Abstract
Genomic profiling of immortalized human mammary epithelial (hTERT-HME1) cells identified several metabolic genes, including the membrane glutamate transporter, SLC1A1, as 1,25-dihydroxyvitamin D3 (1,25D) regulated. In these studies we have surveyed the effects of 1,25D on known glutamate transporters and evaluated its impact on cellular glutamate handling. We confirm that expression of SLC1A1 and all of its known transcript variants are significantly upregulated in hTERT-HME1 cells following 1,25D treatment. Expression of the full-length cognate protein, EAAT3 is correspondingly increased in 1,25D treated hTERT-HME1 cells. Under the same conditions, the expression of two other glutamate transporters - SLC1A6 (EAAT4) and SLC1A2 (EAAT2 or GLT-1) - is enhanced by 1,25D while that of SLC1A3 (EAAT1 or GLAST) and SLC7A11 (xCT) is decreased. Glutamate is not essential for growth of hTERT-HME1 cells, and supplemental glutamate (up to 0.5 mM) does not abrogate the growth inhibitory effects of 1,25D. These data suggest that extracellular glutamate is not a major contributor to cellular energy metabolism in hTERT-HME1 cells under basal conditions and that the growth inhibitory effects of 1,25D are not secondary to its effects on glutamate handling. Instead, the effects of 1,25D on glutamate transporters translated to a decrease in cellular glutamate concentration and an increase in media glutamate concentration, suggesting that one or more of these transporters functions to export glutamate in response to 1,25D exposure. The reduced cellular glutamate concentration may also reflect its incorporation into the cellular glutathione (GSH) pool, which is increased upon 1,25D treatment. In support of this concept, the expression of GCLC (which codes for the rate-limiting enzyme in GSH synthesis) and genes which generate reducing equivalents in the form of NADPH (ie, G6PD, PGD, IDH2) are elevated in 1,25D treated cells. Taken together, these data identify 1,25D as a physiological regulator of multiple membrane glutamate transporters that impacts on overall cellular glutamate handling.
Keywords: Mammary cells, glutamate, glutamine, vitamin D, glutathione
INTRODUCTION
Glutamate, a negatively-charged non-essential amino acid, has been extensively studied for its role as a neurotransmitter in the central nervous system. Aberrant glutamate metabolism and transport has been linked to several neurological conditions including Alzheimer’s disease, schizophrenia, obsessive compulsive disorder and depression (1, 2). Recently, an emerging role for glutamate signaling and/or metabolic flux in cancer has been suggested by in vitro studies. Blocking binding of extracellular glutamate to ionotrophic glutamate receptors (GluRs) inhibits growth of several cancer types, including breast carcinomas, through induction of apoptosis, inhibition of cell division, and reduction of cell motility (3). Furthermore, intracellular glutamate can be metabolized to produce ATP and macromolecules to support cancer cell proliferation (4) and is essential for synthesis of glutathione (GSH). Extensive research has focused on glutamine uptake and catabolism (via glutaminase) as the major source of intracellular glutamate in cancer cells. The metabolic flux of glutamine to glutamate is regulated by oncogenes such as MYC and RAS, which are often altered in breast cancer (5). Despite the interest in glutamine-glutamate flux as a mediator of the metabolic switch in breast cancer, there is limited data on the expression and function of glutamate transporters in normal or cancerous mammary cells.
We recently reported that 1,25-dihydroxyvitamin D3 (1,25D), the ligand for the Vitamin D Receptor (VDR) enhances expression of the glutamate transporter SLC1A1 in two immortalized normal human mammary epithelial cell lines (hTERT-HME1 and HME) as well as in DCIS.com cells (a model of ductal carcinoma in situ). SLC1A1 codes for the excitatory amino acid transporter 3 (EAAT3), a membrane transporter with high specificity for glutamate and cysteine (6). Although not well-studied in mammary gland or human breast cancer, increased expression of SLC1A1 is correlated with differentiation of glioma cells, and SLC1A1-knockout mice exhibit increased oxidative stress due to GSH depletion (7, 8). Additionally, mTOR signaling upregulates SLC1A1 in Xenopus laevis oocytes (9). Interestingly, the induction of SLC1A1 by 1,25D observed in hTERT-HME1, HME and DCIS.com cells was abrogated in MCF10A cells (which have MYC amplification) and in breast cancer cells MCF7 and Hs578T (10). In addition, induction of SLC1A1 gene expression by 1,25D was blunted in HME cells expressing SV-40 (HME-LT cells) and those expressing SV-40 plus oncogenic RAS (HME-PR cells) (11). These studies suggest that 1,25D enhances SLC1A1 expression in normal mammary epithelial cells but that expression of this gene and its regulation by 1,25D is often abrogated in breast cancer cells. Based on these previous studies, we hypothesized that 1,25D mediated up-regulation of SLC1A1 would alter glutamate handling in hTERT-HME1 cells. Our results confirm that 1,25D increases expression of the EAAT3 transporter in mammary epithelial cells and support the concept that this membrane transporter regulates cellular glutamate handling in response to 1,25D exposure.
MATERIALS AND METHODS
Cell lines and cell culture
hTERT-HME1 cells were originally purchased from Clontech as the Infinity™ Human Mammary Epithelial Cell Line (currently available from ATCC). This line was derived from non-tumorigenic mammary epithelial cells immortalized by retroviral transfection of the human telomerase reverse transcriptase (TERT). Cells were maintained in serum free M171 media plus mammary epithelial growth supplement (MEGS, Life Technologies, Grand Island, NY) in a 37°C and 5% CO2 incubator and passaged every 3–4 days. For experiments in which glutamate or glutamine concentrations were varied, cells were plated in M171 media (which contains 0.1 mM glutamate) and grown for 24h before switching to custom glutamine and glutamate free mammary epithelial cell growth medium (PromoCell, Heidelberg, DE) plus MEGS to which glutamate and/or glutamine were added back (Sigma Aldrich, St. Louis, MO).
Cell density assays
Cells were plated in M171 media in 24-well plates at a density of 10,000 cells per well. Twenty-four hours post-attachment, cells were switched to Promocell custom media containing 0–0.5 mM glutamate and/or glutamine in the presence or absence of 100 nM 1,25D (Sigma Aldrich, St. Louis, MO). After 96h, media was removed and adherent cells were fixed with 1% glutaraldehyde. After removal of glutaldehyde, monolayers were stained with 0.1% crystal violet, solution was removed and plates were air-dried overnight. After suspension of stained cell monolayers in 0.2% Triton X-100, absorbance was read at 590 nm on a Victor 3 Perkin Elmer Multilabel Plate Reader (Waltham, MA). In some experiments cells were treated 24h post-attachment with the EAAT inhibitor DL-threo-β-Benzyloxyaspartic acid (DL-TBOA; Tocris Bioscience, Bristol, UK) at 200 uM in the presence or absence of 100nM 1,25D.
Gene expression analysis
For quantitative PCR, cells were plated in 100 mm dishes and treated with 100 nM 1,25D or ethanol vehicle 24h after attachment. Twenty-four hours post-treatment, RNA was isolated with the Qiagen RNeasy kit (Qiagen, Valencia, CA) and purity and concentration were measured on a Nanodrop 1000 Spectrophotometer. TaqMan Reverse Transcriptase reagents (Life Technologies, Grand Island, NY) were used to prepare cDNA which was analyzed in duplicate using SYBR Green PCR Master Mix (ABgene – Thermo Scientific, Pittsburgh, PA) on an ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). Primer sequences for SLC1A1, GCLC, G6PD, PGD, IDH2 and other glutamate transporters were from Origene (Rockville, MD) whereas primers for detection of specific SLC1A1 variants were as reported in Porton et al (12). All primers were synthesized by IDTDNA (Coralville, IA) and sequences are listed in Supplemental Table 1. Data were calculated using the ΔΔCt method, with normalization of CT values for gene-specific primers to that of 18S. Values from 1,25D-treated cells were expressed relative to values from control cells.
Western blotting
Cells were plated in 100 mm dishes and treated with 100 nM 1,25D or vehicle 24 h post-attachment. Whole cell lysates were collected by scraping after 48 h treatment and sonicated in 2× Laemmli Buffer. Protein concentrations were measured using the Pierce BCA Protein Assay (Thermo Scientific, Rockford, IL). Samples containing 50 ug of protein were separated on SDS-PAGE gels, transferred to PVDF membranes using semi-dry transfer, blocked for 1 h in 5% skim milk/PBS and incubated at 4°C overnight with primary antibody directed against EAAT3, the protein product of the SLC1A1 gene (Epitomics, Cambridge, MA) at a dilution of 1:12,000 in 5% skim milk/PBS/0.1% Tween. After 1 h incubation with an anti-rabbit ECL HRP-linked secondary antibody (GE Healthcare, Buckinghamshire, UK) at a dilution of 1:5,000 in 5% skim milk/PBS/0.1% Tween, membranes were developed with Pierce ECL 2 Western Blotting Substrate (Thermo Scientific, Waltham, MA) and imaged on a Storm 860 Molecular Imaging System (GE Healthcare, Pittsburgh, PA). Blots were stripped with acetonitrile and re-probed with a GAPDH primary antibody (AbD Serotec, Raleigh, NC) diluted 1:16,000, followed by incubation with an anti-mouse ECL HRP-linked secondary antibody (GE Healthcare, Buckinghamshire, UK) diluted 1:5,000. Specific bands recognizing EAAT3 and GAPDH were quantified with ImageQuant software and data for EAAT3 expression were normalized to that of GAPDH after background correction.
Glutamate Assay
For cellular glutamate measurements, 106 cells were plated in 100 mm dishes and cultured for 24 h prior to treatment with 100 nM 1,25D, 200uM L-TBOA or vehicle. After an additional 48 h, cells were trypsinized and counted. For assay, 106 cells from each treatment condition were pelleted, resuspended in glutamate assay buffer (BioVision, Milpitas, CA), sonicated and centrifuged. Supernatants, representing clarified cell lysates, were collected for glutamate assay. For measurement of media glutamate content, 100,000 hTERT-HME1 cells per well were plated in 12-well plates and treated exactly as above. After 48h treatment, media was removed for glutamate assay and adherent cells were fixed and stained with crystal violet for assessment of cell density as described above. Glutamate content of lysates and media was determined in a 30 minute reaction with the glutamate colorimetric assay kit as recommended by manufacturer (BioVision, Milpitas, CA). Absorbance was read at 450 nm on a Victor 3 Perkin Elmer Multilabel Plate Reader (Waltham, MA). Intracellular glutamate content was expressed per 106 cells and media content was expressed relative to cell density.
Glutathione Assay
hTERT-HME1 cells (106 cells per 100 mm dish) were treated 24 h after plating with 100 nM 1,25D or vehicle. After 48 h treatment, 106 cells were counted and deproteinated using 5% metaphosphoric acid. Samples diluted in NaPO4/EDTA buffer were analyzed for glutathione concentrations with the Bioxytech GSH/GSSG-412 nm assay (Percipio Biosciences, Burlingame, CA). Briefly, 5,5’-Dithiobis-(2-nitrobenzoic acid) (DTNB) reacts with GSH to form 2TNB, a product that is detectable at 412 nm. Additionally, any oxidized glutathione is recycled into GSH by addition of glutathione reductase and NADPH allowing for measurement of total GSH. Final GSH content was expressed per 106 cells.
Statistical Analysis
Graphpad Prism software (La Jolla, CA) was utilized for statistical analysis. All data is expressed as mean ± standard deviation. For comparison of experiments with more than two groups, significance was determined by one-way ANOVA followed by Tukey post-test. In graphs, mean values that are significantly different (p <0.05) are indicated by different letters above each bar. For experiments with two conditions, significance was measured by Student’s t-test and mean values that are significantly different (p <0.05) are indicated by asterisks above the bars.
RESULTS
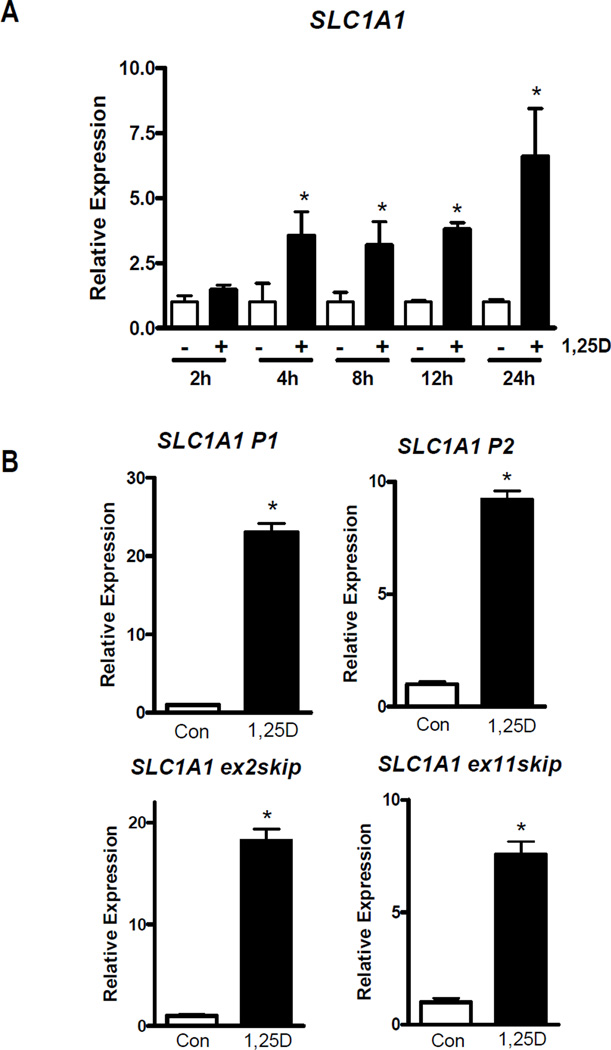
Previous microarray studies identified SLC1A1 as an up-regulated gene in non-transformed mammary cells treated with 100 nM 1,25D for 24 h (11, 13). To determine the kinetics of this response, we measured SLC1A1 gene expression by qPCR in hTERT-HME1 cells treated with 100 nM 1,25D or vehicle for 2, 4, 6, 12 and 24 hours. Compared to time-matched vehicle-treated cells, SLC1A1 was upregulated 3-fold within 4 h of 1,25D-treatment (Figure 1A) suggesting that SLC1A1 might be a direct target of the 1,25D-VDR complex. By 24 h, SLC1A1 was upregulated more than 6-fold compared to vehicle-treated cells. We also examined whether 25-hydroxyvitamin D (25D), a circulating metabolite that is internalized and converted to 1,25D in hTERT-HME1 cells (14, 15), could mimic the effect of 1,25D on SLC1A1. Cells treated with 100 nM 25D for 24 h exhibited 3-fold up-regulation of SLC1A1 compared to control cells (Supplemental Figure 1). These data indicate that SLC1A1 gene expression is induced in mammary epithelial cells by the VDR-ligand, 1,25D, and by the physiologically relevant circulating vitamin D metabolite, 25D.
Figure 1. Effect of 1,25D on SLC1A1 gene expression in hTERT-HME1 cells.
A, hTERT-HME1 cells were treated with 100 nM 1,25D or ethanol vehicle for 2, 4, 8, 12 or 24 h after which RNA was isolated and analyzed by qPCR with primers that detect all SLC1A1 transcript variants. B, RNA isolated from cells treated with vehicle (Con) or 100 nM 1,25D for 24 h was analyzed with primers specific for transcripts arising from promoter 1 (P1), promoter 2 (P2) or those lacking exon 2 (ex2skip) or exon 11 (ex11skip). PCR data were normalized to 18S and expressed relative to control values at each time point which were set to 1. Each bar represents mean ± SD of three independent replicates analyzed in duplicate. *p < 0.05 as measured by one-tailed, unpaired student t-test.
The SLC1A1 gene encodes several variants, all of which are detected by the primer utilized for the data shown in Figure 1A. Since these transcripts may have distinct functions, we examined whether 1,25D selectively altered the expression of transcripts derived from the primary promoter (SLC1A1 P1), the newly-described internal promoter (SLC1A1 P2) or variants lacking exon 2 (SLC1A1 exon2skip) or exon 11 (SLC1A1 exon11skip) (12). After 24 h 100 nM 1,25D treatment, expression of all four SLC1A1 transcripts was upregulated between 7–20 fold relative to vehicle-treated cells (Figure 1B). These data suggest that 1,25D is an important regulator of all known SLC1A1 transcript variants in non-transformed mammary epithelial cells.
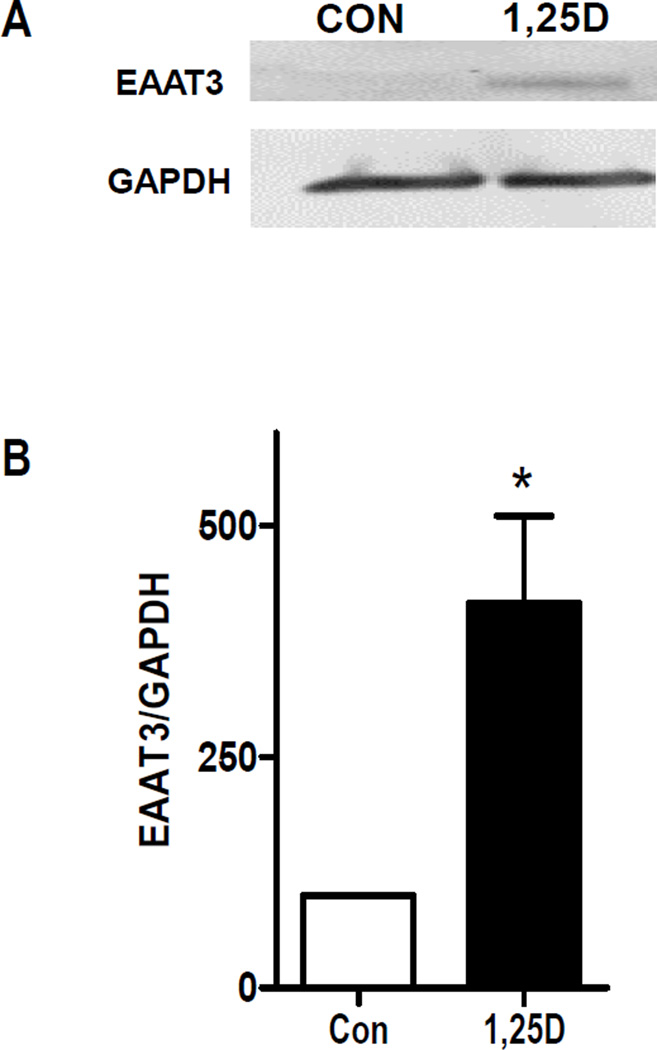
To test whether the upregulated transcription at the SLC1A1 gene locus after 1,25D exposure was sufficient to alter its protein product, we examined the levels of the cognate protein, excitatory amino acid transporter 3 (EAAT3), by western blot. Lysates from hTERT-HME1 cells treated with 100 nM 1,25D or vehicle for 48 h were blotted with an antibody detected against EAAT3. As shown in Figure 2A, western blotting detected specific antibody binding in 1,25D treated cells at approximately 63kDa, the MW at which glycosylated full length EAAT3 runs on SDS-PAGE (16). Little to no EAAT3 protein was detected under basal conditions, suggesting that SLC1A1 gene expression is low in proliferating mammary epithelial cells. Quantitation of triplicate blots (Figure 2B) indicated that 1,25D up-regulated expression of full length EAAT3 approximately 4-fold relative to control cells. No other bands were induced by 1,25D (not shown) suggesting that the variant SLC1A1 transcripts may not be translated in hTERT-HME1 cells.
Figure 2. Expression of EAAT3 in 1,25D-treated hTERT-HME1 cells.
A, hTERT-HME1 cells were treated with 100 nM 1,25D or ethanol vehicle (Con) for 48 h. Whole cell lysates were analyzed by Western blotting with antibodies directed against EAAT3 or GAPDH. Blot represents one of three biological replicates. B, Quantitation of western blot data. Each bar represents mean ± SD of triplicates. *p <0.05 as measured by one-tailed, unpaired student t-test.
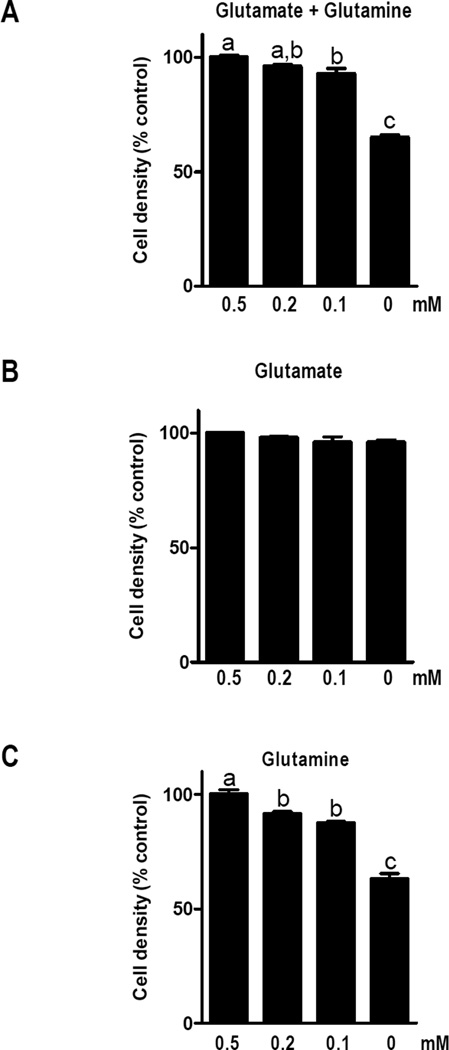
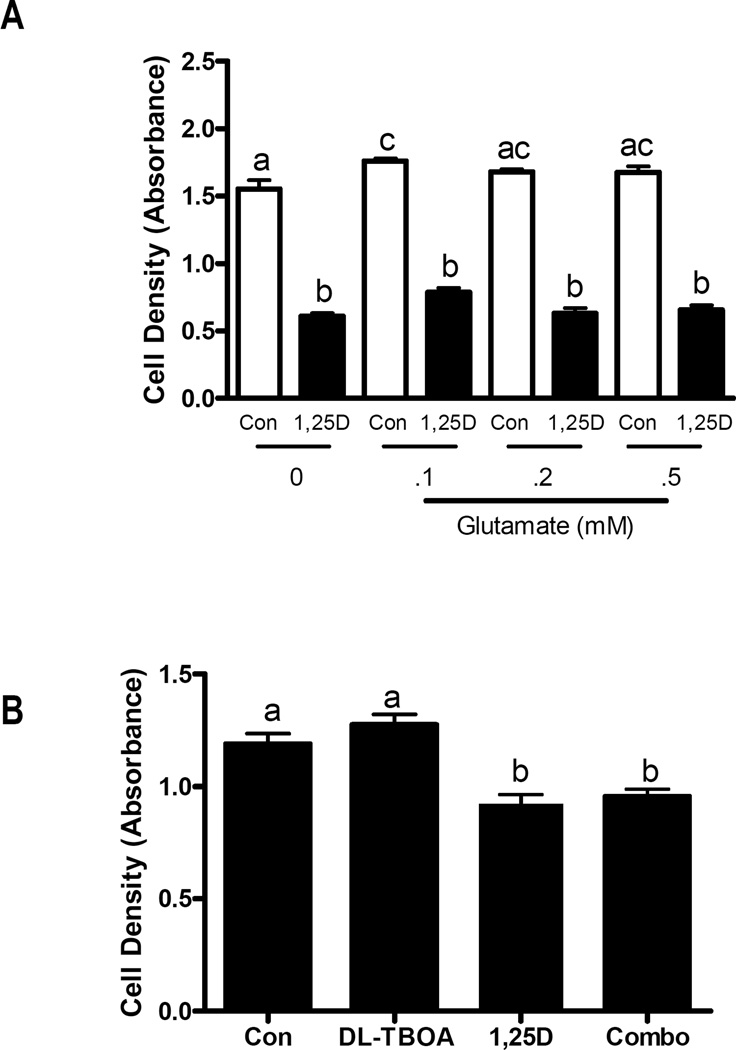
As 1,25D is known to induce growth arrest and differentiation in hTERT-HME1 cells, we tested whether increases in EAAT3 could be mechanistically linked to cell proliferation. We first evaluated whether growth of hTERT-HME1 cultures was dependent on exogenous glutamate. After attachment in full media, cells were switched to custom media devoid of both glutamate and glutamine, and the ability of either amino acid to support growth was evaluated. In the absence of both glutamate and glutamine, cell growth was approximately 60% that of cells grown in the presence of both amino acids (Figure 3A). Interestingly, growth of hTERT-HME1 cells was not significantly reduced in the absence of glutamate (Figure 3B), but was sensitive to glutamine deprivation (Figure 3C). These data indicate that exogenous glutamate is not required for survival or proliferation of hTERT-HME1 cells under basal conditions (ie, when EAAT3 expression is low). To determine if exogenous glutamate influenced the cellular response to 1,25D, cultures were treated with 100 nM 1,25D or vehicle in media containing 0 – 0.5 mM glutamate (in the presence of 0.5 mM glutamine) and density was measured after 96 h. As expected, density of 1,25D treated cultures was significantly decreased compared to control cells, but this effect was not rescued by glutamate supplementation nor enhanced by glutamate deprivation (Figure 4A). These data suggest that even in the presence of 1,25D (when EAAT3 is induced) extracellular glutamate does not alter growth of hTERT-HME1 cells. We also used DL-TBOA, a competitive and non-transportable inhibitor of EAATs (17) to determine whether inhibition of glutamate transport alone would alter cell growth. DL-TBOA did not alter growth of hTERT-HME1 cells under basal conditions or in the presence of 1,25D (Figure 4B).
Figure 3. Effects of glutamate and glutamine on hTERT-HME1 cell density.
Cells grown in complete media M171 for 24 h were switched to custom media containing indicated concentrations of glutamate and/or glutamate. Cell density was measured by crystal violet assay 96 h after media switch. Bars represent the mean ± SD of triplicates. Significance is indicated by letters above the bars: those bars annotated with different letters are significantly different (p<0.05) as assessed by one-way ANOVA and Tukey post-test.
Figure 4. Effects of 1,25D and DL-TBOA on hTERT-HME1 cell density.
A, Cells grown in complete media M171 for 24 h were switched to custom media containing indicated concentrations of glutamate (0, 0.1, 0.2, or 0.5 mM) plus 100nM 1,25D or ethanol vehicle (Con). Cell density was measured after 96 h by crystal violet staining. Data is expressed as absorbance at 590nm which is proportional to cell density. B, Attached cells grown in complete media 171 for 24 h were treated with ethanol vehicle (Con), 100 nM 1,25D, 200 uM DL-TBOA, or the combination of 1,25D and DL-TBOA. After 96 h treatment, culture density was measured after 96 h by crystal violet staining. Data is expressed as absorbance at 590nm which is proportional to cell density. For both A and B, bars represent mean ± SD of triplicates. Significance is indicated by letters above the bars: those bars annotated with different letters are significantly different (p<0.05) as assessed by one-way ANOVA and Tukey post-test.
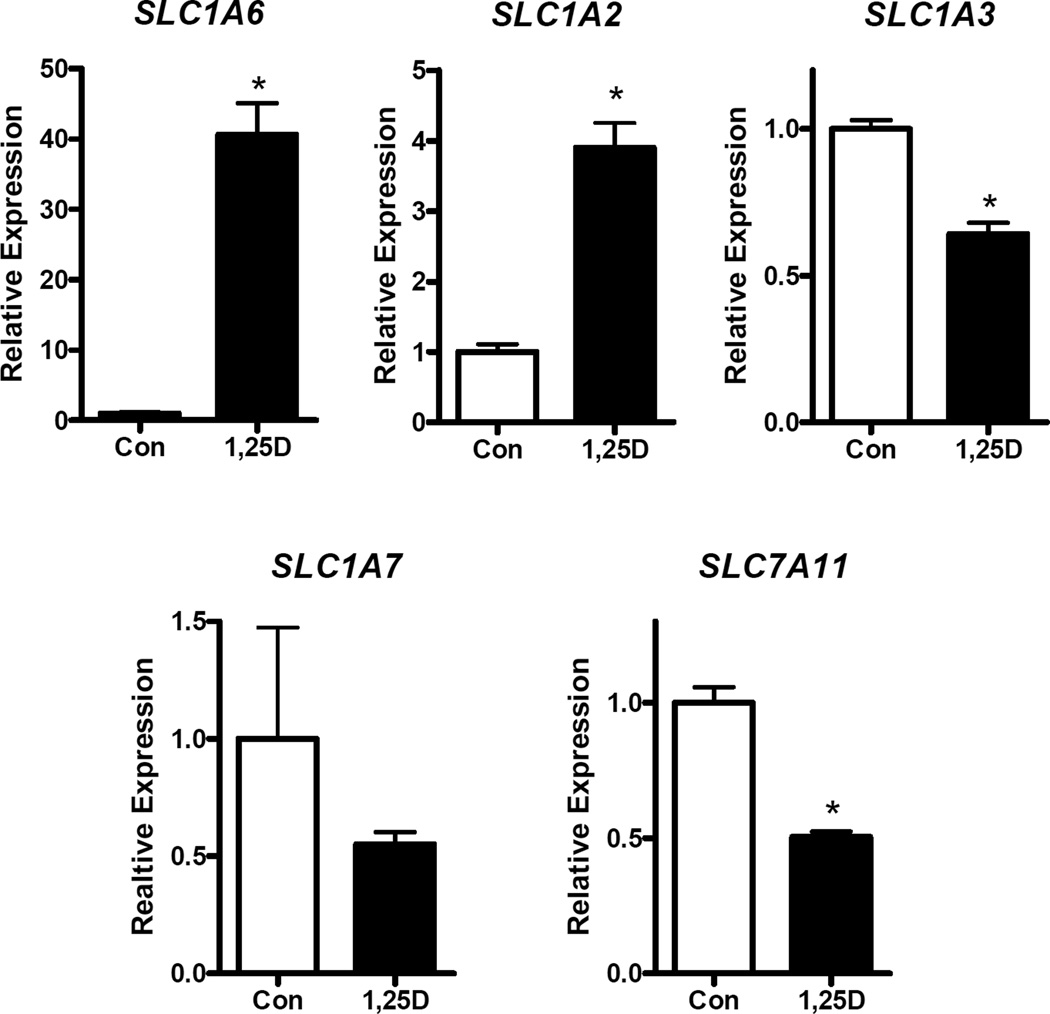
Since there are multiple transporters in the SLC1A family which code for EAATs with distinct transport properties, we measured the expression of other glutamate importers and exporters under basal conditions and after 1,25D treatment. Cells were treated with 100 nM 1,25D for 24 h and expression of SLC1A2 (EAAT2 or GLT-1), SLC1A3 (EAAT1 or GLAST), SLC1A6 (EAAT5), SLC1A7 (EAAT5) and SLC11A7 (xCT) was quantified. Under basal conditions, CT values in the 33–35 range indicated that most of the SLC1A family genes are expressed at low abundance in hTERT-HME1 cells. The exception was SLC1A3 which was robustly expressed (mean CT values of 27). Following 1,25D treatment (Figure 5), significant upregulation of SLC1A2 (4-fold) and SLC1A6 (40-fold) was observed. In contrast, SLC1A7 was not significantly altered and both SLC1A3 and SLC7A11 were down-regulated in the presence of 1,25D. SLC7A11, which encodes the well-studied glutamate exporter xCT, was robustly expressed in hTERT-HME1 cells (mean CT values of 22) and its expression was significantly down-regulated by 1,25D. These data indicate that normal breast epithelial cells in culture express several glutamate transporters that are regulated by 1,25D, with SLC1A1, SLC1A3 and SLC7A11 likely to play biologically significant roles based on their abundance.
Figure 5. Effect of 1,25D on expression of additional glutamate transporters in hTERT-HME1 cells.
RNA isolated from cells treated with vehicle (Con) or 100 nM 1,25D for 24 h was analyzed with primers specific for SLC1A6, SLC1A2, SLC1A3, SLC1A7 and SLC7A11. Data were normalized to 18S and expressed relative to control values which were set to 1. Each bar represents mean ± SD of three independent biological replicates analyzed in duplicate. *p <0.05 as measured by one-tailed, unpaired student t-test.
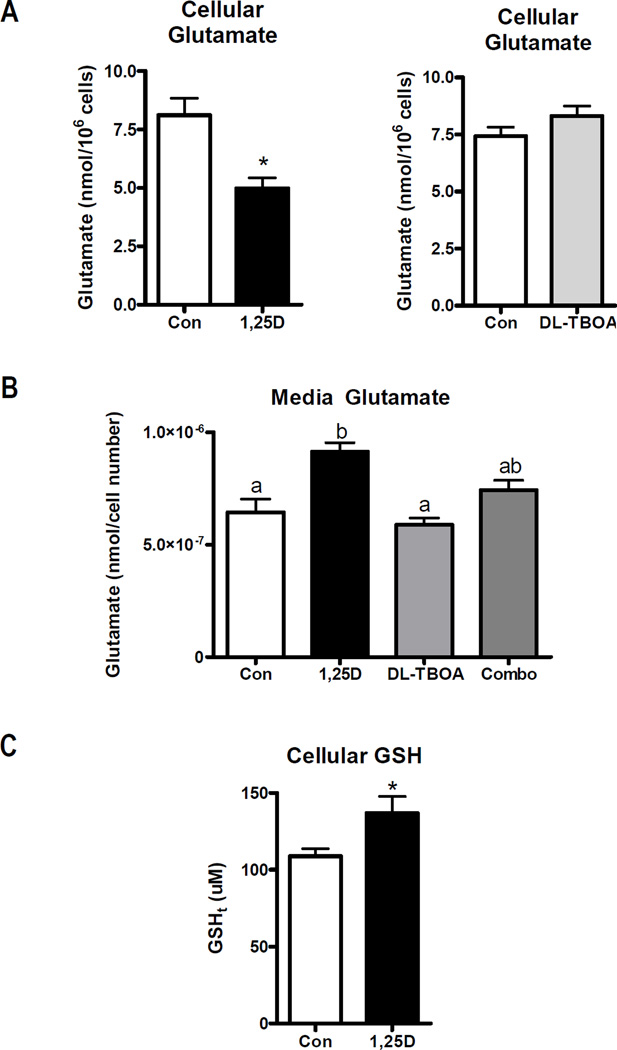
To determine whether the alterations in glutamate transporter expression after 1,25D treatment affected glutamate handling, we assessed both cellular and media glutamate concentrations. hTERT-HME1 cells were treated with 100 nM 1,25D or vehicle for 48 h in media 171 containing glutamate and glutamine. Under these conditions, 1,25D unexpectedly decreased cellular glutamate content whether expressed per 106 cells (as shown in Figure 6A) or as absolute values (49.7 ± 7.7 nmolar in control cells and 24.5 ± 3.8 nmolar in 1,25D-treated cells). Concomitantly, the glutamate content of the media was increased in 1,25D treated cultures (Figure 6A). Interestingly, treatment with the EAAT inhibitor DL-TBOA did not alter cellular or media glutamate content under basal conditions (Figure 6B), possibly due to the relatively low expression of most glutamate transporters in the absence of 1,25D. However, DL-TBOA blunted the increase in media glutamate content induced by 1,25D. This data suggests that one or more DL-TBOA sensitive transporters function to export glutamate in response to 1,25D exposure.
Figure 6. Effect of 1,25D on glutamate and GSH content.
A, Cellular glutamate was measured in lysates from hTERT-HME1 cells treated with vehicle, 100 nM 1,25D or 200 uM DL-TBOA for 48 h. Cell numbers were significantly altered after 48 h of 1,25D treatment (Con, 6.1±0.4 × 106 cells; 1,25D, 4.9±0.5 × 106 cells; p<0.05), therefore data was normalized to 106 cells. B, Media collected from cells treated with 100 nM 1,25D ± 200 uM DL-TBOA for 48 h was analyzed for glutamate content which was expressed relative to culture density evaluated by crystal violet staining of the adherent cells. For both A and B, glutamate was determined with a kinetic colorimetric assay kit as described in methods. C, GSH content was measured in hTERT-HME1 cells treated with 100 nM 1,25D or ethanol vehicle (Con) for 48 h. 106 cells were deproteinated and analyzed with the Bioxytech GSH/GSSG-412 nm assay. For all graphs, bars represent the mean ± SD of triplicates. In A and C, significance was evaluated by one-tailed, unpaired student t-test; * p<0.05. In B, significance was evaluated by one-way ANOVA and Tukey post-test and is indicated by letters above the bars: those bars annotated with different letters are significantly different (p<0.05).
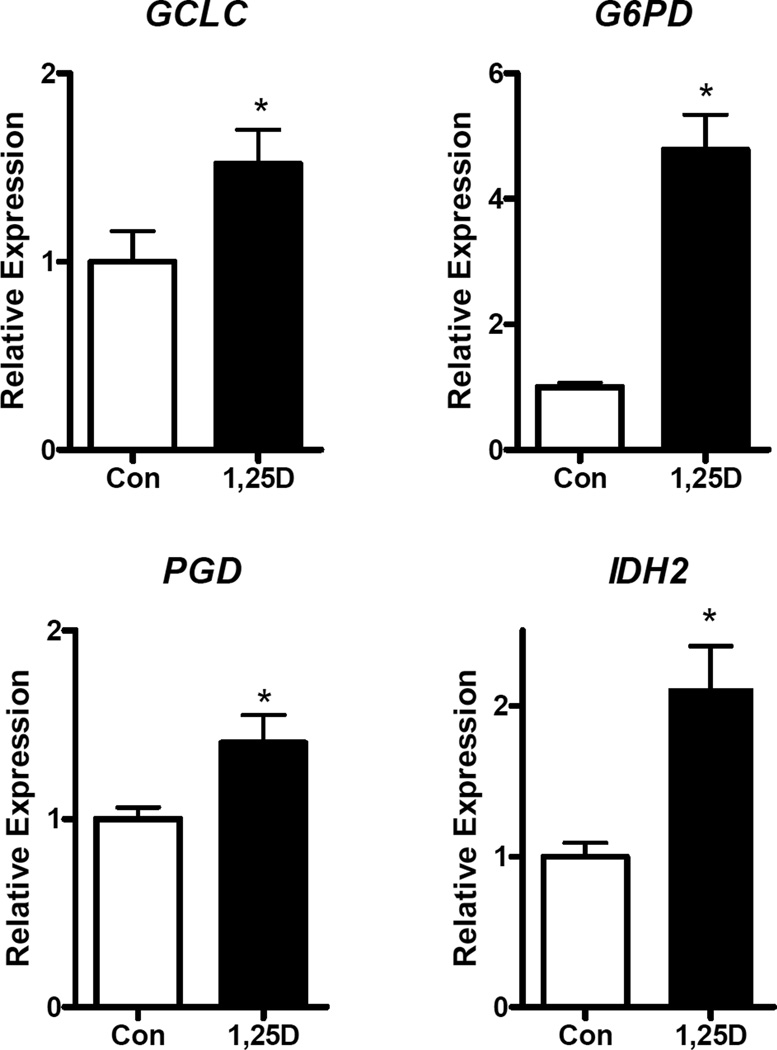
In addition to export, we considered that the reduced content of intracellular glutamate in 1,25D-treated cells could reflect its shunting into the GSH pool. Glutamate is an essential component of glutathione (GSH) and a role for EAAT3 in supplying glutamate for GSH synthesis is supported by studies in Slc1a1-knockout mice which exhibit increased oxidative stress due to low GSH (8). As shown in Figure 6C, 48 h treatment with 1,25D of hTERT-HME1 cells in glutamate-containing media significantly enhanced GSH concentration by ~30%, supporting the idea that intracellular glutamate may be shunted into GSH rather than accumulating as free glutamate. To further explore this concept, we examined the effect of 1,25D on the expression of GSH-related genes including GCLC, which encodes the catalytic subunit of glutamate-cysteine ligase (the rate-limiting enzyme in GSH synthesis) and G6PD (glucose-6-phosphate dehydrogenase), PGD (phosphogluconate dehydrogenase) and IDH2 (isocitrate dehydrogenase 2), metabolic genes that generate NADPH which is essential for maintaining glutathione in its reduced form. As shown in Figure 7, GCLC, G6PD, PGD and IDH2 were all significantly upregulated within 24 h of 1,25D treatment, supporting the concept that vitamin D signaling promotes synthesis and reduction of GSH in hTERT-HME1 cells.
Figure 7. Effect of 1,25D on expression of GSH-related genes in hTERT-HME1 cells.
RNA isolated from cells treated with vehicle (Con) or 100 nM 1,25D for 24 h was analyzed with primers specific for GCLC, G6PD, PGD and IDH2. Data were normalized to 18S and expressed relative to control values which were set to 1. Each bar represents mean ± SD of three independent biological replicates analyzed in duplicate. *p <0.05 as measured by one-tailed, unpaired student t-test.
DISCUSSION
These studies have identified 1,25D, the ligand for the VDR, as a regulator of multiple glutamate transporters in normal human mammary epithelial cells. Although glutamate transporters have been well-studied in the central nervous system due to the role of glutamate as an excitatory neurotransmitter (18–22), the components for glutamate transport and signaling are widely expressed in non-neural systems. However, little is known about the functions of glutamate in these other cell types. The SLC1 family of plasma membrane transporters regulates glutamate influx to maintain optimal intracellular glutamate concentrations for metabolism and GSH synthesis and also can function in the reverse direction to mediate glutamate secretion (16). Recently, accumulation of glutamate has been linked to the epithelial mesenchymal transition (EMT) of mammary epithelial cells (23) and to the aggressiveness of breast cancers (5). Furthermore, deregulation of glutamine to glutamate interconversions has been implicated as a driver of the metabolic switch in breast cancer and this metabolic hub represents a promising therapeutic target (24). Our data demonstrate that 1,25D mediated changes in glutamate transporters are associated with altered cellular glutamate content, suggesting that regulation of glutamate distribution via the SLC1 family of transporters may be mechanistically relevant with respect to the anti-cancer effects of 1,25D.
Using isoform specific PCR primers, we found that 1,25D significantly increased the expression of all four SLC1A1 isoforms described by Porton et al (12). SLC1A1 has previously been identified as a 1,25D regulated gene in neurons and osteoblasts (25, 26), but our work (10, 11) is the first to report such a link in epithelial cells. Here we have explored the relevance of SLC1A1 expression and regulation by 1,25D in mammary cells. The full-length SLC1A1 gene product (EAAT3) was undetectable in untreated cells but clearly induced in response to 1,25D treatment. We found no evidence for expression of the variant SLC1A1 transcripts in untreated or 1,25D treated cells, but further studies will be necessary to confirm this. Intriguingly, 1,25D also up-regulated genes encoding two related transporters SLC1A2 (EAAT2, GLT-1) and SLC1A6 (EAAT4), but exerted opposite effects on SLC1A3 (EAAT1 or GLAST) and SLC7A11 (xCT). Although the specific functional consequences of these transcriptional changes have yet to be precisely defined, the net effect of 1,25D treatment was a reduction in intracellular glutamate content. The observation that media glutamate concentration was concomitantly increased after 1,25D treatment suggests net glutamate secretion, possibly due to SLC1A1, SLC1A2 and/or SLC1A6 encoded transporters functioning in the reverse direction. Although the main function of EAATs is to transport glutamate into the cell, studies have observed reversed glutamate transport, accompanied by an increase in media glutamate concentration (27–29). More detailed measurements of glutamate flux in 1,25D-treated cells will be necessary to test this suggestion. If confirmed, studies to explore how glutamate secretion, which has been demonstrated in breast cancer cells (30), impacts disease progression will be warranted. Although the physiological relevance of glutamate secretion from normal mammary cells has yet to be defined, glutamate release by tumor cells has been shown to act on metabotropic glutamate receptors to alter proliferation, survival and angiogenesis and has also been linked to metastatic bone destruction (31–33).
The reduction in intracellular glutamate may also be secondary to accelerated metabolism or incorporation into GSH pools. In support of the latter, 1,25D treatment increased GSH content and up-regulated GCLC (which codes for the catalytic subunit of Glutamate-Cysteine Ligase, the rate-limiting enzyme in GSH synthesis). The increased GSH content in 1,25D-treated cells is consistent with the known role of EAAT3 in maintenance of intracellular glutamate and cysteine content (34), as well as reports that Slc1a1-null mice exhibit enhanced oxidative stress due to reduced GSH content (8). It is worth noting that 1,25D also transcriptionally up-regulated a set of genes that produce NADPH (G6PD, PGD, IDH2), which provide reducing equivalents for GSH’s function in antioxidant defense. Collectively, our data provides evidence that the reduced intracellular glutamate in 1,25D-treated cells reflects both its secretion into the media and its incorporation into GSH.
Since 1,25D is known to inhibit proliferation of mammary epithelial cells through undefined mechanisms (15), we initially hypothesized that 1,25D-mediated changes in the SLC1 transporters and glutamate flux would alter cellular energy metabolism sufficiently to impact cell proliferation. However, our data indicate that neither glutamate deprivation nor treatment with DL-TBOA (a pan-inhibitor of the EAAT glutamate transporters) altered growth of hTERT-HME1 cultures. Furthermore, although treatment with 1,25D markedly reduced cell growth as expected, this growth inhibition was not rescued by increased exogenous glutamate nor was it abrogated in the presence of DL-TBOA. Therefore, the regulation of glutamate transporters and cellular glutamate handling by 1,25D appears mechanistically unrelated to its growth inhibitory effects in mammary epithelial cells.
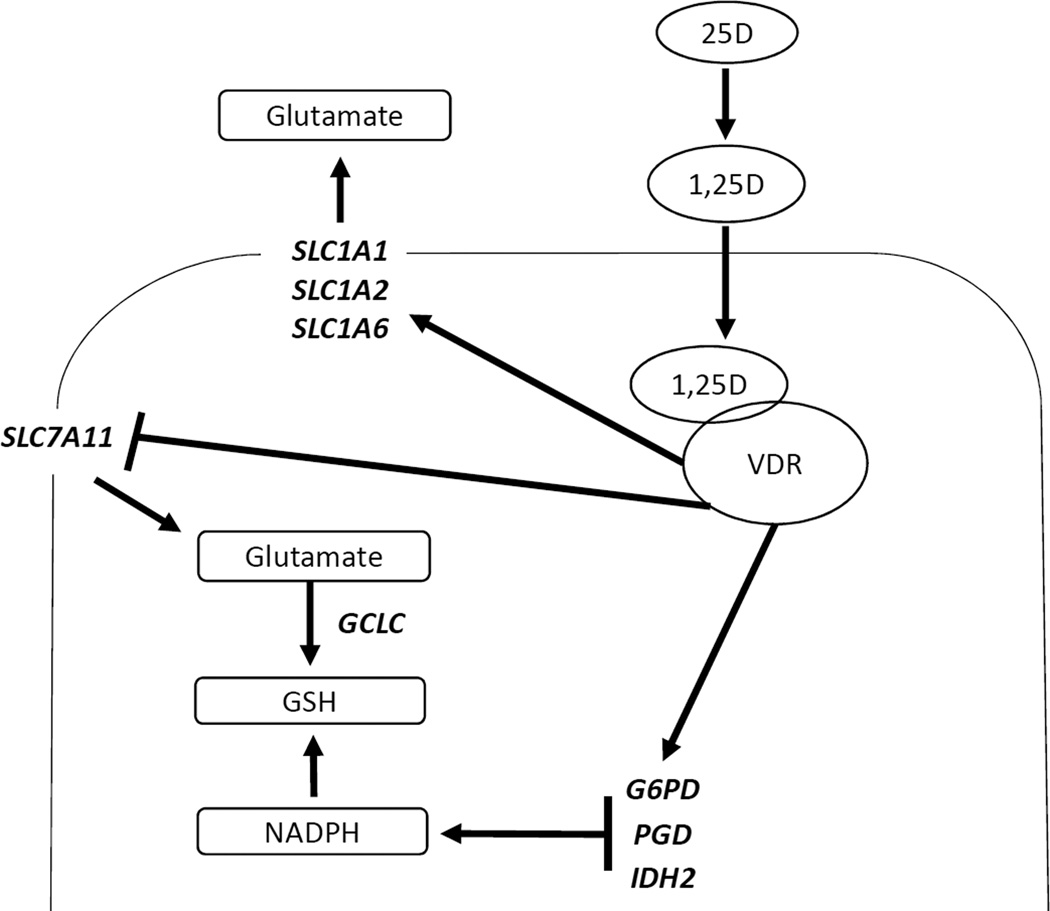
As shown in our model (Figure 8), 1,25D significantly up-regulates three glutamate transporters (SLC1A1, SLC1A2, and SLC1A6) in mammary epithelial cells. Although these transcriptional changes do not appear to directly contribute to the anti-proliferative effects of 1,25D, they are associated with altered cellular content of glutamate and GSH and net glutamate secretion. 1,25D also enhances expression of GCLC, G6PD, PGD, and IDH2, genes which function to increase synthesis of GSH and NADPH for protection against oxidative stress. The links between these cellular changes and the anti-cancer effects of vitamin D signaling warrant continued investigation.
Figure 8. Model depicting effects of 1,25D on glutamate metabolism in hTERT-HME1 cells.
Circulating 25D is internalized and converted to 1,25D which binds VDR to trigger up-regulation of multiple plasma membrane glutamate transporters (SLC1A1, SLC1A2 and SLC1A6) and GSH and NADPH-producing metabolic enzymes (GCLC, G6PD, PGD, IDH2). Concomitantly, other glutamate exchangers (SLC7A11, SLC1A3) are down-regulated. The net result of these transcriptional changes is reduction of intracellular glutamate content, accumulation of glutamate in media and increased total glutathione (GSH) pool.
Supplementary Material
Highlights.
1,25D upregulates glutamate transporters SLC1A1, SLC1A2 & SLC1A6
1,25D reduces cellular glutamate and enhances net glutamate secretion
Growth of mammary epithelial cells is not dependent on exogenous glutamate
Supplemental glutamate does not abrogate growth inhibitory effects of 1,25D
1,25D upregulates GSH content and expression of GCLC, G6PD, PGD and IDH2
Acknowledgments
This work was supported by grant R21CA166434 to JW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Chakrabarty K, Bhattacharyya S, Christopher R, Khanna S. Glutamatergic dysfunction in OCD. Neuropsychopharmacology. 2005 Sep;30(9):1735–1740. doi: 10.1038/sj.npp.1300733. [DOI] [PubMed] [Google Scholar]
- 2.Hynd MR, Scott HL, Dodd PR. Glutamate-mediated excitotoxicity and neurodegeneration in Alzheimer's disease. Neurochem Int. 2004 Oct;45(5):583–595. doi: 10.1016/j.neuint.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Rzeski W, Turski L, Ikonomidou C. Glutamate antagonists limit tumor growth. Proc Natl Acad Sci U S A. 2001 May 22;98(11):6372–6377. doi: 10.1073/pnas.091113598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassago A, Ferreira AP, Ferreira IM, Fornezari C, Gomes ER, Greene KS, et al. Mitochondrial localization and structure-based phosphate activation mechanism of Glutaminase C with implications for cancer metabolism. Proc Natl Acad Sci U S A. 2012 Jan 24;109(4):1092–1097. doi: 10.1073/pnas.1112495109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budczies J, Pfitzner BM, Gyorffy B, Winzer KJ, Radke C, Dietel M, et al. Glutamate enrichment as new diagnostic opportunity in breast cancer. Int J Cancer. 2015 Apr 1;136(7):1619–1628. doi: 10.1002/ijc.29152. [DOI] [PubMed] [Google Scholar]
- 6.Jensen AA, Fahlke C, Bjorn-Yoshimoto WE, Bunch L. Excitatory amino acid transporters: recent insights into molecular mechanisms, novel modes of modulation and new therapeutic possibilities. Curr Opin Pharmacol. 2015 Feb;20C:116–123. doi: 10.1016/j.coph.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Bianchi MG, Gazzola GC, Tognazzi L, Bussolati O. C6 glioma cells differentiated by retinoic acid overexpress the glutamate transporter excitatory amino acid carrier 1 (EAAC1) Neuroscience. 2008 Feb 19;151(4):1042–1052. doi: 10.1016/j.neuroscience.2007.11.055. [DOI] [PubMed] [Google Scholar]
- 8.Aoyama K, Suh SW, Hamby AM, Liu J, Chan WY, Chen Y, et al. Neuronal glutathione deficiency and age-dependent neurodegeneration in the EAAC1 deficient mouse. Nat Neurosci. 2006 Jan;9(1):119–126. doi: 10.1038/nn1609. [DOI] [PubMed] [Google Scholar]
- 9.Almilaji A, Pakladok T, Guo A, Munoz C, Foller M, Lang F. Regulation of the glutamate transporter EAAT3 by mammalian target of rapamycin mTOR. Biochem Biophys Res Commun. 2012 May 4;421(2):159–163. doi: 10.1016/j.bbrc.2012.03.109. [DOI] [PubMed] [Google Scholar]
- 10.Beaudin SG, Robilotto S, Welsh J. Comparative regulation of gene expression by 1,25-dihydroxyvitamin D3 in cells derived from normal mammary tissue and breast cancer. J Steroid Biochem Mol Biol. 2015 Apr;148:96–102. doi: 10.1016/j.jsbmb.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmons KM, Beaudin SG, Narvaez CJ, Welsh J. Gene Signatures of 1,25-Dihydroxyvitamin D Exposure in Normal and Transformed Mammary Cells. J Cell Biochem. 2015 Mar 3; doi: 10.1002/jcb.25129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porton B, Greenberg BD, Askland K, Serra LM, Gesmonde J, Rudnick G, et al. Isoforms of the neuronal glutamate transporter gene, SLC1A1/EAAC1, negatively modulate glutamate uptake: relevance to obsessive-compulsive disorder. Transl Psychiatry. 2013;3:e259. doi: 10.1038/tp.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beaudin SG, Robilotto S, Welsh J. Comparative regulation of gene expression by 1,25-dihydroxyvitamin D in cells derived from normal mammary tissue and breast cancer. The Journal of steroid biochemistry and molecular biology. 2014 Sep 16; doi: 10.1016/j.jsbmb.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowling MJ, Kemmis CM, Taffany DA, Welsh J. Megalin-mediated endocytosis of vitamin D binding protein correlates with 25-hydroxycholecalciferol actions in human mammary cells. J Nutr. 2006 Nov;136(11):2754–2759. doi: 10.1093/jn/136.11.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemmis CM, Salvador SM, Smith KM, Welsh J. Human mammary epithelial cells express CYP27B1 and are growth inhibited by 25-hydroxyvitamin D-3, the major circulating form of vitamin D-3. J Nutr. 2006 Apr;136(4):887–892. doi: 10.1093/jn/136.4.887. [DOI] [PubMed] [Google Scholar]
- 16.Kanai Y, Clemencon B, Simonin A, Leuenberger M, Lochner M, Weisstanner M, et al. The SLC1 high-affinity glutamate and neutral amino acid transporter family. Mol Aspects Med. 2013 Apr-Jun;34(2–3):108–120. doi: 10.1016/j.mam.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Shimamoto K. Glutamate transporter blockers for elucidation of the function of excitatory neurotransmission systems. Chem Rec. 2008;8(3):182–199. doi: 10.1002/tcr.20145. [DOI] [PubMed] [Google Scholar]
- 18.Watkins JC, Jane DE. The glutamate story. Br J Pharmacol. 2006 Jan;147(Suppl 1):S100–S108. doi: 10.1038/sj.bjp.0706444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krnjevic K. When and why amino acids? J Physiol. 2010 Jan 1;588(Pt 1):33–44. doi: 10.1113/jphysiol.2009.176990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skerry TM, Genever PG. Glutamate signalling in non-neuronal tissues. Trends Pharmacol Sci. 2001 Apr;22(4):174–181. doi: 10.1016/s0165-6147(00)01642-4. [DOI] [PubMed] [Google Scholar]
- 21.Nedergaard M, Takano T, Hansen AJ. Beyond the role of glutamate as a neurotransmitter. Nat Rev Neurosci. 2002 Sep;3(9):748–755. doi: 10.1038/nrn916. [DOI] [PubMed] [Google Scholar]
- 22.Hinoi E, Takarada T, Ueshima T, Tsuchihashi Y, Yoneda Y. Glutamate signaling in peripheral tissues. Eur J Biochem. 2004 Jan;271(1):1–13. doi: 10.1046/j.1432-1033.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 23.Bhowmik SK, Ramirez-Pena E, Arnold JM, Putluri V, Sphyris N, Michailidis G, et al. EMT-Induced Metabolite Signature Identifies Poor Clinical Outcome. Oncotarget. 2015 Jul 30; doi: 10.18632/oncotarget.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meadows AL, Kong B, Berdichevsky M, Roy S, Rosiva R, Blanch HW, et al. Metabolic and morphological differences between rapidly proliferating cancerous and normal breast epithelial cells. Biotechnol Prog. 2008 Mar-Apr;24(2):334–341. doi: 10.1021/bp070301d. [DOI] [PubMed] [Google Scholar]
- 25.Tarroni P, Villa I, Mrak E, Zolezzi F, Mattioli M, Gattuso C, et al. Microarray analysis of 1,25(OH)(2)D(3) regulated gene expression in human primary osteoblasts. J Cell Biochem. 2012 Feb;113(2):640–649. doi: 10.1002/jcb.23392. [DOI] [PubMed] [Google Scholar]
- 26.Nissou MF, Brocard J, El Atifi M, Guttin A, Andrieux A, Berger F, et al. The transcriptomic response of mixed neuron-glial cell cultures to 1,25-dihydroxyvitamin D3 includes genes limiting the progression of neurodegenerative diseases. J Alzheimers Dis. 2013;35(3):553–564. doi: 10.3233/JAD-122005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jabaudon D, Scanziani M, Gahwiler BH, Gerber U. Acute decrease in net glutamate uptake during energy deprivation. Proc Natl Acad Sci U S A. 2000 May 9;97(10):5610–5615. doi: 10.1073/pnas.97.10.5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Billups B, Attwell D. Modulation of non-vesicular glutamate release by pH. Nature. 1996 Jan 11;379(6561):171–174. doi: 10.1038/379171a0. [DOI] [PubMed] [Google Scholar]
- 29.Szatkowski M, Barbour B, Attwell D. Non-vesicular release of glutamate from glial cells by reversed electrogenic glutamate uptake. Nature. 1990 Nov 29;348(6300):443–446. doi: 10.1038/348443a0. [DOI] [PubMed] [Google Scholar]
- 30.Willard SS, Koochekpour S. Glutamate signaling in benign and malignant disorders: current status, future perspectives, and therapeutic implications. Int J Biol Sci. 2013;9(7):728–742. doi: 10.7150/ijbs.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banda M, Speyer CL, Semma SN, Osuala KO, Kounalakis N, Torres Torres KE, et al. Metabotropic glutamate receptor-1 contributes to progression in triple negative breast cancer. PLoS One. 2015;9(1):e81126. doi: 10.1371/journal.pone.0081126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fazzari J, Lin H, Murphy C, Ungard R, Singh G. Inhibitors of glutamate release from breast cancer cells; new targets for cancer-induced bone-pain. Sci Rep. 2015;5:8380. doi: 10.1038/srep08380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Speyer CL, Hachem AH, Assi AA, Johnson JS, DeVries JA, Gorski DH. Metabotropic glutamate receptor-1 as a novel target for the antiangiogenic treatment of breast cancer. PLoS One. 2014;9(3):e88830. doi: 10.1371/journal.pone.0088830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanai Y, Hediger MA. The glutamate/neutral amino acid transporter family SLC1: molecular, physiological and pharmacological aspects. Pflugers Arch. 2004 Feb;447(5):469–479. doi: 10.1007/s00424-003-1146-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.