Significance
Protein O-glycosylation is an important process in all cells. Substrates are often modified at multiple Ser/Thr residues, but how a glycosyltransferase can act on a continuously changing substrate is unknown. Here, we have analyzed the mechanism by which the cytosolic O-glycosyltransferase GtfA/B of Streptococcus gordonii modifies the Ser/Thr-rich repeats of adhesin, a protein that mediates the attachment of the bacterium to host cells. GtfA/B is a tetramer, with two molecules of GtfA and GtfB. The GtfB subunit of the glycosyltransferase provides the primary polypeptide-binding site, whereas GtfA performs catalysis. GtfB binds unmodified substrate when conformationally constrained by GtfA and binds modified adhesin molecules when in a relaxed conformation. This model explains how the glycosyltransferase can modify a progressively changing substrate molecule.
Keywords: O-glycosylation, bacterial adhesin, crystal structure, Ser/Thr-rich repeats, enzymatic mechanism
Abstract
O-glycosylation of Ser and Thr residues is an important process in all organisms, which is only poorly understood. Such modification is required for the export and function of adhesin proteins that mediate the attachment of pathogenic Gram-positive bacteria to host cells. Here, we have analyzed the mechanism by which the cytosolic O-glycosyltransferase GtfA/B of Streptococcus gordonii modifies the Ser/Thr-rich repeats of adhesin. The enzyme is a tetramer containing two molecules each of GtfA and GtfB. The two subunits have the same fold, but only GtfA contains an active site, whereas GtfB provides the primary binding site for adhesin. During a first phase of glycosylation, the conformation of GtfB is restrained by GtfA to bind substrate with unmodified Ser/Thr residues. In a slow second phase, GtfB recognizes residues that are already modified with N-acetylglucosamine, likely by converting into a relaxed conformation in which one interface with GtfA is broken. These results explain how the glycosyltransferase modifies a progressively changing substrate molecule.
The glycosylation of proteins at Ser and Thr residues (O-glycosylation) is a ubiquitous and important process (1). This kind of modification is found in all organisms and cells, both in the cytosol and in organelles of the secretory pathway. For example, many eukaryotic intracellular proteins are modified with N-acetylglucosamine (GlcNAc) in the cytosol by O-linked N-acetylglucosamine transferase (OGT), a modification that is thought to counteract phosphorylation of the same residues (2, 3). This modification is also of importance for the function of several nuclear pore proteins (3, 4). Prominent examples of secreted O-glycosylated proteins are the mucins, which are exported from epithelial cells and form gels that serve as lubricants and chemical barriers (5). In many cases, proteins are modified at multiple Ser/Thr residues. For example, substrates of the cytosolic OGT are often modified at Ser/Thr/Pro repeats (2, 3), and secreted mucin proteins are modified at numerous Ser and Thr residues by UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases (ppGalNAcTs) (6).
The mechanisms of all O-glycosylation reactions are only poorly understood. For example, although the recognition signal for N-glycosylation at Asn residues is well established (an Asn-X-Ser/Thr sequence) (7), it is unclear how Ser/Thr residues are selected for O-glycosylation. In cases where Ser/Thr-rich repeats are modified, the glycosyltransferases face the additional problem that the substrate changes during the reaction, being initially unmodified, but becoming progressively modified at an increasingly larger number of Ser/Thr residues. How the same enzyme can recognize and modify a continuously changing substrate molecule remains largely unknown.
O-glycosylation plays a prominent role in the pathogenicity of Gram-positive bacteria and mycobacteria (8, 9). Specifically, O-glycosylation is required for the biogenesis and function of adhesins of streptococci and staphylococci bacteria (9). These adhesins contain serine-rich repeats (SRR) that are heavily modified. The glycoproteins are exported from the cell, but remain associated with the cell wall and allow the bacteria to attach to the host cells and their surrounding extracellular matrix (10–17). In addition, SRR glycoproteins may also mediate interactions between bacteria, facilitating biofilm formation and bacterial colonization (18). The SRR-containing adhesins are a major contributor to bacterial infections, including infective endocarditis, pneumococcal pneumonia, neonatal sepsis, and meningitis (19). In view of their roles in a broad spectrum of infections, these adhesins and their biogenesis machinery are major potential targets for novel antibacterial agents.
SRR-containing adhesins have a peculiar pathway of biosynthesis: They are first O-glycosylated in the cytosol and then exported by a dedicated “accessory Sec system” (20, 21). In Streptococcus gordonii, the adhesin GspB contains two Ser/Thr-rich domains (Ser and Thr account for ∼60% of all amino acids). The Ser/Thr residues are first modified with GlcNAc by a primary glycosyltransferase, the activity of which requires two proteins (GtfA and GtfB) (22). Homologs of GtfA and GtfB are found in many other bacterial species (20, 21). Deletion of GtfA or GtfB abolishes adhesin secretion in Streptococcus gordonii and Streptococcus parasanguinis, and deletion of GtfA reduces the adhesion of bacteria to host cells (23, 24). Structural studies on GtfA from Streptococcus pneumoniae show that it belongs to the GT-B family of glycosyltransferases and contains a binding site for UDP-GlcNAc (25). According to the CAZy classification, GtfA is a member of the GT4 family and retains the stereochemistry of the anomeric bond of the sugar during the enzymatic reaction (26, 27). GtfB has been proposed to be a chaperone for GtfA (28), but its exact function is unknown.
Here, we have analyzed the mechanism of the primary GtfA/B glycosyltransferase of S. gordonii. Crystal structures and biochemical experiments show that the enzyme is a tetramer containing two molecules each of GtfA and GtfB. GtfA contains the active site, whereas GtfB provides the major substrate-binding site. During a first phase of glycosylation, the conformation of GtfB is restrained by GtfA, allowing the binding of substrate molecules containing unmodified Ser/Thr residues. In a slow second phase, GtfB changes into a relaxed conformation that can recognize Ser/Thr residues already modified with GlcNAc. Our results explain how the glycosyltransferase can modify a continuously changing substrate molecule. The GtfA/B enzyme shows interesting similarities and differences to other O-glycosylation enzymes.
Results
Structure of the GtfA/B Glycosyltransferase.
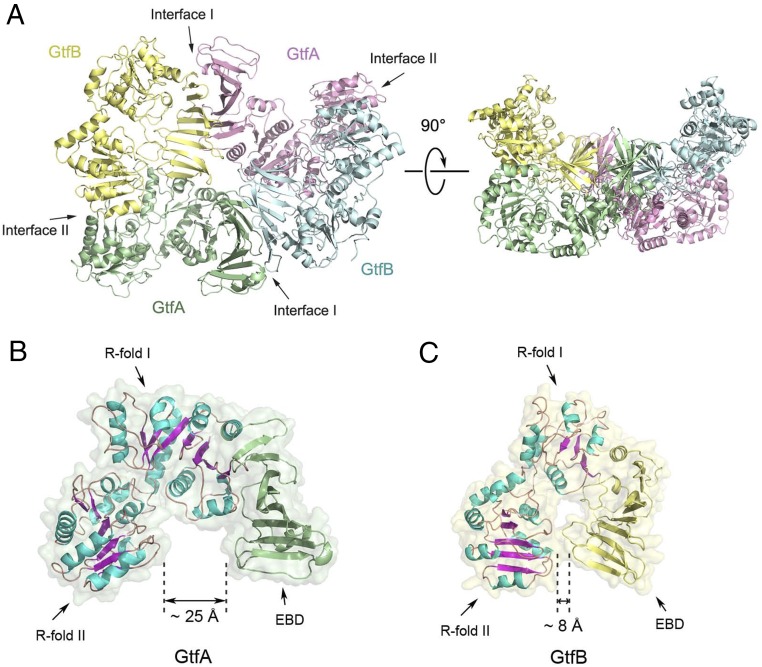
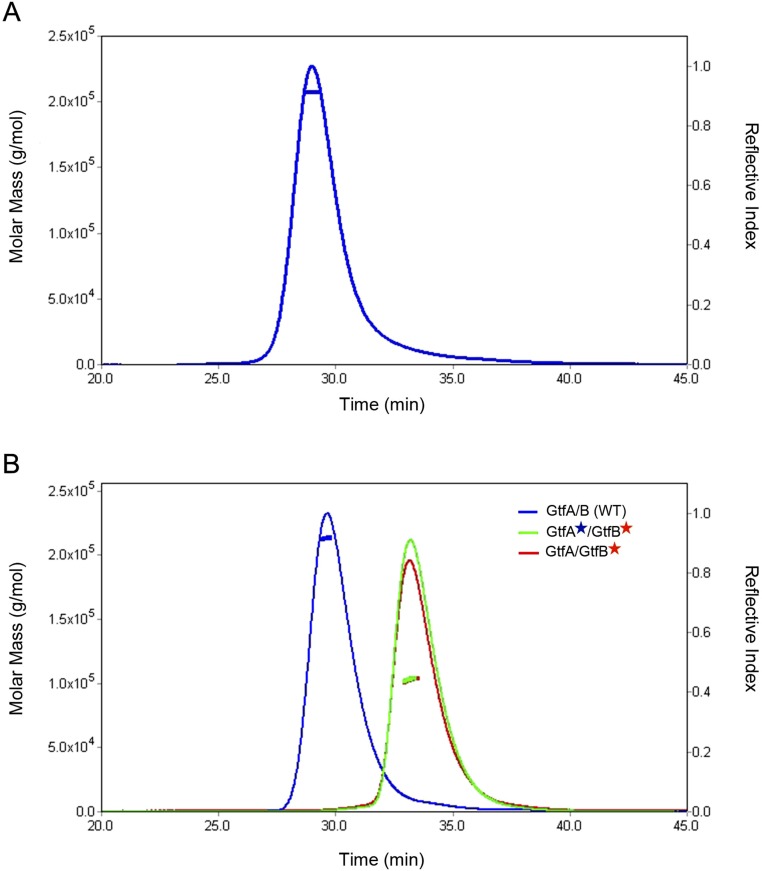
To determine the structure of the active GtfA/B glycosyltransferase, we coexpressed GtfA and GtfB in Escherichia coli and purified the complex. A crystal structure of the GtfA/B complex was determined at 2.92 Å resolution (Table S1). The structure shows a tetramer with two molecules each of GtfA and GtfB (Fig. 1A). A tetramer is likely the oligomeric state of the complex in solution, as demonstrated by size-exclusion chromatography coupled with multiangle light scattering (SEC-MALS) (Fig. S1A). As reported (25), GtfA has a GT-B structure with two Rossmann-like folds (R folds I and II) and an extended β-sheet domain (EBD; also called DUF1975), which together give the molecule a horseshoe-like shape (Fig. 1B). Its structure is similar to that of TarM from Staphylococcus aureus, a teichoic acid α-glycosyltransferase (29, 30).
Table S1.
Data collection and refinement statistics
| Parameters | S. gordonii GtfAB | S. gordonii GtfAB bound to UDP and GlcNAc |
| Data collection | ||
| Resolution limit, Å | 2.92 | 3.84 |
| Space group | I 2 2 2 | P 21 21 21 |
| Cell dimensions | ||
| a, b, c, Å | 155.93, 196.41, 220.86 | 148.98, 196.00, 215.67 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 90 |
| Total reflections | 4,336,858 | 448,209 |
| Unique reflections | 73,613 | 60,983 |
| Rmerge | 0.239 (>1) | 0.385 (>1) |
| CC1/2 | 0.998 (0.774) | 0.990 (0.581) |
| CC* | 0.999 (0.915) | 0.996 (0.87) |
| Mean I/σI | 23.13 (1.90) | 5.28 (1.02) |
| Completeness, % | 99.9 (99.9) | 99.9 (99.9) |
| Multiplicity | 58.9 (60.9) | 7.3 (7.5) |
| Wavelength, Å | 0.9792 | 0.9792 |
| Refinement | ||
| Resolution range, Å | 122.1–2.92 | 94.48–3.84 |
| Rwork/Rfree, % | 21.70/27.50 | 26.89/33.68 |
| No. of nonhydrogen atoms | 15,474 | 30,948 |
| Protein | 15,438 | 30,864 |
| Ligand/ion | 2 | 84 |
| Solvent | 34 | 0 |
| Average B factor, Å2 | 68 | 115 |
| Protein | 68 | 115 |
| Ligand/ion | 17 | 125 |
| Solvent | 33 | 0 |
| rms deviations | ||
| Bond lengths, Å | 0.010 | 0.005 |
| Bond angles, ° | 1.35 | 0.91 |
| Ramachandran analysis, % | ||
| Favored | 95 | 93 |
| Allowed | 5 | 6.5 |
| Outliers | 0.2 | 0.2 |
Statistics for the highest-resolution shell are shown in parentheses. Rmerge = ΣhklΣi|Ii(hkl)-I(hkl)|/ΣhklΣi|Ii(hkl)|, where I(hkl) is the integrated intensity of the reflection. R = Σ|Fobs-Fcalc|/ ΣFobs, where Fobs and Fcalc are observed and calculated structure factors, respectively. Rfree was calculated with 5% of the reflections. CC1/2 is defined as the correlation coefficient between intensities from random half datasets. CC* describes the CC of the full dataset against the true intensities.
Fig. 1.
Crystal structure of the GtfA/B complex. (A) Tetrameric assembly of the GtfA/B complex. The two GtfA subunits are shown as cartoons in green and pink, and the two GtfB subunits in yellow and cyan. Left shows a top view, with GtfA/B interfaces I and II indicated. Right shows a side view. (B) Structure of GtfA in the tetrameric complex, shown as a cartoon inside a surface presentation (in light green). The helices and β-strands in the two R folds are shown in blue and magenta, respectively, and the EBD is in green. (C) As in B, but for GtfB. The surface presentation is in light yellow. The two R folds are shown as in B, the EBD in yellow. Note that the opening of the horseshoe is significantly smaller than in GtfA.
Fig. S1.
Molecular mass of GtfA/B complexes. (A) Shown is the analysis of the purified GtfA/B complex on a Superdex 200 10/300 gel filtration column, coupled with multiangle light scattering (SEC-MALS). The curve shows the reflective index, and the horizontal line is the average molecular mass of the eluted protein. The size corresponds to a tetramer. (B) As in A, but the analysis was also performed with purified complexes in which Gtf proteins carried mutations in interface II. The size of the mutant complexes corresponds to a dimer.
Surprisingly, GtfB has a similar structure as GtfA (Fig. 1C), although this similarity is not apparent from the primary sequence. The two ends of the horseshoe of GtfB are closer together than in GtfA (8 Å versus 25 Å; Fig. 1 B and C). Each copy of GtfA contacts both copies of GtfB in the complex, and each copy of GtfB contacts both GtfA molecules. This arrangement generates two kinds of GtfA-B dimer interfaces, one between the EBDs of GtfA and GtfB (interface surface I: ∼1,560 Å2) and the other between the R folds of GtfA and the ends of the horseshoe of GtfB (interface surface II: ∼960 Å2). PISA interface analysis (Protein Data Bank in Europe; refs. 31 and 32) predicts that interface I is significantly more stable than interface II. This interface is consistent with previous mutational analysis (28, 33).
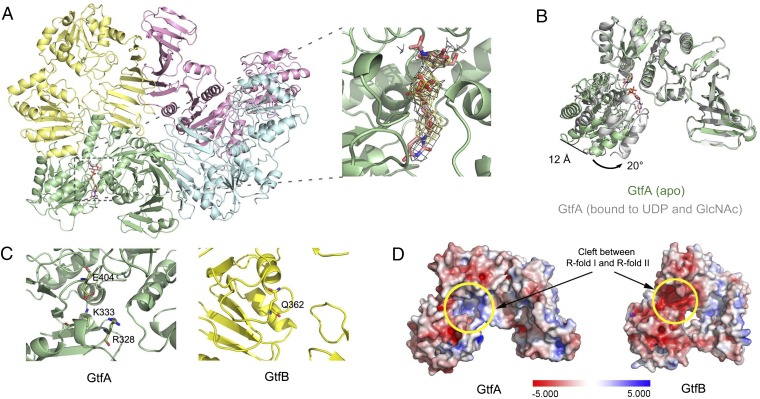
To determine the binding sites for the UDP-GlcNAc substrate in the GtfA/B complex, we cocrystallized the enzyme with UDP and GlcNAc. The structure was refined to a resolution of 3.84 Å (Table S1). Density for UDP and GlcNAc was seen in one copy of GtfA of the tetrameric complex (Fig. 2A). As expected for a GT-B fold glycosyltransferase and observed in a structure of ligand-bound GtfA alone (25, 34, 35), UDP and GlcNAc reside in the cleft between the two R folds. Compared with the apo structure of GtfA, the R-fold II rotates toward R-fold I by ∼20°, thereby embracing the sugar and nucleotide and narrowing the cleft between the R folds (Fig. 2B). The other GtfA molecule in the tetramer remained in the open conformation and did not contain ligands. This molecule was probably prevented from conformational changes by crystal contacts. Both GtfB molecules also remained unchanged. The structure of the tetramer shows that the two active sites of the GtfA molecules point in opposite directions, suggesting that they act independently.
Fig. 2.
Structure of GtfA/B with bound UDP and GlcNAc. (A) Top view of GtfA/B in cartoon presentation with colors as in Fig. 1A, Left. UDP and GlcNAc (shown in stick presentation) are bound to one copy of GtfA. Right shows a magnified view of the active site of GtfA together with a 2mFo-DFc electron density map (in gray) for UDP and GlcNAc after final refinement (at σ = 1.5). The occupancy level of refinement is 1.0. Also shown is an omit map (in yellow), calculated without UDP and GlcNAc (at σ = 3.0). (B) Comparison of GtfA conformations with (gray) and without (apo; green) bound UDP/GlcNAc. (C) Active site residues in GtfA are indicated in stick presentation (Left). GtfB lacks these residues (Right). The essential residue E404 in GtfA is replaced by Q362 in GtfB. (D) The active site in GtfA is positively charged (Left; yellow circle), whereas the corresponding region in GtfB is negatively charged (Right). The electrostatic surface was calculated with the Adaptive Poisson-Boltzmann Solver (55), as implemented in Pymol, using a scale from −5.000 to 5.000 (Bottom).
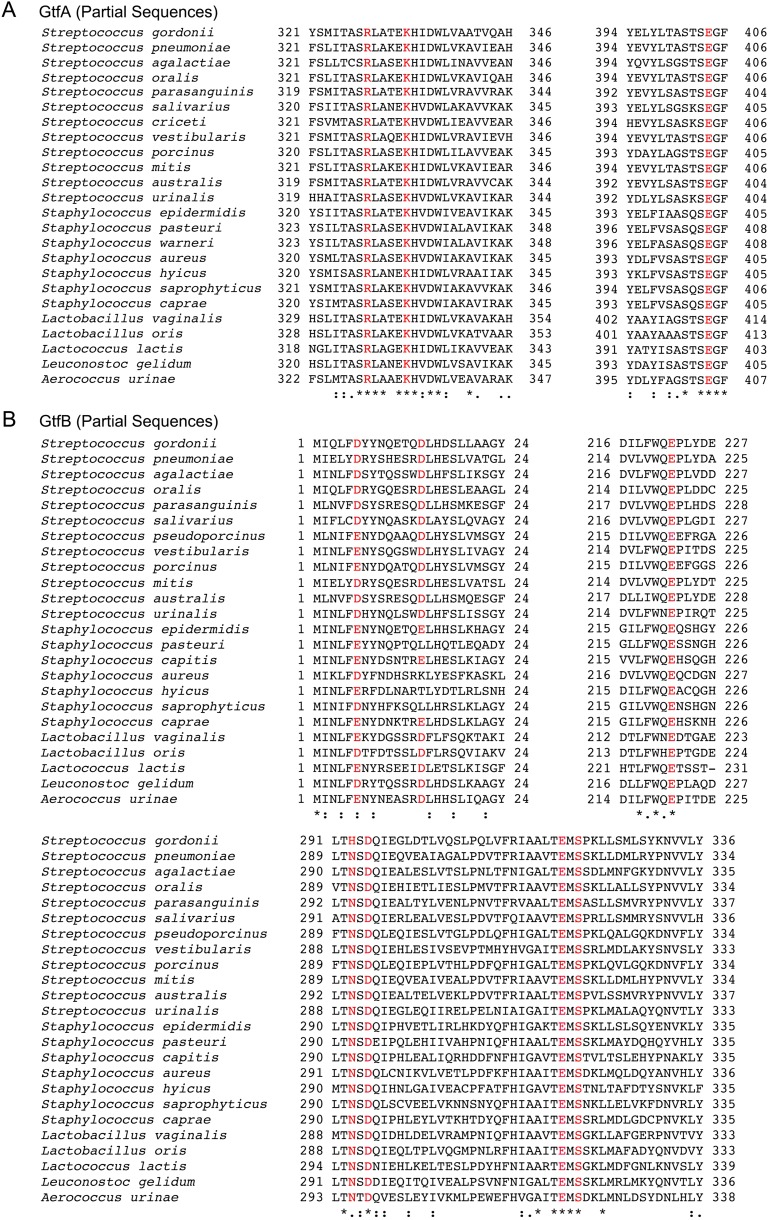
GtfA contains the residues in the active site that are typical for GT-B fold glycosyltransferases, including E404 that is needed for catalysis and K333 and R328 that are involved in binding the phosphates of the UDP-GlcNAc substrate (25, 34, 35) (Fig. 2C). These three residues are evolutionarily conserved (Fig. S2A). The entire binding pocket for the nucleotide sugar is positively charged (Fig. 2D). In contrast, GtfB lacks these residues, because the position of E404 is occupied by Q362, and the positive-charged residues are lacking (Fig. 2C). In fact, the interface between the two R folds has the opposite charge, generated by a number of negatively charged amino acids (Fig. 2D). These features suggest that GtfB does not bind nucleotide-charged sugars or functions as a glycosyltransferase.
Fig. S2.
Conservation of residues involved in GtfA/B function. (A) GtfA proteins from 24 different species were aligned. Regions corresponding to residues 321–346 and residues 394–406 of S. gordonii are shown. The conserved residues corresponding to R328, K333, and E404 of S. gordonii GtfA are colored in red. (B) GtfB proteins from 24 different species were aligned. Regions corresponding to residues 1–24, 216–227, and 291–336 of S. gordonii are shown. The conserved residues corresponding to D6, D14, E222, H293, D295, E319, and S321 of S. gordonii GtfB are colored in red.
GtfA/B Glycosylates the Adhesin GspB in Two Phases.
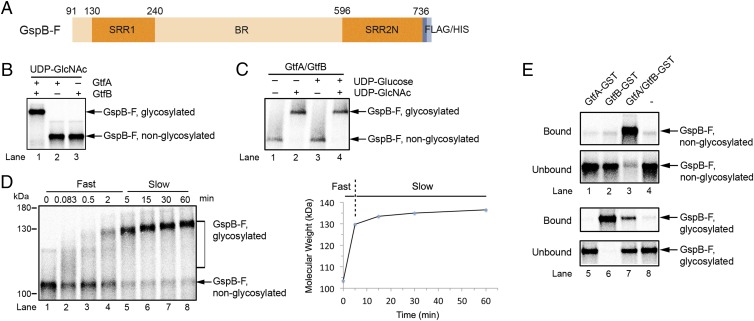
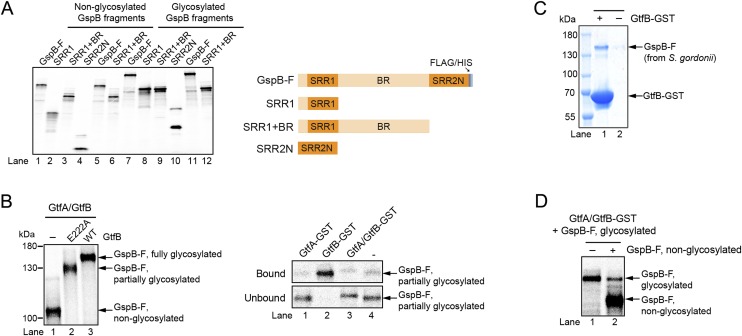
To study the molecular mechanism of the GtfA/B glycosyltransferase, we developed an assay in which substrate is generated by in vitro translation in a reticulocyte lysate system in the presence of [35S]methionine. We used as a substrate a fragment of the adhesin GspB containing residues 91–736 (GspB-F; Fig. 3A). This fragment lacks the signal sequence but includes the first Ser/Thr-rich domain (SRR1), an intervening sequence that normally binds to host cells (binding region; BR), and the N-terminal part of the second Ser/Thr-rich domain (SRR2N). The substrate also contains C-terminal FLAG and His6 tags. GspB-F with the signal sequence is glycosylated in S. gordonii cells and secreted with the same efficiency as full-length adhesin (36).
Fig. 3.
Adhesin glycosylation and binding by GtfA/B. (A) Scheme of the adhesin GspB-F construct used for in vitro experiments. The Ser/Thr-rich region 1 (SRR1), the binding region (BR), and the N-terminal part of Ser/Thr-rich region 2 (SRR2) are indicated. GspB-F contains C-terminal FLAG and His6 tags. (B) GspB-F was synthesized in vitro in reticulocyte lysate in the presence of [35S]methionine. GtfA and GtfB were added as indicated together with UDP-GlcNAc. The samples were incubated for 60 min and subjected to SDS/PAGE followed by autoradiography. (C) As in B, but GspB-F was purified by ammonium sulfate precipitation and Ni-NTA chromatography before in vitro glycosylation in the presence of GtfA and GtfB. Where indicated, UDP-GlcNAc or UDP-Glucose were added. (D) As in B, but GspB-F was purified by ammonium sulfate precipitation before glycosylation by GtfA/B. The reaction was followed over time. Right gives the change in molecular mass of the major GspB-F species. (E) Nonglycosylated and glycosylated GspB-F were partially purified and incubated with GftA-GST, GtfB-GST, or GtfA/GtfB-GST complex. After incubation with GSH beads, the bound and unbound fractions were analyzed by SDS/PAGE and autoradiography.
In vitro translation of GspB-F resulted in a single band visualized after SDS/PAGE and autoradiography. This band corresponds to the nonglycosylated protein (Fig. 3B). The addition of a 1:1 molar ratio of GtfA and GtfB, as well as UDP-GlcNAc, generated fully glycosylated GspB-F, migrating as a single band of much lower mobility in SDS/PAGE (Fig. 3B, lane 1). Similar results were obtained with GspB constructs containing only SRR1, only SRR2N, or SRR1 plus BR (Fig. S3A). GtfA or GtfB alone were totally inactive (Fig. 3B, lanes 2 and 3). When UDP-GlcNAc was not added, the glycosylation reaction with GtfA/B proceeded with only slightly lower efficiency, suggesting that the reticulocyte lysate contains nucleotide-sugar precursor. Indeed, when GspB-F was purified after in vitro translation, no modification by GtfA/B was seen unless UDP-GlcNAc was added (Fig. 3C, lanes 1 and 2). No modification was seen in the presence of UDP-glucose (lanes 3 and 4). These results confirm that O-glycosylation requires both Gtf subunits and that GlcNAc is the primary sugar added to Ser/Thr residues.
Fig. S3.
In vitro glycosylation of GspB fragments and substrate binding of GtfA/B complexes. (A) Various fragments of GspB (Right) were synthesized in vitro in reticulocyte lysate in the presence of [35S]methionine (lanes 1–6). They were then incubated with GtfA/B complex and UDP-GlcNAc for 60 min (lanes 7–12). The samples were analyzed by SDS/PAGE and autoradiography. Lanes 5 and 6 are repetitions of lanes 1 and 3, respectively, and lanes 11 and 12 are repetitions of lanes 7 and 9, respectively. (B) Partially glycosylated GspB-F was generated by using a complex of GtfA and GtfB E222A mutant (Left, lane 2). This intermediate was purified and added to GftA-GST, GtfB-GST, or GtfA/GtfB-GST complex. After incubation with GSH beads, the bound and unbound fractions were analyzed by SDS/PAGE and autoradiography (Right). (C) GspB-F was purified from S. gordonii cells and incubated with GtfB-GST. After incubation with GSH beads, the bound fraction was analyzed by SDS/PAGE and staining with Coomassie blue. A control was performed without GtfB-GST. (D) Glycosylated GspB-F was synthesized in vitro. After purification, a complex of GtfA and GtfB-GST was added, followed by addition of GSH beads. After extensive washing, the beads were incubated in the absence or presence of nonglycosylated GspB-F (at the same concentration as glycosylated GspB-F in the original binding assay), and the bound material was analyzed by SDS/PAGE and autoradiography.
A time course of the glycosylation reaction with GtfA/B showed that the reaction proceeds in two distinct phases, even when the gel mobility shifts are converted into molecular mass changes (Fig. 3D). During a fast phase (up to 5 min for the chosen concentration of GtfA/B in Fig. 3D), unmodified GspB-F was converted into a heterogeneous mixture of glycosylated species, which ran as a smear in SDS gels. In the second phase, a single species appeared, the size of which slowly increased with time. No size change occurred after 60 min, suggesting that, at this time point, all available Ser/Thr residues are modified. The distinct glycosylation phases suggest that GtfA/B recognizes in different ways unmodified substrate and substrate that already contains modified Ser/Thr residues.
Substrate Interaction with GtfA and GtfB.
To better understand the roles of the two subunits of the glycosyltransferase, we performed binding experiments with partially purified in vitro synthesized substrates. Nonglycosylated GspB-F was enriched after in vitro translation by ammonium sulfate precipitation. Fully glycosylated GspB-F was generated by addition of a mixture of GtfA and GST-tagged GtfB (GtfB-GST) after in vitro translation, and subsequent removal of the GtfA/B–GST complex with a GSH resin. The labeled substrates were incubated with GST-tagged GtfA (GtfA-GST), GtfB-GST, or a complex of GtfA and GtfB-GST. After incubation with glutathione resin, the bound and unbound material was analyzed by SDS/PAGE and autoradiography (Fig. 3E). The results show that nonglycosylated GspB-F interacts with GtfA/B, but not with GtfA or GtfB alone (lane 3 versus lanes 1 and 2), explaining why glycosylation requires both subunits. However, glycosylated GspB-F binds strongly to GtfB (lane 6), weakly to GtfA/B (lane 7), and not at all to GtfA (lane 5). Strong binding of GtfB was also observed with GspB-F in which only a subset of Ser/Thr residues were glycosylated in vitro (Fig. S3B), or with glycosylated GspB-F purified from the cytosol of S. gordonii cells (Fig. S3C). Taken together, these results show that the complex of GtfA/B binds more strongly to unmodified than modified substrate, a conclusion that was confirmed in competition experiments (Fig. S3D). However, GtfB alone has no affinity for unmodified substrate, but binds strongly after glycosylation.
GtfB Mediates Substrate Binding.
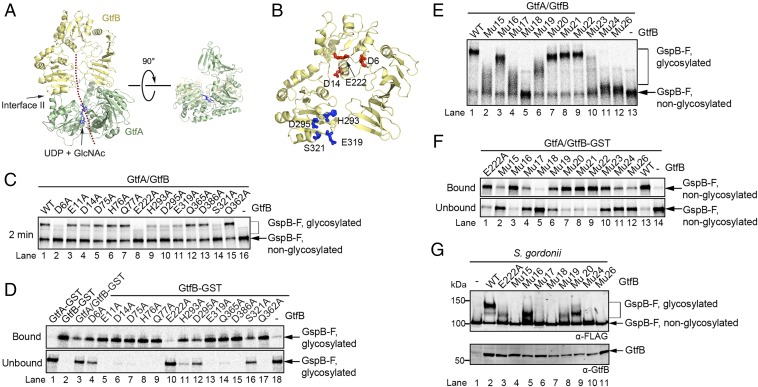
Because GtfB likely lacks enzymatic activity and can interact with glycosylated GspB-F, we wondered whether it contains the major substrate-binding site. The crystal structure indicates a continuous groove across interface II of a GtfA/B dimer, which could accommodate the adhesin substrate (dotted line in Fig. 4A). In this model, the polypeptide substrate would be located in GtfA on top of UDP-GlcNAc in the active site, as seen for human O-GlcNAc transferase (37). From there, the peptide would continue into the opening of the horseshoe of GtfB and exit at the acidic patch.
Fig. 4.
Mutational analysis of GtfB’s substrate binding site. (A) Cartoon of the GtfA/B dimer (GtfA in green, GtfB in yellow) with a polypeptide (dotted red line) in the postulated binding groove (Left). The polypeptide would sit on top of UDP-GlcNAc (in stick presentation), and cross interface II between GtfA and GtfB into a groove of GtfB. Right shows a side view of the groove. (B) Amino acid residues in GtfB’s groove affecting substrate binding. Residues in red and blue are in the acidic patch and R-fold II, respectively. (C) In vitro glycosylation of GspB-F was tested by mixing wild-type GtfA with wild-type GtfB or the indicated GtfB mutants. The samples were incubated for 2 min before analysis by SDS/PAGE and autoradiography. Later time points are shown in Fig. S4A. (D) Purified in vitro glycosylated GspB-F was mixed with GST-tagged wild-type or mutant GtfB. After incubation with GSH beads, the bound and unbound fractions were analyzed by SDS/PAGE and autoradiography. (E) As in C, but with GtfB mutants carrying multiple mutations in the binding groove. Mutant 15: E222A, D6A; Mutant 16: H293A, D295A, E319A, S321A; Mutant 17: E222A, S321A; Mutant 18: E222A, D6A, D14A; Mutant 19: D6A, S321A; Mutant 20: H293A, S321A; Mutant 21: D295A, S321A; Mutant 22: D295A, E319A; Mutant 23: D6A, D14A; Mutant 24: H293A, D295A, E319A, S321A, E222A; Mutant 26: H293A, D295A, E319A, S321A, E222A, D6A. (F) Nonglycosylated GspB-F was mixed with a complex of GtfA and either wild-type or mutant GtfB-GST. After incubation with GSH beads, the bound and unbound fractions were analyzed by SDS/PAGE and autoradiography. (G) The glycosylation of GspB-F was tested in a S. gordonii strain lacking GtfB. Where indicated, the cells were transformed with a plasmid that expresses either wild-type GtfB or the indicated GtfB mutants. The material secreted into the medium was analyzed by SDS/PAGE, followed by immunoblotting with FLAG antibodies. The cell pellet was analyzed by SDS/PAGE and immunoblotting with GtfB antibodies.
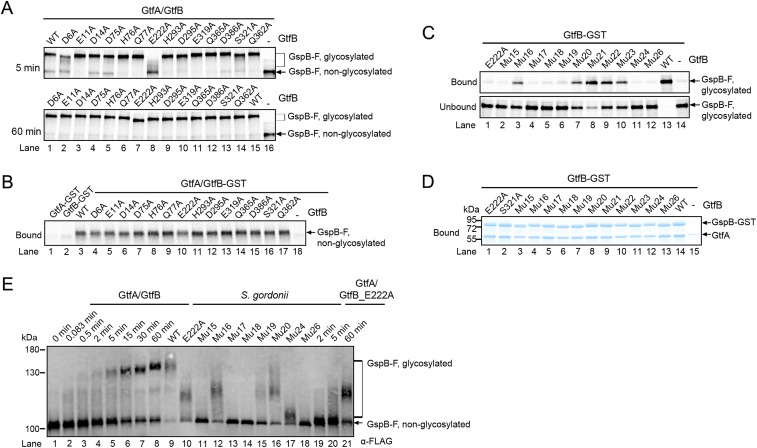
To test the proposed binding model, we first generated single Ala mutations in the acidic patch residues, in R-fold II, and in the EBD (some of these positions are shown in Fig. 4B). The GtfB mutant proteins were purified and tested together with wild-type GtfA for glycosylation of in vitro-synthesized GspB-F (Fig. 4C and Fig. S4A). Two mutations in the acidic patch (E222A and D6A) showed a strong glycosylation defect at early time points. Smaller defects were seen with another mutant in the acidic patch (D14), or when residues were altered in the R-fold II (H293A, D295A, E319A, S321A), which line one side of the postulated binding groove. All these residues are conserved among different bacterial species (Fig. S2B). No effect was seen with a mutant in a nonconserved acidic patch residue (E11A) or with mutants in the EBD (D75, H76, and Q77) that affect the other side of the groove. As expected, mutation of residues in the R-fold II that point away from the groove (Q362A, Q365A, D386A) did not inhibit glycosylation. At later time points, full glycosylation was seen with all mutants, except E222A, which remained underglycosylated (Fig. S4A). Pull-down experiments showed that all mutant GtfA/B complexes interacted well with nonglycosylated GspB-F (Fig. S4B), explaining why none of the mutants was completely defective in glycosylation. However, most GtfB mutants with reduced glycosylation activity also had a weaker affinity for glycosylated GspB-F (Fig. 4D). For example, the E222A mutant was most severely affected in glycosylation (Fig. 4C) and almost completely lost affinity for glycosylated GspB-F (Fig. 4D). These results indicate that the binding to the GtfB groove has more stringent requirements for glycosylated than for unmodified substrate.
Fig. S4.
Analysis of GtfB mutations in the peptide-binding groove. (A) In vitro glycosylation of GspB-F was tested by mixing GtfA with wild-type GtfB or the indicated GtfB mutants. The samples were incubated for 5 or 60 min before analysis by SDS/PAGE and autoradiography. An earlier time point is shown in Fig. 4C. (B) In vitro synthesized, nonglycosylated GspB-F was mixed with a complex of GtfA and either wild-type or mutant GtfB-GST. After incubation with GSH beads, the bound fraction was analyzed by SDS/PAGE and autoradiography. (C) In vitro synthesized, glycosylated GspB-F was purified and mixed with GST-tagged wild-type or mutant GtfB. After incubation with GSH beads, the bound and unbound fractions were analyzed by SDS/PAGE and autoradiography. (D) GtfA was mixed with GST-tagged wild-type or mutant GtfB. After incubation with GSH beads, the bound complex was analyzed by SDS/PAGE and Coomassie blue staining. (E) In vitro synthesized GspB-F was mixed with UDP-GlcNAc and wild-type GtfA/B complex (lanes 1–8) or a complex of GtfA and GtfB E222A mutant (lanes 19–21). The reactions were followed over time and analyzed by SDS/PAGE, followed by immunoblotting with FLAG antibodies. GspB-F secreted by S. gordonii cells was analyzed in parallel (lanes 9–18).
Because single mutations did not completely abolish glycosylation, we introduced multiple mutations into the potential binding surface of GtfB. A mutant in which three residues of the acidic patch were altered (mutant 18; E222A, D6A, D14A) was almost inactive when combined with wild-type GtfA (Fig. 4E). The same is true for mutants in which four residues of the R-fold II and one or two of the acidic patch residues were changed (mutant 24: H293A, D295A, E319A, S321A, E222A; mutant 26: H293A, D295A, E319A, S321A, E222A, D6A). The glycosylation defects of these mutants correlated well with their reduced affinity for nonglycosylated (Fig. 4F) and glycosylated GspB-F (Fig. S4C). Again, the defects were more pronounced with glycosylated substrate. Other mutants had less severe glycosylation defects, although all had a stronger phenotype than those in which only one residue was changed (Fig. 4 E and F). It should be noted that all GtfB mutants behaved like wild-type proteins in gel filtration experiments; they were also indistinguishable in their association with GtfA, as shown by pull-down experiments (Fig. S4D). Taken together, these results show that GtfB is a major contributor to the binding of both nonglycosylated and glycosylated adhesin.
Consistent with the in vitro experiments, with the exception of E222A, single mutations of GtfB had only small effects on the glycosylation of secreted GspB-F in S. gordonii cells, whereas GtfB variants with multiple mutations showed clear defects (Fig. 4G). In general, there was a good correlation between the in vitro and in vivo results. Some mutants generated partially glycosylated GspB-F, which ran on SDS/PAGE at approximately the same position as the material produced with these mutants in vitro (Fig. S4E).
GtfA/B Is a Nonprocessive Enzyme.
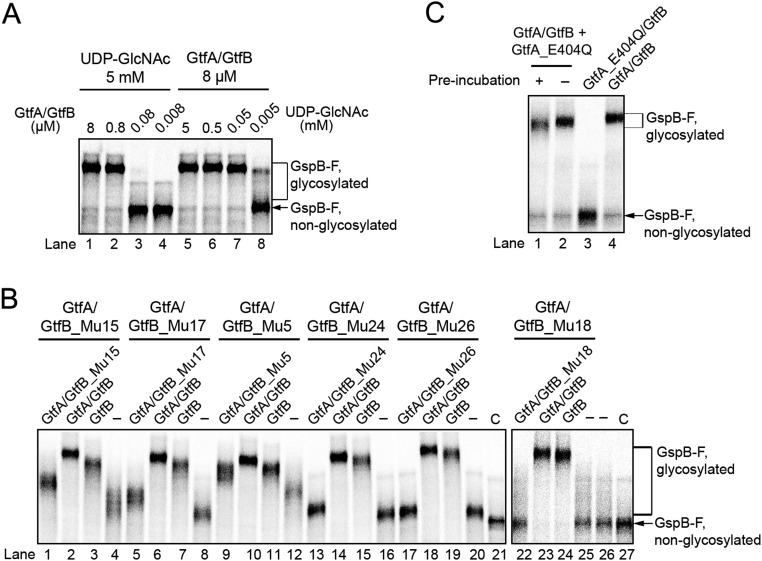
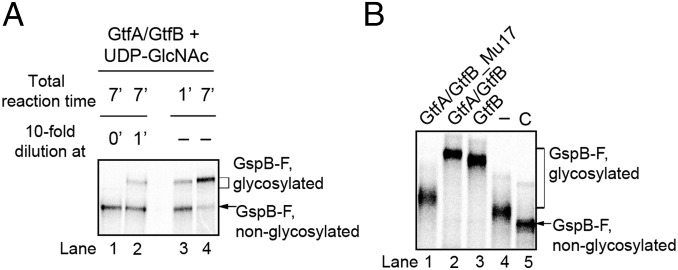
GtfA/B could either be a processive enzyme, i.e., remain bound to the peptide substrate during repeated cycles of sugar attachment to Ser/Thr residues, or it could dissociate from, and rebind to, the substrate during successive modification events. In a first test to distinguish between these possibilities, we incubated GtfA/B with in vitro-synthesized GspB-F for 1 min and then diluted the sample 10-fold, a concentration at which complex formation between free enzyme and substrate is immeasurably slow (Fig. S5A). During a subsequent 6-min incubation, no further modification of GspB-F was observed (Fig. 5A, lane 2 versus 3 and 4), indicating that the enzyme/adhesin complex dissociated. A second test for processivity used the GtfA/GtfB_Mu17 mutant complex. As the concentration of this complex was increased, the adhesin was more completely modified (Fig. 5B, lane 1 versus 4). Thus, partial modification by the mutant complex is likely due to its increased dissociation from the substrate (see also Fig. 4). Finally, dissociation of the enzyme–substrate complex is also supported by the observation that addition of wild-type GtfA/B after glycosylation with GtfA/GtfB_Mu17 or any of the other GtfB mutant complexes resulted in complete glycosylation of the substrate (Fig. 5B, lane 2 and Fig. S5B). Addition of wild-type GtfB alone had almost the same effect (Fig. 5B, lane 3). These results confirm that GtfA/B is a nonprocessive enzyme. The adhesin molecule is released either during each modification cycle or, less likely, after several cycles. In addition, it seems that the GtfA/B tetramer dissociates, allowing mutant GtfB to be replaced with wild-type GtfB.
Fig. S5.
The GtfA/B tetramer dissociates into dimers. (A) In vitro glycosylation reactions were performed with GtfA/B complex and UDP-GlcNAc at the indicated concentrations. After 60-min incubation, the samples were analyzed by SDS/PAGE and autoradiography. (B) To test for tetramer dissociation, glycosylation reactions were performed for 30 min with a complex of GtfA and the indicated GtfB mutants defective in adhesin binding (Fig. 4F). Samples in lanes marked with (-) were analyzed immediately. Where indicated, the same amount of GtfA–GtfB mutant complex, wild-type GtfA/B complex, or GtfB alone were added, and the incubation continued for 30 min before analysis. Lanes marked with “C” show controls without enzyme. (C) A 10-fold excess of catalytically inactive GtfA_E404Q was either preincubated with wild-type GtfA/B complex at 37 °C for 30 min (lane 1) or added immediately at the beginning of a glycosylation reaction (lane 2). Controls were performed with wild-type GtfA/B complex alone (lane 4) or with a complex of GtfA E404Q and wild type GtfB (lane 3).
Fig. 5.
GtfA/B is a nonprocessive enzyme. (A) To test whether GtfA/B is processive, in vitro synthesized GspB-F was incubated with GtfA and GtfB in the presence of UDP-GlcNAc for 1 min. The sample in lane 3 was analyzed immediately, and the one in lane 2 was diluted 10-fold and incubated for additional 6 min (0.08 μM final concentration of GtfA/B). The sample in lane 4 was incubated for an additional 6 min without dilution. Lane 1 shows a sample diluted before incubation. All samples were analyzed by SDS/PAGE and autoradiography. (B) To test for tetramer dissociation, a glycosylation reaction was performed for 30 min with a complex of GtfA and a GtfB mutant (Mu17) (GtfA/GtfB_Mu17) defective in adhesin binding (Fig. 4F). The sample in lane 4 was analyzed immediately. To the samples in lanes 1–3, the same amount of GtfA/GtfB_Mu17 complex, wild-type GtfA/B complex, or GtfB alone was added, and the incubation continued for 30 min before analysis. Lane 5 shows a control without enzyme.
Dissociation of the GtfA/B Tetramer into Dimers.
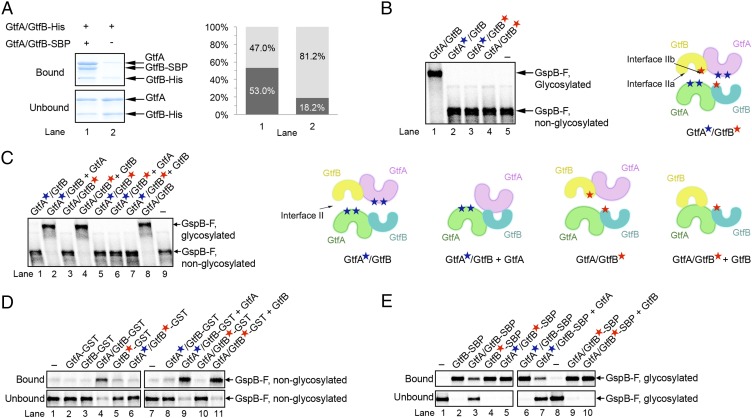
To test whether the GtfA/B tetramer dissociates, we analyzed whether GtfB subunits can be exchanged between different GtfA/B complexes. We generated a complex of GtfA with streptavidin-binding peptide (SBP)-tagged GtfB (GtfB-SBP) and incubated it at 37 °C with a complex of GtfA and His6-tagged GtfB (GtfB-His). After incubation with streptavidin resin, the bound and unbound fractions were analyzed by SDS/PAGE and Coomassie blue staining (Fig. 6A). We found that indeed a significant percentage of the GtfB subunits were exchanged between the GtfA/B complexes (∼35% compared with the 50% theoretical limit). These results confirm that the GtfA/B tetramer can spontaneously dissociate. Consistent with these observations, addition of an excess of a catalytically inactive GtfA mutant (E404Q; Fig. S5C, lane 3) to wild-type GtfA/B resulted in less complete glycosylation (lane 2 versus 4). The effect was even more pronounced after preincubation (lane 1 versus 2). The fast phase of glycosylation was not affected, suggesting that the dissociation of the GtfA/B tetramers is enhanced during the slow phase of glycosylation.
Fig. 6.
Dissociation of the GtfA/B tetramer into dimers. (A) A complex of GtfA and GtfB-SBP was incubated at 37 °C with a complex of GtfA and GtfB-His. After incubation with streptavidin beads, the bound and unbound fractions were analyzed by SDS/PAGE and Coomassie blue staining. The percentage of bound and unbound GtfB-His was quantitated (dark and light gray columns at Right, respectively). (B) Mutations were introduced into interface II of GtfA (blue stars), GtfB (red stars), or both (the cartoon at Right shows the location of the mutations). The resulting GtfA/B dimers were tested for in vitro glycosylation of GspB-F. Controls were performed with wild-type GtfA/B tetramer (lane 1) and in the absence of enzyme (lane 5). (C) As in B, but the reactions with dimerization-defective GtfA or GtfB mutants were complemented with wild-type GtfA or GtfB as indicated. Controls were performed with wild-type GtfA/B tetramer (lane 8) and in the absence of enzyme (lane 9). Cartoons of the tested GtfA/B complexes are shown at Right. (D) The binding of nonglycosylated GspB-F was tested with the indicated combinations of GtfA, GtfB, and GST fusions, with and without dimerization-defective mutations. In lanes 9 and 11, the samples were complemented with wild-type GtfA or GtfB, respectively. Lanes 1 and 7 show controls without added Gtf proteins. Shown are fractions bound and unbound to GSH beads, analyzed by SDS/PAGE and autoradiography. (E) The binding of purified in vitro glycosylated GspB-F was tested with the indicated combinations of GtfA, GtfB, and SBP fusions, with and without dimerization-defective mutations. The samples shown in lanes 7 and 10 were complemented with wild-type GtfA or GtfB, respectively. Lanes 1 and 8 show controls without added Gtf proteins. Shown are fractions bound and unbound to streptavidin beads, analyzed by SDS/PAGE and autoradiography.
We considered it likely that the GtfA/B tetramer dissociates into dimers by disruption of interface II; interface I is extensive, making it unlikely that the other possible dimer or even free GtfB could be generated. We therefore introduced mutations at dimer interface II of either GtfA or GtfB. The GtfA mutations were chosen to disrupt the interaction of both ends of the horseshoe structure of GtfB (interface IIa and IIb), whereas the GtfB mutations affected the interaction of only one end of the horseshoe [interface IIb (the EBD side); Fig. 6B, Right]. When these mutants were mixed with one another or combined with wild-type partner subunits, the complexes were indeed dimers in gel filtration and light scattering experiments (Fig. S1B). These results also show that interface IIb provides most of the binding energy between GtfA and GtfB across interface II.
All constitutive GtfA/B dimers were catalytically inactive (Fig. 6B, Left), supporting the idea that the tetramer is the active species. Dimers containing GtfA interface mutations could be rescued by addition of wild-type GtfA (Fig. 6C, lane 2 versus 1). Similarly, the activity of dimers containing GtfB interface mutations could be restored with wild-type GtfB (lane 4 versus 3). However, dimers containing mutations in both GtfA and GtfB could not be rescued (lanes 5–7). These experiments show that an interaction between GtfA and GtfB across interface II is essential for glycosyltransferase activity, consistent with the postulated continuous substrate-binding groove (Fig. 4A).
All constitutive dimers did not interact with unmodified GspB-F, consistent with their lack of glycosyltransferase activity (Fig. 6D, lanes 5, 6, 8, and 10). As seen before in enzymatic assays, the binding to unmodified substrate could be restored by wild-type subunits if only one side of the interface was mutated (lanes 7–11). Interestingly, all dimers strongly interacted with glycosylated GspB-F (Fig. 6E, lanes 5, 6, and 9). This interaction was as strong as with wild-type or dimerization-defective GtfB alone (lanes 2 and 4). Upon addition of wild-type GtfA, dimers containing GtfA interface mutations showed reduced binding to glycosylated GspB-F (lane 7 versus 6). In contrast, dimers containing GtfB interface mutations retained their strong affinity for glycosylated GspB-F when wild-type GtfB was added (lane 10 versus 9). These results suggest that GtfB undergoes a conformational change. In the tetramer, the conformation of GtfB is constrained by interaction with GtfA across interface II, allowing GtfB to bind nonglycosylated substrate. In the dimer, R-fold II of GtfB is unconstrained and could move outwards, as seen in other glycosyltransferases (38). The resulting relaxed conformation would allow interaction with substrate that already contains modified Ser/Thr residues. The tetrameric complex is not only required for the enzymatic reaction, but also for the release of the glycosylated product.
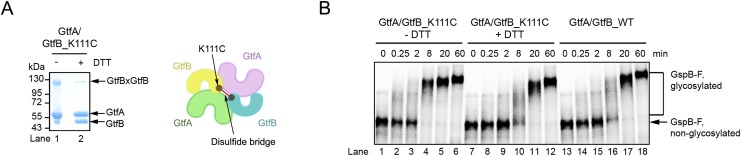
To further test this model, we generated a cross-linked tetramer. A cysteine was introduced into the EBD side of the horseshoe structure of GtfB; it allowed spontaneous disulfide bridge formation between the two GtfB copies in the tetramer (Fig. S6A). The disulfide bridge prevents the dissociation of GtfA and GtfB across interface IIb, but does not affect interface IIa and, thus, the movement of R-fold II of GtfB. The cross-linked tetramer was even more efficient in GspB-F glycosylation (Fig. S6B; compare the 8-min time points, lane 4 versus 10 and 16), supporting the idea that complete dissociation of the tetramer into dimers is not essential and that breaking interface IIa is sufficient to convert GtfB into a conformation that can interact with glycosylated substrate.
Fig. S6.
Glycosylation reactions with cross-linked GtfA/B tetramers. (A) A Cys residue was introduced into position 111 of GtfB (GtfB_K111C). A spontaneous disulfide cross-link forms between the two GtfB molecules in the tetramer (see scheme at Right). In a nonreducing SDS-gel, approximately 50% of all GtfB molecules are cross-linked (lane 1). The cross-linked band disappears after reduction of the disulfide bridge with DTT (lane 2). (B) An in vitro glycosylation reaction was performed with a cross-linked complex of GtfA and GtfB_K111C for different time periods (-DTT). Controls were performed with DTT-treated complex and with wild-type GtfA/B complex.
Discussion
Our results provide insight into the molecular mechanism of an O-glycosyltransferase, the GtfA/B enzyme from S. gordonii. Based on structural and biochemical data, we propose a model that explains how the enzyme deals with a continuously changing polypeptide substrate, which initially is unmodified, but with time contains an increasing number of glycosylated Ser/Thr residues.
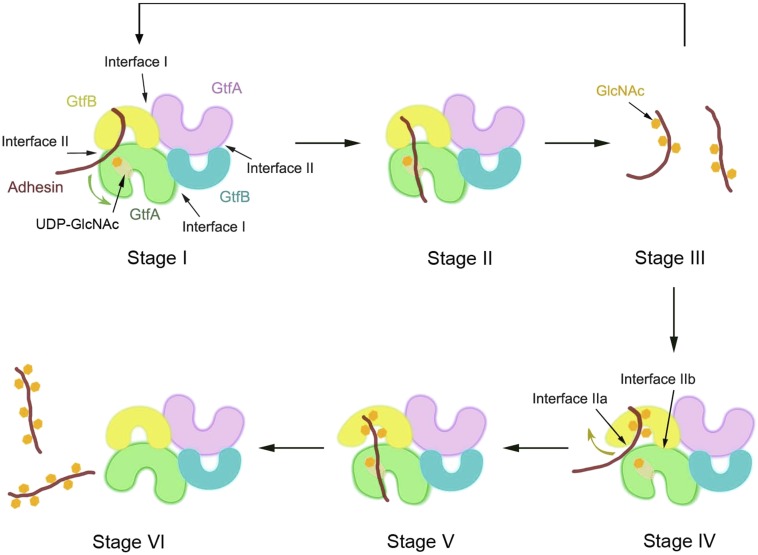
In the first step, the tetrameric GtfA/B complex binds to unmodified adhesin (Fig. 7; stage I). Most of the interaction is provided by a binding groove in GtfB that contains critical residues in an acidic patch and its R-fold II. Mutations in these residues completely abolish substrate binding to the GtfA/B tetramer. However, GtfB alone cannot bind unmodified substrate; rather it is forced into a constrained, interacting conformation by binding to GtfA across interface II. The other substrate of the glycosyltransferase, UDP-GlcNAc, binds to the active site of GtfA into a cleft that is closed by the movement of R-fold II toward R-fold I. It remains unclear whether there is a defined order between sugar binding to GtfA and peptide binding to GtfB. In the next step (Fig. 7, stage II), the adhesin likely binds on top of UDP-GlcNAc in the active site of GtfA, as such an arrangement is observed in human OGT (37) and is consistent with the reported order of substrate binding and product release in related glycosyltransferases (39, 40). After modification of a Ser or Thr residue with GlcNAc, the GtfA/B tetramer dissociates from the substrate (Fig. 7, stage III). As long as there are long stretches of unmodified Ser/Thr residues in the substrate, stages I–III are repeated with fast kinetics. We speculate that the initial, rapid attachment of sugars may prevent the degradation of the adhesin in the cytosol.
Fig. 7.
Scheme of the molecular mechanism of GtfA/B-mediated O-glycosylation. The scheme shows different stages during the glycosylation reaction by GtfA/B. In stage I, the tetrameric GtfA/B complex binds unmodified adhesin (GspB) to GtfB and UDP-GlcNAc to GtfA. R-fold II of GtfA moves toward R-fold I to embrace UDP-GlcNAc (green arrow). In stage II, unmodified adhesin is positioned on top of UDP-GlcNAc in the active site of GtfA. After the enzymatic reaction, modified adhesin is released (stage III). Stages I–III are repeated with fast kinetics as long as there are long stretches of unmodified Ser/Thr residues in adhesin. GtfB can bind to glycosylated Ser/Thr residues when interface IIa is broken and R-fold II of GtfB moves outwards (stage IV, yellow arrow). As in stage II, Ser/Thr residues are modified when positioned on top of UDP-GlcNAc in the active site of GtfA (stage V). Glycosylated adhesin is released upon reassociation of GtfA and GtfB across interface IIa (stage VI).
Eventually during the glycosylation reaction, modified Ser/Thr residues need to be accommodated in the GtfB groove. This interaction is accomplished by breaking interface IIa in the GtfA/B tetramer, releasing the constraint imposed by GtfA on GtfB (Fig. 7, stages IV and V). The resulting conformational change in GtfB probably widens the binding groove so that the bulky sugar residues on Ser/Thr can be accommodated. The widening of the groove likely occurs by an outward movement of R-fold II, because such a conformational change has been observed in GtfA by us (Fig. 2B) and has been reported in other GT-B family members (38). In the GtfA/B dimer, GtfB binds to glycosylated substrate as strongly as in the absence of GtfA (a probably nonphysiological situation), consistent with R-fold II being unconstrained. Our results show that the GtfA/B tetramer can dissociate into dimers during the enzymatic reaction, which requires that both interfaces II are broken at the same time. However, our experiments with cross-linked GtfA/B indicate that complete dissociation into dimers is not required for interaction of the enzyme with glycosylated substrate. It also remains uncertain whether dimers are formed in vivo, and whether both interfaces IIa are broken at the same time.
The last step in the reaction is the release of the glycosylated product, which likely requires the reassociation of GtfA and GtfB across interface IIa (Fig. 7, stage VI). At the end of the glycosylation reaction, there may be an equilibrium between completely modified, unbound adhesin, and a small percentage of modified substrate bound to GtfA/B. This equilibrium might be affected by downstream events during the secretion of the adhesin, such as further glycosylation by the accessory glycosyltransferases (Nss or Gly), or by the binding to proteins involved in secretion (20, 22).
In summary, our results show that there is labor division between the two subunits of the glycosyltransferase: GtfA is the active enzyme, whereas GtfB is inactive, but provides the primary polypeptide-binding site. By changing its conformation, the GtfB subunit can recognize both unmodified and modified adhesin molecules. Our results show that GtfA/B is a nonprocessive enzyme. This mechanism is in contrast to polysaccharide-synthesizing enzymes, such as cellulose synthase (41). In this case, there is directed synthesis through the membrane, which prevents the release of the growing substrate into the cytosol.
GtfA/B is the only known member of the GT-B family of O-glycosyltransferases that requires two subunits for its activity. All other members have only one subunit that binds both the sugar precursor and polypeptide substrate (34, 35). How a polypeptide is recognized in this case is unclear. Although GtfA/B thus seems to use a unique mechanism, some aspects are shared by other O-glycosyltransferases. For example, labor division between two domains is seen in the eukaryotic cytosolic O-GlcNAc transferase (OGT) (37). This enzyme consists of a GT-B fold that contains the active site and a tetratricopeptide (TPR) domain that binds the unmodified polypeptide substrate. The TPR domain contains a narrow peptide-binding tunnel, so it is unlikely to accommodate glycosylated peptide segments. How multiple glycosylation events in Ser/Thr-rich sequences would occur is therefore unclear. In mucin-synthesizing ppGalNAcT enzymes, an active domain with a GT-A fold is able to modify unmodified substrates without the help of any other protein/domain. However, the additional attachment of sugars to modified segments requires a sugar-binding lectin domain (42–44). Thus, different O-glycosylation enzymes have found distinct ways to cope with the problem of a progressively changing substrate structure.
A noncatalytic binding domain, similar to GtfB in the GtfA/B complex, is also found in many glycoside hydrolases. These domains bind to substrate on their own and recruit it to the enzymatically active domain (45). In the case of cellulase 9B (cellulose 1,4-β-endoglucanase) from Cellulomonas, both carbohydrate-binding domains have a similar fold as bacterial 1,3-1,4-β-glucanases, but lack catalytic residues (46–48), reminiscent of the situation with GtfB. We propose that catalytically inactive subunits/domains may have evolved from active enzymes to facilitate substrate recruitment of carbohydrate-modifying enzymes.
Materials and Methods
Protein Purification.
Genes encoding S. gordonii GtfA/B were amplified from S. gordonii genomic DNA by PCR and cloned into the pBAD vector. GtfA contains 6xHis, followed by the sequence GMAS at its N terminus. E. coli BL21 (DE3) cells were transformed with this plasmid, and the expression of recombinant protein was induced with 0.2% (wt/vol) arabinose. Protein was purified from the soluble fraction of a cell lysate by Ni-NTA affinity (QIAGEN) and ion-exchange (HiTrap Q FF; GE Healthcare) chromatography. For crystallization, the proteins were further purified by gel filtration (Superdex 200 10/300 GL; GE Healthcare). Selenomethionine (Se-Met)-derivatized GtfA/B complex was purified from cells grown in M9 minimal medium (Sigma). Se-Met (Anatrace) was added to the medium before induction of expression. Five millimolar DTT (Sigma) was included during the purification steps.
GtfA and GtfB were also overexpressed on their own after cloning into the pET21b vector. C-terminal 6xHis, GST, or SBP tags were added as indicated. Expression was induced with 0.25 mM isopropyl thiogalactopyranoside. Individually expressed GtfA and GtfB proteins were purified by affinity resins followed by gel filtration chromatography. Mutations were introduced into GtfB by QuikChange mutagenesis, and proteins were purified in the same way as the wild-type protein.
Tetramerization-defective GtfA and GtfB mutants (GtfA: Q226A, N249A, D263A, and T267A; GtfB: N62A, D83A, and E86A) were overexpressed and purified as described for the wild-type proteins. GtfA/B dimers were assembled by mixing proteins in a 1:1 molar ratio. The complex was further purified by gel filtration.
Protein Crystallization and Structure Determination.
Crystallization of native and Se-Met substituted 6xHis-GtfA/B complex was performed by the hanging-drop vapor-diffusion method at 22 °C. Optimal crystals were grown in 24-well plates (Hampton Research) by mixing 1 μL of 15 mg/mL protein solution with an equal volume of well solution, containing 100 mM Tris⋅HCl, pH 8.0, 200 mM MgCl2•6H2O, and 10% (wt/vol) PEG 4000. Tetragonal rods appeared after 2–3 d and grew to full size within a week. To crystallize GtfA/B with ligands, GtfA/B was incubated with 10 mM UDP and 10 mM GlcNAc at 4 °C overnight before setting up crystallization trials. Crystals appeared under the same conditions as above. Optimal crystals were obtained by including 30 mM glycyl-glycyl-glycine in the crystallization drops. All crystals were equilibrated in well solution plus 25–30% glycerol (vol/vol) and were flash-frozen in liquid nitrogen stream. Both native and selenium single anomalous diffraction (SAD) datasets were collected at beamline 24ID-E at the Argonne National Laboratory and processed with XDS (49).
Four SAD datasets obtained with Se-Met–containing crystals at the peak wavelength for Se were merged and scaled together. Because the dataset was slightly anisotropic, an anisotropy correction was applied during scaling. The high multiplicity of the merged dataset also helped to improve the quality of the data. Molecular replacement was performed with PHASER in the PHENIX software suite (50), using a published S. pneumoniae GtfA structure (PDB ID code 4PQG), lacking the R-fold II, as the initial search model (25). Two copies of GtfA were identified in each asymmetric unit. Using this partial model, the Se positions were determined and phases were recalculated by AutoSol Wizard in PHENIX. This map allowed visualization of electron density for GtfB. A model was built in Coot (51), facilitated by the positions of Se-Met, and refined with Phenix.refine (Table S1) (52). The final refined atomic model contains two copies of GtfA (an N-terminal Ser derived from the tag and residues 2–503 of GtfA) and two copies of GtfB. One of the GtfB copies comprises residues 1–447 and the other residues 1–445.
The structure of GtfA/B complex bound to UDP and GlcNAc was determined by molecular replacement using PHASER in PHENIX, with the GtfA/B apo structure lacking the R-fold II, as the initial search model. The presence of UDP and GlcNAc in the active site of one copy of GtfA was confirmed in a Fo-Fc difference map calculated with model phases. The model was modified in Coot and refined with Phenix.refine (Table S1). The final refined atomic model contains the same residues as the model for the apo complex, except that the GtfB copies contain residues 1–445 and 1–446, respectively.
Figures showing structures were prepared in PyMOL (Version 1.5.0.4; Schrödinger). All software packages were accessed through SBGrid (53).
SEC-MALS.
One hundred microliters of 1.2 mg/mL protein solution was applied to a Superdex 200 10/300 GL column connected to an ÄKTA Purifier system (GE Healthcare) that is coupled with a multiangle light-scattering instrument (Wyatt Technology). The latter consists of a DAWN EOS detector and an Optilab rEX refractive index detector. The light scattering data were recorded and analyzed by using Astra V software (Wyatt Technology).
In Vitro Glycosylation Assays.
A DNA segment encoding GspB-F was amplified by PCR, using a 5′ end forward primer containing the T7 promoter. GspB-F was synthesized in vitro by TnT Quick Coupled Transcription/Translation System (Promega) in the presence of [35S]methionine. Labeled GspB-F was analyzed by SDS/PAGE by using 7.5% (wt/vol) or 4–20% (wt/vol) acrylamide gels (Bio-Rad). The gels were fixed, dried, and analyzed with a Phosphoimager (Bio-Rad).
To remove UDP-GlcNAc after in vitro translation, the samples were precipitated with 2 M ammonium sulfate and resuspended in a buffer containing 20 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, and 3 mM DTT. For in vitro glycosylation, 0.8 μM GtfA/GtfB complex was mixed with 2 μL of GspB-F solution in 5 μL of final volume. The reaction was started with 0.5 mM UDP-GlcNAc, followed by incubation at 37 °C. Where indicated, 0.5 mM UDP-Glucose was added to 35S-labeled GspB-F purified by an additional Ni-NTA affinity step.
In some experiments, unmodified GspB-F was first incubated with a complex of wild-type GtfA and mutant GtfB (each at 0.8 μM) in the presence of 0.5 mM UDP-GlcNAc at 37 °C for 30 min. The reaction was either stopped by adding SDS/PAGE loading buffer, or supplemented with 0.8 μM of wild-type GtfA/B, GtfA/GtfB_mutant, or GtfB alone, and incubated at 37 °C for additional 30 min.
Purification of Glycosylated 35S-GspB-F.
In vitro-synthesized GspB-F was glycosylated by incubation with 0.8 μM preassembled GtfA/GtfB-GST complex and 0.5 mM UDP-GlcNAc at 37 °C for 60 min. The GtfA/GtfB-GST complex was then removed by incubation with glutathione resin at 4 °C for 2 h. The depletion was repeated twice with fresh resin. The final sample lacked GtfA/B complex, as demonstrated that it was unable to glycosylate in vitro-synthesized 35S-GspB-F.
Partially glycosylated GspB-F was generated by incubating 8 μM preassembled complex of GtfA and GST-tagged GtfB E222A mutant with in vitro synthesized GspB-F and 0.5 mM UDP-GlcNAc. The GtfA/GtfB_E222A-GST complex was then removed by incubation with glutathione resin at 4 °C for 2 h. The depletion was repeated with fresh glutathione resin.
Purification of Glycosylated GspB-F from S. gordonii.
S. gordonii strain carrying GspB-F in place of wild-type GspB was cultured in Todd Hewitt Broth (BD Biosciences) at 37 °C for 5 h. The strain contains endogenous GtfA/B but carries a deletion comprising the genes for the Gly and Nss glycosyltransferases, SecY2, and Asp1-3. The cells were harvested and lysed by sonication. Glycosylated GspB-F was enriched from the clarified cell lysate with an affinity resin containing succinylated wheat germ agglutinin and further purified by gel filtration.
Pull-Down Experiments.
To test the binding to GspB-F, 8 μM Gtf proteins with either GST or SBP tag on one of the proteins were incubated with 3 μL of 35S-labeled GspB-F solutions at 4 °C for 2 h. Magnetic affinity resins (either glutathione or streptavidin beads) were added to the mixtures, and the samples were incubated at 4 °C (glutathione resin) for 2–3 h or at room temperature (streptavidin resin) for 1 h. Bound and unbound fractions were separated by removing the beads with a magnet and analyzed by SDS/PAGE and autoradiography.
To test the binding of glycosylated GspB-F purified from S. gordonii, 2 μM GspB-F was mixed with 2.5 μM GtfB-GST at 4 °C for 2 h before adding the magnetic beads. The bound fraction was analyzed by SDS/PAGE and Coomassie blue staining.
To test the association of GtfA with GtfB mutants, 5 μM GtfA was incubated with 5 μM GST fusion of a GtfB mutant at 4 °C for 2 h. Magnetic glutathione resins were added, and the samples were incubated at 4 °C for 2 h. The bound fractions were analyzed by SDS/PAGE and Coomassie blue staining.
In Vivo Glycosylation of GspB-F by GtfB Mutants.
A nonpolar mutation in gtfB was generated in S. gordonii M99 strain expressing GspB-F (PS1225) by allelic exchange using the pORF4 knockout vector as described (23), creating strain PS3382 (ΔgtfB deletion strain). Different gtfB mutants were subcloned from pET21b into the nisin-inducible Gram-positive expression vector pMSP3545 and introduced into PS3382 by natural transformation (54). After induction with nisin (10 μg/mL) for 4 h, supernatants were collected and probed for secreted GspB-F by using anti-Flag antibodies, whereas protoplasts were treated with mutanolysin and probed for GtfB expression by using anti-GtfB antibodies (Covance).
Cross-Linking of GtfB in the Tetramer.
A cysteine was introduced at position K111 of GtfB, which faces K111 of the other GtfB molecule in the GtfA/B tetramer. GtfA and GtfB_K111C were coexpressed in E. coli BL21 (DE3), and the complex was purified as described for the wild-type complex, but in the absence of any reducing reagent.
Acknowledgments
We thank the staff at the Advanced Photon Source of the Northeastern Collaborative Access Team (NE-CAT) beamline for help with data collection; Phil Jeffrey (Princeton University), Surajit Banerjee (NE-CAT), and Simon Jenni (Harvard Medical School) for help with crystallographic data merging and anisotropy correction; and Stephen Harrison for advice on initial backbone tracing. NE-CAT is supported by National Institute of General Medical Sciences Grant P41 GM103403 from the National Institutes of Health (NIH). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. Y.C. was supported by a Howard Hughes Medical Institute (HHMI)-Helen Hay Whitney Foundation fellowship. The work in the T.A.R. laboratory was supported by NIH Grant GM052586. T.A.R. is an HHMI Investigator. P.M.S. is supported by the Department of Veterans Affairs, NIH Grants R01-AI041513 and R01-AI106987, and the Northern California Institute for Research and Education.
Footnotes
The authors declare no conflict of interest.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5E9T and 5E9U).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600494113/-/DCSupplemental.
References
- 1.Van den Steen P, Rudd PM, Dwek RA, Opdenakker G. Concepts and principles of O-linked glycosylation. Crit Rev Biochem Mol Biol. 1998;33(3):151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- 2.Bond MR, Hanover JA. A little sugar goes a long way: The cell biology of O-GlcNAc. J Cell Biol. 2015;208(7):869–880. doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li B, Kohler JJ. Glycosylation of the nuclear pore. Traffic. 2014;15(4):347–361. doi: 10.1111/tra.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corfield AP. Mucins: A biologically relevant glycan barrier in mucosal protection. Biochim Biophys Acta. 2015;1850(1):236–252. doi: 10.1016/j.bbagen.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Tian E, Ten Hagen KG. Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj J. 2009;26(3):325–334. doi: 10.1007/s10719-008-9162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz F, Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. 2011;21(5):576–582. doi: 10.1016/j.sbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Iwashkiw JA, Vozza NF, Kinsella RL, Feldman MF. Pour some sugar on it: The expanding world of bacterial protein O-linked glycosylation. Mol Microbiol. 2013;89(1):14–28. doi: 10.1111/mmi.12265. [DOI] [PubMed] [Google Scholar]
- 9.Zhou M, Wu H. Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology. 2009;155(Pt 2):317–327. doi: 10.1099/mic.0.025221-0. [DOI] [PubMed] [Google Scholar]
- 10.Samen U, Eikmanns BJ, Reinscheid DJ, Borges F. The surface protein Srr-1 of Streptococcus agalactiae binds human keratin 4 and promotes adherence to epithelial HEp-2 cells. Infect Immun. 2007;75(11):5405–5414. doi: 10.1128/IAI.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bensing BA, López JA, Sullam PM. The Streptococcus gordonii surface proteins GspB and Hsa mediate binding to sialylated carbohydrate epitopes on the platelet membrane glycoprotein Ibalpha. Infect Immun. 2004;72(11):6528–6537. doi: 10.1128/IAI.72.11.6528-6537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siboo IR, Chambers HF, Sullam PM. Role of SraP, a Serine-Rich Surface Protein of Staphylococcus aureus, in binding to human platelets. Infect Immun. 2005;73(4):2273–2280. doi: 10.1128/IAI.73.4.2273-2280.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King NP, et al. UafB is a serine-rich repeat adhesin of Staphylococcus saprophyticus that mediates binding to fibronectin, fibrinogen and human uroepithelial cells. Microbiology. 2011;157(Pt 4):1161–1175. doi: 10.1099/mic.0.047639-0. [DOI] [PubMed] [Google Scholar]
- 14.Sheen TR, et al. Serine-rich repeat proteins and pili promote Streptococcus agalactiae colonization of the vaginal tract. J Bacteriol. 2011;193(24):6834–6842. doi: 10.1128/JB.00094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kukita K, et al. Staphylococcus aureus SasA is responsible for binding to the salivary agglutinin gp340, derived from human saliva. Infect Immun. 2013;81(6):1870–1879. doi: 10.1128/IAI.00011-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shivshankar P, Sanchez C, Rose LF, Orihuela CJ. The Streptococcus pneumoniae adhesin PsrP binds to Keratin 10 on lung cells. Mol Microbiol. 2009;73(4):663–679. doi: 10.1111/j.1365-2958.2009.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo HS, et al. Characterization of fibrinogen binding by glycoproteins Srr1 and Srr2 of Streptococcus agalactiae. J Biol Chem. 2013;288(50):35982–35996. doi: 10.1074/jbc.M113.513358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez CJ, et al. The pneumococcal serine-rich repeat protein is an intra-species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS Pathog. 2010;6(8):e1001044. doi: 10.1371/journal.ppat.1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lizcano A, Sanchez CJ, Orihuela CJ. A role for glycosylated serine-rich repeat proteins in gram-positive bacterial pathogenesis. Mol Oral Microbiol. 2012;27(4):257–269. doi: 10.1111/j.2041-1014.2012.00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bensing BA, Seepersaud R, Yen YT, Sullam PM. Selective transport by SecA2: An expanding family of customized motor proteins. Biochim Biophys Acta. 2014;1843(8):1674–1686. doi: 10.1016/j.bbamcr.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feltcher ME, Braunstein M. Emerging themes in SecA2-mediated protein export. Nat Rev Microbiol. 2012;10(11):779–789. doi: 10.1038/nrmicro2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takamatsu D, Bensing BA, Sullam PM. Four proteins encoded in the gspB-secY2A2 operon of Streptococcus gordonii mediate the intracellular glycosylation of the platelet-binding protein GspB. J Bacteriol. 2004;186(21):7100–7111. doi: 10.1128/JB.186.21.7100-7111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takamatsu D, Bensing BA, Sullam PM. Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol Microbiol. 2004;52(1):189–203. doi: 10.1111/j.1365-2958.2004.03978.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhou M, Peng Z, Fives-Taylor P, Wu H. A conserved C-terminal 13-amino-acid motif of Gap1 is required for Gap1 function and necessary for the biogenesis of a serine-rich glycoprotein of Streptococcus parasanguinis. Infect Immun. 2008;76(12):5624–5631. doi: 10.1128/IAI.00534-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi W-W, et al. Structure of a novel O-linked N-acetyl-D-glucosamine (O-GlcNAc) transferase, GtfA, reveals insights into the glycosylation of pneumococcal serine-rich repeat adhesins. J Biol Chem. 2014;289(30):20898–20907. doi: 10.1074/jbc.M114.581934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carbohydrate Active Enzymes database ( www.cazy.org/). Accessed November 8, 2015.
- 27.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu R, Wu H. A molecular chaperone mediates a two-protein enzyme complex and glycosylation of serine-rich streptococcal adhesins. J Biol Chem. 2011;286(40):34923–34931. doi: 10.1074/jbc.M111.239350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sobhanifar S, et al. Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid α-glycosyltransferase. Proc Natl Acad Sci USA. 2015;112(6):E576–E585. doi: 10.1073/pnas.1418084112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koç C, et al. Structural and enzymatic analysis of TarM glycosyltransferase from Staphylococcus aureus reveals an oligomeric protein specific for the glycosylation of wall teichoic acid. J Biol Chem. 2015;290(15):9874–9885. doi: 10.1074/jbc.M114.619924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. “Protein interfaces, surfaces and assemblies” service PISA at the European Bioinformatics Institute. ( www.ebi.ac.uk/pdbe/prot_int/pistart.html). Accessed June 16, 2015.
- 32.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Bu S, et al. Interaction between two putative glycosyltransferases is required for glycosylation of a serine-rich streptococcal adhesin. J Bacteriol. 2008;190(4):1256–1266. doi: 10.1128/JB.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gloster TM. Advances in understanding glycosyltransferases from a structural perspective. Curr Opin Struct Biol. 2014;28:131–141. doi: 10.1016/j.sbi.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: Structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 36.Bensing BA, Takamatsu D, Sullam PM. Determinants of the streptococcal surface glycoprotein GspB that facilitate export by the accessory Sec system. Mol Microbiol. 2005;58(5):1468–1481. doi: 10.1111/j.1365-2958.2005.04919.x. [DOI] [PubMed] [Google Scholar]
- 37.Janetzko J, Walker S. The making of a sweet modification: Structure and function of O-GlcNAc transferase. J Biol Chem. 2014;289(50):34424–34432. doi: 10.1074/jbc.R114.604405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vetting MW, Frantom PA, Blanchard JS. Structural and enzymatic analysis of MshA from Corynebacterium glutamicum: Substrate-assisted catalysis. J Biol Chem. 2008;283(23):15834–15844. doi: 10.1074/jbc.M801017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang D-M, et al. Glycosyltransferases: Mechanisms and applications in natural product development. Chem Soc Rev. 2015;44(22):8350–8374. doi: 10.1039/c5cs00600g. [DOI] [PubMed] [Google Scholar]
- 40.Lee SS, et al. Mechanistic evidence for a front-side, SNi-type reaction in a retaining glycosyltransferase. Nat Chem Biol. 2011;7(9):631–638. doi: 10.1038/nchembio.628. [DOI] [PubMed] [Google Scholar]
- 41.McNamara JT, Morgan JLW, Zimmer J. A molecular description of cellulose biosynthesis. Annu Rev Biochem. 2015;84:895–921. doi: 10.1146/annurev-biochem-060614-033930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hassan H, et al. The lectin domain of UDP-N-acetyl-D-galactosamine: Polypeptide N-acetylgalactosaminyltransferase-T4 directs its glycopeptide specificities. J Biol Chem. 2000;275(49):38197–38205. doi: 10.1074/jbc.M005783200. [DOI] [PubMed] [Google Scholar]
- 43.Fritz TA, Raman J, Tabak LA. Dynamic association between the catalytic and lectin domains of human UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase-2. J Biol Chem. 2006;281(13):8613–8619. doi: 10.1074/jbc.M513590200. [DOI] [PubMed] [Google Scholar]
- 44.Ten Hagen KG, Fritz TA, Tabak LA. All in the family: The UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 2003;13(1):1R–16R. doi: 10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- 45.Shoseyov O, Shani Z, Levy I. Carbohydrate binding modules: Biochemical properties and novel applications. Microbiol Mol Biol Rev. 2006;70(2):283–295. doi: 10.1128/MMBR.00028-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson PE, Joshi MD, Tomme P, Kilburn DG, McIntosh LP. Structure of the N-terminal cellulose-binding domain of Cellulomonas fimi CenC determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1996;35(45):14381–14394. doi: 10.1021/bi961612s. [DOI] [PubMed] [Google Scholar]
- 47.Brun E, et al. Structure and binding specificity of the second N-terminal cellulose-binding domain from Cellulomonas fimi endoglucanase C. Biochemistry. 2000;39(10):2445–2458. doi: 10.1021/bi992079u. [DOI] [PubMed] [Google Scholar]
- 48.Hahn M, Olsen O, Politz O, Borriss R, Heinemann U. Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-beta-glucanase. J Biol Chem. 1995;270(7):3081–3088. doi: 10.1074/jbc.270.7.3081. [DOI] [PubMed] [Google Scholar]
- 49.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morin A, et al. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bensing BA, Sullam PM. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol Microbiol. 2002;44(4):1081–1094. doi: 10.1046/j.1365-2958.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- 55.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]