination,” would allow a spore to take advantage of conditions that permit growth but do not include specific inducing molecules that stimulate germination (Figure 1A, “spontaneous”).
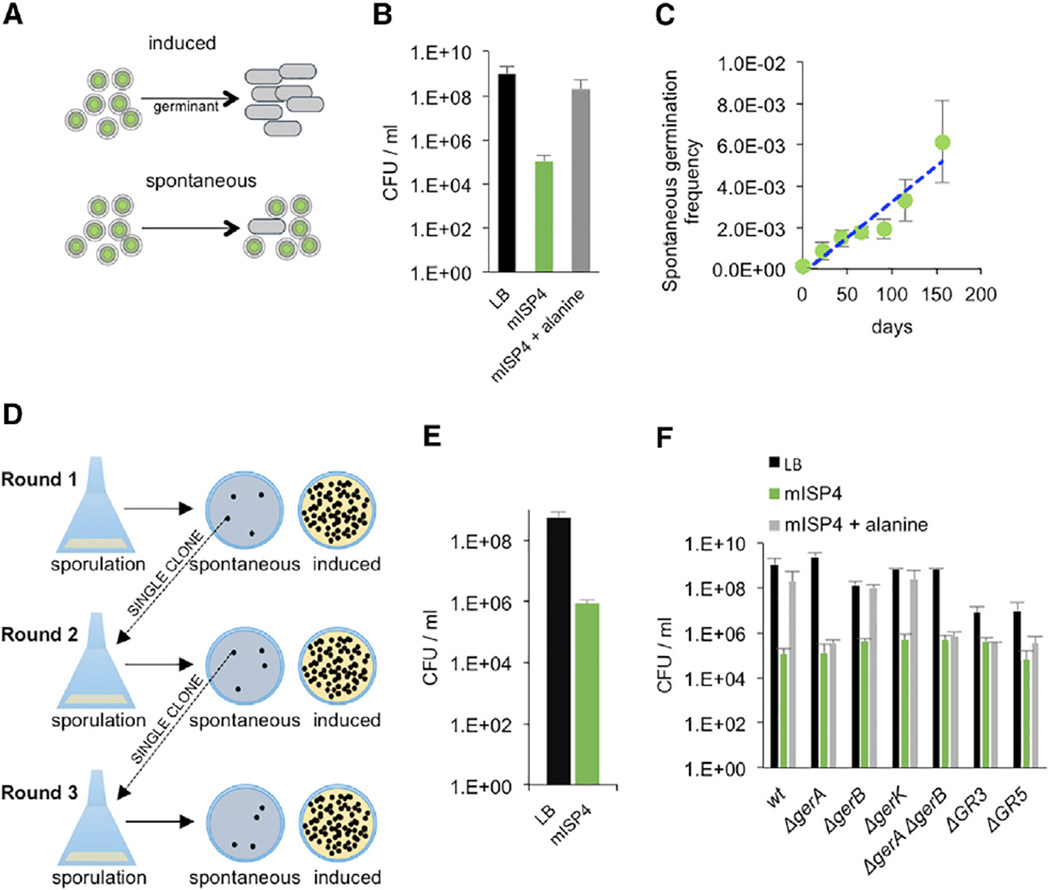
Figure 1. Spontaneous Germination of B. subtilis Spores.
(A) Spores germinate at high frequencies in response to a germinant such as alanine (“induced”) or at lower frequencies in the absence of any specific environmental signal (“spontaneous”).
(B) Spores of B. subtilis (JDB1772) were heat treated and then plated on LB (black), mISP4 (green), and mISP4 supplemented with 10 mM alanine (gray) to determine the fraction of germinating spores. CFU (mISP4) versus CFU (LB), p = 0.004; CFU (mISP4) versus CFU (mISP4ala), p = 0.008 (Mann-Whitney test). Shown is the mean, and error bars represent the SD.
(C) Measurement of the spontaneous germination rate. Wild-type spores (JDB1772, ~1.0 × 109) maintained in distilled water at 37°C were plated at the indicated time points on mISP4, and the number of spontaneously germinated spores was determined. The regression line is: y = 3 × 10−5x; R2 = 0.8963. Shown is the mean, and error bars represent the SD.
(D) Schema of experiment to demonstrate that colony formation on mISP4 is not due to mutation. B. subtilis spores are plated on mISP4 and a single colony from that plate is sporulated by exhaustion in DSM, and the resulting spores are plated on both mISP4 and LB to determine the frequency of spontaneous germination. This process is repeated several times.
(E) Single wild-type B. subtilis colonies (JDB1772) were cycled through three rounds of sporulation and successive spontaneous germination. The descendants after round 3 were plated on LB (black bar) and mISP4 (green bar) to determine the final spontaneous germination rate.
(F) Germination on mISP4 with heat-treated spores of wild-type strain (JDB1772) and strains lacking a subset or all of the five germination receptor mutants (“ΔgerA,” JDB2919; “ΔgerB,” JDB3118; “ΔgerK,” JDB3615; “ΔgerA ΔgerB,” JDB1366; “ΔGR3,” JDB3614; “ΔGR5,” JDB1914) were plated on LB (black), mISP4 (green), or mISP4 + 10 mM alanine (gray). Shown is the mean, and error bars represent the SD.
Spores efficiently germinate in Luria-Bertani (LB), a nutrient-rich medium (Figure 1B, black), since the colony-forming units (CFUs) resulting from plating spores on LB are similar to the CFUs resulting from plating the culture prior to initiation of sporulation (data not shown). To assess spontaneous germination, we used a minimal medium (mISP4) that uses starch as it serves as a carbon source but does not induce germination. Addition of 10 mM alanine to mISP4 stimulates germination to levels similar to that observed in LB, indicating that mISP4 can support efficient germination in the presence of known germinants (Figure 1B, gray). However, even in the absence of added alanine, germination on mISP4 was observed (Figure 1B, green bar). The difference (~1 × 104) between the frequency of germination on mISP4 and on mISP4 with added alanine indicates that a fraction of spores can germinate even in the absence of a known inducer. A quantitatively similar response was observed in different Bacilli, indicating that it is conserved (Figure S1, green bars). The magnitude of this effect is consistent with previous measurements of non-nutrient germination [11].
We determined the rate of spontaneous germination by sampling a population of ~108 spores at regular intervals over 150 days and plating them on mISP4. Under these conditions, ~104 spores germinated per day over the duration of the experiment (Figure 1C), consistent with the frequency obtained previously (Figure 1B). The approximate constancy of this rate suggests that spontaneous germination is not limited to a single sub-population and that every spore is eventually capable of germinating in the absence of a specific inducer.
Spontaneous Germination Is a Result of Phenotypic Variation
Spontaneous germination could be due to either genetic variation (i.e., a mutation) or phenotypic variation. If an acquired mutation is responsible, then spores of bacteria originating from spontaneously germinating spores should have significantly higher levels of spontaneous germination. To test this possibility, we produced spores from cultures derived from single colonies of spontaneously germinated spores and measured their germination on mISP4 and on LB to confirm total spore number (Figure 1D). We measured the frequency on mISP4 to be ~4 × 105, approximately 1,000-fold less than on LB (Figure 1B). We then repeated this procedure twice, and similar frequencies of spontaneous germination were observed in all three rounds, consistent with the absence of an acquired mutation (round 2: ~8 × 105; round 3: ~7 × 105) (Figure 1E). The constancy of these frequencies over sequential passages indicates that spontaneous germination most likely depends on phenotypic variation and not genotypic variation.
Induced germination in response to alanine requires a germination receptor complex encoded by the gerA operon [10]. Variability in the levels of one protein component of this complex, GerAA, has been proposed to affect the differential sensitivity of spores to germination by alanine [12]. To determine whether this variability was responsible for spontaneous germination, we examined spores derived from a strain (ΔgerA) that lacks GerAA. Since this mutant strain had the same rate of germination as a wild-type strain on mISP4, the absence of GerA did not affect the rate of spontaneous germination (Figure 1F, “ΔgerA”). B. subtilis contains four gerA orthologs that in some cases individually mediate germination in response to amino acids other than alanine [10]. However, strains that lack one or more of these germination receptors (including the strain ΔGR5 that lacks all five identified nutrient germination receptors) germinated on mISP4 indistinguishably from the wild-type (Figure 1F, green), indicating that spontaneous germination is independent of any specific nutrient germination receptor.
Spontaneous Germination Is a Result of Variation in the Levels of a Transcription Factor
Another possible source of the phenotypic variation is the spore coat, the proteinaceous multilayer structure that surrounds the spore and provides much of its resistance to environmental stressors [13]. During sporulation, an asymmetric division produces a mother cell and a smaller cell that after engulfment will become the metabolically inactive spore (Figure 2A) [14]. Assembly of the spore coat involves a network of transcription factors operating in the mother cell that sequentially regulate the transcription of genes encoding the ~80 proteins that comprise the coat [13]. Variations in the behavior of this network could lead to variations in the structure of the coat and thereby affect spore germination. An important component of the regulatory network underlying spore coat assembly is the GerE transcription factor that, in association with the mother-cell-specific σK factor, affects the expression of >50 genes, many of which encode coat proteins [15] (Figure 2A). While GerE is not strictly essential for spore formation, spores lacking gerE exhibit changes to their surfaces [16]. Thus, variation in GerE levels may have phenotypic consequences for the spore.
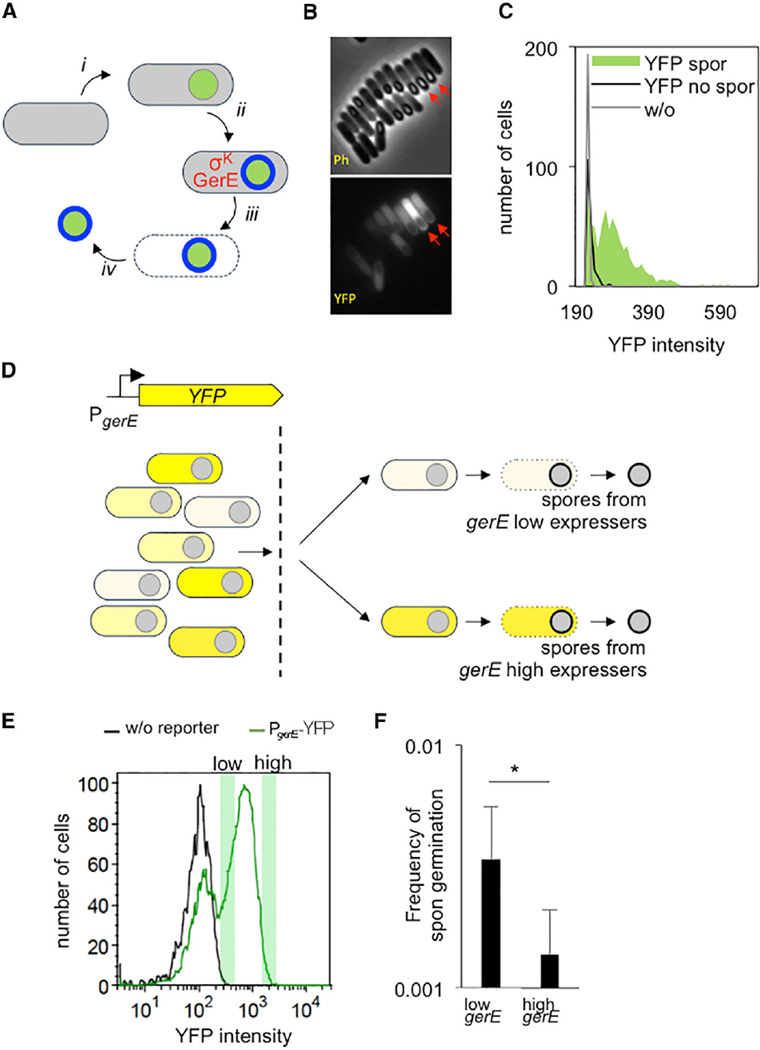
Figure 2. Phenotypic Variation in Spontaneous Germination.
(A) In sporulating cells, the forespore (green) becomes engulfed within the mother cell (gray) (i); the transcription factors σK and GerE act in the mother cell and together control the expression of genes encoding proteins of the spore coat (blue) (ii); lysis of the mother cell (iii) results in release of the mature spore into the environment (iv).
(B) A PgerE-YFP strain was sporulated by re-suspension, and fluorescence was monitored at 5 hr following initiation of sporulation (Ph, phase; YFP, fluorescence). Note the variation in the total cellular fluorescence of individual cells that appeared to be at a similar stage in spore formation (red arrows).
(C) Quantification of (B). The YFP intensity of ~725 sporulating cells (“YFP spor,” green area), ~175 non-sporulating cells (“YFP no spor,” black line), and~200 cells lacking the reporter (“w/o,” gray line) was measured.
(D) Sporulating PgerE-YFP cells are separated into populations expressing either high levels or low levels of YFP. These populations are then allowed to complete sporulation separately.
(E) Cells analyzed by fluorescence-activated cell sorting (FACS) at hour 5 following initiation of sporulation by re-suspension exhibited a bimodal fluorescence distribution. Fractions taken from the right green peak (green shaded boxes) associated with “high YFP” (intensity between 1 × 103 and 2 × 103 a.u.) and “low YFP” (intensity between 275 and 450 a.u.) were identified and separated. The left green peak represents non-PgerE-YFP-expressing cells since they overlap with the fluorescence of cells of a strain that lacked the PgerE-YFP reporter (black) and are predominantly non-sporulating cells as visualized by microscopy.
(F) Spores that originated from distinct fractions deriving from either high or low gerE-expressing cells that were identified in (E) were plated on mISP4 and LB. The ratio of CFUs (“Frequency of spontaneous germination”) was determined; asterisk (*) denotes that p = 0.03 (Mann Whitney test). Shown is the mean, and error bars represent the SD.
Using a PgerE-YFP reporter, we observed that 5 hr after the initiation of sporulation, the YFP fluorescence of individual sporulating cells varied (Figure 2B, “YFP”; Figure 2C, green). The cells appeared to be relatively synchronized in sporulation, as assayed by microscopy (Figure 2B, “Ph”), suggesting that a lack of synchrony was likely not sufficient to cause this variation. To demonstrate that these differences in GerE levels in individual cells resulted in phenotypic variation, we separated isogenic sporulating cells containing PgerE-YFP into high and low YFP-expressing fractions by flow cytometry (Figure 2D). We took advantage of the observation that sporulating cells become committed to the completion of sporulation after ~2 hr, even if the environment changes back to conditions that do not support sporulation [17]. Following sorting of cells at hour 5 of sporulation, the fractions associated with the top ~5% and the bottom ~5% of the YFP signal (Figure 2E) were allowed to complete sporulation separately for ~24 hr. The resulting spores were plated on mISP4, and the frequency of spontaneous germination was determined. A significant difference between the fractions was observed, with the cells expressing higher levels of gerE having a lower rate of spontaneous germination (Figure 2F). This is also supported by higher rates of spontaneous germination when the scaffold protein CotE in the spore coat was deleted (Figure S2). Thus, GerE likely directly represses spontaneous germination, and GerE-mediated variations in the makeup of the spore coat may be at least partially responsible for the frequency of spontaneous germination.
Recombination of σK during Sporulation Affects the Rate of Spontaneous Germination
We next addressed whether this variation in GerE levels is subject to modulation. The gerE gene is transcribed by a sporulation-specific σK-containing RNAP [18]. σK is subject to complex regulation involving transcription of its gene by an earlier cell-specific transcription factor and the subsequent proteolytic processing of the encoded protein [19]. In addition, σK is the product of a disrupted gene. The two open reading frames (ORFs) encoding σK, spoIVCB and spoIIIC, are separated by an intervening sequence of ~48 kb, called the skin element (Figure 3A). The recombinase SpoIVCA excises the skin element in the mother cell and joins spoIVCB and spoIIIC, allowing transcription of a gene encoding full-length σK [20]. We examined whether this recombination affects gerE expression using a mutant strain called skinless where spoIVCB and spoIIIC are already joined and therefore the genomic rearrangement is not required [20]. GerE levels in the population were lower in the skinless mutant (Figure 3B), indicating that gerE expression is affected by the recombination event. The removal of the skin element also affected the expression of earlier genes in sporulation, suggesting a global perturbation of the regulatory system during sporulation (Figure S3).
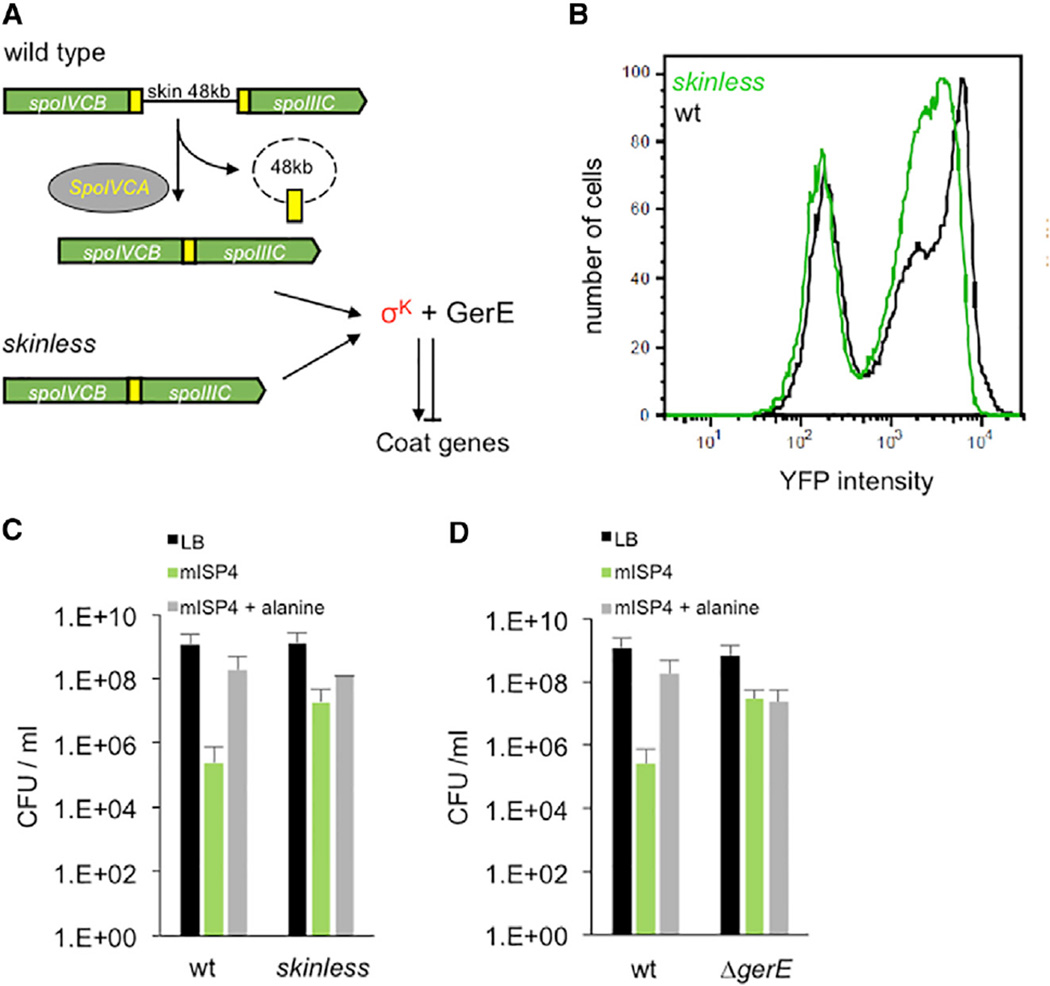
Figure 3. Regulation of Spontaneous Germination by Chromosomal Rearrangement.
(A) In wild-type cells, spoIIIC and spoIVCB are normally separated by the ~48 kb skin element that is excised by the SpoIVCA recombinase. This allows the transcription of a spoIIIC-spoIVCB gene fusion and synthesis of σK, which in association with GerE, regulates the expression of spore coat genes. In skinless cells, spoIVCB and spoIIIC are already fused, allowing synthesis of σK without chromosomal rearrangement.
(B) Fluorescence distributions of cells from either wild-type (black) or skinless (green) strains expressing PgerE-YFP at 5 hr after initiation of sporulation in re-suspension media. The right peak (fluorescence > 5 × 102) of the wild-type strain (black) has a mean intensity of 8 × 103, and the skinless strain (green) has a mean intensity of 3 × 103. The left green and black peaks (fluorescence < 5 × 102) likely represent non-PgerE-YFP-expressing cells since they are predominantly non-sporulating as determined by microscopy.
(C) Germination of wild-type or skinless spores on LB (black), mISP4 (green) or mISP4 + 10mM alanine (gray). For mISP4, CFU wild-type versus CFU skinless; p = 0.028 (unpaired t test, two-tailed).
(D) Germination of wild-type and ΔgerE (JDB3577) spores on LB (black), mISP4 (green), or ISP4+10 mM alanine (gray). For mISP4, CFU wild-type versus CFU ΔgerE; p = 0.005, unpaired t test, two tailed. Shown is the mean, and error bars represent the SD.
The inverse relationship between GerE levels and the frequency of spontaneous germination (Figure 2F) suggests that skinless spores should have a higher frequency of spontaneous germination. Consistent with this hypothesis, we observed a ~100-fold higher frequency of spontaneous germination of a skinless strain (Figure 3C). An implication of this observation is that variation in the timing of the skin recombination event may be an important factor in setting the frequency of spontaneous germination in wild-type spores. Finally, this inverse relationship also suggests that spores derived from a ΔgerE strain, which has the lowest levels of GerE, should have higher levels of spontaneous germination. We confirmed this prediction by observing that ΔgerE spores spontaneously germinate at a significantly higher frequency than the wild-type and, in fact, at a frequency similar to that observed with wild-type spores in response to alanine (Figure 3D). Thus, GerE plays a key role in establishing the phenotypic diversity underlying spontaneous germination. Interestingly, a ΔgerE strain is significantly impaired in nutrient germination [16], in contrast with spontaneous germination, thereby providing further evidence that the two processes are independent.
Physiological Consequences of Spontaneous Germination
We used two assays to examine the consequences of the increased frequency of spontaneous germination of the skinless mutant. First, wild-type and skinless spores had no obvious differences in their appearance by light microscopy, with a preponderance of phase-bright spores (Figure 4A, i and ii; Figure 4B). When the spores were incubated for 6 days in mISP4 with xylose as the carbon source, since starch interfered with the imaging, the skinless spores had largely formed rod-shaped cells, indicating that they had germinated and re-initiated growth. In contrast, wild-type spores remained phase bright (Figure 4A, iii and iv; Figure 4B). Importantly, these spores germinated and re-initiated growth when alanine was present, indicating that they were still competent to germinate (Figure 4A, v; Figure 4B). Thus, even though wild-type spores fail to efficiently germinate under this particular growth-supportive condition, their ability to spontaneously germinate allows them to take advantage of growth-promoting conditions. That is, a population of spores can “respond” even if the individual cells are unable to detect the presence of such a condition.
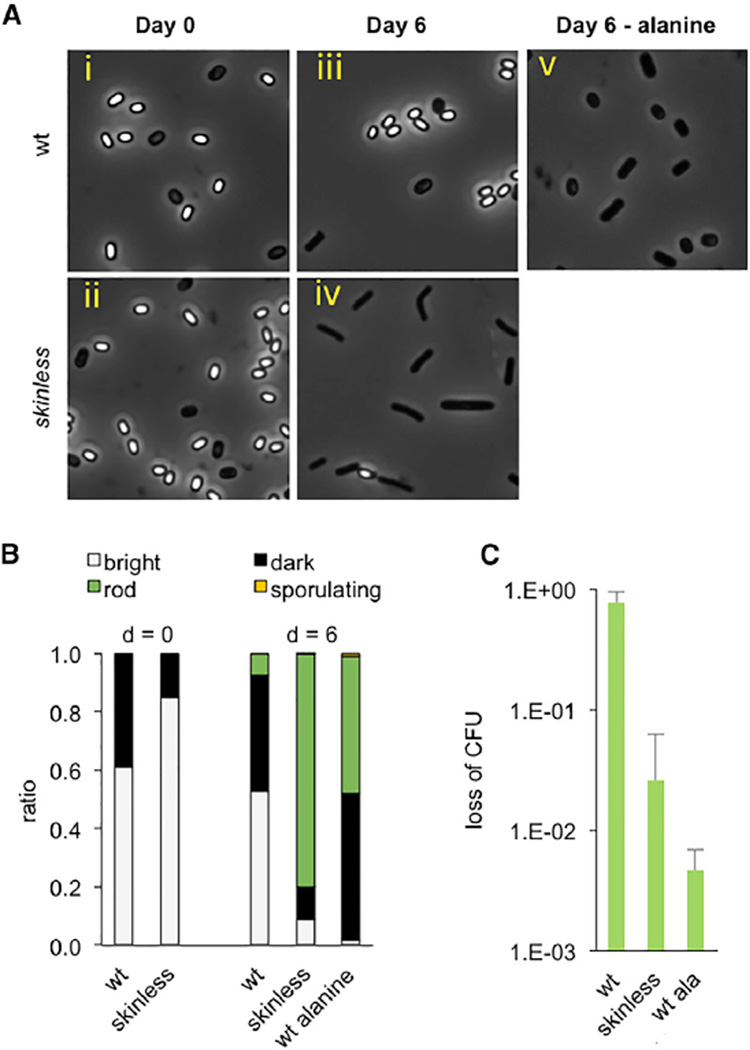
Figure 4. Consequences of Spontaneous Germination in Favorable and Unfavorable Environments.
(A) Spontaneous germination of wild-type and skinless spores in liquid mISP4. Spores were generated by exhaustion, re-suspended in mISP4 with xylose, and incubated at room temperature. Samples taken at day 0 and day 6 following initiation of incubation were analyzed by light microscopy.
(B) Quantification of (A) with fraction of phase-bright spores (white), phase-dark (black, considered to be less dormant than phase-bright spores) cells, vegetative cells, rods (green), and sporulating cells (yellow), as determined by observation using phase-contrast microscopy.
(C) Killing of skinless spores. Following incubation in mISP4 + xylose for 6 days, wild-type and skinless spores were subjected simultaneously to chloramphenicol (10 µg/ml), ampicillin (100 µg/ml), and kanamycin (50 µg/ml) for 120 min and plated on mISP4 to determine CFUs. As a control, wild-type spores were incubated with 10 mM alanine for 60 min before exposure to antibiotics. The ratio of CFUs before and after treatment with antibiotics “loss of CFU” is reported. For wild-type versus skinless, p = 0.03; for wild-type versus wild-type + alanine, p = 0.03 (unpaired t test, two tailed). Shown is the mean, and error bars represent the SD.
In a second assay, we subjected wild-type and skinless spores to treatment with antibiotics following incubation in liquid mISP4 for 6 days. Whereas wild-type spores remained viable, as measured by CFUs, the skinless spores exhibited a sharp drop in viability (Figure 4C). This loss in viability is also seen when wild-type spores are exposed to alanine, consistent with the known sensitivity of germinated spores to antibiotics. Thus, a high frequency of spontaneous germination can render a population sensitive to bactericidal compounds.
DISCUSSION
B. subtilis spores have been a model system for studying microbial dormancy since their initial characterization in 1876 [21]. Germination of these spores occurs in response to high levels of certain amino acids via stimulation of germination receptors [9]. Here, we demonstrate that they also germinate at a reduced frequency (~1 in 104) in the absence of any known inducing signal. This mechanism produces individuals at a low rate that will either grow under favorable conditions or die under unfavorable conditions. Thus, spontaneous germination allows the population to respond to environmental changes, thereby overcoming the impaired ability of individual dormant spores to accurately sense environmental signals. The advantages of such a mechanism are apparent given that other bacteria with growth-limited phenotypes such as E. coli persisters [22] and Mycobacterium tuberculosis [23] also appear to exhibit stochastic exit from quiescence.
Spontaneous germination, at least in part, results from variation in the levels of the transcription factor GerE, which is responsible for regulating the expression of many of the proteins that comprise the coat of the spore (Figures 2F and 3D). Cell-to-cell variation or noise in gene expression can lead to alternate phenotypic states within an isogenic population [24] and thereby facilitate bet-hedging, a strategy particularly useful under conditions of environmental variability [25]. This cell-to-cell variation can be subject to genetic control [26]; for example, reducing the noise in the expression of the transcriptional regulator of genetic competence in B. subtilis affects the magnitude of phenotypic competence [27]. Here, we demonstrate that, similarly, spontaneous germination is under genetic control since a mutant strain that lacks proper regulation of gerE expression and exhibits changes in the levels of GerE (Figure 3B) has a greatly increased frequency of spontaneous germination (Figure 3C).
A central issue in microbial physiology is understanding how bacteria detect the many potential metabolic substrates present in the environment [28]. This question is particularly relevant for dormant bacteria since they need to reliably assess whether the environment supports growth before exiting dormancy [29]. Here, we have shown that the minimal medium mISP4 does not stimulate spore germination, despite the fact that it supports growth. However, the ability of spores to spontaneously germinate on mISP4 allows the spores to take advantage of these growth-promoting conditions. Thus, the population can assess whether a given milieu supports growth without requiring detection of any particular substrate.
Microbes can exist in metabolically quiescent states that facilitate their survival in stressful environments. However, quiescence is likely to impair proper functioning of their sensory apparatus. For example, the energetic cost of operating the chemotaxis sensing machinery is comparable to that required for producing its components [30], suggesting that the metabolic requirements of sensory processing are significant. Thus, spontaneous exit from dormancy may be advantageous to a variety of organisms because it does not require any sensory input. The relatively low frequency of spontaneous germination in the population may reflect both positive and negative evolutionary pressures that produce a bet-hedging strategy similar to that observed in other decision-making scenarios [31]. Specifically, a relatively high frequency may enhance the population’s ability to take advantage of favorable growth conditions, but it also renders the population sensitive to growth-inhibiting conditions. The relatively low rate of spontaneous germination that we measure under our experimental conditions (Figure 1C; ~104/day) would allow a population of 109 spores (as can be found in 1 g soil [32]) to last for >100 years before exhaustion. A population of dormant cells could therefore regularly assess the environment for long periods of time, thereby promoting survival of the population during extended periods of austerity and contributing to microbial diversity [33].
EXPERIMENTAL PROCEDURES
Detailed experimental procedures are described in Supplemental Information.
Supplementary Material
Highlights.
A sub-population of Bacillus subtilis bacterial spores germinate spontaneously
Phenotypic variability underlies spontaneous germination
The phenotypic variability of spontaneous germination is under genetic control
Acknowledgments
We thank Peter Setlow, Michael Delay, and members of our lab for helpful discussions. We acknowledge D. Higgins for first identifying ISP4 as a non-germination- inducing B. subtilis growth medium and M. Elowitz for providing the PgerE-YFP. This work was supported by an EMBO fellowship to A.S. J.D. is a Burroughs-Welcome Investigator in the Pathogenesis of Infectious Disease and an Ellison Medical Foundation Senior Scholar in Aging.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, three figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2015.07.018.
AUTHOR CONTRIBUTIONS
A.S. conducted the experiments. A.S. and J.D. designed the experiments and wrote the paper.
REFERENCES
- 1.Rittershaus ES, Baek SH, Sassetti CM. The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe. 2013;13:643–651. doi: 10.1016/j.chom.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 3.Acar M, Mettetal JT, van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nat. Genet. 2008;40:471–475. doi: 10.1038/ng.110. [DOI] [PubMed] [Google Scholar]
- 4.Lidstrom ME, Konopka MC. The role of physiological heterogeneity in microbial population behavior. Nat. Chem. Biol. 2010;6:705–712. doi: 10.1038/nchembio.436. [DOI] [PubMed] [Google Scholar]
- 5.Choi PJ, Cai L, Frieda K, Xie XS. A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science. 2008;322:442–446. doi: 10.1126/science.1161427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon AJ, Halliday JA, Blankschien MD, Burns PA, Yatagai F, Herman C. Transcriptional infidelity promotes heritable phenotypic change in a bistable gene network. PLoS Biol. 2009;7:e44. doi: 10.1371/journal.pbio.1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epstein SS. Microbial awakenings. Nature. 2009;457:1083. doi: 10.1038/4571083a. [DOI] [PubMed] [Google Scholar]
- 8.Buerger S, Spoering A, Gavrish E, Leslin C, Ling L, Epstein SS. Microbial scout hypothesis, stochastic exit from dormancy, and the nature of slow growers. Appl. Environ. Microbiol. 2012;78:3221–3228. doi: 10.1128/AEM.07307-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paredes-Sabja D, Setlow P, Sarker MR. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 2011;19:85–94. doi: 10.1016/j.tim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Setlow P. Germination of spores of Bacillus species: what we know and do not know. J. Bacteriol. 2014;196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paidhungat M, Setlow P. Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 2000;182:2513–2519. doi: 10.1128/jb.182.9.2513-2519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart KA, Setlow P. Numbers of individual nutrient germinant receptors and other germination proteins in spores of Bacillus subtilis. J. Bacteriol. 2013;195:3575–3582. doi: 10.1128/JB.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenney PT, Driks A, Eichenberger P. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 2013;11:33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 2012;36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS, Losick R. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2004;2:e328. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh S, Setlow B, Wahome PG, Cowan AE, Plomp M, Malkin AJ, Setlow P. Characterization of spores of Bacillus subtilis that lack most coat layers. J. Bacteriol. 2008;190:6741–6748. doi: 10.1128/JB.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dworkin J, Losick R. Developmental commitment in a bacterium. Cell. 2005;121:401–409. doi: 10.1016/j.cell.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 18.Cutting S, Mandelstam J. The nucleotide sequence and the transcription during sporulation of the gerE gene of Bacillus subtilis. J. Gen. Microbiol. 1986;132:3013–3024. doi: 10.1099/00221287-132-11-3013. [DOI] [PubMed] [Google Scholar]
- 19.Konovalova A, Søgaard-Andersen L, Kroos L. Regulated proteolysis in bacterial development. FEMS Microbiol. Rev. 2014;38:493–522. doi: 10.1111/1574-6976.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkel B, Losick R, Stragier P. The Bacillus subtilis gene for the development transcription factor sigma K is generated by excision of a dispensable DNA element containing a sporulation recombinase gene. Genes Dev. 1990;4:525–535. doi: 10.1101/gad.4.4.525. [DOI] [PubMed] [Google Scholar]
- 21.Cohn F. Untersuchungen ueber Bakterien. IV. Beitraege zur Biologie der Bacillen. Beitraege zur Biologie der Planzen. 1876;2:249–276. [Google Scholar]
- 22.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 23.Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, McKinney JD. Dynamic persistence of antibiotic- stressed mycobacteria. Science. 2013;339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 24.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaumont HJ, Gallie J, Kost C, Ferguson GC, Rainey PB. Experimental evolution of bet hedging. Nature. 2009;462:90–93. doi: 10.1038/nature08504. [DOI] [PubMed] [Google Scholar]
- 26.Casadesús J, Low DA. Programmed heterogeneity: epigenetic mechanisms in bacteria. J. Biol. Chem. 2013;288:13929–13935. doi: 10.1074/jbc.R113.472274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torsvik V, Øvreås L. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 2002;5:240–245. doi: 10.1016/s1369-5274(02)00324-7. [DOI] [PubMed] [Google Scholar]
- 29.Dworkin J, Shah IM. Exit from dormancy in microbial organisms. Nat. Rev. Microbiol. 2010;8:890–896. doi: 10.1038/nrmicro2453. [DOI] [PubMed] [Google Scholar]
- 30.Govern CC, Ten Wolde PR. Optimal resource allocation in cellular sensing systems. Proc. Natl. Acad. Sci. USA. 2014;111:17486–17491. doi: 10.1073/pnas.1411524111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 2008;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 32.Brandes Ammann A, Kölle L, Brandl H. Detection of bacterial endospores in soil by terbium fluorescence. Int. J. Microbiol. 2011;2011:435281. doi: 10.1155/2011/435281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones SE, Lennon JT. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. USA. 2010;107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.