Abstract
Identifying and annotating distal regulatory enhancers is critical to understand the mechanisms that control gene expression and cell-type-specific activities. Next-generation sequencing techniques have provided us an exciting toolkit of genome-wide assays that can be used to predict and annotate enhancers. However, each assay comes with its own specific set of analytical needs if enhancer prediction is to be optimal. Furthermore, integration of multiple genome-wide assays allows for different genomic features to be combined, and can improve predictive performance. Herein, we review the genome-wide assays and analysis schemes that are used to predict and annotate enhancers. In particular, we focus on three key computational topics: predicting enhancer locations, determining the cell-type-specific activity of enhancers, and linking enhancers to their target genes.
Keywords: Enhancer prediction, Epigenomics, Enhancer–gene linking, Enhancer activity, Epigenetics
1. Introduction
During animal development, a single cell divides many times to give rise to a great variety of cell-types and tissues. Each of an individual’s cell-types has its own specific set of characteristics, yet they are all constructed from the same “blueprints,” as they possess the same genome sequence. The genome defines all the genes that are expressed in an individual; however, only a specific subset of genes is expressed in any given cell-type. Thus, cell-type-specific gene expression must be tightly controlled throughout development. Furthermore, incorrect patterns of gene expression can result in diseases, such as cancers.
The expression of genes is controlled by RNA polymerase II (RNA Pol II), which transcribes DNA into RNA. The initiation of transcription occurs at the transcription start site (TSS). Adjacent to the TSS is the gene promoter, which contains cis-regulatory elements that are bound by transcription factors (TFs) and regulate gene expression. Enhancers, which are distal regulatory sites bound by TFs, interact with promoters through DNA looping and further tune gene expression [1,2]. The looping increases the local concentration of TFs that recruits RNA Pol II to initiate the transcriptional process [3,4]. While gene-coding regions and promoters have been well annotated, identification of enhancers remains a great challenge, as they can be located hundreds of kilobases (kb) to millions of bases from their interacting genes and function independently of their location and/or orientation relative to the TSS [4,5].
Initial genome-wide enhancer identification strategies relied on properties of the DNA sequence, such as clusters of TF binding sites [6,7] and highly conserved genomic regions [8–11]. However, these approaches may not be accurate enough and lack information about the cell-type specificity of the identified enhancers. More recently, next-generation sequencing technologies have given rise to numerous genome-wide assays that allow the cell-type-specific measurement of genomic properties. These approaches have started to be applied en masse, especially by projects like ENCODE [12] and the Roadmap Epigenomics Project [13]. Herein, we review the use of genome-wide assays, particularly computational strategies to identify enhancers on a genome-wide level and to link enhancers to their target genes.
2. Identifying the location of enhancers
In this section we discuss approaches that use epigenomic data to predict the genomic positions of enhancers. Important assays and predictive features they provide are discussed and brought into context, such as enhancer sequence patterns, ChIP-seq, chromatin signatures, and DNA methylation.
The genome-wide mapping and annotation of enhancers is a critical step towards a comprehensive understanding of the underlying principles of mammalian gene regulation. An initial genome-wide approach to enhancer discovery relies on non-coding regions of the genome that are conserved across multiple species. The assumption is that functional regions evolve under constraints and thus at a lower rate than non-functional regions. This approach has been used extensively in the past decade, when genome sequences of multiple species became available for comparison. While most commonly used to predict functional TF binding sites, several studies used this approach to identify enhancers [7–11]. However, recent studies showed that enhancers may not be conserved across species and that conservation alone is insufficient to identify cell-type-specific activity of enhancers [14,15]. Deletion of conserved regions of the mouse genome resulted in viable mice showing that these regions are not always crucial [16]. Moreover, conserved regions of the genome may have non-enhancer functions, such as attaching to the cellular matrix [17]. Consequently, additional tissue-specific information is needed for more accurate enhancer prediction and annotation (Fig. 1).
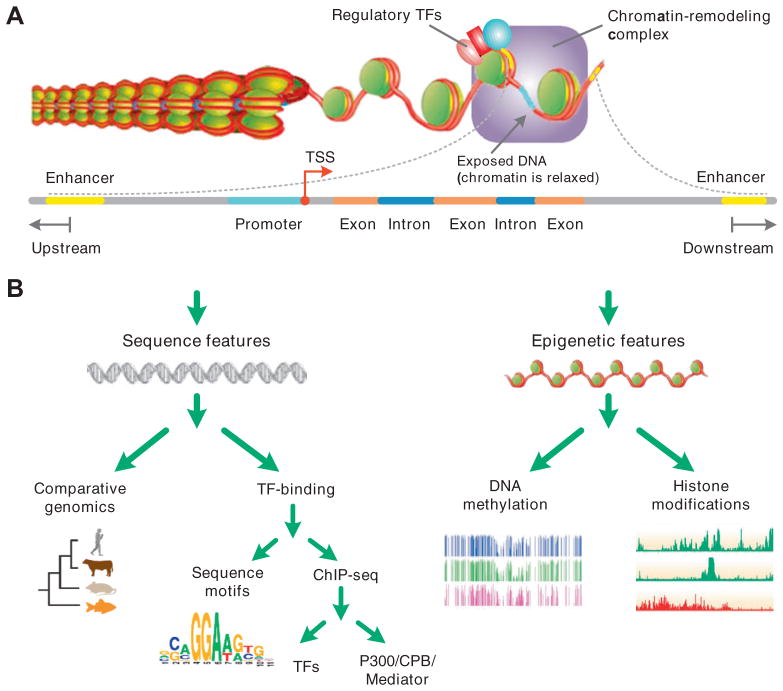
Fig. 1.
Transcriptional controls and enhancers features in use for computational prediction. (A) Schematic representation of the transcriptional activity. Regulatory TFs recruit chromatin-remodeling complex (coactivators) and histone acetyltransferases (HATs). After decondensation of chromatin, regulatory TFs recruit basal transcription complex and RNA Pol II to form the initial complex and begin the transcription. (B) A classification of features used in computational models for enhancer prediction. Sequence features are those mainly relevant to TF binding regions while epigenetic features are relevant to modifications of chromatin structure.
Since enhancer activity is dependent on sequence-specific DNA-binding of TFs, which in turn recruits coactivators to initiate gene transcription, particular sequence signatures associated with TF binding sites can be exploited to predict enhancer locations. Sequence features, typically corresponding to cis-regulatory elements, can be detected using known-TF motifs or de novo motif discovery methods. Several recent studies have demonstrated the usefulness of predicting enhancers from combinations of cell-type-specific sequence motifs [18–20]. Sets of cell-type-specific enhancers and/or promoters might be regulated through common mechanisms, and therefore, sequence motifs might be shared between the sets. Several recent studies used DNA oligomers of a specific length, referred to as k-mers, to form length k motifs from a training set of enhancer sequences; then a statistical model was applied to learn and generalize the rules to discriminate enhancers from non-functional DNA sequences [21–23].
Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) is a powerful method to identify cell-type-specific binding sites of TFs [24,25]. These binding sites have been used in combination with machine learning methods to predict the locations of enhancers [6,26]. Such methods are limited as many TF ChIP-seq binding sites are not functional [27,28] and any specific TF will only bind to a subset of a cell-type-specific enhancers.
Sequence-specific binding TFs often recruit cofactor proteins, such as chromatin-modifying enzymes, for example: histone acetyltransferase p300/CBP, BRG1 complex and Mediator complex [29,30]. The binding of cofactors facilitates chromatin remodeling and DNA looping to form crucial enhancer–promoter interaction [31,32]. Therefore, genome-wide profiling of cofactor occupancy provides a general strategy for detecting enhancers [33,34]. For instance, Visel et al. used a transgenic mouse assay to show that 87% of enhancers identified from p300 ChIP-seq in three tissues were reproducibly active [33].
Nucleosome positioning and dynamics (assembly, mobilization and disassembly of nucleosomes) also influence gene transcription [35]. Furthermore, enhancer activity is associated with characteristic chromatin signatures that consist of histone tail modifications, including H3 lysine 4 monomethylation (H3K4me1), H3K4me3 and H3K27ac [36–38]; such chromatin signatures can be identified by clustering analysis of histone modification ChIP-seq data [39,40] (Fig. 2A). As an example, in human CD4+ T cells, 39 histone modifications have been mapped and several combinations of histone modifications were found to mark enhancers; however, no single histone modification was associated with more than 35% of enhancers [41]. These results suggested that histone modifications are likely to act cooperatively to mark enhancers. This complication suggests that statistical models must consider multiple histone modifications when predicting enhancers.
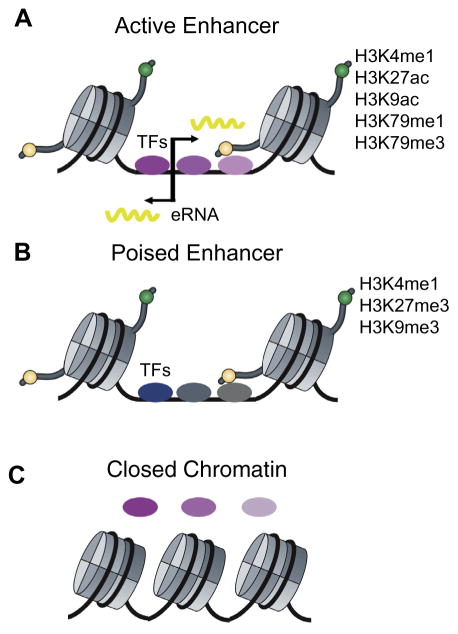
Fig. 2.
Epigenomic features that mark active and poised enhancers. (A) Generally active enhancers are marked by H3K4me1, H3K27ac, H3K9ac, H3K79me1, and H3K79me3. They are also bi-directionally transcribed, producing eRNAs that are 1– 2 kb in length. (B) Poised enhancers are not active but instead are primed for activation during development and are marked by H3K4me1, H3K27me3, and H3K9me3. (C) Closed chromatin is not bound by TFs. Binding of pioneer TFs often induces the transition from “closed” to “open” chromatin.
Sophisticated computational methods have been developed to predict enhancer locations from histone modifications and the majority fit into two categories: discriminative and generative models (Table 1). The discriminative category is inherently supervised and requires a large training set, usually collected from coactivator binding sites, such as p300. Examples of computational tools in this category are: CSI–ANN [42], ChromaGenSVM [43], and RFECS [44]. CSI–ANN first applies a Particle Swarm Optimization technique to train a time-delay neural network whose optimal structure is determined by testing different numbers of hidden layer nodes and delays. The model then slides a 2.5 kb window across the genome to determine if regions match the profile of enhancers. ChromaGenSVM trains a support vector machine (SVM) to recognize the histone modification profiles associated with enhancers. It integrates a genetic algorithm to automatically select the types of histone marks and the window size of the epigenomic profiles that best characterize enhancer regions. For example, from 38 distinct ChIP-seq chromatin marks in human CD4+ T cells, ChromaGenSVM picked out a set of only five epigenomic marks (H3K4me1, H3K4me3, H3R2me2, H3K8ac, and H2BK5ac) that best characterize active enhancers. Furthermore, it was determined that the optimal window size for ChIP-chip data was 5 kb but this dropped to 1 kb with ChIP-seq. RFECS is a Random Forest based method that trains a forest of binary decision trees, in which the Fisher discriminative approach is used as a linear classifier at each tree branch. Each feature is a multi-dimensional vector of 100 bp bins forming a window of 2 kb along the enhancers, and the final enhancer prediction is determined by votes from all the trees in the forest. These three methods have a similar workflow of training and prediction but apply different statistical models. These statistical methods also provide a systematic way to evaluate the contribution of individual histone marks to enhancer location prediction. For example, RFECS identified H3K4me1 and H3K4me3 as the most important features when predicting enhancer locations [44]. It is worth of noting that the optimal set of histone marks to predict enhancer locations may not be unique due to the functional redundancy of these marks (see below). Comparing the performance of these methods should also be interpreted with caution as no gold standard set of enhancer locations currently exists. Furthermore, commonly used evaluation criteria, such as overlapping predicted enhancers with DNase I hypersensitivity site (DHS) or p300 binding sites, only provide indirect evidence and represent only a subset of enhancers.
Table 1.
Computational tools for enhancer identification.
| Category | Approach | Statistical model | Program/Note | Ref. |
|---|---|---|---|---|
| Sequence motif | De novo motifs | Support vector machine | kmer-SVM | [21–23] |
| TF-binding motifs | n/a | EEL-enhancer element locator | [6] | |
| ChIP-seq | p300/Mediator | n/a | Peak profile scanning | [33,34,51] |
| DNA methylation | n/a | Support vector machine | SVMmap | [61] |
| Genomic window based | methylSeekR | [58,60] | ||
| Histone modification | Discriminative models | Time-delay neural network | CSI–ANN | [42] |
| Support vector machine | ChromaGenSVM | [43] | ||
| Random Forest | RFECS | [44] | ||
| Generative models | Dynamic Bayesian network | Segway | [48] | |
| Hidden Markov model | ChromHMM | [47] | ||
| Modular Hidden Markov model | ChroModule | [45,49] |
In the generative category, multiple methods use hidden Markov model (HMM) or dynamic bayesian network (DBN), including: Chromia [45], ChromHMM [46,47], Segway [48], and ChroModule [49]. Both Chromia and ChroModule are supervised learning HMMs; Chromia focuses on predicting promoters and enhancers while ChroModule uses a modularHMMto segment the entire genome into five categories: promoters, enhancers, transcribed, repressed and background. Chromatin modifications surrounding regulatory elements often form characteristic shapes, such as bimodal H3K4me3 peaks at active promoters [36,39,40,50,51]. Robust methods are needed to represent these profiles as they vary in terms of length, magnitude and pattern of epigenomic modification. Both Chromia and ChroModule use a mixture of Gaussians to flexibly represent the diverse signal patterns associated with regulatory elements and capture the signature patterns. For example, the enhancer module in ChroModule can model enhancers either with or without a nucleosome free region using a Gaussian mixture model. Therefore, ChroModule can capture novel signal patterns and combination of epigenomic modifications associated with regulatory elements. Alternatively, ChromHMM puts read counts into 200 bp bins that are discretized [52]. In order to associate the genomic locations with HMM states, a posterior probability distribution over the state of each interval (bin) is computed. Segway exploits a DBN model that works with the full data matrix at 1 bp genomic resolution [48]. An advantage of the DBN framework is that it can handle heterogeneous missing data. The method uses a single Gaussian distribution to represent the sequencing signals. When applying ChromHMM and Segway, a critical step is to select the optimal number of states that best fits the data, which is normally achieved by testing a range of states and picking the best performing one. The unsupervised learning strategy in ChromHMM and Segway allows these methods to learn unknown combination of chromatin signatures, which may correspond to novel biology. In order to interpret the segmentation results of these methods and annotate each state, additional information such as TSS and p300 binding sites are needed to associate the identified chromatin states to functional elements like promoters and enhancers.
Methods in the discriminative category aim to predict enhancers while generative methods segment the epigenome and enhancers are annotated as a part of segmentation. Choice of method depends on the purpose of the analysis and the availability of data, as illustrated by a comparison between ChromHMM and Segway annotations [47].
Besides histone modification, DNA methylation – the addition of a methyl group to the nucleotide cytosine – is another epigenomic feature that can predict enhancer locations. Different cell-types and tissues display distinct patterns of DNA methylation [53–55] and specific changes in DNA methylation are associated with development of cancers and autoimmune diseases, such as rheumatoid arthritis [56,57]. Enhancers have methylation levels between 10 and 50% while promoters have methylation levels between 0 and 10% and the rest of the genome is marked by higher levels of DNA methylation (hypermethylation) [58]. In the Stadler et al. study, the characteristic DNA methylation levels are modeled by a three-state HMM model to compartmentalize the genome into cell-type-specific enhancer and promoter regions [58]. Similar to segmentation of chromatin modification data, HMM models have been developed to segment methylomes to systematically uncover the regulatory regions associated with characteristic DNA methylation levels [59,60]. Additionally, these methods can also reveal regions with consecutive DNA methylation levels; such as partially methylated domains (PMD), which are broad and inactive regions of the genome. Furthermore, changes in DNA methylation at enhancers correlate with changes in the expression of distal genes that may be regulated by the enhancers. Therefore, given the gene expression levels and DNA methylation profiles in different tissues it is possible to simultaneously identify enhancers and link them to their target genes (see below) [61]. For example, Aran et al. first identified differentially expressed genes across 58 human cell-types. To train a statistical model they created two sets of genomic regions that represent positive and negative examples. The positive set was the correlation between CpG methylation in the promoters and the expression levels of their corresponding genes, these were taken as the positive examples. The background consisted of randomly selected CpG-gene pairs that are located in different chromosomes. Then they trained a support vector machine (SVM), known as SVMmap (Table 1), to identify CpG-gene pairs within a set of genomic window. These distal methylation site and gene pairs were found to be enriched with enhancer-associated histone modifications and significantly overlapped those detected using 5C (see below).
In summary, various genomic and epigenomic properties have been exploited to predict genome-wide enhancer positions. Owing to the complex nature of development and cell-type-specificity, any single modification or factor used in isolation is often not the best predictor of enhancer locations. Instead the integration of multiple layers of genomic/epigenomic information is more powerful for producing a comprehensive annotation and understanding of enhancers. Following this trend, several recent studies have started to explore integrative models to identify potential enhancers [62,63]. Furthermore, there are recent advances in high-throughput enhancer identification assays (e.g. STARR-seq [64] or using bidirectional expression of short transcripts measured by GRO-seq [65]). These techniques should become even more powerful once they are combined with computational methodologies. When a complete set of enhancers are cataloged, much work will be required to examine spatiotemporal activity of enhancers in a high-throughput, unbiased, and dynamic way, especially in the context of multiple developmental stages.
2.1. Predicting the cell-type-specific activity of enhancers
Since the activity signatures of enhancers are diverse and complex, it is important to chart their developmental and cell-type specificity. We discuss the techniques that are available and how they can be used to determine enhancer activity on a spatiotemporal level (location and developmental stage specificity).
As discussed above, enrichment of H3K4me1 [36], hypersensitivity to nuclease digestion [66], and sequence conservation between species [8,9,67] have been exploited to identify enhancers (Fig. 2A). However, not all enhancers exhibiting these properties are functionally active in a specific cell-type (Fig. 2B and C). The histone acetyltransferase p300 was initially used to measure enhancer activity [33] but follow-up studies showed that only a fraction of enhancers bound by p300 modulate transcription in a given cell-type [37,38]. H3K4me1, by itself, is not sufficient to distinguish active enhancers from inactive ones [37,38], while H3K27ac, in combination with H3K4me1, were shown to be a more robust indicator of enhancer activity [37,38]. For example, genome-wide analysis in mouse embryonic stem cells (ESCs) and four other cell-types demonstrated that enhancers marked by H3K27ac are associated with genes with higher levels of expression [37]. In addition to H3K27ac, several other epigenomic signatures have also been associated with enhancer activity: H3K4me3, which is enriched in active promoters, was found to reflect enhancer activity in T cells [68]; H3K79me3 and RNA Pol II are also significantly enriched for active enhancers in Drosophila melanogaster embryos [69]. Together, these results suggest that there is not just one epigenomic modification associated with enhancer activity [69]. Indeed, enhancers marked by H3K4me1 but not by H3K27ac in human ESCs were shown to be active in other tissues [37]. This suggests that these enhancers, termed poised enhancers, are in a primed state and become active upon differentiation or stimuli. Furthermore, poised enhancers are associated with enrichment of H3K27me3 [38] and H3K9me3 [70].
A more direct measure of enhancer activity is the transcription of RNAs at the enhancer site. While transcription at enhancers was first observed more than 20 years ago [71], only recently has evidence been found that enhancer transcription is genome-wide and indicative of enhancer activity. Kim et al. identified a large number of short (<2 kb), bi-directionally transcribed, and non-polyadenylated RNA transcripts at enhancer sites [72]. These RNA transcripts are termed as enhancer RNAs (eRNAs). Interestingly, eRNA transcription levels were found to be correlated with the expression levels of their target genes. Moreover, eRNAs are produced only in the presence of functional target promoters. These findings suggest that eRNA transcription is associated with enhancer activity [72]. Wang et al. further confirmed that eRNA transcription is a robust indicator of enhancer activity [73]. Computational methods have been developed to predict enhancer activity from eRNA transcription levels measured by global nuclear run-on sequencing (GRO-seq) [65]. For example, Melgar et al. extracted GRO-seq signals at enhancers and in nearby windows, then used a Bayesian model to predict enhancer activity [65]. Furthermore, knockdown experiments showed that eRNAs are functional consequence of enhancers, rather than by-products of gene transcription [74].
Although eRNA transcription is a direct indicator of enhancer activity, its abundance is low and its accurate measurement requires higher sequencing depth than mRNA. Thus, integration of chromatin signatures and eRNA transcription becomes a promising approach to predict enhancer activity. Zhu et al. used a logistic regression model to learn the relationship between chromatin signatures and eRNA production [75]. They demonstrated that four histone modification marks are sufficiently accurate to predict eRNA transcription. Interestingly, many combinations of four modifications can achieve superior predictions, which is consistent with other studies showing that histone marks redundantly mark enhancers. They then used the luciferase reporter assay to confirm that their model predicted enhancer activity more accurately than using H3K27ac in isolation.
Both chromatin modifications and eRNAs exhibit high cell-type specificity during development, differentiation and homeostasis, indicating enhancers have a crucial role in the fine tuning cell-type-specific gene expression. In vivo mapping of p300 binding sites in mouse embryonic forebrain, midbrain, limb, and heart identified enhancers that recapitulate tissue-specific gene expression in transgenic mouse assays [33]. H3K4me1-defined enhancers are also cell type-specific and are correlated with gene expression patterns [36]. H3K27ac and eRNA production are even more dynamic than the universal mark H3K4me1 [37,38,70]. Genome-wide mapping of H3K27ac and eRNA production reveals highly dynamic enhancer activity during embryogenesis [76], embryonic limb development [77] and at estrogen receptor binding sites [78]. Interestingly, a significant portion of H3K4me1-defined enhancers are not enriched with H3K27ac and do not produce bidirectional eRNA transcripts. These poised enhancers are likely not regulatory active but may become active in other cell-types, especially those in the same developmental lineage, or following stimulation [37].
A recent study used CAGE sequencing to identify active enhancers via eRNA identification [79]. However, only 43,011 active enhancers were identified in 808 human cell-types and tissues, suggesting that eRNA alone is still limited to predict enhancers due to the low eRNA abundance that makes detection difficult. This recent observation further highlights the importance of overcoming this hurdle by training computational models to capture the characteristic patterns of multiple chromatin modifications based on the top active enhancers with the most abundant eRNAs to more exhaustively identify enhancers.
2.2. Linking enhancers with their target genes
Once a putative enhancer is identified it is important to confidently identify the genes it is regulating. In this section we will discuss methods that can be used to link enhancers and genes. In particular, methods used to analyze chromosome conformation capture methods, such as Hi-C and ChIA-PET.
To determine the regulatory functions of enhancers they must be linked to the genes whose expression they control. This task is very important, as enhancer activity is critical in controlling cell-type-specific patterns of gene expression. Knowing enhancer–gene interactions permits the identification of the regulatory targets of non-promoter TF binding sites; such as those identified with ChIP-seq or using DNA-binding motifs. Furthermore, these interactions can be identified en masse allowing transcriptional regulatory network to be constructed [46,80]. However, linking enhancers to their target genes is very challenging since enhancers and target genes: (i) may be very distant [81], (ii) may be located on different chromosomes [82], (iii) may have many-to-many relationships [83], and (iv) have cell-type-specific interactions [51]. Thus, to accurately link enhancers to their target genes a method must be able to evaluate a huge number of potential enhancer–gene combinations in a cell-type-specific manner.
The simplest method is to assign enhancers to the closest TSS. However, this method can generate many false positives and a 5C (see below) study has shown as little as 7% of enhancer interactions are with the closest TSS [84]. An advancement on the closest TSS approach is to take into account of genomic domains created by the insulator protein, CTCF [85], which is assumed to block promoter- enhancer interactions [51,80]. However, the assignment of promoter-enhancer pairs by either of these two methods has been shown to be only slightly better than the random control [86]. Comparing promoter and enhancer activity between different cell-types can achieve more accurate assignment of promoter-enhancer pairs. The idea is that interacting enhancers and promoters display similar patterns of activity across cell-types. A logistic regression classifier coupled with transcription factor (TF) motifs and expression levels was used to assign promoter-enhancer interactions and classify TFs into activators or repressors [46]. However, this approach is based on modeling one-to-one promoter-enhancer relationships and is biased towards the closest TSS. To better model the many-to-many interaction relationship that exists between promoters and enhancers, the genome can be divided into domains of co-regulated promoters and enhancers, called enhancer promoter units (EPUs) [86]. Chromatin modification data from 19 mouse tissues/cell-types was used to define EPUs; there were an average of 5.67 enhancers per TSS and the domains are highly correlated with 3D chromatin domains identified using a technique called Hi-C (see below).
There are a number of experimental techniques that use variants of chromosome conformation capture (3C) [87] to identify interactions between enhancers and their target genes. In 3C experiments, regions of genomic DNA are cross-linked using formaldehyde, forming stable complexes between regions of the genome that are nearby and potentially interacting. These DNA region complexes can be measured using PCR primers that are designed to measure the level of interaction between two regions of interest, such as a gene and an enhancer [88]. Other variants of 3C, such as 4C and 5C [84,89], increase the throughput and reduce biases by allowing interactions between multiple regions to be tested at one time. These techniques are more powerful when coupled with Hi-C and high-throughput sequencing, allowing for genome-wide maps of interacting regions [90] (Fig. 3A). Recently, Hi-C was used to uncover over one million long-range chromatin interactions at 5–10 kb resolution [91]; however, this study used 3.4 billion uniquely mapped reads making it very costly to experimentally determine high-resolution enhancer–gene interactions in other cell-types or under other conditions. An alternative technology, chromatin interaction analysis by paired-end tag sequencing (ChIA-PET), measures interactions at a specific factor of interest [92] that requires less sequencing depth (Fig. 3B). In particular, ChIA-PET is powerful when targeting a general factor that is enriched at interacting enhancer–promoter pairs, such as CTCF [93] or pre-initiation complexes of RNA Pol II [94]. However, ChIA-PET requires a high-quality antibody and interactions not involving the factor of interest are missed. Similar to the observation that not all binding sites of a TF detected by ChIP-seq are functional, it is worth of noting that the interactions defined by Hi-C or ChIA-PET may not be functional enhancer–gene pairs but rather physical contacts due to nearby functional interactions.
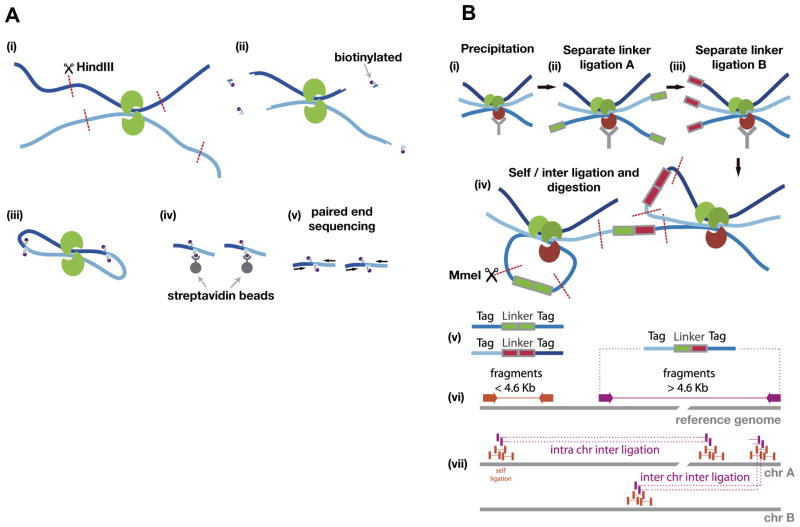
Fig. 3.
Overview of Hi-C and ChIA-PET. (A) A schematic shows the Hi-C protocol. (i) Formaldehyde cross-linking of the cells (Proteins in green, Chromatin dark blue and light blue), followed by digestion (HindIII) of the chromatin. (ii) The restriction site is used to attach biotinylated nucleotides (purple). (iii) Ligation of the open ends. (iv) Streptavidin beads are used to isolate the biotinylated molecules, followed by (v) paired-end sequencing. (B) Schematic of ChIA-PET analysis: The chromatin is prepared by formaldehyde cross-linking, fragmentation (not shown) and (i) precipitation, followed by (ii), (iii) separate linker ligation A and B. Then, the separate probes are mixed to allow for proximity (inter) and self-ligation, which is followed by (iv) MmeI restriction enzyme digestion. After sequencing, the resulting tag-linker products (v) are mapped to the genome (vi). The linker can be used to categorize between self and inter ligation, which allows for clustering of self-ligation and long-range chromatin interactions (vii).
Within the nucleus, genomic loci are organized in functional compartments, which interact on a level correlated to gene expression. Hi-C aims to find these interactions by interrogating all possible loci as it is not focused on pre-defined interaction sites. However, Hi-C data may incorporate many false positive signals and include biases brought about by random and self-polymer looping, technical and experimental inaccuracies, and biases during the experiment [95]. Hence, when using Hi-C to search for locus interactions, the first step should be to filter the interactions, to adjust for and remove biases. An initial filtering of contacts with a mixture Poisson regression model, 23,337,830 potential inter-loci connections were reduced to 96,137 [96]. A more accurate filtering might be achieved by removing biases through comparison of multiple Hi-C data sets [97]. The method is motivated by the idea that equivalent biases for detecting contacts between two regions exist within multiple datasets. To do this, an iterative correction procedure was applied to mapped reads to uncover relative contact probabilities. An iterative normalization process then calculates contact probabilities between two pairs of regions for the complete set of probability-pairs, which helps to eliminate biases.
Some Hi-C analysis methods have explicitly attempted linking enhancers and genes. One recent method used a geometric distribution- based model to form hotspots (or clusters) of adjacent Hi-C reads that represent potential sites of DNA–DNA interaction [98]. This first step was done without any consideration of read pairing that provides connection in Hi-C. Next interaction hotspots are extended by ~3 kb to account for Hi-C reads being enriched at the cut sites of the restriction enzymes used in the assay. Finally, high confidence enhancer–gene links are identified between hotspot pairs where one contains a DHS and enhancer-related histone modifications and a second is an annotated promoter. The two hotspots must also be connected by 2 or more Hi-C reads. The predicted enhancer–gene links were enriched with p300 binding sites and enhancer binding TFs. Furthermore, the target promoters were significantly bound by RNA Pol II. More recently, a statistical approach to distinguish between random loops, technical or experimental biases in the dataset and true mid-range contacts was proposed [99]. It extended an existing method [100] via replacing the discrete binning approach originally used to sort out random looping events, by a spline-fitting procedure to achieve an enhanced estimate of contact probability. By using this approach, 6–46% more contacts were found compared to the original method. Validation of the method was done using a ChIA-PET contact catalog, resulting in 77% matching enhancer–promoter interactions, which also outperformed the original binning approach. Furthermore, predictions were successfully compared with 3C-validated contacts.
Alternatively, to remove noise in the contacts identified in Hi-C, it is also possible to combine the approach with cross-species conservation data (Fig. 4A) [101]. For example, Lu et al. compared the presence/absence of genomic regions across 45 species and established a phylogenetic profile for each gene and enhancer in human, in which the predictions were being made. Enhancer–gene interactions supported by high Hi-C reads have an average correlation of 0.6 between their phylogenetic profiles while an average of 0.35 was observed in the background. Based on this observation, they next determined the cut-offs of correlation coefficient and Hi-C read counts for selecting enhancer–gene pairs that resulted in enhancer–gene links that were more reproducible between replica cell lines. This method allows the prediction of many-to-many enhancer–gene links. They observed that genes regulated by the same enhancers were functionally related and co-expressed, and links could help to explain the role of a disease-causing SNP. However, this method is limited by the sequencing depth of Hi-C (only 2 reads were required to connect an enhancer and gene) and newly evolved enhancer–gene links maybe be missed as the two are unlikely to share cross-species conservation patterns.
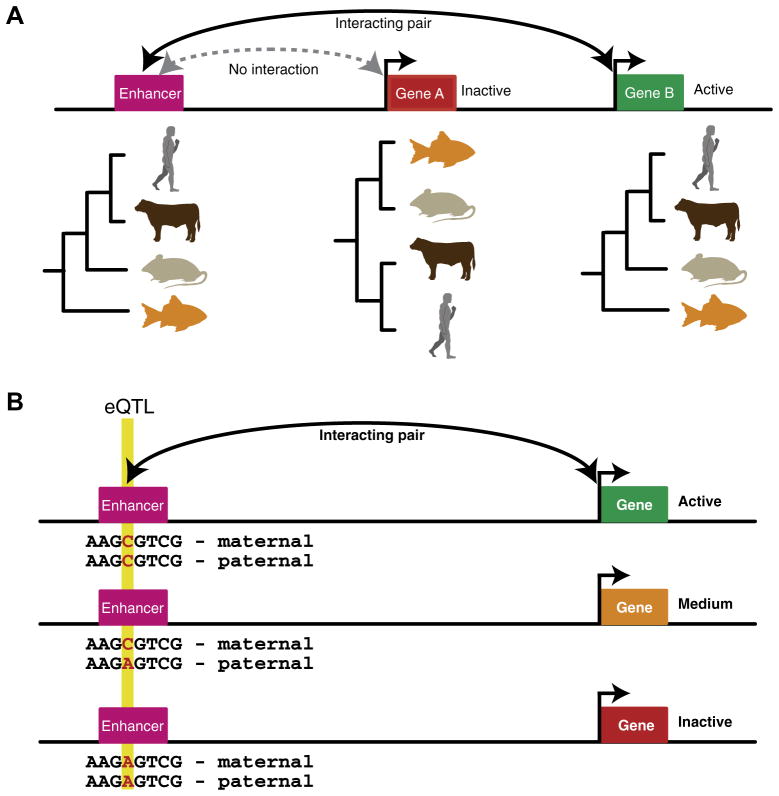
Fig. 4.
Concepts used to link enhancers to their target genes. (A) A schematic shows the phylogenetic profiles of two genes and an enhancer. The gene and enhancer pair that share the same phylogenetic profile are shown to interact resulting in the expression of ‘Gene B’ while ‘Gene A’ does not interact with the enhancer and is inactive. (B) A schematic shows an eQTL located within an enhancer that interacts with the shown gene. The genotype ‘A’ has a negative effect on enhancer activity resulting in a reduction in gene expression. Correlating these changes over multiple genotypes allows enhancers and genes to be linked.
Owing to the great financial cost involved in experimentally generating genome-wide high-resolution maps for enhancer and gene interactions from Hi-C, there is a great need for computational methodologies that can identify interactions from other types of data. Integrative computational methodologies combine cell-type-specific genome-wide experiments with non-cell-type-specific information to link enhancers with their target genes. For example, ChIP-seq for the histone acetyltransferase p300, which localizes to enhancers, was combined with four general features: (i) genomic distance from an enhancer to its target gene, (ii) conservation of genes and enhancers across species, (iii) distance in a protein– protein interaction network between TFs binding to the enhancer and the target gene, and (iv) Gene Ontology (GO) similarities between regulators and the putative target genes [102]. The protein– protein interaction and GO features were motivated by the fact that auto-regulatory loops are common in gene regulatory networks, and therefore, enhancer-binding proteins might be relatively proximal to their target genes in the network and have similar GO classifications. These features were combined in a Random Forest classifier to achieve a greater than 2-fold improvement over any single feature. However, this method can only link enhancers to genes within 2000 kb and identify one-to-one relationships. Furthermore, they assumed that all differentially expressed genes were positive targets of an enhancer, which is not always the case.
Mapping studies have identified expression quantitative trait loci (eQTLs), which link alterations in gene expression to SNPs (Fig. 4b). To identify eQTLs, both SNP variants and gene expression are measured in multiple genotypes of the same cell-type. Then the two are correlated allowing links between the SNPs and alterations in gene expression. When eQTLs lay outside of genes and promoters, they likely correspond to enhancers. Thus, eQTLs provide a way of linking enhancers to target genes [103,104]. Combined with enhancer predictions made from epigenomic data, eQTLs can be used to computationally predict enhancer–gene links; such as an integrative Random Forest model that used the following features: (i) genomic distance, (ii) co-occurrence of TF ChIP-seq peaks at enhancers and promoters, (iii) co-expression of target gene and TFs with ChIP-seq peaks at the promoter, (iv) DHS at the enhancer, (v) similarity in the GO-terms of the target gene and TFs with ChIP-seq peaks at the promoter, (iv) the strength of any CTCF peaks located between the enhancer and target gene [105]. Using all the features the method achieved a great improvement in performance, an area under the receiver operator curve (AUC) of 0.9 compared to 0.75 when only a single feature was used. Furthermore, when enhancers and genes are separated by >150 kb, the genomic distance feature becomes uninformative while the other features remain good predictors and the overall model performance only decreases slightly.
3. Conclusions
The advent of next-generation sequencing has brought with it a plethora of genome-wide assays that have revolutionized our ability to interrogate the genome. Owing to projects like ENCODE [106], Roadmap Epigenomics Project [13] and BLUEPRINT [107], the catalogue of human enhancers and its annotation has risen rapidly in recent years. To accompany these advances there have been many sophisticated enhancer prediction and annotation methods developed. In particular, integrative analysis strategies that use multiple genome-wide assays are becoming powerful; continuous development on the experimental and computational technologies is likely to further improve the prediction accuracy of enhancers and enhancer-target gene linkage.
There are still many challenges ahead. Firstly, large sets of functionally confirmed enhancers in different cell-types, at various developmental stages and under diverse cellular conditions are needed for developing novel computational methods and further interrogating genome-wide measurements. Secondly, better understanding of the mechanistic relationship between epigenomic modifications, eRNA transcription, and enhancer activity is critical for advancing prediction and annotation of enhancers in the human genome. Thirdly, an emerging need is to develop computational methods that can model and infer the dynamics of enhancer activity and enhancer–gene interactions in a biological context-dependent manner. With the fast advancement of genomic and computational technologies, overcoming these hurdles may come much sooner than expected.
References
- 1.Ong CT, Corces VG. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith E, Shilatifard A. Nat Struct Mol Biol. 2014;21:210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- 3.Krivega I, Dean A. Curr Opin Genet Dev. 2012;22:79–85. doi: 10.1016/j.gde.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulger M, Groudine M. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calo E, Wysocka J. Mol Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 7.Blanchette M, Bataille AR, Chen X, Poitras C, Laganiere J, Lefebvre C, Deblois G, Giguere V, Ferretti V, Bergeron D, Coulombe B, Robert F. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Nat Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 10.Allende ML, Manzanares M, Tena JJ, Feijoo CG, Gomez-Skarmeta JL. Methods. 2006;39:212–219. doi: 10.1016/j.ymeth.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Poulin F, Nobrega MA, Plajzer-Frick I, Holt A, Afzal V, Rubin EM, Pennacchio LA. Genomics. 2005;85:774–781. doi: 10.1016/j.ygeno.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 12.ENCODE Project Consortium. PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Wang J, Ward L, Sarkar A, Quon G, Kheradpour P, Heravi-Moussavi A, Coarfa C, Harris AR, Ziller M, Schultz M, Eaton M, Pfenning A, Wang X, Polak P, Karlic R, Amin V, Wu Y-C, Sandstrom RS, Whitaker JW, Elliott G, Lowdon R, Beaudet Arthur E, Boyer Laurie, Farnham P, Fisher S, Haussler D, Jones S, Li W, Marra M, Sunyaev S, Thomson JA, Tlsty TD, Tsai L-H, Wang W, Waterland RA, Zhang M, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosalvjevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Nature. 2014 submitted for publication. [Google Scholar]
- 14.Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Bristow J, Ren B, Black BL, Rubin EM, Visel A, Pennacchio LA. Nat Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meireles-Filho AC, Stark A. Curr Opin Genet Dev. 2009;19:565–570. doi: 10.1016/j.gde.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glazko GV, Koonin EV, Rogozin IB, Shabalina SA. Trends Genet. 2003;19:119–124. doi: 10.1016/S0168-9525(03)00016-7. [DOI] [PubMed] [Google Scholar]
- 18.Narlikar L, Sakabe NJ, Blanski AA, Arimura FE, Westlund JM, Nobrega MA, Ovcharenko I. Genome Res. 2010;20:381–392. doi: 10.1101/gr.098657.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taher L, Smith RP, Kim MJ, Ahituv N, Ovcharenko I. Genome Biol. 2013;14:R117. doi: 10.1186/gb-2013-14-10-r117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitaker JW, Chen Z, Wang W. Nat Methods. doi: 10.1038/nmeth.3065. In press. http://dx.doi.org/10.1038/nmeth.3065, http://www.nature.om/nmeth/journal/vaop/ncurrent/abs/nmeth.3065.html. [DOI] [PMC free article] [PubMed]
- 21.Fletez-Brant C, Lee D, McCallion AS, Beer MA. Nucleic Acids Res. 2013;41:W544–556. doi: 10.1093/nar/gkt519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorkin DU, Lee D, Reed X, Fletez-Brant C, Bessling SL, Loftus SK, Beer MA, Pavan WJ, McCallion AS. Genome Res. 2012;22:2290–2301. doi: 10.1101/gr.139360.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee D, Karchin R, Beer MA. Genome Res. 2011;21:2167–2180. doi: 10.1101/gr.121905.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson DS, Mortazavi A, Myers RM, Wold B. Science. 2007;316:1497– 1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 25.Farnham PJ. Nat Rev Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yip KY, Cheng C, Bhardwaj N, Brown JB, Leng J, Kundaje A, Rozowsky J, Birney E, Bickel P, Snyder M, Gerstein M. Genome Biol. 2012;13:R48. doi: 10.1186/gb-2012-13-9-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher WW, Li JJ, Hammonds AS, Brown JB, Pfeiffer BD, Weiszmann R, MacArthur S, Thomas S, Stamatoyannopoulos JA, Eisen MB, Bickel PJ, Biggin MD, Celniker SE. Proc Natl Acad Sci U S A. 2012;109:21330– 21335. doi: 10.1073/pnas.1209589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo CL. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borggrefe T, Yue X. Semin Cell Dev Biol. 2011;22:759–768. doi: 10.1016/j.semcdb.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 30.Roeder RG. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Conaway RC, Conaway JW. Curr Opin Genet Dev. 2011;21:225–230. doi: 10.1016/j.gde.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Afzal V, Simpson PC, Rubin EM, Black BL, Bristow J, Pennacchio LA, Visel A. Nat Genet. 2012;44:89–93. doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henikoff S. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 36.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 37.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hon G, Ren B, Wang W. PLoS Comput Biol. 2008;4:e1000201. doi: 10.1371/journal.pcbi.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hon G, Wang W, Ren B. PLoS Comput Biol. 2009;5:e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Firpi HA, Ucar D, Tan K. Bioinformatics. 2010;26:1579–1586. doi: 10.1093/bioinformatics/btq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez M, Miranda-Saavedra D. Nucleic Acids Res. 2012;40:e77. doi: 10.1093/nar/gks149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajagopal N, Xie W, Li Y, Wagner U, Wang W, Stamatoyannopoulos J, Ernst J, Kellis M, Ren B. PLoS Comput Biol. 2013;9:e1002968. doi: 10.1371/journal.pcbi.1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Won KJ, Chepelev I, Ren B, Wang W. BMC Bioinf. 2008;9:547. doi: 10.1186/1471-2105-9-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffman MM, Ernst J, Wilder SP, Kundaje A, Harris RS, Libbrecht M, Giardine B, Ellenbogen PM, Bilmes JA, Birney E, Hardison RC, Dunham I, Kellis M, Noble WS. Nucleic Acids Res. 2012;41:827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffman MM, Buske OJ, Wang J, Weng Z, Bilmes JA, Noble WS. Nat Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Won KJ, Zhang X, Wang T, Ding B, Raha D, Snyder M, Ren B, Wang W. Nucleic Acids Res. 2013;41:4423–4432. doi: 10.1093/nar/gkt143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, Mieczkowski P, Lieb JD, Zhao K, Brown M, Liu XS. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ernst J, Kellis M. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. Nat Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, Yang H, Wang T, Lee AY, Swanson SA, Zhang J, Zhu Y, Kim A, Nery JR, Urich MA, Kuan S, Yen CA, Klugman S, Yu P, Suknuntha K, Propson NE, Chen H, Edsall LE, Wagner U, Li Y, Ye Z, Kulkarni A, Xuan Z, Chung WY, Chi NC, Antosiewicz-Bourget JE, Slukvin I, Stewart R, Zhang MQ, Wang W, Thomson JA, Ecker JR, Ren B. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, Gnirke A, Meissner A. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakano K, Whitaker JW, Boyle DL, Wang W, Firestein GS. Ann Rheum Dis. 2013;72:110–117. doi: 10.1136/annrheumdis-2012-201526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitaker JW, Shoemaker R, Boyle DL, Hillman J, Anderson D, Wang W, Firestein GS. Genome Med. 2013;5:40. doi: 10.1186/gm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schubeler D. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 59.Schroeder DI, Lott P, Korf I, LaSalle JM. Genome Res. 2011;21:1583–1591. doi: 10.1101/gr.119131.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burger L, Gaidatzis D, Schubeler D, Stadler MB. Nucleic Acids Res. 2013;41:e155. doi: 10.1093/nar/gkt599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aran D, Sabato S, Hellman A. Genome Biol. 2013;14:R21. doi: 10.1186/gb-2013-14-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen CY, Morris Q, Mitchell JA. BMC Genomics. 2012;13:152. doi: 10.1186/1471-2164-13-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Podsiadło A, Wrzesień M, Paja W, Rudnicki W, Wilczyński B. BMC Syst Biol. 2013;7:S16. doi: 10.1186/1752-0509-7-S6-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 65.Melgar MF, Collins FS, Sethupathy P. Genome Biol. 2011;12:R113. doi: 10.1186/gb-2011-12-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 68.Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, Holota H, Imbert J, Andrau JC, Ferrier P, Spicuglia S. EMBO J. 2011;30:4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE. Nat Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 70.Zentner GE, Tesar PJ, Scacheri PC. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tuan D, Kong S, Hu K. Proc Natl Acad Sci U S A. 1992;89:11219–11223. doi: 10.1073/pnas.89.23.11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu XD. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu Y, Sun L, Chen Z, Whitaker JW, Wang T, Wang W. Nucleic Acids Res. 2013;41:10032–10043. doi: 10.1093/nar/gkt826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bogdanovic O, Fernandez-Minan A, Tena JJ, de la Calle-Mustienes E, Hidalgo C, van Kruysbergen I, van Heeringen SJ, Veenstra GJ, Gomez-Skarmeta JL. Genome Res. 2012;22:2043–2053. doi: 10.1101/gr.134833.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cotney J, Leng J, Oh S, DeMare LE, Reilly SK, Gerstein MB, Noonan JP. Genome Res. 2012;22:1069–1080. doi: 10.1101/gr.129817.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jorgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Muller F, Forrest AR, Carninci P, Rehli M, Sandelin A, de Hoon MJ, Haberle V, Kulakovskiy IV, Lizio M, Schmeier S, Dimont E, Schmid C, Schaefer U, Medvedeva YA, Plessy C, Vitezic M, Severin J, Semple CA, Young RS, Francescatto M, Alam I, Albanese D, Altschuler GM, Arakawa T, Archer JA, Arner P, Babina M, Rennie S, Balwierz PJ, Beckhouse AG, Pradhan-Bhatt S, Blake JA, Blumenthal A, Bodega B, Bonetti A, Briggs J, Brombacher F, Califano A, Cannistraci CV, Carbajo D, Chierici M, Ciani Y, Clevers HC, Dalla E, Davis CA, Detmar M, Diehl AD, Dohi T, Drablos F, Edge AS, Edinger M, Ekwall K, Endoh M, Enomoto H, Fagiolini M, Fairbairn L, Fang H, Farach-Carson MC, Faulkner GJ, Favorov AV, Fisher ME, Frith MC, Fujita R, Fukuda S, Furlanello C, Furuno M, Furusawa J, Geijtenbeek TB, Gibson AP, Gingeras T, Goldowitz D, Gough J, Guhl S, Guler R, Gustincich S, Ha TJ, Hamaguchi M, Hara M, Harbers M, Harshbarger J, Hasegawa A, Hasegawa Y, Hashimoto T, Herlyn M, Hitchens KJ, Ho Sui SJ, Hofmann OM, Hori F, Huminiecki L, Iida K, Ikawa T, Jankovic BR, Jia H, Joshi A, Jurman G, Kaczkowski B, Kai C, Kaida K, Kaiho A, Kajiyama K, Kanamori-Katayama M, Kasianov AS, Kasukawa T, Katayama S, Kato S, Kawaguchi S, Kawamoto H, Kawamura YI, Kawashima T, Kempfle JS, Kenna TJ, Kere J, Khachigian LM, Kitamura T, Klinken SP, Knox AJ, Kojima M, Kojima S, Koseki H, Koyasu S, Krampitz S, Kubosaki A, Kwon AT, Laros JF, Lee W, Lipovich L, Mackay-sim A, Manabe R, Mar JC, Marchand B, Mathelier A, Mejhert N, Meynert A, Mizuno Y, de Lima Morais DA, Morikawa H, Morimoto M, Moro K, Motakis E, Motohashi H, Mummery CL, Murata M, Nagao-Sato S, Nakachi Y, Nakahara F, Nakamura T, Nakamura Y, Nakazato K, van Nimwegen E, Ninomiya N, Nishiyori H, Noma S, Nozaki T, Ogishima S, Ohkura N, Ohmiya H, Ohno H, Ohshima M, Okada-Hatakeyama M, Okazaki Y, Orlando V, Ovchinnikov DA, Pain A, Passier R, Patrikakis M, Persson H, Piazza S, Prendergast JG, Rackham OJ, Ramilowski JA, Rashid M, Ravasi T, Rizzu P, Roncador M, Roy S, Rye MB, Saijyo E, Sajantila A, Saka A, Sakaguchi S, Sakai M, Sato H, Satoh H, Savvi S, Saxena A, Schneider C, Schultes EA, Schulze-Tanzil GG, Schwegmann A, Sengstag T, Sheng G, Shimoji H, Shimoni Y, Shin JW, Simon C, Sugiyama D, Sugiyama T, Suzuki M, Suzuki N, Swoboda RK, t Hoen PA, Tagami M, Takahashi N, Takai J, Tanaka H, Tatsukawa H, Tatum Z, Thompson M, Toyoda H, Toyoda T, van de Wetering M, van den Berg LM, Verardo R, Vijayan D, Vorontsov IE, Wasserman WW, Watanabe S, Wells CA, Winteringham LN, Wolvetang E, Wood EJ, Yamaguchi Y, Yamamoto M, Yoneda M, Yonekura Y, Yoshida S, Zabierowski SE, Zhang PG, Zucchelli S, Summers KM, Hide W, Freeman TC, Lenhard B, Bajic VB, Taylor MS, Makeev VJ. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Won KJ, Xu Z, Zhang X, Whitaker JW, Shoemaker R, Ren B, Xu Y, Wang W. Nucleic Acids Res. 2012;40:8199–8209. doi: 10.1093/nar/gks584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vokes SA, Ji H, Wong WH, McMahon AP. Genes Dev. 2008;22:2651– 2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dorsett D. Curr Opin Genet Dev. 1999;9:505–514. doi: 10.1016/s0959-437x(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 83.Ferretti E, Cambronero F, Tumpel S, Longobardi E, Wiedemann LM, Blasi F, Krumlauf R. Mol Cell Biol. 2005;25:8541–8552. doi: 10.1128/MCB.25.19.8541-8552.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sanyal A, Lajoie BR, Jain G, Dekker J. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Phillips JE, Corces VG. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dekker J, Rippe K, Dekker M, Kleckner N. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 88.Weedon MN, Cebola I, Patch AM, Flanagan SE, De Franco E, Caswell R, Rodriguez-Segui SA, Shaw-Smith C, Cho CH, Lango H. Nat Genet. 2014;46:61–64. doi: 10.1038/ng.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, Wong E, Sheng J, Zhang Y, Poh T, Chan CS, Kunarso G, Shahab A, Bourque G, Cacheux-Rataboul V, Sung WK, Ruan Y, Wei CL. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Y, Wong CH, Birnbaum RY, Li G, Favaro R, Ngan CY, Lim J, Tai E, Poh HM, Wong E, Mulawadi FH, Sung WK, Nicolis S, Ahituv N, Ruan Y, Wei CL. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dekker J, Marti-Renom MA, Mirny LA. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lan X, Witt H, Katsumura K, Ye Z, Wang Q, Bresnick EH, Farnham PJ, Jin VX. Nucleic Acids Res. 2012;40:7690–7704. doi: 10.1093/nar/gks501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hwang YC, Zheng Q, Gregory BD, Wang LS. Nucleic Acids Res. 2013;41:4835–4846. doi: 10.1093/nar/gkt188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ay F, Bailey TL, Noble WS. Genome Res. 2014;24:999–1011. doi: 10.1101/gr.160374.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu Y, Zhou Y, Tian W. Nucleic Acids Res. 2013;41:10391–10402. doi: 10.1093/nar/gkt785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rodelsperger C, Guo G, Kolanczyk M, Pletschacher A, Kohler S, Bauer S, Schulz MH, Robinson PN. Nucleic Acids Res. 2011;39:2492–2502. doi: 10.1093/nar/gkq1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, Ingle C, Beazley C, Gutierrez M. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang D, Rendon A, Wernisch L. Nucleic Acids Res. 2013;41:1450–1463. doi: 10.1093/nar/gks1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. Nature. 2012;489:57–74. [Google Scholar]
- 107.Adams D, Altucci L, Antonarakis SE, Ballesteros J, Beck S, Bird A, Bock C, Boehm B, Campo E, Caricasole A, Dahl F, Dermitzakis ET, Enver T, Esteller M, Estivill X, Ferguson-Smith A, Fitzgibbon J, Flicek P, Giehl C, Graf T, Grosveld F, Guigo R, Gut I, Helin K, Jarvius J, Kuppers R, Lehrach H, Lengauer T, Lernmark A, Leslie D, Loeffler M, Macintyre E, Mai A, Martens JH, Minucci S, Ouwehand WH, Pelicci PG, Pendeville H, Porse B, Rakyan V, Reik W, Schrappe M, Schubeler D, Seifert M, Siebert R, Simmons D, Soranzo N, Spicuglia S, Stratton M, Stunnenberg HG, Tanay A, Torrents D, Valencia A, Vellenga E, Vingron M, Walter J, Willcocks S. Nat Biotechnol. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]