Significance
Growing experimental evidence suggests that the pathological spreading of alpha-synuclein aggregates in Parkinson’s disease is mediated through a process of templated seeding whereby aggregates catalyze the conversion of soluble protein molecules into their aggregated forms. A molecular-level understanding of this process is still lacking. Here, we determine the concentrations and numbers of aggregates necessary for the effective seeding of alpha-synuclein, thus providing a quantitative framework to understand the conditions when its seeded propagation is favorable. We find that high concentrations of aggregates are needed for seeding yet that aggregates cause cytotoxicity at significantly lower concentrations. This suggests that templated seeding is unlikely to be the main mechanism of spreading in Parkinson’s disease but occurs together with oligomer-induced cellular stress.
Keywords: amyloid aggregation, kinetic analysis, templated seeding, prion-like propagation, neurodegeneration
Abstract
The protein alpha-synuclein (αS) self-assembles into small oligomeric species and subsequently into amyloid fibrils that accumulate and proliferate during the development of Parkinson’s disease. However, the quantitative characterization of the aggregation and spreading of αS remains challenging to achieve. Previously, we identified a conformational conversion step leading from the initially formed oligomers to more compact oligomers preceding fibril formation. Here, by a combination of single-molecule fluorescence measurements and kinetic analysis, we find that the reaction in solution involves two unimolecular structural conversion steps, from the disordered to more compact oligomers and then to fibrils, which can elongate by further monomer addition. We have obtained individual rate constants for these key microscopic steps by applying a global kinetic analysis to both the decrease in the concentration of monomeric protein molecules and the increase in oligomer concentrations over a 0.5–140-µM range of αS. The resulting explicit kinetic model of αS aggregation has been used to quantitatively explore seeding the reaction by either the compact oligomers or fibrils. Our predictions reveal that, although fibrils are more effective at seeding than oligomers, very high numbers of seeds of either type, of the order of 104, are required to achieve efficient seeding and bypass the slow generation of aggregates through primary nucleation. Complementary cellular experiments demonstrated that two orders of magnitude lower numbers of oligomers were sufficient to generate high levels of reactive oxygen species, suggesting that effective templated seeding is likely to require both the presence of template aggregates and conditions of cellular stress.
Neurodegenerative disorders such as Parkinson’s disease (PD) and Alzheimer’s disease are becoming increasingly common as a result of increasing longevity in the population of the modern world, and there are no effective disease-modifying therapies to date (1, 2). The main characteristics of these disorders are the deposition and spreading of aggregated proteins causing neuronal loss that is accompanied with motor and cognitive deficits (3–5).
Alpha-synuclein (αS) is a small (14.5 kDa) intrinsically disordered protein expressed in neurons and presynaptic nerve terminals (6). It is abundant in the neuronal cytosol of a healthy brain, and its function is thought to be associated with axonal transport (7, 8). The assembly of monomeric αS into amyloid fibrils that form Lewy bodies (LBs) and Lewy neurites is a hallmark of PD (9).
The deposition of αS inclusions in PD follows a common pattern that correlates with clinical symptoms (10). Furthermore, transplanted embryonic neurons in patients with PD developed LB deposits, suggesting that these aggregates can spread (11). Experiments in wild-type and transgenic mice showed that injection of fibrils of recombinant αS could lead to the aggregation of the endogenous protein, supporting this concept (12–14), and the selective fate of the aggregate-containing neurons was demonstrated (15). Although the pathological spreading of αS has now been reproduced in many laboratories, the molecular mechanism of the observed phenomenon is not fully understood and this is important for rational development of therapies. One emerging explanation is that it occurs by prion-like propagation of αS aggregates (16, 17). However, the prion-like role of αS in the process of its pathological spreading is still under debate (18), and alternative hypotheses exist to explain the observations (16). At the molecular level, prion-like propagation is linked to the process of templated seeding, in which the aggregates of αS enter a cell and act as templates to promote the misfolding and aggregation of cellular proteins, resulting in an increased number of aggregates that can then spread to neighboring cells (19). To date, there is a lack of quantitative investigations of how many aggregates, either oligomers or fibrils, are needed to promote efficiently the aggregation of soluble αS, to determine and evaluate the conditions when the templated seeding of αS might be favorable.
To access the quantitative information on the templated seeding requirements for αS, it is important to achieve a comprehensive understanding of its aggregation pathway. A concentration-dependence study can reveal reaction orders of the key steps in a detailed molecular-level mechanism of αS aggregation that can be ultimately used to make predictions of αS seeding behavior. To date, evidence has been obtained about the mechanism of αS aggregation in solution (20–22) and in the presence of lipid membranes that are thought to play an important role in vivo (23–25). Single-molecule studies combined with detailed kinetic analysis have provided us with an opportunity to define the sequence of events during the aggregation process in great detail (26–28). Our related studies using single-molecule fluorescence techniques to follow the aggregation of αS identified a slow conversion step from the initially formed, proteinase (endopeptidase) K-sensitive oligomers to more compact, proteinase K-resistant oligomers. We determined the apparent rates for this process and showed that the converted oligomers caused the highest damage to neuronal cells (27) and were stable with respect to the changes in buffer conditions (28).
In the present study, we have extended this approach to analyze the kinetics of aggregation over a 280-fold range of αS concentrations, from 0.5 µM to 140 µM, and used these data to develop an expanded kinetic model for the aggregation of αS and to determine the rate constants for the individual steps of the reaction. The availability of this model, as well as of data acquired over a wide concentration range, allows us to make quantitative predictions of the aggregation behavior under a variety of predefined conditions and calculate the numbers of oligomers and fibrils required for the templated seeding of αS. This in vitro analysis allows us to assess specifically the propensity of αS protein to undergo a templated seeding process and reveals that this process will bypass the slow nucleation step only under conditions where a high number of aggregates are present within volumes of the order of the size of a living cell. Additional quantitative cellular assays reveal that cytotoxicity occurs at lower concentrations than those required for seeding, suggesting that the spreading of αS under more complex in vivo conditions is likely to involve templated seeding in combination with complementary cell-mediated processes.
Results
Single-Molecule Förster Resonance Energy Transfer Measurements Show That αS Oligomers Are Formed at 0.5- to 140-µM Concentrations Within Several Hours.
All of the single-molecule experiments were performed with full-length (140 residues) αS with an alanine to cysteine mutation at residue 90 for fluorophore incorporation (A90C). The attachment of Alexa Fluor dyes to cysteine 90 was demonstrated to have no significant effect on the kinetics of fibril formation in our previous studies (27, 28), a result attributable to the fact that this residue is at the periphery of the nonamyloid component (NAC) region, which is a key constituent of the αS β-sheet fibril core. In addition, attachment at this position results in the fluorophores becoming in close proximity on the formation of β-sheet structure during aggregation, enabling the conversion process to be followed by intermolecular Förster resonance energy transfer (FRET) between dyes located on different monomers. Therefore, as has been shown in our previous work (27, 28), lower FRET efficiencies are observed for initially formed oligomers lacking significant persistent structure, whereas higher FRET efficiencies are identified for more compact β-sheet-rich oligomers.
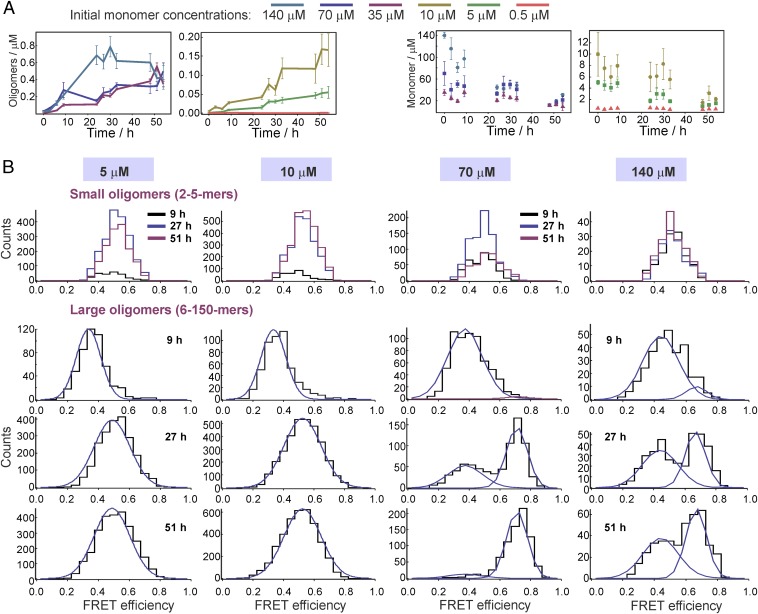
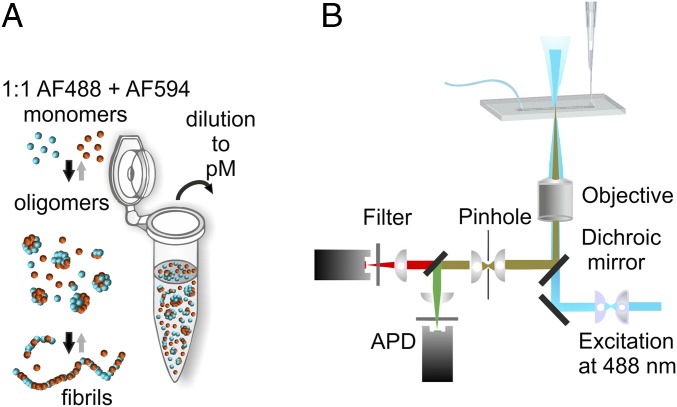
We measured αS oligomer formation in solution at physiologically related pH at 37 °C, with initial monomer concentrations of 0.5 µM, 5 µM, 10 µM, 35 µM, 70 µM, and 140 µM, using the single-molecule FRET (sm-FRET) technique to follow the changes in the numbers of oligomers in the samples within the first 54 h (see Fig. 2). In these experiments, using αS labeled with Alexa Fluor 488 (αS-AF488) and Alexa Fluor 594 (αS-AF594), equimolar quantities of αS-AF488 and αS-AF594 were mixed to give the chosen starting concentration and allowed to aggregate (Fig. 1A), and aliquots were withdrawn for the measurements at the times indicated. Upon withdrawal, the solutions were immediately diluted for single-molecule analysis and continuously passed through a microfluidic channel to reduce the sampling time (28, 29) (Fig. 1B). A 488-nm (blue) laser beam was focused in the center of the channel, to excite the AF488 dye directly, and the resulting fluorescence was simultaneously collected in both AF488 (donor) and AF594 (acceptor) channels. AF594-labeled monomers passing through the confocal volume are undetectable, whereas AF488-labeled monomers give rise to single bursts in the donor channel, enabling the level of monomeric αS to be monitored during the aggregation reaction. As oligomers typically contain both types of label, they are detected as simultaneous intense bursts in the donor and the acceptor channels, due to the emission from both the directly excited AF488 and the nonradiatively excited AF594 via FRET. In this manner, oligomers can be distinguished from monomers despite the fact that the latter are found at much higher concentrations; and the oligomer number and fraction in the solution can be quantified as a function of the reaction time to give kinetic profiles of oligomer formation. The analyses of the experimental data are detailed in Methods and the results are shown in Fig. 2. In addition, the intensities recorded in both donor and acceptor channels for each oligomer were used to determine its FRET efficiency value (Eq. 1 in Methods), and the FRET efficiency values of all oligomers detected in each sample are shown as a FRET efficiency histogram (Fig. 2B), which contains information on both the number of oligomers in the sample and their structural characteristics.
Fig. 2.
Results of the sm-FRET experiment. (A) Kinetic profiles of oligomer formation and monomer depletion, plotted against the incubation time (SEM, N = 6, N is a separate sample). Of note, the increase in the oligomer concentrations for 0.5-µM solutions is present and the resulting species are in the low-nanomolar range, which is not readily visible on the scale in A. Magnification is shown in SI Appendix, Fig. S7A. (B) Representative FRET efficiency histograms, resulting from sm-FRET aggregation experiments with initial protein concentrations of 5 µM, 10 µM, 70 µM, and 140 µM, detected over 400 s. The data were split into two apparent size groups: small (2–5 monomer units) and large (6–150 monomer units) oligomers. For the large oligomers, illustrative fits to Gaussian functions are shown in blue (SI Appendix, Eq. S1), and the resulting mean FRET efficiency values, E, were as follows: for 5 µM, 9 h, E = 0.34, and 27 h and 51 h, global E = 0.49; for 10 µM, 9 h, E = 0.33, and 27 h and 51 h, global E = 0.52; for 70 µM, global E(low-FRET) = 0.37 and global E(high-FRET) = 0.71; for 140 µM, global E(low-FRET) = 0.44 and global E(high-FRET) = 0.67. Further details of the fitting functions and the resulting average parameters (N = 6) are given in SI Appendix, section 1.1.
Fig. 1.
Schematic representation of sm-FRET experiment to probe the aggregation of alpha-synuclein (αS). (A) A 1:1 stoichiometric ratio of αS monomer labeled with Alexa Fluor 488 and Alexa Fluor 594, shown as blue and orange spheres, was combined at a given initial concentration and allowed to aggregate. During the process, the monomeric protein assembles into oligomers, the main focus of the present experiments, and then amyloid fibrils. Aliquots were withdrawn, diluted, and analyzed using a single-molecule microscope. (B) Schematic of the setup used for the single-molecule FRET experiments.
Variations in the Number of Oligomers and Their FRET Histograms as a Function of the Starting Protein Concentration.
Within 54 h of the initiation of the aggregation reaction, oligomer formation was observed for all concentrations of αS, and the highest concentrations of oligomers were formed in the samples with the highest initial protein concentrations and varied as a function of the initial monomer concentration (Fig. 2A). We performed control experiments to verify that we monitor the formation of oligomeric aggregates of αS, using TEM imaging and sm-FRET measurements with removal of fibrils at late incubation times, as detailed in SI Appendix, Figs. S1–S3. The FRET efficiency histograms of the oligomers probed at various time points (Fig. 2B) indicated the existence of two distinct populations, the initially formed, disordered “low-FRET” and the more compact, “high-FRET” oligomers, in agreement with our previous studies that were carried out at the 70-µM concentration of αS (27, 28). In these studies, the high-FRET oligomers were found to be more proteinase K-resistant, more cytotoxic, and more stable toward the changes in buffer conditions compared with the low-FRET species, confirming that the changes in the FRET efficiency represent changes in αS oligomer structure. The experimental data, as described previously, were divided into groups with different oligomer apparent sizes, based on the number of peaks that were resolvable in the FRET efficiency histograms, as shown in SI Appendix, Fig. S4. The groups included “small” oligomers containing 2–5 monomer units (-mers) which showed one peak in the FRET histograms, and “large” oligomers containing 6–150-mers where two peaks could be identified in the FRET histograms. Species composed of more than 150-mers, and species occupying neighboring time bins, typically observed in the samples past the first day of incubation, were assumed to arise from fibrils and excluded from the measurements (SI Appendix, Fig. S3). The apparent size estimation was based on oligomer brightness, as discussed in detail in SI Appendix, and the overall distributions are shown in SI Appendix, Fig. S8. This shows that the majority of the species detected at all times in the aggregation reaction were smaller than 10-mers. We also performed additional total internal reflection fluorescence microscopy (TIRFM) measurements to confirm these results, as detailed in SI Appendix, Fig. S9. In the case of the measurements made on samples with the initial concentrations of 35–140 µM, one FRET peak was observed in the histograms of the small oligomers at all aggregation times, which could be fitted to a single Gaussian distribution (SI Appendix, Eq. S1) centered at a FRET efficiency value (E) of 0.5 (SI Appendix, section 1.1). For large oligomers, two populations could be distinguished, particularly at late aggregation times, and the histograms were globally fitted to double Gaussian functions (SI Appendix, Eq. S1), with E values of 0.4 and 0.6 (Fig. 2B and SI Appendix, section 1.1). These two populations were assessed to have distinctly different kinetic profiles, with the low-FRET population appearing and increasing within the first 24 h of the experiment, but the high-FRET population reaching a maximum value at longer times. They were assigned to the two types of αS oligomers reported previously (27), the low-FRET and the high-FRET oligomers, and the time dependence of these two populations was monitored separately. The kinetic traces for these low- and high-FRET species separately are shown in Fig. 3B. Examination of the histograms obtained for the lowest-concentration samples (0.5 µM) showed that only the low-FRET population could be observed for the studied aggregation period (SI Appendix, Fig. S7B). For the 5-µM and 10-µM samples, the low-FRET distribution was clearly detectable during the first 9 h of the measurements, whereas at later times the histograms showed a much broader single distribution, and the overall oligomer concentrations were extracted without separation into the low- and high-FRET subpopulations (Fig. 2B).
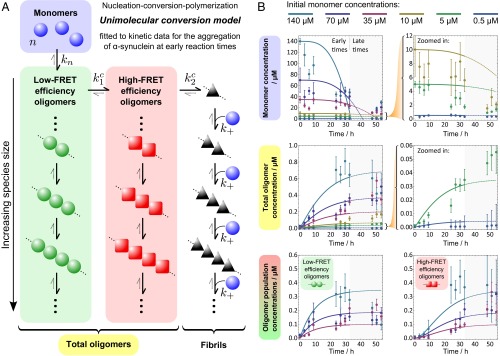
Fig. 3.
Modeling the kinetics of αS aggregation. (A) The model considers a coarse-grained conversion reaction between oligomeric populations as a whole, with no size dependence, and allows fibrils to grow once formed. Here, unimolecular conversions with no monomer dependence are assumed between populations (an alternative model with bimolecular conversions is presented in SI Appendix, Fig. S12A). (B) The resulting nucleation–conversion–polymerization model is used to describe the observed populations, whereby monomer units form low-FRET efficiency oligomers with rate constant kn and an average reaction order of n. These oligomers can then convert to ordered high-FRET efficiency oligomers via a first-order reaction with rate constant k1c, with a subsequent final first-order conversion to fibrils with rate constant k2c. Fibrils can then recruit single-monomer units to grow in a succession of elongation steps, with a length-independent rate constant k+. At early reaction times, reverse reactions can be neglected, and conversion constants were fixed as equal such that k1c = k2c ≡ kc .The resulting simplified model, with four free parameters, was fitted globally to early-time (up to 33 h) kinetic data showing changes with time in monomeric and oligomeric populations for a range of initial monomer concentrations. The resulting nucleation reaction order was found to be n = 0.90 ± 0.1 with rate constants kn = (4.0 ± 2.0) × 10−4 µM1−n h−1, kc = (9.5 ± 5.0) × 10−2 h−1, and k+ = (9.0 ± 7.0) × 10−2 µM−1 h−1.
Reactive Oxygen Species Measurements.
We previously reported that oligomers can be taken up rapidly by neurons and astrocytes and that the high-FRET oligomers promote the production of reactive oxygen species (ROS) when applied to primary neuronal cultures (27). To check whether the oligomers of αS generated in these experiments were able to cause cellular damage, we performed ROS assays and found that the oligomers prepared at either 70 µM or 5 µM initial protein concentration induced the production of ROS, as detailed in SI Appendix, section 1.8 and Fig. S15. Subsequently, we performed a series of ROS measurements to determine the lowest concentration of aggregates needed to be added to produce detectable ROS (SI Appendix), and this concentration was around 50 pM, which corresponds to 30 oligomers per volume approximately corresponding to a single cell (10 µm)3.
The Critical Aggregation Concentration of αS Is Submicromolar.
We measured the critical aggregation concentration (CαS) of αS under the conditions used in this study, by estimating the total concentration of both monomers and oligomers that were released from αS fibrils after prolonged incubation of the fibrils in pure buffer solution. TEM images of the samples of fibrils after the incubation in buffer are in SI Appendix, Fig. S10, and confirm oligomer release from the fibrils, in agreement with the observations in our previous work (27). The CαS value corresponds to the total concentration of monomer present in equilibrium with fibrillar aggregates and was measured in supernatants after removal of fibrillar pellets by ultracentrifugation. This approach yielded a value of 0.7 ± 0.2 µM. The method used for the CαS measurement is fully described in SI Appendix, section 1.7. This CαS value is lower than the result in an earlier report, 28 µM, which was measured using quantitative amino acid analysis (20). It is closer to a more recently reported value of ca. 2.7 µM, which was obtained from absorption measurements of denatured supernatants and subsequent extrapolation to the situation exploring the absence of denaturant (30).
Kinetic Analysis.
Two simple models are consistent with the kinetic data.
In our previous study of the aggregation of αS (27), we identified a conversion from relatively disordered low-FRET oligomers to more compact high-FRET oligomers and were able to perform the kinetic analysis of these data to provide the rate of this conversion. In light of the previously unidentified experimental information on the different protein species formed during the aggregation of the protein at different concentrations, we have been able to expand the model obtained in the previous study. This yielded an explicit and, importantly, predictive aggregation model. Similar to the earlier analysis, we replace the simplified nucleated polymerization models (31–33), in which monomeric units are in direct equilibrium with fibrillar structures, by nucleation–conversion–polymerization models (34), which introduce a series of conformational conversion steps before fibril formation. Such a class of models can be solved analytically for early reaction times, allowing a global fit to both monomeric and oligomeric data. The simplest such model considers a general mechanism whereby low-FRET oligomers are formed from monomeric units in solution. These low-FRET oligomers then convert to high-FRET oligomers, which in turn convert into fibrils (Fig. 3A). Fibrils then grow by monomer addition as has been inferred from the observation that the elongation of the fibrils is initially linear in monomer concentration (22). The assumption of the on-pathway nature of the oligomers is supported by multiple observations. The presence of the lag phase in the formation of high-FRET oligomers relative to that of the low-FRET oligomers suggests that these species originate from the rearrangement of their preceding low-FRET species, as an off-pathway relationship cannot generate such an effect. This is further confirmed by the oligomer release upon fibril disaggregation (SI Appendix, Fig. S10), which was previously demonstrated to occur with the high-FRET oligomers being released at early times, followed by the low-FRET species upon longer incubations (27). By the principle of microscopic reversibility this shows that both high- and low-FRET species are on the pathway to form fibrils. In addition, we have recently demonstrated a correlation between the rates of the oligomer and fibril formation for αS and its pathological mutants, particularly the inhibition of both of these processes for the A30P variant, further supporting the oligomer on-pathway nature (35). Additionally, from the TEM imaging experiments of aggregation and disaggregation (SI Appendix, Figs. S2 and S10), the diameters of oligomers and fibrils generated in these experiments appear comparable. Finally, we can directly observe that oligomers can elongate in the presence of freshly added αS monomer, using TIRFM imaging (SI Appendix, Fig. S11). Because sm-FRET data suggest that oligomers account for a very small percentage of total system mass throughout the aggregation reaction, this allowed a coarse-grained treatment of each oligomer population (described in Methods). The dependence of these conversion processes on oligomer size has only a very minor effect on the overall conversion kinetics and thus does not enter the analysis and we consider only an average overall flux between populations. These considerations do not, however, discriminate between different possible reaction orders of the conversion reactions, and so both a monomer-independent unimolecular conversion model (Fig. 3A) and a monomer-dependent bimolecular conversion model (SI Appendix, Fig. S12A) were considered. Finally, both models were simplified by setting the conversion rate constant for the low-FRET to high-FRET oligomer conversion step equal to that for the high-FRET oligomer to fibril conversion, because allowing these rate constants to differ introduces an extra free parameter to the fitting procedure in each case, yet makes no significant difference to the quality of the fits. The kinetic equations and method used are discussed in further detail in Methods.
We were thus able to use both the unimolecular and the bimolecular conversion models to fit the kinetic data over the entire concentration range, particularly the oligomer concentrations (Fig. 3B and SI Appendix, Fig. S12B), to extract rate constants and a nucleation reaction order. A summary of the results is given in Fig. 3, and the obtained values are in agreement with our earlier studies (27, 28). In both cases we obtain a nucleation reaction order close to one, suggesting that the initial nucleation step governed by is a nonelementary step, for example a change in conformation, that can be resolved in principle into a series of steps by more detailed modeling.
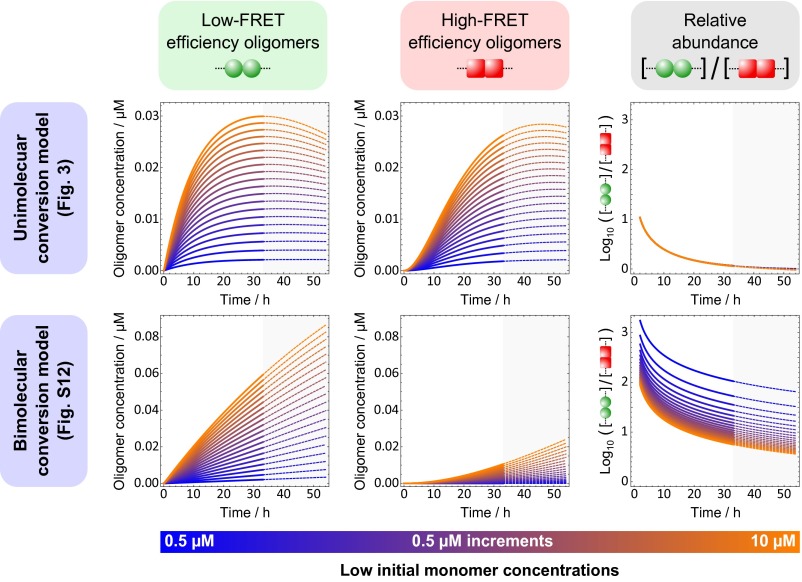
A unimolecular conversion model explains the observed intermediate-FRET histograms at low concentrations.
To distinguish between a unimolecular conversion mechanism and a bimolecular conversion mechanism, both models were used with their fitted rate constants to predict the numbers of low-FRET and high-FRET oligomers with reaction time at concentrations below 10 µM, and these predictions are shown in Fig. 4. A unimolecular conversion model predicts similar and stable relative abundance of low-FRET and high-FRET oligomers up to 54 h during aggregation reactions under these conditions, whereas a bimolecular model predicts much larger variations with time between and within each population. The former finding is consistent with the observation of blurred single-FRET peaks at these concentrations, shown in Fig. 2B; the observed FRET behavior is intermediate between the low-FRET and high-FRET extremes, whereas a bimolecular model would predict significant dominance of the low-FRET population’s contribution to the overall FRET behavior. A unimolecular conversion model is thus preferred and retained for further study below. The unimolecular conversion model gives a rate constant for each conversion step of 9.5 × 10−2 h−1, giving a half-life of each of the conversion steps of the aggregation reaction, outlined in Fig. 3A, of about 7 h. The overall characteristic reaction time, measured by an inflection in the production of fibril mass with time, is given by this model as 34 h for 140 μM and 183 h for 0.5 μM. The value of k+ obtained here, 25 M−1 s−1, for the addition of monomer to a short fibril is slower than that obtained for the addition of monomer to large fibrils under similar but not identical conditions (22). This suggests that the short fibrils formed early in the aggregation process still differ in structure from mature fibrils.
Fig. 4.
Predictions of αS aggregation kinetics at low concentrations by nucleation–conversion–polmerization models. After fitting to the available kinetic data, both unimolecular (Fig. 3) and bimolecular (SI Appendix, Fig. S12) conversion models were used to predict how the concentrations of low-FRET efficiency oligomers and high-FRET efficiency oligomers vary over time at low initial monomer concentrations; the fitting was carried out by considering only the total concentration of oligomers at these low monomer concentrations. As the fitting was carried out over a reaction time of 33 h and overlaid over 54 h, a similar time range was used for the predictions. From the first two columns, it is clear that a unimolecular conversion model alone predicts similar concentrations of both types of oligomers and furthermore predicts a ratio of concentrations that is very stable with changing initial monomer concentration. These predictions are consistent with the single-peak FRET histograms at low concentrations shown in Fig. 2B.
Seeding predictions from the unimolecular conversion model.
In templated seeding, the addition of preformed aggregates to the protein solution causes the acceleration of the aggregation reaction (36). Seeding by αS fibrils has been efficient in vitro (22, 37). However, determining whether oligomers of αS can also take part in the process of seeding has been difficult experimentally, because they are present in low concentrations during the aggregation process, their different types are hard to isolate and stabilize, and the precise quantification of these seeds before their addition remains challenging. Experimental evidence exists for both the promotion (38) and the inhibition (39) of aggregation reaction induced by various αS oligomers.
Having established the extended kinetic model that is consistent with all of the experimental data, we used this model to explore the impact of seeding with either high-FRET oligomers or small fibrils. Low-FRET oligomers were not considered in this analysis owing to their demonstrated lower stability (27, 28). We determined the concentrations of either high-FRET oligomers or small fibrils that are required to double the initial aggregation rate of αS over a 1010-fold range of the different initial concentrations of αS (SI Appendix, Fig. S13). According to the predictions, lower concentrations of fibrils than of oligomers are required to double the aggregation rate of αS, meaning that fibrils will be more prone to seed this reaction if present at equal concentrations. In addition, we assessed how sensitive the system is to seeding over the explored αS monomer concentration range, by looking at the ratio of the monomer concentration to seed concentration, termed “effectiveness” (SI Appendix, Fig. S13). The results reveal that the impact of seeding will vary, depending not only on the nature of the seeds, but also on the initial αS monomer concentration (34); below 10 nM of monomeric αS seeding by either type of aggregates will be relatively ineffective, whereas it will become effective between 10 nM and 1 µM and particularly prominent at the concentrations above 1 µM.
To assess the role of seeds in αS aggregation at more physiologically related conditions, we estimated the numbers of oligomers or fibrils that would need to be introduced to a volume approximately corresponding to a single cell (10 µm)3 to double the aggregation rate in the presence of a set of chosen initial concentrations of αS, which include the range of reported in vivo concentrations of αS (40, 41). According to the kinetic model, above the CαS the seeding will result in the formation of fibrils, whereas below the CαS it will lead to faster production of oligomers. Table 1 summarizes these predictions, and the first column lists the initial concentrations of αS, followed by columns containing the numbers of either high-FRET oligomers or fibrils that are required to be added to these initial concentrations to double the aggregation rate. In addition, Table 1 includes the concentrations of αS monomer that would be needed to prepare the required oligomer seed numbers during the aggregation process. Following these predictions, if the monomer concentration is 2 µM, 10,000 fibrils are needed, or 16,000 high-FRET oligomers, and an approximately five times higher (9.4 µM) concentration of αS monomer would be required for the production of this number of oligomers. We investigated how the resulting seed numbers vary upon either lowering or increasing the initial concentration of αS, as illustrated in Table 1, and the required seed numbers remained in the order of thousands of species. For example, at 2 nM concentration, which is the lowest αS concentration considered in this analysis, 2,700 high-FRET oligomers or 2,400 fibrils are needed to double the aggregation rate. The seed numbers at 3 µM and 4 µM of initial αS, mimicking the conditions of the protein overproduction (42, 43), are also included (Table 1).
Table 1.
Summary of predicted seed concentrations and numbers required to double the initial aggregation rate
| Total concentration/µM | High-FRET oligomer seeding required to double initial rate/µM | No. of high-FRET oligomers implied in a (10 μm)3 cell-like volume | Initial monomer concentration required to produce this concentration of high-FRET oligomers during the aggregation reaction/µM | Fibril seeding required to double initial rate/µM | No. of fibrils implied in a (10 μm)3 cell-like volume |
| 2 | 0.0271 | 16,000 | 9.4 | 0.0174 | 10,000 |
| 0.2 | 0.0098 | 5,900 | 2.9 | 0.0079 | 4,700 |
| 0.02 | 0.0048 | 2,900 | 1.3 | 0.0042 | 2,500 |
| 0.002 | 0.0045 | 2,700 | 1.2 | 0.0039 | 2,400 |
| 3 | 0.0326 | 20,000 | 11.8 | 0.0199 | 12,000 |
| 0.003 | 0.0044 | 2,600 | 1.1 | 0.0038 | 2,300 |
| 4 | 0.0373 | 22,000 | 13.8 | 0.0218 | 13,000 |
| 0.004 | 0.0043 | 2,600 | 1.1 | 0.0038 | 2,300 |
To investigate the effects of the elevated concentration of the initial monomeric αS on the aggregate production, we used the kinetic model to estimate the number of oligomers that would be formed in a 48-h period at the initial αS monomer concentration of 2 μM in the (10 µm)3 volume and determined how this number changes if the concentration is 3 μM instead (SI Appendix, Fig. S14). This analysis shows that the simulated cell-like volume would contain about 104 oligomers in a period of 48 h, if there was no inhibition of the aggregation reaction. This number is again large, of the same order of magnitude as the numbers of species required for seeding, although, for example, the number of proteasomes available for the degradation in a biological cell was estimated to be about 1 million (44). However, if the oligomers were degraded significantly slower than monomers, then oligomer formation could reduce the overall rate of proteasomal degradation in a cell. Interestingly, the consequence of the increased initial monomer concentration of αS from 2 μM to 3 μM in this simulation is that higher concentrations of both oligomers and fibrils are produced at all times up to 48 h (SI Appendix, Fig. S14). Considering the ability of oligomers to cause cellular damage, demonstrated in this and other studies (45), this observation is consistent with the established correlation between an increase in the concentration of αS and pathology.
Discussion
Here, we have exploited single-molecule FRET microscopy to study the initial steps of αS aggregation and explored systematically the formation of oligomers over a range of the starting protein concentrations from 0.5 μM to 140 μM. We then developed an extended kinetic model to analyze the experimental data over the full range of the examined αS concentrations, which allowed an estimation to be made of the rate constants of the main microscopic steps of the reaction shown in Fig. 3A, and making predictions of the number of αS seeds required to increase the aggregation rate, providing an insight into the conditions when the process of templated seeding might be favorable.
Following our predictions, templated seeding by fibrils will enable the aggregation reaction to proceed to growth of more fibrils by monomer addition, without the steps dependent on the formation and conversion of oligomers. Seeding is expected to be particularly effective at minimum concentrations of αS above 1 µM (SI Appendix, Fig. S13), supporting the importance of αS concentration elevation in the disease. To quantify the number of seeds required to affect the aggregation rate of αS in a volume approximately corresponding to a single cell (10 µm)3, we estimated how many oligomers or fibrils would need to be introduced to this volume to double the aggregation rate of αS (Table 1). Clearly, these calculations do not aim to address various complex factors of the cellular environment, among which are, for example, the presence of cellular organelles and lipid surfaces, or altered salt content and pH that could increase the number of aggregates (22, 25), or molecular chaperones (46) and protein degradation systems (47), which prevent aggregate formation and remove aggregates once formed. Nevertheless, the predicted seed numbers based on our analysis provide a quantitative insight into the requirements to observe effective templated seeding of αS in vitro, which is significant because quantification of this process is currently lacking due to experimental difficulties in its elucidation. The predicted numbers of aggregates are large, in the order of 104 species per cell-like volume, values that correspond to micromolar concentrations of the aggregates, requiring multiple species to enter this volume at the same time. These requirements for the templated seeding of αS are relatively high in comparison with, for example, recently reported results for tau K18 using a similar approach (26). It is interesting to note that our kinetic analysis can be used to predict some of the conditions when a small number of oligomers or fibrils may be effective at seeding the aggregation reaction, and hence templated seeding might readily occur. Two factors appear to be important for this process to be favorable. The first determining factor is the nucleation rate, and if the nucleation rate is slow then a small number of oligomers or fibrils are most effective. For example, if the nucleation rate of αS is reduced in our model by a factor of 103, which is close to the published nucleation rate for the tetrapeptide repeat of tau (26), but all of the other rates remain the same, then the number of high-FRET oligomers required to double the initial aggregation rate at 2 μM in the (10 μm)3 volume would be reduced ∼39-fold, from 16,000 to 410. For a single high-FRET oligomer to be required for templated seeding, the nucleation rate constant of αS must be reduced by a markedly larger factor of 108. The other determining factor is the initial protein concentration and whether it is above or below the critical aggregation concentration, which at our conditions was found to be 0.7 μM for αS. At 2 μM, above the critical aggregation concentration, 16,000 oligomers are needed whereas below it, at 0.2 μM, only 5,900 oligomers will be sufficient. These oligomers, if added to a hypothetical cell, will lead to the faster formation of more oligomers, but not fibrils. Therefore, templated seeding will be more effective at low concentrations of free monomer.
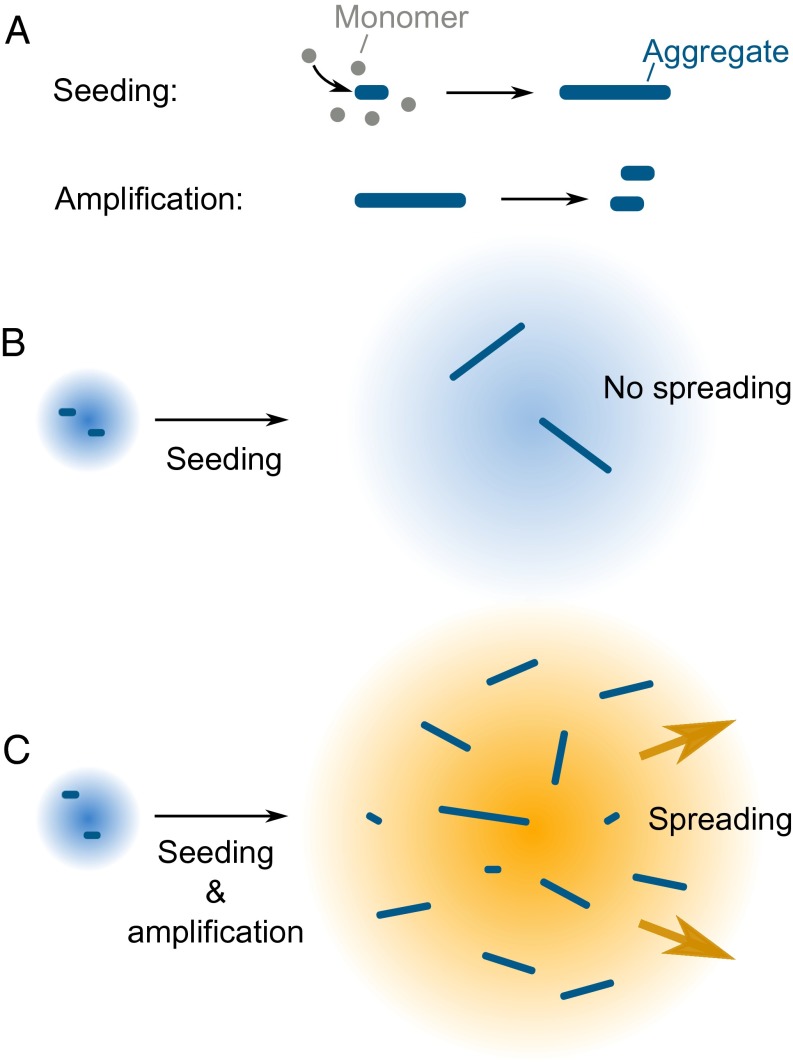
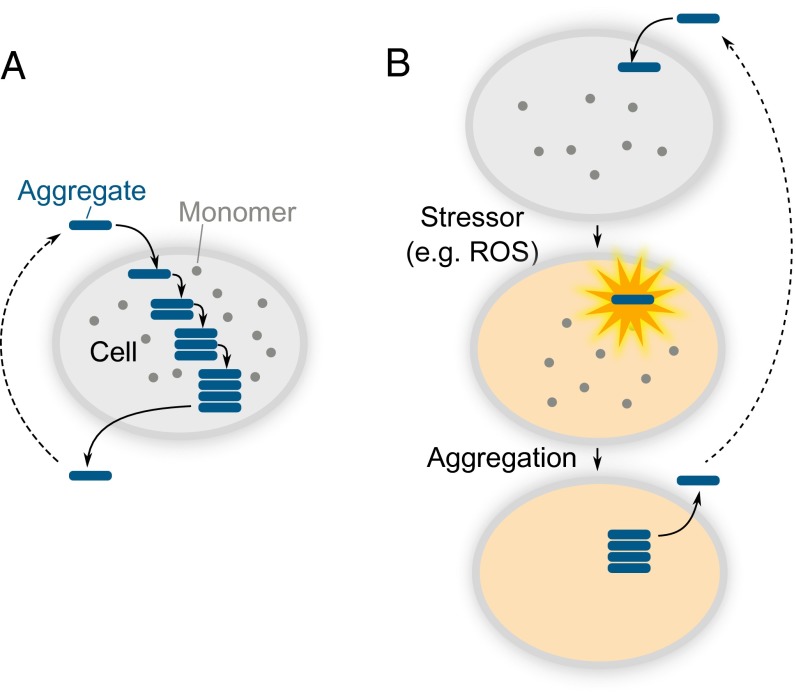
It is interesting to discuss our seeding predictions in the context of prion-like propagation of αS, because the processes of templated seeding and prion-like spreading have been frequently linked in the literature (16). However, despite occurring at the same time, the processes can be distinguished at the molecular level, as is schematically illustrated in Fig. 5, and the templated seeding mechanism constitutes only a part of the spreading process. Templated seeding results in an accelerated production of protein aggregates. In isolation, this would lead to the formation of aggregates and be followed by their passive diffusion (Fig. 5B). To achieve sustainable aggregate-driven spreading and prevent dilution of aggregates as they propagate from cell to cell, a process of aggregate amplification is essential in addition to templated seeding (Fig. 5C). In combination, these two processes would create a positive feedback loop, involving aggregate production and multiplication, resulting in the aggregate-driven (prion-like) spreading of αS, schematically shown in Fig. 6A. Therefore, effective prion-like spreading of αS in vivo requires a combination of conditions favoring both the templated seeding and the aggregate amplification processes. However, although αS aggregate amplification was demonstrated in vitro at low pH (22), we detected no amplification at neutral pH in these experiments and thus additional cellular-based processes are needed, not included in the kinetic model, to achieve the aggregate amplification required for sustained spreading.
Fig. 5.
Schematic representation of the processes required for αS spreading. (A) Seeding results in the formation of aggregates from monomeric protein, and amplification involves the multiplication of existing aggregates. (B) The seeding process alone would not lead to efficient spreading due to the possibility that the formed aggregates would be diluted out. (C) The combination of seeding and amplification can lead to continuous aggregate spreading.
Fig. 6.
Schematic outline of two simplified models of αS aggregate spreading in a cellular environment. (A) Aggregate-driven, or prion-like, propagation of aggregated species. The primary role of the aggregate upon entering a cell is to induce the aggregation of monomeric protein by the mechanism of templated seeding. (B) Cell-driven model. The initial role of the aggregate is to induce cellular stress, which disrupts the homeostasis and creates conditions where protein aggregation becomes favorable.
We have found that the oligomers of αS produced during the aggregation process are neurotoxic, which is consistent with our previous work (27). Using ROS measurements, we estimated that the numbers of oligomers required to promote the production of ROS in neuronal cells under our experimental conditions are in the order of tens of oligomers, which is similar to earlier results reported for amyloid-beta peptide, where cellular damage was found to occur when a single oligomer entered a recipient cell (48). The numbers of aggregates required for ROS production are therefore two orders of magnitude lower than what is required for the templated seeding, and this difference suggests that the templated seeding by the oligomers occurs less readily than the cellular damage caused by the aggregates themselves. This idea is consistent with the long-established link between oxidative stress and neurodegeneration (49) and the fact that the oxidative reactions can promote the aggregation of αS (50), and it is corroborated by experimentally observed correlation between αS seeding and cellular toxicity (15). Based on our findings, we hypothesize that for small aggregates in vivo, if templated seeding occurs, it is under conditions of raised levels of ROS, which in turn may promote αS aggregation, resulting in the aggregate spreading in a cell-driven way that does not strongly depend on the seeding effectiveness, as illustrated in Fig. 6B. Because our results suggest that cellular stress is required for sustained spreading, and larger αS fibrils were previously found less effective at exerting it (27), this implies that a larger number of fibrils than of oligomers will be required for the spreading to occur. Taken together, our quantitative analysis suggests that the mechanism of templated seeding by oligomers or fibrils is unlikely to solely drive the spreading of αS aggregates in PD, because this will always occur under conditions of cellular stress. Ultimately, this suggests that reducing cellular stress may be a possible therapeutic strategy to prevent the spread of disease through the brain.
Summary
We have characterized the early stages of αS aggregation, using in vitro single-molecule experiments and kinetic analysis. Our proposed model treats the initially formed low-FRET oligomers, the more compact high-FRET oligomers, and the fibrils of αS as distinct species, requiring successive conversion steps. The combination of single-molecule measurements and kinetic analysis used in this study has provided a quantification of αS aggregates in seeding the aggregation process, which is important in the context of PD, and the approach is applicable to other peptides and proteins that are likely to be involved in neurodegenerative diseases.
Methods
αS Sample Preparation for sm-FRET.
Monomeric full-length A90C and wild-type αS were expressed and purified according to a previously described protocol (51). A90C was labeled with maleimide-linked Alexa Fluor 488 (AF488) or Alexa Fluor 594 (AF594) (Life Technologies) and separated from the free dyes according to the previously reported protocol (27, 28, 52). Aliquots were flash-frozen, stored at –80 °C, and thawed once before use. For sm-FRET aggregation experiments, a 1:1 molar ratio of AF488- and AF594-labeled monomeric αS was combined in Tris buffer (25 mM Tris, 0.1 M NaCl, pH 7.4) with 0.01% NaN3, up to a starting protein concentration of 0.5 µM, 5 µM, 10 µM, 35 µM, 70 µM, or 140 µM and a sample volume of 300 µL. It is established that αS aggregation in vitro can be promoted by constant agitation (53); therefore the solutions were incubated in the dark at 37 °C with constant agitation at 200 rpm (New Brunswick Scientific Innova 43), and aliquots were withdrawn at regular intervals for sm-FRET experiments. The purity of the starting material was confirmed by sm-FRET measurements of the samples before the incubation.
sm-FRET Data Acquisition.
Aliquots from the dual-labeled aggregating samples were diluted by a serial dilution of 103- to 105-fold in Tris buffer at room temperature immediately before the measurement, a concentration suitable for single-molecule analysis, keeping the multiple-occupancy events negligible. The analyzed solution was introduced into an inlet of a straight-channeled microfluidic device (PDMS; 25 µm height, 100 µm length) via a gel-loading tip and passed through the channel at a constant rate of 2 cm/s by a syringe pump (PHD2000; Harvard Apparatus), according to the previously reported method (29).
The setup used for single-molecule measurements (Fig. 1B) is analogous to one previously described (54). For the FRET measurement, a collimated 488-nm laser beam (Spectra Physics NewPort Cyan) was directed through a back port of an inverted microscope (Nikon Eclipse Ti-U) at 2 mW power (measured at the back port of the microscope), where it was reflected by a dichroic mirror (Semrock DiO1 R405/488/594) and sent through an oil immersion objective (Plan Apo VC 60 x, NA 1.40; Nikon) to be focused 10 µm into the center of the microfluidic channel. Fluorescence signal was collected by the same objective, imaged onto a 100-µm pinhole (Thorlabs), and separated into two channels by a dichroic mirror (Horiba 585DRLP). Donor fluorescence was filtered by a long-pass (Edge Basic 514) and a band-pass filter (535AF45; Omega Filters) before being focused onto an avalanche photodiode (APD) (Perkin-Elmer). Acceptor fluorescence was directed through a long-pass filter (610ALP; Horiba) and a band-pass filter (BrightLine 629/53) before being focused onto a second APD. Synchronous output from the APDs was collected by a custom-implemented field-programmable gate array (FPGA) (Celoxica RC10). Data were acquired for 400 s (80 frames, 100,000 bins per frame, 50 µs bin width) per aliquot and consisted of time-binned photon bursts in the donor and the acceptor channel (SI Appendix, Fig. S5). It was verified that the chosen experimental conditions and the detection time led to the stable rate of coincident events (SI Appendix, Fig. S6), suggesting the absence of oligomer dissociation during the measurements.
sm-FRET Data Analysis.
Data were analyzed using custom-written Igor Pro (Wavemetrics) code, according to a previously reported method (28). Time bins with intensities greater than 15 photons per bin in the donor (emission from AF488) and the acceptor (AF594) channel simultaneously (the AND criterion) (55) were assigned to be due to oligomeric events and selected for the analysis. The donor counts that did not fit the criterion but were above the applied threshold were saved separately and assigned as monomeric αS bursts. The threshold of 15 photons per bin for both channels was determined using the previously established optimized threshold selection method (56). The values of the photon bursts were corrected for the crosstalk and the autofluorescence from the donor to acceptor channels according to , where ID is the modified intensity in the donor channel, D is the original intensity in the donor channel, AD is the autofluorescence in the donor channel (1.6 photons per bin, the average signal from buffer only), and , where IA is the modified intensity in the acceptor channel, A is the original intensity in the acceptor channel, AA is the autofluorescence in the acceptor channel (1.3 photons per bin), and C is the crosstalk from donor to acceptor channel (13%). The crosstalk was negligible from the acceptor to donor channel.
For every simultaneous oligomeric burst, the FRET efficiency was calculated as
| [1] |
where ID is the donor intensity in the presence of an acceptor, IA is the acceptor intensity, and is the gamma factor specific to the instrument (0.99), which accounts for the relative detection efficiencies of the dyes and their quantum yield.
The FRET efficiency values were binned into histograms with bin width of 0.05 (Fig. 2).
Subsequently, the data were split into two size groups: small (2–5-mers) and large (6–150-mers). Large species, either occupying consecutive time bins or greater than 150-mers, were excluded from the analysis as detailed in Horrocks et al. (29). At small sizes, one peak was observed in the FRET efficiency histograms in all measured samples (SI Appendix, Fig. S4). At large sizes, either one (0.5–10 µM) or two (35–140 µM) FRET efficiency peaks could be distinguished. The two distinguishable peaks were assigned to be due to low-FRET oligomers and high-FRET oligomers. The FRET efficiency histograms were integrated to give oligomer kinetic traces, as detailed in SI Appendix, section 1.1. For 0.5- to 10-µM samples, the overall change in oligomer populations was obtained. For 35- to 140-µM samples, the separate kinetic traces for low-FRET and high-FRET oligomers were resolved.
Kinetic Analysis.
The following kinetic moment equations were used to model the aggregation of αS, neglecting the relatively slow reverse conversions and depolymerization reactions.
For the monomer-independent unimolecular conversion model,
| [2] |
For the monomer-dependent bimolecular conversion model,
where P1 is the number concentration of low-FRET oligomers, P2 is the number concentration of high-FRET oligomers, P3 is the number concentration of fibrils, M1 is the mass concentration of low-FRET oligomers (concentration of monomer residues involved), M2 is the mass concentration of high-FRET oligomers (concentration of monomer residues involved), M3 is the mass concentration of fibrils (concentration of monomer residues involved), m(t) is the concentration of free monomer units, mtot is the total concentration of monomer in the system, k1+ is the rate constant governing growth of low-FRET oligomers, and k2+ is the rate constant governing growth of high-FRET oligomers. All other symbols are defined in Fig. 3 and SI Appendix, Fig. S12.
These moment equations are derived by taking sums of the system’s kinetic master equations over the length distribution of each species, as previously described (31–34). Both models are first linearized for early times by taking (discarding the last equation in each case) and then solved analytically using the Mathematica 10.0 software package, to obtain closed-form expressions for each population. Initial conditions were chosen according to the seeding scenario explored (for data fitting, unseeded conditions are represented by and . The resulting expressions were then globally fitted to experimental data up to 33 h for a range of values for via a weighted least-squares Levenberg–Marquardt algorithm, leaving all rate constants and the nucleation reaction order free (Fig. 3 and SI Appendix, Fig. S12). The negligible mass of oligomers observed throughout the aggregation reaction suggests that the consumption of monomer mass by growth of oligomers can be neglected; terms involving and were thus neglected in both models, with no adverse effect observed on the quality of the resulting fits. Furthermore, and were equated into a single parameter in each model, as described in Kinetic Analysis.
The overall characteristic reaction time in the unimolecular model, measured by the time at which an inflection is observed in the fibril mass concentration without linearization of the equations, is given to a good approximation by as described previously (34). For seeding simulations, a doubling in the initial reaction rate was quantified by considering a halving in the 10th time of the reaction upon seeding, that is, the time taken for the fibril mass concentration to reach , which lies within the range of validity of the linearized early time solution described above.
Supplementary Material
Acknowledgments
We thank Nadia Shivji and Beata Blaszczyk for αS protein expression, Peter Jönsson for help with preliminary total internal reflection fluorescence microscopy (TIRFM) imaging experiments, Chris Taylor for help with preliminary autodilution experiments, and Michel Goedert for critical reading of the manuscript. M.I. is funded by a Tayyeb-Hussain Scholarship. G.A.G. is funded by the Schiff Foundation. S.G. is funded through a Wellcome Trust Intermediate Clinical Fellowship. Funding from the Frances and Augustus Newman Foundation, the European Research Council, and the Biothechnology and Biophysical Sciences Research Council is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. V.N.U. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1524128113/-/DCSupplemental.
References
- 1.Dorsey ER, et al. Projected number of people with Parkinson disease in the most populous nations, 2005 through 2030. Neurology. 2007;68(5):384–386. doi: 10.1212/01.wnl.0000247740.47667.03. [DOI] [PubMed] [Google Scholar]
- 2.Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer’s disease. Alzheimers Dement. 2007;3(3):186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- 3.Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 4.Prusiner SB. Cell biology. A unifying role for prions in neurodegenerative diseases. Science. 2012;336(6088):1511–1513. doi: 10.1126/science.1222951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 6.George JM. The synucleins. Genome Biol. 2002;3(1):S3002. doi: 10.1186/gb-2001-3-1-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonini NM, Giasson BI. Snaring the function of alpha-synuclein. Cell. 2005;123(3):359–361. doi: 10.1016/j.cell.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Iwai A, et al. The precursor protein of non-A beta component of Alzheimer’s disease amyloid is a presynaptic protein of the central nervous system. Neuron. 1995;14(2):467–475. doi: 10.1016/0896-6273(95)90302-x. [DOI] [PubMed] [Google Scholar]
- 9.Spillantini MG, et al. Filamentous alpha-synuclein inclusions link multiple system atrophy with Parkinson’s disease and dementia with Lewy bodies. Neurosci Lett. 1998;251(3):205–208. doi: 10.1016/s0304-3940(98)00504-7. [DOI] [PubMed] [Google Scholar]
- 10.Braak H, et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24(2):197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 11.Li JY, et al. Lewy bodies in grafted neurons in subjects with Parkinson’s disease suggest host-to-graft disease propagation. Nat Med. 2008;14(5):501–503. doi: 10.1038/nm1746. [DOI] [PubMed] [Google Scholar]
- 12.Luk KC, et al. Intracerebral inoculation of pathological α-synuclein initiates a rapidly progressive neurodegenerative α-synucleinopathy in mice. J Exp Med. 2012;209(5):975–986. doi: 10.1084/jem.20112457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luk KC, et al. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338(6109):949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuda-Suzukake M, et al. Prion-like spreading of pathological α-synuclein in brain. Brain. 2013;136(Pt 4):1128–1138. doi: 10.1093/brain/awt037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osterberg VR, et al. Progressive aggregation of alpha-synuclein and selective degeneration of lewy inclusion-bearing neurons in a mouse model of parkinsonism. Cell Reports. 2015;10(8):1252–1260. doi: 10.1016/j.celrep.2015.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brundin P, Li JY, Holton JL, Lindvall O, Revesz T. Research in motion: The enigma of Parkinson’s disease pathology spread. Nat Rev Neurosci. 2008;9(10):741–745. doi: 10.1038/nrn2477. [DOI] [PubMed] [Google Scholar]
- 17.Golde TE, Borchelt DR, Giasson BI, Lewis J. Thinking laterally about neurodegenerative proteinopathies. J Clin Invest. 2013;123(5):1847–1855. doi: 10.1172/JCI66029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chauhan A, Jeans AF. Is Parkinson’s disease truly a prion-like disorder? An appraisal of current evidence. Neurol Res Int. 2015;2015:345285. doi: 10.1155/2015/345285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy J. Expression of normal sequence pathogenic proteins for neurodegenerative disease contributes to disease risk: ‘Permissive templating’ as a general mechanism underlying neurodegeneration. Biochem Soc Trans. 2005;33(Pt 4):578–581. doi: 10.1042/BST0330578. [DOI] [PubMed] [Google Scholar]
- 20.Wood SJ, et al. alpha-synuclein fibrillogenesis is nucleation-dependent. Implications for the pathogenesis of Parkinson’s disease. J Biol Chem. 1999;274(28):19509–19512. doi: 10.1074/jbc.274.28.19509. [DOI] [PubMed] [Google Scholar]
- 21.Fink AL. The aggregation and fibrillation of alpha-synuclein. Acc Chem Res. 2006;39(9):628–634. doi: 10.1021/ar050073t. [DOI] [PubMed] [Google Scholar]
- 22.Buell AK, et al. Solution conditions determine the relative importance of nucleation and growth processes in α-synuclein aggregation. Proc Natl Acad Sci USA. 2014;111(21):7671–7676. doi: 10.1073/pnas.1315346111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu M, Li J, Fink AL. The association of alpha-synuclein with membranes affects bilayer structure, stability, and fibril formation. J Biol Chem. 2003;278(41):40186–40197. doi: 10.1074/jbc.M305326200. [DOI] [PubMed] [Google Scholar]
- 24.Auluck PK, Caraveo G, Lindquist S. α-Synuclein: Membrane interactions and toxicity in Parkinson’s disease. Annu Rev Cell Dev Biol. 2010;26:211–233. doi: 10.1146/annurev.cellbio.042308.113313. [DOI] [PubMed] [Google Scholar]
- 25.Galvagnion C, et al. Lipid vesicles trigger α-synuclein aggregation by stimulating primary nucleation. Nat Chem Biol. 2015;11(3):229–234. doi: 10.1038/nchembio.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shammas SL, et al. A mechanistic model of tau amyloid aggregation based on direct observation of oligomers. Nat Commun. 2015;6:7025. doi: 10.1038/ncomms8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cremades N, et al. Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell. 2012;149(5):1048–1059. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horrocks MH, et al. Fast flow microfluidics and single-molecule fluorescence for the rapid characterization of α-synuclein oligomers. Anal Chem. 2015;87(17):8818–8826. doi: 10.1021/acs.analchem.5b01811. [DOI] [PubMed] [Google Scholar]
- 29.Horrocks MH, et al. Single molecule fluorescence under conditions of fast flow. Anal Chem. 2012;84(1):179–185. doi: 10.1021/ac202313d. [DOI] [PubMed] [Google Scholar]
- 30.Baldwin AJ, et al. Metastability of native proteins and the phenomenon of amyloid formation. J Am Chem Soc. 2011;133(36):14160–14163. doi: 10.1021/ja2017703. [DOI] [PubMed] [Google Scholar]
- 31.Oosawa F, Asakura S. Thermodynamics of the Polymerization of Protein. Academic; New York: 1975. [Google Scholar]
- 32.Cohen SIA, et al. Nucleated polymerization with secondary pathways. I. Time evolution of the principal moments. J Chem Phys. 2011;135(6):065105. doi: 10.1063/1.3608916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen SIA, Vendruscolo M, Dobson CM, Knowles TPJ. Nucleated polymerization with secondary pathways. II. Determination of self-consistent solutions to growth processes described by non-linear master equations. J Chem Phys. 2011;135(6):065106. doi: 10.1063/1.3608917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia GA, Cohen SIA, Dobson CM, Knowles TPJ. Nucleation-conversion-polymerization reactions of biological macromolecules with prenucleation clusters. Phys Rev E Stat Nonlin Soft Matter Phys. 2014;89(3):032712. doi: 10.1103/PhysRevE.89.032712. [DOI] [PubMed] [Google Scholar]
- 35.Tosatto L, et al. Single-molecule FRET studies on alpha-synuclein oligomerization of Parkinson’s disease genetically related mutants. Sci Rep. 2015;5:16696. doi: 10.1038/srep16696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lansbury PT., Jr Structural neurology: Are seeds at the root of neuronal degeneration? Neuron. 1997;19(6):1151–1154. doi: 10.1016/s0896-6273(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 37.Nonaka T, Watanabe ST, Iwatsubo T, Hasegawa M. Seeded aggregation and toxicity of alpha-synuclein and tau: Cellular models of neurodegenerative diseases. J Biol Chem. 2010;285(45):34885–34898. doi: 10.1074/jbc.M110.148460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danzer KM, Krebs SK, Wolff M, Birk G, Hengerer B. Seeding induced by alpha-synuclein oligomers provides evidence for spreading of alpha-synuclein pathology. J Neurochem. 2009;111(1):192–203. doi: 10.1111/j.1471-4159.2009.06324.x. [DOI] [PubMed] [Google Scholar]
- 39.Lorenzen N, et al. The role of stable α-synuclein oligomers in the molecular events underlying amyloid formation. J Am Chem Soc. 2014;136(10):3859–3868. doi: 10.1021/ja411577t. [DOI] [PubMed] [Google Scholar]
- 40.Kellie JF, et al. Quantitative measurement of intact alpha-synuclein proteoforms from post-mortem control and Parkinson’s disease brain tissue by intact protein mass spectrometry. Sci Rep. 2014;4:5797. doi: 10.1038/srep05797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mollenhauer B, et al. α-Synuclein and tau concentrations in cerebrospinal fluid of patients presenting with parkinsonism: A cohort study. Lancet Neurol. 2011;10(3):230–240. doi: 10.1016/S1474-4422(11)70014-X. [DOI] [PubMed] [Google Scholar]
- 42.Ahn TB, et al. alpha-Synuclein gene duplication is present in sporadic Parkinson disease. Neurology. 2008;70(1):43–49. doi: 10.1212/01.wnl.0000271080.53272.c7. [DOI] [PubMed] [Google Scholar]
- 43.Singleton AB, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302(5646):841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 44.Princiotta MF, et al. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003;18(3):343–354. doi: 10.1016/s1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 45.Chen SW, et al. Structural characterization of toxic oligomers that are kinetically trapped during α-synuclein fibril formation. Proc Natl Acad Sci USA. 2015;112(16):E1994–E2003. doi: 10.1073/pnas.1421204112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daturpalli S, Waudby CA, Meehan S, Jackson SE. Hsp90 inhibits α-synuclein aggregation by interacting with soluble oligomers. J Mol Biol. 2013;425(22):4614–4628. doi: 10.1016/j.jmb.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Hao R, et al. Proteasomes activate aggresome disassembly and clearance by producing unanchored ubiquitin chains. Mol Cell. 2013;51(6):819–828. doi: 10.1016/j.molcel.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narayan P, et al. Rare individual amyloid-β oligomers act on astrocytes to initiate neuronal damage. Biochemistry. 2014;53(15):2442–2453. doi: 10.1021/bi401606f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bowling AC, Beal MF. Bioenergetic and oxidative stress in neurodegenerative diseases. Life Sci. 1995;56(14):1151–1171. doi: 10.1016/0024-3205(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 50.Hashimoto M, et al. Oxidative stress induces amyloid-like aggregate formation of NACP/alpha-synuclein in vitro. Neuroreport. 1999;10(4):717–721. doi: 10.1097/00001756-199903170-00011. [DOI] [PubMed] [Google Scholar]
- 51.Hoyer W, et al. Dependence of alpha-synuclein aggregate morphology on solution conditions. J Mol Biol. 2002;322(2):383–393. doi: 10.1016/s0022-2836(02)00775-1. [DOI] [PubMed] [Google Scholar]
- 52.Thirunavukkuarasu S, Jares-Erijman EA, Jovin TM. Multiparametric fluorescence detection of early stages in the amyloid protein aggregation of pyrene-labeled alpha-synuclein. J Mol Biol. 2008;378(5):1064–1073. doi: 10.1016/j.jmb.2008.03.034. [DOI] [PubMed] [Google Scholar]
- 53.Serpell LC, Berriman J, Jakes R, Goedert M, Crowther RA. Fiber diffraction of synthetic alpha-synuclein filaments shows amyloid-like cross-beta conformation. Proc Natl Acad Sci USA. 2000;97(9):4897–4902. doi: 10.1073/pnas.97.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orte A, Clarke R, Balasubramanian S, Klenerman D. Determination of the fraction and stoichiometry of femtomolar levels of biomolecular complexes in an excess of monomer using single-molecule, two-color coincidence detection. Anal Chem. 2006;78(22):7707–7715. doi: 10.1021/ac061122y. [DOI] [PubMed] [Google Scholar]
- 55.Ying L, Wallace M, Balasubramanian S, Klenerman D. Ratiometric analysis of single-molecule fluorescence resonance energy transfer using logical combinations of threshold criteria: A study of 12-mer DNA. J Phys Chem B. 2000;104(21):5171–5178. [Google Scholar]
- 56.Clarke RW, Orte A, Klenerman D. Optimized threshold selection for single-molecule two-color fluorescence coincidence spectroscopy. Anal Chem. 2007;79(7):2771–2777. doi: 10.1021/ac062188w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.