Abstract
Altered expressions of glycans is associated with cell-cell signal transduction and regulation of cell functions in the bone marrow microenvironment. Studies of this microenvironment often use two human bone marrow stromal cell lines, HS5 and HS27a, co-cultured with myeloid cells. We hypothesized that differential protein glycosylation between these two cell lines may contribute to functional differences in in vitro co-culture models. In this study, we applied an integrated strategy using genomic, proteomic, and functional glycomic techniques for global expression profiling of N-glycans and their related genes and enzymes in HS5 vs. HS27a cells. HS5 cells showed significantly enhanced levels of bisecting N-glycans (catalyzed by MGAT3), whereas HS27a cells showed enhanced levels of Galβ1, 4GlcNAc (catalyzed by β4GalT1). This integrated strategy provides useful information regarding the functional roles of glycans and their related glycogenes and glycosyltransferases in the bone marrow microenvironment, and a basis for future studies of crosstalk among stromal cells and myeloma cells in co-culture.
Keywords: bone marrow stromal cells, glycans, gene microarray, lectin microarray, SILAC
INTRODUCTION
The bone marrow microenvironment consists of a specialized population of cells that play essential roles in regulation, self-renewal, and differentiation of adult stem cells. The microenvironment supports maturation of hematopoietic stem cells (HSCs) and hematopoietic progenitor cells (HPCs) and their release into the vascular system [1]. Mesenchymal/stromal cells that represent integral components of the microenvironment contribute to the regulation and release of HPCs via adhesion molecules, extracellular matrix (ECM), and soluble factors, including cytokines and chemokines [2, 3]. HS5 and HS27a, two bone marrow stroma cell lines, both derived from the same healthy marrow donor [4], express strikingly different functions [5, 6]. HS5 has a fibroblastic appearance and secretes high levels of granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage CSF (GM-CSF), macrophage CSF (M-CSF), interleukin-6 (IL-6), IL-8, and IL-11, and supports the proliferation of later stages of co-cultured hematopoietic cells [4, 7]. HS27a secretes low levels of growth factors but express high levels of glycoproteins such as ICAM-1(CD54) and MCAM (CD146), and supports the formation of “cobblestone” areas [4]. These two cell lines have been widely used in in vitro studies as representative components of the bone marrow microenvironment and as partners in the crosstalk between marrow stroma cells and co-cultured hematopoietic cells [5, 8, 9]. For example, apoptosis-resistant clonal myelodysplastic syndrome (MDS) progenitor cells from patients with advanced MDS acquired sensitivity to apoptosis induced by TNF-α following stromal contact [10–12]. HS5 and HS27a cells were also used for establishing the xenotransplantation murine model of MDS [13]. Kerbauy et al. observed engraftment of distinct clonal MDS-derived hematopoietic precursors when stromal cells (HS5 and HS27a cells combined) were co-injected via an intramedullary route [14]. Li et al. recently reported that intravenous co-administration of HS27a cells (but not HS5 cells) with HPCs from MDS patients facilitated engraftment of clonal CD34+ cells of any karyotype [15]. Their findings suggest that HS27a cells are more effective than HS5 cells in supporting primitive clonal MDS precursors.
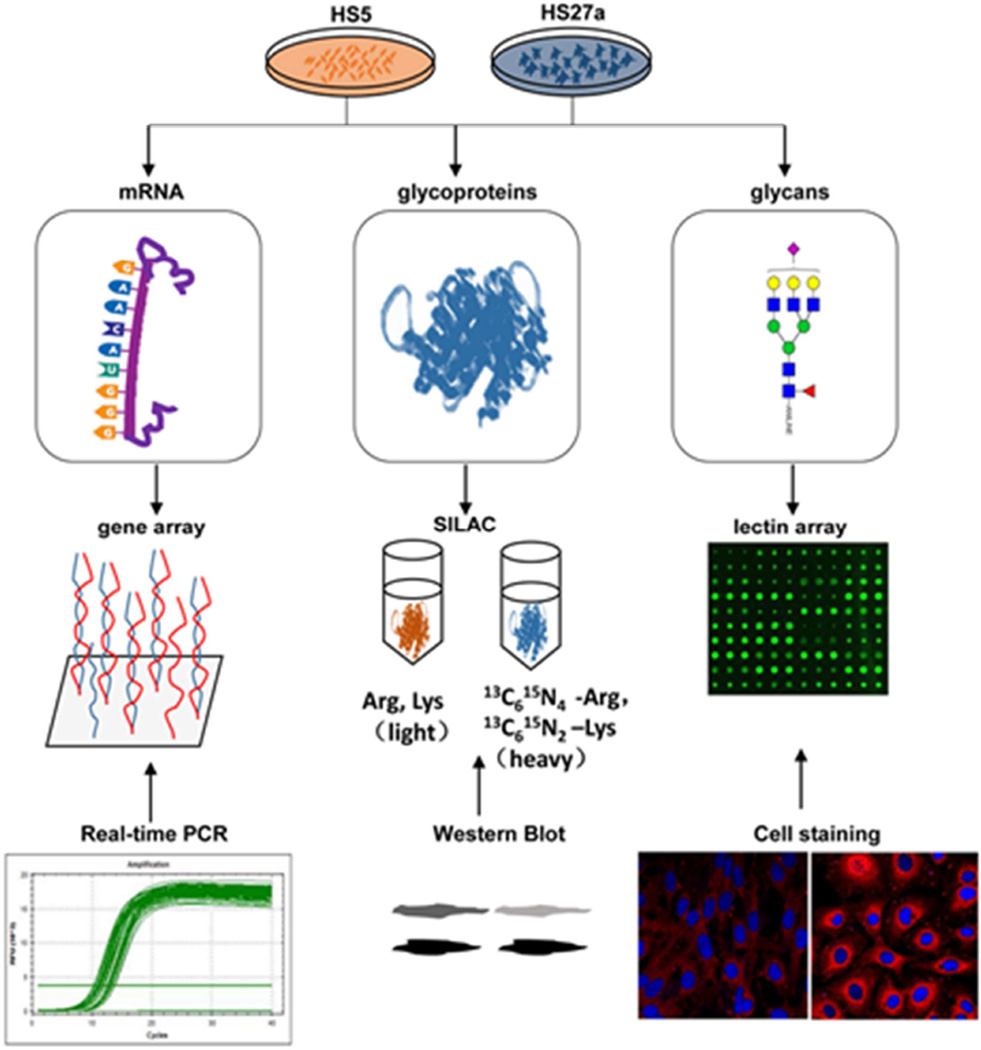
Glycosylation modification plays crucial roles in cell adhesion, differentiation, proliferation, apoptosis, and signal transduction [16–19]. Mice with knockout (Fut8−/−) of the gene that encodes the α1, 6-fucosyltransferase enzyme showed abnormal pro-B cell to pre-B cell transition and reduction of peripheral blood B cells and immunoglobulin production [20]. Cell surface antigens such as CD133, a 120-kDa glycosylated polypeptide used as an HSC biomarker, are often glycosylated [21, 22]. Differences of glycosylation expression between stromal HS5 and HS27a cells have not been addressed in any study to date. We used an integrated strategy that combines genomic, proteomic, and functional glycomic techniques for comparative profiling of glycans, their related genes, and proteins in HS5 vs. HS27a cells. Expression levels of glycosyltransferases and glycosidases were quantitatively analyzed by the Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) method, and glycans were subjected to lectin microarray analysis. This integrated strategy (summarized in Figure 1) was useful in revealing global differences in glycan expression between these two cell lines, and can be extended to similar comparative studies of other cell lines.
Figure 1.
The integrated strategy (schematic) used in this study.
MATERIALS AND METHODS
Cell Lines and Culture
Human marrow stromal cell lines HS5 and HS27a, originally derived from marrow of a healthy subject and immortalized by transduction with human papilloma virus E6/E7 constructs were were maintained as previously described [23]. Multiple aliquots from early passages were cryopreserved for later use.
SILAC was performed as described previously [24, 25]. SILAC reagents and media were from Thermo Scientific (San Jose, CA, USA); final concentration of arginine (Arg) and lysine (Lys) was 100 µg/mL. HS27a cells were cultured in SILAC medium containing 13C6 15N4 Arg and 13C6 Lys (heavy). HS5 cells were cultured in SILAC medium containing 12C6 14N4 Arg and 12C6 Lys (light). Culture medium was replaced every other day until cells were 70–80% confluent. Cells were grown for 5 or 6 passages. Labeling efficiency was checked to ensure an incorporation rate >95%.
Glycogene Microarray Analysis
Differential gene expression in HS5 and HS27a cells was analyzed by B. Torok-Storb's group (Fred Hutchinson Cancer Research Center; Seattle, WA, USA) as described previously [5]. Open-access data at http://www.ncbi.nlm.nih.gov/sites/GDSbrowser (GEO accession: GSE463) were downloaded. Glycogenes listed on the GlycoV4 oligonucleotide microarray (covering 1260 human glycogenes) were extracted, and analyzed as described [26, 27]. Raw values were normalized using the robust multichip average (RMA) expression summary. Data were processed using R program software and Bioconductor project (GEOquery 2.23.2, www.r-project.org/). Fold changes were estimated by fitting a linear model for the genes, and linear modeling was performed with the Limma package in the R program software for differential expression analysis. Transcripts differentially expressed in HS5 vs. HS27a samples were compared using thresholds of fold change >1.5, fold change <0.67, adjusted p-value <0.05.
RNA Isolation
Total RNA from cultured HS5 and HS27a cells was isolated using an RNApure Tissue Kit (CWBiotech; Beijing, China) according to the manufacturer’s instructions. Highly purified RNA (A260/A280 >1.8) was used.
Quantitative Real-Time PCR (RT-PCR)
Primers were designed using the DNAMAN program V. 6.0.3 (Lynnon Biosoft; Vaudreuil, Quebec, Canada). Total RNA was converted into cDNA using a ReverTra Ace-α First-strand cDNA Synthesis Kit (Toyobo; Osaka, Japan). Quantitative RT-PCR was performed by Light Cycler based SYBR Green I dye detection with UltraSYBR Mixture (CWBiotech). mRNA levels of target genes were normalized to expression of β-actin and quantified using the 2−ΔΔCT method [28].
Total Protein Extraction and Western Blot Analysis
Unlabeled and labeled cells with 75–90% confluence were lysed with T-PER Tissue Protein Extraction Reagent (Thermo Scientific, Hudson, NH, USA) according to the manufacturer’s instructions. In brief, cells were trypsinized, resuspended in 1×PBS (0.01 mol/L phosphate buffer containing 0.15 mol/L NaCl, pH 7.2), added with an appropriate amount of T-PER Reagent containing 0.1% aprotinin, incubated on ice for 30 min, and homogenized. The sample was centrifuged for 15 min at 12,000 rpm (4 °C), and the supernatant was collected and stored at −80 °C. Protein content was determined by BCA assay (Beyotime; Shanghai, China).
Proteins from each sample were separated by 10% SDS-PAGE and transferred onto PVDF membranes (Bio-Rad; Hercules, CA, USA) by the Trans-Blot Turbo Transfer System (Bio-Rad). Membranes were soaked in 5% (w/v) skim milk in TBST (20 mM Tris-HCl, 150 mM NaCl, 0.05% Tween 20, pH 8.0) for 2 hr at 37 °C, probed with primary antibodies against MGAT3 (Santa Cruz Biotechnology; Santa Cruz, CA, USA) and β4GalT1 (Abcam; Cambridge, MA, USA) overnight at 4 °C, and incubated with appropriate HRP-conjugated secondary antibody. Bands were visualized using enhanced chemiluminescence detection kit Westar Nova (Cyanagen; Bologna, Italy) with imaging by ChemiDoc™ XRS+ (Bio-Rad).
Quantitative Analysis of Proteins Associated with Glycan Biosynthesis by SILAC
Cells with various labeling were lysed by T-PER Reagent. Proteins were mixed at equivalent ratios, reduced (10 mM DTT, 45 min), and alkylated (30 mM IAM, 45 min at room temperature [RT] in the dark). Sequencing grade modified trypsin (Promega; Madison, WI, USA) was added to 1:100 (w/w), and the mixture was incubated at 37 °C for 24 hr [29]. 2D-LC-MS was performed using LTQ Orbitrap MS (Thermo Fisher Scientific; Waltham, MA, USA) as described previously [30, 31] (see supplemental data). Data were analyzed using MaxQuant software program V. 1.4.1.2 [31, 32].
Lectin Microarray Analysis and Data Analysis
Lectin microarray analysis was performed as described previously [33]. In brief, 37 commercial lectins from Vector Laboratories (Burlingame, CA, USA), Sigma-Aldrich, and Calbiochem Merck (Darmstadt, Germany) were immobilized on a solid support at high spatial density. Glycoprotein samples labeled with fluorescent dye Cy3 (GE Healthcare; Buckinghamshire, UK) were applied and scanned with a GenePix 4000B confocal scanner (Axon Instruments; Union City, CA, USA). Raw values less than the average background were omitted. The median for each lectin was globally normalized to the sum of the medians of all valid data for the 37 lectins [33]. Differences between the two data sets were evaluated by Student's t-test applied to each lectin signal (fold change >1.5, fold change <0.67, p-value <0.05).
Lectin Staining
HS5 and HS27a cells were cultured in 24-well plates with sterilized glycergel (DakoCytomation; Carpinteria, CA, USA) to 60–70% confluence. Culture medium was discarded and 2% fresh paraformaldehyde was added to fix cells. Cells were immobilized for 15 min at RT, washed with 1×PBS, permeabilized with 0.2% Triton X-100 in 1×PBS for 2 min at RT, and blocked with 5% BSA in 1×PBS overnight at 4 °C. Fixed cells were incubated with 15–20 µg/mL Cy3 fluorescein-labeled lectins in 5% BSA for 3 hr in the dark at RT, washed with PBS, stained with 4 µg/mL DAPI in 1×PBS for 10 min at RT, washed again with PBS, and observed by laser confocal fluorescence microscopy (model Eclipse Ti-U; Nikon; Tokyo, Japan).
RESULTS
Expression of Glycan-Related Genes in HS5 and HS27a Cells
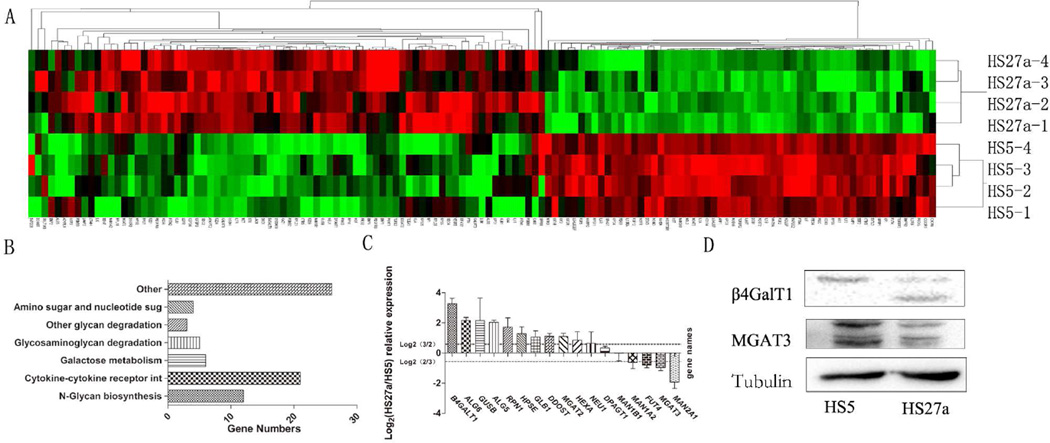
Of ~17,000 genes in the microarray, 130 glycogenes were differentially expressed and were visualized as a “heatmap” using the Cluster and Tree View software program (http://www.eisenlab.org/eisen/?page_id=42) (Figure 2A). On the basis of DAVID software analysis (http://david.abcc.ncifcrf.gov/), 66 glycogenes were annotated and classified into the following eight groups based on their functions: N-glycan synthesis (12 genes), glycosaminoglycan degradation (5), other glycan degradation (3), cytokine-cytokine receptor interaction (21), amino and nucleotide sugar metabolism (4), Galactose metabolism (6), and other functions (26) (Figure 2B; Table S1).
Figure 2.
Comparative glycogene expression of HS5 vs. HS27a cells by glycogene microarray analysis.
(A) "Heatmap" representation of differentially expressed genes involved in metabolism of glycosphingolipids, glycoproteins, and glycosaminoglycans. Red: genomic activation. Green: genomic inhibition. Black: no clear link. Gray: missing data.
(B) Annotations of genes by DAVID software program were classified, and category distribution numbers are shown as a bar chart. Categories and numbers: N-Glycan biosynthesis, 12. Glycosaminoglycan degradation, 5. Other glycan degradation, 3. Cytokine-cytokine receptor interaction, 21. Amino and nucleotide sugar metabolism, 4. Galactose metabolism, 6. Other, 26.
(C) Gene expression of ALG5, ALG6, β4GalT1, DPAGT1, DDOST, FUT4, GLB1, GUSB, HEXA, HPSE, MAN1B1, MAN1A2, MAN2A1, MGAT2, MGTA3, NEU1, and RPN1 was analyzed by RT-PCR, as described in M&M. Experiments were performed in biological triplicate. Relative expression was analyzed by the 2−ΔΔCt method and presented as Log2 relative expression for HS5 vs. HS27a, with Log2(3/2) and Log2(2/3) as threshold values. Values above Log2(3/2) and below Log2(2/3) indicate significant up-regulation and down-regulation, respectively.
(D) Western blot analysis of MGAT3 and β4GalT1, with β-tubulin expression as protein loading control.
Among the 15 N-glycan related genes (including glycan degradation and N-glycan synthesis), 12 (ALG5, ALG6, β4GALT1, DPAGT1, DDOST, MAN1A2, MAN1B1, MGAT2, RPN1, GLB1, HEXA, NEU1) showed increased expression and 3 (β4GalT3, MGAT3, MAN2A1) showed reduced expression in HS27a compared with HS5. Among the glycosaminoglycan degradation group, 4 genes (GLB1, GUSB, HPSE, HEXA) were up-regulated and 1 (HS3ST3B1) was down-regulated in HS27a. These findings are summarized in Table 1 (p-value <0.05 for all arrays).
Table 1.
Differential Expression of Glycan-Related Genes in HS27a vs. HS5 Cells (Fold change =HS27a/HS5)
| gene symbol | accession ID | p-value | Fold change |
gene description | ||
|---|---|---|---|---|---|---|
| A | B | c | ||||
| ALG5 | NM_013338 | 0.005 | 2.010 | dolichyl-phosphate β-glucosyltransferase | ||
| ALG6 | NM_013339 | 0.009 | 1.545 | α-1,3-glucosyltransferase | ||
| B4GALT1 | NM_001497 | 0.000 | 2.723 | βGlcNAc β 1,4- galactosyltransferase, polypeptide 1 | ||
| B4GALT3 | NM_003779 | 0.032 | 0.765 | βGlcNAc β 1,4- galactosyltransferase, polypeptide 3 | ||
| DDOST | NM_005216 | 0.025 | 1.417 | dolichyl-diphosphooligosaccharide protein glycosyltransferase subunit | ||
| DPAGT1 | NM_001382 | 0.002 | 1.565 | dolichyl-phosphate N-acetylglucosaminephosphotransferase 1 | ||
| MAN1A2 | NM_006699 | 0.000 | 1.605 | mannosidase, α, class 1A, member 2 | ||
| MAN1B1 | NM_016219 | 0.002 | 1.718 | mannosidase, α, class 1B, member 1 | ||
| MGAT2 | NM_002408 | 0.004 | 1.613 | monoacylglycerol O-acyltransferase 2 | ||
| MGAT3 | NM_002409 | 0.000 | 0.262 | mannosyl-glycoprotein β-1,4-N-acetylglucosaminyltransferase | ||
| RPN1 | NM_002950 | 0.002 | 1.661 | ribophorin I | ||
| MAN2A1 | NM_002372 | 0.000 | 0.182 | mannosidase, α, class 2A, member 1 | ||
| GLB1 | NM_000404 | 0.005 | 1.492 | galactosidase, β1 | ||
| HEXA | NM_000520 | 0.006 | 2.140 | hexosaminidase A (α polypeptide) | ||
| NEU1 | NM_000434 | 0.001 | 2.231 | sialidase 1 (lysosomal sialidase) | ||
| GLB1 | NM_000404 | 0.005 | 1.492 | galactosidase, β 1 | ||
| GUSB | NM_000181 | 0.000 | 2.614 | glucuronidase | ||
| HS3ST3B1 | NM_006041 | 0.000 | 0.135 | heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 | ||
| HPSE | NM_006665 | 0.009 | 3.330 | heparanase | ||
| HEXA | NM_000520 | 0.017 | 2.140 | (α polypeptide) | ||
Group A: N-Glycan biosynthesis related genes.
Group B: Glycosaminoglycan degradation related genes.
Identifiers, names, and functional descriptions are from information available in public databases, primarily the National Center for Biotechnology Information (NCBI) UniGene database and GenBank.
Average value in gene expression of HS27a (n=4) was divided by average value in gene expression of HS5 (n=4).
The genes listed in Table 1 were defined with significant change of expression either fold change >1.5 or <0.67. The enzyme MGAT3 (encoded by MGAT3 gene), which produces bisecting GlcNAc by catalyzing transfer of GlcNAc residues in β1,4 linkage to the β1,4-mannose residue in the core region of N-glycans [34–36] had lower expression in HS27a than in HS5 (fold change =0.262). β1,4-galactosyltransferase (encoded by β4GalT1), which catalyzes addition of UDP-Gal to terminal GlcNAc in β1,4 linkage, was higher in HS27a than in HS5 (fold change =2.723). MGAT2 (encoding monoacylglycerol acyltransferase 2) was higher in HS27a (fold change =1.613). HEXA, which encodes a ganglioside-binding protein that degrades GM2 in association with GM2 activator [37], was higher in HS27a (fold change =2.140).
The findings from gene microarray analysis were validated by RT-PCR (Figure 2C). Among the 18 N-glycan synthesis genes, β4GalT1, MAN1A2, ALG6, GUSB, ALG5, RPN1, HPSE, GLB1, DDOST, MGAT2, and HEXA showed increased expression whereas FUT4, MGAT3, MAN2A1, and HS3ST3B1 showed reduced expression in HS27a compared with HS5, consistently with microarray analysis results.
Quantitation of Glycan Biosynthesis-Related Proteins by SILAC and Western Blot Analysis
Data from gene microarray analysis and RT-PCR revealed differential expression at the transcriptional level of glycan-related genes in HS5 vs. HS27a cells. Labeled proteins isolated from the two cell lines were mixed (1:1), digested, and analyzed by ultrahigh-resolution liquid chromatography-tandem mass spectrometry (nLC-ESI-MS/MS). Among 4257 proteins detected in two experiments, 10 enzymes involved in glycan biosynthesis and degradation (ALG6, β4GALT1, GLB1, HEXA, MAN1A2, MAN1B1, MAN2A1, MGAT2, RPN1, DDOST) were detected (Table 2). Among these 10 glycan-related enzymes, four glycosyltransferases (DDOST, β4GalT1, MAN1A2, MGAT2) showed increased expression in HS27a (HS27a/HS5 ratio >1.50) and one (MAN2A1) showed reduced expression (ratio <0.67), consistently with gene microarray analysis results. Protein expression levels were confirmed by Western blot analysis. Protein level of β4GalT1 in HS27a is shown in Figure 2D. Both microarray and RT-PCR analyses showed a significant reduction of MGAT3 mRNA level in HS27a (Figure 2A, C). However, a reduced MGAT3 protein level in HS27a was revealed only by Western blot, not by SILAC (Figure 2D). Taken together, integrated findings from microarray analysis, RT-PCR, SILAC, and Western blot analysis indicate differential expression of both mRNA and protein levels of glycan genes in HS5 vs. HS27a cells.
Table 2.
Differential Expression of 10 Glycan Biosynthesis Related Proteins (Glycosyltransferases and Glycosidases)
| Gene symbol |
UniProt | peptide | MS | PEP | SCORE | HS27a/HS5 ratio |
|---|---|---|---|---|---|---|
| ALG6 | Q9Y672 | FINPDWIALHTSR | 1568.81 | 0.000205 | 80.31 | 0.708 |
| β4GALT1 | Q86XA6 | LPQLVGVSTPLQGGSNSAAAIGQSSGELR | 2793.46 | 1.35E-06 | 106.28 | 4.322 |
| GLB1 | C9J539 | HHLGDDVVLFTTDGAHK | 1860.91 | 1.22E-12 | 128.95 | 0.999 |
| HEXA | H3BU85 | HYLPLSSILDTLDVMAYNK | 2192.12 | 4.82E-15 | 147.07 | 1.340 |
| MAN1A2 | O60476 | NPGVFLIHGPDEHR | 1586.80 | 0.000321 | 123.10 | 1.655 |
| MAN1B1 | B3KQC5 | VPSGGYSSINNVQDPQKPEPR | 2268.11 | 0.000328 | 77.51 | 1.205 |
| MAN2A1 | Q16706 | WWDIIDIQK | 1215.63 | 0.000755 | 114.97 | 0.387 |
| MGAT2 | Q10469 | IFHAGDCGMHHK | 1408.61 | 0.001631 | 75.82 | 1.708 |
| RPN1* | P04843 | ISVIVETVYTHVLHPYPTQITQSEK | 1479.77 | 1.42E-08 | 86.36 | 1.180 |
| DDOST* | E7EWT1 | TAVIDHHNYDISDLGQHTLIVADTENLLK | 3244.64 | 1.24E-17 | 151.19 | 1.328 |
Gene symbol and UniProt number are taken from in NCBI and UniProt databases.
Protein encoded by the gene was detected twice by LTQ Orbitrap MS, and PEP and SCORE are derived from the more reliable experiment. HS27a/HS5 ratio is the average from the two experiments. Proteins without * were detected once by Orbitrap MS.
Glycopattern Analysis
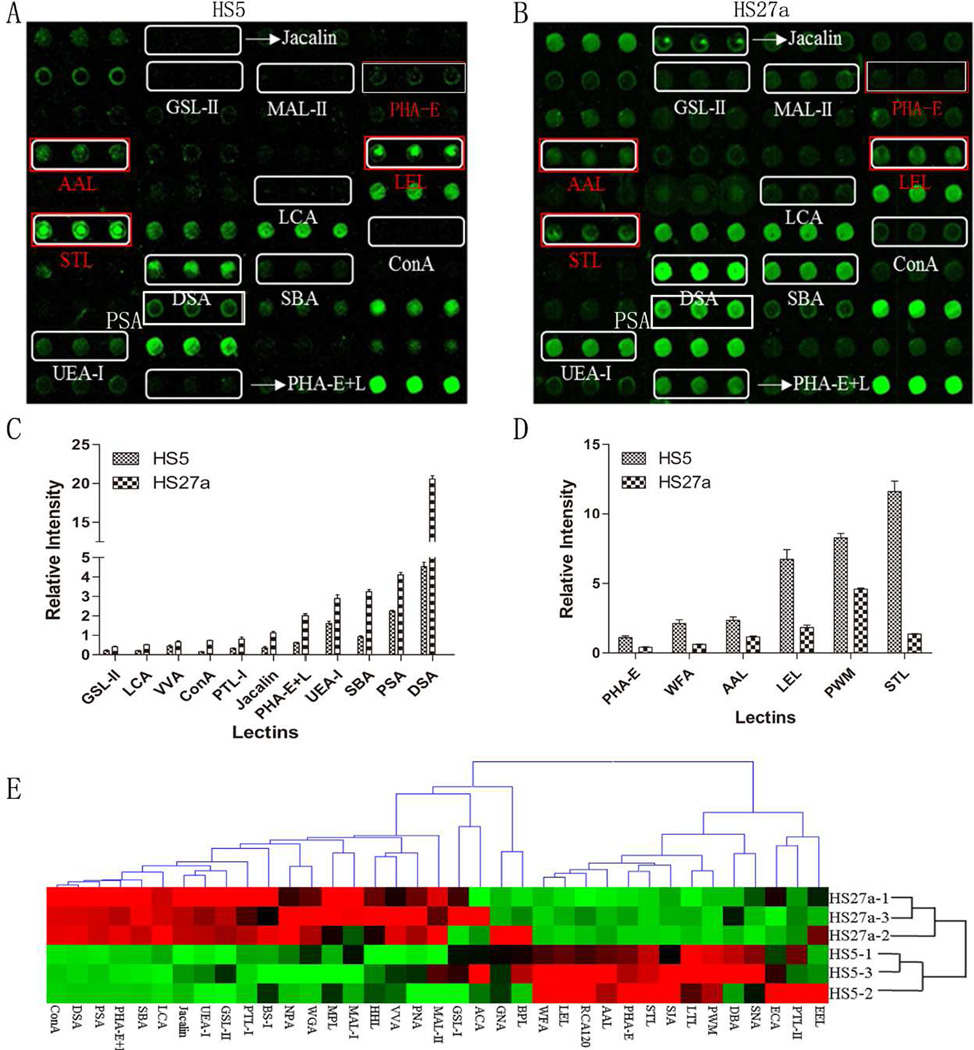
Patterns of glycoproteins reflect the expression, function, and metabolism of oligosaccharides in cells. We used lectin microarrays containing 37 lectins (Table S2), two negative controls (BSA), and one positive control (Cy3-BSA) (Figure S1) to analyze fine glycan structures of glycoproteins in HS5 vs. HS27a. Significant differences (>1.5 fold change, <0.67 fold-change, p <0.05) of glycans recognized by 18 different lectins were observed between the two cell lines (Figure 3A, B). HS27a showed increased expression of 11 glycan structures (recognized by lectins ConA, DSA, SBA, Jacalin, PHA-E+L, LCA, PTL-I, GSL-II, PSA, UEA-I, and VVA) and reduced expression of seven glycan structures (recognized by PWM, AAL, PHA-E, WFA, LEL, PTL-II, and STL) (Figure 3C; Table 3). Hierarchical clustering analysis and visualization were performed, allowing classification of lectin signal patterns (Figure 3D).
Figure 3.
Glycan profiling of HS5 and HS27a by lectin microarray analysis.
(A) Differentially expressed glycans in scanned image for HS5 are indicated.
(B) Differentially expressed glycans in scanned image for HS27a are indicated. Twelve glycan structures (recognized by lectins ConA, DSA, SBA, Jacalin, PHA-E+L, LCA, PTL-I, GSL-II, PSA, UEA-I, and VVA) showed higher expression in HS-27a than in HS5. Eight glycan structures (recognized by PWM, AAL, PHA-E, WFA, LEL, and STL) showed lower expression in HS27a.
(C), (D) Normalized fluorescence intensity of lectins with altered signals (HS27a/HS5 ratio >1.5 or <0.6, p <0.05; see Table 3).
(E) Variation of expression of glycans recognized by 37 lectins, presented as a heatmap. Red: fluorescence signal activation. Green: signal inhibition. Black: no clear link. Gray: missing data.
Table 3.
Differential Glycopatterns of HS5 and HS27a Cells Determined by Lectin Microarray Analysis
| Lectin Abbreviation |
Average Intensity (HS5) |
Average Intensity (HS27a) |
HS27a/ HS5 ratio |
Specificity |
|---|---|---|---|---|
| ConA | 0.154 | 0.734 | 4.783 | branched and terminal mannose, terminal GlcNAc |
| DSA | 4.537 | 20.572 | 4.535 | GlcNAc |
| SBA | 0.918 | 3.243 | 3.525 | Terminal GalNAc (especially GalNAcα1-3Gal) |
| Jacalin | 0.348 | 1.129 | 3.260 | Galβ1-3GalNAcα-Ser/Thr(T) and GalNAcα- Ser/Thr(T) |
| PHA-E+L | 0.625 | 2.014 | 3.235 | Bisecting GlcNAc and biantennary N-glycans and tetra-antennary complex-type N-glycan |
| PTL-I | 0.208 | 0.517 | 2.466 | αGalNAc and Gal |
| LCA | 0.325 | 0.802 | 2.460 | Fucα-1,6GlcNAc (core fucose) |
| GSL-II | 0.212 | 0.413 | 1.968 | GlcNAc and galactosylated N-glycans |
| PSA | 2.243 | 4.117 | 1.835 | Fucα-1,6GlcNAc (core fucose) |
| UEA-I | 1.609 | 2.898 | 1.801 | Fucα1-2Galβ1-4Glc(NAc) |
| VVA | 0.433 | 0.666 | 1.539 | GalNAc and GalNAcα-Ser/Thr (Tn) |
| PWM | 8.283 | 4.591 | 0.554 | GlcNAc |
| AAL | 2.346 | 1.150 | 0.490 | Terminal Fucα-1,6GlcNAc, Fucα-1,3Galβ-1,4GlcNAc |
| PHA-E | 1.102 | 0.410 | 0.372 | Bisecting GlcNAc and biantennary N-glycans |
| WFA | 2.119 | 0.624 | 0.294 | GalNAcα/β1-3/6Gal |
| LEL | 6.727 | 1.826 | 0.271 | Poly-LacNAc and (GlcNAc)n |
| STL | 11.602 | 1.360 | 0.117 | Oligomers of GlcNAc |
Average value in HS27a glycopatterns (n=3) was divided by average value of HS5 expression (n=3).
In brief, decreased fluorescence intensity of PHA-E indicated down-regulation of bisecting GlcNAc N-glycan structure in HS27a. Decreased AAL fluorescence intensity indicated low fucosylation of Fucα1, 3GlcNAc and Fucα1, 6Galβ1, 4GlcNAc in HS27a. Increased MAL-II fluorescence intensity indicated high sialylation in HS27a. PTL-I (recognizing αGalNAc and Gal), GSL-II (recognizing GlcNAc and galactosylated N-glycans), and MAL-I (recognizing Galβ-1,4GlcNAc) showed higher fluorescence intensities in HS27a (Table S2). Fluorescence intensity of LEL (recognizing poly-LacNAc and (GlcNAc)n) was lower in HS27a than in HS5 (Figure 3C).
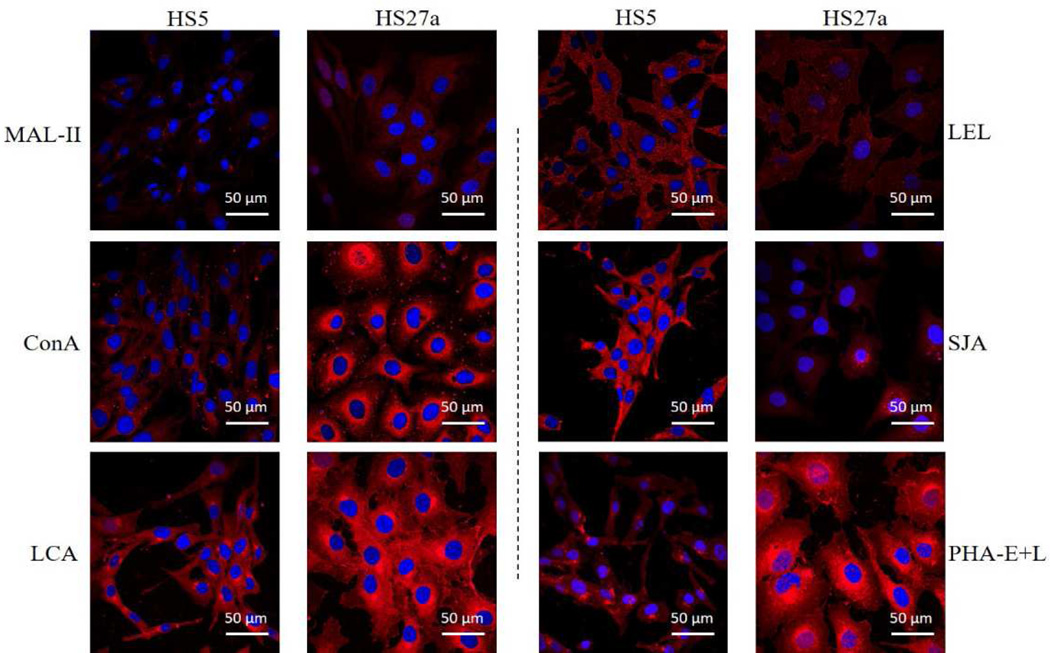
The differential glycopatterns in HS5 vs. HS27a were confirmed by lectin staining with Mal-II, LEL, PHA-E+L, LCA, SJA, and ConA. HS27a showed increased fluorescence signal intensities of LCA, ConA, Mal-II, and PHA-E+L but reduced signal intensities of SJA and LEL, consistently with lectin microarray results (Figure 4).
Figure 4.
Altered expression of glycans evaluated by lectin staining.
Six lectins (MAL-II, ConA, LCA, LEL, SJA, PHA-E+L) labeled with Cy3 were applied, and cell staining was performed as described in M&M. Panels show signals from merge images of Cy3-conjugated lectins and DAPI staining of nuclei in HS5 (left) and HS27a (right). Objective magnification 60×.
DISCUSSION
Changes of oligosaccharide structures on proteins are associated with numerous physiological and pathological events. Altered levels of N-glycans, O-glycans, and other glycoconjugates have been reported in many types of cancer [38, 39]. Increased mutation frequency of glycophorin A (GPA), the major cell surface sialoglycoprotein of human erythrocytes, is often detected in patients with MDS, aplastic anemia (AA), or paroxysmal nocturnal hemoglobinuria (PNH) [40]. Proteoglycan biosynthesis was previously studied in two phenotypically distinct murine bone, MS3-2A and D2XRII [41]. Human marrow stromal cell lines HS27a and HS5 have been used in many MDS studies, but global expression profiling of their glycans and related genes has not been performed to date.
We characterized glycan expression levels in HS5 and HS27a using an integrated strategy of gene microarray, proteomic, and lectin microarray analysis. Our findings were confirmed by Western blot analysis, RT-PCR, and lectin histochemistry. In HS27a, MGAT3 expression was greatly reduced at the mRNA level, with consequent suppression of MGAT3 products, bisecting GlcNAc N-glycans, as recognized by PHA-E in lectin microarray. Significant differential expression of MGAT3 in HS5 vs. HS27a was detected by Western blotting but not by SILAC, perhaps because of sensitivity limitation of SILAC. MGAT3 and its products (bisecting GlcNAc N-glycans) are involved in biosynthesis of complex-type and hybrid-type oligosaccharides, and may inhibit cell migration through alteration of N-glycans in ECM and adhesion molecules such as E-cadherin, laminin, and integrin [42, 43]. MGAT3 overexpression inhibited epithelial-mesenchymal transition (EMT) induced by TGF-β1 in epithelial cell lines [44]. MGAT3 expression is enhanced in hepatocytes during hepatocarcinogenesis [45]. The observed up-regulation of MGAT3 in brains of Alzheimer's disease (AD) patients may represent an adaptive response to protect brain cells from additional β-amyloid production, which may be responsible for much of the pathology of AD [46]. The finding that levels of MGAT3 and bisecting GlcNAc N-glycans are much lower in HS27a than in HS5a, provides useful information regarding the altered microenvironment created by co-culture of HS5 or HS27a with myeloid cells.
β4GalT1 was also differentially expressed in the two cell lines. LacNAc, the product of β4GalT1, is attached to O-glycans, N-glycans, and glycolipids, and plays a crucial role in a variety of biological processes, including morphogenesis, brain development, cellular adhesion [47–49], cell-cell interaction, cell-ECM interaction, and metastatic capacity [50]. Aberrant glycosylation, particularly overexpression of lacto-series type 1 and type 2 structures (often in the form of poly-LacNAc) with various types of fucosylation and sialylation, is observed in many human cancers [51, 52]. In the present study, expression of β4GalT1 at both the mRNA and protein levels was markedly higher in HS27a than in HS5. Fluorescence intensity of lectin MAL-I which could recognize Galβ1, 4GlcNAc structure, was stronger in HS27a (Table S2). Galactosylated N-glycans, recognized by PTL-I and GSL-II (Table 3), were also enhanced in HS27a. On the other hand, fluorescence intensity of LEL, which could also recognized Galβ1,4GlcNAc, was lower in HS27a. This mainly because that the glycan-binding specificities of many lectins remain to be fully explored [26], and differences in structure need to be confirmed by mass spectroscopic and other techniques. LacNAc synthesis is part of the pathway leading to the terminal capping group sialyl Lewis X (Lex) [53, 54]. Sialyl Lex and sialyl Lea are both ligands of the cell adhesion molecule ELAM-1 [55], and display greatly increased levels in a variety of cancer cells [56]. Our understanding of the bone marrow microenvironment will be improved by further studies of sialyl Lex and sialyl Lea function.
Glycosylation patterns and their changes in transformed cells have received steadily increasing attention from cancer researchers during recent decades. However, it is often difficult to relate cancer cell phenotypes to glycosylation of specific proteins and their encoding genes [57]. We developed and applied an integrated strategy with genomic, proteomic, and functional glycomic techniques for global expression profiling of human stromal HS5 and HS27a cells. The present findings provide a useful basis for our future studies of crosstalk among stromal cells and myeloma cells in co-culture system.
Supplementary Material
Highlights.
Integrated strategy was used to profile N-glycan and glycogenes in two human bone marrow cells.
Bisecting N-glycans, catalyzed by MGAT3, were enhanced in HS5 cells.
Glycan structure of Galβ1,4GlcNAc, catalyzed by β4GalT1, was significantly increased in HS27a cells.
Acknowledgments
This study was supported by the National Science Foundation of China (No. 81470294, 31400691), the Natural Science Foundation of Jiangsu Province, China (No. BK20140169), Jiangsu Province Recruiting Plan for High-level, Innovative and Entrepreneurial Talents, the Fundamental Research Funds for the Central Universities (No. JUSRP51319B), Jiangsu Province "Six Summit Talent" Foundation (2013-SWYY-019), and the 111 Project (No. 111-2-06). The authors are grateful to Dr. S. Anderson for English editing of the manuscript. The authors also thank Dr. Li Zheng (Northwestern University, China) for assistance with the lectin microarray analysis and Dr. Xue Peng (Chinese Academy of Sciences, Beijing, China) for assistance with the LTQ Orbitrap mass analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fliedner TM. The role of blood stem cells in hematopoietic cell renewal. Stem Cells. 1998;16(Suppl 1):13–29. doi: 10.1002/stem.160361. [DOI] [PubMed] [Google Scholar]
- 2.Mendez-Ferrer S, Michurina TV, Ferraro F, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatia R, McGlave PB, Dewald GW, Blazar BR, Verfaillie CM. Abnormal function of the bone marrow microenvironment in chronic myelogenous leukemia: Role of malignant stromal macrophages. Blood. 1995;85:3636–3645. [PubMed] [Google Scholar]
- 4.Roecklein BA, Torok-Storb B. Functionally distinct human marrow stromal cell lines immortalized by transduction with the human papilloma virus E6/E7 genes. Blood. 1995;85:997–1005. [PubMed] [Google Scholar]
- 5.Graf L, Iwata M, Torok-Storb B. Gene expression profiling of the functionally distinct human bone marrow stromal cell lines HS-5 and HS-27a. Blood. 2002;100:1509–1511. doi: 10.1182/blood-2002-03-0844. [DOI] [PubMed] [Google Scholar]
- 6.Iwata M, Sandstrom RS, Delrow JJ, Stamatoyannopoulos JA, Torok-Storb B. Functionally and phenotypically distinct subpopulations of marrow stromal cells are fibroblast in origin and induce different fates in peripheral blood monocytes. Stem Cells Dev. 2014;23:729–740. doi: 10.1089/scd.2013.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garrido SM, Appelbaum FR, Willman CL, Banker DE. Acute myeloid leukemia cells are protected from spontaneous and drug-induced apoptosis by direct contact with a human bone marrow stromal cell line (hs-5) Experimental Hematology. 2001;29:448–457. doi: 10.1016/s0301-472x(01)00612-9. [DOI] [PubMed] [Google Scholar]
- 8.Lichtman MA. The ultrastructure of the hemopoietic environment of the marrow: A review. Experimental Hematology. 1981;9:391–410. [PubMed] [Google Scholar]
- 9.Iwata M, Awaya N, Graf L, Kahl C, Torok-Storb B. Human marrow stromal cells activate monocytes to secrete osteopontin, which down-regulates NOTCH1 gene expression in CD34+ cells. Blood. 2004;103:4496–4502. doi: 10.1182/blood-2004-01-0256. [DOI] [PubMed] [Google Scholar]
- 10.Bhagat TD, Spaulding E, Sohal D, et al. MDS marrow stroma is characterized by epigenetic alterations. Blood. 2008;112(1243):3635. [Google Scholar]
- 11.Mhyre AJ, Marcondes AM, Spaulding EY, Deeg HJ. Stroma-dependent apoptosis in clonal hematopoietic precursors correlates with expression of PYCARD. Blood. 2009;113:649–658. doi: 10.1182/blood-2008-04-152686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prinetti A, Aureli M, Illuzzi G, et al. Gm3 synthase overexpression results in reduced cell motility and in caveolin-1 upregulation in human ovarian carcinoma cells. Glycobiology. 2010;20:62–77. doi: 10.1093/glycob/cwp143. [DOI] [PubMed] [Google Scholar]
- 13.Elias HK, Schinke C, Bhattacharyya S, Will B, Verma A, Steidl U. Stem cell origin of myelodysplastic syndromes. Oncogene. 2014;33:5139–5150. doi: 10.1038/onc.2013.520. [DOI] [PubMed] [Google Scholar]
- 14.Kerbauy DMB, Lesnikov V, Torok-Storb B, Bryant E, Deeg HJ. Engraftment of distinct clonal MDS-derived hematopoietic precursors in NOD/SCID-·2microglobulin-deficient mice after intramedullary transplantation of hematopoietic and stromal cells. Blood. 2004;104:2202–2203. doi: 10.1182/blood-2004-04-1518. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Marcondes AM, Ragoczy T, Telling A, Deeg HJ. Effect of intravenous coadministration of human stroma cell lines on engraftment of long-term repopulating clonal myelodysplastic syndrome cells in immunodeficient mice. Blood Cancer Journal. 2013;3:e113. doi: 10.1038/bcj.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neeper M, Schmidt AM, Brett J, et al. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. The Journal of Biological Chemistry. 1992;267:14998–15004. [PubMed] [Google Scholar]
- 17.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 18.Brownlee M. The pathobiology of diabetic complications: A unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 19.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Ishihara K, Yokota T, et al. Reduced α4β1 integrin/vcam-1 interactions lead to impaired pre-b cell repopulation in α 1,6-fucosyltransferase deficient mice. Glycobiology. 2008;18:114–124. doi: 10.1093/glycob/cwm107. [DOI] [PubMed] [Google Scholar]
- 21.Freeze H, Elbein A. In: Essentials of Glycobiology. Varki A, editor. Vol. 66. 2009. [PubMed] [Google Scholar]
- 22.Zhou F, Cui C, Ge Y, et al. α2,3-sialylation regulates the stability of stem cell marker CD133. Journal of Biochemistry. 2010;148:273–280. doi: 10.1093/jb/mvq062. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Marcondes AM, Gooley TA, Deeg HJ. The helix-loop-helix transcription factor TWIST is dysregulated in myelodysplastic syndromes. Blood. 2010;116:2304–2314. doi: 10.1182/blood-2009-09-242313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ong SE. Stable isotope labeling by amino acids in cell culture, silac, as a simple and accurate approach to expression proteomics. Molecular & Cellular Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 25.Yang G, Tan Z, Lu W, et al. Quantitative glycome analysis of N-glycan patterns in bladder cancer vs normal bladder cells using an integrated strategy. Journal of Proteome Research. 2015;14:639–653. doi: 10.1021/pr5006026. [DOI] [PubMed] [Google Scholar]
- 26.Tan Z, Lu W, Li X, et al. Altered N-glycan expression profile in epithelial-to-mesenchymal transition of nmumg cells revealed by an integrated strategy using mass spectrometry and glycogene and lectin microarray analysis. Journal of Proteome Research. 2014;13:2783–2795. doi: 10.1021/pr401185z. [DOI] [PubMed] [Google Scholar]
- 27.Guan F, Schaffer L, Handa K, Hakomori SI. Functional role of gangliotetraosylceramide in epithelial-to-mesenchymal transition process induced by hypoxia and by TGF-β. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24:4889–4903. doi: 10.1096/fj.10-162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nature Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 30.Olsen JV, de Godoy LM, Li G, et al. Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a C-trap. Molecular & Cellular Proteomics : MCP. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Cox J, Matic I, Hilger M, et al. A practical guide to the maxquant computational platform for SILAC-based quantitative proteomics. Nature Protocols. 2009;4:698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- 32.Cox J, Mann M. Maxquant enables high peptide identification rates, individualized PPB-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 33.Yu H, Zhu M, Qin Y, et al. Analysis of glycan-related genes expression and glycan profiles in mice with liver fibrosis. J Proteome Research. 2012;11:5277–5285. doi: 10.1021/pr300484j. [DOI] [PubMed] [Google Scholar]
- 34.Narasimhan S. Control of glycoprotein synthesis. Udp-GlcNAc: Glycopeptide β 4-N-acetylglucosaminyltransferase iii, an enzyme in hen oviduct which adds glcnac in β 1–4 linkage to the β-linked mannose of the trimannosyl core of N-glycosyl oligosaccharides. Journal of Biological Chemistry. 1982;257:10235–10242. [PubMed] [Google Scholar]
- 35.Priatel JJ, Sarkar M, Schachter H, Marth JD. Isolation, characterization and inactivation of the mouse Mgat3 gene: The bisecting N-acetylglucosamine in asparagine-linked oligosaccharides appears dispensable for viability and reproduction. Glycobiology. 1997;7:45–56. doi: 10.1093/glycob/7.1.45. [DOI] [PubMed] [Google Scholar]
- 36.Song Y, Aglipay JA, Bernstein JD, Goswami S, Stanley P. The bisecting GlcNAc on N-glycans inhibits growth factor signaling and retards mammary tumor progression. Cancer Research. 2010;70:3361–3371. doi: 10.1158/0008-5472.CAN-09-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamanaka S, Johnson MD, Grinberg A, et al. Targeted disruption of the Hexa gene results in mice with biochemical and pathologic features of Tay-Sachs disease. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:9975–9979. doi: 10.1073/pnas.91.21.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saldova R, Royle L, Radcliffe CM, et al. Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and igG. Glycobiology. 2007;17:1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 39.De Leoz ML, Young LJ, An HJ, et al. High-mannose glycans are elevated during breast cancer progression. Molecular & Cellular Proteomics : MCP. 2011 doi: 10.1074/mcp.M110.002717. 10:M110 002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hattori H, Machii T, Ueda E, Shibano M, Kageyama T, Kitani T. Increased frequency of somatic mutations at glycophorin a loci in patients with aplastic anaemia, myelodysplastic syndrome and paroxysmal nocturnal haemoglobinuria. British Journal of Haematology. 1997;98:384–391. doi: 10.1046/j.1365-2141.1997.2233037.x. [DOI] [PubMed] [Google Scholar]
- 41.Kirby SL, Bentley SA. Proteoglycan synthesis in two murine bone marrow stromal cell lines. Blood. 1987;70:1777–1783. [PubMed] [Google Scholar]
- 42.Miyagawa S, Ueno T, Nagashima H, Takama Y, Fukuzawa M. Carbohydrate antigens. Current Opinion in Organ Transplantation. 2012;17:174–179. doi: 10.1097/MOT.0b013e3283508189. [DOI] [PubMed] [Google Scholar]
- 43.Isaji T, Gu J, Nishiuchi R, et al. Introduction of bisecting GlcNAc into integrin α5β1 reduces ligand binding and down-regulates cell adhesion and cell migration. Journal of Biological Chemistry. 2004;279:19747–19754. doi: 10.1074/jbc.M311627200. [DOI] [PubMed] [Google Scholar]
- 44.Xu Q, Isaji T, Lu Y, et al. Roles N-acetylglucosaminyltransferase iii in epithelial-to-mesenchymal transition induced by transforming growth factor β1 (TGF-β1) in epithelial cell lines. The Journal of Biological Chemistry. 2012;287:16563–16574. doi: 10.1074/jbc.M111.262154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyoshi E, Nishikawa A, Ihara Y, et al. N-acetylglucosaminyltransferase iii and v messenger rna levels in lec rats during hepatocarcinogenesis. Cancer Research. 1993;53:3899–3902. [PubMed] [Google Scholar]
- 46.Akasaka-Manya K, Manya H, Sakurai Y, et al. Protective effect of N-glycan bisecting GlcNAc residues on β-amyloid production in alzheimer's disease. Glycobiology. 2010;20:99–106. doi: 10.1093/glycob/cwp152. [DOI] [PubMed] [Google Scholar]
- 47.Shen A, Wang H, Zhang Y, Yan J, Zhu D, Gu J. Expression of β-1,4-galactosyltransferase II and V in rat injured sciatic nerves. Neuroscience Letters. 2002;327:45–48. doi: 10.1016/s0304-3940(02)00381-6. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura N, Yamakawa N, Sato T, Tojo H, Tachi C, Furukawa K. Differential gene expression of β-1,4-galactosyltransferases I, II and V during mouse brain development. Journal of Neurochemistry. 2001;76:29–38. doi: 10.1046/j.1471-4159.2001.00004.x. [DOI] [PubMed] [Google Scholar]
- 49.Vadaie N, Hulinsky RS, Jarvis DL. Identification and characterization of a drosophila melanogaster ortholog of human β1,4-galactosyltransferase VII. Glycobiology. 2002;12:589–597. doi: 10.1093/glycob/cwf074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castronovo V, Luyten F, van den Brûle F, Sobel ME. Identification of a 14-kda laminin binding protein (HLBP14) in human melanoma cells that is identical to the 14-kDa galactoside binding lectin. Arch Biochem Biophys. 1992;297:132–138. doi: 10.1016/0003-9861(92)90650-l. [DOI] [PubMed] [Google Scholar]
- 51.Hakomori S. Aberrant glycosylation in tumors and tumor-associated carbohydrate antigens. Advances in Cancer Research. 1989;52:257–331. doi: 10.1016/s0065-230x(08)60215-8. [DOI] [PubMed] [Google Scholar]
- 52.Hakomori S. Aberrant glycosylation in cancer cell membranes as focused on glycolipids: Overview and perspectives. Cancer Research. 1985;45:2405–2414. [PubMed] [Google Scholar]
- 53.Sperandio M, Gleissner CA, Ley K. Glycosylation in immune cell trafficking. Immunological Reviews. 2009;230:97–113. doi: 10.1111/j.1600-065X.2009.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ujita M, Misra AK, McAuliffe J, Hindsgaul O, Fukuda M. Poly-N-acetyllactosamine extension in N-glycans and core 2-and core 4-branchedo-glycans is differentially controlled by i-extension enzyme and different members of the β1, 4-galactosyltransferase gene family. Journal of Biological Chemistry. 2000;275:15868–15875. doi: 10.1074/jbc.M001034200. [DOI] [PubMed] [Google Scholar]
- 55.Takada A, Ohmori K, Yoneda T, et al. Contribution of carbohydrate antigens sialyl Lewis A and sialyl Lewis X to adhesion of human cancer cells to vascular endothelium. Cancer Research. 1993;53:354–361. [PubMed] [Google Scholar]
- 56.Kannagi R. Molecular mechanism for cancer-associated induction of sialyl Lewis X and sialyl Lewis A expression-the warburg effect revisited. Glycoconjugate Journal. 2004;20:353–364. doi: 10.1023/B:GLYC.0000033631.35357.41. [DOI] [PubMed] [Google Scholar]
- 57.Hakomori S. Glycosylation defining cancer malignancy: New wine in an old bottle. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:10231–10233. doi: 10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.