Abstract
Although subtypes of pancreatic ductal adenocarcinoma (PDAC) were described, this malignancy is clinically still treated as a single disease. Here, we present patient-derived models representing the full spectrum of previously identified quasi-mesenchymal (QM-PDA), classical and exocrine-like PDAC subtypes, and identify two markers—HNF1A and KRT81—that enable stratification of tumors into different subtypes by immunohistochemistry. Individuals bearing tumors of these subtypes show significant differences in overall survival and their tumors differ in drug sensitivity, with the exocrine-like subtype being resistant to tyrosine kinase inhibitors and paclitaxel. Cytochrome P450 3A5 (CYP3A5) metabolizes these compounds in tumors of the exocrine-like subtype, and pharmacological or shRNA-mediated CYP3A5 inhibition sensitizes tumor cells to these drugs. Whereas hepatocyte nuclear factor 4 alpha (HNF4A) controls basal expression of CYP3A5, drug-induced CYP3A5 upregulation is mediated by the nuclear receptor NR1I2. CYP3A5 also contributes to acquired drug resistance in QM-PDA and classical PDAC, and is highly expressed in several additional malignancies. These findings designate CYP3A5 as predictor of therapy response and as a tumor cell-autonomous detoxification mechanism that must be overcome to prevent drug resistance.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a highly aggressive disease with dismal prognosis1. In both Europe and the USA pancreatic cancer is the fourth leading cause of cancer death2,3. Treatment with gemcitabine4, FOLFIRINOX scheme5 or the albumin-paclitaxel conjugate nab-paclitaxel6 only offer a modest increase in overall survival. Despite extensive testing of targeted therapies in clinical trials, thus far all of the examined compounds confer little or no survival benefit in unselected cohorts of PDAC patients1,7,8.
Although patient stratification according to molecular characteristics has not yet been performed in clinical trials for PDAC, transcriptional profiling of whole tumor tissues suggested the existence of subtypes of PDAC that differ in patient survival and tumor metastasis 9,10. Additionally, three PDAC subtypes were described based on gene expression profiling of laser capture microdissected epithelial tumors; these subtypes were termed classical, quasi-mesenchymal (QM-PDA) and exocrine-like11. However, in a larger panel of human and mouse PDAC cell lines, only the classical and the QM-PDA subtype were identified11, suggesting that currently used PDAC cell lines inadequately represent the heterogeneity of human PDAC. In addition, the classical and QM-PDA subtypes were suggested to differ in response to a range of chemotherapeutics, but the drug sensitivity of the exocrine-like subtype has yet to be determined11.
Although resistance of PDAC to therapy is well described1, little is known about the molecular mechanisms mediating it. Members of the cytochrome P450 (CYP) enzyme family have been previously only investigated with regard to a role in systemic drug metabolism12,13 or their up- or down-regulation in solid tumors compared to normal tissues14. Thus, the functional role and impact of CYPs on tumor-cell autonomous drug resistance remains largely unknown14,15.
Here, we show that the exocrine-like PDAC subtype is resistant towards the small molecule drugs dasatinib, erlotinib and paclitaxel, and that this resistance is mediated by a cell autonomous CYP3A5-dependent drug detoxification mechanism. CYP3A5 also contributes to acquired drug resistance in other subtypes of PDAC and in other malignancies.
Results
Establishment of PDAC models including the exocrine-like subtype
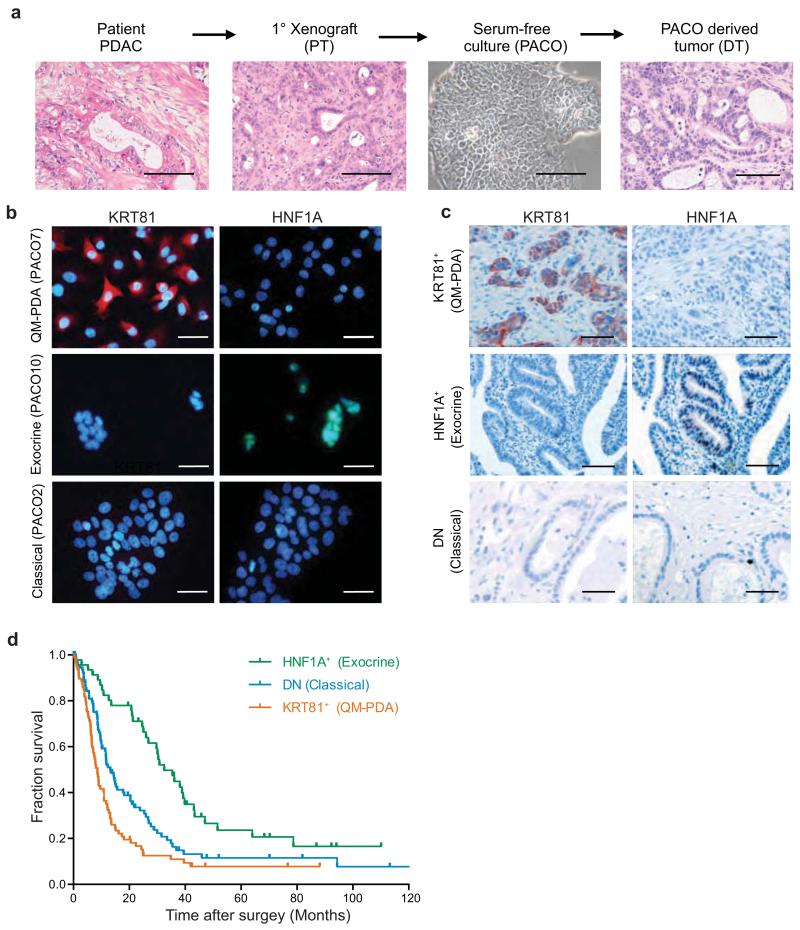
First, we established patient-derived PDAC models to provide an in vitro and in vivo platform for functional studies. Patient-derived PDAC specimens were surgically grafted onto the pancreas of immune-deficient NOD.Cg-Prkdcscid Il2rgtm1Wjl (NSG) mice. Tumors from primary xenografts (PT) were then used to propagate primary PDAC cell lines (PACO) (Fig. 1a and Supplementary Table 1). Comparison of the resulting PACO derived tumors (DT) with the original xenografts (PT) showed conservation of histomorphological characteristics (Fig. 1a) and of RNA expression profiles (Supplementary Table 2). Matching recent genomic profiling data16-18, all eight analyzed PACO lines harbored mutations in KRAS and six out of eight in TP53 (Supplementary Table 3).
Figure 1.
Subtype stratification of PDAC models and patients by two markers.
(a) Schematic overview of the experimental workflow used to generate orthotopic xenografts and PACO cells. H&E staining of a human PDAC tumor, the corresponding first passage xenograft (PT), phase contrast image of the derived cell line (PACO10) and the respective derived xenograft (DT). Scale bar, 100 μM. (b) KRT81 and HNF1A immunofluorescence staining on PACO lines from the three different subtypes (n = 3). Scale bar, 50 μM. (c) KRT81 and HNF1A immunostaining on sections from a TMA of individuals with PDAC (n = 241). Scale bar, 100 μM. (d) Kaplan-Meier analysis of overall survival of subjects with PDAC (n = 217). Tumor sections on the TMA were retrospectively subtyped into three groups based on KRT81 and HNF1A expression as determined by immunostaining (HNF1A+: n = 46; DN: n = 92; KRT81+: n = 79). P value was determined by log-rank test.
Next, we determined which PDAC subtypes are represented in our PDAC models. To this end, we used the PDAssigner genes11 to subtype eight PACO lines as well as the respective PT and DT xenografts by Gene Set Enrichment Analysis (GSEA). All three subtypes, including the exocrine-like subtype, are represented (Supplementary Fig. 1a,b). Specifically, the gene-expression profiles of two of our PACO lines and xenografts were enriched for the classical subtype (PACO2 and 17), three for the exocrine-like (PACO10, 14, 18) and three for the QM-PDA (PACO7, 9, 19) subtype. Taken together, these results demonstrate that our models faithfully preserve histomorphological characteristics of the originating tumors, and for the first time enable the study of functional differences between all three PDAC subtypes.
Prognostic value of HNF1A and KRT81
Given that histopathology supplemented by immunohistochemistry is still currently the standard method for tumor subtyping, we sought to identify surrogate protein markers for each of these three PDAC subtypes to facilitate clinical patient stratification. We subjected PACO cell lines to array-based gene-expression profiling. A list of genes showing strong (> 5 fold, P < 0.05) differential expression between the different PACO subtypes was further filtered for candidates that showed heterogeneous expression across PDAC specimens in the Protein Atlas database19 to generate a candidate biomarker list. Additionally, GSEA of transcription factor activity genesets on the original expression profiles revealed an enrichment for transcripts with binding-sites for the transcription factor HNF1A in the exocrine-like subtype, suggesting HNF1A as a putative marker for this subtype (data not shown). We stained PACO lines and xenografts for all marker candidates, evaluated signal intensity and subtype-specificity (Supplementary Table 4). We excluded markers that stained only weakly or subtype unspecific. This analysis identified nuclear positivity for HNF1A to be specific for exocrine-like PDAC, while staining for cytokeratin 81 (KRT81) was specific for the QM-PDA subtype (Fig. 1b). Additionally, in the TCGA pancreatic adenocarcinoma (PAAD) cohort, KRT81 expression also inversely correlated with HNF1A (Supplementary Fig. 1c). None of the candidate markers for the classical subtype showed a reliable and exclusive staining in this subtype. Nevertheless, the specificity of the KRT81 and HNF1A allowed us to define classical subtype specimens as double-negative (DN). Hence, we defined surrogate markers for the three subtypes as KRT81+HNF1A− for the QM-PDA subtype, KRT81−HNF1A+ for the exocrine-like subtype and KRT81−HNF1A− for the classical subtype.
We verified the association of our marker-defined subtypes with the PDAssigner signatures in an independent validation cohort of primary PDAC xenografts (Supplementary Table 5). Principal component analysis (PCA) demonstrated that transcriptional profiles clustered according to KRT81 and HNF1A defined subtypes (Supplementary Fig. 1d). Hierarchical clustering using the PDAssigner genes further showed a separation into three groups, revealing a high concordance with our marker-defined groups (Supplementary Fig. 1e). GSEA of the marker-defined groups revealed enrichment of the QM-PDA signature in the KRT81+ cases and of the exocrine-like signature in the HNF1A+ cases. The DN cases were enriched for all three signatures and could not be unequivocally assigned (Supplementary Fig. 1f), suggesting that further refinement of the PDAssigner could lead to a more robust classification. Collectively, our surrogate markers separate the validation cohort into three distinct groups, of which the KRT81+ and HNF1A+ cases are enriched in the respective PDAssigner-defined subtypes11.
Next, we tested whether subtype stratification of a cohort of 231 individuals with PDAC by using these two markers is associated with clinical outcome. In a retrospective study using immunohistochemistry we designated these PDAC tumors as 45% DN, 35% KRT81+ and 20% HNF1A+ (Fig. 1c,d and Supplementary Table 6). We also identified 14 KRT81+HNF1A+ double positive specimens and excluded them from the analysis. Log-rank analysis revealed significant differences in overall survival of individuals with PDAC of different subtypes (P < 0.001) (Fig. 1d). Subjects with HNF1A+ tumors had the best mean survival (43.5 months), followed by the DN subtype (26.3 months) and the KRT81+ subtype (16.5 months). Moreover, Cox proportional hazards multivariate regression analysis revealed that the survival impact of the subtype classification is independent of established conventional prognostic factors such as stage, grade and age20,21 (Supplementary Table 7). Subtype was associated with grade, as the HNF1A+ cases were more differentiated (24% G3), the KRT81+ samples tended to be less differentiated (50.6% G3) and the DN cases ranged in between (41.5% G3) (Supplementary Table 8). While this association was significant (P = 0.01), grade alone was insufficient to predict subtype. Hence, HNF1A and KRT81 can be used to stratify patients into subtypes of PDAC that differ in overall survival.
Exocrine-like PDAC cells are resistant to tyrosine kinase inhibitors
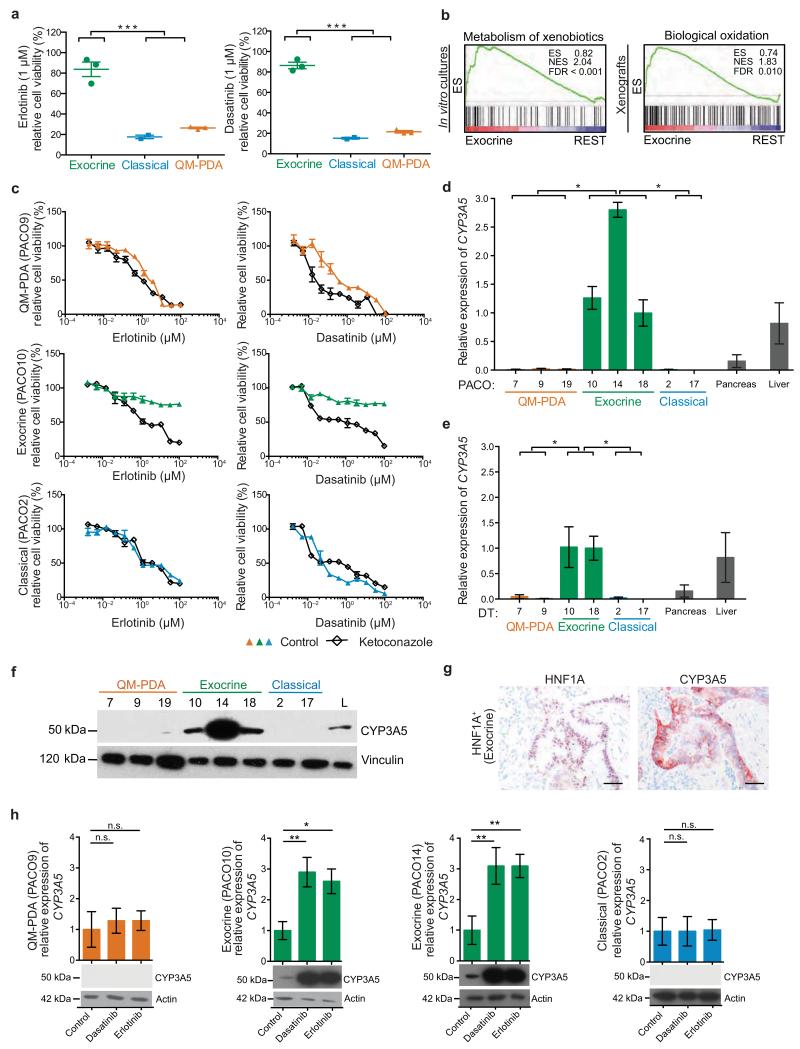
To address whether the subtypes differ in drug sensitivity, PACO lines were treated with the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) erlotinib and the SRC/ABL1 selective TKI dasatinib, which are approved or under investigation for treatment of PDAC, respectively22-24. We treated PACO lines of each subtype with 1 μM (Fig. 2a) or 10 μM (Supplementary Fig. 2a) erlotinib and dasatinib, corresponding to 0.3–3 and 6–60 fold peak plasma concentrations reported in humans, respectively25,26. Analysis after 48 hours revealed that the classical and QM-PDA were sensitive, whereas the exocrine-like cells were almost completely resistant. To exclude that the observed resistance was due to varying proliferation rates, we treated the PACO lines as described above for 7 days, which confirmed the difference in drug response (Supplementary Fig. 2b,c). To identify the mechanisms underlying the observed drug resistance we used GSEA to compare the exocrine-like PACO lines and xenografts with the classical and the QM-PDA PACO lines and xenografts. This analysis revealed an enrichment of signatures comprising genes involved in xenobiotic biotransformation in the exocrine-like PDAC models (Fig. 2b and Supplementary Fig. 2d,e). For validation we analyzed an independent dataset generated from laser micro-dissected PDAC11, confirming the up-regulation of similar gene-sets in exocrine-like PDAC samples (Supplementary Fig. 2f). Thus, xenobiotic biotransformation might contribute to the observed drug resistance in the exocrine-like PDAC subtype.
Figure 2.
Exocrine-like PDAC, which express CYP3A5, are resistant to TKIs
(a) PACO line specific drug sensitivities to 1 μM erlotinib or dasatinib after 48 h. Bars depict mean ± SD (n = 2; ***P < 0.001; grouped one-way ANOVA). (b) Gene set enrichment analysis (GSEA) of the exocrine-like subtype compared to the classical and QM-PDA subtype (REST), using the indicated gene signatures. Left panel: PACO cell lines. Right panel: PT + DT xenografts. Statistical significance was assessed using 10,000 permutations. ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate. (c) PACO lines treated with erlotinib or dasatinib for 48 h post ketoconazole (100 nM) or vehicle pre-treatment for 2 h (n = 3). (d, e) CYP3A5 expression, as measured by qRT-PCR, in PACO lines (d) and PACO derived xenografts (e) compared to pancreas and liver mRNA. Values are relative to PACO18 mRNA expression and depict mean ± SEM (n = 3; *P < 0.05; grouped one-way ANOVA). (f) Anti-CYP3A5 immunoblot of PACO cell lines. Vinculin was used as loading control. L = liver protein lysate. (g) CYP3A5 and HNF1A immunostainings on PDAC sections from HNF1A+ individuals (n = 217). Scale bar, 100 μM. (h) CYP3A5 expression analysis by qRT-PCR and immunoblot at basal level and in response to 10 μM dasatinib or erlotinib of QM-PDA, exocrine-like and classical PACO lines. Values are relative to untreated controls and depict mean ± SEM (n = 3; *P < 0.05; **P < 0.01; n.s. = not significant, Student’s T-test). Actin was used as loading control for immunoblots.
Enzymes of the cytochrome P450 family (CYP) systemically metabolize small molecule drugs by oxidation, resulting in a potential inactivation27,28. To test if xenobiotic biotransformation is indeed involved in the observed drug resistance, we pre-treated cells of each subtype with the pan-CYP inhibitor ketoconazole14. One PACO line of each subtype was pre-treated with 100 nM ketoconazole or vehicle for 2 hours, followed by the addition of serial dilutions of erlotinib or dasatinib. Relative cell viability was determined after 48 hours. To compare TKI effects across independent experiments, we calculated activity areas as previously described29. Ketoconazole pretreatment significantly increased sensitivity exclusively in the exocrine-like PDAC cells, rendering their drug response comparable to the other subtypes (Fig. 2c and Supplementary Fig. 2g). These results suggest that CYPs contribute to drug resistance in the exocrine-like PDAC subtype.
CYP3A5 is expressed and inducible in exocrine-like PDAC
Members of the CYP3A subfamily are major contributors to xenobiotic biotransformation of small molecule drugs in the liver12. We thus tested expression of all three CYP3A family members CYP3A4, CYP3A5 and CYP3A7, in both PACO cell lines and derived xenografts (DT). qRT-PCR revealed that CYP3A5 is exclusively expressed in the exocrine-like subtype at comparable or even higher levels than in normal liver and pancreas (Fig. 2d,e). In contrast, expression of CYP3A4 and CYP3A7 was low or absent (Supplementary Fig. 2h–k). The exocrine-like specific expression of CYP3A5 was also confirmed at the protein level in PACO lines (Fig. 2f and Supplementary Fig. 2l) and in specimens of individuals with HNF1A+ tumors (Fig. 2g). The marker-defined exocrine-like xenografts of the validation cohort of primary PDAC xenografts also expressed significantly more CYP3A5 than the other two subtypes (Supplementary Fig. 2m). In line with these findings, expression of CYP3A5 correlated positively with HNF1A and inversely with KRT81 in the PAAD dataset (Supplementary Fig. 2n).
Enzymes involved in xenobiotic biotransformation can be induced in response to their substrates30. To test whether this regulatory mechanism is also functional in PDAC cells, we measured CYP3A5 mRNA and protein in PACO lines at steady state and in response to 10 μM dasatinib or erlotinib. Exposure to either drug boosted CYP3A5 expression in the exocrine-like but not in the classical and QM-PDA PACO lines (Fig. 2h). No induction of CYP3A4 or CYP3A7 was observed (Supplementary Fig. 2o). Taken together, these data reveal that CYP3A5 is highly expressed and further inducible in the exocrine-like subtype in vitro and in vivo.
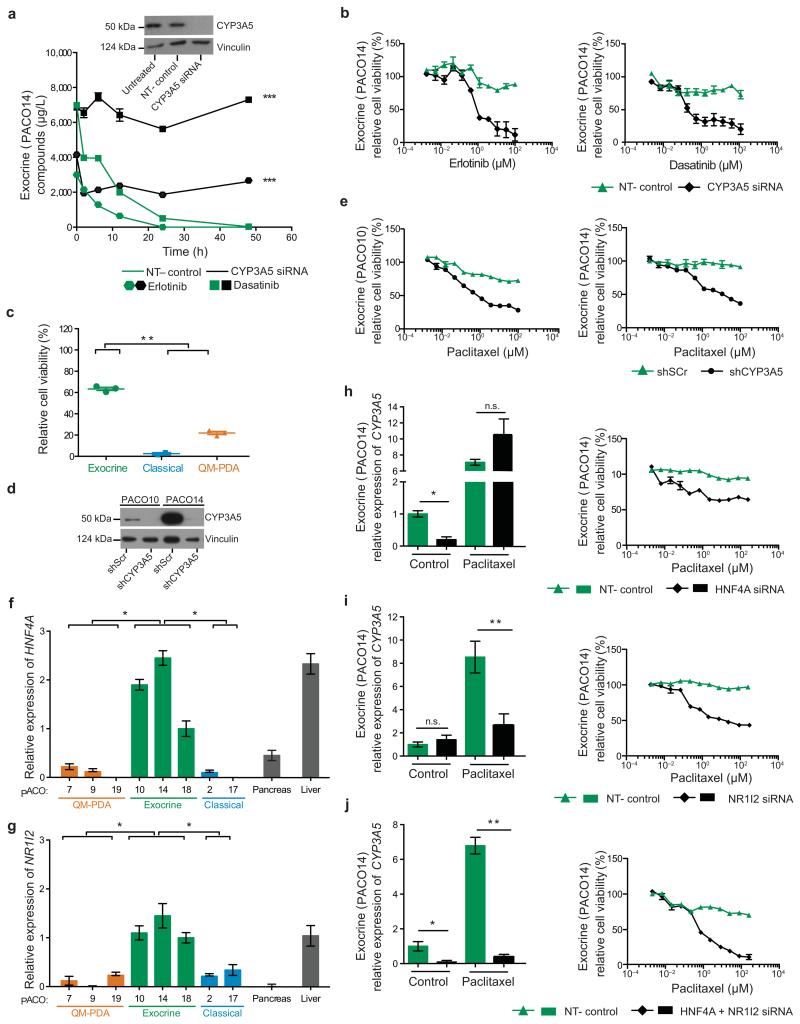
CYP3A5 mediates drug resistance in exocrine-like PDAC cells
To test if CYP3A5 metabolizes erlotinib and dasatinib in exocrine-like PDAC cells, we measured their chemical modification in two different exocrine-like PACO lines transfected with non-targeting (NT–control) or CYP3A5 siRNA. Quantitative mass spectrometric analysis (LC-MS/MS) analysis27,28 revealed a rapid conversion of erlotinib and dasatinib in NT-control cells, as illustrated by loss of their unmodified forms from the supernatant; this did not occur in CYP3A5 siRNA cells (Fig 3a and Supplementary Fig. 3a,b). Chemical modifications by CYP enzymes can have a neutral effect, activate or inactivate small molecule inhibitors31. If CYP3A5 inactivates these compounds, its expression would explain the observed resistance towards erlotinib and dasatinib in exocrine-like PDAC cells. Indeed, siRNA knockdown of CYP3A5 significantly and exclusively sensitized the exocrine-like PACO cells to these drugs (Fig. 3b and Supplementary Fig. 3c,d). As microtubule-targeting taxanes are also substrates for CYP3A family members32,33, we next asked whether CYP3A5 expression impacts the recently introduced paclitaxel-based treatment for PDAC5,6. Treatment of PACO lines with paclitaxel indeed revealed that the exocrine-like subtype was significantly more resistant compared to the other two subtypes (Fig. 3c and Supplementary Fig. 3e–g). To verify the role of CYP3A5 in this context, we established control (shScr) or stable knockdown of CYP3A5 (shCYP3A5) in two exocrine-like PACO lines (Fig. 3d). As observed for erlotinib and dasatinib (Supplementary Fig. 3h,i), knockdown of CYP3A5 sensitized the exocrine-like PACO cells to paclitaxel (Fig. 3e).
Figure 3.
CYP3A5 mediates drug resistance and is regulated by HNF4A and NR1I2 in exocrine-like cells in vitro
(a) Anti-CYP3A5 immunoblot of untreated, non-targeting (NT–control) and CYP3A5 siRNA transfected exocrine-like cells. Vinculin was used as loading control. Compound concentrations in the supernatant of exocrine-like cells transfected with CYP3A5 or NT–control siRNA, followed by treatment with erlotinib or dasatinib (10 μM). Concentrations were determined by LC-MS/MS (n = 6; ***P < 0.001; two-way ANOVA). (b) Exocrine-like cells treated with erlotinib or dasatinib for 48 h, post CYP3A5 or NT–control siRNA transfection (n = 3). (c) PACO line specific sensitivities to 1 μM paclitaxel after 48 h. Bars depict mean ± SD (n = 2; **P < 0.01; grouped one-way ANOVA). (d) Anti-CYP3A5 immunoblot comparing shCYP3A5 with shScr exocrine-like cells. Vinculin was used as loading control. (e) CYP3A5 knockdown or control exocrine-like cells treated with paclitaxel for 48 h (n = 3). (f, g) HNF4A (f) and NR1I2 (g) expression, as measured by qRT-PCR, in PACO lines compared to pancreas and liver mRNA. Values are relative to PACO18 mRNA expression and depict mean ± SEM (n = 3; *P < 0.05; grouped one-way ANOVA). (h-j) CYP3A5 expression in response to 10 μM paclitaxel or DMSO (control) after 48 h of HNF4A- (h), NR1I2- (i) and HNF4A-/NR1I2- double knockdown (j) exocrine-like cells. Values are relative to untreated, NT–control mRNA expression and depict mean ± SEM (n = 3; *P < 0.05; **P < 0.01; n.s. = not significant; Student’s T-test). Exocrine-like cells treated with paclitaxel for 48 h post of HNF4A- (h), NR1I2- (i) and HNF4A-/NR1I2- double knockdown (j) (n = 3).
The strong upregulation of CYP3A5 in response to erlotinib, dasatinib and paclitaxel (Fig. 2h and Supplementary Fig. 3j) suggests a major contribution of CYP3A5 induction to the observed drug resistance. Of the transcription factors known to regulate the expression of CYP3A family members34-37, HNF4A and NR1I2 (PXR) are selectively expressed in exocrine-like PACO cells at levels comparable to those in normal liver (Fig. 3f,g). While HNF4A-dependent transcription is activated by its ubiquitous ligand linoleic acid38, NR1I2 initiates transcription in response to xenobiotics such as erlotinib, dasatinib and paclitaxel39,40. We performed individual or combined siRNA knockdowns of these two transcription factors to test their contribution to basal and induced expression of CYP3A5 (Supplementary Fig. 3k). Basal expression of CYP3A5 was significantly reduced by knockdown of HNF4A but not NR1I2 (Fig. 3h,i and Supplementary Fig. 3l,m). In contrast, induction of CYP3A5 expression by erlotinib, dasatinib and paclitaxel was significantly impaired by knockdown of NR1I2 but not HNF4A (Fig. 3h,i and Supplementary Fig. 3l,m). Combined knockdown of HNF4A and NR1I2 significantly decreased both basal and drug-induced CYP3A5 expression (Fig. 3j and Supplementary Fig. 3n,o). Next, we tested the contribution of both factors to drug resistance. Knockdown of either HNF4A or NR1I2 rendered exocrine-like PACO cells susceptible to all tested drugs (Fig. 3h,i and Supplementary Fig. 3l,m). The combined knockdown of HNF4A and NR1I2 rendered the cells even more sensitive as achieved by the individual knockdowns (Fig. 3j and Supplementary Fig. 3n).
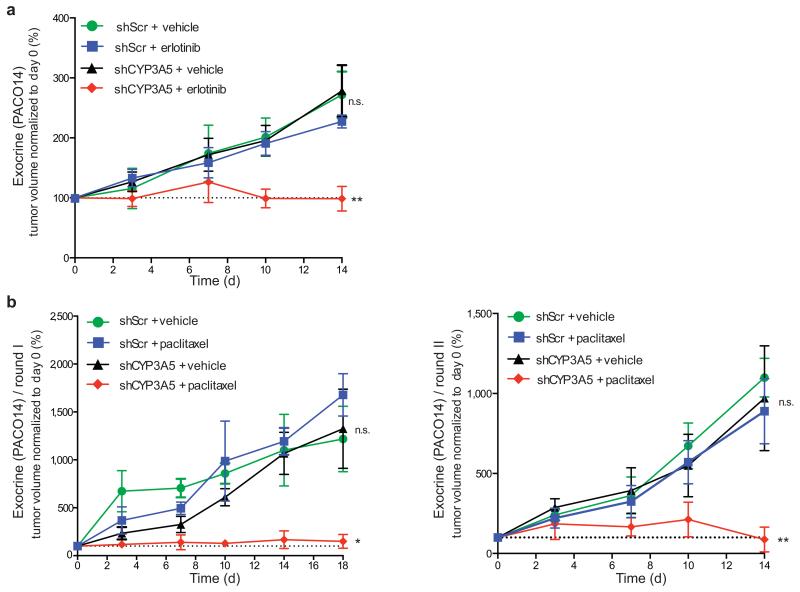
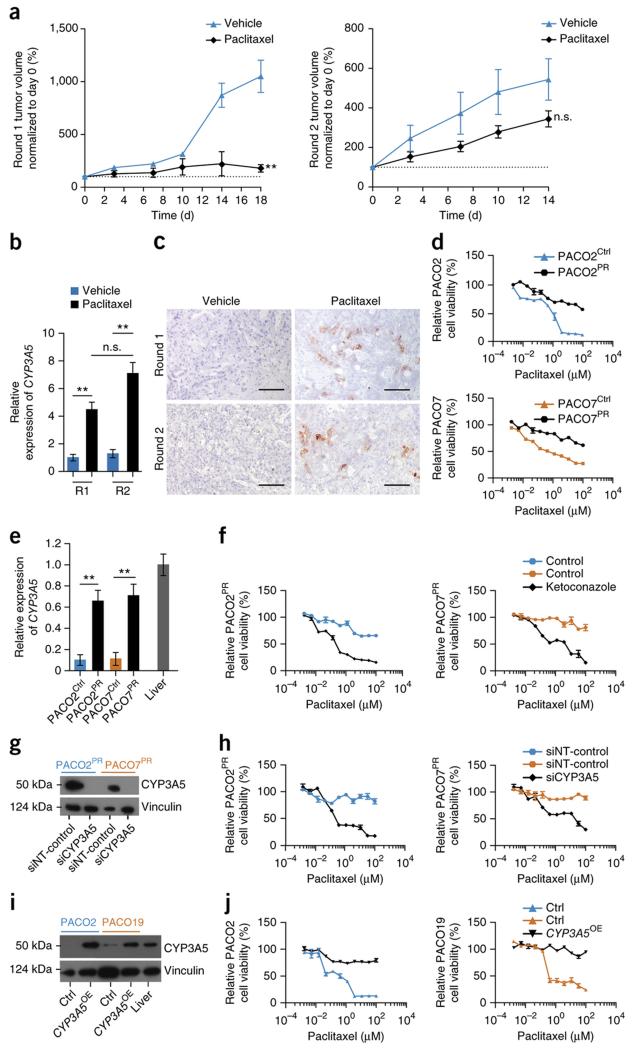
We next asked if ablation of CYP3A5 expression could sensitize established tumors to small molecule drugs in vivo. We thus established subcutaneous tumors from shScr or shCYP3A5 exocrine-like PACO cells in NSG mice. Once tumors reached an average volume of 200 mm3, mice were treated with erlotinib or vehicle by oral gavage for five consecutive days, following two days of rest for a total of 14 days. Whereas treatment with erlotinib had no significant effect on the growth rate of the shScr tumors, growth of the shCYP3A5 tumors was significantly inhibited (Fig. 4a and Supplementary Fig. 4a). Additionally, CYP3A5 expression was significantly increased in the erlotinib but not vehicle treated shScr tumors (Supplementary Fig. 4b). Knockdown of CYP3A5 also significantly enhanced the response to paclitaxel (Fig. 4b and Supplementary Fig 4c). To extend the in vivo treatment period we re-injected cells recovered after treatment round one into secondary mice (round two). Even in round two, paclitaxel significantly suppressed growth of shCYP3A5 tumors (Fig. 4b and Supplementary Fig. 4c) and CYP3A5 expression was significantly higher in paclitaxel- compared to vehicle-treated shScr groups (Supplementary Fig. 4d). We conclude that long-term suppression of CYP3A5 in exocrine-like xenografts does not lead to induction of alternative resistance pathways.
Figure 4.
CYP3A5 mediates drug resistance in exocrine-like PDAC cells in vivo
(a) Growth curves of PDAC xenografts from exocrine-like shScr and shCYP3A5 cells, treated for two cycles of 5 days with erlotinib (100 mg/kg) and 2 days recovery. (b) Growth curves of PDAC xenografts from exocrine-like shScr and shCYP3A5 cells treated with two cycles of 5 days paclitaxel (2 mg/kg) and 2 days recovery followed by 4 days of paclitaxel (left panel, round I). Cells from one xenograft per treatment group were re-injected and treated for two cycles of 5 days paclitaxel and 2 days recovery (right panel, round II). Tumor volume was measured with a digital caliper. Shown are tumor volumes normalized to baseline (day 0) and depict mean ± SEM. P values were determined at the end point using one-sided Mann-Whitney U test. (n = 6 mice per treatment group; *P < 0.05; **P < 0.01; n.s. = not significant).
CYP3A5 contributes to acquired resistance in QM-PDA and classical PDAC cells
The development of secondary resistance limits the efficacy of drug treatment in PDAC1. We thus asked if CYP3A5 also contributes to acquired resistance. To this end, tumors of the classical subtype were treated with paclitaxel for two rounds for a total of 32 days. Paclitaxel treatment significantly inhibited tumor growth of classical xenografts during the first treatment round, while longer-term treatment led to a marked development of paclitaxel resistance (Fig. 5a). CYP3A5 mRNA and protein expression significantly increased after paclitaxel treatment (Fig. 5b,c). Since CYP3A4 and CYP3A7 remained absent (Supplementary Fig. 5a), these data suggest a role of CYP3A5 in acquired drug resistance.
Figure 5.
CYP3A5 contributes to acquired resistance in QM-PDA and classical PDAC cells
(a) Growth curves of PDAC xenografts derived from classical cells treated as described for (Fig. 4b) round I (left panel, round I). Cells from one xenograft per group were re-injected and treated as described for (Fig. 4b), round II (right panel, round II). (n=6 mice per treatment group; **P < 0.01; n.s. = not significant). (b) CYP3A5 expression, as measured by qRT-PCR, in tumors after paclitaxel or vehicle treatment after the first (RI) and after the second (RII) treatment round. Values are relative to RI vehicle control and depict mean ± SEM (n = 3; **P < 0.01; Student’s T-test). (c) CYP3A5 immunostainings on PACO17 xenograft sections post paclitaxel or vehicle treatment after the first (Round I) and the second (Round II) treatment round (n = 3). Scale bar, 100μM. (d) Parental (PACO2Ctrl, PACO7Ctrl) and paclitaxel-resistant (PACO2PR, PACO7PR) classical and QM-PDA cell lines treated with paclitaxel for 48 h (n = 3). (e) CYP3A5 expression in PACO2Ctrl and PACO7Ctrl cells compared to PACO2PR and PACO7PR cells. Values are relative to liver mRNA and depict mean ± SEM (n = 3; **P < 0.01; Student’s T-test). (f) PACO2PR and PACO7PR cells treated with paclitaxel for 48 h post ketoconazole (100 nM) or vehicle pre-treatment for 2 h (n = 3). (g) Anti-CYP3A5 immunoblot of NT–control or CYP3A5 siRNA transfected PACO2PR and PACO7PR cells. Vinculin was used as loading control. (h) PACO2PR and PACO7PR cells treated with paclitaxel for 48 h post transfection with CYP3A5 or non-targeting (NT-control) siRNA (n = 3). (i) Anti-CYP3A5 immunoblot of classical and QM-PDA cell lines transduced with CYP3A5_OX or Ctrl vectors. Vinculin was used as loading control. (j) CYP3A5_OX- or Ctrl- transduced classical and QM-PDA cell lines treated with paclitaxel for 48 h (n = 3).
To functionally explore this hypothesis, we generated paclitaxel resistant classical (PACO2PR) and QM-PDA (PACO7PR) PACO lines (Fig. 5d). In line with the findings from the in vivo treatment, CYP3A5 expression was significantly increased in these PACO sublines compared to the parental control lines (Fig. 5e), whereas expression of CYP3A4 and CYP3A7 remained absent (Supplementary Fig. 5b). Similar results were obtained for dasatinib and erlotininb resistant lines (Supplementary Fig. 5c,d). Inhibition of CYP3A5 with ketoconazole (Fig. 5f) or CYP3A5 knockdown (Fig. 5g,h) restored drug sensitivity in the paclitaxel resistant sublines to levels comparable to that of the parental lines. Furthermore, ectopic expression of CYP3A5 in non-exocrine-like cells conferred drug-resistance (Fig. 5i,j and Supplementary Fig. 5e,f), confirming that CYP3A5 upregulation is a primary mechanism responsible for the observed acquired resistance.
CYP3A5 contributes to drug resistance in other malignancies
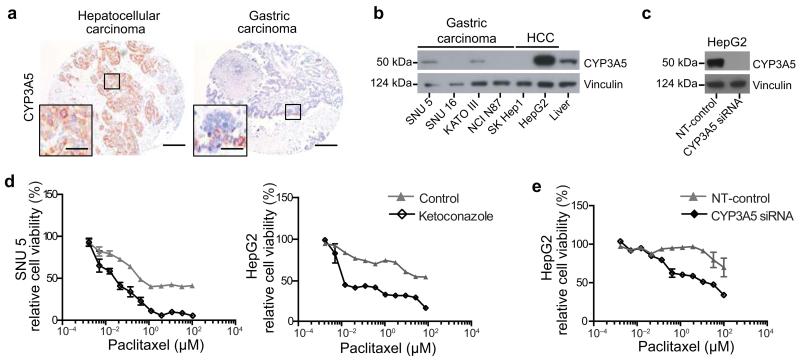
Expression of CYP family members has been described in a range of tumors14,41. To address whether CYP3A5 mediates resistance in tumor entities other than PDAC, we stained a TMA comprising 438 individual tissue samples of 33 distinct tumor entities for CYP3A5 (Fig. 6a and Supplementary Fig. 6a). Samples from 10 out of 33 tumor entities expressed detectable CYP3A5. Particularly high CYP3A5 expression was found in the majority of hepatocellular carcinoma, gastric carcinoma, cervical carcinoma, adrenal gland cortical carcinoma and biliary tract cancers, indicating that CYP3A5 may mediate drug resistance in a considerable fraction of solid tumor entities (Supplementary Table 9). To begin to test this hypothesis we screened a number of gastric and hepatocellular carcinoma cell lines for CYP3A5 expression. The gastric cancer cell line SNU 5 and the hepatocellular carcinoma cell line HepG2 expressed CYP3A5 at levels comparable to normal liver and were selected for further experiments (Fig. 6b and Supplementary Fig. 6b). Upon paclitaxel exposure CYP3A5 expression was further induced (Supplementary Fig. 6c). Moreover, pre-treatment with ketoconazole sensitized both cell lines to paclitaxel treatment (Fig. 6d). A similar sensitization was observed by CYP3A5 knockdown in HepG2 cells (Fig. 6c,e and Supplementary Fig. 6d), suggesting that CYP3A5 expression contributes to drug resistance in tumor types in addition to PDAC.
Figure 6.
CYP3A5 contributes to drug resistance in other malignancies
(a) CYP3A5 immunostainings of human hepatocellular and gastric carcinoma paraffin sections from a tissue microarray containing various tumor entities (n = 16). Scale bar, 100 μM; Scale bar, 5 μM. (b) Anti-CYP3A5 immunoblot of four gastric and two hepatocellular (HCC) carcinoma cell lines, compared to total normal liver lysate. Vinculin was used as loading control. (c) Anti-CYP3A5 immunoblot of CYP3A5 NT-control siRNA transfected HepG2 cells. Vinculin was used as loading control. (d) SNU 5 and HepG2 cells treated with paclitaxel for 48 h, post ketoconazole (100 nM) or vehicle pre-treatment for 2 h. (e) HepG2 cells treated with paclitaxel for 48 h, post CYP3A5 or NT-control siRNA transfection (n = 3).
Discussion
We here confirm the existence of three reported PDAC subtypes11 and identify two surrogate markers, HNF1A and KRT81, for tumor stratification by immunohistochemistry. Our finding that individuals with resectable HNF1A+ exocrine-like PDAC have the best survival might be perceived contradictory at first. However, patient survival is not only determined by drug response; growth rate of the primary tumor as well as the propensity for and the pattern of metastasis also influence survival38. In fact, exocrine-like PACO cells are slowly expanding in culture and have a delayed onset in xenograft formation compared to the classical and QM-PDA subtypes (data not shown). This suggests that exocrine-like tumors, despite showing treatment resistance, are the least aggressive PDAC subtype.
Drug response in patients is influenced by hepatic CYPs that mediate systemic drug metabolism, whereas only minor amounts of these enzymes are expressed in other tissues42,43. Although a role for CYPs in tumor cell autonomous drug detoxification has been postulated34,44-55, this concept has never been functionally demonstrated. We now demonstrate that CYP3A5 contributes to both basal and acquired resistance to small molecule drugs in PDAC. Since CYP3A5 is dispensable for normal physiology56,57, its inhibition in cancers is a promising therapeutic option. Designing a CYP3A5-specific inhibitor may be challenging due to the structural similarities amongst CYP3A family members, although a highly selective CYP3A4 inhibitor has been reported58. Enzymes of the CYP family are frequently induced by their substrates12,37. We show that basal and substrate-induced expression of CYP3A5 is differentially regulated by HNF4A and NR1I2. Interfering with these regulatory mechanisms may provide an alternative approach to suppress the CYP3A5 pathway, thus overcoming basic and acquired drug-resistance in PDAC.
The described CYP3A5 mediated resistance mechanism is not limited to PDAC, since expression and functional analyses suggest that subsets of other cancer entities may employ the same resistance strategy. Consequently, CYP3A5 expression should be considered in the interpretation of drug trials, as targets of this enzyme likely have decreased efficacy in CYP3A5-expressing tumors.
Online Methods
Human tissue specimens
Tissue samples were obtained from patients admitted to the Department of General, Visceral and Transplantation Surgery, University of Heidelberg (Prof. Dr. Markus W. Büchler). The study was approved by the ethical committee of the University of Heidelberg (case number 301/2001) and conducted in accordance with the Helsinki Declaration; written informed consent was obtained from all patients. Patient and tumor characteristics are summarized in Supplementary Table 1. The PDAC validation cohort consists of a subset of the HIPO-015 study, for which xenografts were readily available. The xenografts were also derived from surgical removed specimens from individuals with PDAC that received partial pancreatoduodenectomy at the University Hospital Heidelberg. The study was approved by the ethical committee of the University of Heidelberg (case number S-206/2011) and conducted in accordance with the Helsinki Declaration; written informed consent was obtained from all patients. Patient and tumor characteristics of the PDAC xenograft validation cohort are summarized in Supplementary Table 5.
Xenografts of primary tumor specimens and PACO cell lines
To establish primary xenografts, tumors were cut into pieces of 1-2 mm3 and implanted onto the pancreas of female NOD.Cg-Prkdcscid Il2rgtm1Wjl (NSG) mice at the age of 8 to 12 weeks, which were bred in the animal facility of the German Cancer Research Center. For the generation of xenografts from the PACO lines, a suspension of 105 -106 cultured cells in Matrigel (2 mg/ml) (BD) was injected into the pancreas of NSG mice. Engraftment of tumors and subsequent growth was monitored by regular palpation of the implantation site. Orthotopically grafted tumors were surgically removed and subsequently analyzed by immunohistochemistry and gene-expression profiling, or used for the generation of PACO cultures. Animal care and all procedures followed the German legal regulations and were previously approved by the governmental review board of the state of Baden-Wuerttemberg, Regierungspraesidium Karlsruhe authorization numbers G64-10, G39-13, G105-14 and G80-15.
Cell Culture
For the generation of PACO cultures, primary xenografts were resected after attaining a volume of approximately 1 cm3. Tumor pieces were first minced using sterile scalpels and dissociated into single cells by incubation with 1 μg/ml collagenase IV (Sigma) for 2 h at 37 °C. The resulting suspension was filtered through a 100 μM mesh, and cell debris and dead cells were removed by density centrifugation (FiColl Paque Plus, Amersham). Remaining erythrocytes were removed using ACK Buffer (Lonza). For establishing PACO cultures, single cells (5 × 106) were seeded in T75 flasks (Primaria, BD) in serum-free medium (referred to as PACO medium) as described before59. Adherent monolayer cultures were maintained at 37 °C and 5% CO2. After the outgrowth of tumor cells, contaminating fibroblasts were removed by trypsinization. We obtained SNU 5, SNU 16, KATO III, NCI N87, SK Hep1 and HepG2 cells from the American Type Culture Collection.
Erlotinib-, dasatinib- and paclitaxel-resistant PACO cells were generated by continuously exposing cells to the individual compounds (10 nM or 20 nM) for two months. In brief, medium supplemented with the respective drugs was replaced every 4 days. At a confluency of 70%, cells were passaged and allowed to recover for 24 h until treatment was continued. Erlotinib, dasatinib or paclitaxel resistances were confirmed by dose-response studies as described below.
All cell lines used were monthly authenticated by Single Nucleotide Polymorphism profiling and tested for mycoplasma contaminations (both Multiplexion).
Sanger sequencing
Genomic DNA was prepared from PACO cells using DNAeasy Blood & Tissue Kit (Qiagen). Genomic DNA regions containing KRAS and TP53 mutations were amplified by PCR using Q5 hot start high-fidelity master mix (New England Biolabs Inc.) according to manufacturer’s instructions. PCR primer pairs used are summarized in Supplementary Table 10. PCR products were purified using the High Pure PCR Product Purification Kit (Roche). Sanger sequencing was subsequently performed (GATC) and analyzed using ApE software, Version 2.0.49. Variant positions are relative to the reference sequences NM_004985 (KRAS) and NM_00546 (TP53).
Gene-expression analysis
Total RNA was isolated from different PACO lines at early and late passages at 80% confluency, or from 50 mg of tumor tissue using the miRNeasy kit (Qiagen) according to manufacturer’s instructions. Gene expression analysis was performed using Illumina HumanHT12v4 BeadChips at the Genomics and Proteomics Core Facility of the German Cancer Research Center (GPCF DKFZ, Heidelberg). Correlation plots, Spearman’s rank correlation coefficients and significance (two-tailed) of log2 gene expression from the PACO datasets (PACO lines, primary (PT) and secondary (DT) xenografts) were generated using Graph Pad Prism 6.0b software.
Gene Set Enrichment Analysis (GSEA) was conducted on quantile-normalized data from the PACO datasets and the validation cohort. In order to assign the corresponding PDAC subtype to the individual PACO samples, the previously described PDAssigner signatures were used to derive genesets for each individual subtype11. We computed P-values using 1,000 or 10,000 permutations for each geneset and adjusted them with the FDR method 60. We performed subtype assignment for the initial eight samples by comparing each individual sample against the remaining seven (REST) for each geneset. Samples were assigned to the signature and respective subtype with the lowest FDR (QM-PDA: PACO7, PACO9, PACO19; exocrine-like: PACO10, PACO14, PACO18; classical: PACO2, PACO17). GSEA was then repeated using the stratified groups for comparison. This initial cohort was used to identify surrogate protein markers for each subtype and subsequent PDAC subtype classifications were marker-based by immunohistochemistry.
RSEM normalized RNA-sequencing expression data of 183 primary PDAC tumors were obtained from the Cancer Genome Atlas (TCGA) PAAD data sets available online (Broad Institute). Spearman’s rank correlation coefficients and significance (two-tailed) were calculated using Graph Pad Prism 6.0b software for each pair of genes.
Gene expression data of the HIPO-015 xenograft validation cohort were quantile-normalized and corrected for unwanted variation using the fsva function of the surrogate variable analysis (SVA) package61,62. Specifically, first, the number of latent factors (“surrogate variables”) was determined by the function n.sv(). Then, the surrogate variables were estimated using the function sva() (with the subtypes as known covariates) and finally the function fsva() was used to regress out the surrogate variables and to obtain the corrected gene expression data. Probes mapping to multiple genes were excluded and the first probe per gene was retained from the remaining set. Unsupervised principal component analysis (PCA) was then computed from the glog2 transformed data using the 500 genes with highest variability across samples63. Calculations were performed by R Version 3.2.2.
The Significance Analysis of Microarray (SAM)64 was used to identify differentially regulated genes at a FDR < 0.05 with a fold change of > 2. Additionally, differential expression of CYP3A5 was validated based on log2 transformed mRNA expression of the annotated xenograft samples from the validation cohort. Intensities of the probe with the highest average intensity per gene were retrieved from quantile-normalized microarray data. Unpaired t-test (two-tailed) was used to compute statistical significance (P < 0.05). Calculations were performed by R Version 3.2.2.
Hierarchical clustering analysis was performed on quantile-normalized, SVA-corrected, glog2-transformed gene expression data of the validation cohort using R Version 3.2.2. Probes mapping to multiple genes were excluded and the first probe per gene was retained from the remaining set. The intersection between the set of gene expression symbols and the previously determined PDAssigner contained the variables used for hierarchical clustering with Manhattan distances and single- linkage.
Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-4029. The results shown in Supplementray Figure 1c and 2n are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/
LC-MS/MS analysis
(S)-(−)Propranolol hydrochloride (Internal Standard) was purchased from Sigma. Acetonitrile was from Bernd Kraft (Duisburg, Germany), ammonium acetate, formic acid from Merck, methanol from VWR International and dimethylsulfoxide from Applichem. 500 μl of reaction media was quenched with 1,000 μl of acetonitrile at each time point and mixed. After centrifugation, clear supernatants were pre-diluted with PACO media and acetonitrile at a ratio 1 : 25. 100 μl of the sample were transferred into a new vial, followed by addition of 10 μl (S)-(−) (S)-(−) Propranolol hydrochloride solution (105 μg/L) and finally vigorously mixed. 10 μl were injected onto column. Calibration and quality control samples were prepared by spiking either dasatinib or erlotinib to the PACO media. The sample was injected onto a PerfectSil Target ODS-3 HPLC column (3 μm, 100 × 2.1 mm, MZAnalysentechnik), using an Agilent 1100 (Agilent) binary pump and degasser, with a CTC PAL sampler (CTC Analytics). Chromatographic separation was performed by gradient elution at a constant flow rate of 250 μl/ min for 15 min. The gradient consisted of 20 mM NH4OAc plus 0.1% formic acid (mobile phase A) and 400 mM NH4OAc/methanol/acetonitrile 5 : 5 : 90 plus 0.1% formic acid (mobile phase B). The gradient applied was 0.0 min, 70% A/30% B; 1.5 min 70% A/ 30% B; 3.0 min 5% A/ 95% B; 11.0 min 5% A/ 95% B; 11.5 min 70% A/ 30% B and 15 min 70% A/ 30% B. From 4 to 8 min runtime, the eluate was directed to a QTrap 5500 mass spectrometer (SCIEX) equipped with an electrospray ion source. Mass transitions of 488.1 to 401.1 for dasatinib, 394.0 to 278.1 for erlotinib and 260.1 to 116.1 for (S)-(−) Propranolol were monitored. Ionization was achieved at 5.5 kV and a temperature of 400 °C. Nitrogen was applied as curtain, collision and drying gas. De-clustering potentials, collision energy and collision exit potential was as follows: 26 V, 39 V and 12 V for dasatinib, 16 V, 43 V and 26 V for erlotinib and 61 V, 23 V and 14 V for (S)-(−) Propranolol.
Tissue Microarray
The tissue microarray (TMA) was constructed from individuals that received partial pancreatoduodenectomy for PDAC between 1991 and 2006 at the Charité University Hospital Berlin. The use of this tumor cohort for biomarker analysis has been approved by the Charité University ethics committee (EA1/06/2004). Formalin-fixed and paraffin-embedded tissue samples were used to generate tissue microarrays as previously described65. Briefly, each PDAC included was represented by three different tissue cores, measuring 1 mm in diameter, chosen by a board of certified pathologists as being representative for the respective tumor. From the defined regions, tissue cylinders of 1.5 mm diameter were punched from each donor sample and arrayed into a new ‘recipient’ paraffin block using a semiautomated tissue microarrayer (Beecher Instruments). The human various cancers high density TMA, which is composed of VA2-SBC, VB2-SBC and VC2-SBC (n = 438), was purchased from Super Bio Chips (BioCat).
Immunohistochemistry and marker-based stratification
For a list of all marker candidates tested and a summary of the results obtained see Supplementary Table 4. Tumor specimens were fixed in 10% formalin overnight and embedded in paraffin. For immunohistochemistry, slides were deparaffinized and rehydrated. Antigen retrieval was enhanced by boiling in a steam pot at pH 6 in Dako target retrieval solution (Dako) for 15 min, followed by cooling for 30 min and washing in distilled water. Nonspecific binding was blocked using the Linaris Avidin/Biotin blocking Kit (Vector Labs) according to the manufacturers’ instructions. Slides were incubated with primary antibodies for 30 min, rinsed in PBS-T (PBS with 0.5% Tween-20), incubated for 20 min with the appropriate secondary antibody using the Dako REAL Detection System and rinsed in PBS-T. After blocking of endogenous peroxidase and incubation with Streptavidin HRP (20 min at room temperature (RT)), slides were developed with AEC (Dako) and counterstained with Hematoxylin. Primary antibodies and dilutions are described in Supplementary Table 11. All antibodies were diluted in Dako antibody diluent.
Three pathologists evaluated all sections independently; discordant cases were discussed using a multiheaded microscope until consensus was achieved. The study was carried out blinded to the identity of the specimens. A case was considered positive for a given marker (CYP3A5, KRT81, HNF1A), if the tumor cells in the respective tissue microarray spots showed a detectable staining regardless of the strength of the signal or the number of positive cells. However, in those instances, in which staining of tumor cells was detectable for any of the markers the respective staining was usually strong. Hence, if any tumor cell was found positive for KRT81 or HNF1A in any of the cores, the tumor was defined as QM-PDA or exocrine-like, respectively. Stromal cells were negative in all instances; normal acinar pancreatic cells (when present) expressed HNF1A homogenously to a moderate degree but were consistently negative for the other two markers.
As whole tissue slides were used in the validation cohort of xenografts the scoring system was adapted to account for a higher level of heterogeneity observed in the KRT81 and HNF1A stainings. Specifically, a cut off of at least 10% KRT81-positive tumor cells was introduced to consider a case QM-PDA. Positivity of a single tumor cell did not justify an allocation to a specific biological subtype. Additionally, the evaluation of HNF1A stainings was adapted by only considering cases with moderate or strong nuclear staining reactions exocrine-like. Few cases with an extremely light nuclear staining reaction of HNF1A were observed, which was not considered to represent a biologically relevant HNF1A expression.
Immunofluorescence
PACO cells were seeded on T75 flasks (Primaria, BD) and grown to 60-70% confluency. Cells were fixed in 4% freshly depolymerized formaldehyde for 15 min, permeabilized with 0.25% (v/v) Triton X-100 (Sigma) in PBS for 45 min and blocked with 1% BSA in PBS for 1 h. Primary antibodies (Supplementary Table 11) were incubated O/N at 4 °C, and detected by fluorescence using secondary antibodies coupled to fluorochromes (Life Technologies) diluted 1:1,000 for 1 h in the dark. Isotype-matched secondary antibodies conjugated with Alexa-Fluor-488 or PE were incubated for 1 h at RT. Slides were mounted using ProLong Antifade GOLD with DAPI (Life Technologies) as described by the manufacturer.
Western blot analysis
Whole cell lysates of PACO cells were prepared using RIPA buffer (Cell Signaling Technology), 1 mM PMSF (Sigma), 1 mM EDTA and Halt Protease/Phosphatase Inhibitor Cocktail (Pierce). Protein lysates were resolved on 4-12% Bis/Tris NuPage gels with MOPS running buffer (Life Technologies) and blotted on nitrocellulose membranes (Amersham International). Membranes were blocked for 1 h in TBS containing 0.1% (v/v) Tween-20 with 20% (w/v) non-fat dry milk powder (blocking solution). Primary antibodies (Supplementary Table 11) were incubated O/N at 4 °C in blocking solution. Secondary HRP-coupled antibodies (Southern Biotech) were diluted 1:10,000 in blocking solution and incubated for 1 h at RT. Membranes were washed in 0.1% TBSTween and immunocomplexes were detected using the ECL kit (Amersham International). As positive control recombinant CYP3A5, CYP3A4, CYP3A7 (Abnova) and human liver lysates (Novus) were used.
Real-time quantitative PCR
Total RNA was extracted using the miRNeasy mini kit (Qiagen) and reverse transcribed using the high capacity cDNA reverse transcription kit (Applied Biosystems). cDNA corresponding to 10 ng of starting RNA was used for relative RNA quantification (qRT-PCR). TaqMan probes (Applied Biosystems) for CYP3A5 (HS00241417_m1), CYP3A4 (HS0060406_m1), CYP3A7 (Hs00426361_m1), HNF4A (Hs00230853_m1) NR1I2 (Hs01114267_m1), PPIA (HS04194521_s1) and GAPDH (HS9999905_m1) were used to acquire expression data with the Viia 7 Real-Time PCR System (Applied Biosystems). The ViiA 7 software 1.1 was used for data acquisition and analysis. As positive control, RNA from total normal liver and pancreas were used (Novus).
siRNA transfection of PACO cells
PACO cells were grown to 80% confluency. The transfection reagent Dharmafect 4 (Thermo Scientific), non-targeting (NT-control) and CYP3A5, HNF4A or NR1I2 siRNA (On-Target plus SMARTpool Thermo Scientific, Supplementary Table 12) were pre-incubated at RT for 5 min at a ratio of 1:4 in IMDM culture medium (Gibco). For the HNF4A and NR1I2 double knockdown, the individual siRNAs were pre-incubated together at a ratio of 1:8 in IMDM culture medium (Gibco). Dharmafect 4 was then combined with the siRNAs and incubated for further 20 min at RT. The mixture was then added to the PACO culture medium. The culture medium was aspirated and the transfection agent-RNA complex mixture was added to the monolayer. Flasks were incubated at 37 °C for 72 h until further analysis.
Generation of stable knockdown cells
Stable shRNA-mediated knockdown of CYP3A5 was achieved by targeting the sequence TTGATTTCAACATCTTTCT (shCYP3A5) in a pGIPZ vector (GE healthcare, Thermo Scientific; Supplementary Table 12). In addition the non-silencing control pGIPZ vector (shScr) was used as negative control (GE healthcare, Thermo Scientific). Lentiviral particles were produced in HEK 293T cells. Viral particles were concentrated and PACO cells were transduced at a multiplicity of infection of 1 to 5. Successfully transduced cells were selected by cell sorting for GFP. qRT-PCR and Western blotting confirmed knockdown efficiency.
Stable expression of CYP3A5
PACO cells were stably transduced with the expression vector pLenti-GIII-CMV-RFP-2A-Puro (Applied Biological Materials Inc.) containing the full CYP3A5 open reading frame (CYP3A%_OX) or empty control vector (Control). Lentiviral particles were produced in HEK 293T cells. Viral particles were concentrated and PACO cells were transduced at a multiplicity of infection of 1 to 5. Successfully transduced cells were selected by cell sorting for RFP. qRT-PCR and Western blotting confirmed CYP3A5 expression.
Drug treatment assays
Dasatinib, erlotinib and paclitaxel (LC Laboratories) were dissolved in water-free DMSO. For the determination of relative cell viability, 10 μM and 1 μM or serial dilutions of the three drugs were screened in quadruplicates. In brief, 8000 cells per well were seeded in 96-well plates 24 h prior to the addition of the individual compounds. For the co-treatment experiments, either siRNA transfection was carried out as described, or cells were pretreated with 100 nM ketoconazole for 2 h and then treated in the presence of ketoconazole. After incubation for 48 h or 7 days, cell viability was assessed using CellTiterBlue (Promega) following manufacturer’s instructions. Vehicle (DMSO) was used as negative control. Treatment with 20 μM staurosporine (LC Laboratories) was used as positive control. Responses were normalized to DMSO- and staurosporine- treated controls. Relative cell viability curves were plotted using Graph Pad Prism 6.0b software.
In vivo drug treatment
Tumors were established by subcutaneously injecting 5 × 105 shCYP3A5 or shScr exocrine-like (PACO10, PACO14) and classical (PACO17) cells into female NSG mice at the age of 8 to 12 weeks, using Matrigel (2 mg/ml) in a total injection volume of 100 μl. After the tumors reached a size of approximately 200 mm3, mice were randomly assigned to the respective treatment groups (n = 6 per group) for drug administration. Erlotinib was prepared in 0.5% methylcellulose, 0.1% Tween 80 and 99.4% water for injection (WFI). Erlotinib (100 mg/kg) or vehicle were administrated by oral gavage at 5 consecutive days followed by 2 days of rest, for a duration of 14 days. Paclitaxel was prepared in 50% Cremophor EL (Sigma) and 50% absolute ethanol (Sigma) to a concentration of 6 mg/ml. Before administration, paclitaxel was further diluted in 0.9% NaCl (Braun) to a final concentration of 0.4 mg/ml. Paclitaxel (2 mg/kg) or vehicle were then administrated by interperitoneal injection for two cycles of 5 days paclitaxel (2 mg/kg) and 2 days recovery followed by 4 days of paclitaxel. Cells from one xenograft per treatment group were re-injected, and treated for two cycles of 5 days paclitaxel and 2 days recovery. Tumor volumes were determined blinded twice weekly by calliper measurements and calculated according to the formula (length × height × width) × (π/6) at the end of the experiments. Tumor growth was calculated for each individual tumor by normalizing to the tumor volume at day 0. After 14 or 18 days of treatment, mice were sacrificed and tumors were resected for further analysis. Mice were excluded if they had to be sacrificed before the treatment started, or due to exceeding the maximum allowable tumor size during the treatment experiment.
Statistical analysis
For all in vitro experiments at least three biological replicates were used or grouped analyses were carried out. For all in vivo experiments at least six mice per treatment group were used. Hence, for the reported differences, the sample size used gave sufficient power to call them reliable. Quantitative results were analyzed by one-way analysis of variance (one-way ANOVA), one-sided Mann-Whitney U test and Student’s t test using Graph Pad Prism 6.0b software. Survival analysis was performed using Mantel-Cox log-rank test as well as Cox proportional hazards multivariate regression analysis using the Statistical Package for the Social Sciences (IBM SPSS software). Additionally, Pearson chi-square test was used for comparative data analysis, using SPSS. We considered P < 0.05 (two-sided) as statistically significant. For GSEA a false discovery rate (FDR) of < 0.2 was considered statistically significant. In vitro treatment data were evaluated by determining the activity area29 from each dose response curve by adding max (100 – mean response, 0) for every concentration. Activity areas were compared by paired t-test. Calculations were performed in R Version 3.1.066. Estimation of variation within each group was determined by standard error of the mean or standard deviation.
Supplementary Material
Acknowledgements
We thank E. Soyka, S. Bauer and A. Hieronymus for excellent technical assistance. We also thank the Microarray and the NGS unit of the Genomics & Proteomics Core Facility, German Cancer Research Center (DKFZ), for providing expression profiling, NGS and related services and all members of the flow cytometry core facility for excellent support. We thank the DKFZ-Heidelberg Center for Personalized Oncology (DKFZ-HIPO) for technical support and funding through HIPO-015. This work was in part supported by the Dietmar Hopp Foundation and the BioRN Spitzencluster “Molecular and Cell based Medicine” (E.M.N., C.E., E.E., C.K., V.V., W.N., C.R., J.E., F.M.Z., A.T., M.R.S.), the German Bundesministerium für Bildung und Forschung (BMBF) e:Med PANC-STRAT program (grant 01ZX1305 (A.T., M.R.S., M.S., R.E., N.A.G., T.H., O.S., A.S., A.M., W.W.)), the Helmholtz pre-clinical comprehensive cancer center (E.E., A.T., M.R.S.) and the DKFZ-NCT program NCT3.0 (A.T., M.R.S., O.E., M.S., R.E., N.A.G., T.H., O.S.). A.S. was supported by a fellowship of the National Center for Tumor Diseases (NCT) - Heidelberg School of Oncology (HSO). E.E. is recipient of an EMBO long-term fellowship (ALTF 344-2013). The collection and processing of the specimens via PancoBank was supported by Heidelberger Stiftung Chirurgie (M.W.B.), BMBF (grant 01GS08114 (M.W.B.)) and Biomaterial Bank Heidelberg/BMBH (BMBF grant 01EY1101).
Footnotes
Accession codes
Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-4029.
Conflict of Interest Statement: The authors declare no competing financial interests
References
- 1.Hidalgo M. Pancreatic cancer. The New England journal of medicine. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 2.Malvezzi M, Bertuccio P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2014. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2014 doi: 10.1093/annonc/mdu138. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 4.Burris HA, 3rd, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 5.Conroy T, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. The New England journal of medicine. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 6.Von Hoff DD, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. The New England journal of medicine. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore MJ, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 9.Biankin AV, Maitra A. Subtyping Pancreatic Cancer. Cancer Cell. 2015;28:411–413. doi: 10.1016/j.ccell.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 10.Kim S, et al. Identifying molecular subtypes related to clinicopathologic factors in pancreatic cancer. Biomedical engineering online. 2014;13(Suppl 2):S5. doi: 10.1186/1475-925X-13-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collisson EA, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011;17:500–U140. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guengrich FP. Cytochrome P450 Enzymes. In: McQueen CA, editor. Comprehensive Toxicology (Second Edition) Vol. 9. Elesvier Ltd.; 2010. pp. 43–76. [Google Scholar]
- 13.Rochat B. Role of cytochrome P450 activity in the fate of anticancer agents and in drug resistance: focus on tamoxifen, paclitaxel and imatinib metabolism. Clinical pharmacokinetics. 2005;44:349–366. doi: 10.2165/00003088-200544040-00002. [DOI] [PubMed] [Google Scholar]
- 14.Bruno RD, Njar VCO. Targeting cytochrome P450 enzymes: A new approach in anti-cancer drug development. Bioorganic & Medicinal Chemistry. 2007;15:5047–5060. doi: 10.1016/j.bmc.2007.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michael M, Doherty MM. Drug metabolism by tumours: its nature, relevance and therapeutic implications. Expert Opinion on Drug Metabolism & Toxicology. 2007;3:783–803. doi: 10.1517/17425255.3.6.783. [DOI] [PubMed] [Google Scholar]
- 16.Yachida S, Iacobuzio-Donahue CA. Evolution and dynamics of pancreatic cancer progression. Oncogene. 2013;32:5253–5260. doi: 10.1038/onc.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhlen M, et al. Towards a knowledge-based Human Protein Atlas. Nature biotechnology. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 20.Wolfgang CL, et al. Recent progress in pancreatic cancer. CA: a cancer journal for clinicians. 2013;63:318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hruban RH, Pitman MB, Klimstra DS. Tumors of the pancreas. American Registry of Pathology in collaboration with the Armed Forces Institute of Pathology; Washington, D.C.: 2007. [Google Scholar]
- 22.Hong DS, et al. A phase 1 study of gemcitabine combined with dasatinib in patients with advanced solid tumors. Investigational new drugs. 2013;31:918–926. doi: 10.1007/s10637-012-9898-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.George TJ, Jr., Trevino JG, Liu C. Src inhibition is still a relevant target in pancreatic cancer. The oncologist. 2014;19:211. doi: 10.1634/theoncologist.2013-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trevino JG, et al. Inhibition of SRC expression and activity inhibits tumor progression and metastasis of human pancreatic adenocarcinoma cells in an orthotopic nude mouse model. The American journal of pathology. 2006;168:962–972. doi: 10.2353/ajpath.2006.050570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling J, et al. Metabolism and excretion of erlotinib, a small molecule inhibitor of epidermal growth factor receptor tyrosine kinase, in healthy male volunteers. Drug metabolism and disposition: the biological fate of chemicals. 2006;34:420–426. doi: 10.1124/dmd.105.007765. [DOI] [PubMed] [Google Scholar]
- 26.Christopher LJ, et al. Metabolism and disposition of dasatinib after oral administration to humans. Drug metabolism and disposition: the biological fate of chemicals. 2008;36:1357–1364. doi: 10.1124/dmd.107.018267. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Zhao M, He P, Hidalgo M, Baker SD. Differential metabolism of gefitinib and erlotinib by human cytochrome P450 enzymes. Clin Cancer Res. 2007;13:3731–3737. doi: 10.1158/1078-0432.CCR-07-0088. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, et al. Identification of the human enzymes involved in the oxidative metabolism of dasatinib: an effective approach for determining metabolite formation kinetics. Drug metabolism and disposition: the biological fate of chemicals. 2008;36:1828–1839. doi: 10.1124/dmd.107.020255. [DOI] [PubMed] [Google Scholar]
- 29.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding X, Zhang QY. 4.02 - Enzyme Regulation. In: McQueen CA, editor. Comprehensive Toxicology (Second Edition) Elsevier; Oxford: 2010. pp. 9–29. [Google Scholar]
- 31.Janne PA, Gray N, Settleman J. Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nature reviews. Drug discovery. 2009;8:709–723. doi: 10.1038/nrd2871. [DOI] [PubMed] [Google Scholar]
- 32.Haaz MC, Rivory L, Riche C, Vernillet L, Robert J. Metabolism of irinotecan (CPT-11) by human hepatic microsomes: participation of cytochrome P-450 3A and drug interactions. Cancer research. 1998;58:468–472. [PubMed] [Google Scholar]
- 33.Sonnichsen DS, Relling MV. Clinical pharmacokinetics of paclitaxel. Clinical pharmacokinetics. 1994;27:256–269. doi: 10.2165/00003088-199427040-00002. [DOI] [PubMed] [Google Scholar]
- 34.Burk O, et al. The induction of cytochrome P450 3A5 (CYP3A5) in the human liver and intestine is mediated by the xenobiotic sensors pregnane X receptor (PXR) and constitutively activated receptor (CAR) The Journal of biological chemistry. 2004;279:38379–38385. doi: 10.1074/jbc.M404949200. [DOI] [PubMed] [Google Scholar]
- 35.Burk O, Wojnowski L. Cytochrome P450 3A and their regulation. Naunyn-Schmiedeberg's Arch Pharmacol. 2004;369:105–124. doi: 10.1007/s00210-003-0815-3. [DOI] [PubMed] [Google Scholar]
- 36.Tirona RG, et al. The orphan nuclear receptor HNF4alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat. Med. 2003;9:220–224. doi: 10.1038/nm815. [DOI] [PubMed] [Google Scholar]
- 37.Tompkins LM, Wallace AD. Mechanisms of cytochrome P450 induction. Journal of biochemical and molecular toxicology. 2007;21:176–181. doi: 10.1002/jbt.20180. [DOI] [PubMed] [Google Scholar]
- 38.Yuan X, et al. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PloS one. 2009;4:e5609. doi: 10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koutsounas I, Theocharis S, Patsouris E, Giaginis C. Pregnane X receptor (PXR) at the crossroads of human metabolism and disease. Curr Drug Metab. 2013;14:341–350. doi: 10.2174/1389200211314030009. [DOI] [PubMed] [Google Scholar]
- 40.Wu B, Li S, Dong D. 3D structures and ligand specificities of nuclear xenobiotic receptors CAR, PXR and VDR. Drug Discov Today. 2013;18:574–581. doi: 10.1016/j.drudis.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Michael M, Doherty MM. Tumoral drug metabolism: Overview and its implications for cancer therapy. Journal of Clinical Oncology. 2005;23:205–229. doi: 10.1200/JCO.2005.02.120. [DOI] [PubMed] [Google Scholar]
- 42.Pavek P, Dvorak Z. Xenobiotic-induced transcriptional regulation of xenobiotic metabolizing enzymes of the cytochrome P450 superfamily in human extrahepatic tissues. Curr Drug Metab. 2008;9:129–143. doi: 10.2174/138920008783571774. [DOI] [PubMed] [Google Scholar]
- 43.Ding X, Kaminsky LS. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annual review of pharmacology and toxicology. 2003;43:149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- 44.Downie D, et al. Profiling cytochrome P450 expression in ovarian cancer: Identification of prognostic markers. Clinical Cancer Research. 2005;11:7369–7375. doi: 10.1158/1078-0432.CCR-05-0466. [DOI] [PubMed] [Google Scholar]
- 45.Hukkanen J, et al. Induction and regulation of xenobiotic-metabolizing cytochrome P450s in the human A549 lung adenocarcinoma cell line. American journal of respiratory cell and molecular biology. 2000;22:360–366. doi: 10.1165/ajrcmb.22.3.3845. [DOI] [PubMed] [Google Scholar]
- 46.Murray GI, Patimalla S, Stewart KN, Miller ID, Heys SD. Profiling the expression of cytochrome P450 in breast cancer. Histopathology. 2010;57:202–211. doi: 10.1111/j.1365-2559.2010.03606.x. [DOI] [PubMed] [Google Scholar]
- 47.Bergheim I, et al. Cytochrome P450 levels are altered in patients with esophageal squamous-cell carcinoma. World journal of gastroenterology : WJG. 2007;13:997–1002. doi: 10.3748/wjg.v13.i7.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dhaini HR, et al. Cytochrome P450 CYP3A4/5 expression as a biomarker of outcome in osteosarcoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2003;21:2481–2485. doi: 10.1200/JCO.2003.06.015. [DOI] [PubMed] [Google Scholar]
- 49.Gharavi N, El-Kadi AOS. Expression of cytochrome P450 in lung tumor. Current Drug Metabolism. 2004;5:203–210. doi: 10.2174/1389200043489045. [DOI] [PubMed] [Google Scholar]
- 50.McFadyen MC, Melvin WT, Murray GI. Cytochrome P450 CYP1B1 activity in renal cell carcinoma. Br J Cancer. 2004;91:966–971. doi: 10.1038/sj.bjc.6602053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oyama T, et al. Expression of cytochrome P450 in tumor tissues and its association with cancer development. Frontiers in bioscience : a journal and virtual library. 2004;9:1967–1976. doi: 10.2741/1378. [DOI] [PubMed] [Google Scholar]
- 52.Oyama T, et al. Cytochrome P450 in non-small cell lung cancer related to exogenous chemical metabolism. Frontiers in bioscience (Scholar edition) 2012;4:1539–1546. doi: 10.2741/s350. [DOI] [PubMed] [Google Scholar]
- 53.Schmidt R, et al. CYP3A4, CYP2C9 and CYP2B6 expression and ifosfamide turnover in breast cancer tissue microsomes. Br J Cancer. 2004;90:911–916. doi: 10.1038/sj.bjc.6601492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugawara M, et al. Expressions of Cytochrome P450, UDP-Glucuronosyltranferase, and Transporter Genes in Monolayer Carcinoma Cells Change in Subcutaneous Tumors Grown As Xenografts in Immunodeficient Nude Mice. Drug Metabolism and Disposition. 2010;38:526–533. doi: 10.1124/dmd.109.030668. [DOI] [PubMed] [Google Scholar]
- 55.Basseville A, et al. Irinotecan induces steroid and xenobiotic receptor (SXR) signaling to detoxification pathway in colon cancer cells. Molecular cancer. 2011;10:80. doi: 10.1186/1476-4598-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuehl P, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nature genetics. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 57.Westlind-Johnsson A, et al. Comparative analysis of CYP3A expression in human liver suggests only a minor role for CYP3A5 in drug metabolism. Drug Metabolism and Disposition. 2003;31:755–761. doi: 10.1124/dmd.31.6.755. [DOI] [PubMed] [Google Scholar]
- 58.Walsky RL, et al. Selective mechanism-based inactivation of CYP3A4 by CYP3cide (PF-04981517) and its utility as an in vitro tool for delineating the relative roles of CYP3A4 versus CYP3A5 in the metabolism of drugs. Drug metabolism and disposition: the biological fate of chemicals. 2012;40:1686–1697. doi: 10.1124/dmd.112.045302. [DOI] [PubMed] [Google Scholar]
Methods-only references
- 59.Vermeulen L, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parker HS, Corrada Bravo H, Leek JT. Removing batch effects for prediction problems with frozen surrogate variable analysis. PeerJ. 2014;2:e561. doi: 10.7717/peerj.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 64.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stenzinger A, et al. High SIRT1 expression is a negative prognosticator in pancreatic ductal adenocarcinoma. BMC cancer. 2013;13:450. doi: 10.1186/1471-2407-13-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.