Abstract
Purpose
Uveal melanoma (UM) can be classified by gene expression profiling (GEP) into Class 1 (low metastatic risk) and Class 2 (high metastatic risk), the latter being strongly associated with mutational inactivation of the tumor suppressor BAP1. Nevertheless, a small percentage of Class 1 tumors give rise to metastatic disease. The purpose of this study was to identify biomarkers of metastasis in Class 1 tumors.
Experimental Design
389 consecutive patients with UM were assigned to Class 1 or 2 using a prospectively validated 12-gene prognostic classifier. Selected tumors were further analyzed using global GEP and SNP microarrays. PRAME mRNA expression was analyzed in 64 Class 1 tumors by qPCR.
Results
Among Class 1 UMs, the most significant predictor of metastasis was PRAME mRNA expression (P=0.0006). The 5-year actuarial rate of metastasis was 0% for Class1PRAME−, 38% for Class1PRAME+, and 71% for Class 2 tumors. Median metastasis-free survival for Class1PRAME+ patients was 88 months, compared to 32 months for Class 2 patients. Findings were validated using three independent datasets, including one using disomy 3 to identify low-risk UM. Chromosome copy number changes associated with Class1PRAME+ tumors included gain of 1q, 6p, 8q, and 9q and loss of 6q and 11q. PRAME expression was associated with larger tumor diameter (P=0.05) and SF3B1 mutations (P=0.003).
Conclusions
PRAME is an independent prognostic biomarker in UM that identifies increased metastatic risk in patients with Class 1 or disomy 3 tumors. This finding may further enhance the accuracy of prognostic testing and precision medicine for UM.
Keywords: Uveal melanoma, metastasis, prognosis, gene expression profiling, PRAME
INTRODUCTION
Uveal melanoma (UM) is the most common primary cancer of the eye and has a propensity for fatal hematogenous metastasis to the liver (1). The molecular landscape of primary UM has been well characterized (2). UMs can be categorized by gene expression profiling (GEP) into two molecular classes associated with metastatic risk: Class 1 (low metastatic risk) and Class 2 (high metastatic risk)(3). Class 1 tumors retain a differentiated melanocytic phenotype and transcriptome, whereas Class 2 tumors have a de-differentiated phenotype and stem cell-like transcriptome (4). The Class 2 signature is strongly associated with mutations in BAP1 on chromosome 3p21, usually accompanied by loss of the other copy of chromosome 3, consistent with the “two hit” model of tumor suppressor gene inactivation (5, 6). Four other frequently mutated genes have been identified in UM. Hemizygous mutations in the G-protein subunits GNAQ and GNA11 are early or initiating events in UM (7–9). GNAQ and GNA11 mutations are mutually exclusive, and one or the other is found in ~83% of UMs (9). Hemizygous mutations in SF3B1 and EIF1AX are virtually mutually exclusive with each other, and with BAP1 mutations, and they have been linked to good prognosis (10, 11).
GEP-based assignment of UMs to Class 1 or Class 2 with a 12-gene expression classifier has been validated in a prospective, multi-center study and is now routinely performed for clinical use in many centers (12, 13). While the vast majority of UM metastases occur in patients with Class 2 tumors, a small proportion of Class 1 tumors also give rise to metastasis. Based on a retrospective analysis of expression data from the 12-gene classifier on Class 1 tumors that metastasized, a provisional sub-grouping of Class 1 tumors into “1A” and “1B” based on the expression of two of these genes (CDH1 and RAB31) has been used as a provisional indicator of Class 1 patients who may be at increased risk of metastasis (14). Class 1A tumors have low CDH1/RAB31 expression, and Class 1B tumors have high CDH1/RAB31 expression. To identify additional and potentially more accurate biomarkers for metastasis in Class 1 tumors, we conducted a genome-wide integrated transcriptomic and chromosomal analysis in our cohort of Class 1 tumors. We identified the cancer-testis antigen PRAME (preferentially expressed antigen in melanoma) as a biomarker for increased metastatic risk in Class 1 tumors. This finding has important implications for precision management in patients with UM and may aid in the stratification of patients for clinical trials.
MATERIAL AND METHODS
Clinical samples
This study was conducted with the approval of the Institutional Review Boards of Washington University and the University of Miami. Tumor samples were obtained at enucleation or by fine need biopsy between November 1998 and May 2015 from patients with UMs arising from the ciliary body and/or choroid. Samples were snap frozen and stored at -80°C. Baseline clinical information, metastatic status, and final outcome were recorded for each patient. Prognostic molecular class assignments (Class 1 or Class 2) were obtained using a prospectively validated 12-gene classifier as previously reported (13). Mutation status for GNAQ, GNA11, BAP1, SF3B1, and EIF1AX were obtained by Sanger sequencing as previously described (5, 7, 10, 11). Tumor samples from Leiden University were obtained between 1999 and 2008 by enucleation and clinical and histopathologic features were retrieved from patient charts and histology reports. Data on survival were obtained from medical records and the Integral Cancer Center West, which updates patient follow-up information, including date and cause of death, on a yearly basis. Under Dutch law, tumor material may be used for research purposes, and patients signed an informed consent for genetic testing. The tenets of the Declaration of Helsinki were followed in all studies.
RNA expression analysis
Total RNA was isolated using TRIzol and the Qiagen RNeasy kit. RNA quality was assessed on the Agilent Bioanalyzer 2100. RNA from enucleated samples was hybridized to Illumina Human HT12v4 BeadChip arrays, and resulting data have been deposited in the NCBI’s Gene Expression Omnibus (15) and are accessible through GEO Series accession number GSE73652. Raw data were normalized by cubic spline using Illumina GenomeStudio. Significance Analysis of Microarrays (SAM version 4.0) was used to generate a ranked list of differentially expressed genes based on false discovery rate and fold change (16). Gene Set Enrichment Analysis (GSEA version 2.0.4) was used to identify significantly enriched functional gene sets (17). Differentially up-regulated and down-regulated genes from SAM were input into GSEA and analyzed using the Molecular Signatures Database (MSigDB) to identify chromosomal position and transcription factor target gene sets. A circular plot of gene expression data was generated using the Perl-based Circos graphical program (18). Expression data of specific chromosomal regions were plotted using the R package GViz. Principal component analysis (PCA) and 3D visualization using the 12-gene expression classifier were performed using Partek Genomics Suite. Unsupervised PCA and 3D visualization using global transcriptomic data were performed on the top 20% most variable genes across all samples using the stats and rgl packages in R.
PRAME mRNA expression was analyzed using real-time quantitative PCR (qPCR). RNA was converted to cDNA using the Applied Biosystems High Capacity cDNA Reverse Transcription Kit, and gene expression was quantified using the Applied Biosystems 7900HT Real-Time PCR System with TaqMan® primers and Gene Expression Master Mix following the manufacturer’s protocol. The control genes for the 12-gene expression classifier (MRPS21, RBM23 and SAP130) were included. TaqMan® primers were Hs01022301_m1 (PRAME), Hs00230458_m1 (MRPS21), Hs00216503_m1 (RBM23), and Hs00368617_m1 (SAP130). Ct values were calculated using the manufacturer’s software, and mean Ct values were calculated for all triplicate sets. ΔCt values were calculated by subtracting the mean Ct of each discriminating gene from the geometric mean of the mean Ct values of the three endogenous control genes, as previously described (19).
For Kaplan-Meier actuarial analysis, PRAME mRNA expression data were scaled from 0 to 1 and plotted from lowest to highest expression. A Loess model was constructed to represent the best fit of the data (2nd degree, span=0.5, family=Gaussian, fitting by least-squares), and the differences in predicted values of the Loess model were plotted to estimate slope change (Supplementary Fig. 1A). Samples with minimal differences (blue dots) were coded as PRAME−, whereas those with significant differences (red dots) were coded as PRAME+.
For validation, three independent gene expression datasets and associated clinical annotations were analyzed. Two of these datasets were obtained from GEO (15), and tumor samples were stratified into Class 1 or 2 based on the 12-genes in the UM classifier. Dataset #GSE22138 was performed on the Affymetrix Human Genome U133 Plus 2.0 Array platform and dataset #GSE44295 was performed on the Illumina Ref8 platform. These datasets consisted of 20 Class1PRAME− and 31 Class1PRAME+ tumors. The third dataset contained 25 tumors classified as low risk based on disomy 3, which is significantly associated with the Class 1 signature (6, 13), and was performed at Leiden University on the Illumina Human HT12v4 BeadChip Array platform. For these additional datasets, PRAME gene expression was scaled from 0 to 1 for all samples and categorized as PRAME+ or PRAME−, as described above.
Immunohistochemistry
Immunohistochemistry (IHC) for PRAME was performed with the SIGMA Prestige anti-PRAME antibody (HPA045153) at 4 μg/ml on formalin-fixed, paraffin-embedded (FFPE) slides using the VECTASTAIN® UNIVERSAL Elite ABC Kit (Vector Laboratories, PK-6200) and the ImmPACT™ SG Peroxidase Substrate Kit (Vector Laboratories, SK-4705) according to the manufacturer’s recommendation, with the following changes: Primary antibody was incubated overnight at 4°C with addition of 0.3% Triton X-100. Antigen retrieval was performed in Sodium Citrate Buffer (10 mM, 0.05% Tween 20, pH 6.0) in an autoclave at 121°C for 10 min. Slides were slowly cooled down to 80°C for 25 min, and additionally for 10 min at room temperature. The blocking step was changed to 1 h. Incubation with secondary antibody was increased to 1 h with addition of 0.1% Triton X-100. Incubation with VECTASTAIN Elite ABC reagent was increased to 1 h. ImmPACT™ SG reagent was applied for 20 min, with changing to fresh substrate after 7 and 14 min. Nuclear fast red counter-stain (Vector Laboratories, H-3403) was applied for 10 minutes. Negative controls included tumors known to be lacking PRAME mRNA expression and samples tested with secondary antibody only. Two independent investigators assessed the IHC slides for PRAME nuclear staining in a blinded manner. Samples were graded by the percentage of cells demonstrating nuclear staining. The scores from the independent graders were averaged.
Chromosome copy number analysis
Genomic DNA was isolated using the Qiagen DNeasy kit. For the Class1met+ samples, loss of heterozygosity for chromosome 3 was determined using a single nucleotide polymorphism (SNP) assay as previously described (20). For PRAME+ and PRAME− samples, SNP analysis was performed using the Affymetrix Genome-Wide Human SNP 6.0 array and data were analyzed for copy number gains and losses as described in the Affymetrix Genotyping Console software user manual and then plotted with Integrative Genomics Viewer. For the Leiden dataset, monosomy 3 was determined using Affymetrix 250K_SNP and Cytoscan HD microarrays as previously described with a cutoff value of < 1.9 for chromosomal loss (6).
Statistical analysis
Statistical analysis was determined using Medcalc® version 14.10.2. Statistical significance was determined using the Mann-Whitney test for continuous variables, Fisher exact test for discrete variables, Cox proportional hazards regression for evaluation of prognostic factors for survival, Spearman’s rank correlation coefficient for comparison of continuous non-parametric data, and Kaplan-Meier method with log-rank test and right censoring for comparing survival between groups.
RESULTS
Clinical characteristics of metastasizing Class 1 uveal melanomas
Our study design is summarized in Supplementary Table 1. Among 389 consecutive patients with UM involving the ciliary body and/or choroid who underwent GEP prognostic testing, the molecular class assignment was Class 1 in 216 (56%) patients and Class 2 in 173 (44%) patients. Class 2 GEP was associated with markedly greater metastatic risk than Class 1 GEP, with metastatic disease being detected in 12/216 (6%) Class 1 cases versus 63/173 (36%) Class 2 cases (log rank test, P<0.0001)(Supplementary Fig. 1B).
We wished to determine whether any clinicopathologic features differed between Class 1 tumors with metastasis (Class1met+) and Class 2 tumors with metastasis (Class2met+). Class2met− tumors were not included in this analysis because the high metastatic rate and limited follow-up in some cases would have led to the erroneous inclusion of tumors that would later metastasize. Pairwise comparisons of clinicopathologic features are summarized in Supplementary Table 2. Detailed clinicopathologic and molecular features of all the Class1met+ tumors are summarized in Supplementary Table 3. Among 52 patients in whom metastasis was detected and the sites of metastasis were available, the liver was affected in 6/12 (50%) patients with a Class1met+ tumor compared to 36/40 (90%) patients with a Class2met+ tumor (Fisher exact test, P=0.02). Other metastatic sites in Class 1 tumors included lung in 4, bone in 3, and stomach in 2 cases. Compared to Class2met+ tumors, Class1met+ tumors were associated with younger patient age and less frequent ciliary body involvement (Mann-Whitney test, P=0.04 and P=0.02, respectively).
Similarly, we compared the characteristics of the Class 1 tumors with and without metastatic disease. Pairwise comparisons of clinicopathologic features are summarized in Supplementary Table 2. There were no significant differences between Class1met+ and Class1met− tumors with regard to age, sex, tumor diameter or thickness, ciliary body involvement or cell type. For Class1met+ tumors, mutations were identified in BAP1 in none of 8, EIF1AX in none of 6, GNAQ in 5 of 8, GNA11 in 1 of 7 and SF3B1 in 4 of 8 samples in which DNA was still available for sequencing (Supplementary Table 3).
Comparison of gene expression profiles among Class 1 tumors
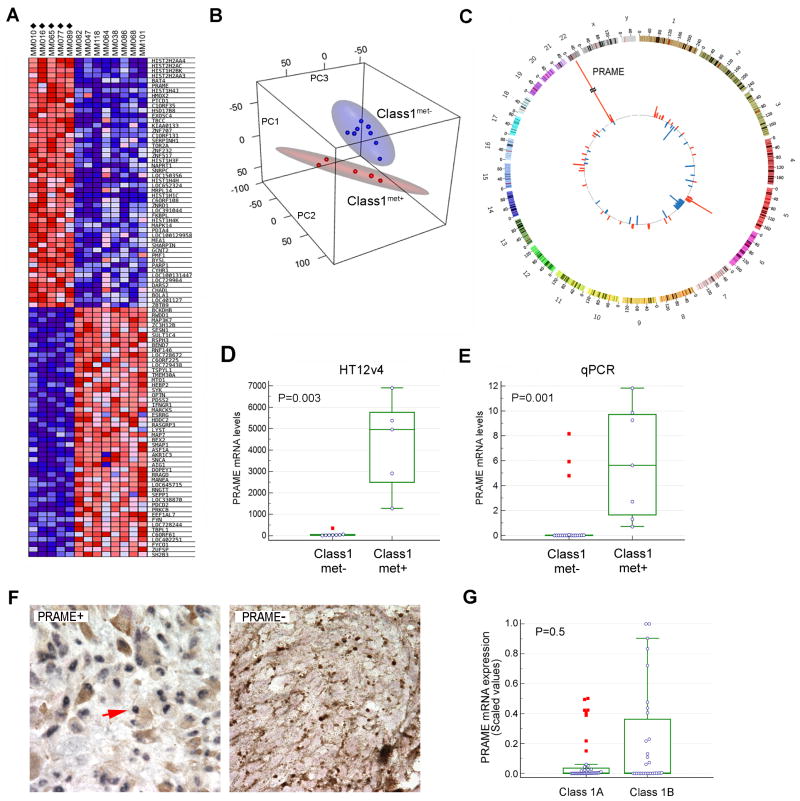
Among the 5 known UM driver mutations, SF3B1 mutations were most strongly associated with Class 1 metastasis, but since these mutations were only present in 50% of metastatic cases, it is not an adequate biomarker for Class 1 metastasis. Thus, we turned to transcriptomic profiling. Initially, we analyzed 108 Class 1 tumors by PCA using the mRNA expression of genes from the 12-gene classifier. PCA organized the samples into two clusters corresponding to their metastatic status (Supplementary Fig. 1C), indicating that there were potentially meaningful transcriptomic differences between Class1met+ and Class1met− tumors. To identify genes that were differentially expressed between Class1met+ and Class1met− tumors, we performed a genome-wide transcriptomic analysis of 5 Class1met+ tumors and 8 Class1met− tumors with at least 1 year of follow-up using the Illumina Human HT12v4 Expression BeadChip array. Using SAM, differentially expressed genes were identified at a significance level of FDR<0.01 (Fig. 1A and Supplementary Table 4). Using unsupervised PCA, tumor samples once again clustered into two distinct groups that corresponded to their metastatic status, with no overlap in their 95% confidence ellipsoids (Fig. 1B). By far, the most highly overexpressed gene in Class1met+ tumors was PRAME (Mann-Whitney test, P=0.003) (Fig. 1C–D).
Figure 1.
Gene expression profiling of Class 1 primary uveal melanomas based on metastatic status. (A) Heatmap showing the 50 most highly up-regulated and down-regulated genes in 5 Class 1 primary uveal melanomas that metastasized (Class1met+ tumors, indicated by diamonds) versus 8 that did not (Class1met− tumors). Gene expression profiles were obtained using Illumina Human HT12v4 BeadChip arrays and analyzed with Significance Analysis of Microarrays (SAM). Blue = decreased gene expression, red = increased gene expression in Class1met+ versus Class1met− tumors. (B) Unsupervised principal component analysis of the same dataset, showing differential clustering of Class1met+ (red) and Class1met− (blue) tumors. Red and blue ellipsoids represent 95% confidence intervals for each respective group. (C) Circos plot of the same dataset demonstrating genes that are significantly up-regulated (red) or down-regulated (blue) in Class1met+ compared to Class1met− tumors with respect to chromosomal location. (D) Box-and-whisker plots of PRAME mRNA expression in Class1met+ versus Class1met− tumors ≥1 year of follow-up using the Illumina Human HT12v4 BeadChip array dataset. (E) Box-and-whisker plots of PRAME mRNA expression in Class1met+ versus Class1met− tumors with ≥3 years of follow-up using qPCR. (F) Representative examples of immunohistochemical staining for PRAME in Class1PRAME+ and Class1PRAME− tumors, 100x magnification. Blue, PRAME nuclear staining (arrow); brown, intrinsic melanin pigment; red, nuclear counterstain. (G) Analysis of PRAME mRNA expression with respect to Class 1A versus Class 1B designation. In box-and-whiskers plots, the central box represents the 25th to 75th percentiles, the middle line represents the median, the horizontal bars represent the minimum and maximum values, except for “far out” values, indicated by red boxes and defined as values larger than the upper 75% plus 3 times the interquartile range.
We then analyzed PRAME expression by qPCR in 26 Class 1 tumors, 7 with metastasis and 19 without metastasis with even longer follow-up of >3 years. These samples included 17 that were not previously analyzed. Consistent with the array data, 16/19 (84%) Class1met− tumors showed minimal PRAME expression (Class1PRAME−), whereas 7/7 (100%) of Class1met+ tumors showed elevated PRAME expression (Class1PRAME+, Mann-Whitney test, P=0.001) (Fig. 1E). We performed IHC for PRAME protein on 6 of the Class 1 tumors in which original FFPE blocks were still available, and as expected, PRAME mRNA expression correlated with nuclear protein expression (Fig. 1F, Supplementary Fig. 1D). There was no significant association between PRAME expression status and the currently used Class “1A/1B” designation (Mann-Whitney test, P=0.5) (Fig. 1G). However, among Class 1 tumors in which both PRAME and 1A/1B data were available, all 6 Class 1 tumors that metastasized were PRAME+ and Class 1B.
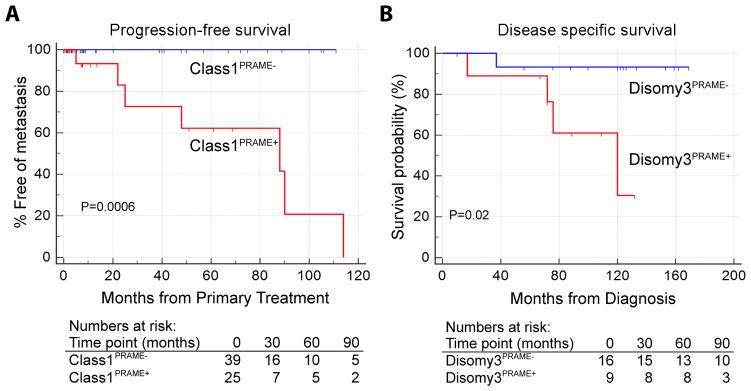
To obtain a more accurate assessment of the prevalence of PRAME−expressing Class 1 tumors, and to obtain actuarial estimates of metastatic risk associated with PRAME expression in Class 1 tumors, we performed qPCR on an additional 38 samples, for a total of 64 Class 1 tumors analyzed by qPCR. The overall median follow-up was 8.2 months (mean 31.5 months, interquartile range 3.3–57.1 months). Using the stratification procedure described in Materials and Methods, 39 (61%) tumors were categorized as Class1PRAME− and 25 (39%) as Class1PRAME+. Using Kaplan-Meier survival analysis, PRAME status was strongly associated with metastasis (log rank test, P=0.0006). Indeed, no metastatic events occurred among Class1PRAME− cases (Fig. 2A). The 5-year actuarial probability of metastasis was 15% for Class 1 tumors overall, 0% for Class1PRAME− tumors and 38% for Class1PRAME+ tumors, compared to 71% for Class 2 tumors. The median metastasis-free survival for patients with Class1PRAME+ tumors was 88 months, compared to 32 months for Class 2 tumors. PRAME expression showed a significant association with larger basal tumor diameter (Spearman correlation, P = 0.04) and with SF3B1 mutations (Mann-Whitney test, P = 0.003) (Supplementary Fig. 1E).
Figure 2.
Survival analysis in patients with Class 1 primary uveal melanoma. (A) Kaplan-Meier plot showing metastasis-free survival in 39 patients with Class1PRAME− (blue) and 25 patients with Class1PRAME+ (red) uveal melanomas. (B) Kaplan-Meier plot showing metastasis-free survival in 16 patients with Disomy3PRAME− (blue) and 9 patients with Disomy3PRAME+ (red) uveal melanomas.
To further validate the association of high PRAME levels with metastasis, we combined two independent published datasets with long follow-up (GSE22138 and GSE44295), which included 20 Class1PRAME− and 31 Class1PRAME+ cases (median follow-up 68.0 months, mean 80.4 months, interquartile range 41.4–101.5 months). By Kaplan-Meier survival analysis, PRAME expression was significantly associated with metastasis (log rank test, P = 0.05) (Supplementary Fig. 1F). The 5-year actuarial probability of metastasis was very similar to our dataset: 21% for Class 1 tumors overall, 5% for Class1PRAME− tumors, and 31% for Class1PRAME+ tumors.
We analyzed a third dataset with long follow-up from Leiden University that defined low risk UM by the presence of disomy 3. This dataset of 64 cases included 16 Disomy3PRAME− tumors and 9 Disomy3PRAME+ tumors, with a median follow-up of 121.0 months (mean, 107.7 months; interquartile range, 85.0–138.0 months) and 76.0 months (mean, 83.8 months; interquartile range, 72.0–109.0 months), respectively. Comparable to the other datasets, PRAME expression in these tumors was associated with increased metastatic risk (log rank test, P = 0.02) (Fig. 2B). Similar to our previous findings, PRAME expression in these tumors was associated with larger basal tumor diameter (P = 0.001).
Functional analysis of the Class1PRAME+ transcriptome
Genes that were differentially expressed between Class1PRAME− and Class1PRAME+ tumors were analyzed for chromosomal location, biological function and transcription factor binding sites using GSEA and MSigDB to gain insights into tumor biology. Genes that were up-regulated in Class1PRAME+ tumors showed a significant enrichment in chromosomal location at 1q21, 1q42, 6p21-22 and 8q24, whereas down-regulated genes showed enrichment across most of chromosome 6q (FDR<0.01 for all)(Supplementary Table 5). At FDR=0.02, enrichment of down-regulated genes was also observed at 11q23-24.
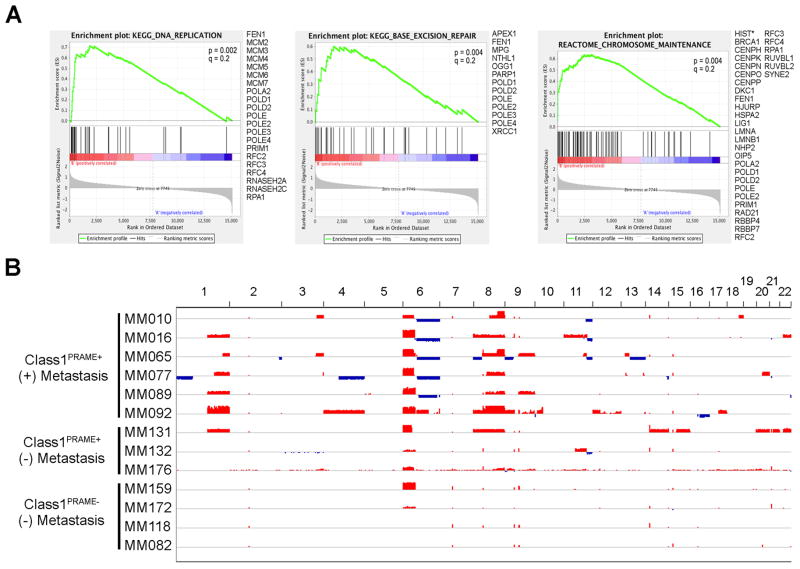
Genes that were up-regulated in Class1PRAME+ tumors were also enriched for biological functions related to chromosome maintenance, DNA replication and base excision repair (FDR≤0.2 for all)(Fig. 3A and Supplementary Table 6). Analysis of genes up-regulated in Class1PRAME+ tumors for transcription factor binding sites within the promoter region (± 2 Kb of the transcription start site) showed enrichment for two known transcription factors: NFY and SP1 (FDR<0.01 for both)(Supplementary Table 5). Interestingly, PRAME associates with NFY at its CCAAT binding site in promoter regions to induce transcriptional activation (21), and many of the up-regulated genes with NFY sites in their promoters were located within regions of chromosomal gain on 1q and 6p in Class1PRAME+ tumors (Supplementary Fig. 2).
Figure 3.
Functional and chromosome copy number analyses in Class 1 primary uveal melanoma. (A) Gene Set Enrichment Analysis (GSEA) of annotated gene sets exhibiting significant enrichment for genes that were up-regulated in Class1PRAME+ tumors relative to Class1PRAME− tumors. (B) High density Affymetrix Genome-Wide Human SNP 6.0 Array analysis of Class1PRAME+ and Class1PRAME− tumors with respect to metastatic status. Blue indicates copy number loss, and red, copy number gain.
Chromosomal alterations in Class1PRAME+ tumors
The correlation between mRNA expression of PRAME and of genes involved in chromosome maintenance suggested that Class1PRAME+ tumors may demonstrate increased chromosomal gains and losses relative to Class1PRAME− tumors. To test this hypothesis, we used high density Affymetrix® Genome-Wide Human SNP 6.0 arrays to analyze 9 Class1PRAME+ tumors (6 with metastasis and 3 without metastasis) and 4 Class1PRAME− tumors (all without metastasis). Indeed, there was a correlation between PRAME expression and specific chromosomal gains and losses, even in this limited set of cases (Fig. 3B). Consistent with our transcriptomic analysis (Supplementary Table 5), the most common chromosome copy number changes specific to Class1PRAME+ tumors included 1q gain (5 cases, all with metastasis), 6p gain with 6q loss (5 cases, all with metastasis) and 8q gain (8 cases, 6 with metastasis and 2 without metastasis). Less common but potentially important changes that were specific to Class1PRAME+ tumors included 9q gain (3 cases, all with metastasis) and 11q loss (4 cases, 3 with metastasis and 1 without metastasis). 6p gain was only specific to Class1PRAME+ tumors when accompanied by 6q loss, consistent with isochromosome 6p (22). In the Leiden dataset, gain or amplification of 8q was observed in 6/16 Disomy3PRAME− and in 8/9 Disomy3PRAME+ tumors (Fisher exact test, P = 0.03). Although chromosome 6 status did not show a significant difference between Disomy3PRAME− and Disomy3PRAME+ tumors (data not shown), 8/9 Disomy3PRAME+ tumors displayed a gain of 6p. We elected not to pursue further chromosomal analyses since PRAME met our objective for this study, which was to identify a strong biomarker for Class 1 metastasis.
DISCUSSION
The clinical and molecular features associated with metastasis in Class 1 tumors were clearly distinct from those associated with metastasis in Class 2 tumors. Class 1 UMs that metastasized tended to occur in younger patients, less frequently involved the ciliary body, less frequently metastasized to the liver, lacked BAP1 mutations, frequently had SF3B1 mutations, and exhibited a distinct GEP. The transcriptomes of Class 1 and Class 2 tumors were so dramatically different that the comparatively subtle differences between Class1met− and Class1met+ were masked until Class 2 tumors were first removed from the analysis. When this was done, however, the gene expression differences between these Class 1 subgroups were clearly evident, and PRAME was the single most differentially expressed gene that distinguished Class 1 tumors based on their metastatic status. When we expanded the analysis to a larger number of tumor samples and stratified the tumors based on the presence or absence of PRAME expression measured by qPCR, Kaplan-Meier survival analysis confirmed the strong association of PRAME with metastasis in Class 1 tumors. This finding was validated in three independent datasets, including one that used disomy 3 to define “low risk” UM. As expected, we also found that PRAME mRNA expression correlated with PRAME nuclear protein expression by IHC. A rigorous analysis of PRAME IHC staining was beyond the scope of this study but needs to be performed in a large number of FFPE samples to confirm whether this has similar prognostic value as PRAME mRNA expression.
The 12-gene classifier is a prospectively validated clinical prognostic test for UM (13), and we anticipate that the addition of PRAME will further enhance the classifier’s accuracy. There was no significant correlation between PRAME status and the provisional “1A/1B” system that the classifier currently uses. As such, we are initiating a multi-center study to compare these two predictors, and we anticipate that PRAME will likely be the more accurate biomarker for metastasis in Class 1 UMs since it was identified in an unsupervised manner from a global genomic analysis and was independently validated.
PRAME was initially discovered as a tumor antigen recognized by cytolytic T cells in cutaneous melanoma (23). PRAME expression is a marker of poor clinical outcome in a variety of cancers (24), and PRAME mRNA expression was recently found to be an important biomarker for differentiating benign nevi from malignant melanoma of the skin (25). PRAME may promote tumor progression, at least in part, by inhibiting differentiation, growth arrest, and apoptosis induced by retinoic acid signaling (26). PRAME binds the retinoic acid receptor (RAR) and recruits EZH2 to form a heterotrimeric complex that represses transcription of genes containing RAR binding sites. Consistent with the possibility that PRAME may inhibit retinoic acid signaling in Class1PRAME+ tumors, we found that 16% of genes down regulated in Class1PRAME+ tumors had a retinoic acid response element within ±10 Kb of the transcription start site or gene end, including RARB itself (FDR ≤ 0.05, Supplementary Table 5). The cooperation between PRAME, RAR, and EZH2 suggests a potential therapeutic role for EZH2 inhibitors in patients with metastatic Class 1 tumors (27), particularly since EZH2 has been shown to repress immune signaling in uveal melanoma cells (28). Additionally, combination therapy with retinoic acid and an HDAC inhibitor may have therapeutic benefit by synergistically inhibiting PRAME to induce differentiation and cell cycle exit (29). Interestingly, UM cells treated with retinoic acid become sensitive to killing by cytolytic T cells and NK cells (30), suggesting a potential role for immunotherapy in Class 1 metastasis. Vaccines against PRAME in cutaneous melanoma and other cancers are currently in clinical trials (Trial Nos. NCT01149343 and NCT01853878), and other immunotherapies directed against PRAME−expressing cancers are in development (31).
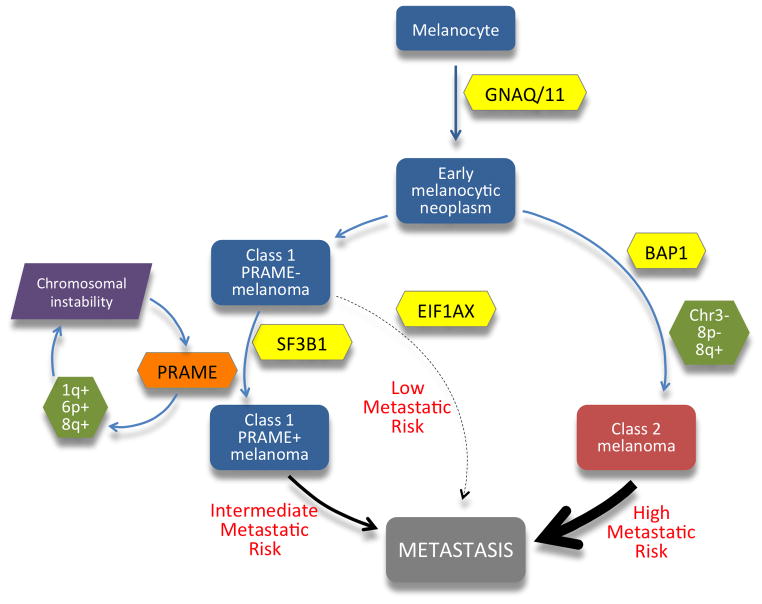
PRAME also associates with the transcription factor NFY at transcriptionally active promoters and enhancers containing the NFY binding motif CCAAT (21). NFY is a highly conserved, sequence-specific, nucleosome-like, trimeric complex that interacts with DNA in a sequence-specific manner (32). Though PRAME does not directly bind to NFY, its association at the NFY CCAAT binding motif is required for NFY-mediated transcriptional activation, whereas lack of PRAME is associated with repression (21). Intriguingly, many of the genes that were up-regulated in Class1PRAME+ tumors contain NFY sites in their promoter and are located within the regions of 1q and 6p gain in Class1PRAME+ tumors. Further, many of these genes play a role in meiotic recombination, telomere and chromosome maintenance, DNA replication and base excision repair, all of which have been implicated in chromosomal instability (33). Moreover, NFYA, the component of the NFY trimeric complex that contains the CCAAT DNA-binding motif, is also located on 6p and is significantly up-regulated (FDR < 0.05). These findings suggest that PRAME up-regulation may cooperate with copy number gains on 1q and 6p to facilitate chromosomal instability associated with tumor progression (Fig. 4).
Figure 4.
Molecular hypothesis of tumor progression in uveal melanoma, highlighting the role of PRAME reported herein. Increased PRAME mRNA expression in Class 1 uveal melanomas is associated with transcriptional up-regulation of key genes involved in chromosome maintenance and stability, many of which are located on chromosome 1q and 6p and contain regulatory elements for transcription factors that interact with PRAME, including the retinoic acid receptor and NFY complexes. These observations suggest a possible feed-forward mechanism in which progressively increasing PRAME expression and specific chromosomal gains mutually reinforce one another to promote Class 1 tumor progression. There was a significant association of this pathway with SF3B1 mutations, which were mutually exclusive with EIF1AX mutations, suggesting a bifurcated pathway. This Class 1 metastatic pathway was distinct from the more common pathway leading to metastasis through the bi-allelic loss of BAP1 and acquisition of the Class 2 gene expression profile.
Mutations in SF3B1 and EIF1AX are associated with Class 1 GEP and have been reported to be good prognostic factors in UM, whereas BAP1 mutations are associated with Class 2 GEP and poor prognosis (10, 11). While this study affirms that SF3B1 mutations are associated with better prognosis than BAP1 mutations, we found that SF3B1 mutations were associated with increased metastatic risk among Class 1 tumors, which almost never have BAP1 mutations. Similarly, another group recently reported that patients with disomy 3 have a worse disease-free survival when an SF3B1 mutation is present (34). In the same way, 6p gain is a good prognostic factor relative to Class 2 GEP (and monosomy 3), 6p gain should not be thought of as “protective” against metastasis because it is strongly associated with metastasis in Class 1 tumors. On the other hand, EIF1AX mutations were not found in any of the metastasizing Class 1 tumors and were almost always mutually exclusive with SF3B1 and BAP1 mutations, suggesting that this mutation may have value as a favorable prognostic factor. Larger numbers of cases will need to be evaluated to determine the prognostic value of these mutations.
A limitation of this study was the small number of Class1met+ cases, but this is a reflection of the infrequency of metastasis in Class 1 tumors. To overcome this limitation, we validated the prognostic significance of PRAME in three independent datasets, which all had long (> 6 years) median follow-up data. Further, we are planning a prospective, multicenter study to validate these findings, establish the optimal PRAME expression threshold for maximizing positive and negative predictive value, and determine whether PRAME has prognostic value in Class 2 tumors. It is possible that some Class1met− tumors might have metastasized if they had been followed for a longer period of time. Indeed, rare metastatic events can occur even decades after primary ocular treatment (35). However, it is difficult to obtain such long-term data, and our findings are of practical value for the vast majority of patients for whom 5-year outcome projections are most important for personal and clinical decision-making.
For Class 1 tumors, the inclusion of PRAME expression may further augment the prognostic accuracy of the clinically available 12-gene classifier for UM. This finding allows patients with UM to be stratified into Class1PRAME− (low metastatic risk), Class1PRAME+ (intermediate metastatic risk) and Class 2 (high metastatic risk) for purposes of metastatic surveillance and clinical trials of targeted adjuvant therapies.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Uveal melanoma (UM) can be stratified into Class 1 (low metastatic risk) and Class 2 (high metastatic risk) using a prospectively validated 12-gene expression profile. While the vast majority of metastases occur in Class 2 tumors, a small percentage of Class 1 tumors also metastasize. This study revealed that metastasis in Class 1 tumors is strongly associated with expression of the PRAME oncogene. This finding was validated in three independent datasets, including one using disomy 3 as a surrogate marker for low risk UM. Adding PRAME to the 12-gene expression classifier will likely enhance its prognostic accuracy by identifying Class 1 tumors with intermediate metastatic risk. Some cancers expressing PRAME have been shown to be recognized by cytotoxic T cells, suggesting that immunotherapy may have a role in the precision management of patients with PRAME+ UM.
Acknowledgments
Financial support: This work was supported by grants to J.W.H. from the National Cancer Institute (R01 CA125970 and CA161870), Research to Prevent Blindness, Inc. Senior Scientific Investigator Award, and Melanoma Research Alliance, to J.W.H. and M.G.F. from the Melanoma Research Foundation, to M.G.F. from the Sheila and David Fuente Graduate Program in Cancer Biology, Sylvester Comprehensive Cancer Center, to S.K. by the 2015 AACR-Ocular Melanoma Foundation Fellowship, in honor of Robert C. Allen, MD, Grant Number 15-40-39-KURT, and to the Bascom Palmer Eye Institute from NIH Core Grant P30EY014801, Research to Prevent Blindness Unrestricted Grant, and Department of Defense Grant #W81XWH-09-1-0675. The funding organizations had no role in the design or conduct of this research.
Footnotes
Conflict of Interest: Dr. Harbour is the inventor of intellectual property used in the study and receives royalties from its commercialization. He is a paid consultant for Castle Biosciences, licensee of intellectual property presented in this article. No other authors disclosed any potential conflicts of interest.
Author contributions
Conception and design: J. William Harbour
Development of methodology: J. William Harbour
Acquisition of data: Matthew G. Field, Christina L. Decatur, Kaleigh Kozak, Gulcin Gezgin, Pieter van der Velden, Martine J. Jager, and Stefan Kurtenbach
Analysis and interpretation of data: Matthew G. Field, Kaleigh Kozak, Gulcin Gezgin, Pieter van der Velden, Martine J. Jager, Stefan Kurtenbach and J. William Harbour
Administrative, technical and material support: Christina L. Decatur
Study supervision: J. William Harbour
References
- 1.Ramaiya KJ, Harbour JW. Current management of uveal melanoma. Expert Review of Ophthalmology. 2007;2:939–46. [Google Scholar]
- 2.Harbour JW, Chao DL. A molecular revolution in uveal melanoma: implications for patient care and targeted therapy. Ophthalmology. 2014;121:1281–8. doi: 10.1016/j.ophtha.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64:7205–9. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang SH, Worley LA, Onken MD, Harbour JW. Prognostic biomarkers in uveal melanoma: evidence for a stem cell-like phenotype associated with metastasis. Melanoma Res. 2008;18:191–200. doi: 10.1097/CMR.0b013e3283005270. [DOI] [PubMed] [Google Scholar]
- 5.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Essen TH, van Pelt SI, Versluis M, Bronkhorst IH, van Duinen SG, Marinkovic M, et al. Prognostic parameters in uveal melanoma and their association with BAP1 expression. Br J Ophthalmol. 2014;98:1738–43. doi: 10.1136/bjophthalmol-2014-305047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onken MD, Worley LA, Long MD, Duan S, Council ML, Bowcock AM, et al. Oncogenic mutations in GNAQ occur early in uveal melanoma. Invest Ophthalmol Vis Sci. 2008;49:5230–4. doi: 10.1167/iovs.08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45:133–5. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin M, Masshofer L, Temming P, Rahmann S, Metz C, Bornfeld N, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45:933–6. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010;12:461–8. doi: 10.2353/jmoldx.2010.090220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onken MD, Worley LA, Char DH, Augsburger JJ, Correa ZM, Nudleman E, et al. Collaborative Ocular Oncology Group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012;119:1596–603. doi: 10.1016/j.ophtha.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castle Biosciences Inc. DecisionDx-UM Summary. c2015 [Internet] [cited 2014 December 10]; Available from: http://www.myuvealmelanoma.com/health-care-professionals/decisiondx-um-summary/
- 15.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harbour JW. A prognostic test to predict the risk of metastasis in uveal melanoma based on a 15-gene expression profile. Methods Mol Biol. 2014;1102:427–40. doi: 10.1007/978-1-62703-727-3_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onken MD, Worley LA, Person E, Char DH, Bowcock AM, Harbour JW. Loss of heterozygosity of chromosome 3 detected with single nucleotide polymorphisms is superior to monosomy 3 for predicting metastasis in uveal melanoma. Clin Cancer Res. 2007;13:2923–7. doi: 10.1158/1078-0432.CCR-06-2383. [DOI] [PubMed] [Google Scholar]
- 21.Costessi A, Mahrour N, Tijchon E, Stunnenberg R, Stoel MA, Jansen PW, et al. The tumour antigen PRAME is a subunit of a Cul2 ubiquitin ligase and associates with active NFY promoters. EMBO J. 2011;30:3786–98. doi: 10.1038/emboj.2011.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aalto Y, Eriksson L, Seregard S, Larsson O, Knuutila S. Concomitant loss of chromosome 3 and whole arm losses and gains of chromosome 1, 6, or 8 in metastasizing primary uveal melanoma. Invest Ophthalmol Vis Sci. 2001;42:313–7. [PubMed] [Google Scholar]
- 23.Ikeda H, Lethe B, Lehmann F, van Baren N, Baurain JF, de Smet C, et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6:199–208. doi: 10.1016/s1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- 24.Epping MT, Bernards R. A causal role for the human tumor antigen preferentially expressed antigen of melanoma in cancer. Cancer Res. 2006;66:10639–42. doi: 10.1158/0008-5472.CAN-06-2522. [DOI] [PubMed] [Google Scholar]
- 25.Clarke LE, Bryan Warf M, Flake DD, 2nd, Hartman AR, Tahan S, Shea CR, et al. Clinical Validation of a Gene Expression Signature that Differentiates Benign Nevi from Malignant Melanoma. J Cutan Pathol. 2015;42:244–52. doi: 10.1111/cup.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122:835–47. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Crea F, Fornaro L, Bocci G, Sun L, Farrar WL, Falcone A, et al. EZH2 inhibition: targeting the crossroad of tumor invasion and angiogenesis. Cancer Metastasis Rev. 2012;31:753–61. doi: 10.1007/s10555-012-9387-3. [DOI] [PubMed] [Google Scholar]
- 28.Holling TM, Bergevoet MW, Wilson L, Van Eggermond MC, Schooten E, Steenbergen RD, et al. A role for EZH2 in silencing of IFN-gamma inducible MHC2TA transcription in uveal melanoma. J Immunol. 2007;179:5317–25. doi: 10.4049/jimmunol.179.8.5317. [DOI] [PubMed] [Google Scholar]
- 29.Epping MT, Wang L, Plumb JA, Lieb M, Gronemeyer H, Brown R, et al. A functional genetic screen identifies retinoic acid signaling as a target of histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2007;104:17777–82. doi: 10.1073/pnas.0702518104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vertuani S, Dubrovska E, Levitsky V, Jager MJ, Kiessling R, Levitskaya J. Retinoic acid elicits cytostatic, cytotoxic and immunomodulatory effects on uveal melanoma cells. Cancer Immunol Immunother. 2007;56:193–204. doi: 10.1007/s00262-006-0185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amir AL, van der Steen DM, van Loenen MM, Hagedoorn RS, de Boer R, Kester MD, et al. PRAME−specific Allo-HLA-restricted T cells with potent antitumor reactivity useful for therapeutic T-cell receptor gene transfer. Clin Cancer Res. 2011;17:5615–25. doi: 10.1158/1078-0432.CCR-11-1066. [DOI] [PubMed] [Google Scholar]
- 32.Nardini M, Gnesutta N, Donati G, Gatta R, Forni C, Fossati A, et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell. 2013;152:132–43. doi: 10.1016/j.cell.2012.11.047. [DOI] [PubMed] [Google Scholar]
- 33.Silva AG, Graves HA, Guffei A, Ricca TI, Mortara RA, Jasiulionis MG, et al. Telomere-centromere-driven genomic instability contributes to karyotype evolution in a mouse model of melanoma. Neoplasia (New York, NY) 2010;12:11–9. doi: 10.1593/neo.91004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kilic E, Koopmans AE, Yavuzyigitoglu S, Vaarwater J, van Ijcken WF, Paridaens D, et al. SF3B1 and EIF1AX mutations in uveal melanoma: a protective factor, or not? [abstract] Acta Ophthalmol. 2014;92 [Abstracts from the 2014 European Association for Vision and Eye Research Conference] [Google Scholar]
- 35.Midena E, de Belvis V, Dei Tos AP, Antonini C. Isolated brain metastasis of malignant choroidal melanoma 27 years after enucleation. Arch Ophthalmol. 1999;117:1553–6. doi: 10.1001/archopht.117.11.1553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.