Abstract
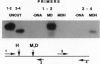
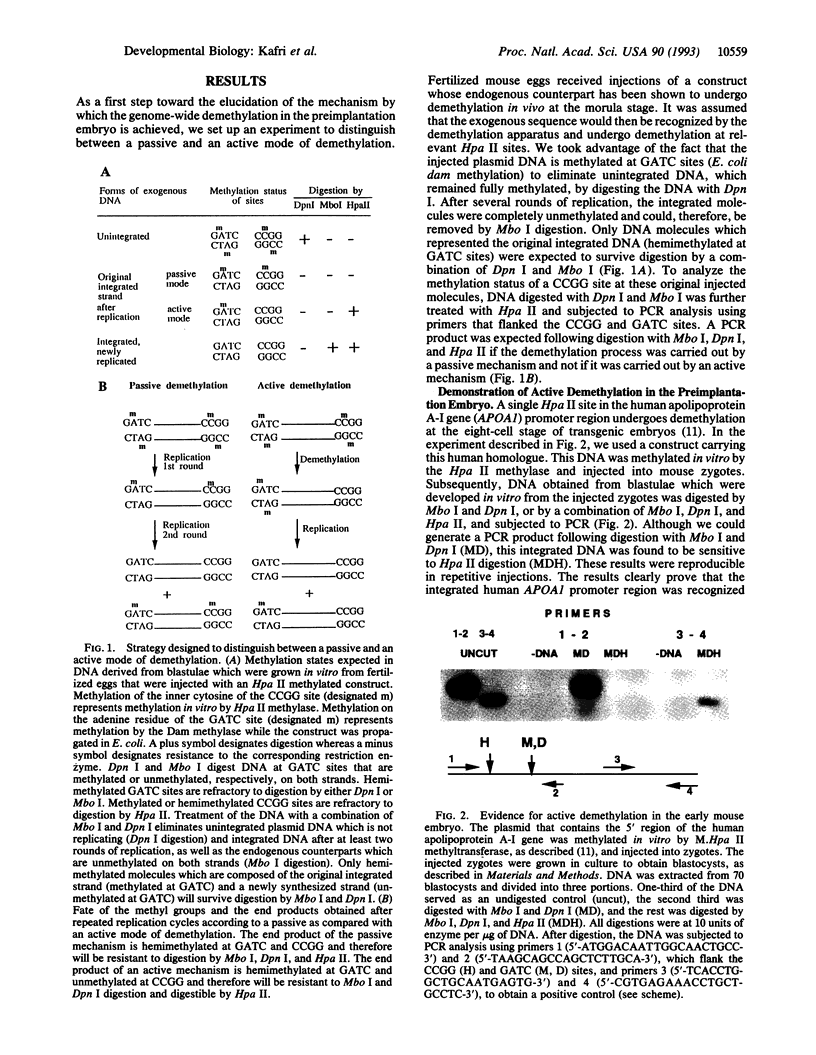
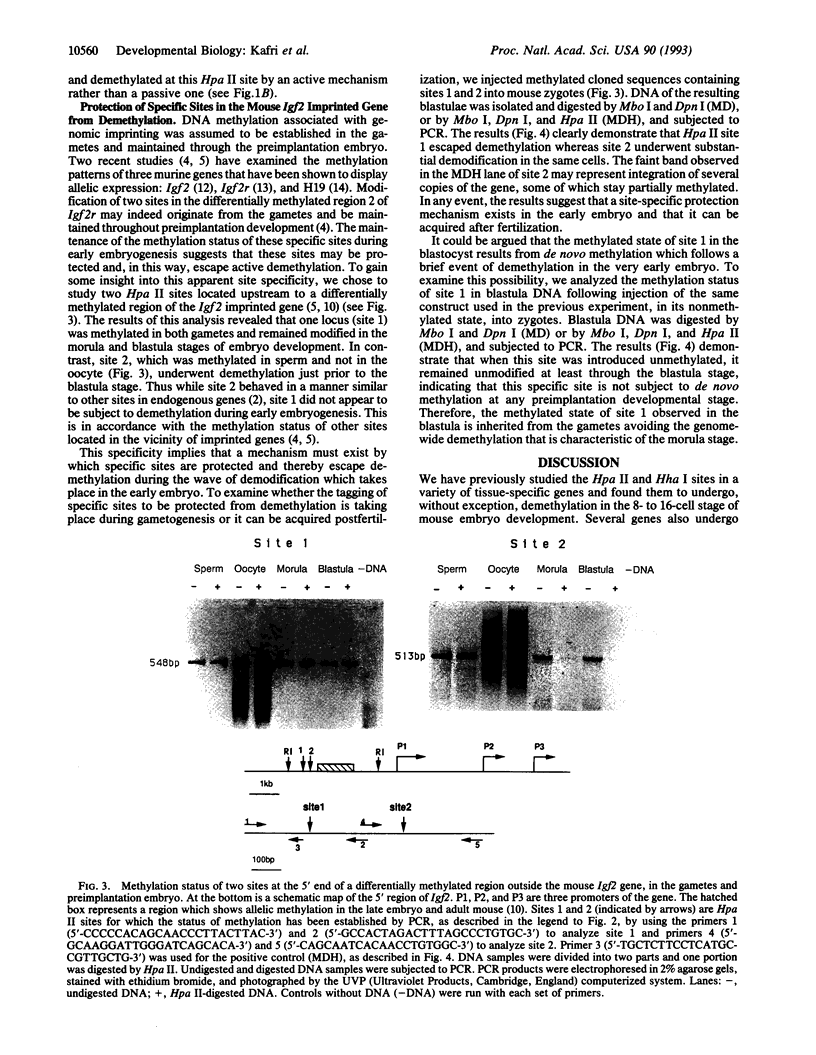
Gene-specific methylation patterns in mammals play a role in a variety of biological processes in the embryo and adult tissues. These patterns are established during embryo development by a process that involves genome-wide demethylation in the morula and de novo methylation in the pregastrula. To elucidate the mechanism of demethylation in the early mouse embryo, we have injected mouse zygotes with gene sequences that were methylated in vitro by Hpa II methylase and analyzed the methylation status of specific sites in blastocyst DNA. Because it had been propagated in Escherichia coli, the DNA used for these injections was also methylated at adenine residues in GATC sites. This allowed us to eliminate fully methylated, unintegrated DNA by Dpn I digestion and fully unmethylated, integrated DNA that underwent several rounds of replication by Mbo I digestion. The integrated, originally injected DNA strands were in a hemimethylated state and survived this treatment. The methylation status of Hpa II sites in these molecules was analyzed by Hpa II digestion of the genomic DNA isolated from blastocysts, followed by PCR amplification using appropriate primers. The results demonstrate that demethylation is achieved by an active mechanism and that specific sites in imprinted genes escape demethylation, maintaining a methylated state throughout preimplantation development.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barlow D. P., Stöger R., Herrmann B. G., Saito K., Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991 Jan 3;349(6304):84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- Bartolomei M. S., Zemel S., Tilghman S. M. Parental imprinting of the mouse H19 gene. Nature. 1991 May 9;351(6322):153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- Brandeis M., Kafri T., Ariel M., Chaillet J. R., McCarrey J., Razin A., Cedar H. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 1993 Sep;12(9):3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson L. L., Page A. W., Bestor T. H. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 1992 Dec;6(12B):2536–2541. doi: 10.1101/gad.6.12b.2536. [DOI] [PubMed] [Google Scholar]
- DeChiara T. M., Robertson E. J., Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991 Feb 22;64(4):849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- Howlett S. K., Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991 Sep;113(1):119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- Jost J. P. Nuclear extracts of chicken embryos promote an active demethylation of DNA by excision repair of 5-methyldeoxycytidine. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4684–4688. doi: 10.1073/pnas.90.10.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri T., Ariel M., Brandeis M., Shemer R., Urven L., McCarrey J., Cedar H., Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992 May;6(5):705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- Monk M., Adams R. L., Rinaldi A. Decrease in DNA methylase activity during preimplantation development in the mouse. Development. 1991 May;112(1):189–192. doi: 10.1242/dev.112.1.189. [DOI] [PubMed] [Google Scholar]
- Monk M., Boubelik M., Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987 Mar;99(3):371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Jones P. A., Chaillet J. R., Ferguson-Smith A. C., Barton S. C., Reik W., Surani M. A. Parental imprinting: potentially active chromatin of the repressed maternal allele of the mouse insulin-like growth factor II (Igf2) gene. Genes Dev. 1992 Oct;6(10):1843–1856. doi: 10.1101/gad.6.10.1843. [DOI] [PubMed] [Google Scholar]
- Shemer R., Kafri T., O'Connell A., Eisenberg S., Breslow J. L., Razin A. Methylation changes in the apolipoprotein AI gene during embryonic development of the mouse. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11300–11304. doi: 10.1073/pnas.88.24.11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer R., Walsh A., Eisenberg S., Breslow J. L., Razin A. Tissue-specific methylation patterns and expression of the human apolipoprotein AI gene. J Biol Chem. 1990 Jan 15;265(2):1010–1015. [PubMed] [Google Scholar]
- Stöger R., Kubicka P., Liu C. G., Kafri T., Razin A., Cedar H., Barlow D. P. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993 Apr 9;73(1):61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]