Summary
Multi-dimensional mass spectrometry-based shotgun lipidomics (MDMS-SL) is a powerful technology platform among current lipidomics practices due to its high efficiency, sensitivity, and reproducibility, as well as its broad coverage. This platform has been broadly used to determine the altered lipid profiles induced by diseases, injury, genetic manipulations, drug treatments, and aging, among others. Herein, we summarized the principles underlying this platform and presented a protocol for analysis of many of the lipid classes and subclasses covered by MDMS-SL directly from lipid extracts of brain samples. We believe that this protocol could aid the researchers in the field to determine the altered lipid patterns in neurodegenerative diseases and brain injury.
Keywords: Alzheimer’s disease, brain injury, lipidome, mass spectrometry, neurodegeneration, shotgun lipidomics
1. Introduction
Lipidomics, defined as the large-scale study of cellular lipids, is a rapidly expanding research field (1–3). Numerous new discoveries and advances have been made in recent years (3–12). Its essential roles in identifying the biochemical mechanisms of lipid metabolism, investigating the functions of an individual gene of interest, identifying novel biomarkers, and evaluating drug efficacy, among others are becoming increasingly appreciated. One important task in lipidomics is the large-scale identification and quantitation of individual lipid molecular species in each cellular lipidome.
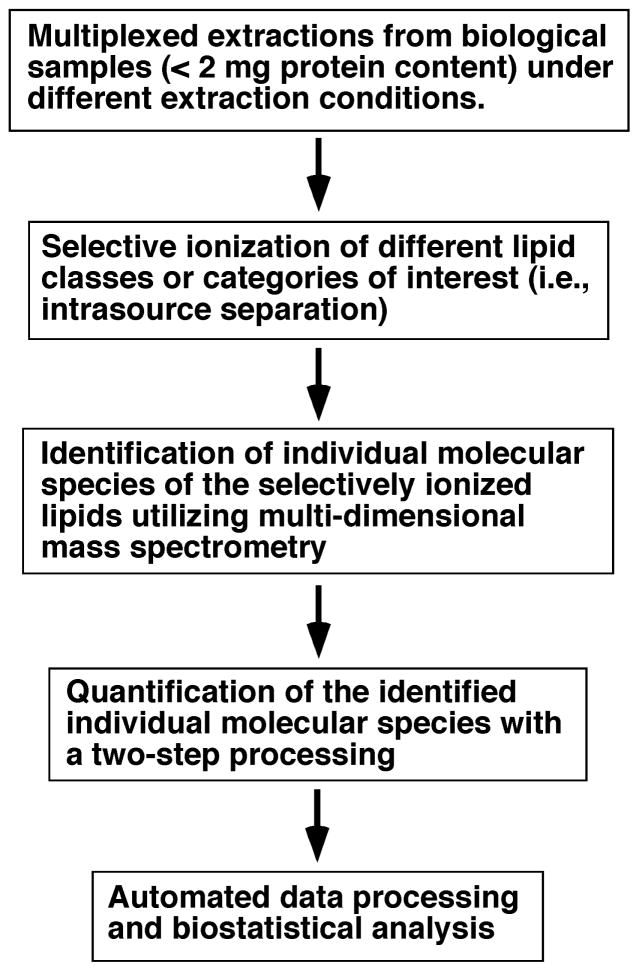
One of the major new developments in current lipidomics is the multi-dimensional mass spectrometry (MS)-based shotgun lipidomics (MDMS-SL) (4, 13, 14). The principle underlying the direct infusion-based MDMS-SL technology is to maximally exploit the unique chemical and physical properties of lipid classes in combination with the special advantages inherited in MS for lipid analysis, thereby achieving maximal separation and ionization, and minimal ion suppression. This principle and the comparison of its differences with other approaches have been extensively described in our review article (14). The workflow of this platform is schematically illustrated (Figure 1).
Figure 1.
A workflow of multi-dimensional mass spectrometry-based shotgun lipidomics.
In brief, the lipids of each biological sample (commonly containing less than 2 mg of protein content from cell, tissue or biologic fluid samples) can be extracted by solvent(s) under acidic, basic, and/or neutral conditions (i.e., multiplexed extractions). The differential solubility of the different lipid classes in various solvents under varying pH conditions is exploited in this step which is a critical step to maximally separate and enrich the lipid class(es) of interest. For example, many lipid classes (e.g., sphingosine-1-phosphate, lysophosphatidic acid (LPA), acylcarnitine, etc) can be efficiently extracted under acidic conditions (4) or recovered from aqueous phase (Han, unpublished data). Gangliosides and acyl-CoA are highly soluble in polar solvents and are partitioned into the aqueous phase during chloroform extraction (15–17). Thus, these lipid classes can be reverse-extracted by using butanol or other solvents under acidic conditions. Moreover, very hydrophobic lipid classes (e.g., cholesterol and its esters, triacylglycerol (TAG), non-esterified fatty acid (NEFA), etc) can be extracted and enriched with hexane. Fluorenylmethoxylcarbonyl (Fmoc) chloride can be added to quickly tag the amine-containing lipids and increase the sensitivity for analysis of these lipids through neutral-loss scanning of the tagged Fmoc moiety (18). Base hydrolysis of all ester-linked glycerolipids could be exploited for isolation and enrichment of sphingoid-backbone containing lipids (19). In contrast, vinyl ether-linked lipid species (i.e., plasmalogens) are labile under acidic conditions. This chemical instability could be used to unambiguously identify the presence of plasmalogen species with a comparison of the mass spectra acquired before and after acid treatment (20).
The electrospray ion source behaves like an electrophoretic cell and can selectively separate different charged moieties under high electrical potential (typically ~ 4 kV) (21, 22). Since different lipid classes possess different electrical properties, largely depending on the nature of their polar head groups (1, 4), the electrospray ion source can be used to resolve lipid classes in a crude lipid extract based upon the intrinsic electrical properties of each lipid class (now termed “intrasource separation of lipids”) (4, 13, 23). In shotgun lipidomics, the differential acidic or basic properties of lipid classes in a lipid solution prepared under a specific pH condition are exploited to selectively ionize different lipid classes in the positive- or negative-ion modes and to achieve a maximal ionization sensitivity (24). Therefore, the lipid classes containing phosphate (e.g., anionic phospholipids, ethanolamine glycerophospholipid (PE), acyl-CoA, and sphingosine-1-phosphate), sulfate (e.g., sulfatide), and carboxylate (e.g., gangliosides and NEFA) can be selectively ionized in the negative-ion mode, for some classes under basic conditions (i.e., in the presence of NH4OH or LiOH in a concentration of 50% of PE concentration), whereas lipid classes containing amine (e.g., acylcarnitine) can be readily ionized in the positive-ion mode under acidic conditions (4). Molecular species of other lipid classes can be ionized as either alkaline or anion (e.g., chloride, acetate, or formate) adducts in the positive- or negative-ion mode, respectively, as discussed previously in details (4).
Finding a sensitive and unique fragment after collision induced-dissociation, which is specific to a class or a group of lipids of interest, is the third key step for successfully identifying, profiling, and quantifying individual molecular species in the class or the group. Specifically, either neutral-loss scanning (NLS) or precursor-ion scanning (PIS) at the mass or m/z ratio of the fragment of interest, respectively, can be performed to “isolate” a given class or a group of lipids from which individual lipid molecular species can be identified in a multi-dimensional array analysis fashion (13, 14). Here, each of these fragments represents a building block of the class or the group of lipids and all the building blocks of each lipid class together constitute an additional dimension to the molecular ions present in the survey scan, which is referred to as the first dimension (7, 13). For example, three moieties linked to the hydroxyl groups of glycerol can be recognized as three individual building blocks and if each building block is identified then each individual glycerol-derived lipid molecular species in a given sample can be determined (13).
Finally, quantitation by shotgun lipidomics is performed in a two-step procedure (13, 25, 26). First, the abundant and non-overlapping molecular species of a class are quantified by comparing the ion peak intensity of each individual identified molecular species to that of the pre-selected internal standard of the class after 13C de-isotoping (4, 27) from a survey scan. Next, some or all of these determined molecular species of the class (plus the pre-selected internal standard) are used as standards to determine the content of other low-abundance or over lapping molecular species using one or multiple NLS and/or PIS scans which are specific to the building blocks (e.g., headgroup) of the lipid class of interest (see above). Multiple standards are necessary in this second step since the fragmentation kinetics of different molecular species may be different (28, 29). It should be pointed out that such an approach by using tandem MS spectrum along with at least two internal standards for quantitation has been broadly employed in the field (29–32). Through this second step in the quantitation process, the linear dynamic range of quantitation can be dramatically extended by eliminating background noise and by filtering the overlapping molecular species through a multi-dimensional mass spectrometric approach (4).
Through lipid class-selective intrasource ionization and subsequent multi-dimensional MS analyses, shotgun lipidomics, at its current stage, enables us to fingerprint and quantify individual molecular species of most major and many minor lipid classes in cellular lipidomes, which collectively represent > 95% of the total lipid mass (composed of hundreds to thousands of molecular species), directly from their CHCl3 extracts after multiplexed sample preparation. These classes of lipids include choline glycerophospholipid (PC), PE, phosphatidylinositol (PI), phosphatidylglycerol (PG), phosphatidylserine (PS), phosphatidic acid (PA), sphingomyelin (SM), monohexosylceramide (i.e., galactosylceramide and/or glucosylceramide, HexCer), sulfatide, NEFA, TAG, lysoPC, lysoPE, lysoPA, acylcarnitine, cholesterol and cholesteryl esters, and ceramide (Cer) (including dihydroceramide). Special methodologies for cardiolipin (CL) (33), 4-hydroxyalkenal (34), sphingosine-1-phosphate (35), sulfatide (36), and sphingosine, psychosine, and lysoSM (19) have also been developed based on their chemical properties.
In this chapter, the protocol for identification and quantitation of some representative lipid classes for research of neurodegenerative diseases and brain injury are described. Although we believe that the MDMS-SL technology platform is powerful for comprehensive analysis of the majority of lipid classes present in cellular lipidomes and the described protocol is readily applicable to other studies, the specific difference of the lipidomic research on neurodegenerative diseases and brain injury from the majority of other studies should be recognized and the solution to address those concerns should be provided. Up to date, only MDMS-SL has addressed this concern and provides an instant criterion to direct a representative sampling during the analysis.
The major biological materials used for lipidomic analysis in the research of neurodegeneration and brain injury are the brain tissues. One of the concerns in lipidomic analysis of these kinds of samples is their homogeneity of cell populations present in different samples. For example, neurons are enriched in gray matter whereas oligodendrocytes are mainly present in white matter. One complication present in any study with brain samples, particularly those from large brains, e.g., human, is the varying degree of different cell populations in the sampled tissues. Differences in the ratio of co-existing gray to white matter represent an unpredictable variable which may overshadow real differences between the samples from disease states relative to controls.
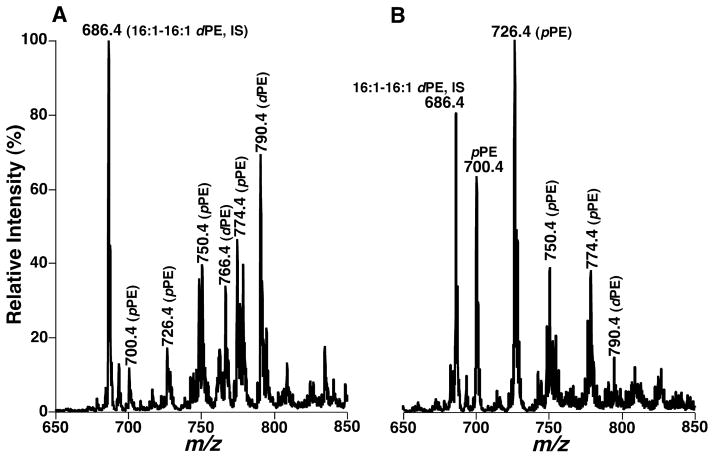
MDMS-SL analysis of human brain samples demonstrates the presence of very distinct lipid profiles of PE molecular species in gray and white matter samples (Figure 2). Specifically, ESI-MS analysis of lipid extracts of cortex gray matter from post-mortem subjects has demonstrated multiple predominant deprotonated ion peaks corresponding to PE species (Figure 2A) in which over 80 mol% of PE species and 55–60% of plasmalogen PE (pPE) species contain polyunsaturated fatty acyl chains at the sn-2 position (37). In contrast, ESI-MS analysis of lipid extracts of white matter from different brain regions has revealed the presence of one predominant peak at m/z 726.4 containing monounsaturated acyl chain (18:1–18:1 pPE) which represents over 85 mol% of the total pPE (Figure 2B). Accordingly, the distinct PE molecular species profiles between brain gray and white matters provide an important criterion to distinguish gray and white matters or determine the degree of cross contamination from co-existing gray and white matters. This degree of the cross-contamination can be accurately determined based upon the peak intensity ratios of ions at m/z 726.4 (18:1–18:1 pPE) and 790.4 (18:0–22:6 PE) following determination of a disease state with the entire profiles (Figure 2).
Figure 2.
Distinct profiles of ethanolamine glycerophospholipid molecular species in lipid extracts of cognitively normal human occipital gray (Panel A) and white matter (Panel B). Plasmenylethanolamine and phosphatidylethanolamine are abbreviated as “pPE” and “dPE”, respectively. “IS” denotes internal standard. (Reproduced with permission from reference (52) from Elsevier B.V., Copyright 2010).
2. Materials
2.1 Equipment (see Note 1)
Nano-ESI source device (TriVersa NanoMate, Advion Bioscience Ltd., Ithaca, NY)
Mass spectrometers (Thermo TSQ VANTAGE, San Jose, CA; ABSciex 4800 MALDI TOF/TOF Analyzer, Framingham, MA)
2.2 Reagents
In addition to the commonly used reagents, the majority of lipid internal standards including 1,2-dimyristoleoyl-sn-glycero-3-phosphocholine (di14:1 PC), 1,2-dipalmitoleoyl-sn-glycero-3-phosphoethanolamine (di16:1 PE), 1,2-dipentadecanoyl-sn-glycero-3-phosphoglycerol (sodium salt) (di15:0 PG), 1,2-dimyristoyl-sn-glycero-3-phosphoserine (sodium salt) (di14:0 PS), 1,2-dimyristoyl-sn-glycero-3-phosphate (sodium salt) (di14:0 PA), 1,1′,2,2′-tetramyristoyl cardiolipin (T14:0 CL), 1-heptadecanoyl-2-hydroxy-sn-glycero-3-phosphocholine (17:0 lysoPC), N-lauroryl sphingomyelin (N12:0 SM), N-heptadecanoyl ceramide (N17:0 Cer), N-lauroyl sulfatide (N12:0 sulfatide) are purchased from Avanti Polar Lipids, Inc., Alabaster, AL. except noted). Other lipid internal standards are used including 7,7,8,8-d4-palmitic acid (d4-16:0 NEFA) (Cambridge Isotope Laboratories, Andover, MA), triheptadecenoin (T17:1 TAG) (Nu Chek, Inc., Elysian, MN), and N-pentadecanoyl galactosylceramide (N15:0 GalCer) (Matreya, Inc., Pleasant Gap, PA).
3. Methods
3.1 Cellular lipid extraction and sample preparation
Overall, tissue samples are homogenized in 10 times diluted phosphate buffered saline (PBS). An appropriate amount of internal standard premixture is spiked into the homogenates or other biofluid samples. After a modified Bligh and Dyer extraction procedure as previously described (38), the generated lipid extract is analyzed by ESI-MS and ESI-MS/MS.
Keep the brain sample (~ 25 mg) in a 1.5-mL Eppendorf tube. Add 300 μL of 10 times diluted PBS in the tube. The samples are separately homogenized for 1 min by using a disposable soft tissue homogenizer with an up-and-down dabbing motion (see Note 2). Pipet an aliquot of 25 μL for determination of protein content.
Protein assay is carried out by using a 96-well microplate and following the manufacturer’s instruction using bovine serum albumin as standard.
Accurately transfer individual homogenate of the tissue samples in step 1 to a disposable culture borosilicate glass tube (16 × 100 mm) and record the transferred volume. Add a certain amount of the premixture of internal standards based on the protein content of the transferred homogenate (see Note 3).
For liquid samples (e.g., plasma, serum, or cerebrospinal fluid) for research of neurodegeneration and brain injury, individual sample is accurately transferred into a disposable culture borosilicate glass tube (16 × 100 mm) and the transferred volume of the sample is recorded. Add a certain amount of the premixture of internal standards based on the volume of the transferred sample (see Notes 3 and 4).
Prepare extraction solvent, chloroform/methanol (1/1; v/v) (solvent A), and 10 and 50 mM lithium chloride solutions.
Add 4 mL solvent A to the glass tube for extraction (Step 3 or 4), and an appropriate volume of 50 mM LiCl to bring the aqueous phase to a final volume of 2 mL. Cap the tubes and vortex them for 20 s. The samples are then centrifuged at 2700 rpm for 10 min.
Collect the bottom layer to a new borosilicate glass tube (see Note 5). Add 2 mL chloroform to individual glass tubes with the residual top layer. Cap the tubes and vortex them for 20 s. The samples are again centrifuged at 2700 rpm for 10 min.
Collect the bottom layer and combine it with that collected in Step 7 (see Note 5). Evaporate the combined bottom layer under a stream of nitrogen with a nitrogen-evaporator until totally dried.
Resuspend individual residue in Step 8 with 4 mL of solvent A, and add 2 mL of 10 mM LiCl. Cap the tubes and vortex them for 20 s. The samples are centrifuged at 2700 rpm for 10 min. Repeat Steps 7 and 8.
Resuspend individual lipid extract residue from Step 9 with solvent A in a volume of 200 μL/mg protein or 1 mL/mL original liquid samples. The lipid extracts are flushed with nitrogen, capped, and stored at −20 °C for MS analysis.
A quarter of each individual lipid extract (Step 10) is transferred to a conic centrifuge glass test tube and the solvent is evaporated under the stream of nitrogen. A small volume of ice-cold LiOMe solution (1 M, 50 μL) in MeOH is added to the test tube at 0 °C. The reaction mixture is vortexed for 15 s, stood in an ice bath for 1 h, and quenched with 2 mL of 0.4% acetic acid solution. The pH of the quenched reaction solution should be adjusted to 4 – 5 by addition of acetic acid if necessary. The aqueous phase is washed with hexane (2 mL x 3 times) and discarded. The lipids in the aqueous phase are extracted by the modified Bligh and Dyer method as described in Step 9. The combined CHCl3 phase was dried under a stream of nitrogen. Each individual extract is reconstituted in 100 μL of solvent A, flushed with nitrogen, capped, and stored at −20 °C for analysis of sphingolipidome (19).
3.2 Mass spectrometric analysis of lipids
Lipid classes present in the prepared lipid samples with or without hydrolysis with lithium methoxide are analyzed in three different modes: negative-ion ESI, negative-ion ESI plus lithium hydroxide, and positive-ion ESI plus lithium hydroxide. Negative-ion ESI-MS analysis of brain PE species in the presence of LiOH is first performed to determine the cell populations in an individual sample as previously described (37, 39). In the case of that the criteria for the purity of cell population are not met, cellular lipid extraction and sample preparation (Section 3.1) have to be repeated.
Dilute each lipid extract solution prepared to < 50 μM of total lipids with chloroform/methanol/isopropanol (1/2/4; v/v/v) with or without LiOH (2% to 5%) in a chemically-resistant 96-well microplate (see Note 6).
Set the ionization voltage of the nanospray ionization source at 1.2 kV in the positive-ion mode, −1.2 kV in the negative-ion mode, and gas pressure at 2.0 psi. Nanospray ionization for each sample is performed by a customized sequence subroutine operated under the Chipsoft software (see Note 7).
For mass spectrometric analysis, collect 2-min duration of signal averaging in the profile mode for each survey MS scan (see Note 8). For tandem mass spectrometric analysis, set collision gas pressure at 1.0 mTorr, vary collision energy with the class of lipids, and collect a 5-min period of signal averaging in the profile mode for each tandem MS spectrum, including PIS and NLS, which are sensitive and specific to the lipid class or the category of lipid classes of interest, as show in Table 1. All of the mass spectra are automatically acquired by a customized sequence subroutine operated under Xcalibur software. An alternative method for analysis of sulfatide species by using matrix-assisted laser desorption/ionization time-of-flight/time-of-flight (MALDI TOF/TOF) mass spectrometer could be employed (see Note 9).
Table 1.
Summary of the specific scans in each lipid class used to identify and quantify individual molecular speciesa
| Lipid class (ref) | Ion format | Scans for class specific prescreen | Scans for identification of acyl chain and/or regioisomers | Preliminary scans for the second step quantitation |
|---|---|---|---|---|
| PC (43) | [M+Li]+ | NLS189.1, −35eV | NLS(59.0+FA), −40eV | NLS183.1, −35eV for polyunsaturated acyl chain containing species NLS59.0, −24eV for plasmalogen species NLS189.1, −35eV for all the other species |
| lysoPC (43) | [M+Na]+ | NLS59.0, −22eV NLS205.0, −34eV |
PIS104.1, −34eV PIS147.1, −34eV |
NLS59.0, −22eV NLS205.0, −34eV |
| PE, lysoPE (18) | [M-H]− [M-H+Fmoc]− ([M+C15H9O2]−) |
PIS196.1, 50eV for [M-H]− NLS222.2, 30eV |
PIS(FA-H), 30eV | NLS222.2, 30eV for [M-H+Fmoc]− |
| PI, lysoPI (23) | [M-H]− | PIS241.1, 45eV | PIS(FA-H), 47eV | PIS241.1, 45eV |
| PS, lysoPS (23) | [M-H]− | NLS87.1, 24eV | PIS(FA-H), 30eV | NLS87.1, 24eV |
| PG, PA, lysoPG, lysoPA (23) | [M-H]− | PIS153.1, 35eV | PIS(FA-H), 30eV | PIS153.1, 35eV |
| CL, mono-lysoCL (33) | [M-2H]2− | Full MS at high resolution | PIS(FA-H) at high resolution, 25eV; NLS(FA-H2O) at high resolution, 22eV | |
| TAG (27) | [M+Li]+ | NLS(FA), −35eV | ||
| Sphingomyelin (43) | [M+Li]+ | NLS213.2, −50eV | NLS(neutral fragments from sphingoid backbone) | NLS213.2, −50eV |
| Ceramide (44) | [M-H]− | NLS(neutral fragments from sphingoid backbone), (e.g., NLS256.2, 32eV for d18:1 non-hydroxyl species) | NLS(neutral fragments from sphingoid backbone), (e.g., NLS256.2, 32eV for d18:1 non-hydroxyl species) | NLS(neutral fragments from sphingoid backbone), (e.g., NLS256.2, 32eV for d18:1 non-hydroxyl species) |
| Hexosyl ceramide (45, 46) | [M+Li]+ | NLS162.2, −50eV | NLS(neutral fragments from sphingoid backbone) | NLS162.2, −50eV |
| Sulfatide (47) | [M-H]− | PIS 97.1, 65eV | NLS(neutral fragments from sphingoid backbone) | PIS97.1, 65eV |
| Sphingoid base-1-phosphate (48) | [M-H]− | PIS79.1, 24eV | PIS79.1, 24eV | |
| Sphingoid base (19) | [M+H]+ | NLS48.0, −18eV | NLS48.0, 18eV | |
| Psychosine (49) | [M+H]+ | NLS180.0, −24eV | NLS180.0, −24eV | |
| Cholesterol (50) | [cholesteryl methoxyacetate +MeOH+Li]+ | PIS97.1, −22eV | PIS97.1, −22eV | |
| Acyl carnitine (51) | [M+H]+ | PIS85.1, −30eV | PIS85.1, −30eV for all species; PIS145.1, −30eV for hydroxyl species | PIS85.1, −30eV |
| Acyl-CoA (16) | [M-H]−, [M-2H]2− [M-3H]3− |
PIS134.0, 30eV | PIS134.0, 30eV | PIS134.0, 30eV |
| 4-hydroxyalkenal (34) | [M+carnosine+H]+ | NLS71.2, −28eV NLS117.2, −26eV |
NLS71.2, −28eV NLS117.2, −26eV |
NLS and PIS stand for neutral-loss scan and precursor-ion scan, respectively. FA and (FA-H) denote free fatty acid and fatty acyl carboxylate anion, respectively. The abbreviations of phospholipid classes are given in the text.
3.3 Mass spectrometric data analysis
Data processing of MS analyses including ion peak selection, data transferring, baseline correction, peak intensity comparison, and quantification is conducted by self-programmed MicroSoft Excel macros (40). The principles of the macros are summarized as follows:
3.3.1 Establishment of the database of lipid classes and individual molecular species
The building block concept regarding lipid molecular structures is extensively employed to build the database of the program (40). On the basis of the differences of these building blocks, the majority of lipid classes present in the mammalian cellular lipidomes are classified into five categories, including glycerophospholipids, glycerolipids, sphingolipids, sterols, and metabolites (see Note 10).
All lipid classes are defined by backbones and building blocks. For example, PC is one class of lipids in the glycerophospholipid category. It has glycerol as its backbone with three building blocks connected the three hydroxy groups. The phosphocholine head group at the sn-3 position specifies the class. The oxygen atom of glycerol at the sn-1 position is connected to an aliphatic chain through an ester, ether, or vinyl ether bond, which defines the phosphatidyl-, plasmanyl-, and plasmenyl- subclasses of PC, according to the IUPAC nomenclature (see Note 11). The oxygen atom at the sn-2 position is connected to the other aliphatic chain through an ester bond.
The building blocks of fatty acyl chain vary with the number of carbon atoms and the number of double bonds, as well as the location of the double bonds in the aliphatic chains. The variations of the carbon atom number and double bond number in the aliphatic chains composite of the entire lipid class, such as PC, in the database of lipid classes for MDMS-SL analysis (see Note 12).
3.3.2 Automated data processing to identify and quantify individual molecular species of a class of interest present in a cellular lipidome
Tabular raw data from the mass spectra are transferred by self-programmed software directly from the Xcalibur platform.
The baseline levels of the tabular raw data from the mass spectra are determined based on the fact that an accelerated intensity change exists from noise to signal (41). The precisely determined baseline level is deduced from the raw data for identification and quantification of analytes.
An ion peak list of the molecular species in a lipid class of interest present in the analyzed lipid extract is generated by matching the m/z values of the detected ion peaks after baseline correction in the specific scan (i.e., PIS or NLS, Table 1) with those of the candidate species in the established database of the lipid class of interest. This peak list represents all the detectable species of the specific class including isomeric species, and contains information about the total number of carbon atoms and the total number of double bonds of the aliphatic chain(s) from the lipid database of the program.
Identification of acyl chain moieties is achieved by loading all PIS or NLS data specific for acyl chain information. The combination of the paired aliphatic chains is determined by the restriction of the total number of carbon atoms and the total number of double bonds present in the acyl chains identified for each individual species.
Before direct quantification of the class of lipid molecular species of interest, the effects of 13C isotope need to be considered (4, 27). There exist two types of these effects. The first type of effect comes from the carbon number difference between a given molecular species and the selected internal standard. The second type of effect is because of the overlapping of the ion peak of the species of interest (m/z = M) with the 13C isotope peak of another species containing an additional double bond (m/z = M-2) (see Note 13).
Quantification of the identified individual molecular species is performed in a two-step procedure (40). An algorithm first determines whether there exist overlapping or low-abundance peaks in the peak list of interest. The step one quantification is performed for the abundant and non-overlapping peaks by direct ratiometric comparison to the ion peak intensity of the selected internal standard of the class in the survey MS scan after baseline correction and removal of 13C isotope effects.
The determined non-overlapping and abundant species plus the exogenously added internal standard are the candidate standards for the second step of quantification. The corrected ion peak intensities of the overlapping and/or low-abundance species from the class-specific PIS or NLS (Table 1) are used to quantify these species with ratiometric comparison to the ion-peak intensities of the candidate standards (see Note 14).
Acknowledgments
This work was partially supported by National Institute of General Medical Sciences Grant R01 GM105724 and intramural institutional research funds. Special thanks are expressed to Ms. Imee Tiu for her editorial assistance.
Footnotes
The nanospray source is controlled by Chipsoft 8.3.1 software. All MS or tandem MS analyses are operated under the Xcalibur software. Additional equipments and supplies needed include analytical balance (readability 0.01 mg), multi-sample bio-pulverizer (12 wells, capacity 10–100mg per well), cryogenic vials (2.0 mL), Branson digital sonifier 450, vortex shaker and mixer, razor blade or scissors, tissue tearor, 1.5-mL Eppendorf tubes, 1.5 mL polypropylene pestles (disposable soft tissue homogenizer) with handheld pellet pestle motor, disposable culture borosilicate glass tubes (16 × 100 mm), 5.75″ disposable borosilicate glass Pasteur pipets, Drummond pipet-aids, table top centrifuge, analytical nitrogen evaporator, 96-well microplates (transparent for protein assay and chemical resistance for preparing lipid samples for direct infusion).
During sonification or homogenization, samples are kept in an ice bath to keep cold.
The internal standards mixture includes di14:1 PC, di16:1 PE, di15:0 PG, di14:0 PS, di14:0 PA, T14:0 CL, d4-16:0 NEFA, 17:0 lysoPC, T17:1 TAG, N12:0 SM, N17:0 Cer, N12:0 sulfatide, N15:0 GalCer, etc. The stock solution of individual internal standard is prepared in either solvent A or pure chloroform with a concentration approximately 1 mg/mL. The amount of each individual lipid species in the premixture is prepared based on the abundance of the corresponding lipid class in the samples. The molecular species of internal standards are selected because they represent < 0.1% of the endogenous cellular lipid mass levels as predetermined by ESI-MS lipid analysis.
Alternatively, liquid samples can also be normalized to their protein contents. In this case, an aliquot of the liquid sample is used to determine the protein content prior to addition of the internal standard premixture.
In order to eliminate the contamination from the top layer (aqueous phase) to the bottom phase, insert the glass Pasteur pipet into the upper layer by slowly air bubbling until the pipet inserts into the bottom layer, which could avoid the upper layer liquid going into the pipet. After taking the glass pipet out from the upper layer, remove the aqueous contaminant outside of the pipet tip by gently swirling the tip on the edge of the glass tube and quickly transfer the bottom layer to a clean glass tube.
The total lipid concentration of a lipid extract can be estimated on the basis of the protein content or previous experience (38). This knowledge is useful for estimation of the concentrations of total lipids to prevent lipid aggregation during analysis. The lithium hydroxide is made of 200-time dilution of a saturated methanol solution.
Since sample ionization and spectra collection are operated with two separate software programs (i.e., ChipSoft and Xcalibur, respectively), the ionization polarity and time controlled by the ChipSoft should be matched to those of the mass spectrometer. The mass spectrometer will be triggered to start collecting spectra with the start of the nanospray.
For the triple-quadrupole mass spectrometer, the first and third quadrupoles are used as independent mass analyzer with a mass resolution of 0.7 Th, and the second quadrupole serves as a collision cell for tandem mass spectrometry. For the analysis of cardiolipin, the mass resolution is set at 0.3 Th to detect its doubly-charged ions (33).
Alternatively, MALDI TOF-MS by using 9-aminoacridine as matrix could be used to selectively desorption/ionization of sulfatide species over other examined anionic lipids present in lipid extracts of biological samples (36). The structure of individual sulfatide species can be elucidated through product ion analysis by employing MALDI TOF/TOF-MS in this case.
MDMS-SL allows us to analyze lipid molecular species of a class of interest in a non-targeted approach. Therefore, the database should be as broad and flexible as possible. The former covers all possible natural lipid molecular species, whereas the latter allows to modify or expand the database as necessary.
To date, the plasmanyl and plasmenyl subclasses have been identified only in choline, ethanolamine, and serine glycerophospholipids in mammalian lipidomes (42).
The molecular species that are included in our database are approximately 6,500 glycerophospholipid species, 3,200 glycerolipid species, 26,000 sphingolipid species, 100 sterol lipids, and 410 metabolites (40). Therefore, a total of over 36,000 molecular species, not counting regioisomers, oxidized lipids, or other covalently modified entities, are included in the initial construction of the database. Moreover, by modifying the general chemical formulas, the constructed databases can easily be extended to cover any new species or subclasses in each lipid class when the sensitivity of mass spectrometers is further improved or any unusual lipid profiles are analyzed from a biological sample.
The isotope effects from other atoms, such as hydrogen, nitrogen, or phosphorus, are usually neglected due to extremely low abundance of its isotope or no difference between the species and the selected internal standard.
An algorithm could be generated based on two variables (i.e., the differences in the number of total carbon atoms and the number of total double bonds present in fatty acyl chains of each individual species from those of the selected standards) with multivariate least-square regression to determine the correction factors for each individual molecular species for the second-step quantification (40). With this second step, the linear dynamic range of quantification is extended dramatically to quantify overlapping and/or low-abundance species with one or more MS/MS scans to reduce background noise, increase S/N ratios of low-abundance species, and filter the overlapping molecules with class-specific PIS or NLS.
References
- 1.Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res. 2003;44:1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Lagarde M, Geloen A, Record M, Vance D, Spener F. Lipidomics is emerging. Biochim Biophys Acta. 2003;1634:61. doi: 10.1016/j.bbalip.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 4.Han X, Gross RW. Shotgun lipidomics: Electrospray ionization mass spectrometric analysis and quantitation of the cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 5.Walker JM, Krey JF, Chen JS, Vefring E, Jahnsen JA, Bradshaw H, Huang SM. Targeted lipidomics: fatty acid amides and pain modulation. Prostaglandins Other Lipid Mediat. 2005;77:35–45. doi: 10.1016/j.prostaglandins.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Serhan CN. Mediator lipidomics. Prostaglandins Other Lipid Mediat. 2005;77:4–14. doi: 10.1016/j.prostaglandins.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Han X. Neurolipidomics: challenges and developments. Front Biosci. 2007;12:2601–2615. doi: 10.2741/2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanova PT, Milne SB, Forrester JS, Brown HA. LIPID arrays: New tools in the understanding of membrane dynamics and lipid signaling. Mol Interv. 2004;4:86–96. doi: 10.1124/mi.4.2.6. [DOI] [PubMed] [Google Scholar]
- 9.Welti R, Shah J, Li W, Li M, Chen J, Burke JJ, Fauconnier ML, Chapman K, Chye ML, Wang X. Plant lipidomics: discerning biological function by profiling plant complex lipids using mass spectrometry. Front Biosci. 2007;12:2494–2506. doi: 10.2741/2250. [DOI] [PubMed] [Google Scholar]
- 10.Schiller J, Suss R, Fuchs B, Muller M, Zschornig O, Arnold K. MALDI-TOF MS in lipidomics. Front Biosci. 2007;12:2568–2579. doi: 10.2741/2255. [DOI] [PubMed] [Google Scholar]
- 11.Albert CJ, Anbukumar DS, Monda JK, Eckelkamp JT, Ford DA. Myocardial lipidomics. Developments in myocardial nuclear lipidomics. Front Biosci. 2007;12:2750–2760. doi: 10.2741/2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, Thiele C, Ekroos K, Shevchenko A. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal Chem. 2006;78:6202–6214. doi: 10.1021/ac060545x. [DOI] [PubMed] [Google Scholar]
- 13.Han X, Gross RW. Shotgun lipidomics: multi-dimensional mass spectrometric analysis of cellular lipidomes. Expert Rev Proteomics. 2005;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- 14.Han X, Yang K, Gross RW. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom Rev. 2012;31:134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsui ZC, Chen QR, Thomas MJ, Samuel M, Cui Z. A method for profiling gangliosides in animal tissues using electrospray ionization-tandem mass spectrometry. Anal Biochem. 2005;341:251–258. doi: 10.1016/j.ab.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 16.Kalderon B, Sheena V, Shachrur S, Hertz R, Bar-Tana J. Modulation by nutrients and drugs of liver acyl-CoAs analyzed by mass spectrometry. J Lipid Res. 2002;43:1125–1132. doi: 10.1194/jlr.m200060-jlr200. [DOI] [PubMed] [Google Scholar]
- 17.Golovko MY, Murphy EJ. An improved method for tissue long-chain acyl-CoA extraction and analysis. J Lipid Res. 2004;45:1777–1782. doi: 10.1194/jlr.D400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Han X, Yang K, Cheng H, Fikes KN, Gross RW. Shotgun lipidomics of phosphoethanolamine-containing lipids in biological samples after one-step in situ derivatization. J Lipid Res. 2005;46:1548–1560. doi: 10.1194/jlr.D500007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X, Cheng H, Yang K, Gross RW, Han X. Alkaline methanolysis of lipid extracts extends shotgun lipidomics analyses to the low abundance regime of cellular sphingolipids. Anal Biochem. 2007;371:135–145. doi: 10.1016/j.ab.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang K, Zhao Z, Gross RW, Han X. Shotgun lipidomics identifies a paired rule for the presence of isomeric ether phospholipid molecular species. PLoS ONE. 2007;2:e1368. doi: 10.1371/journal.pone.0001368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikonomou MG, Blades AT, Kebarle P. Electrospray-ion spray: a comparison of mechanisms and performance. Analytical Chemistry. 1991;63:1989–1998. [Google Scholar]
- 22.Gaskell SJ. Electrospray: principles and practice. J Mass Spectrom. 1997;32:677–688. [Google Scholar]
- 23.Han X, Yang J, Cheng H, Ye H, Gross RW. Towards fingerprinting cellular lipidomes directly from biological samples by two-dimensional electrospray ionization mass spectrometry. Anal Biochem. 2004;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Han X, Yang K, Yang J, Fikes KN, Cheng H, Gross RW. Factors influencing the electrospray intrasource separation and selective ionization of glycerophospholipids. J Am Soc Mass Spectrom. 2006;17:264–274. doi: 10.1016/j.jasms.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Han X, Cheng H, Mancuso DJ, Gross RW. Caloric restriction results in phospholipid depletion, membrane remodeling and triacylglycerol accumulation in murine myocardium. Biochemistry. 2004;43:15584–15594. doi: 10.1021/bi048307o. [DOI] [PubMed] [Google Scholar]
- 26.Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzchalia T, Shevchenko A. Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Analytical Chemistry. 2006;78:585–595. doi: 10.1021/ac051605m. [DOI] [PubMed] [Google Scholar]
- 27.Han X, Gross RW. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal Biochem. 2001;295:88–100. doi: 10.1006/abio.2001.5178. [DOI] [PubMed] [Google Scholar]
- 28.Han X, Gross RW. Structural determination of picomole amounts of phospholipids via electrospray ionization tandem mass spectrometry. J Am Soc Mass Spectrom. 1995;6:1202–1210. doi: 10.1016/1044-0305(95)00568-4. [DOI] [PubMed] [Google Scholar]
- 29.Brugger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci USA. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehmann WD, Koester M, Erben G, Keppler D. Characterization and quantification of rat bile phosphatidylcholine by electrospray-tandem mass spectrometry. Anal Biochem. 1997;246:102–110. doi: 10.1006/abio.1996.9941. [DOI] [PubMed] [Google Scholar]
- 31.Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X. Profiling membrane lipids in plant stress responses. Role of phospholipase Da in freezing-induced lipid changes in Arabidopsis. J Biol Chem. 2002;277:31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- 32.Welti R, Wang X. Lipid species profiling: a high-throughput approach to identify lipid compositional changes and determine the function of genes involved in lipid metabolism and signaling. Curr Opin Plant Biol. 2004;7:337–344. doi: 10.1016/j.pbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Han X, Yang K, Yang J, Cheng H, Gross RW. Shotgun lipidomics of cardiolipin molecular species in lipid extracts of biological samples. J Lipid Res. 2006;47:864–879. doi: 10.1194/jlr.D500044-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang M, Fang H, Han X. Shotgun lipidomics analysis of 4-hydroxyalkenal species directly from lipid extracts after one-step in situ derivatization. Anal Chem. 2012;84:4580–4586. doi: 10.1021/ac300695p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang X, Han X. Characterization and direct quantitation of sphingoid base-1-phosphates from lipid extracts: A shotgun lipidomics approach. J Lipid Res. 2006;47:1865–1873. doi: 10.1194/jlr.D600012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng H, Sun G, Yang K, Gross RW, Han X. Selective desorption/ionization of sulfatides by MALDI-MS facilitated using 9-aminoacridine as matrix. J Lipid Res. 2010;51:1599–1609. doi: 10.1194/jlr.D004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han X, Holtzman DM, McKeel DW., Jr Plasmalogen deficiency in early Alzheimer’s disease subjects and in animal models: molecular characterization using electrospray ionization mass spectrometry. J Neurochem. 2001;77:1168–1180. doi: 10.1046/j.1471-4159.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 38.Christie WW, Han X. Lipid Analysis: Isolation, Separation, Identification and Lipidomic Analysis. 4. The Oily Press; Bridgwater, England: 2010. [Google Scholar]
- 39.Cheng H, Wang M, Li JL, Cairns NJ, Han X. Specific changes of sulfatide levels in individuals with pre-clinical Alzheimer’s disease: an early event in disease pathogenesis. J Neurochem. 2013;126 doi: 10.1111/jnc.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang K, Cheng H, Gross RW, Han X. Automated lipid identification and quantification by multi-dimensional mass spectrometry-based shotgun lipidomics. Anal Chem. 2009;81:4356–4368. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang K, Fang X, Gross RW, Han X. A practical approach for determination of mass spectral baselines. J Am Soc Mass Spectrom. 2011;22:2090–2099. doi: 10.1007/s13361-011-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vance DE, Vance JE. Biochemistry of Lipids, Lipoproteins and Membranes. 5. Elsevier Science B.V; Amsterdam: 2008. [Google Scholar]
- 43.Yang K, Zhao Z, Gross RW, Han X. Systematic analysis of choline-containing phospholipids using multi-dimensional mass spectrometry-based shotgun lipidomics. J Chromatogr B. 2009;877:2924–2936. doi: 10.1016/j.jchromb.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han X. Characterization and direct quantitation of ceramide molecular species from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal Biochem. 2002;302:199–212. doi: 10.1006/abio.2001.5536. [DOI] [PubMed] [Google Scholar]
- 45.Han X, Cheng H. Characterization and direct quantitation of cerebroside molecular species from lipid extracts by shotgun lipidomics. J Lipid Res. 2005;46:163–175. doi: 10.1194/jlr.D400022-JLR200. [DOI] [PubMed] [Google Scholar]
- 46.Hsu FF, Turk J. Structural determination of glycosphingolipids as lithiated adducts by electrospray ionization mass spectrometry using low-energy collisional-activated dissociation on a triple stage quadrupole instrument. J Am Soc Mass Spectrom. 2001;12:61–79. doi: 10.1016/S1044-0305(00)00194-X. [DOI] [PubMed] [Google Scholar]
- 47.Hsu FF, Bohrer A, Turk J. Electrospray ionization tandem mass spectrometric analysis of sulfatide. Determination of fragmentation patterns and characterization of molecular species expressed in brain and in pancreatic islets. Biochim Biophys Acta. 1998;1392:202–216. doi: 10.1016/s0005-2760(98)00034-4. [DOI] [PubMed] [Google Scholar]
- 48.Jiang X, Han X. Characterization and direct quantitation of sphingoid base-1-phosphates from lipid extracts: A shotgun lipidomics approach. J Lipid Res. 2006;47:1865–1873. doi: 10.1194/jlr.D600012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang X, Yang K, Han X. Direct quantitation of psychosine from alkaline-treated lipid extracts with a semi-synthetic internal standard. J Lipid Res. 2009;50:162–172. doi: 10.1194/jlr.D800036-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng H, Jiang X, Han X. Alterations in lipid homeostasis of mouse dorsal root ganglia induced by apolipoprotein E deficiency: A shotgun lipidomics study. J Neurochem. 2007;101:57–76. doi: 10.1111/j.1471-4159.2006.04342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su X, Han X, Mancuso DJ, Abendschein DR, Gross RW. Accumulation of long-chain acylcarnitine and 3-hydroxy acylcarnitine molecular species in diabetic myocardium: identification of alterations in mitochondrial fatty acid processing in diabetic myocardium by shotgun lipidomics. Biochemistry. 2005;44:5234–5245. doi: 10.1021/bi047773a. [DOI] [PubMed] [Google Scholar]
- 52.Han X. Multi-dimensional mass spectrometry-based shotgun lipidomics and the altered lipids at the mild cognitive impairment stage of Alzheimer’s disease. Biochim Biophys Acta. 2010;1801:774–783. doi: 10.1016/j.bbalip.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]