Abstract
As in many polarized cells, spindle alignment in yeast is essential and cell cycle regulated. A key step that governs spindle alignment is the selective binding of the Kar9 protein to only one of the two spindle pole bodies (SPBs). It has been suggested that cyclin-dependent kinase Cdc28, in complex with cyclin Clb4, associates only with the SPB in the mother cell and so prevents Kar9 binding to this SPB. However, here we show that the nonoverexpressed Clb4 associates with the budward-directed SPB through Kar9. Cdc28-Clb4 then uses Kar9 as a carrier to move from this SPB to the plus ends of astral microtubules, where Cdc28-Clb4 regulates the interactions between microtubule ends and subdomains of the bud cortex. In the absence of Cdc28-Clb4 activity (G1/S phase), astral microtubules interact with the bud tip in a manner dependent on actin, Myo2, and Kar9. Coincidentally with reaching the bud cortex in S phase, Cdc28-Clb4 facilitates the dissociation of the microtubule bud tip interaction and their capture by the bud neck. This transition prevents the preanaphase spindle from becoming prematurely pulled into the bud. Thus, Cdc28-Clb4 facilitates spindle alignment by regulating the interaction of astral microtubules with subdomains of the bud cortex.
Keywords: Adenomatous polyposis coli, APC, cell polarity, Kar9, spindle orientation
The budding yeast Saccharomyces cerevisiae is a highly polarized model organism that divides perpendicular to the mother-bud axis at the predetermined site of bud emergence. This mode of division makes the alignment of the mitotic spindle along the mother-bud axis essential for survival. A precise temporal program of microtubule (MT)-cortex interactions ensures the correct positioning of the spindle. In G1/S phase of the cell cycle, astral MTs, which are organized by the yeast spindle pole body (SPB), are directed into the growing bud (the daughter cell; Byers and Goetsch 1975). Astral MT interactions with the bud tip position the nucleus in the mother cell body close to the bud neck (Segal et al. 2002).
The SPB then duplicates in early S by a conservative mechanism creating a newly formed and a pre-existing (“old”) SPB (Byers and Goetsch 1975; Pereira et al. 2001). At this stage of the cell cycle, the Bim1-Kar9 complex has a key role in the alignment of the assembling spindle along the mother-bud axis. Bim1 is an EB1-like MT-binding protein (Schwartz et al. 1997), and Kar9 shows weak similarities to the adenomatous polyposis coli (APC) tumor suppressor (Miller and Rose 1998; Bienz 2001). The Bim1-Kar9 complex binds to only the old, but not the new, SPB and becomes transported from the old SPB to MT plus ends, where Kar9 ensures the association of MTs with the bud cortex (Liakopoulos et al. 2003; Maekawa et al. 2003). Because only the astral MTs organized by the old SPB have Bim1-Kar9 attached to it, and Kar9 interacts with the budward-directed actin motor Myo2, the astral MTs of the old SPB will be pulled into the bud, whereas the MTs of the new SPB fail to show persistent bud cortex interactions (Beach et al. 2000; Korinek et al. 2000; Lee et al. 2000; Miller et al. 2000; Yin et al. 2000; Liakopoulos et al. 2003). This positions the old SPB close to the mother-bud junction (bud neck), whereas the new SPB becomes centered in the mother cell body (Pereira et al. 2001).
Coincident with the assembly of the bipolar spindle in S phase, astral MTs establish new interactions with the bud neck. These interactions require a functional septin ring and depend upon Kar9 function (Segal et al. 2000; Kusch et al. 2002). During this phase of the cell cycle, astral MTs also depolymerize, which in turn further assists in the orientation of the preanaphase spindle along the mother-bud axis and positions the nucleus adjacent to the bud neck (Kusch et al. 2002).
In budding yeast the actin-dependent spindle orientation occurs in a restricted period that extends from G1 phase to shortly before anaphase (Theesfeld et al. 1999). With anaphase onset, when the spindle extends into the bud, a second dynein-dependent pathway provides the force for nuclear movement (Yeh et al. 1995; Carminati and Stearns 1997). Genetic data indicate that the Kar9 and dynein pathways partially compensate for each other in spindle orientation. Failure of one pathway leads to defects in spindle alignment and chromosome instability, but mutants are viable. However, simultaneous loss of both pathways is lethal (Miller and Rose 1998; Miller et al. 1998).
Although these data indicate that the astral MT-cortex interactions are highly coordinated throughout the cell cycle, little is known about their regulation. However, it is clear that yeast cyclin-dependent kinase, Cdc28, is associated with SPBs and MTs. In early S phase, a fraction of Cdc28 is transported, together with Kar9, from the old SPB to MT ends directed into the bud. In contrast, Cdc28 is not associated with MTs organized by the new SPB (Maekawa et al. 2003). The role of Cdc28 in the regulation of the localization of the Bim1-Kar9 complex with the SPB and MTs is still a matter of debate. Although one study suggests that Cdc28 regulates the transport of Bim1 and Kar9 (Maekawa et al. 2003), another report, in which overexpressed Clb4-GFP was used to define the localization of Clb4 to the new SPB, concludes that Cdc28-Clb4 prevents Bim1-Kar9 from binding to the new SPB (Liakopoulos et al. 2003). Here, we have found that nonoverexpressed Clb4 selectively associates with the old SPB in a Kar9-dependent manner. The Cdc28-Clb4 complex becomes transported from the SPB to astral MT plus ends, where, in S phase, Clb4 facilitates the dissociation of MT ends from the bud tip region and their capture by the bud neck. Thus, Clb4 controls the interaction of astral MT plus ends with subdomains of the bud cortex.
Results
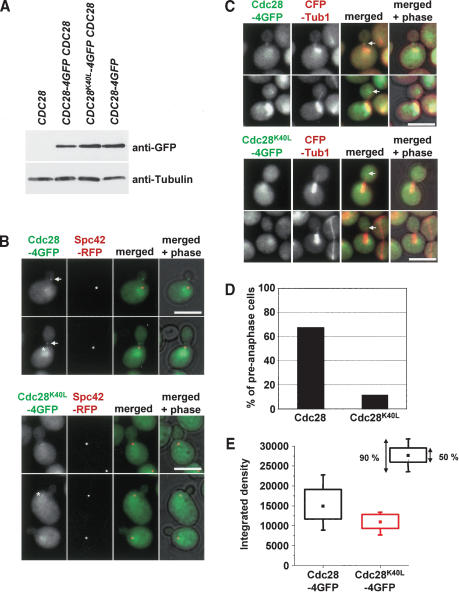
Kinase activity is required for efficient association of Cdc28 with the SPB
Previously, we showed that the yeast cyclin-dependent kinase, Cdc28, binds to the SPB in early S phase and is subsequently transported, along with Kar9, to astral MT plus ends (Maekawa et al. 2003). It is unclear whether Cdc28 has to be in an active state for this transportation event. We addressed the requirement for Cdc28 activity for SPB loading and MT transport with the Cdc28K40L mutant, which lacks phosphorylation activity due to the failure to bind ATP (Agarwal and Cohen-Fix 2002). Cdc28-4GFP and Cdc28K40L-4GFP expressed from the CDC28 promoter were each integrated into the genome of separate strains, which also contained wild-type CDC28. CDC28-4GFP and CDC28K40L-4GFP proteins were expressed to a similar degree (Fig. 1A). As reported (Maekawa et al. 2003), Cdc28-4GFP localized to the nucleus, the SPB (Fig. 1B, asterisks), and as a dot close to the bud neck (Fig. 1B, arrows). The neck signal resulted from astral MT plus ends, which contact the bud neck in preanaphase (Fig. 1C). Inactivation of Cdc28 activity only mildly affected SPB association of Cdc28K40L-4GFP (Fig. 1B, asterisk). However, in CDC28K40L-4GFP cells the signal at the MT plus end was diminished to a striking degree. Although 67.4% of CDC28-4GFP cells in preanaphase showed a bud neck signal, this was only the case for 11% of CDC28K40L-4GFP cells (Fig. 1B-D). In addition, in these 11% of cells the Cdc28K40L-4GFP signal was only ∼0.5 of the intensity of that seen in cells expressing Cdc28-4GFP (Fig. 1E). As a control, we established that the untagged Cdc28K40L did not have a dominant-negative effect on astral MT plus end localization of Cdc28-4GFP. About 73%-74% of preanaphase cells showed Cdc28-4GFP at astral MT plus ends independently of whether CDC28 or CDC28K40Lwere coexpressed. Thus, it is likely that Cdc28 has to be in an active state for the efficient transport to astral MT plus ends.
Figure 1.
Cdc28 has to be in an active state for efficient localization to astral microtubule plus ends. (A) CDC28-4GFP and CDC28K40L-4GFP accumulated to a similar degree. Immunoblots of yeast cell extracts with anti-GFP and anti-Tub2 antibodies. (B-E) Cdc28K40L bound only slightly reduced to SPBs, but its association with astral MT plus ends was strongly reduced. (B) Colocalization of Cdc28-4GFP and Cdc28K40L-4GFP with the SPB marker Spc42-RFP (asterisks). In contrast to Cdc28-GFP (arrows), Cdc28K40L-4GFP fails to bind to the bud neck in the majority of cells. (C) Cdc28-4GFP colocalizes as a dot with astral MT plus ends (CFP-Tub1). Such a localization was observed only rarely with Cdc28K40L-4GFP. The arrow marks the plus end of astral MTs. (D) Quantification of C. The percentage of preanaphase cells with a clear dot-like Cdc28-4GFP or Cdc28K40L-4GFP signal at astral MT plus ends directed into the bud is shown. (E) The signal intensity of the Cdc28-4GFP or Cdc28K40L-4GFP dots of C was quantified. Bars, 5 μm.
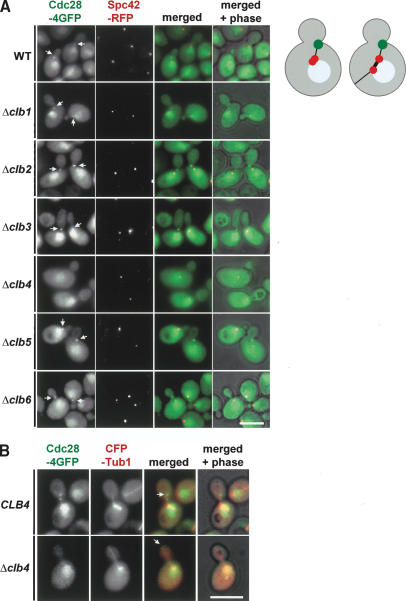
Cdc28 localization with astral MTs requires Clb4
Cyclin-dependent kinases have to associate with a cyclin subunit to be active (Dorée and Hunt 2002). The observation that Cdc28 kinase has likely to be in an active state for efficient MT localization (Fig. 1) suggests that Cdc28 is associated with a cyclin when it is transported to astral MT plus ends. Deletion of this cyclin may affect MT binding of Cdc28. Because Cdc28 does not associate with SPBs and MTs in G1 phase of the cell cycle (Maekawa et al. 2003), it is unlikely that G1-type cyclins are important for the SPB and MT binding of Cdc28. Budding yeast has six B-type cyclins, namely, Clb1-6, with diverse functions in S phase and mitosis. None of the B-type cyclins are essential for viability of yeast cells (Andrews and Measda 1998). We could therefore test cyclin deletion mutants for MT association of Cdc28. Deletion of CLB1, CLB2, CLB3, CLB5, and CLB6 did not affect colocalization of Cdc28 with astral MT ends that contacted the bud neck (Fig. 2A, arrows). However, the bud neck signal of Cdc28-4GFP was completely eliminated in Δclb4 cells (Fig. 2A). Further analysis confirmed that Cdc28-4GFP did not associate with astral MT plus ends of Δclb4 cells (n > 100; Fig. 2B). When taken together, this indicates that Cdc28 association with astral MT plus ends is dependent upon Clb4.
Figure 2.
Localization of Cdc28 with astral MT plus ends is dependent on Clb4. (A) The indicated cell types with CDC28-4GFP SPC42-RFP were analyzed for Cdc28-4GFP bud neck localization (arrows). Cdc28 was strongly enriched inside the nucleus (Maekawa et al. 2003). The cartoon cells on the right show the SPB (red), Cdc28 at astral MT plus ends (green), astral MTs (thin black line), nuclear MTs (thick black line), and the nucleus (blue). (B) In 100% of Δclb4 cells (n > 100) Cdc28-4GFP failed to associate with astral MT plus ends (arrows). Bars, 5 μm.
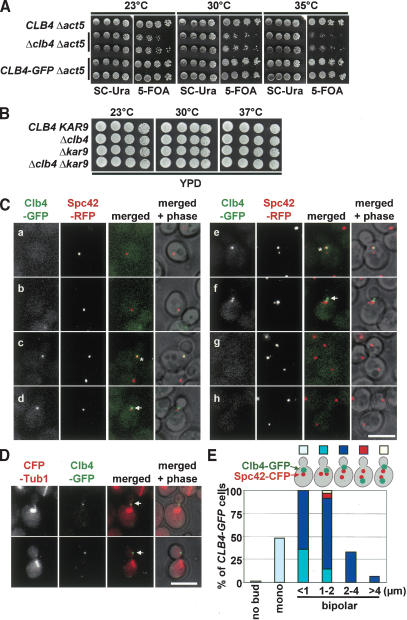
Clb4 moves from the budward-directed SPB to astral MT plus ends
The Clb4 dependency suggests that it could be the B-type cyclin that associates with Cdc28 at astral MT ends. However, a previous study concluded that Clb4 is selectively associated with the motherward-directed new SPB, but not with the old SPB that migrates into the bud in anaphase (Liakopoulos et al. 2003). Because Liakopoulos et al. (2003) used overexpressed Gal1-CLB4-GFP, we re-evaluated the Clb4 localization by using CLB4-GFP, which was the only source of Clb4 activity and was expressed at the natural locus from the native CLB4 promoter. Based on two criteria, the CLB4-GFP construct was functional. First, ACT5 encodes an actin-related protein that is part of the dynactin complex (Li et al. 1994). Δclb4 was synthetically toxic when combined with Δact5 (Fig. 3A, 35°C). CLB4-GFP Δact5 cells grew as Δact5 cells, indicating that CLB4-GFP was functional. The synthetically toxic phenotype of Δact5 Δclb4 also suggests that Clb4 functions predominately in the early Kar9 but not the late dynein/dynactin spindle alignment pathway. Consistently, Δclb4 Δkar9 cells did not show an enhanced growth defect when compared with Δclb4 or Δkar9 cells (Fig. 3B). Second, astral MTs of preanaphase CLB4 and CLB4-GFP cells interact with the cell cortex in very similar manner, whereas in Δclb4 cells they were mainly directed toward the bud tip (Supplementary Fig. 1A). This finding further supports the notion that CLB4-GFP is fully functional.
Figure 3.
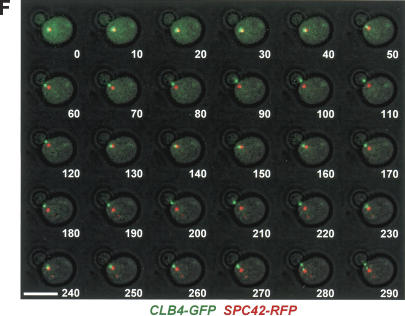
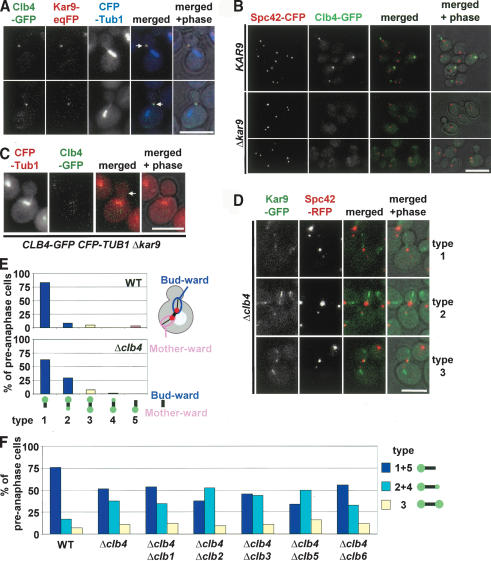
Clb4 is transported from the SPB to astral MT plus ends. (A) CLB4-GFP is functional. A growth spot test of the indicated cell types with the URA3-based pRS316-ACT5 plasmid on selective plates (SC-Ura) and 5-FOA plates. 5-FOA selects for the cells that spontaneously lost the pRS316-ACT5 plasmid. The fact that Δclb4 Δact5 cells grew worse than CLB4 Δact5 and CLB4-GFP Δact5 cells indicates that the CLB4-GFP construct is functional. (B) Growth test of the indicated cell types at 23°C, 30°C, and 37°C. (C) Cell cycle-dependent localization of Clb4. Cells with CLB4-GFP SPC42-RFP in G1 (panels a,b), early S (panels c,d), S/G2 (panels e,f), and anaphase (panels g,h) were analyzed for Clb4-GFP localization. Because of the slow folding of the RFP molecule, the pre-existing old SPB is more strongly stained by RFP than the newly formed SPB (Pereira et al. 2001). The arrows point toward Clb4-GFP signals at the bud neck. Asterisks indicate Clb4 associating with the SPB. (D) Clb4 is concentrated at astral MT plus ends. Preanaphase CFP-TUB1 CLB4-GFP cells were analyzed for Clb4-GFP localization. Arrows point toward the plus end of MTs. (E) Preanaphase CLB4-GFP SPC42-CFP cells were fixed and analyzed as indicated for SPB association of Clb4. Quantification of Supplementary Figure 1B. n = 180. (F) Single-plane time-lapse analysis of CLB4-GFP SPC42-RFP cells. The merged Clb4-GFP (in green), Spc42-RFP signals (red), and phase contrast images were taken every 10 sec. Bars, 5 μm.
The localization of Clb4-GFP was analyzed in exponentially growing cells carrying Spc42-RFP as SPB marker. The Spc42-RFP protein discriminates between the old and newly formed SPB: On SPB separation in S phase, the RFP signal of the old SPB was stronger than that seen at the newly formed SPB (Pereira et al. 2001). No Clb4-GFP signal was observed in unbudded G1 cells (Fig. 3C, panels a,b) when Clb4 is degraded by the APC/C-proteasome (Visintin et al. 1997). In most early S-phase cells, Clb4-GFP associated with the single SPB signal (Fig. 3C, panel c, asterisk). In some cells the Clb4-GFP signal did not colocalize with Spc42-RFP but instead associated with the bud neck (Fig. 3C, panel d, arrow). On SPB separation in S phase, Clb4-GFP was selectively associated with the budward-directed old SPB (Fig. 3C, panel e, asterisk; Pereira et al. 2001) but not with the SPB located toward the mother cell body (Figs. 3C, 4B). Clb4-GFP was also observed as a dot-like signal at the bud cortex (Fig. 3C, panel f, arrow). This signal reflects astral MT plus ends that at this stage of the cell cycle interact with the bud neck (Fig. 3D, arrows). Clb4-GFP was not detectable in anaphase cells, probably because of the degradation of the protein (Fig. 3C, panels g,h).
Figure 4.
Kar9 directs Clb4 to SPBs. (A) Colocalization of Clb4 and Kar9 at astral MTs. Fixed CLB4-GFP KAR9-eqFP611 CFP-TUB1 cells were analyzed by fluorescence microscopy. (B) SPB localization of Clb4 is dependent on Kar9. Localization of Clb4-GFP in KAR9 SPC42-CFP and Δkar9 SPC42-CFP cells. (C) Clb4 fails to associate with astral MT plus ends in Δkar9 cells. The arrow points toward the plus end of an astral MT. (D) Kar9 localization in Δclb4 cells. Live images of preanaphase cells were examined. The three major cell types are shown. In type 1 cells, Kar9 is only associated with the budward-directed old SPB and/or astral MTs organized by this SPB. In type 2 cells, the majority of Kar9-GFP signal (>70%) is associated with the old SPB. A weaker signal is at the new SPB in the mother cell. In type 3 cells, both SPBs and associated astral MTs have an equal Kar9-GFP signal. (E) Quantification of D, n > 100. The cartoon indicates the regions that were considered as budward- and motherward-directed signals. (F) SPB localization of Kar9-GFP in mutants (S288c background) lacking two B-type cyclins. Experiment as in D and E. (G) The distribution of Kar9-GFP at SPBs, along astral MTs and at astral MT plus ends in preanaphase CLB4 and Δclb4 cells (both with KAR9-GFP CFP-TUB1). n > 100. (H) In Δclb4 cells, Kar9-GFP is frequently dispersed along the length of astral MTs. In preanaphase CLB4 KAR9-GFP CFP-TUB1 cells, Kar9 is predominately associated with the plus end of astral MTs. In preanaphase Δclb4 cells, a clear Kar9 signal is seen at astral MT plus ends. However, an additional Kar9 signal is dispersed along the length of the astral MTs. Bars, 5 μm.
RFP forms tetramers (Baird et al. 2000). Spc42-RFP could therefore alter the properties of SPBs, thereby affecting SPB binding of Clb4. To exclude this possibility, we re-evaluated the localization of Clb4 in CLB4-GFP SPC42-CFP cells. In 77% of cells with a short bipolar spindle (1-2 μm), Clb4 was associated only with the bud-ward-directed SPB (Fig. 3E; Supplementary Fig. 1B). Only ∼6% of cells showed Clb4 at the motherward-directed SPB, and ∼3% had a Clb4 signal at both SPBs (Fig. 3E, 1-2 μm). In the residual cells, the spindle was not yet aligned along the mother-bud axis. In these cells Clb4 was associated with only one of the two SPBs. When these data are taken together, the functional, nonoverexpressed Clb4-GFP associated in the vast majority of preanaphase cells with the budward-directed SPB.
The localization pattern of nonoverexpressed Clb4-GFP is reminiscent of the distribution of Cdc28-4GFP, which is transported from the SPB to astral MT plus ends (Maekawa et al. 2003). This was also the case for Clb4, as indicated by time-lapse video microscopy of living CLB4-GFP cells (Fig. 3F). The Clb4-GFP signal colocalized with the SPB (Spc42-RFP) in small-budded S-phase cells and then progressively moved away from the SPB toward the bud neck. A similar behavior was observed for the Cks1 protein, which is a subunit of the Cdc28 protein kinase complex (Supplementary Fig. 2A-D; Hadwiger et al. 1989). Thus, Clb4 and Cks1 as Cdc28 are transported from SPBs to astral MT plus ends.
SPB and MT localization of Clb4 are dependent on Kar9; however, Kar9 associates with astral MTs in Δclb4 cells
The cotransport of Cdc28-Clb4 and Bim1-Kar9 from the SPB to the plus end of astral MTs and the phosphorylation of Kar9 by Cdc28 in vitro and in vivo (Liakopoulos et al. 2003; Maekawa et al. 2003) suggest that all four proteins are part of a larger complex during the transport process. Colocalization of Kar9 and Clb4 (Fig. 4A) and of Clb4 and Cdc28 (data not shown) further supported this notion. One of the two subcomplexes could use the other as a carrier to reach and regulate MT plus ends. To test this possibility, we analyzed Clb4 localization in Δkar9 cells and Kar9 localization in Δclb4 cells. In most (∼90%) Δkar9 cells, a Clb4-GFP signal was undetectable at either SPBs or astral MT ends (Fig. 4B,C). A very weak Clb4-GFP signal colocalized in only <10% of Δkar9 cells with the SPB marker Spc42-CFP (Fig. 4B). Because Clb4 levels were unchanged in Δkar9 cells (data not shown), it was not the degradation of Clb4 in Δkar9 cells that lead to the absence of a Clb4 signal, but instead Kar9 directed Clb4 to SPBs and MTs. Similarly, deletion of Kar9 abolished Cks1 association with astral MT plus ends (Supplementary Fig. 2E). In contrast, Cdc28-4GFP was still associated with SPBs in Δkar9 cells (Maekawa et al. 2003). This is because Cdc28 in complex with other B-type cyclins probably interacts with SPB components such as Spc42 (Bailly et al. 2003; Maekawa et al. 2003). Thus, Kar9 is essential for the SPB and astral MT binding of Clb4 and Cks1.
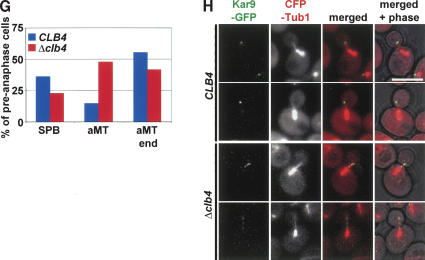
Deletion of CLB4 did not affect the SPB localization of Kar9 in G1 phase cells; 89.2% of wild type and 92.3% of Δclb4 cells had a Kar9-GFP signal that colocalized with the SPB marker Spc42-RFP. However, after SPB separation the strong polar localization of Kar9 with the bud-ward-directed SPB seen in wild-type cells was mildly disturbed in preanaphase Δclb4 cells (Liakopoulos et al. 2003; Maekawa et al. 2003). In ∼27% of Δclb4 cells, a weak Kar9 signal (less than one-third of the signal at the budward-directed SPB) was also observed at the mother-ward-directed SPB (Fig. 4D,E). This effect was slightly enhanced by the additional deletion of other B-type cyclins, namely, CLB2 and CLB5 (Fig. 4F). This result was not dependent on our S288c strain background but was also seen in W303 cells (cf. Fig. 4E [S288c] and Supplementary Fig. 3A [W303]). Thus, Cdc28-Clb activity is moderately important for the polar association of Kar9 with SPBs.
A marked difference between Δclb4 cells and wild-type cells was the increased association of Kar9 along the length of astral MTs in Δclb4 cells (Fig. 4D). Despite this expanded MT association of Kar9 in Δclb4 cells, Kar9 still showed a clear association with astral MT plus ends (Fig. 4G,H; Supplementary Fig. 3B). Clb4 is therefore required to restrict Kar9 MT localization to the plus end of MTs.
In Δclb4 cells, Kar9 is strongly underphosphorylated (Supplementary Fig. 4A; Liakopoulos et al. 2003), and this reduced phosphorylation could account for the altered localization of Kar9. However, mutated Kar9 proteins in which two or all six Cdk1 consensus sites were mutated and which failed to become hyperphosphorylated (Supplementary Fig. 4B), did not show the strongly dispersed localization along astral MTs that is typical of Δclb4 cells (Supplementary Fig. 4C). In addition, these Kar92A and Kar96A proteins had only a weak effect upon the polar association with the budward-directed SPB independent of the strain background (Supplementary Fig. 4D,E). It is therefore unlikely that Cdc28-Clb4 regulates Kar9 localization solely through Kar9 phosphorylation.
Kar9 anchors astral MTs to the bud cortex in the absence of Cdc28-Clb activity
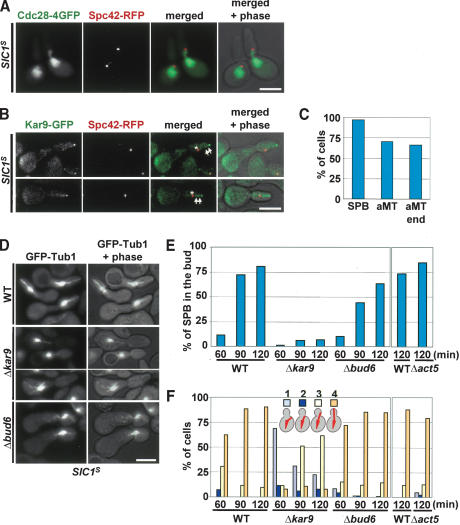
Our data suggest that a considerable fraction of Kar9 becomes transported to astral MT plus ends, even in the absence of Cdc28-Clb4 activity (Fig. 4).To exclude the possibility that another B-type cyclin partially complements for the role of Clb4 in Kar9 transport, Cdc28-Clb was completely inhibited by overexpression of stabilized versions of the inhibitor SIC1 (SIC1-ΔNT or SIC1T5V,T33V,S76A; Schwob et al. 1994). Both constructs gave similar results, and they are referred to as SIC1S. Cells with overexpressed SIC1S arrested in late G1 phase with duplicated but unseparated SPBs without detectable Cdc28-Clb activity (Drury et al. 1997; Desdouets et al. 1998). In these G1 cells, Cdc28-4GFP was nuclear and did not associate with the SPB (Spc42-RFP) or astral MTs (Fig. 5A). This is in agreement with the absence of Cdc28-4GFP from SPBs in α-factor arrested cells, which also lack Cdc28-Clb activity (Maekawa et al. 2003). In contrast, the kinase dead Cdc28K40L-4GFP (Fig. 1) was able to bind to SPBs of cycling S-phase cells (the cell cycle is driven by CDC28), probably because of the formation of Cdc28K40L-Clb complexes.
Figure 5.
Kar9 anchors astral MTs to the bud cortex in the absence of Cdc28-Clb activity. (A) Cdc28-4GFP localization in SIC1S arrested cells. Cells were released from α-factor block, and cell cycle progression was arrested by the overexpression of SIC1T5V,T33V,S76A. (B) Synchronized KAR9-GFP SPC42-RFP cells were arrested in late G1 through overexpression of Gal1-SIC1-ΔNT. Kar9-GFP associated dot-like with the SPB (Spc42-RFP, asterisks), astral MT plus ends, and along astral MTs (arrows). (C) Kar9 localization of cells of B was determined by fluorescence microscopy. n > 100. (D) In pMet25-SIC1T5V,T33V,S76A-HA cells, astral MT association with the bud tip is dependent on KAR9 but not BUD6. G1 cells of indicated cell types with GFP-Tub1 arrested by α-factor were released into medium lacking methionine, allowing the expression of SIC1T5V,T33V,S76A. (E) SPBs were frequently pulled into the bud in SIC1T5V,T33V,S76A cells. The same experiment as D. Δact5 cells were analyzed in parallel to the corresponding wild type in a separate experiment. Numbers refer to minutes after release from the α-factor block. The position of the SPB (the intersection of astral and nuclear MTs) was determined for each time point in ∼100 cells. (F) Experiment of E but the astral MT organization was scored as belonging to one of four categories (see cartoon cells). (1) Cells lacking astral MTs touching either the bud neck or the bud cortex; (2) cells with astral MTs contacting the bud neck; (3) cells with astral MT contacting the bud cortex (excluding the bud tip region); (4) cells with astral MTs contacting the bud tip. n > 100.
In virtually all SIC1S cells, a fraction of Kar9 did associate with the SPB despite the absence of Cdc28 at this location (Fig. 5B, asterisks). In ∼70% of cells, Kar9 localized along astral MTs (Fig. 5B, arrows) and strongly associated with astral MT plus ends (Fig. 5B,C). This suggests that Kar9 binds to astral MT plus ends even in the absence of Cdc28-Clb activity.
We were now able to address whether Kar9 is active in anchoring MT ends to the bud cortex when Cdc28-Clb activity is absent. In SIC1S arrested wild-type cells, the SPB was frequently pulled into the bud (Fig. 5B,D). This probably arose from the stable association of astral MT ends with bud tip that continued to grow. In contrast, in SIC1S Δkar9 cells the SPB was no longer pulled into the bud (Fig. 5D,E). This was because of the strongly reduced bud tip interactions of MTs ends in Δkar9 cells. Although ∼90% of astral MT ends interacted with the bud tip in SIC1S wild-type cells, this number was reduced to ∼8% in Δkar9 cells (Fig. 5F, 120 min SIC1S-induction). Thus, in late G1 phase, in the absence of Cdc28-Clb activity, Kar9 anchors astral MT ends to the bud tip.
We also investigated the requirement for the dynein/dynactin complex and the protein Bud6. Bud6 has been implicated in the Kar9-independent anchoring of astral MTs to the bud cortex early in the cell cycle (Segal et al. 2002). However, in SIC1S cells deletion of BUD6 had only a weak effect on the bud localization of the SPB (Fig. 5D,E) and on the bud tip anchoring of astral MT ends (Fig. 5F). Similarly, deletion of the dynactin subunit ACT5 hardly had any effect upon astral MTs in SIC1S cells (Fig. 5F). Thus, in cells lacking Cdc28-Clb activity, Kar9 is the main determinant that anchors astral MTs to the bud cortex.
Clb4 regulates the association of astral MTs with cortical domains of the bud
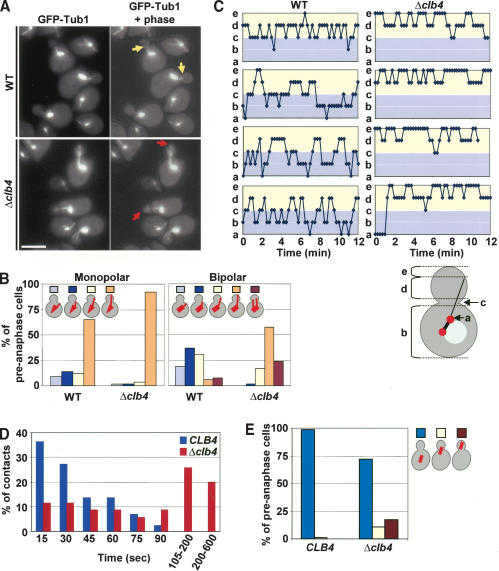
Cdc28-Clb activity is not essential for the establishment of Kar9-dependent astral MT interactions with the bud tip (Fig. 5). However, Cdc28-Clb4 may affect the turnover of established interactions. To address this possibility, we analyzed the interaction of astral MT ends with the bud cortex of Δclb4 cells. Wild-type CLB4 and Δclb4 cells were synchronized by an established α-factor block and release procedure. In wild-type CLB4 and Δclb4 cells in G1 or early S phase (as indicated by the monopolar spindle), most astral MTs interacted with the bud tip region (Fig. 6A,B; Segal et al. 2002). In wild-type cells, astral MT cortex interactions changed dramatically with formation of a bipolar spindle in late S phase: In most S/G2 cells, astral MTs interacted with the bud neck region (37%) or the central bud cortex (31%), whereas in 19% of cells no clear interaction with the bud neck or bud cortex was observed. Only ∼8% of astral MTs remained associated with the bud tip (Fig. 6B). This switch toward more central cortical domains was not observed in Δclb4 cells, in which the bud tip interactions persisted in most S/G2 cells (58%; Fig. 6B). An additional phenotype of preanaphase Δclb4 cells (∼25%) was the simultaneous interaction of astral MT bundles organized by the two SPBs with the bud cortex (Fig. 6B; Liakopoulos et al. 2003). This phenotype is probably caused by the partial symmetric association of Kar9 with both SPBs, which occurred in ∼27% of Δclb4 cells (Fig. 4E).
Figure 6.
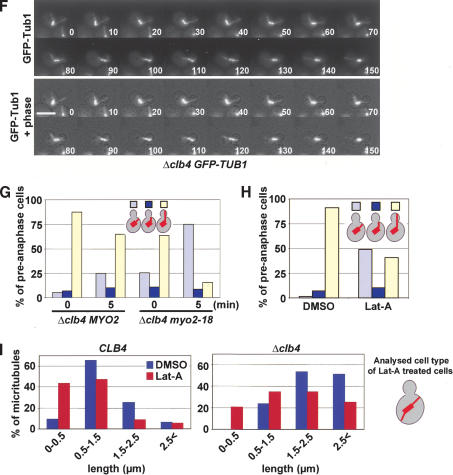
Clb4 regulates the binding of astral MTs with cortical domains of the bud. (A) MT organization of α-factor synchronized CLB4 and Δclb4 cells (both with GFP-TUB1). The yellow arrows in preanaphase CLB4 cells point toward astral MTs touching the bud neck. In contrast, in most Δclb4 cells, the astral MTs were directed toward the bud tip (red arrows). Shown are Z-series projections. (B) Quantification of astral MT bud cortex interactions of cells in A. The thick bar in the cartoon cells indicates the monopolar (late G1 or early S phase) or short bipolar nuclear spindle (S/G2 phase cells). The astral MTs are shown as thin lines. Astral MT bud cortex interactions were scored as in Figure 5F. An additional category was cells with both astral MT bundles directed into the bud. n > 100. (C) Analysis of astral MT interactions with the cell cortex by three-dimensional time-lapse video microscopy using an Ultraview spinning-disk confocal microscope. The position of astral MT plus ends were as outlined in the cartoon shown on the bottom of the subfigure: (a) astral MT end at or near SPB; (b) astral MT end in mother cell body; (c) astral MT end contacting the bud neck; (d) astral MT end contacting the bud cortex; (e) astral MT end contacting the bud tip. Fourteen wild-type and Δclb4 cells were analyzed >12 min each. Four representative examples are shown for each cell type. (D) Budward-directed astral MTs interact in Δclb4 cells longer with the cortex than in CLB4 wild-type cells. Duration of bud cortex interaction of cells imaged in C was determined and grouped as outlined in the figure. CLB4 wild-type cells, n = 44; Δclb4 cells, n = 35. (E) Synchronized CLB4 and Δclb4 cells expressing GFP-TUB1 were analyzed for the position of the preanaphase spindle as indicated. (F) Time-lapse analysis of Δclb4 GFP-TUB1 cells. The movie shows that the short spindle was virtually entirely pulled into the bud after 110 sec. The short spindle then reversed back through the mother-bud neck (130-150 sec). The length of the spindle remained relatively constant at ∼2 μm throughout the sequence. Supplementary Movie 3 shows another example. (G) Astral MT interactions with the bud tip in Δclb4 cells are dependent on MYO2. The indicated cell types (with GFP-TUB1) were shifted for 5 min from 23°C (t = 0 min) to 36°C (t = 5 min). (H) Logarithmically growing GFP-TUB1 cells were treated with Lat-A or DMSO for 10 min. Astral MT cell cortex interactions of fixed cells were determined by fluorescent microscopy. (I) Length of astral MTs in wild-type and Δclb4 cells after Lat-A treatment. Both cell types were treated as in H. The length of budward-directed astral MTs was determined in DMSO-treated cells (n > 60). In Lat-A-treated cells, the length of astral MTs originating from the budward-directed SPB but not contacting the bud cortex was determined (see cartoon on the left; n > 60). Bars, 5 μm.
Is the Δclb4 phenotype due to a failure of MTs to bind to the septin ring of the bud neck, or a failure to release MT plus ends from the bud tip? We excluded the possibility that the reduced interaction of astral MT ends with the bud neck in Δclb4 cells was caused by a septin defect by analyzing the localization of the septin subunits Cdc3, Cdc11, and Cdc12 (data not shown). Consistently, inactivating the septin ring in cdc12-6 cells (Kusch et al. 2002) did not significantly increase the bud tip interaction of astral MTs (Supplementary Fig. 3C). This suggests that the bud tip interaction of MTs in Δclb4 cells is not because of a defective septin ring.
Thus, CLB4 regulates the way astral MTs interact with the bud cortex. If Cdc28-Clb4 activity is absent, as is the case in G1 of wild-type cells or in Δclb4 cells, astral MTs preferentially interact with the bud tip region. With the accumulation of Cdc28-Clb4 activity in S phase of wild-type cells, these MT interactions switch from the bud tip to the bud neck.
Clb4 regulates the duration of astral MTs with the bud cortex
To evaluate the model outlined above, we analyzed astral MT bud cortex interactions of preanaphase wild-type and Δclb4 cells by time-lapse video microscopy (Supplementary Movies 1, 2). In wild-type cells, astral MTs interacted with the bud neck region (Fig. 6C, c) and then moved along the bud cortex to the middle of the bud (Fig. 6C, d). Astral MTs then reversed back to the mother cell (Fig. 6C, b) or even depolymerized back to the SPB (Fig. 6C, a). Bud tip interactions were rarely observed in preanaphase wild-type cells (Fig. 6C, e). In contrast, in Δclb4 cells, astral MT ends showed extended periods of interactions with the bud tip region (Fig. 6C, e). Astral MTs suddenly shrank back toward the middle of the bud cortex (Fig. 6C, d) and then re-established associations with the bud tip. Bud neck interactions were not frequent in Δclb4 cells (Fig. 6C, c). This result suggests that in an unperturbed cell cycle, Clb4 facilitates the switching of astral MT interactions from the bud tip region toward the bud neck.
The result above indicates that astral MT plus ends remain attached to the bud tip region in Δclb4 cells. This more persistent binding could be the result of a reduced turnover of astral MT plus ends with binding sites at the bud cortex. If this is the case, we would expect that the astral MTs of Δclb4 cells interact longer with the bud cortex than those in wild-type cells. We determined the duration of a continuous contact of astral MT plus ends with the bud cortex from the same time-lapse sequences (Fig. 6D). Although in wild-type cells, bud cortex interactions were maintained for 35.1 sec in average, this duration time was dramatically increased to 129 sec in Δclb4 cells. Thus, astral MT plus ends interact in Δclb4 cells approximately four times longer with the bud cortex than in wild-type cells.
If the interactions between astral MT ends and the bud tip persist and the MTs shorten, then we would expect the preanaphase spindle to be pulled into the bud. Indeed, such migration into the bud occurred in 28.2% of the preanaphase Δclb4 cells but rarely in wild-type (0.8%) cells (Fig. 6E). Time-lapse analysis of live Δclb4 cells confirmed that the short preanaphase spindle, which was correctly positioned along the mother-bud axis, was pulled into the bud (Fig. 6F, t = 100; Supplementary Movie 3). This spindle then reversed back into the mother cell body (Fig. 6F, t = 150). Thus, the Clb4-regulated astral MT bud cortex interactions ensure the correct positioning of the preanaphase spindle.
In Δclb4 cells the interaction of astral MT ends with the bud tip is dependent on Kar9, Myo2, and actin
In Δclb4 cells, astral MT plus ends interact preferentially with the bud tip region (Fig. 6C). To understand the proteins involved in this capture, the binding of astral MT ends to the bud cortex was analyzed in Δclb4 cells. Kar9 interacts with the myosin V motor Myo2, which thereby mediates interactions with the bud cortex (Yin et al. 2000; Hwang et al. 2003). It has also been described that overexpressed Kar9 binds to the bud tip region in a manner that is dependent on the status of Bud6, the polarity determinants Spa2 and Pea2, and the yeast formin Bni1 (Miller et al. 1999).
Bud tip contact of astral MTs in Δclb4 cells was strongly dependent on Kar9 (Supplementary Fig. 3D). This Kar9 dependency could mean that the reduced phosphorylation of Kar9, observed in Δclb4 cells (Supplementary Fig. 4A; Liakopoulos et al. 2003), is responsible for the more stable interaction of MT ends with the bud tip. However, a comparison of the phenotypes of Δclb4, Δkar9, KAR9AA, and KAR96A cells excluded this possibility (Supplementary Fig. 4F).
We next investigated the role of Myo2 in the bud tip association of astral MTs in Δclb4 cells. For this experiment we used the conditional lethal myo2-18 allele, which is specifically defective in its interaction with Kar9 (Yin et al. 2000). In Δclb4 cells in S/G2, ∼88% astral MTs, organized by the budward-oriented SPB, were directed toward the bud tip region at 23°C (Fig. 6G, 0 min). A short incubation at 36°C decreased the bud tip orientation by 26% (Fig. 6G, 5 min). In Δclb4 myo2-18 cells the bud tip interaction was strongly reduced by 75% upon shifting the cells to 36°C (Fig. 6G). Thus, Myo2 plays an important role in the anchorage of astral MTs to the bud tip region in Δclb4 cells.
Depolymerization of F-actin by Lat-A also affected bud cortex interactions of astral MTs in Δclb4 cells (Fig. 6H). In addition, we noticed that the budward-directed astral MTs of Δclb4 cells were about twice as long as those of wild-type cells (2.04 ± 0.62 μm vs. 1.11 ± 0.54 μm). Lat-A treatment reduced the length of these MTs in both cell types by ∼40% (Fig. 6I; 1.34 ± 0.91 μm in Δclb4 cells and 0.68 ± 0.67 μm in wild-type cells). This means that even without bud cortex interactions, astral MTs of Δclb4 cells were about twice as long as in wild-type cells.
Deletion of SPA2, PEA2, BNI1, or BUD6 did not affect astral MT orientation in Δclb4 cells (Supplementary Fig. 3E). These data suggest that in the absence of Clb4, astral MT ends interact with the bud cortex in a manner that is dependent upon the function of the Kar9-Myo2 complex and actin.
Astral MT dynamics is altered in Δclb4 cells
Cdc28-Clb4 could modulate the dynamic behavior of MTs. The dynamic properties of astral MTs directed into the bud of preanaphase wild type and Δclb4 cells were determined from time-lapse data. In budding yeast, astral MTs turn over only at their plus end (Maddox et al. 2000). Measuring the length of astral MTs over time reflects changes that occur at the plus end. Although the catastrophe and rescue frequencies were similar in CLB4 and Δclb4 cells, the growth and shrinkage rates were moderately increased by 25% and 20% in Δclb4 cells (Table 1; Supplementary Fig. 5). As observed for fixed cells (Fig. 6I), the average length of budward-directed astral MTs were twice as long in living Δclb4 cells as in wild-type cells. As may be expected from the longer duration of single interactions of MTs with the bud cortex (Fig. 6D), astral MTs of Δclb4 cells interacted for longer per time interval with the bud cortex (87.6% of time) in comparison to wild-type cells (60.9% of time). Thus, Clb4 moderately affects growth and shrinking rates of budward-directed astral MTs, whereas catastrophe and rescue frequencies were not significantly different.
Table 1.
Astral microtubule dynamics in CLB4 and Δclb4 cells
| CLB4 (n = 14) | Δclb4 (n = 14) | ||
|---|---|---|---|
| Rates (μm/min) | |||
| Growing | 1.25 ± 0.35 | 1.56 ± 0.38 | p < 0.0001 |
| Shrinking | 1.91 ± 0.68 | 2.28 ± 0.93 | p < 0.0001 |
| Frequencies (event/sec) | |||
| Catastrophe | 0.0183 ± 0.0036 | 0.0198 ± 0.0046 | p = 0.36 |
| Rescue | 0.0224 ± 0.0057 | 0.0233 ± 0.0036 | p = 0.59 |
| Event duration (min) | |||
| Growing | 0.88 ± 0.34 | 0.95 ± 0.35 | p = 0.17 |
| Shrinking | 0.64 ± 0.19 | 0.69 ± 0.25 | p = 0.21 |
| Pausing | 0.58 ± 0.12 | 0.56 ± 0.13 | p = 0.83 |
| Event length changes (μm) | |||
| Growing | 1.10 | 1.48 | |
| Shrinking | 1.23 | 1.57 | |
| Total times (%) | |||
| Growing | 53.3 | 54.2 | |
| Shrinking | 38.1 | 43.9 | |
| Pausing | 8.6 | 1.9 | |
| Average length (μm) | 0.98 ± 0.13 | 1.84 ± 0.38 | p < 0.0001 |
| aMT-cortex contact (% of time) | 60.9 | 87.6 |
For CLB4 and Δclb4 cells, 14 time-lapse sequences of 12,045 or 10,125 sec were obtained with the spinning-disk confocal microscope. Frequencies of catastrophes and rescues were calculated in each time-lapse sequence, and the averages of them were shown (Kosco et al. 2001). Event length changes equal mean event rates times mean event durations. P values were by unpaired t test.
Discussion
It is now well established that the cyclin-dependent kinase Cdc28 plays important roles in regulating the orientation of the mitotic spindle. The Cdc28-Clb5 complex restricts astral MT organization in S phase to the old SPB by an unknown mechanism (Segal et al. 1998, 2000). In addition, Cdc28 becomes transported together with Kar9 from the budward-directed SPB along astral MTs to the site where astral MT ends interact with the bud cortex (Maekawa et al. 2003). Another report proposed that the Cdc28-Clb4 complex associates selectively with the SPB in the mother cell body, where it prevents the binding of the Bim1-Kar9 complex to this SPB (Liakopoulos et al. 2003).
In this study we have re-examined the role of Cdc28 at SPBs and astral MTs. Several observations suggest that Cdc28, in complex with the B-type cyclin Clb4 and Cks1, gets transported from SPBs along astral MTs to astral MT plus ends. First, time-lapse analysis of Cdc28-4GFP (Maekawa et al. 2003), the nonoverexpressed, functional Clb4-GFP (Fig. 3A,F; Supplementary Fig. 1A), and Cks1-4GFP (Supplemenatary Fig. 2D) showed that the proteins were transported from the budward-directed old SPB to astral MT plus ends directed into the bud. Whether this transport is through a MT-dependent motor activity or the binding of the Bim1-Kar9 and Cdc28-Clb4 to growing MT plus ends requires further experiments. Second, the association of Cdc28 and Cks1 with astral MT ends was entirely dependent on Clb4 but not on any of the other five B-type cyclins (Fig. 2; data not shown). Third, colocalization of Clb4 and Cdc28 (data not shown) and Cks1 and Cdc28 (Supplementary Fig. 2A) is consistent with the interaction of the three proteins at MTs plus ends. Fourth, as has been reported for Cdc28 (Maekawa et al. 2003), the association of Clb4 and Cks1 with SPBs and MT ends was strongly dependent upon Kar9 function (Fig. 4B; Supplementary Fig. 2E).
What is the function of the Cdc28-Clb4 complex at astral MTs? In agreement with published data (Liakopoulos et al. 2003), we observed a role of CLB4 in the selective binding of Kar9 to the budward-directed SPB (Fig. 4E). However, in our experiments deletion of CLB4 or the introduction of mutations in Kar9, which prevented its phosphorylation by Cdc28 (Supplementary Fig. 4B-D), only mildly affected Kar9 SPB asymmetry (Fig. 4E). The additional deletion of other B-type cyclins, namely, CLB2 and CLB5, only slightly increased Kar9 at the motherward-directed SPB (Fig. 4F). The more pronounced effect observed by Liakopoulos et al. (2003) is probably explained by the use of the MT depolymerizing drug nocodazole that directs Kar9 to both SPBs even in wild-type cells (cf. Fig. 4B,C in Liakopoulos et al. 2003 and our Supplementary Fig. 3F). Interestingly, our experiments suggest that the kinetics of Kar9 symmetrization in the presence of nocodazole is slightly increased in Δclb4 cells compared with wild-type cells (Supplementary Fig. 3F). The reason for this difference between the two cell types is unknown but could be due to the altered permeability of the cell wall for the drug.
Liakopoulos et al. (2003) proposed, in part based on the selective localization of overexpressed Clb4-GFP with the motherward-directed SPB, that Cdc28-Clb4 activity at this SPB locally phosphorylates Kar9. This phosphorylation event would then lead to the disassembly of the Bim1-Kar9 complex, whereas Bim1-Kar9 could still bind to the budward-directed SPB not carrying the Cdc28-Clb4 complex. However, the selective localization of the nonoverexpressed, functional Clb4-GFP to the budward-directed SPB (Figs. 3C,E, 4B) makes this model less likely. Instead, Cdc28-Clb activity could influence the amount of free cytoplasmic Kar9. Previously, it has been shown that an increase in Kar9 levels targets the protein to both SPBs (Miller et al. 2000). Alternatively, phosphorylated Kar9 may have a higher affinity for the budward-directed old SPB, which is biochemically distinct from the new SPB positioned in the mother cell body (Pereira et al. 2001).
Our data show that the Cdc28-Clb4-Cks1 complex is transported in a “piggy-back style” through Kar9 from SPBs to astral MT plus ends (Fig. 4A,B; Supplementary Fig. 2E; Maekawa et al. 2003). Deletion of Kar9 completely inhibits transport of Cdc28-Clb4 (Fig. 4B), whereas in Δclb4 cells Kar9 is still transported to MT plus ends (Fig. 4G,H). However, in the absence of Cdc28-Clb4 activity, the Kar9 signal was stretched along astral MTs, whereas in preanaphase wild-type cells it was restricted to MT plus ends (Figs. 4G,H, 5B,C). This observation is most consistent with a role of Cdc28-Clb4 in the transport of Kar9 along astral MTs or during timely SPB loading. Cdc28-Clb4 could regulate the speed of transport through modification of a kinesin motor protein. Alternatively, Cdc28-Clb4 may restrict the period during which the Bim1-Kar9 complex can be loaded onto SPBs to G1 phase of the cell cycle. Lack of Cdc28-Clb4 would then result in a constant loading of the Bim1-Kar9 complex onto SPBs and transport to MT ends.
Even in the absence of Cdc28-Clb4 activity, Kar9 became concentrated at astral MT plus ends and functioned in mediating the interaction of MT ends with the bud cortex (Fig. 5E,F; Supplementary Fig. 3D). This, together with the result outlined in Fig. 4E, indicates that the main function of Cdc28-Clb4 is not in targeting Kar9 to astral MT plus ends or in the establishment of Kar9 SPB asymmetry but instead in regulating the way MT ends interact with the bud cortex. In G1/S phase of wild-type (no Cdc28-Clb4 activity) and Δclb4 cells, astral MTs interact predominantly with the bud tip region (Fig. 6A,B). Bud tip binding was also observed in SIC1S cells, which lack Cdc28-Clb activity (Fig. 5). In wild-type cells, astral MTs were eventually released from the bud tip region in S phase and realigned to interact with the bud neck (Fig. 6B). This transition did not occur in Δclb4 cells (Fig. 6B), suggesting that the Cdc28-Clb4 complex has a key role in regulating this switch of astral MT bud cortex interactions. Time-lapse analysis confirmed this conclusion. In preanaphase wild-type cells, MT ends moved rapidly from the bud neck to the central region of the bud cortex and finally retreated to the bud neck or even the cortex of the mother cell (Fig. 6C; Supplementary Movie 1). In contrast, in preanaphase Δclb4 cells most MT plus ends contacted the bud tip region (Fig. 6B,C; Supplementary Movie 2).
What is regulated by Cdc28-Clb4 that leads to the switch in astral MT bud cortex interactions? The most pronounced difference between preanaphase wild-type and Δclb4 cells was the four times increase in the time of a continuous contact of astral MT plus ends with the bud cortex in Δclb4 cells (Fig. 6D). This prolonged interaction in Δclb4 cells was not caused by a decrease in astral MT plus end dynamics. In fact, in Δclb4 cells, growing and shrinking rates of astral MTs were even slightly increased, whereas catastrophe and rescue frequencies were very similar to wild-type cells (Table 1). Moreover, in Δclb4 cells no obvious correlation between the length of an astral MT and the duration of a bud cortex contact was observed (data not shown). Conversely, even when long astral MTs contacted the bud tip in wild-type cells, this interaction was short lived (Fig. 6C). The longer duration of an interaction in Δclb4 cells therefore suggests that astral MTs interact more firmly with attachment sites at the bud cortex. This conclusion is consistent with the observation that interactions of astral MT ends with the bud cortex in Δclb4 cells were strongly dependent on the attachment proteins Myo2 and F-actin (Fig. 6G,H). In addition, in ∼28% of Δclb4 cells the preanaphase spindle became prematurely pulled into the bud (Fig. 6E,F; Supplementary Movie 3). Such a phenotype is expected when shortening astral MTs remain stably anchored to the bud cortex. When take together, these data suggest that Cdc28-Clb4 promotes the turnover of MT plus ends with attachment sites at the bud cortex.
A second difference was the longer astral MTs in Δclb4 cells (Table 1; Fig. 6I). Short treatment of cells with Lat-A that impairs attachments of astral MT ends with the bud cortex (Theesfeld et al. 1999) decreased the length of astral MTs in wild-type and Δclb4 cells. However, these detached astral MTs were still twice as long in Δclb4 than in wild-type cells (Fig. 6I), indicating that Clb4, independently of bud cortex interactions, directly affects the properties of plus ends of astral MTs possibly through the Cdc28-Clb4 dependent phosphorylation of MT-associated proteins.
Recently, it has been shown that the MT-associated Kar9 interacts with the motor Myo2, which then pulls MT ends into the bud (Yin et al. 2000; Hwang et al. 2003). It is unclear what exactly happens when Myo2 reaches the end of the actin cables near the bud tip. Our data now suggest that MT ends remain attached to the bud tip region until S phase, in a manner dependent on Kar9, F-actin, and Myo2 (Fig. 6G,H; Supplementary Fig. 3D). The Cdc28-Clb4 complex then resolves this connection (Fig. 6B,C). Considering that the Cdc28-Clb4 complex uses Kar9 as a means of transport (Fig. 4B; Maekawa et al. 2003), we propose that as soon as Cdc28-Clb4 reaches astral MT plus ends, it modifies proteins at the bud cortex or the MT end. This then causes the release of MT ends from the bud tip and allows their capture at the bud neck. However, phosphorylation of Kar9 is not necessary to release astral MT ends from the bud tip region (Supplementary Fig. 4F). Thus, other proteins in the Kar9-Myo2 complex or at the bud cortex are likely the targets of Cdc28-Clb4. At the bud neck MTs become depolymerized and so position the preanaphase nucleus close to the bud neck (Kusch et al. 2002).
In many cell types, mitotic spindle orientation is essential for the position of the division plane, the development of cell-type diversity, cell cycle progression, and accurate chromosome segregation. As in yeast cells, cortical APC and the centrosome are also required for mitotic spindle orientation in asymmetric stem cell division in Drosophila testis (Yamahita et al. 2003). Thus, the Kar9/APC-dependent Cdk-cyclin B transport from centrosomes to MT ends and the regulation of the interaction of astral MT plus ends with domains of the cell cortex may be conserved features of the spindle alignment program. In fact, from yeast to human cells, Cdk1 is found on spindles, spindle poles, and astral MTs (Bailly et al. 1989; Alfa et al. 1990), and spindle alignment in Drosphila neuroblasts depends on Cdk1 activity (Tio et al. 2001).
Materials and methods
Yeast strains and plasmids
Strains and plasmids are listed in Supplementary Table 1. Unless indicated, yeast strains were derivatives of YPH499 (Sikorski and Hieter 1989). Yeast strains were constructed by PCR-based methods (Knop et al. 1999). Plasmid pSM1023 (4GFP-KanMX6) was used for the construction of 4GFP, pCT101 for eqFP611 (Wiedenmann et al. 2002), pYM46 and pHM43 for RFP, pSM571 and pYM26 for GFP, and pHM82 and pHM84 for CFP (Knop et al. 1999). CDC28 and CDC28K40L on URA3-based integration plasmids (pHM1 and pHM98) were digested at unique BsaBI site (upstream of the K40L mutation) and then integrated into the CDC28-4GFP-kanMX6 locus to generate CDC28 CDC28-4GFP-kanMX6 or CDC28 CDC28K40L-4GFP-kanMX6. For CDC28-4GFP CDC28 and CDC28-4GFP CDC28K40L, pHM1 and pHM98 were digested at AflII site that locates downstream of the mutation. MTs were labeled by using the pRS306-CFP-TUB1 or pAFS125-GFP-TUB1 constructs. A Δact5::kanMX6 pRS316-ACT5 shuffle strain was used to test the functionality of CLB4-GFP. Only cells, which spontaneously lost the pRS316-ACT5 URA3-based plasmid (Sikorski and Hieter 1989), can grow on 5′-fluoroorotic acid (5-FOA) plates.
Yeast cell extract and immunoblotting
Extract of yeast cells was prepared by trichloroacetic acid lysis. Cell extracts were analyzed by immunoblotting using a standard protocol (Knop et al. 1999).
Cell cycle analysis and growth conditions
Yeast strains were grown in yeast extract, peptone, and dextrose medium containing 100 mg/L adenine (YPAD medium; Sherman 1991). Cells for live cell imaging were grown in SC medium. pMet25-SIC1T5V,T33V,S76A-HA cells were grown in SD medium. Cells were arrested with 10 μg/mL mating pheromone α-factor for 2.5 h at 30°C. Then SIC1T5V,T33V,S76A expression was induced by incubating the cells in the same medium but without methionine and cysteine for 30 min. Cells were released from the block by washing with SC-methionine/cysteine medium. Gal1-SIC1-ΔNT cells were grown in YP medium with 3% raffinose and arrested with α-factor for 2.5 h at 30°C. SIC1-ΔNT expression was induced by the addition of 2% galactose for 30 min, and then cells were released in YP medium containing 3% raffinose and 2% galactose.
Microscopy and image analysis
Yeast cells with RFP-, eqFP611-, CFP-, or GFP-tagged proteins were analyzed by fluorescence microscopy after fixation with 4% paraformaldehyde in 150 mM phosphate buffer (pH 6.5) for 10 min or in live cells. Images were acquired in Z-series 13-18 planes 0.3 or 0.35 μm apart. All planes and projected images were examined. Images shown in figures are projections of all planes with signals in focus, except for Figure 3F and Supplementary Figure 2D. The Cdc28-4GFP and Cdc28K40L signals in Figure 1E were quantified with Metamorph software (Universal Imaging Corporation). For live cell imaging, cells were on slabs of 25% gelatin containing SD medium supplemented with the required amino acids. Single-plane time-lapse analysis was as described (Pereira et al. 2001). For GFP-TUB1 cells, two independent time-lapse series were analyzed. The first time-lapse series was acquired in a Zeiss 200M inverted microscope (Carl Zeiss) equipped with 63× 1.4NA Plan-Apochromat Objective. Images were acquired at 15-sec intervals at 30°C. The 2 × 2 binned images were acquired in Z-series 14 planes 0.4 μm apart. The second time-lapse series were acquired with Ultraview spinning-disk confocal microscope (PerkinElmer Life Sciences) at 15-sec intervals at room temperature (∼25°C). The 2 × 2 binned images were acquired in Z-series 23-25 planes 0.4 μm apart. At each time point, the length of MT was measured with Imaris software (Bitplane). Growth, shrinkage, pauses, catastrophes, and rescues were defined and calculated as described (Kosco et al. 2001). Frequencies of catastrophe and rescue and average length are mean values of individual cells. Variations around mean values are given as SDs. Comparisons of statistical significance were by unpaired t-test. Figure 6D and E, Table 1, and Supplementary Movies 1-3 were from the second time lapse series; Figure 6G was from the first series. The bud tip region was defined as one-third of the bud cortex closest to the bud tip.
Acknowledgments
We thank O. Cohen-Fix, M. Knop, K. Nasmyth, A. Murray, and T. Huffaker for yeast strains and plasmids. We are grateful to S. Bagley for the help with microscopy and I. Hagan, K. Labib, and members of Schiebel lab for helpful discussions. The work of E.S. is supported by a core grant of Cancer Research UK.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.298704.
References
- Agarwal R. and Cohen-Fix, O. 2002. Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes & Dev. 16: 1371-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfa C.E., Ducommun, B., Beach, D., and Hyams, J.S. 1990. Distinct nuclear and spindle pole body populations of cyclincdc2 in fission yeast. Nature 347: 680-682. [DOI] [PubMed] [Google Scholar]
- Andrews B. and Measda, V. 1998. The cyclin family of budding yeast: Abundant use of a good idea. Trends Genet. 14: 66-72. [DOI] [PubMed] [Google Scholar]
- Bailly E., Doree, M., Nurse, P., and Bornens, M. 1989. p34cdc2 is located in both nucleus and cytoplasm; part is centrosomally associated at G2/M and enters vesicles at anaphase. EMBO J. 8: 3985-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly E., Cabantous, S., Sondaz, D., Alain Bernadac, A., and Simon, M.-N. 2003. Differential cellular localization among mitotic cyclins from Saccharomyces cerevisiae: A new role for the axial budding protein Bud3 in targeting Clb2 to the mother-bud. J. Cell Sci. 116: 4119-4130. [DOI] [PubMed] [Google Scholar]
- Baird G.S., Zacharias, D.A., and Tsien, R.Y. 2000. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. 97: 11984-11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach D.L., Thibodeaux, J., Maddox, P., Yeh, E., and Bloom, K. 2000. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol. 10: 1497-1506. [DOI] [PubMed] [Google Scholar]
- Bienz M. 2001. Spindles cotton on to junctions, APC and EB1. Nat. Cell. Biol. 3: E1-E3. [DOI] [PubMed] [Google Scholar]
- Byers B. and Goetsch, L. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124: 511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati J.L. and Stearns, T. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138: 629-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdouets C., Santocanale, C., Drury, L.S., Perkins, G., Foiani, M., Plevani, P., and Diffley, J.F. 1998. Evidence for a Cdc6p-independent mitotic resetting event involving DNA polymerase α. EMBO J. 17: 4139-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorée M. and Hunt, T. 2002. From Cdc2 to Cdk1: When did the cell cycle kinase join its cyclin partner? J. Cell Sci. 115: 2461-2464. [DOI] [PubMed] [Google Scholar]
- Drury L.S., Perkins, G., and Diffley, J.F. 1997. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 16: 5966-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger J.A., Wittenberg, C., Mendenhall, M.D., and Reed, S.I. 1989. The Saccharomyces cerevisiae CKS1 gene, a homolog of the Schizosaccharomyces pombe suc1+ gene, encodes a subunit of the Cdc28 protein kinase complex. Mol. Cell. Biol. 5: 2034-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E.S., Kusch, J., Barral, Y., and Huffaker, T.C. 2003. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell. Biol. 161: 483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers, K., Pereira, G., Zachariae, W., Winsor, B., Nasmyth, K., and Schiebel, E. 1999. Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast 15: 963-972. [DOI] [PubMed] [Google Scholar]
- Korinek W.S., Copeland, M.J., Chaudhuri, A., and Chant, J. 2000. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science 287: 2257-2259. [DOI] [PubMed] [Google Scholar]
- Kosco K.A., Pearson, C.G., Maddox, P.S., Wang, P.J., Adams, I.R., Salmon, E.D., Bloom, K., and Huffaker, T.C. 2001. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell. 12: 2870-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch J., Meyer, A., Snyder, M.P., and Barral, Y. 2002. Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes & Dev. 16: 1627-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L., Tirnauer, J.S., Li, J., Schuyler, S.C., Liu, J.Y., and Pellman, D. 2000. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science 287: 2260-2262. [DOI] [PubMed] [Google Scholar]
- Li M.H., Karpova, T.S., and Cooper, J.A. 1994. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell 78: 669-679. [DOI] [PubMed] [Google Scholar]
- Liakopoulos D., Kusch, J., Grava, S., Vogel, J., and Barral, Y. 2003. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112: 561-574. [DOI] [PubMed] [Google Scholar]
- Maddox P.S., Bloom, K.S., and Salmon, E.D. 2000. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2: 36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H., Usui, T., Knop, M., and Schiebel, E. 2003. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 22: 438-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K. and Rose, M.D. 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140: 377-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K., Heller, K.K., and Rose, M.D. 1998. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol. Biol. Cell 9: 2051-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K., Matheos, D., and Rose, M.D. 1999. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J. Cell Biol. 144: 963-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K., Cheng, S.-C., and Rose, M.D. 2000. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell 11: 2949-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Tanaka, T.U., Nasmyth, K., and Schiebel, E. 2001. Modes of spindle pole body inheritance and segregation of the Bfa1p/Bub2p checkpoint protein complex. EMBO J. 22: 6359-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz K., Richards, K., and Botstein, D. 1997. BIM1 encodes a microtubule-binding protein in yeast. Mol. Biol. Cell 8: 2677-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E., Bohm, T., Mendenhall, M.D., and Nasmyth, K. 1994. The B-type cyclin kinase inhibitor p40 (Sic1) controls the G1 to S transition in Saccharomyces cerevisiae. Cell 79: 233-244. [DOI] [PubMed] [Google Scholar]
- Segal M., Clarke, D.J., and Reed, S.I. 1998. Clb5-associated kinase activity is required early in the spindle pathway for correct preanaphase nuclear positioning in Saccharomyces cerevisiae. J. Cell Biol. 143: 135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M., Clarke, D.J., Maddox, P., Salmon, E.D., Bloom, K., and Reed, S.I. 2000. Coordinated spindle assembly and orientation requires Clb5-dependent kinase in budding yeast. J. Cell Biol. 148: 441-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M., Bloom, K., and Reed, S.I. 2002. Kar9p-independent microtubule capture at Bud6p cortical sites primes spindle polarity before bud emergence in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 4141-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194: 3-21. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter, P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theesfeld C.L., Irazoqui, J.E., Bloom, K., and Lew, D.J. 1999. The role of actin in spindle orientation changes during the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 146: 1019-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tio M., Udolph, G., Yang, X., and Chia, W. 2001. cdc2 links the Drosophila cell cycle and asymmetric division machineries. Nature 409: 1063-1067. [DOI] [PubMed] [Google Scholar]
- Visintin R., Prinz, S., and Amon, A. 1997. CDC20 and CDH1: A family of substrate-specific activators of APC-dependent proteolysis. Science 278: 460-463. [DOI] [PubMed] [Google Scholar]
- Wiedenmann J., Schenk, A., Röcker, C., Girod, A., Spindler, K.-D., and Nienhaus, G.U. 2002. A far-red fluorescent protein with fast maturation and reduced oligomerization tendency from Entacmaea quadricolor (Anthozoa, Actinaria). Proc. Natl. Acad. Sci. 99: 11646-11651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamahita Y.M., Jones, D.L., and Fuller, M.T. 2003. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301: 1547-1550. [DOI] [PubMed] [Google Scholar]
- Yeh E., Skibbens, R.V., Cheng, W.J., Salmon, E.D., and Bloom, K. 1995. Spindle dynamic and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130: 687-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Pruyne, D., Huffaker, T.C., and Bretscher, A. 2000. Myosin V orientates the mitotic spindle in yeast. Nature 406: 1013-1015. [DOI] [PubMed] [Google Scholar]