Abstract
Succinylation refers to modification of lysine residues with succinyl groups donated by succinyl-CoA. Sirtuin5 (Sirt5) is a mitochondrial NAD+-dependent deacylase that catalyzes the removal of succinyl groups from proteins. Sirt5 and protein succinylation are conserved across species, suggesting functional importance of the modification. Sirt5 loss impacts liver metabolism but the role of succinylation in the heart has not been explored. We combined affinity enrichment with proteomics and mass spectrometry to analyze total succinylated lysine content of mitochondria isolated from WT and Sirt5−/− mouse hearts. We identified 887 succinylated lysine residues in 184 proteins. 44 peptides (5 proteins) occurred uniquely in WT samples, 289 (46 proteins) in Sirt5−/− samples, and 554 (133 proteins) were common to both groups. The 46 unique proteins in Sirt5−/− heart participate in metabolic processes such as fatty acid β-oxidation (Eci2) and branched chain amino acid catabolism, and include respiratory chain proteins (Ndufa7, 12, 13, Dhsa). We performed label-free analysis of the peptides common to WT and Sirt5−/− hearts. 16 peptides from 9 proteins were significantly increased in Sirt5−/− by at least 30%. The adenine nucleotide transporter 1 showed the highest increase in succinylation in Sirt5−/− (108.4 fold). The data indicate that succinylation is widespread in the heart and enriched in metabolic pathways. We examined whether the loss of Sirt5 would impact ischemia-reperfusion (I/R) injury and we found an increase in infarct size in Sirt5−/− hearts compared to WT littermates (68.5+/− 1.1% Sirt5−/− vs 39.6+/− 6.8% WT) following 20 minutes of ischemia and 90 minutes reperfusion. We further demonstrate that I/R injury in Sirt5−/− heart is restored to WT levels by pretreatment with dimethyl malonate, a competitive inhibitor of succinate dehydrogenase (SDH), implicating alteration in SDH activity as causative of the injury.
Keywords: Sirt5, Ischemia-reperfusion, Cardiac, Succinylation, Succinate
Introduction
An emerging class of post-translational modifications (PTMs) involves modification of lysine side chains with thioester-coenzyme A metabolites [1–5]. Succinylation refers to the modification of lysine residues with succinyl groups that are donated by the intermediate metabolite succinyl-CoA (SucCoA) [4]. The phenomenon of protein succinylation is conserved across species, and has been identified in E. coli, S. cerevisiae, D. melanogaster, M. musculus, and H. sapiens [4, 6–10]. To date, a cellular succinyltransferase has not been identified, and the reaction of SucCoA with lysine side chains is presumed to occur spontaneously [11]. Conditions of the mitochondrial matrix, where pH is relatively high and SucCoA is concentrated, support this spontaneous reaction, thus it is not surprising that succinylated proteins are enriched in mitochondria [7, 9]. Manipulation of cellular SucCoA levels via genetic deletion of TCA enzymes in yeast demonstrates that global protein succinylation positively correlates with SucCoA levels [7]. SucCoA levels reflect the metabolic status of a cell, suggesting that protein succinylation and the intermediate metabolite SucCoA function as a metabolic signaling pathway. The mammalian Sirtuin (Sirt) family of proteins includes seven members, three of which (Sirt3, Sirt4, and Sirt5) are predominantly located in the mitochondria. Sirt5 catalyzes the desuccinylation of lysine residues in a reaction that requires consumption of NAD+ [12]. The study of Sirt5 and its desuccinylase activity has recently been described, and the general recognition that succinylation is a bona fide PTM and the ability of Sirt5 to regulate it in liver was only recently reported [12]. Succinylation can both increase and decrease protein activity. Succinylation of protein components of the pyruvate and succinate dehydrogenase complexes correlates with increased complex activity [9]. Conversely, succinylation of 3-hydroxy-3-methylglutaryl-CoA synthetase 2 (Hmgcs2) decreases activity of the enzyme, and ketogenesis is altered in the Sirt5 knockout mouse liver [10].
The function of Sirt5 in the heart has not been explored. The unrelenting energy needs of the heart underlie a crucial need for proper metabolic tuning in cardiac tissue. Although metabolic remodeling in liver in the Sirt5−/− mouse has been described,[9, 10, 13], there are little or no data on succinylation in the heart. To investigate this, we combined affinity enrichment with proteomics to capture an image of lysine succinylation in cardiac mitochondria. The data demonstrate that protein succinylation occurs at baseline in heart mitochondria and suggests that Sirt5 functions as a desuccinylase. The data in this paper provide the first description of the cardiac succinylome. We find that succinylation is widespread in the heart, and proteins succinylated in cardiac mitochondria participate in the processes of oxidative phosphorylation, fatty acid oxidation, ketogenesis, and branched chain amino acid catabolism, among others. We investigated whether the metabolic alterations in the Sirt5−/− mice might alter the response of the heart to ischemia-reperfusion injury, and we found that the Sirt5 knockout mouse has increase ischemia-reperfusion (I/R) injury.
Methods
Animals
All animals were treated and cared for in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health (NIH), Revised 2011), and protocols were approved by the Institutional Animal Care and use Committee. The sexes and ages of mice at time of experimentation are indicated within particular method descriptions. The Sirt5−/− mice are on a C57B6 background and have been backcrossed more than 10 generations.
Mitochondria isolation from mouse hearts
Mitochondria were isolated by differential centrifugation according to standard procedures [14]. Briefly, hearts were minced in mitochondrial isotonic buffer (“Buffer B”) consisting of 225 mM mannitol, 75 mM sucrose, 5 mM MOPS, 0.5 mM EGTA, and 2 mM taurine, 5 mM nicotinamide, 1 μM trichostatin A (pH 7.25). Minced heart tissue was homogenized by Polytron. To digest contractile proteins, trypsin (0.001g/0.1g wet tissue) in buffer B was added to homogenized hearts for 5 minutes on ice. Homogenates were centrifuged at 500×g for five minutes (4 deg C) and the resulting supernatant was centrifuged at 11,000×g for five minutes (4 deg C) to pellet mitochondria. Final mitochondrial pellets were resuspended in Buffer B.
Immunoblotting
Protein extracts were quantified with BCA protein assay (Pierce Cat #23225) or Bradford protein assay (Sigma Cat# B6916). Proteins were reduced with 50 mM DTT, denatured with LDS (lithium dodecyl sulfate), heated, and equivalent amounts of protein were loaded on NuPage 4–12% Bis-Tris gels (Invitrogen) and electrophoresed in 1× MES buffer. Protein gels were transferred to nitrocellulose membranes (0.2 μM pore size). Membranes were blocked with 5% milk. Primary antibodies were incubated in 5% BSA overnight at 4 deg C. Secondary antibody incubations were performed in 5% milk at room temperature for 30–45 minutes. Blots were developed with reagents from the Amersham ECL Select Western Blotting kit (Amersham, Cat #RPM2235) and signal was collected with chemiluminescence film (Amersham, Cat #28906834). Sirt5 antibody, Mdh2 antibody, and the antibodies designed to detect acetylated lysine residues were obtained from Cell Signaling Technologies (Sirt5, #8779; Mdh2, #8610; AcK #9441; AcK #9681). The antibody designed to detect succinylated lysine residues was obtained from PTM Biolabs (PTM-401).
Affinity purification of peptides containing succinylated lysine residues
To measure succinylation we used an approach that has been used previously to measure acetylation [2, 15]. This approach employs an antibody against succinyl-lysine (SuK) to precipitate SuK containing peptides, which are then identified by mass spectrometry. Hearts were collected from three male wild type and four Sirt5−/− male mice at 6 months of age. Mitochondria were isolated via differential centrifugation in the presence of deacylase inhibitors (5 mM NAM and 1 μM trichostatin A). Isolated mitochondrial pellets (800 ug) were resuspended in 8.0 M guanidine hydrochloride (Sigma, #G9284). Samples were first reduced with 5 mM DTT for 1 hour at 60 deg C, then alkylated with 15 mM iodoacetamide for 30 minutes at room temperature in the dark. Alkylation reactions were quenched with DTT. Samples were diluted with 25 mM TEAB such that guanidine hydrochloride concentration was < 0.8 M. The samples were then digested overnight at 37 deg C with trypsin (Pierce, V5111) at protein:enzyme ratio of approximately 50:1. The tryptic peptides were acidified with formic acid, and desalted on Oasis HLB 1 cc cartridges (Waters #WAT200677) per manufacturer’s instructions. Collected eluants were lyophilized overnight and resolubilized in NETN buffer (50 mM Tris-Cl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.5% NP-40). Tryptic peptides were enriched via immunoprecipitation with anti-succinyl lysine antibody (PTM Biolabs, PTM-401) immobilized on Protein G agarose beads (Life Technologies, 15920-010). Peptides and antibody-beads were incubated overnight at 4 deg C with end-over-end rotation. The beads were washed 3× with 1 mL NETN buffer, then 2× with 1 mL ETN buffer (50 mM Tris-Cl (pH 8.0), 100 mM NaCl, 1 mM EDTA). Bound succinylated peptides were eluted from the agarose with 0.1% TFA. The eluate was cleaned with C18 tips (Varian Omix, A57003100). Peptides were dried and samples resuspended in 0.1% formic acid for mass spec analysis. The samples were analyzed on an LTQ Orbitrap Elite (Thermo Fisher Scientific, San Jose, CA) coupled with an Eksigent nanoLC-Ultra 1D plus system (Dubin, CA). Peptides were separated on a PicoFrit analytical column (250 mM long, ID 75 μM, tip ID 10 μm, packed with BetaBasic 5 μm 300 Å particles, New Objective, Woburn, MA) using a 160-minute linear gradient of 5–35% acetonitrile in 0.1% formic acid at a flow rate of 250 nL/min. Mass analysis was carried out in data-dependent analysis mode, where MS1 scanned full MS mass range from m/z 300 to 2000 at 60,000 mass resolution and 10 CID MS2 scans were sequentially carried out in the Orbitrap and the ion trap, respectively. The LCMS data were searched against the SwissProt database, taxonomy Mus musculus (mouse) using Mascot server (Matrix Science, London, UK; version 2.4). Searching parameters were set as follows: precursor mass tolerance at 20 ppm, fragment ion mass tolerance at 0.8 Da, trypsin enzyme with 4 miscleavages, carbamidomethylation of cysteine as fixed modification, and deamidation of asparagine and glutamine, oxidation of methionine, and succinylation of lysine as variable modifications. Peptides were filtered with 1.0% false discovery rate (FDR). Relative quantification of succinylated peptides were performed using QUOIL (QUantification without Isotope Labeling), an in-house software program designed as a label-free approach to peptide quantification by LC-MS/MS [16].
Pathway Analysis
Pathway analysis was performed with the Ingenuity Pathway Analysis tool (IPA, Ingenuity Systems, www.ingenuity.com). A dataset containing the UniprotKB IDs for all proteins identified as succinylated was uploaded to the application. The entire mouse proteome was used as the reference set. A core analysis was performed to identify protein function and a canonical pathway analysis was used to identify the pathways to which the proteins mapped. Significance is measured within IPA by two means: 1.) Determination of a ratio calculated by dividing the number of differentially expressed genes that map to a particular pathway by the total number of genes in that pathway; 2.) Calculation of a p-value by Fisher’s exact test to determine the probability that the association between the genes in the dataset and the canonical pathway can be explained by chance.
Langendorff Heart Perfusion
Global ischemia/reperfusion was carried out via Langendorff technique with WT and Sirt5−/− mice. Mice were anesthetized with an intraperitoneal injection of 0.10 cc pentobarbital sodium diluted 1:5 in perfusate. The abdominal cavity was exposed with a transverse incision and 0.05 cc heparin was administered to the inferior vena cava. The heart was quickly isolated and briefly (less than 1 minute) placed in ice-cold Krebs-Heinseleit (KH) buffer (in mmol/L: 25 NaHCO3, 120 NaCl, 11 glucose, 4.7 KCl, 1.2 KH2PO4, 1.2 MgSO4, and 1.75 CaCl2) prior to cannulation. The aorta was cannulated on a Langendorff apparatus and the heart was perfused in retrograde fashion with KH buffer at a constant pressure of 100 cm of water at 37°C. All hearts were perfused with KH buffer gassed with 95% O2 and 5% CO2. Hearts were subjected to 30 minutes of equilibration followed by 20 minutes of ischemia. When indicated, hearts were treated with dimethyl malonate for 20 minutes prior to ischemia.
ROS Measurements of Perfused Hearts
The hearts were Langendorff perfused with KH buffer for 30 min of equilibration followed with 20 min of no-flow global ischemia for I/R control. Dimethyl malonate (DMM, 5 mmol/L, Santa Cruz) was infused in the perfusate during the last 20 min of perfusion for the DMM-I/R group. MitoSox red (2.5 μmol/L) and DAPI (1 μg/mL) (Molecular Probes) as indicators of superoxide and nuclei, respectively, were infused with the perfusate at the onset of reperfusion for 5 minutes. The hearts were then removed and ventricles were cut into ~1.0 mm transverse slices for immediate confocal imaging using a Zeiss LSM 780 confocal microscope equipped with a Plan-Apochromat 63×/1.40 Oil DIC M27 immersion objective. The imaging of DAPI and MitoSox red was achieved by excitation at 405 nm, and the emissions were collected at 420 to 480 nm and 560 to 615 nm, respectively. The Z-Stack scan function of the confocal microscope was used to identify the optimal focal plane; the same focal plane and laser gain were used for all the imaging to ensure the signals among groups are comparable.
Lactate Production
Accumulation of lactate was measured in hearts subjected to 20 minutes of global ischemia. Global ischemia was carried out via Langendorff technique in the manner described above. Hearts isolated from 3 female WT and 3 female Sirt5−/− mice, all between 23–28 weeks of age, were analyzed. After the 20 minutes ischemic interval, hearts were snap frozen, minced, and extracted with 3.6% perchloric acid. Samples were centrifuged at 3000×g for 10 minutes at 4 deg C and the supernatant was neutralized with K2CO3 to pH ~ 7.0. Lactate concentration in each sample was determined with a colorimetric Lactate Assay Kit (Biovision, cat# K607-100).
Sample preparation and labeling with tandem mass tags (TMT)
To measure total protein abundance, hearts were isolated from three male WT and three male Sirt5−/− mice, all between 14–16 weeks of age, in the manner described previously in these methods. Hearts were quickly isolated, washed in ice-cold PBS, and minced. 1.0 mL lysis buffer (7 M Urea, 2 M Thiourea, 4% CHAPS) was added to minced heart tissue and Precellys homogenizing beads (Precellys, KT 03961-1-009.2). Homogenization was carried out in a Precellys Homogenizer (Bertin Technologies, chilled with liquid nitrogen. The samples spun at 6500 rpm for two 20-second cycles. Extracted protein was quantified with Bradford assay (Sigma, B6916). 100 ug protein from each heart were reduced with 10 mM TCEP (2 hours at room temperature), then alkylated with 17 mM iodoacetamide (30 minutes at room temperature in the dark). 6 volumes of ice cold acetone were added and protein was precipitated overnight at −20 deg C. Protein was pelleted by centrifugation (8000×g, 10 minutes, 4 deg C), air dried, and resuspended in 100 mM TEAB. Each sample was digested overnight at 37 deg C with 2.5 ug trypsin (Pierce, V5111). Each enzymatic digest of reduced and alkylated proteins was then labeled with one of six TMT tags (Pierce, 90110) according to the manufacturer’s protocol. Briefly, TMT reagents were dissolved in anhydrous acetonitrile at room temperature and one tag was added to each of the trypsinized samples. Labeling occurred at room temperature for 1 hour and were quenched with 5% hydroxylamine for 15 minutes. Samples were subsequently mixed, acidified with formic acid, and desalted on Oasis HLB 1 cc cartridges (Waters #WAT200677) per manufacturer’s instructions, and subsequently separated into 12 fractions using basic reverse phase liquid chromatography. Each fraction was then completely dried in by SpeedVac then resuspended in 0.1% formic acid for mass spec analysis on an Orbitrap Fusion (Thermo Fisher Scientific, San Jose, CA). Relative protein quantitation was calculated based on the intensities of TMT report ions using Scaffold 4.0 (Proteome Software, Portland, OR).
Statistics
The proteomics data were analyzed using QUIOL. For infarct size, recovery of function and lactate measurements we used a t-test to measure differences between 2 groups and ANOVA to compare differences between multiple groups followed by a Tukey post hoc test. Because the Mito Sox data were not normally distributed these data were analyzed using a Kruskal-Wallis ANOVA on Ranks.
Results
Lysine succinylation is increased in heart mitochondria of Sirt5−/− mice
Succinylation of lysine residues is a dynamic PTM that regulates enzyme activity [4, 9, 10, 12]. Previous work identified Sirt5 as a lysine desuccinylase with weak lysine deacetylase activity [12]. As shown in Online Fig 1a, we confirmed that Sirt5 was not present in the Sirt5−/− hearts. To determine if Sirt5 functions as a desuccinylase in heart mitochondria, we analyzed the total succinylated lysine content of mitochondria isolated from mouse hearts. Western blot analysis demonstrates a strong increase in lysine succinylation in Sirt5−/− mitochondria relative to WT (Online Fig 1b), but no significant change in lysine acetylation in Sirt5 KO mitochondria relative to WT (Online Fig. 1b). Taken together, these data are consistent with the hypothesis that Sirt5 functions as a desuccinylase in cardiac mitochondria.
Specific lysine residues are targeted for succinylation
To identify the sites of mitochondrial lysine succinylation (SuK) in mouse heart, we combined affinity enrichment with proteomics (Fig. 1a). Mouse heart mitochondria were isolated from Sirt5−/− and WT littermates. Equal amounts of protein from each sample were digested with trypsin and succinylated peptides were immunoprecipitated with an antibody specific for succinylated lysine residues (PTM Biolabs, #PTM-401). The isolated peptides were analyzed with mass spectrometry and were searched against the mouse proteome with the Mascot search engine. With a false discovery rate (FDR) of <1%, we identified 2787 succinylated peptides. We excluded redundant peptides and peptides that were only detected in 1 sample, which generated a data set that included 887 unique SuK peptides which mapped to 184 proteins (see Supplemental Table 1). Of these, 44 peptides occurred only in the WT samples, 289 only in the Sirt5−/− samples, and 554 were common to both groups (Fig 1b).
Figure 1. Identification of sites of lysine succinylation in cardiac mitochondria.
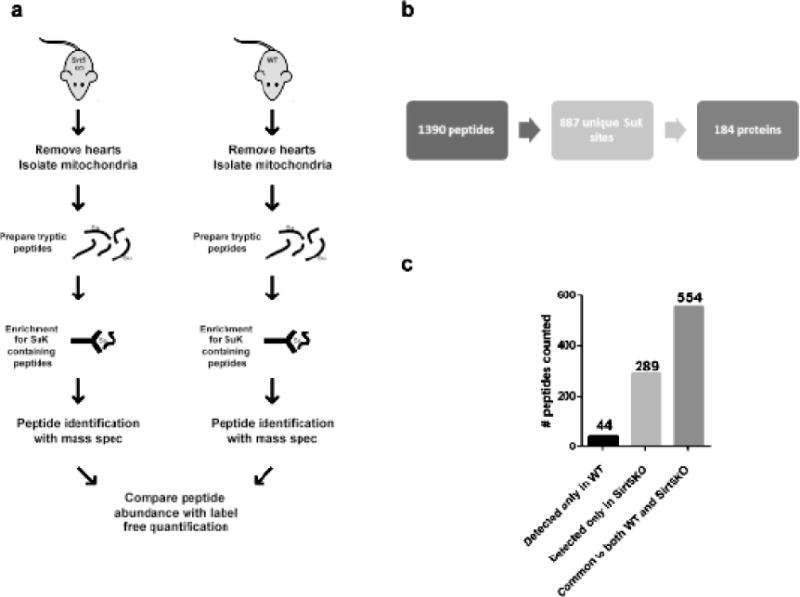
(a) Mouse heart mitochondria were isolated from Sirt5−/− (n=4) and WT (n=3) mice. Each sample was digested with trypsin and succinylated lysine (SuK) containing peptides were isolated by immunoprecipitation with an antibody designed to recognize SuK (PTM Biolabs, PTM-401). Isolated peptides were analyzed with mass spectrometry and label free analysis was used to quantify the relative abundance of peptides common between the sample groups. (b) To be considered for analysis, a peptide must have been detected in at least 2 of the 7 samples. We identified 1390 succinylated (SuK) peptides, which corresponded to 887 unique sites of lysine succinylation. These 887 peptides constitute the cardiac succinylome, and map to 184 proteins. (c) Of the 887 identified SuK peptides, 44 occurred uniquely in WT samples, 289 occurred uniquely in Sirt5 KO samples, and 554 were common to both groups.
We aimed to identify biological processes that are affected by succinylation in the heart. The 184 succinylated proteins identified in at least 2 samples were analyzed with the Ingenuity Pathway Analysis tool (IPA, Ingenuity Systems, www.ingenuity.com) to determine the molecular pathways in which the proteins participate. Using the mouse proteome as reference, we compiled a list of pathways enriched with succinylated proteins. Pathways that passed this analysis with a p-value ≤ 0.05 are presented in Supplemental Table 2. The top 12 pathways, obtained by only including those pathways with a p- value > (−log(p)) = 5.581) are presented in Figure 2. Succinylated cardiac mitochondrial proteins participate in processes such as oxidative phosphorylation, fatty acid oxidation, ketogenesis, and branched chain amino acid catabolism, among others. Many are enzymatic components of the TCA cycle.
Figure 2. Pathways enriched with succinylated proteins.
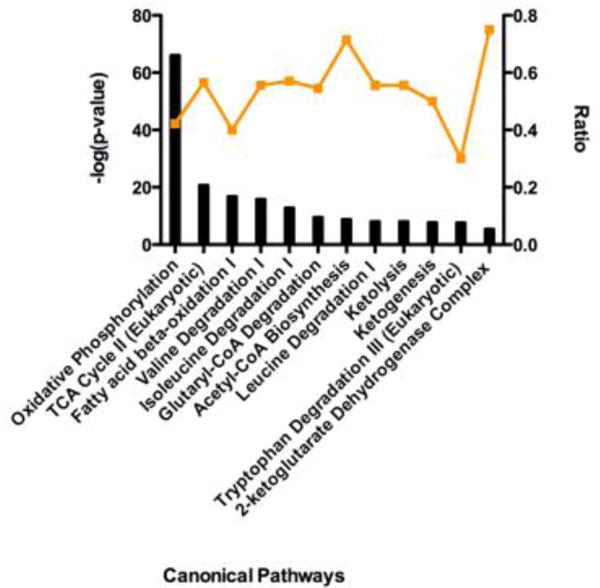
IPA core analysis was used to generate a list of pathways enriched with succinylated proteins. The top 12 scoring canonical pathways are represented. The left y-axis bar represents the [−log (p-value)] of the pathway, calculated by Fisher’s exact test. The right y-axis points (orange line) represent the ratio calculated by dividing the number of succinylated proteins that map to a particular pathway by the total number of proteins in that pathway. The top scoring pathways include oxidative phosphorylation, the TCA cycle, and β-oxidation of fatty acids.
Identification of specific Sirt5 substrates with label-free peptide quantification
We aimed to identify the specific protein substrates of Sirt5. We first defined the 289 peptides that were only found to be succinylated in the Sirt5−/− hearts as Sirt5 substrates (Supplemental Table 3). These peptides mapped to 114 proteins which were organized into functional pathways with the IPA pathway analysis tool (Fig 3a). Sirt5 substrates include proteins that participate in fatty acid metabolism, the TCA cycle, and in oxidative phosphorylation, including subunit A of the succinate dehydrogenase complex (Dhsa). We also applied a label free analysis with an in-house software program (QUantification with Out Isotope Labeling (QUOIL)) to identify common peptides that were enriched in succinylation in the Sirt5−/− hearts [16]. Peptides whose abundance was increased at least 1.3 fold in Sirt5−/− samples relative to WT were assigned as Sirt5 substrates. These 216 peptides mapped to 66 proteins. A complete list of the label-free quantification data is presented (Supplemental Table 4). If we further limited the data to peptides increased at least 1.3 fold with p ≥ 0.05, the list was reduced to 16 peptides which mapped to 9 proteins (Supplemental Table 4). The distribution of fold change among these 16 peptides was inspected and found to be widely varied (Fig 3b). The highest fold change, 108.4, occurred in a peptide that mapped to the ADP/ATP translocase 1 protein (Adt1) while the lowest fold change, 1.87, was detected in a peptide that mapped to an acyl-CoA dehydrogenase, Acadv.
Figure 3. Identification of Sirt5 substrates.

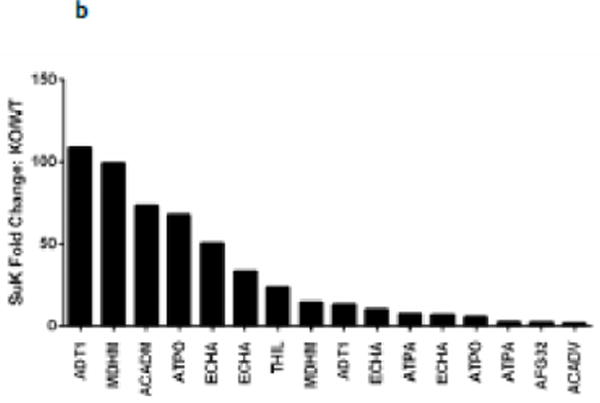
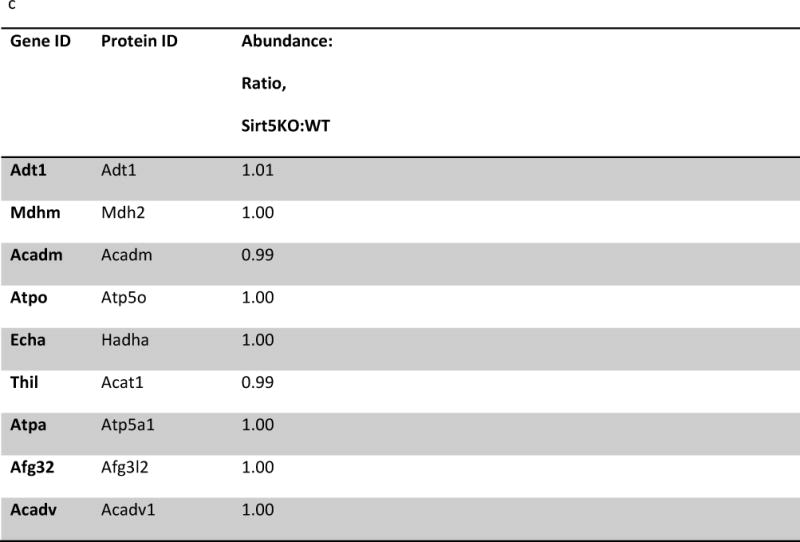
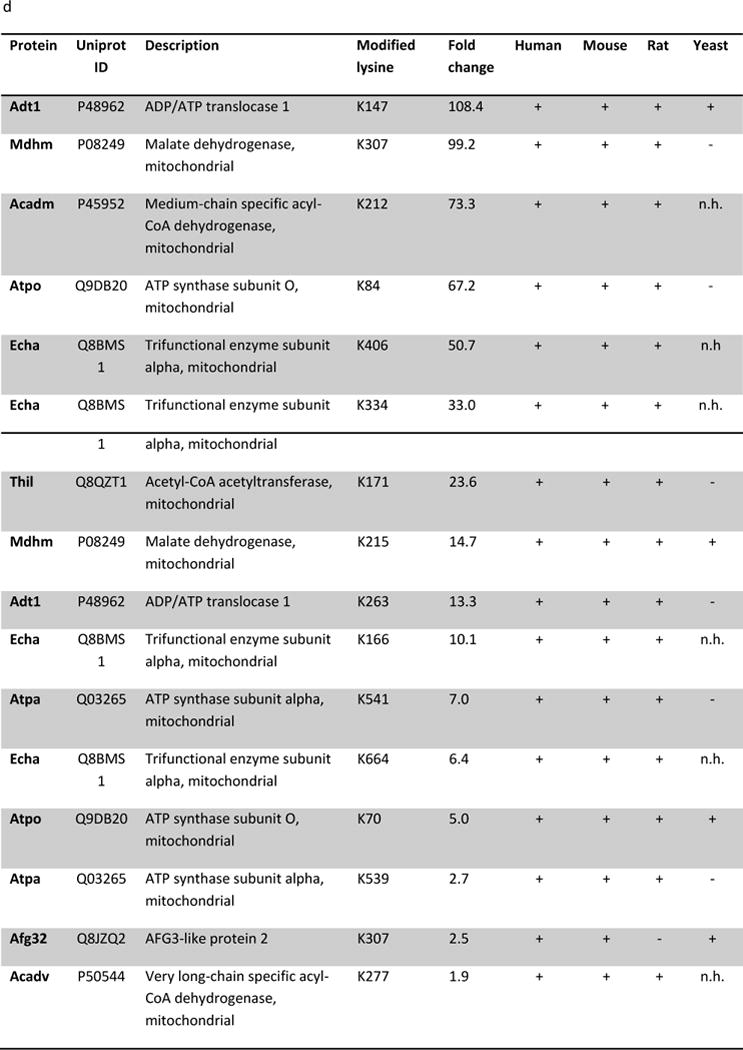
(a) IPA core analysis was used to functionally classify proteins mapped from peptides that were detected only Sirt5−/− samples. (b) A label free analysis program (QUOIL) was used to determine the relative abundance of succinylated peptides detected in both WT and Sirt5−/− samples. Represented here are the 16 peptides from 9 proteins increased by > 30% with p < 0.05 in Sirt5−/− hearts. (c) Relative protein quantitation was calculated based on the intensities of TMT report ions using Scaffold 4.0 (Proteome Software, Portland, OR). The average ratio of protein expression (Sirt5−/−: WT) is expressed numerically. The data demonstrate no significant differences in total protein abundance between Sirt5−/− and WT mice. (d) Conservation of the Sirt5 substrate lysines across human, mouse, rat, and S. Cerevisiae (yeast). Protein alignments were performed with the “Align” tool at www.uniprot.org; “+” indicates a conserved lysine, “−“ indicates that the lysine is not conserved, “n.h.” indicates that a homologous protein is not expressed by the organism.
To be sure that the relative changes in succinylation do not reflect altered protein abundance we used a quantitative labeling approach using tandem mass tags to determine relative protein levels between WT and Sirt5−/− hearts. Tryptic peptides prepared from protein extracted from Sirt5−/− and WT littermate hearts were labeled with tandem mass tags, and relative protein abundance was compared across the samples. As illustrated in figure 3c, the data demonstrate that, with the exception of Sirt5 abundance, there are no significant differences in protein abundance between Sirt5−/− and WT hearts, indicating that the relative changes in peptide abundance reflect the degree of lysine succinylation (Fig 3b). These data also suggest that loss of Sirt5 does not lead to compensatory changes in other proteins. The entire protein abundance data set is presented in the supplement to this paper (Supplemental Table 5).
We inspected the degree to which the Sirt5-targeted lysine residues in the peptides are conserved by evolution. Each of the 16 lysine residues is conserved between human and mouse, while 15/16 are conserved between human, mouse, and rat. We extended the analysis to include the S. cerevisiae genome and found that only 10/16 peptides mapped to proteins with homologues in yeast. Of these, 4 lysine residues are conserved between the organisms (Fig 3d). The strong degree of conservation among these residues suggests potential biological importance of the modification [17].
Sirt5-KO exacerbates ischemia/reperfusion injury
Some of the metabolic alterations that have been reported to occur in Sirt5−/− mice could be beneficial, while others detrimental in the setting of ischemia reperfusion (I/R). We therefore examined the effect of loss of Sirt5 on I/R injury. Three female WT and three female Sirt5−/− perfused hearts were subjected to I/R injury. Post-ischemic recovery of rate pressure product (RPP) and infarct size were measured to assess cell injury. We found that Sirt5−/− mice exhibit an increased susceptibility to I/R injury. Recovery of the RPP after I/R was significantly decreased in Sirt5−/− hearts relative to WT (28.7% vs 50.6% of preischemia RPP) (Fig 4a). Infarct size was measured with TTC staining and data were calculated as percentage of infarct relative to total ventricular area. The data demonstrate that infarct size was significantly increased in Sirt5−/− hearts relative to WT (68.5% vs 40.2% of total ventricular area) (Fig 4b). Taken together, these data suggest that Sirt5 loss negatively impacts the ability of the heart to recover from ischemic injury. We considered that the increased I/R injury evident in the Sirt5−/− heart could be a result of altered anaerobic glycolysis. To investigate this, lactate was quantified in extracts isolated from ischemic WT and Sirt5−/− hearts. However, no significant difference in lactate accumulation was detected between the ischemic hearts (Fig 4c).
Figure 4. Sirt5−/− mice are susceptible to ischemia-reperfusion injury.
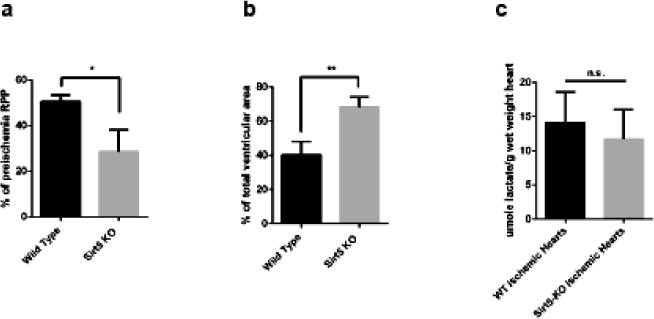
(a) Assessment of the rate pressure product (RPP = heart rate*systolic blood pressure) after ischemia-reperfusion injury in hearts of three female WT and three female Sirt5−/− mice. Baseline RPP priiror to ischemia averaged 54,009+/−4572 (HR*cm of water), and LVDP at baseline was 130+/−9 cm of water. (b) Infarct size in WT and Sirt5−/− mice following 20 minutes of global ischemia and 90 minutes of reperfusion. (c) WT and Sirt5−/− mice show no difference in production of lactate after 20 minutes of global ischemia *p<0.05; **p<0.01; n.s., not significant, measured by t-test. Values are means +/− SEM n=3 in each group.
Accumulation of succinate is reported to be a metabolic signature of ischemic tissue and a driver of cellular injury upon reperfusion [18]. It has been recently reported that that during ischemia succinate accumulates due to generation of fumerate and reversal of succinate dehydrogenase (SDH). Furthermore on reperfusion the accumulated succinate is re-oxidized by SDH and leads to reverse electron flow through complex I generating reactive oxygen species (ROS) leading to I/R injury. Succinylation of protein components of the SDH complex has been shown to increase complex activity [9]. Our study identified 4 succinylated lysine residues in the SDH subunit Dhsa and 1 succinylated lysine in subunit Dhsb. Of these, 2 residues (K179, K335) in Dhsa were classified as Sirt5 targets, as these modifications were only detected in Sirt5−/− hearts (Fig 5a). We considered the possibility that increased succinylation of Dhsa in the Sirt5−/− heart underlies the observed exacerbation of I/R injury in the knockout animals, as succinylation of SDH increases its activity. To test this, perfused hearts isolated from Sirt5−/− and WT animals were treated with 5 mM dimethyl malonate, a competitive inhibitor of SDH, for 20 minutes before ischemia/reperfusion [18]. Consistent with the hypothesis, dimethyl malonate treatment attenuated I/R injury in WT hearts. Strikingly, dimethyl malonate treatment of Sirt5−/− hearts restored RPP recovery to WT levels (Fig 5b). We measured infarct size in Sirt5−/− and WT hearts and show that, consistent with the recovery of function data, there is no significant difference between infarct size when hearts are pretreated with dimethyl malonate. The data are consistent with a model in which loss of Sirt5 activity increases succinylation of Dhsa, promotes SDH activity, and increases oxidation of succinate after ischemia. The increased rate of succinate oxidation exacerbates I/R injury in the Sirt5−/− heart, presumably through increased generation of ROS. To further test this hypothesis we used Mito Sox to measure ROS in Sirt5−/− and WT hearts after 20 min of ischemia followed by 5 min of reperfusion. As shown in Figure 6a, we find a slight but significant increase in ROS in Sirt5−/− hearts compare to WT hearts after ischemia and reperfusion. Furthermore, addition of methyl malonate significantly reduced ROS in Sirt5−/− and WT hearts. Summary data of the changes in fluorescence averaged over four hearts in each group is shown in figure 6b.
Figure 5. Inhibition of succinate dehydrogenase attenuates injury in WT hearts and restores recovery in Sirt5−/− hearts.
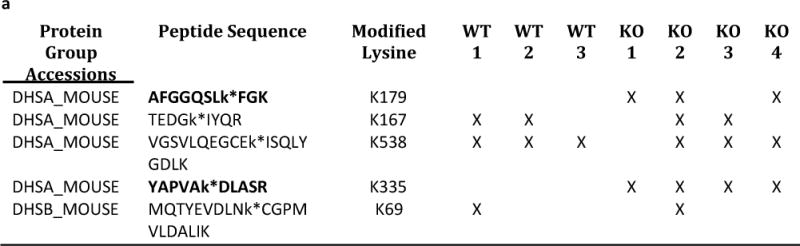
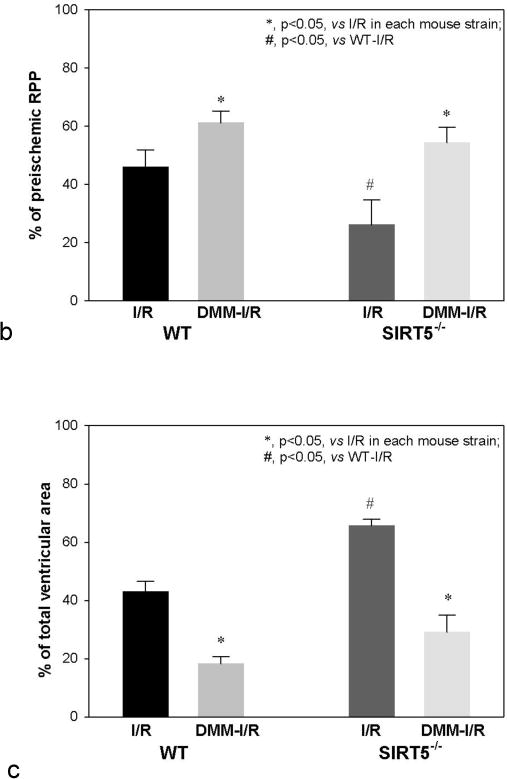
(a) M/S identification of 5 peptides that map to protein components of the SDH complex. 2/4 peptides mapped to SDH subunit Dhsa (bold font) are classified as Sirt5 substrates, as these peptides were only detected in Sirt5−/− samples. (b) Assessment of the rate pressure product (RPP = heart rate*systolic blood pressure) when a competitive inhibitor of SDH, dimethyl malonate, is delivered to WT and Sirt5−/− hearts before ischemia-reperfusion injury. Malonate, which is generated in the cell upon addition of dimethyl malonate attenuates injury in the WT heart and restores recovery in Sirt5−/− hearts to WT levels. Baseline RPP prior to ischemia (or addition of DMM) was 45,585+/−1922, and baseline LVDP was 124+/−4 cm of water. Significance was determined by ANOVA followed by a post hoc Tukey.t-test. Values are means +/− SEM. N=11 for WT I/R (no drug), n=6 for WT I/R treated with dimethyl malonoate (DMM-I/R), n=9 for SIRT5KO (no drug), and n=4 for SIRT5KO + DMM-I/R. Male and female mice were used in this study. There were no sex differences. (c) Infarct size in WT and Sirt5−/− mice after hearts were pretreated with 5 mM methyl malonate for 20 minutes before 20 minutes of global ischemia and 90 minutes of reperfusion. Significance was determined by ANOVA followed by a post hoc. Values are means +/− SEM.N values are the same as 5b.
Figure 6. Mito Sox measurement of superoxide in I/R.
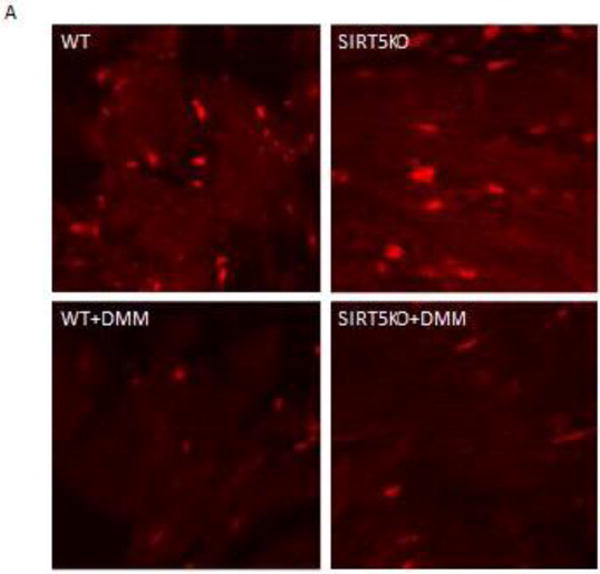
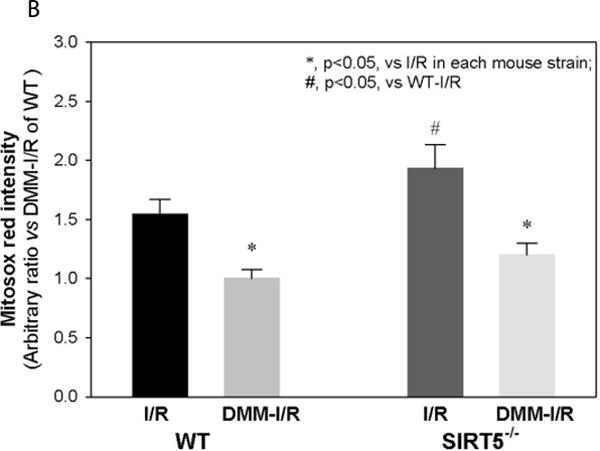
Panel A shows representative images of section of WT, SIRT5-KO, WT + dimethyl malonate (DMM) and SIRT5-KO + DMM hearts treated with Mito Sox following 20 min of ischemia and 5 min of reperfusion. The bright red spots are due to Mito Sox binding to nucleotides as confirmed by co-localization with DAPI (data not shown). The mean and standard error for each group (n=4) are provided in panel B. * indicates Ssignificantly was determined difference between groups as measured by Kruskal-Wallis ANOVA and a Student-Newman Keuls test.
Discussion
Succinylation is a reversible post-translational modification that is conserved across species. Several studies demonstrate that lysine succinylation can result in inhibition or activation of metabolic enzymes in liver [9, 10, 12]. This study reports the first comprehensive proteomic analysis of succinylation in the heart. We identified 184 succinylated proteins in cardiac mitochondria isolated from Sirt5−/− and WT mice, and demonstrate that major metabolic pathways are enriched with these proteins. Succinylated proteins in the heart participate in the processes of oxidative phosphorylation, TCA cycling, branched chain amino acid catabolism, and β-oxidation of fatty acids. Appropriate regulation of these pathways and processes is critical to optimal cardiac function. We applied label-free quantification to our data to determine which peptides were increased in abundance in Sirt5−/− tissue, in order to delineate specific proteins targeted for desuccinylation by Sirt5. We determined that 289 peptides (46 proteins) were detected only in Sirt5−/− mitochondria, and 16 peptides (9 proteins) were significantly hypersuccinylated in Sirt5−/− heart mitochondria. Thus, these findings are consistent with a role for Sirt5 in regulating succinylation in the heart.
Overlap between lysine succinylation and ubiquitination
Negative crosstalk between PTMs describes a scenario in which modification of an amino acid with one modifier precludes its modification with an alternate modifier. Ubiquitination of mitochondrial proteins at lysine residues has been described [19–21]. We examined the degree to which lysine succinylation and lysine ubiquitination overlap. To accomplish this, we compared the 305 Sirt5 substrate lysines identified in our study to a published dataset containing ubiquitinated lysines mapped from mouse heart tissue [22] (Supplemental Table 6). 166 peptides identified as containing at least one ubiquitinated lysine mapped to proteins that are also Sirt5 substrates. We found that 54 lysines overlapped between the two datasets. Thus, 17.7% of Sirt5 substrate lysines can be ubiquitinated at the same position (Fig 7a). Using Phosphosite (Cell Signaling Technologies) [23]), we determined that, of the 54 overlapping sites, 51 (94%) are conserved between mouse and human, and 45 (83.3%) are also acetylated in mouse (Fig 7b). Ubiquitination of mitochondrial proteins has been shown to impact mitochondrial biogenesis, to regulate fission/fusion, to control turnover of individual proteins by the proteasome, and to oversee turnover of mitochondria via mitophagy [24–27]. The data suggest that metabolic derangements that underlie dysregulation of protein succinylation may also impact protein ubiquitination, which may contribute to mitochondrial discord.
Figure 7. Overlap of succinylation and ubiquitination in mouse heart.
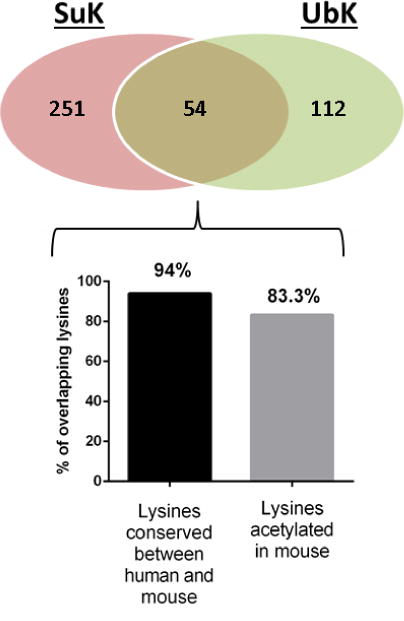
The Venn diagram illustrates the overlap between Sirt5 targeted lysines and ubiquitination. Of the 54 overlapping lysine residues, 51/54 (94%) are conserved between mouse and human. 45/54 (83.3%) are known to be acetylated in mouse.
The biological function of Sirt5 in the heart is not well characterized. The Sirt5−/− mouse used in our study appears normal at baseline; these animals are born with Mendelian ratios, are fertile, and do not demonstrate gross abnormality or decline in heath as they age [28]. We are aware of only two studies that have examined Sirt5 in heart. The first demonstrated that intermittent hypoxia in a hyperbaric chamber for 6 hours per day for 42 days increased Sirt5 protein levels in rat hearts [29]. However it is not clear whether this hypoxic induced increase in Sirt5 is beneficial or detrimental. The second study showed that Sirt5 levels are decreased in H9c2 cells following 12 hours hydrogen peroxide treatment and that siRNA knockdown of Sirt5 reduced apoptosis following hydrogen peroxide treatment. [30]. To better clarify the role of Sirt5 in a model of ischemia and reperfusion we subjected Langendorff perfused hearts from Sirt5−/− mice and their wild type littermates to ischemia and reperfusion and assessed recovery of function and infarct size. We found that loss of Sirt5 and the resultant increase in succinylation leads to an increase in I/R injury. The Sirt5−/− hearts have increased post-ischemic contractile dysfunction and increased infarct size.
A number of metabolic alterations have been reported to be regulated by succinylation; we considered whether any of these modifications might be involved in the increase in I/R injury that we observed. Increased succinylation of enzymes involved in beta-oxidation have been shown to decrease fatty acid oxidation in hepatocytes from Sirt5−/− mice. We considered that this might lead to an increase in glycolysis that could increase lactate production during ischemia and might contribute to the increase in I/R injury. However we found no change in lactate production during ischemia between WT and Sirt5−/− hearts.
It has recently been reported that inhibition of SDH is beneficial in I/R, because there is a build-up of succinate during ischemia which on reperfusion results in reversal of Complex I and generation of ROS [18]. Recent studies in liver reported an increase in succinylation of SDH which increases its activity. Consistent with studies in the liver, we found increased succinylation of SDH in the heart. We treated hearts with an inhibitor of SDH and consistent with the hypothesis, we found that inhibition of SDH rescued the Sirt5−/− from increased I/R injury. We further showed that inhibition of SDH reduced superoxide production in both WT and Sirt5−/− hearts. Although these data suggest that increased I/R injury in Sirt5−/− hearts is mediated, at least in part, by SDH, Sirt5 could have other effects and other targets that might also influence the response to I/R.
In summary, our study is the first to provide global insight into lysine succinylation in the heart. Sirt5 loss leads to an increase in protein succinylation in the heart, and also correlates with increased susceptibility to I/R injury.
Supplementary Material
Highlights.
Identified 184 proteins in the cardiac succinylone
Demonstrated that Sirt5 functions as a desuccinylase in heart
Demonstrated that loss of Sirt5 increases ischemia-reperfusion (I/R) injury
Increase I/R injury in Sirt5−/− hearts depends on succinate dehydrogenase activity
Acknowledgments
This work was funded by the National Heart, Lung, and Blood Institute (NHLBI) Intramural program. We thank Dr. Christian Combs and the NHLBI Light Microscopy Core for their assistance with the Mito Sox measurements
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–4. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–18. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 3.Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10:M111 012658. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–17. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colak G, Xie Z, Zhu AY, Dai L, Lu Z, Zhang Y, et al. Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol Cell Proteomics. 2013;12:3509–20. doi: 10.1074/mcp.M113.031567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinert BT, Scholz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, et al. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–51. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, et al. Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics. 2012;11:100–7. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919–30. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18:920–33. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner GR, Hirschey MD. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell. 2014;54:5–16. doi: 10.1016/j.molcel.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–9. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J, Sadhukhan S, Noriega LG, Moullan N, He B, Weiss RS, et al. Metabolic characterization of a Sirt5 deficient mouse model. Sci Rep. 2013;3:2806. doi: 10.1038/srep02806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J, Morgan M, Shen RF, Steenbergen C, Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res. 2007;101:1155–63. doi: 10.1161/CIRCRESAHA.107.155879. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen TT, Wong R, Menazza S, Sun J, Chen Y, Wang G, et al. Cyclophilin D modulates mitochondrial acetylome. Circ Res. 2013;113:1308–19. doi: 10.1161/CIRCRESAHA.113.301867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang G, Wu WW, Zeng W, Chou CL, Shen RF. Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: Reproducibility, linearity, and application with complex proteomes. J Proteome Res. 2006;5:1214–23. doi: 10.1021/pr050406g. [DOI] [PubMed] [Google Scholar]
- 17.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–4. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chouchani ET, Pell VR, Gaude E, Aksentijevic D, Sundier SY, Robb EL, et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–5. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azzu V, Brand MD. Degradation of an intramitochondrial protein by the cytosolic proteasome. J Cell Sci. 2010;123:578–85. doi: 10.1242/jcs.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azzu V, Mookerjee SA, Brand MD. Rapid turnover of mitochondrial uncoupling protein 3. Biochem J. 2010;426:13–7. doi: 10.1042/BJ20091321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margineantu DH, Emerson CB, Diaz D, Hockenbery DM. Hsp90 inhibition decreases mitochondrial protein turnover. PLoS One. 2007;2:e1066. doi: 10.1371/journal.pone.0001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner SA, Beli P, Weinert BT, Scholz C, Kelstrup CD, Young C, et al. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol Cell Proteomics. 2012;11:1578–85. doi: 10.1074/mcp.M112.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–20. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen MM, Leboucher GP, Livnat-Levanon N, Glickman MH, Weissman AM. Ubiquitin-proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol Biol Cell. 2008;19:2457–64. doi: 10.1091/mbc.E08-02-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu M, St-Pierre P, Shankar J, Wang PT, Joshi B, Nabi IR. Regulation of mitophagy by the Gp78 E3 ubiquitin ligase. Mol Biol Cell. 2013;24:1153–62. doi: 10.1091/mbc.E12-08-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heo JM, Livnat-Levanon N, Taylor EB, Jones KT, Dephoure N, Ring J, et al. A stress-responsive system for mitochondrial protein degradation. Mol Cell. 2010;40:465–80. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos JM, Mishra M, Kowluru RA. Posttranslational modification of mitochondrial transcription factor A in impaired mitochondria biogenesis: implications in diabetic retinopathy and metabolic memory phenomenon. Exp Eye Res. 2014;121:168–77. doi: 10.1016/j.exer.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–14. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu WZ, Wu XF, Zhang Y, Zhou ZN. Proteomic analysis of mitochondrial proteins in cardiomyocytes from rats subjected to intermittent hypoxia. Eur J Appl Physiol. 2012;112:1037–46. doi: 10.1007/s00421-011-2050-9. [DOI] [PubMed] [Google Scholar]
- 30.Liu B, Che W, Zheng C, Liu W, Wen J, Fu H, et al. SIRT5: a safeguard against oxidative stress-induced apoptosis in cardiomyocytes. Cell Physiol Biochem. 2013;32:1050–9. doi: 10.1159/000354505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


