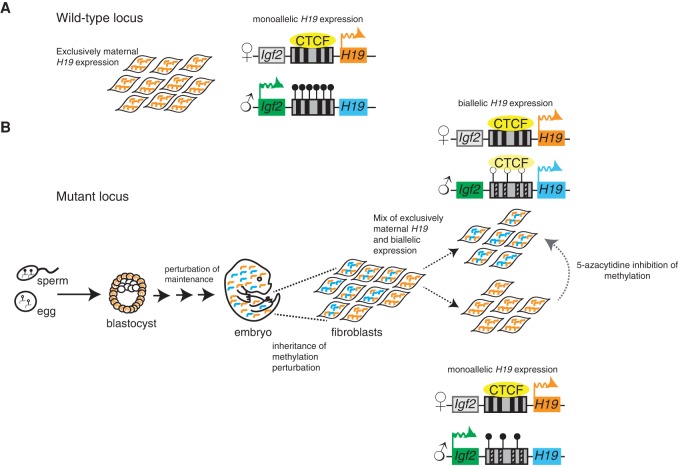
Imprinting defects can lead to inappropriate expression from the normally silenced allele, but it remains unclear whether every cell in a mutant organism follows the population average. Ginart et al. applied a new fluorescence in situ hybridization method that measures allele-specific expression in single cells to address this question in mutants exhibiting aberrant H19/Igf2 imprinting.
Keywords: gene expression, imprinting, single cell
Abstract
Imprinting is a classic mammalian epigenetic phenomenon that results in expression from a single parental allele. Imprinting defects can lead to inappropriate expression from the normally silenced allele, but it remains unclear whether every cell in a mutant organism follows the population average, which would have profound implications for human imprinting disorders. Here, we apply a new fluorescence in situ hybridization method that measures allele-specific expression in single cells to address this question in mutants exhibiting aberrant H19/Igf2 (insulin-like growth factor 2) imprinting. We show that mutant primary embryonic mouse fibroblasts are comprised of two subpopulations: one expressing both H19 alleles and another expressing only the maternal copy. Only in the latter cell population is Igf2 expression detected. Furthermore, the two subpopulations are stable in that cells do not interconvert between the two expression patterns. Combined small input methylation analysis and transcriptional imaging revealed that these two mutant subpopulations exhibit distinct methylation patterns at their imprinting control regions. Consistently, pharmacological inhibition of DNA methylation reduced the proportion of monoallelic cells. Importantly, we observed that the same two subpopulations are also present in vivo within murine cardiac tissue. Our results establish that imprinting disorders can display striking single-cell heterogeneity in their molecular phenotypes and suggest that such heterogeneity may underlie epigenetic mosaicism in human imprinting disorders.
Gene expression in diploid organisms can depend on factors beyond just DNA regulatory sequences and the binding of transcription factors. A classic manifestation of such behavior is when two otherwise indistinguishable maternal and paternal alleles of a gene are expressed differently due to epigenetic regulatory mechanisms. Well-studied examples in mammals include the phenomena of X inactivation, random monoallelic expression, and genomic imprinting (Lee and Bartolomei 2013; Savova et al. 2013). In at least some of these cases, the decision of which allele to express appears to occur at the single-cell level, but the lack of tools for measuring allele-specific expression in single cells has prevented direct observations.
Imprinted gene expression, which occurs predominantly in mammals, refers to genes that are monoallelically expressed exclusively from either the maternal or paternal allele (Lee and Bartolomei 2013). Approximately 150 imprinted genes have been identified in mice, with fewer characterized in humans, and these genes largely reside in 1- to 2-Mb clusters located through the genome. Within these clusters are three to more than a dozen imprinted genes, most of which are regulated by a differentially methylated DNA imprinting control region (ICR) (Lee and Bartolomei 2013). Deletion of the ICR results in loss of imprinting of most genes in the cluster. Smaller ICR deletions and errors in ICR-specific differential DNA methylation or other epigenetic modifications also result in aberrant expression of imprinted genes and can lead to human disease, underscoring the critical nature of the ICR (Kalish et al. 2014).
In certain contexts, expression of imprinted genes can deviate from solely monoallelic expression and display biallelic expression. Such deviations occur either developmentally, as in the case of Kcnq1, which becomes biallelically expressed in the midgestation mouse embryo (Gould and Pfeifer 1998), or tissue-specifically, as in the numerous imprinted genes that exhibit placental-specific imprinting (Tunster et al. 2013). Moreover, loss of imprinted gene expression occurs in certain pathological states, including human imprinting disorders and cancer (Kalish et al. 2014). Importantly, when normally imprinted genes show some degree of biallelic expression in population-based assays, it is unclear whether every cell exhibits the same ratio of allelic expression as the population average, whether individual cells express exclusively either the maternal or paternal allele, or whether individual cells express one or both alleles of a given gene. This uncertainty is because, until recently, it was not possible to assess allele-specific expression in single cells within a population. Moreover, expression patterns may be cell type-specific in complex tissues, but such patterns remain undetected because of the inability to isolate pure cell populations or examine them at the single-cell level. Such information could prove valuable in understanding the mechanisms governing imprinted gene regulation as well as the etiology of loss of imprinting.
The imprinted gene H19 is an ideal system in which to examine imprinting at the single-cell level. H19 is a long noncoding RNA that is normally only expressed from the maternal allele. Studies suggest that H19 regulates growth during development (Gabory et al. 2010), and it is aberrantly expressed in many cancers (Feinberg and Tycko 2004). At the same time, the neighboring gene insulin-like growth factor 2 (Igf2) is transcribed from only the paternal allele. This reciprocal pattern of transcription depends on the ICR, which is unmethylated on the maternal allele, thus allowing shared enhancers to activate H19 alone, and methylated on the paternal allele, thus directing those same enhancers away from H19 and toward Igf2.
In wild-type mammals, only the maternal allele of H19 is transcribed, but, in the human disorder Russell-Silver syndrome (Gicquel et al. 2005), defects in imprinting lead to an overall biallelic H19 expression pattern. This same defect results in decreased Igf2 expression, leading to a reduction in organism size. We previously developed a mouse model of Russell-Silver syndrome in which mutations to the ICR (H19+/DMD-9CG) exhibited a similar biallelic pattern of H19 transcription and reduction in organism size (Engel et al. 2004). However, while these changes in the allelic pattern of expression hold at the level of an entire organism or population of cells, the lack of tools for measuring imprinting in single cells meant that we could not determine whether every cell in the population exhibits the same degree of aberrant biallelic H19 expression or whether individual subpopulations have different allele-specific expression patterns that only match the population average in aggregate. Indications that such subpopulations may exist come from the observation that at least some disorders involving H19 exhibit mosaic phenotypes, with different cells in the organism affected to different extents (Kalish et al. 2013).
Recently, Levesque et al. (2013) and Hansen and van Oudenaarden (2013) described techniques for detecting single-nucleotide polymorphisms (SNPs) at the single-cell and single-molecule level using RNA fluorescent in situ hybridization (FISH). This technique, designated SNP FISH, allowed us to see whether individual mutant cells have different imprinting behavior that deviates from the population average. Using H19 SNP FISH, we show that we can detect allele-specific H19 expression at the single-cell level in both mouse embryonic fibroblasts (MEFs) and cardiac tissue. Upon interrogation of cells from an imprinting mutant mouse, we found that mutant cells formed two subpopulations: one in which cells express H19 biallelically (consistent with the bulk population measurements) and one in which H19 expresses exclusively from the maternal allele, as in the wild type. Moreover, consistent with the enhancer-blocking (insulator) model of imprinting at this locus, only cells with monoallelic H19 expression exhibit transcription of Igf2. We also provide evidence that cells stably maintain their monoallelic or biallelic expression pattern after numerous cell divisions. Monoallelic mutant colonies show methylation patterns similar to that of the wild type, and inhibition of methylation maintenance leads to fewer monoallelic colonies. Thus, these studies demonstrate that defects in parental allele-specific imprinted expression can manifest themselves via profound cell-to-cell heterogeneity, providing a potential explanation for the phenotypic mosaicism often associated with imprinting disorders.
Results
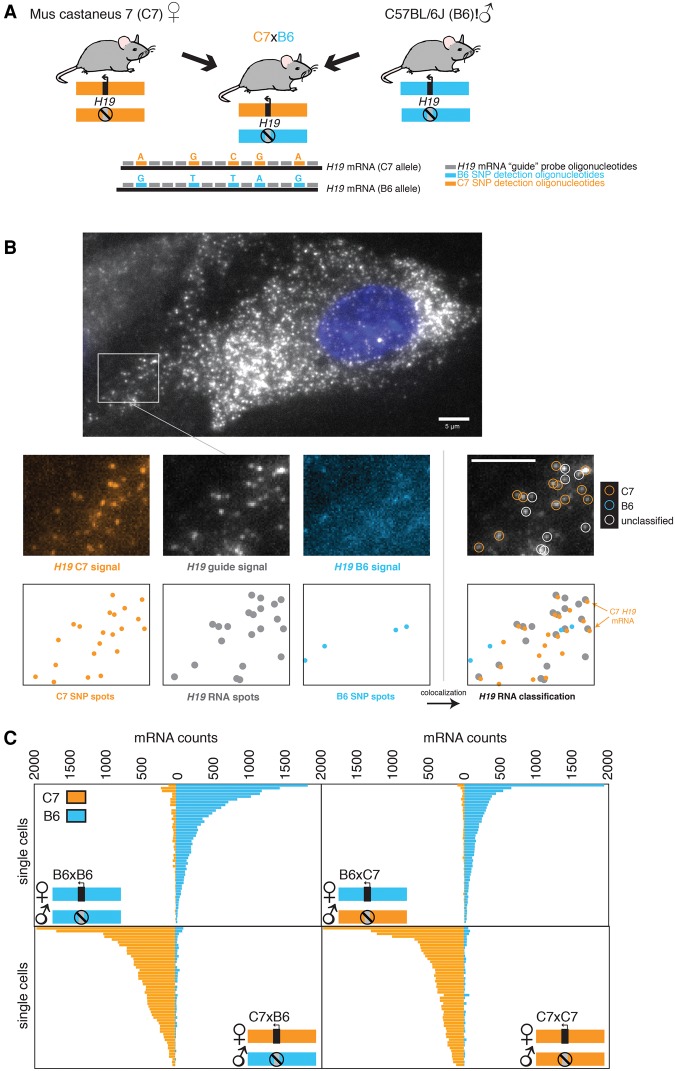
To measure allele-specific expression of the imprinted gene H19 in single cells, we mated two mouse strains (Mus musculus castaneus [C7] and C57BL/6J [B6]) that have five different SNPs in the H19 gene and then performed SNP FISH on primary MEFs isolated from these mice (Fig. 1A). The SNP FISH method works by first using a series of fluorescently labeled oligonucleotides (the “guide” probe) to identify total H19 RNA as fluorescent spots via microscopy (Raj et al. 2008). Next, to discriminate RNA transcribed from the C7 allele from that transcribed from the B6 allele of H19, we used SNP-specific probes targeting each of the five SNPs that vary between the two alleles, with all five of the C7 allele-specific probes labeled with one fluorophore and the B6 allele-specific probes labeled with a different fluorophore (Fig. 1A). Upon performing SNP FISH with both the guide probes and the SNP-specific probes, the guide probes were used to pick out legitimate H19 RNA signals, and then colocalization of these signals with those from either the C7 or B6 allele-specific SNP probes was used to classify the particular H19 RNA as arising from either the C7 or B6 allele (Fig. 1B; Supplemental Fig. 1).
Figure 1.
SNP FISH enables single-cell allele-specific measurements of imprinted gene expression in genetically defined mice. (A) F1 hybrids generated between C7 female with B6 male mice permits detection of parental allele-specific expression of genes on chromosome 7. We designed five SNP FISH probes to detect the SNPs on H19 RNA. (B) Micrograph demonstrating allele-specific detection of B6 and C7 alleles in a representative MEF. Below the large micrograph is a representative region demonstrating (from left to right) the H19 C7 maternal probe, the guide probe, and the B6 paternal probe and the RNA classification demonstrating colocalization. Below each micrograph are the computationally detected spots corresponding to single RNA transcripts. We labeled the guide probes with Cal fluor 610 and the C7- and B6-specific SNP FISH probes targeting the five SNPs between the two alleles with Cy5 and Cy3, respectively. (C) Quantification of allele-specific expression in single MEFs grown for the depicted genotypes. n = 50, randomly subsampled out of a total n = 63 for B6 × B6, n = 80 for B6 × C7, n = 59 for C7 × B6, and n = 76 for C7 × C7. Note that, in all mouse crosses, the maternal allele is written first. Each bar represents the number of H19 RNA classified as either B6 or C7 in an individual cell. Bars, 5 µm.
Using this scheme, we were able to classify 48%–60% of the H19 RNA coming from one allele or the other; the remainder were unclassifiable due to either lack of any SNP FISH probe signal (28%–51%) or the presence of both SNP FISH fluorophores (3%–15%) (Supplemental Fig. 2A), presumably due to cross-hybridization of some subset of the five different SNP-specific probes. We verified the accuracy of our colocalization algorithm by artificially introducing a small, random pixel shift between the guide and SNP FISH probe imaging channels; looking for spurious colocalization; and finding a large decrease in the average rate of total colocalization (Supplemental Fig. 2B). Also, swapping the dye labels on the SNP FISH probes yielded similar results, showing that the specificity of the hybridization does not depend on the chemical properties of the dyes used (Supplemental Fig. 2C). Furthermore, the variability observed in H19 RNA counts was not the result of cell-to-cell variability in detection frequency, which remained roughly constant irrespective of the number of H19 RNA molecules in the cell (Supplemental Fig. 3D).
We validated the specificity of our SNP FISH approach by quantifying allele-specific expression of H19 in MEFs from reciprocal F1 hybrid mice described above (Fig. 1C). In wild-type mice, H19 is expressed exclusively from the maternal allele, and we confirmed that all four breeding combinations (B6 × B6, B6 × C7, C7 × C7, and C7 × B6; note that the maternal allele is listed first) showed that the majority of H19 expression derived from the maternal allele. We found that the percentage of H19 RNA that was misclassified as coming from the paternal allele was ∼5% (Supplemental Fig. 2A). This percentage was similar between the B6 × C7 MEFs, where we could detect slight paternal expression of C7 H19 RNA, and the B6 × B6 MEFs, in which the only possible detection of C7 H19 RNA is through off-target hybridization (Fig. 1C; Supplemental Fig. 2A). Thus, it likely reflects cross-hybridization of the paternally targeted SNP FISH probes to the maternal RNA rather than leaky expression of the paternal allele. Similarly, the degree of off-target hybridization was concordant between the C7 × B6 MEFs and the C7 × C7 MEFs (Fig. 1C; Supplemental Fig. 2A). We observed that the H19 maternal ratio, defined as expression from the maternal allele of H19 divided by total H19 expression, was >80% in most wild-type MEFs, and there is a distribution of that ratio between 78% and 100% (Supplemental Fig. 3A). Based on this, we defined a monoallelic expression threshold as a maternal H19 ratio of >80%.
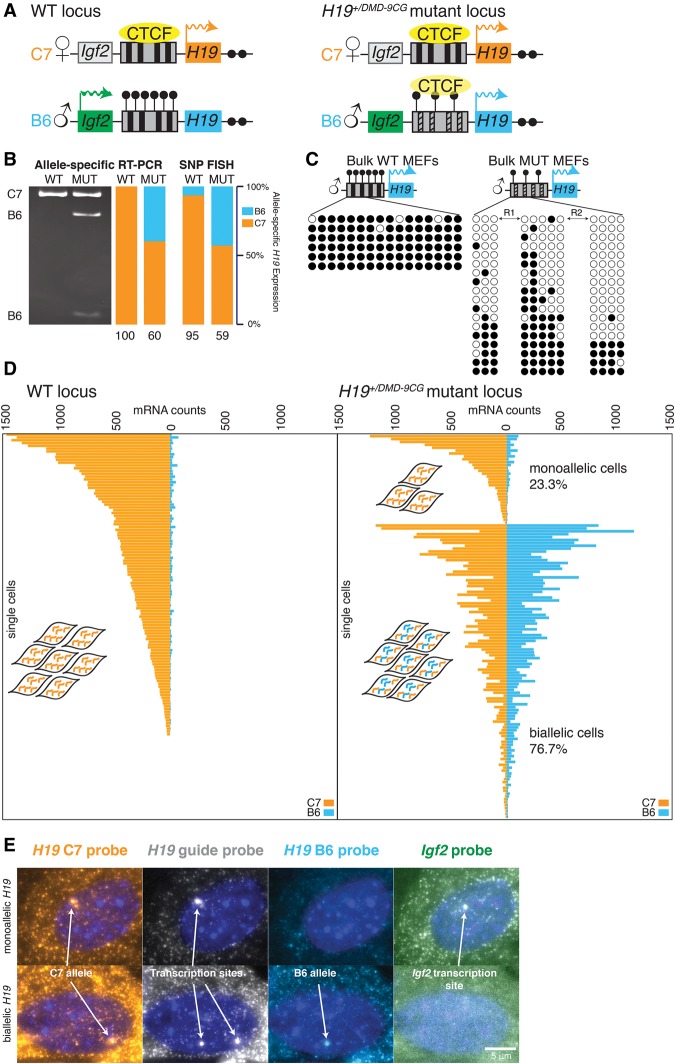
Having established the fidelity of the assay, we next examined H19 expression in mutants with defective imprinting. We used mice with paternally transmitted mutations that alter nine key CG sites in the CTCF-binding sites within the ICR of the H19/Igf2 locus (H19+/DMD-9CG) (Engel et al. 2004). On the paternal wild-type allele (Fig. 2A, left), where H19 is normally inactive, CG sites in the ICR (also known at the differentially methylated domain [DMD]) are methylated, thus blocking the binding of CTCF and hence the recruitment of enhancers to the H19 promoter, thereby repressing expression. In the mutant, nine of these CpG sites are mutated, leading to decreased methylation and aberrant transcription of H19 from the paternal allele (Fig. 2A, right; Engel et al. 2004). We confirmed expression from the paternal allele in bulk MEFs isolated from mutant mice by RT–PCR: After PCR amplification of H19 cDNA, we digested the PCR product with a restriction enzyme that specifically cuts only the paternal (B6) copy of the amplicon, leaving the maternal copy undigested (Fig. 2B). Quantification of maternal versus paternal H19 expression by this assay revealed that ∼60% of H19 RNA expressed from the maternal allele. We also performed bisulfite sequencing to reveal any methylation changes in the ICR. We found that all DNA strands from the paternal allele were essentially fully methylated in the wild-type mice, but H19+/DMD-9CG mutant mice had a mixture of fully methylated and largely unmethylated paternal DNA strands (Fig. 2C).
Figure 2.
SNP FISH reveals the presence of both monoallelically and biallelically expressing cells in mice harboring CG mutations in the CTCF sites in the paternal H19 ICR. (A) Depiction of the wild-type and mutant H19 loci. The mouse H19 locus is regulated by an ∼2-kb ICR (also designated as the DMD), which serves as a CTCF-dependent enhancer blocker on the wild-type maternal allele. On the paternal allele, the ICR is methylated, and CTCF does not bind. Mutations in the CTCF-binding sites, as in the H19+/DMD-9CG mutant mouse, cause a decrease in methylation of the ICR on the paternal allele. (B) Measurement of allele-specific expression in bulk populations of MEFs from C7 × B6 wild-type and H19+/DMD-9CG mice by both RT–PCR (as quantified by degree of DNA digestion specific to the paternal allele) and SNP FISH. The quantification shows the percentage of H19 RNA from the two alleles. (C) Methylation analysis of bulk wild-type and mutant H19+/DMD-9CG MEFs. Each row is an individual DNA strand isolated from the MEFs. Filled black circles indicate methylation at the CpG, and open circles indicate no methylation. R1 and R2 refer to the repeat regions in which several CpGs are mutated in the mutant MEFs. (D) Allele-specific expression of H19 in individual MEFs from the C7 × B6 wild-type mouse (left) and the C7 × B6 H19+/DMD-9CG mutant mouse (right) depicted in A. Each horizontal bar represents an individual cell. (E) Representative micrographs of both monoallelic and biallelic expression in MEFs from a C7 × B6 H19+/DMD-9CG mutant mouse. H19 allele-specific transcription sites are as annotated, and the Igf2 transcription site that is distinct from the H19 site is noted in the far right panel. The nucleus was labeled with DAPI (blue). Bars, 5 µm.
This heterogeneity in the methylation status on individual DNA strands suggested the possibility of transcriptional heterogeneity in the mutant population. We thus sought to distinguish whether the aberrant expression of the paternal H19 allele occurs in every cell or just a subpopulation of cells. Using SNP FISH, we measured both paternal and maternal H19 in individual MEFs isolated from the wild-type (Fig. 2D, left) and H19+/DMD-9CG mutant mice (Fig. 2D, right). Surprisingly, we found a large spread in the ratio of maternal to paternal H19 RNA in individual cells, ranging from 12% to 97%, with an average of 59% (Supplemental Fig. 3B). Importantly, the average expression of 59% by SNP FISH is essentially the same as the 60% maternal expression detected by the RT–PCR assay on populations of cells (Fig. 2B). We observed that 76.7% of the mutant cells exhibited biallelic expression, but the remaining (23.3%) cells contained mostly maternal H19 RNA, similar to the ratio of methylated to nonmethylated control regions seen in the bulk analysis of mutant MEFs (Fig. 2C) and the <80% ratio of maternal to paternal H19 transcripts observed in wild-type MEFs (Supplemental Fig. 3C). Very few cells exhibited a similarly strong paternal bias (an expression ratio <20% maternal RNA expression). Notably, we observed extensive cell-to-cell variability in the overall level of H19 expression, and the cells with the highest levels of H19 tended to be those exhibiting biallelic expression (Supplemental Fig. 3C).
At the population level, biallelic expression of H19 RNA in the H19+/DMD-9CG mutant mice is associated with greatly reduced expression of Igf2 due to decreased methylation at the ICR, leading to an aberrant enhancer-blocking function on the paternal allele (Engel et al. 2004). However, given the variability in the allelic ratio in these mutant MEFs, we wondered whether MEFs exhibiting primarily maternal H19 (as in the wild-type cells) would also express Igf2. To test this, we costained MEFs using RNA FISH probes specific to Igf2 and found that some H19+/DMD-9CG mutant cells and all wild-type MEFs contained Igf2 RNA. Thus, Igf2 is only observed in cells that predominantly expressed maternal H19, as predicted by the enhancer-blocking model governing imprinting at this locus (Fig. 2D; Supplemental Fig. 4). However, Igf2 was not observed in every mutant cell that contained only maternal H19 RNA. Moreover, the expression level of Igf2 in most of these cells was not as high as in the wild-type cells (Supplemental Fig. 4), suggesting that, while some H19+/DMD-9CG cells display monoallelic H19 expression, the mutations in the ICR still lead to abnormal expression of Igf2.
The ability to spatially localize transcripts also allowed us to assess transcriptional activity in single cells by examining accumulations of nascent transcripts at the sites of transcription in the nucleus (Levesque and Raj 2013). In cells that contained only maternal H19 RNA, we observed nascent transcription from only a single chromosome (Fig. 2E, top). In contrast, we observed transcription from both H19 alleles in those cells with both paternal and maternal H19 RNA (Fig. 2E, bottom). Here again, Igf2 was detected only in cells expressing H19 monoallelically. These results show that the lack of paternal transcripts in the monoallelic cells is due to monoallelic transcription rather than other post-transcriptional effects, such as rapid degradation of the paternal transcript, in which case we might still have seen transcription from both alleles but mature transcripts only from the maternal allele.
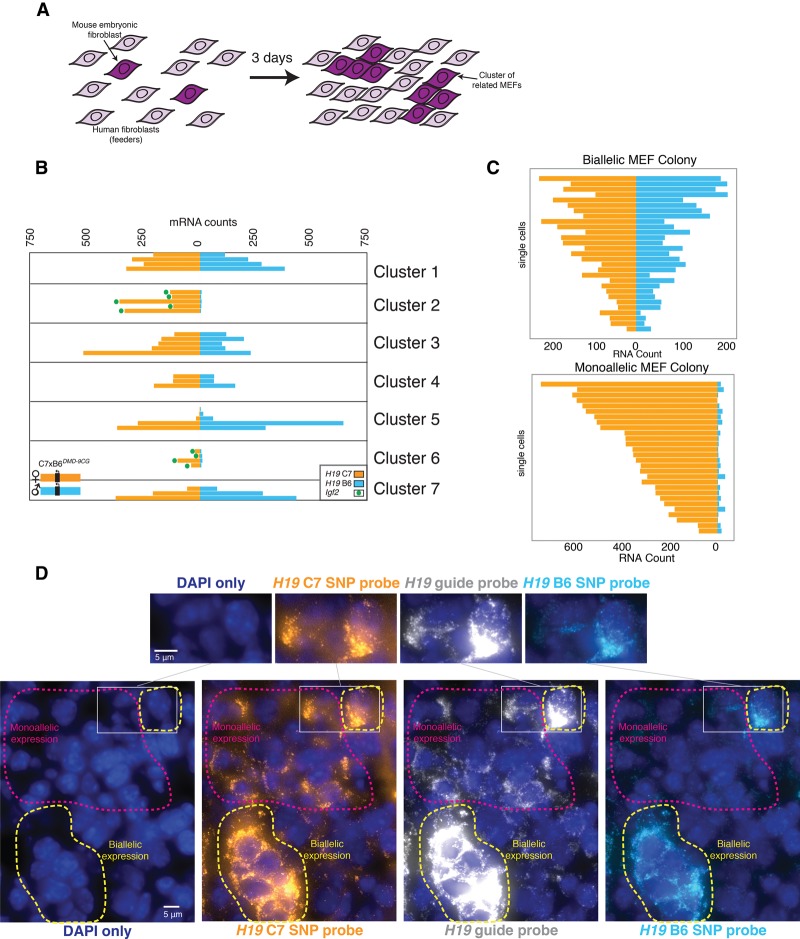
Our observation that there were two distinct subpopulations in the mutant cells—one monoallelic and one biallelic in H19 expression—prompted us to ask whether cells interconverted between these two subpopulations. Specifically, we sought to determine whether cells maintained their allelic expression ratios through cell division. To answer this question, we grew small, isolated clones of MEFs for multiple cell divisions, thus allowing the use of the spatial proximity of cells as an indicator of their relatedness. To culture isolated mutant MEFs, we grew them on a layer of human foreskin fibroblasts that we used as “feeder” cells. Over the course of 72 h, the cells divided one to three times, resulting in small clusters of related mutant MEFs typically containing two to eight cells (Fig. 3A). We found that, while the overall population displayed a heterogeneous mix of monoallelically and biallelically H19-expressing cells, individual clones consisted of exclusively monoallelically or biallelically expressing cells (Fig. 3B,C; Supplemental Fig. 5). To verify that the relatively long half-life of H19 RNA did not give a false impression of heritability in these smaller clusters of cells, we also imaged colonies from cells grown for up to 11 cell divisions, revealing the same heritable allelic expression pattern (Fig. 3C). Thus, our results show that monoallelic or biallelic expression is heritable through several cell divisions in mutant MEFs. Also, while the levels of H19 varied from cell to cell, the overall expression level of H19 was, on average, 50% higher in biallelic cells (Supplemental Fig. 3D). This result is consistent with the known functional role of H19 as an inhibitor of growth: The H19+/DMD-9CG mice are indeed significantly smaller than their wild-type littermates, due in part to reduced expression of the growth factor Igf2 (Engel et al. 2004).
Figure 3.
Monoallelic or biallelic expression behavior is maintained through cell divisions. (A) Depiction of an experiment in which small clusters of H19+/DMD-9CG MEFs were grown in the presence of human foreskin fibroblasts as feeder cells. (B) Allele-specific expression in individual H19+/DMD-9CG MEFs arranged by clusters containing at least three cells. Each bar represents a single cell in which allele-specific H19 mRNA counts are shown, and green dots indicate Igf2 expression in that cell. (C) Allele-specific quantification from representative large (>300 cells) monoallelic and biallelic colonies grown from a single parent MEF preparation. (D) Allele-specific expression in cardiac tissue from H19+/DMD-9CG mice with annotations for biallelic and monoallelic regions of H19 expression. Nuclei were labeled with DAPI (blue). Bars, 5 µm.
The heritability of H19 expression in mutant MEFs raised the possibility that individual tissues from the H19+/DMD-9CG mutant mice may also display mosaic patterns of monoallelic and biallelic H19 expression. We thus performed SNP FISH in cardiac tissue. We selected this tissue because it is relatively homogeneous in terms of number of cell types. Our guide probes revealed that H19 was expressed very highly in tracks of cells (Fig. 3D; Supplemental Fig. 6). Furthermore, within these tracks, we found areas containing only maternal H19 and other areas in which both maternal and paternal H19 RNA were visible (Fig. 3D); the high levels of H19 RNA in these cells allowed the SNP FISH probes to be readily detectable. As in the MEFs, we also found Igf2 expression only in the H19 monoallelic patches of cells (Supplemental Fig. 6). These areas appeared to consist of clusters of cells with the same expression pattern, indicating that the subpopulations with distinct allelic expression patterns detected in mutant MEFs are also observed in vivo.
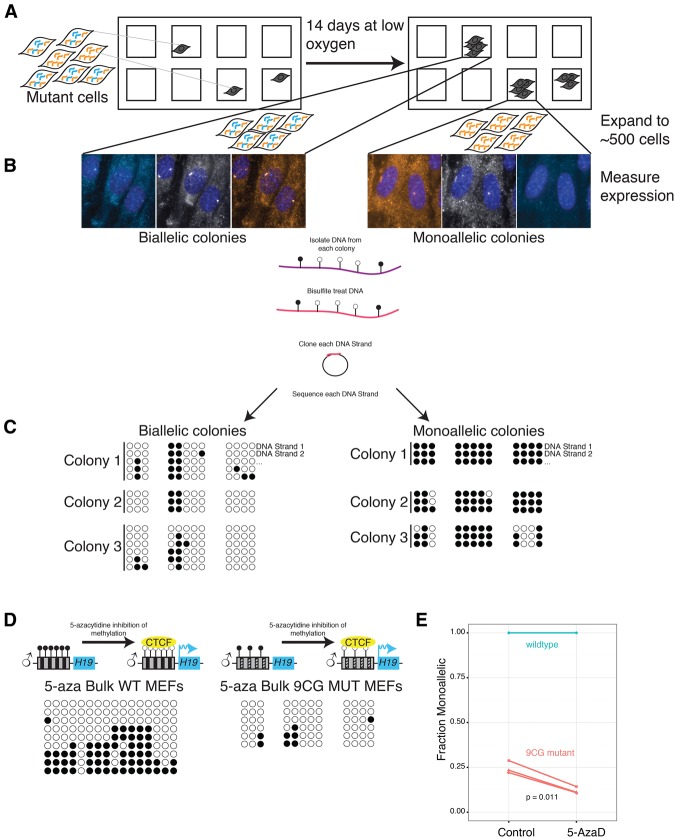
Our bulk methylation analysis in the mutants showed that roughly a quarter of the ICR sequences corresponding to the paternal allele are fully methylated, similar to the fraction of mutant MEFs displaying monoallelic H19 expression by SNP FISH, suggesting that variability in methylation may underlie the heterogeneity in allele-specific transcription. To test this possibility, we sought to measure both transcriptional and methylation heterogeneity within individual cells; however, it is difficult to accurately perform bisulfite sequencing in single cells, especially in combination with RNA FISH. To circumvent this issue, we took advantage of the fact that the allele specificity of expression was heritable and seeded individual wells with single-mutant MEFs and grew them for 14 d under hypoxic conditions (Fig. 4A), allowing the colonies to expand until they reached ∼500 cells, at which point the number of cells was sufficient for methylation analysis. After growth, we fixed the cells and performed SNP FISH to determine whether each individual colony expressed H19 in a monoallelic or biallelic manner. We then extracted DNA from the colony, treated it with bisulfite, and performed a methylation analysis. We found that mutant colonies with biallelic H19 expression showed minimal methylation in the ICR, while mutant colonies with monoallelic H19 expression showed almost complete methylation, similar to wild-type MEFs (Fig. 4C). These results demonstrate that DNA methylation heterogeneity is tightly associated with allele-specific transcriptional heterogeneity.
Figure 4.
Monoallelic or biallelic expression over multiple generations is due to methylation differences. (A) Depiction of an experimental design of colonies. Mutant MEFs were diluted to approximately a single cell per well. They were grown in low oxygen for 14 d, SNP FISH was performed, and the cells were imaged to determine whether they are monoallelic or biallelic in H19 expression. (B) Micrographs of monoallelic and biallelic colonies. After imaging, DNA was isolated from each colony, and the DNA was bisulfite-treated followed by PCR amplification, cloning, and sequencing. From each colony, three separate PCR amplifications were performed. (C) Methylation analysis of monoallelic and biallelic colonies. Each row represents an individual DNA strand from a colony. Closed black circles indicate methylation at that CpG, and open circles indicate no methylation. (D) Methylation analysis after 5-aza-2′-deoxycytidine treatment. (E) Fraction of monoallelic cells before and after treatment with 5-aza-2′-deoxycytidine. The connected lines represent the same MEFs divided into untreated and treated wells. Each line is a separate biological replicate, including two identical wild-type replicates.
To demonstrate that differences in methylation can cause the observed differences in allele-specific expression, we also treated the mutant MEFs with 5-aza-2′-deoxycytidine, which is a DNA methyltransferase inhibitor (Christman 2002). We found that the percentage of monoallelic mutant cells decreased significantly upon addition of 5-aza-2′-deoxycytidine (Fig. 4E). We confirmed the effects of 5-aza-2′-deoxycytidine by performing bulk methylation analysis on treated and untreated cells, finding decreased levels of methylation as expected (Fig. 4D). These results show that altering methylation can cause cells to interconvert between the two observed allele-specific expression patterns and suggest that variability in maintenance of methylation underlies the observed heterogeneity in allele-specific H19 transcription.
Discussion
Imprinting is a prototypical form of epigenetic gene regulation, with wild-type cells and organisms invariably showing correct parent of origin expression. However, mutant organisms with imprinting defects show a broad spectrum of phenotypes and often display pronounced phenotypic mosaicism in that different tissues in the organism will exhibit the mutant phenotype to varying degrees (Thorvaldsen et al. 2002). We investigated the allele-specific expression pattern of H19 in single cells in order to explore whether gene expression heterogeneity in single cells may underlie this mosaicism. Previous bulk population assays have shown that mutations or deletions in the H19 ICR can lead to biallelic expression with varying allelic ratios (Thorvaldsen et al. 2002). An open question in the field is the degree to which individual cells in the population follow the population average. Here, we show that the cells in H19+/DMD-9CG mutant mice that display population-level biallelic expression of H19 can be divided into two subpopulations: one that displays the biallelic expression associated with the mutant phenotype and another that shows monoallelic expression as in the wild-type. Consistent with the enhancer-blocking model of imprinting, only the monoallelically expressing cells have methylated ICRs and transcribe Igf2, thus forming a subpopulation of cells that behave crudely like the wild type. (Notably, the expression of Igf2 is still lower than in wild type, suggesting an incomplete rescue in these cells [Supplemental Fig. 4]). Thus, one can consider this imprinting mutation to be incompletely penetrant at the cellular level (Raj et al. 2010).
Given that all of the cells in the mutant organisms are genetically identical, this variability is most likely of a nongenetic origin. Single-cell analysis has shown that variability in transcript abundance is often due to random bursts of transcription (Golding et al. 2005; Chubb et al. 2006; Raj et al. 2006; Raj and van Oudenaarden 2008; Suter et al. 2011), and one could imagine that the mixed allele-specific ratios that we observed in the mutant population could arise from infrequent bursts of paternal H19 transcription. However, if bursts underlay the transcriptional heterogeneity that we observed, then one would expect rapid interconversion between the biallelic and monoallelic cells (Deng et al. 2014; Reinius and Sandberg 2015). In contrast to this hypothesis, in both our clonal MEF expansions and cardiac tissue from H19+/DMD-9CG mice, we found that monoallelic cells only give rise to other monoallelic cells, and biallelic cells only give rise to biallelic cells. This finding indicates that, once determined, the transcriptional state of the paternal allele of H19 is locked in for subsequent divisions, akin to random monoallelic expression (Gimelbrant et al. 2007; Savova et al. 2013). Our results therefore suggest a model in which the decision to silence the paternal copy is made stochastically at some point during development, after which the cell maintains and propagates that decision (Fig. 5). It is unclear exactly when this stochastic decision takes place, although earlier work with the H19+/DMD-9CG mice showed no defects in the germline establishment of the imprint (Engel et al. 2004), suggesting that the variability may arise during the early maintenance of the imprint. It is also unclear exactly how strong the fidelity of the maintenance of the paternal expression level is. Our MEF expansion results show that this maintenance lasts at least eight to 11 mitotic generations (from a single cell through the low thousands), and it may be that the inheritance lasts even longer than that. It is currently unclear how long the memory lasts in cardiac tissue, where we observed moderately sized clusters. Further experiments will be required to fully characterize the fidelity with which the imprint passes from mother cell to daughter cell and the degree to which that fidelity changes during different stages of development.
Figure 5.
Model depicting a possible mechanism leading to monoallelic and biallelic mutant colonies. (A) Wild-type MEFs express H19 monoallelically. (B) In the H19+/DMD-9CG mouse model, methylation is established normally, as in wild type. Sperm is methylated (closed circles), and the egg is unmethylated (open circles). The blastocyst is largely methylated, and H19 is expressed from the maternal allele. After the blastocyst stage, there is a perturbation of the maintenance of methylation in the mutant embryo, resulting in biallelic H19 expression. Allele-specific SNP FISH allowed us to demonstrate that there are two distinct cell populations—one that is biallelic, showing loss of ICR methylation, and a second that exhibits monoallelic H19 expression, with a wild-type ICR methylation pattern. Disruption of the maintenance of methylation by 5-aza-2′-deoxycytidine led to a decrease in the number of monoallelic cells present in the mutant cell population.
Spatiotemporal mapping of the allele-specific expression pattern of H19 may also reveal more about the dynamics of the phases of imprinting maintenance. A recent study showed that DNA methylation is dynamically maintained by transcription factors in stem states before switching to static propagation through templating in somatic states (Shipony et al. 2014), and a study in mouse embryonic stem cells showed that methylation plays a critical role in maintaining transcriptional heterogeneity (Singer et al. 2014). Prior work has also shown that disrupting imprinting maintenance in the early embryo by knocking down key maintenance proteins can lead to a mosaic pattern later in development (Lorthongpanich et al. 2013). Taken together, these findings provide further support for the possibility of a potential temporal mechanism for the developmental regulation of H19, with a critical period of dynamic methylation during early development underlying the mixed monoallelic and biallelic expression patterns that we observed here.
Consistent with these hypotheses, our results strongly suggest that cell-to-cell variability in methylation at the ICR is responsible for the cell-to-cell variability in allele-specific expression. Not only is it strongly associated at the single-cell level, but inhibiting methylation also reduced the relative abundance of cells that exclusively expressed maternal H19 as predicted, thus showing that it can directly influence transcription. These results suggest that methylation underlies the cellular memory of the transcriptional state, as other studies in mouse embryonic stem cells have found as well (Singer et al. 2014). These findings raise questions about the process that gives rise to the heterogeneity in methylation itself. Interestingly, we noticed that the ICR in the imprinting mutant tended to be either completely methylated or only very sparsely methylated, with few to no cells exhibiting medium levels of methylation. This suggests that the stochastic process underlying the variable methylation may randomly demethylate at some potentially key sites, after which demethylation spreads throughout the entire ICR. Such an “all or none” methylation process would explain why we see such a clear distinction between the monoallelic and biallelic populations of cells. That said, upon treatment with 5-aza-2′-deoxycytosine, wild-type cells display a clear decrease in methylation, with many ICRs showing an almost complete lack of methylation, but we observed no paternal bias in H19 transcription. These results suggest that methylation may not be completely determining and that the mutations in the ICR in the mutant may sensitize the expression of H19 to changes in methylation.
Ultimately, it will be interesting to see whether our results apply equally well to other imprinted genes and loci. The H19+/DMD-9CG mice mimic the methylation changes observed in some Russell-Silver syndrome patients, and the patients have both asymmetry and small size (Kalish et al. 2014). Humans with the inverse disorder, Beckwith-Wiedemann syndrome, display hemihypertrophy and overgrowth, often with profound mosaicism (Kalish et al. 2014). Imprinted gene expression is also critically important for proper brain development (Perez et al. 2015), and these types of mosaic phenotypes may play a role in neurological disease as well. Our results establish that imprinting defects can lead to heritable variability in allele-specific expression, resulting in epigenetic mosaics. Further studies of other imprinted loci may establish the generality of this phenomenon and its underlying mechanisms, perhaps even extending to nonimprinted loci (Yuan et al. 2016).
Materials and methods
Cell culture and fixation
We isolated MEFs from mice at embryonic day 13.5 as previously described (Verona et al. 2008). To determine parent-specific expression of the imprinted gene H19, we used the C57BL/6 (CAST7) strain (C7) (Mann et al. 2003), which possesses chromosome 7 from the M. musculus castaneus strain in a C57BL/6 (B6) (The Jackson Laboratory) background. We isolated MEFs from either B6 mice crossed with B6 mice, C7 mice crossed with C7 mice, C7 mice crossed with B6 mice, or B6 mice crossed with C7 mice. C7 mice were crossed with H19DMD-9CG mice in a B6 background. We grew MEFs in DMEM with GlutaMax (Gibco) with 10% FBS (Sigma) and penicillin/streptomycin. We counted MEFs and plated them at a density of 25,000 cells per well on Lab-Tek chambered coverglass (Thermo Scientific). Twenty-four hours after plating, adherent cells were washed with PBS, fixed with 4% formaldehyde in PBS, washed with PBS twice, and stored in 70% ethanol at 4°C. For coculture of MEFs and primary human foreskin fibroblasts (feeders) (American Type Culture Collection, CRL-2097), we plated 2000 MEFs together with 20,000 feeder cells, allowed them to grow for 72 h, and then fixed them as described above. For colonies from individual MEFs, primary foreskin fibroblasts were plated with 5000 feeder cells per well, and MEFs were diluted to a concentration of 0.5 cells per well. Colonies were grown for 14 d in 5% oxygen and then fixed as above.
5-aza-2′-deoxycytidine methylation inhibition
MEFs were isolated as above and plated at a cell density of 10,000 cells per well on Lab-Tek chambered coverglass slides (Thermo Scientific). Eighteen hours after plating, 1 µM 5-aza-2′-deoxycytidine (Invivogen) was added in cell culture medium. Cells were then cultured for 72 h before standard fixation.
Tissue harvest, sectioning, and fixation
We dissected neonates using standard techniques, and tissues were mounted in Tissue-Plus O.C.T. compound (Fisher Healthcare), flash-frozen in liquid nitrogen, and then stored at −80°C. Tissues were cryosectioned at 7 μm using a Leica CM1850 cryostat. We adhered tissue samples to positively charged Colorfrost plus slides (Fisher Scientific). We fixed and stored slide-mounted sections according to the same protocol as cultured cells with one minor addition. Tissue sections that were imaged with Igf2 were covered with Triton X-100 (Sigma) and then gently shaken in nuclease-free water (Ambion) for 30 min prior to probe hybridization to reduce background.
RNA probe design and synthesis
For each the five SNP positions between the B6 and C7 strains, we designed probes by matching free energies of hybridization as specified in Levesque et al. (2013). We optimized mask oligonucleotides to leave 10-base-pair (bp) overhangs for each of the SNP probes and pooled all five together to act as the complete allele-specific probe. We provide all oligonucleotide sequences in Supplemental Table 1. We coupled the allele-specific probes to the Cy3 or Cy5 fluorophores (GE Healthcare) and purchased H19 guide probes labeled with Cal fluor 610 (Biosearch Technologies). We coupled probes targeting Igf2 mRNA to Atto488.
RNA SNP FISH
We performed RNA SNP FISH as per Levesque et al. (2013) with minor modifications. Briefly, we incubated our cells overnight at 37°C in hybridization buffer (10% dextran sulfate, 2× saline–sodium citrate [SSC], 10% formamide) with 5 nM concentration of the B6 and C7 allele-specific probes and 15 nM concentration of the mask probe, ensuring excess mask for complete hybridization to the B6 and C7 probes. The following morning, we performed two washes in wash buffer (2× SSC, 10% formamide), each consisting of a 30-min incubation at 37°C. After the second wash, we rinsed once with 2× SCC and once with anti-fade buffer. Finally, we mounted the sample for imaging in an anti-fade buffer with catalase and glucose oxidase (Raj et al. 2008) to prevent photobleaching.
We performed RNA FISH on cell culture samples grown on a Lab-Tek chambered coverglass using 50 μL of hybridization solution spread into a thin layer with a coverslip and placed in a parafilm-covered culture dish with a moistened paper towel to prevent excessive evaporation. When performing RNA FISH on tissues, we added an additional clearing step involving the addition of 8% SDS in PBS before adding the hybridization buffer in order to reduce background (Yang et al. 2014).
Imaging
Samples were imaged on a Leica DMI600B automated wide-field fluorescence microscope equipped with a 100× Plan Apo objective, a Pixis 1024BR cooled charge-coupled device camera, and a Prior Lumen 220 light source. We imaged cells by taking a series of Z-stacks spaced by 0.35 µm. We tuned the exposure times depending on the dyes used: 2000 msec for the H19 guide probe, 4000 msec for the C6 and B6 allele-specific probes, and 4000 msec for the Atto488 probe.
Image analysis
We first segmented and thresholded images using a custom Matlab software suite (downloadable at https://bitbucket.org/arjunrajlaboratory/rajlabimagetools/wiki/Home). Segmentation of cells included the nuclear and cytoplasmic region. We fit each spot to a two-dimensional Gaussian profile specifically on the Z-plane on which it occurs in order to ascertain subpixel-resolution spot locations as well as intensity amplitudes. Colocalization took place in two stages: In the first stage, guide spots searched for the nearest-neighbor SNP probes within a 3.0-pixel (360-nm) window. We ascertained the median displacement vector field for each match and subsequently used it to correct for chromatic aberrations. After this correction, we used a more stringent 1.5-pixel (195-nm) radius to make the final determination of colocalization. For the 5-aza-2′-deoxycytidine and large colonies, cells were manually classified as monoallelic or biallelic based on the relative signal intensity in SNP channels. In order to test random colocalization due to spots occurring randomly by chance, we took our images and shifted the guide channel by adding 10 pixels (1.3 µm) to the X and Y coordinates and then performing colocalization.
Our pixel shift control did reveal that the rate of spurious colocalization could be higher in cells with a higher density of RNA molecules; for instance, in C7 × B6 H19+/DMD-9CG mutants MEFs, we observed a false colocalization rate of 15%–20% for cells with >500 H19 RNA in them but 3%–10% for cells with <500 H19 RNA.
DNA methylation analysis
Allele-specific bisulfite sequencing was performed as previously described using nested PCR for the H19 ICR (de Waal et al. 2014). Briefly, bisulfite mutagenesis was performed with 1 µg of isolated genomic DNA from MEFs using the Epitect bisulfite kit (Qiagen). For individual colony analysis with smaller quantities of DNA, the Epitech Plus kit was used. The bisulfite-treated DNA was used for PCR amplification using nested primers, and the amplified products were cloned into a vector using the Strataclone PCR cloning kit (Agilent), transformed into chemically competent Escherichia coli cells, and plated on LB plates with 0.1 mg/mL ampicillin. Three independent nested PCRs were performed from each sample. Recombinant plasmids were isolated and sequenced at the University of Pennsylvania DNA Sequencing Facility. Maternal and paternal alleles were distinguished by using multiple polymorphisms between the B6 and C7 alleles in the F1 hybrids as previously described (Tremblay et al. 1997; Market-Velker et al. 2010).
Allele-specific RT–PCR analysis
Total RNA was extracted from MEFs and reverse-transcribed as previously described (Ideraabdullah et al. 2014). Briefly, 2.5 ng of cDNA was used for all assays. Allele-specific expression was conducted following amplification of H19 using primers HE2 (TGATGGAGAGGACAGAAGGG) and HE4 (TTGATTCAGAACGAGACGGAC) and digestion by restriction enzyme Cac8I specified by polymorphisms between the B6 and C7 alleles as previously described (Thorvaldsen et al. 2006). Digested RT–PCR fragments were resolved on a 12% polyacrylamide gel. The C7 product was uncut and 235 bp, and the B6 product was cut with 173-bp and 62-bp fragments. The band intensities were quantified using ImageJ software (http://rsb.info.nih.gov/ij). Total Igf2 expression was confirmed by quantitative RT–PCR using the same total RNA and primers Igf2f (CGCTTCAGTTTGTCTGTTCG) and Igf2r (GCAGCACTCTTCCACGATG).
Supplementary Material
Acknowledgments
We thank all members of the Bartolomei and Raj laboratory for helpful suggestions and discussions. We particularly thank Long Cai for providing the protocol for background reduction in tissue. We thank Ian Mellis for his help spot-checking data and code. We also thank Dr. Joanne Thorvaldsen, Dr. Gerd Blobel, Dr. Shelley Berger, Dr. Ken Zaret, and Dr. Richard Schultz for helpful comments on the manuscript. P.G., J.M.K., M.S.B., and A.R. acknowledge support from the National Institutes of Health (1R33EB019767). J.M.K. acknowledges support from Alex's Lemonade Stand Foundation, the National Institutes of Health (KL2TR000139 and K08 CA193915), and St. Baldrick's Foundation. P.G. and A.R. acknowledge support from a National Institutes of Health Director's New Innovator award (1DP2OD008514). A.R. also acknowledges support from a Burroughs-Wellcome Fund Career Award at the Scientific Interface, a National Science Foundation CAREER award (1350601), and U01HL129998. M.S.B. also acknowledges support from GM051279.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.275958.115.
References
- Christman JK. 2002. 5-azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21: 5483–5495. [DOI] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH. 2006. Transcriptional pulsing of a developmental gene. Curr Biol 16: 1018–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Ramsköld D, Reinius B, Sandberg R. 2014. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343: 193–196. [DOI] [PubMed] [Google Scholar]
- de Waal E, Mak W, Calhoun S, Stein P, Ord T, Krapp C, Coutifaris C, Schultz RM, Bartolomei MS. 2014. In vitro culture increases the frequency of stochastic epigenetic errors at imprinted genes in placental tissues from mouse concepti produced through assisted reproductive technologies. Biol Reprod 90: 22–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel N, West AG, Felsenfeld G, Bartolomei MS. 2004. Antagonism between DNA hypermethylation and enhancer-blocking activity at the H19 DMD is uncovered by CpG mutations. Nat Genet 36: 883–888. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. 2004. The history of cancer epigenetics. Nat Rev Cancer 4: 143–153. [DOI] [PubMed] [Google Scholar]
- Gabory A, Jammes H, Dandolo L. 2010. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays 32: 473–480. [DOI] [PubMed] [Google Scholar]
- Gicquel C, Rossignol S, Cabrol S, Houang M, Steunou V, Barbu V, Danton F, Thibaud N, Le Merrer M, Burglen L, et al. 2005. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat Genet 37: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. 2007. Widespread monoallelic expression on human autosomes. Science 318: 1136–1140. [DOI] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC. 2005. Real-time kinetics of gene activity in individual bacteria. Cell 123: 1025–1036. [DOI] [PubMed] [Google Scholar]
- Gould TD, Pfeifer K. 1998. Imprinting of mouse Kvlqt1 is developmentally regulated. Hum Mol Genet 7: 483–487. [DOI] [PubMed] [Google Scholar]
- Hansen CH, van Oudenaarden A. 2013. Allele-specific detection of single mRNA molecules in situ. Nat Methods 10: 869–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideraabdullah FY, Thorvaldsen JL, Myers JA, Bartolomei MS. 2014. Tissue-specific insulator function at H19/Igf2 revealed by deletions at the imprinting control region. Hum Mol Genet 23: 6246–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalish JM, Conlin LK, Bhatti TR, Dubbs HA, Harris MC, Izumi K, Mostoufi-Moab S, Mulchandani S, Saitta S, States LJ, et al. 2013. Clinical features of three girls with mosaic genome-wide paternal uniparental isodisomy. Am J Med Genet A 161A: 1929–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalish JM, Jiang C, Bartolomei MS. 2014. Epigenetics and imprinting in human disease. Int J Dev Biol 58: 291–298. [DOI] [PubMed] [Google Scholar]
- Lee JT, Bartolomei MS. 2013. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152: 1308–1323. [DOI] [PubMed] [Google Scholar]
- Levesque MJ, Raj A. 2013. Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nat Methods 10: 246–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MJ, Ginart P, Wei Y, Raj A. 2013. Visualizing SNVs to quantify allele-specific expression in single cells. Nat Methods 10: 865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorthongpanich C, Cheow LF, Balu S, Quake SR, Knowles BB, Burkholder WF, Solter D, Messerschmidt DM. 2013. Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science 341: 1110–1112. [DOI] [PubMed] [Google Scholar]
- Mann MRW, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS. 2003. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod 69: 902–914. [DOI] [PubMed] [Google Scholar]
- Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MRW. 2010. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet 19: 36–51. [DOI] [PubMed] [Google Scholar]
- Perez JD, Rubinstein ND, Fernandez DE, Santoro SW, Needleman LA, Ho-Shing O, Choi JJ, Zirlinger M, Chen S-K, Liu JS, et al. 2015. Quantitative and functional interrogation of parent-of-origin allelic expression biases in the brain. Elife 4: e07860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. 2008. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135: 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. 2006. Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4: e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. 2008. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5: 877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Rifkin SA, Andersen E, van Oudenaarden A. 2010. Variability in gene expression underlies incomplete penetrance. Nature 463: 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinius B, Sandberg R. 2015. Random monoallelic expression of autosomal genes: stochastic transcription and allele-level regulation. Nat Rev Genet 16: 653–664. [DOI] [PubMed] [Google Scholar]
- Savova V, Vigneau S, Gimelbrant AA. 2013. Autosomal monoallelic expression: genetics of epigenetic diversity? Curr Opin Genet Dev 23: 642–648. [DOI] [PubMed] [Google Scholar]
- Shipony Z, Mukamel Z, Cohen NM, Landan G, Chomsky E, Zeliger SR, Fried YC, Ainbinder E, Friedman N, Tanay A. 2014. Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature 513: 115–119. [DOI] [PubMed] [Google Scholar]
- Singer ZS, Yong J, Tischler J, Hackett JA, Altinok A, Surani MA, Cai L, Elowitz MB. 2014. Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol Cell 55: 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. 2011. Mammalian genes are transcribed with widely different bursting kinetics. Science 332: 472–474. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Mann MRW, Nwoko O, Duran KL, Bartolomei MS. 2002. Analysis of sequence upstream of the endogenous H19 gene reveals elements both essential and dispensable for imprinting. Mol Cell Biol 22: 2450–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen JL, Fedoriw AM, Nguyen S, Bartolomei MS. 2006. Developmental profile of H19 differentially methylated domain (DMD) deletion alleles reveals multiple roles of the DMD in regulating allelic expression and DNA methylation at the imprinted H19/Igf2 locus. Mol Cell Biol 26: 1245–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay KD, Duran KL, Bartolomei MS. 1997. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol 17: 4322–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunster SJ, Jensen AB, John RM. 2013. Imprinted genes in mouse placental development and the regulation of fetal energy stores. Reproduction 145: R117–R137. [DOI] [PubMed] [Google Scholar]
- Verona RI, Thorvaldsen JL, Reese KJ, Bartolomei MS. 2008. The transcriptional status but not the imprinting control region determines allele-specific histone modifications at the imprinted H19 locus. Mol Cell Biol 28: 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen C-K, Lubeck E, Shah S, Cai L, Gradinaru V. 2014. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158: 945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Chan GC, Beeler D, Janes L, Spokes KC, Dharaneeswaran H, Mojiri A, Adams WJ, Sciuto T, Garcia-Cardeña G, et al. 2016. A role of stochastic phenotype switching in generating mosaic endothelial cell heterogeneity. Nat Commun 7: 10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.