Abstract
Murine double minute-2 (MDM2) is a negative regulator of P53, and its T309G polymorphism has been suggested as a risk factor for a variety of cancers. Increasing evidence has shown the association of MDM2 T309G polymorphism with head and neck carcinoma (HNC) risk. However, the results are inconsistent. Thus, we performed a meta-analysis to elucidate the association.
The meta-analysis retrieved studies published up to August 2015, and essential information was extracted for analysis. Separate analyses on ethnicity, source of controls, sample size, detection method, and cancer types were also conducted. Odds ratios (ORs) and their 95% confidence intervals (CIs) were used to estimate the association.
Pooled data from 16 case–control studies including 4625 cases and 6927 controls failed to indicate a significant association. However, in the subgroup analysis of sample sizes, an increased risk was observed in the largest sample size group (>1000) under a recessive model (OR = 1.52; 95% CI = 1.08–2.13). Increased risks were also found in the nasopharyngeal cancer in the subgroup analysis of cancer types (GG vs TT: OR = 2.07; 95% CI = 1.38–3.12; dominant model: OR = 1.48; 95% CI = 1.13–1.93; recessive model: OR = 1.76; 95% CI = 1.17–2.65).
The results suggest that homozygote GG alleles of MDM2 SNP309 may be a low-penetrant risk factor for HNC, and G allele may confer nasopharyngeal cancer susceptibility.
INTRODUCTION
Head and neck carcinoma (HNC) is the sixth most frequent type of malignant tumor worldwide; HNC is a group of biologically similar cancers that originate in the head and neck regions, such as oral cavity, pharyngeal cavity, and larynx.1 Nevertheless, the underlying mechanisms of this tumor are unclear. The possible etiological risk factors of HNC include smoking, drinking,2 papilloma virus infection,3 and betel quid chewing,4 as well as exposure to toxic substances.5 However, only a small proportion of the people exposed to these external factors eventually develop HNC, thus indicating that internal factors, such as gene variation, may play a role in its susceptibility.
Previous published meta-analyses assessed the association of several gene variations with HNC risk. Polymorphisms of MTHFR C677T,6 EPHX1 Tyr113His,7 and CYP1B1 Leu432Val8 have been suggested to increase HNC risk. However, a positive association of CCND1 G870A,9 XRCC1 Arg399Gln,10 and XPD Lys751Gln11 with HNC susceptibility was not indicated. Thus, the functions of different gene polymorphisms for HNC risk may differ because of their exact mechanisms.
P53 is an established tumor suppressor that is mutated in a variety of cancers.12 Murine double minute-2 (MDM2) is an important gene and a key negative regulator of P53; this gene is overexpressed in diverse cancer types,13 including HNC.14 MDM2 can interact and suppress P53, leading to P53 degradation through the ubiquitination pathway15; thus, MDM2 is speculated to be a potential target for cancer therapy.16
A MDM2 single nucleotide polymorphism (SNP) at the 309th nucleotide in the first intron (rs2279744), with a T to G change, may increase the affinity for stimulatory protein 1 binding and MDM2 expression, and subsequently activate the P53 pathway.17 The elevated MDM2 protein expression may mediate the ubiquitylation and proteasomal degradation of p53, and the ability of damaged cells is enhanced as a result to escape the cell-cycle checkpoint.18 Therefore, the neoplastic transformation of normal cells may be initiated.
An increasing number of studies have been conducted on the association of MDM2 T309G polymorphism with HNC risk. In 201119 and 2012,20 2 published meta-analyses focused on this association. Nevertheless, their results were inconsistent and conflicting. The meta-analysis of 7 studies published in 2011 showed that MDM2 SNP309 G allele probably functions as an HNC protective factor for Caucasians rather than Asians.19 The meta-analysis of 9 studies published in 2012 suggested that MDM2 SNP309 variation may not be a risk factor for HNC; however, increased risk was found in nasopharyngeal cancer when subgroup analysis was performed.20 Given these findings, this study performed an updated meta-analysis that included the most recent published data up to August 2015 to obtain a more precise estimate of the association.
MATERIALS AND METHODS
Literature Search Strategy
Medline, EMBASE, and Chinese National Knowledge Infrastructure without a language limitation were searched for publications published up to August 2015.
Combinations of the following keywords were used in the search: murine double minute-2 (MDM2), mouth, larynx, pharynx, nasopharynx, head and neck, neoplasm, tumor, cancer, variation, and polymorphism. All relevant studies were retrieved, and the bibliographies were checked for other possible publications.
Ethical approval or patient consent was not needed because this is a meta-analysis in which all data were extracted from published literature.
Inclusion Criteria
The following criteria were used for literature selection: a study on the association of MDM2 T309G polymorphism with HNC risk; case–control or cohort design; and available data on sample size, odds ratios (ORs), and 95% confidence intervals (CIs), as well as genetic distribution or information that readers can infer the results. Accordingly, studies with the following characteristics were excluded: different study design; unavailability of significant information for data collection and analysis; and reviews or duplicate publications. After systematic search and selection, we reviewed all papers in accordance with the criteria mentioned above for further analysis.
Data Extraction
Eligible publications were independently reviewed by 2 authors according to the inclusion criteria. Necessary information was extracted into a database. For any discrepancies, a discussion was made to reach an agreement. During this process, if a conflicting evaluation still existed, another author was consulted to resolve the dispute, and then a final decision was made by the majority of the votes.
Statistical Analysis
The ORs of the association between MDM2 T309G polymorphism and HNC risk were calculated for the included studies. Three genetic models were used for pooling the ORs, namely, a homozygote comparison model (GG vs TT), a dominant model (GG + GT vs TT), and a recessive model (GG vs GT + TT), respectively. The OR and its 95% CI for each study were plotted against the number of participants in order to detect any possible sample size bias. The between-study heterogeneity was estimated by a Chi-square based Q statistic test. A P-value of the Q test less than 0.1 was considered the existence of heterogeneity. In this situation, a random-effect model (DerSimonian and Laird)21 was selected for data pooling. Conversely, a fixed-effect model (Mantel and Haenszel)22 was used if the P-value was more than 0.1. The statistical significance of overall effect size estimate was assessed by a Z test. The Hardy–Weinberg equilibrium (HWE) of the controls was evaluated by Fisher exact test. To assess the potential influences of the publication bias on the results, funnel plots were generated.23 An asymmetrical plot usually indicates the existence of the publication bias. Egger linear regression test24 was performed to evaluate the symmetry of the plot. All statistical analysis was carried out by using the STATA 11.0 software (Stata Corporation, College Station, TX). A P of less than 0.05 was considered statistically significant, except where otherwise specified.
RESULTS
Study Characteristics
Publications were retrieved and screened on the basis of the criteria. As shown in Figure 1, 298 publications were identified, of which 270 irrelevant papers were initially excluded. Afterward, 3 review articles19,20,25 and 2 studies26,27 that focused on other genetic variations rather than SNP309 polymorphism were discarded. Five noncase–control studies28–31 and 2 studies with insufficient data32,33 were further excluded. Consequently, 16 studies were selected for data extraction and assessment.34–49
FIGURE 1.
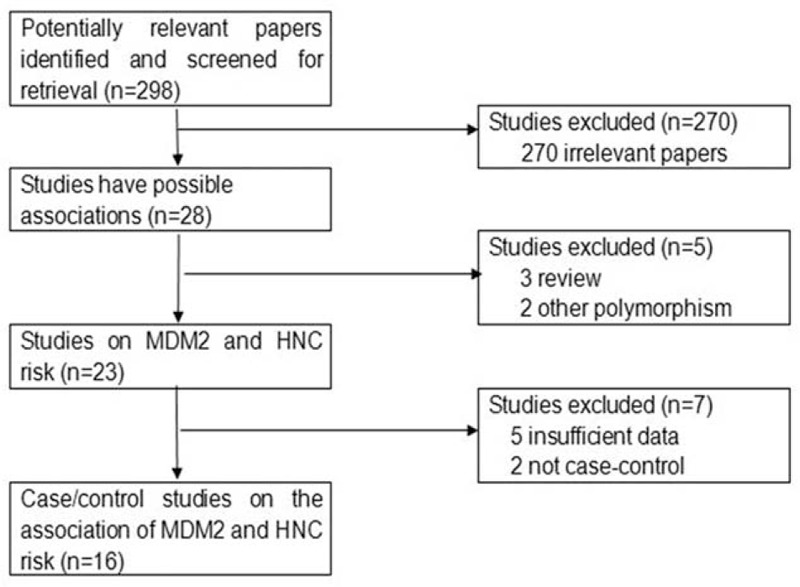
The flow diagram of included/excluded studies.
All of the selected papers were written in English. We established a database according to the extracted information from each study. The necessary information is listed in Table 1, which contains the first author and the number and characteristics of cases and controls for each study as well as other necessary data. The selected studies included 6 groups of Caucasians,34,42–44,46,49 8 groups of Asians,35–41,48 and 2 groups of mixed populations.45,47
TABLE 1.
Characteristics of Studies Included in the Meta-Analysis
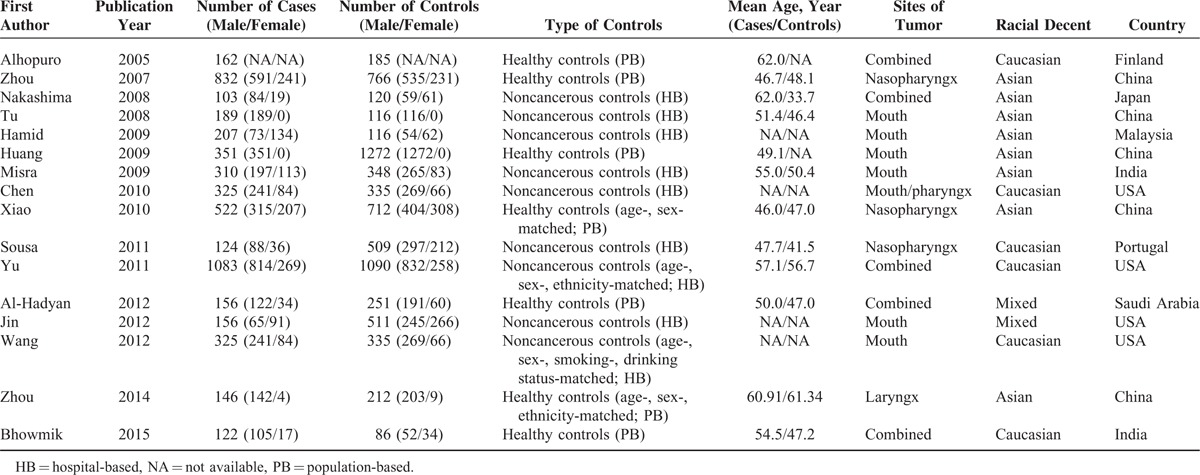
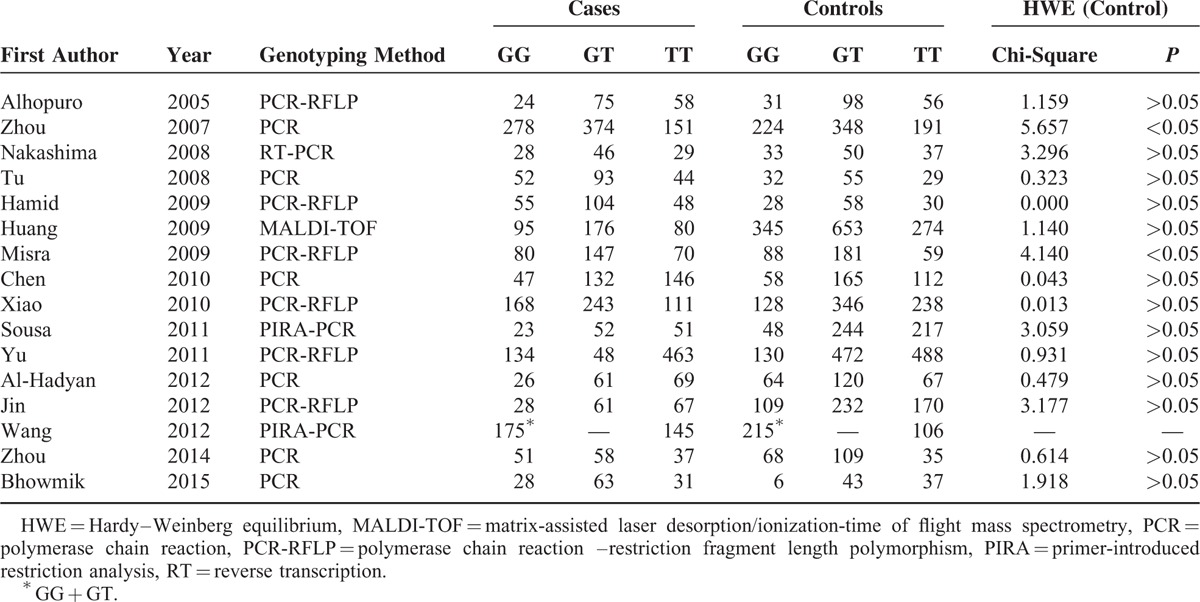
Table 2 presents the distributions of MDM2 T309G genotype and the genotyping methods of the selected studies. The genetic distributions of the control groups in all studies were in accordance with HWE except for 2 studies.35,40 Notably, the distributions of GG and GT were combined as GG + GT in the study of Wang et al46; therefore, the relevant data were included only in the dominant model assessment.
TABLE 2.
Distribution of MDM2 SNP309 Genotype Among HNC Cases and Controls Included in the Meta-Analysis
Meta-Analysis Results
The main results of the meta-analysis are shown in Table 3. Random-effect models were used because an evident heterogeneity was observed in the 3 genetic models. The pooled data based on 4625 cases and 6927 controls in a homozygote comparison (OR = 1.08; 95% CI = 0.81–1.44), and dominant (OR = 0.85; 95% CI = 0.63–1.15) and recessive (OR = 1.20; 95% CI = 0.97–1.48) genetic models failed to demonstrate a marked association of MDM2 T309G polymorphism with HNC risk, indicating that this polymorphism may not confer susceptibility to HNC (Figure 2).
TABLE 3.
Main Results of the Pooled Data in the Meta-Analysis
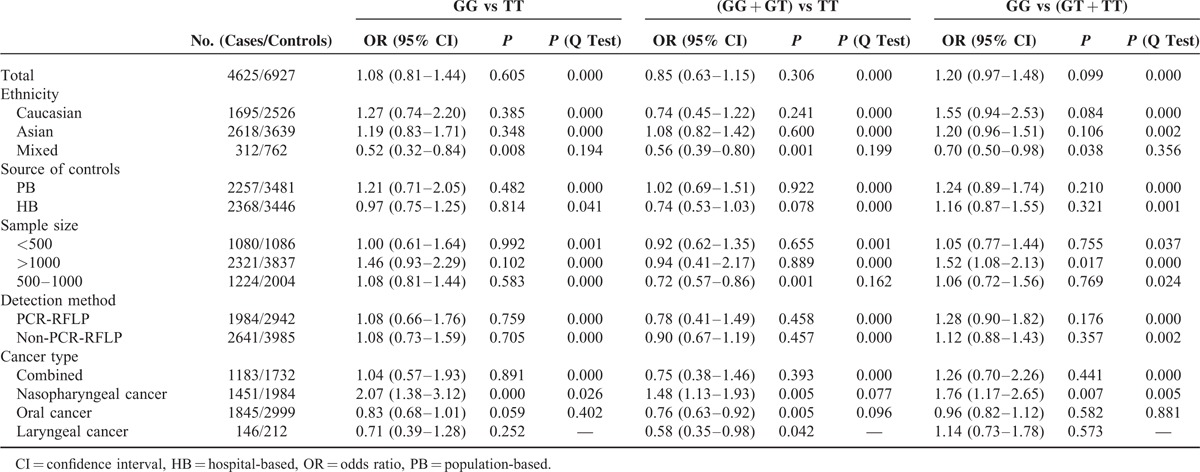
FIGURE 2.
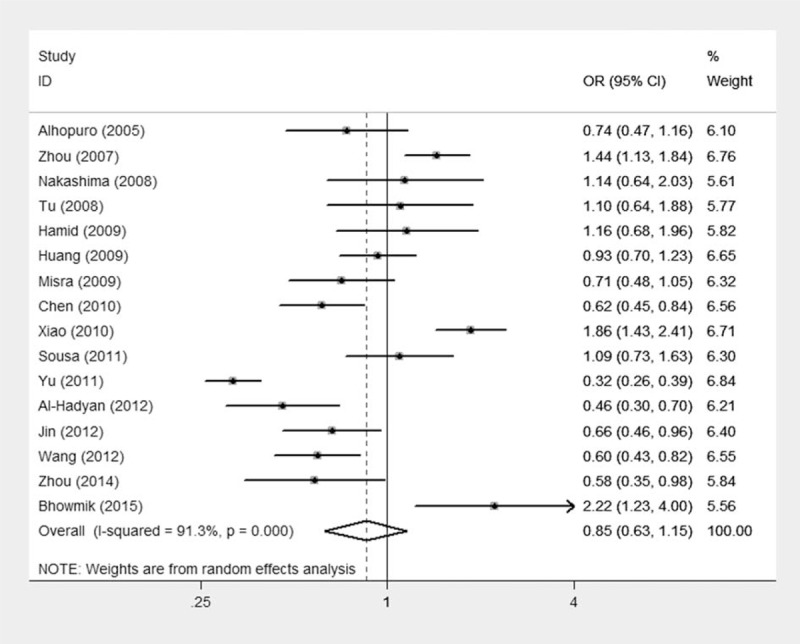
Meta-analysis for the association of HNC risk with MDM2 T309G polymorphism (GG + GT vs TT; overall data).
To address the possible effects of confounding factors on the overall results, we conducted subgroup analyses. When data were divided by ethnicity in the subgroup analyses, no associations were observed among Asians, Caucasians, and mixed ethnicities in accordance with the overall data. Similar results were found in the subgroup analyses of source of controls and detection method. Notably, when the overall data were divided by sample size, an increased cancer risk existed in the group of “greater than 1000” (>1000) under a recessive model (OR = 1.52; 95% CI = 1.08–2.13). This result indicates that individuals carrying GG alleles may have an increased HNC risk compared with those bearing T allele. Similar results could not be achieved in the other 2 subgroups (<500 or 500–1000).
In the subgroup of cancer types, an increased risk was observed in the nasopharyngeal carcinoma group (GG vs TT: OR = 2.07; 95% CI = 1.38–3.12; dominant model: OR = 1.48; 95% CI = 1.13–1.93; recessive model: OR = 1.76; 95% CI = 1.17–2.65) but not in other cancer types under the three genetic models. This finding suggests that individuals who carry variant G allele may have an excess nasopharyngeal cancer risk compared with those who harbor wild-type T allele.
Sensitivity Analysis and Bias Diagnostics
To determine the stability of the results, we conducted sensitivity analysis. We repeated the analysis by removing the studies whose genetic distributions in controls were not in line with HWE and found that the results were not overturned. One-way sensitivity analysis50 was then selected to evaluate the robustness of the data by omitting any single study each time in the repeated analyses. The results also supported the notion that the overall data were stable. Afterward, funnel plots were created to evaluate possible publication bias. The plots appeared to be visually symmetrical, and the symmetries were further confirmed by the Egger linear regression tests (Figure 3), in which the P values were more than 0.05 and all the 95% CIs of the regression curves included the origins (homozygote comparison: t = − 0.79, P = 0.443; dominant model: t = 0.81, P = 0.431; recessive model: t = − 0.91, P = 0.381), indicating that results are not easily affected by publication bias.
FIGURE 3.
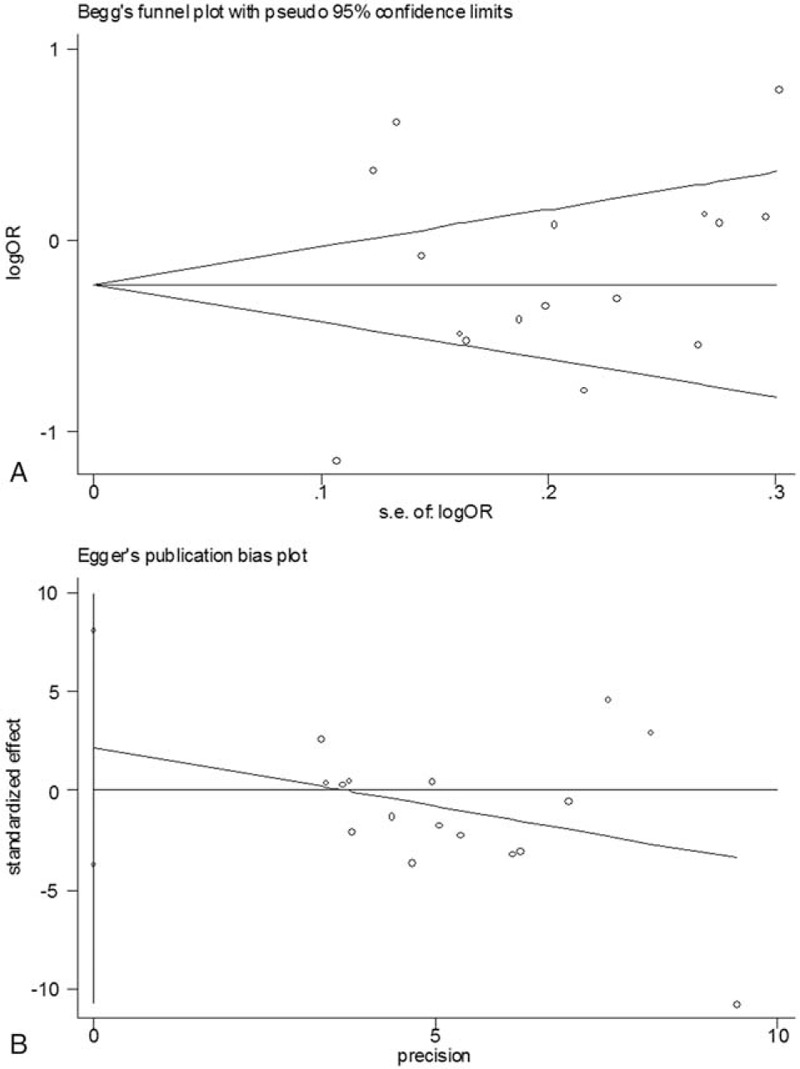
Publication bias test for the overall data (GG + GT vs TT; (A) Funnel plot; (B) Egger linear regression test).
DISCUSSION
We performed an updated meta-analysis that assessed the association of MDM2 T309G with HNC risk. The results showed that GG alleles may increase HNC risk. Subgroup analyses revealed that G allele may increase nasopharyngeal carcinoma risk.
The sample sizes of the present study were markedly higher (5190 cases and 7352 controls) than those of previous published meta-analyses performed in 2011 (1629 cases and 2472 controls)19 and 2012 (2755 cases and 4121 controls).20 The data of the present meta-analysis may be more convincing because of the much larger number of included studies and involved participants. Moreover, more confounding factors were considered in the present meta-analysis, and the sensitivity analysis as well as the publication bias tests indicated the robustness of the results. In a more recent meta-analysis of nasopharyngeal carcinoma published in 2015 by Yang et al,51 their results showed that MDM2 T309G polymorphism may not be associated with nasopharyngeal cancer risk. However, Yang et al51 included an unpublished academic dissertation that had not been peer-reviewed by an open-published journal. An important study on nasopharyngeal carcinoma by Zhou et al35 was also missed. Hence, the data in the present meta-analysis may increase power to obtain a more precise estimate of the association.
In the subgroup analysis of sample sizes, increased cancer risk was observed in the group (>1000) under a recessive model. This finding indicates the function of G allele in increasing cancer susceptibility. The data in this group (>1000) may be more credible than the other groups (<500 or 500–1000) because larger sample size may help statistically increase power for obtaining a precise estimate. Interestingly, the increased risk was also found in nasopharyngeal cancer but not in other cancers when the data were stratified by cancer types. This discrepancy may be attributed to several reasons. First, variant G allele of MDM2 SNP309 is associated with increased MDM2 expression.52 Second, unlike other types of cancer, such as oral cancer and laryngeal cancer, most of the nasopharyngeal cancer cases are poorly differentiated instead of well-differentiated. MDM2 expression has a relationship with advanced T stages and poor cancer differentiation,53 especially with cancer tissues with increased cellularity and atypia.54 Compared with other cancer types, MDM2 appears to be highly expressed in nasopharyngeal cancers with poor differentiation.55 Third, a great proportion of nasopharyngeal cancer cases correlate with Epstein–Barr virus (EBV) infection; MDM2 expression may interact with EBV, and cancer genesis and development are promoted as a result. For instance, EBV encodes EBNA-5 protein, and MDM2 serves as a connection that facilitates the formation of the trimolecular protein EBNA-5–MDM2–P53. Thus, EBNA-5 inhibits P53 polyubiquitination in a concentration-dependent manner and expediently impairs P53 activity.56 Moreover, MDM2 is capable of self-ubiquitination, and it is often degraded by ubiquitin proteasome to maintain the upregulation and downregulation balance of its levels in cells. MDM2 may be augmented by EBV-encoded latent membrane protein 1 and accumulated as ubiquitinated species in cells.57 In addition, EBV may help MDM2 to become resistant to its antagonist.58 The above points may help clarify the reasons why the G allele of MDM2 SNP309 may increase nasopharyngeal carcinoma susceptibility.
Notably, several limitations may be considered for the interpretation of results. First, only studies on Caucasians, Asians, and mixed populations were involved in the present analysis, although the race origin barely affected the overall results. Data on other ethnicities must be evaluated to determine the potential effects of ethnic variation on HNC susceptibility. Second, given that hospital-based (HB) controls were used in several selected studies, nondifferential misclassification bias may inevitably exist. This bias may exist because the individuals may not be a full representative of the whole population. Moreover, the controls in several studies were not well-matched to the cases. However, the subgroup analysis on this subject did not statistically show any difference from the overall result. Third, several important confounding factors, such as age, gender, smoking, drinking, and HPV infection, were not considered for subgroup analyses because relevant information was insufficient in the primary reports. Few included studies reported detailed data about the interaction between MDM2 polymorphism and P53 mutational status, which is an important factor for cancer risk. Thus, subgroup analysis on this subject could not be assessed, and future studies with large sample sizes and more confounding factors are needed to elucidate the association.
In summary, the data in the present meta-analysis indicated that homozygote GG alleles of MDM2 T309G polymorphism may be a risk factor for HNC, and G allele may confer the susceptibility to nasopharyngeal carcinoma. Further investigations are necessary to obtain more precise results.
Footnotes
Abbreviations: CI = confidence interval, EBV = Epstein–Barr virus, HB = hospital-based, HNC = head and neck cancer, HWE = Hardy–Weinberg equilibrium, MDM2 = murine double minute-2, OR = odds ratio, PB = population-based, SNP = single nucleotide polymorphism.
Author contributions: XZ and XZ designed and planned the analysis; ZX and QL wrote the manuscript draft; ZX and HY processed data and prepared the figures and tables. All authors reviewed the manuscript.
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Specenier P, Vermorken JB. Cetuximab: its unique place in head and neck cancer treatment. Biologics 2013; 7:77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toporcov TN, Znaor A, Zhang ZF, et al. Risk factors for head and neck cancer in young adults: a pooled analysis in the INHANCE consortium. Int J Epidemiol 2015; 44:169–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Field N, Lechner M. Exploring the implications of HPV infection for head and neck cancer. Sex Transm Infect 2015; 91:229–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Little MA, Pokhrel P, Murphy KL, et al. The reasons for betel-quid chewing scale: assessment of factor structure, reliability, and validity. BMC Oral Health 2014; 14:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choudhury JH, Ghosh SK. Gene-environment interaction and susceptibility in head and neck cancer patients and in their first-degree relatives: a study of Northeast Indian population. J Oral Pathol Med 2015; 44:495–501. [DOI] [PubMed] [Google Scholar]
- 6.Zhuo X, Song J, Li D, et al. MTHFR C677T polymorphism interaction with heavy alcohol consumption increases head and neck carcinoma risk. Sci Rep 2015; 5:10671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Ge L, Sui Q, et al. Systematic review and meta-analysis of the relationship between EPHX1 polymorphisms and the risk of head and neck cancer. PLoS ONE 2015; 10:e0123347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen M, Hu YY, Hu YK, et al. Quantitative assessment of the influence of CYP1B1 polymorphisms and head and neck squamous cell carcinoma risk. Tumour Biol 2014; 35:3891–3897. [DOI] [PubMed] [Google Scholar]
- 9.Lin H, Fang L, Lin D. Association of cyclin D1 variants with head and neck cancer susceptibility: evidence from a meta-analysis. Asian Pac J Cancer Prev 2014; 15:5645–5651. [DOI] [PubMed] [Google Scholar]
- 10.Liu C, Yin Q, Jiao G, et al. Association between the XRCC1 Arg399Gln polymorphism and head and neck cancer susceptibility: a meta-analysis based on case-control studies. DNA Cell Biol 2014; 33:378–387. [DOI] [PubMed] [Google Scholar]
- 11.Lin H, Lin D, Zheng C. Association of XPD Lys751Gln polymorphism with head and neck cancer susceptibility: evidence from 11,443 subjects. Diagn Pathol 2014; 9:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hainaut P. p53 in metabolism, aging and cancer. Ann Dermatol Venereol 2014; 141 (Suppl. 12):S200–S201. [DOI] [PubMed] [Google Scholar]
- 13.Berberich SJ. Mdm2 and MdmX involvement in human cancer. Subcell Biochem 2014; 85:263–280. [DOI] [PubMed] [Google Scholar]
- 14.Denaro N, Lo Nigro C, Natoli G, et al. The role of p53 and MDM2 in head and neck cancer. ISRN Otolaryngol 2011; 2011:931813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poyurovsky MV, Katz C, Laptenko O, et al. The C terminus of p53 binds the N-terminal domain of MDM2. Nat Struct Mol Biol 2010; 17:982–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q, Zeng SX, Lu H. Targeting p53-MDM2-MDMX loop for cancer therapy. Subcell Biochem 2014; 85:281–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bond GL, Hu W, Bond EE, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 2004; 119:591–602. [DOI] [PubMed] [Google Scholar]
- 18.Meek DW. Regulation of the p53 response and its relationship to cancer. Biochem J 2015; 469:325–346. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Zheng Y, Lei D, et al. MDM2 309T > G polymorphism and risk of squamous cell carcinomas of head and neck: a meta-analysis. Asian Pac J Cancer Prev 2011; 12:1899–1903. [PubMed] [Google Scholar]
- 20.Zhang Y, Bai Y, Guan J, et al. The MDM2 309 T/G polymorphism is associated with head and neck cancer risk especially in nasopharyngeal cancer: a meta-analysis. Onkologie 2012; 35:666–670. [DOI] [PubMed] [Google Scholar]
- 21.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986; 7:177–188. [DOI] [PubMed] [Google Scholar]
- 22.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959; 22:719–748. [PubMed] [Google Scholar]
- 23.Munafo MR, Clark TG, Flint J. Assessing publication bias in genetic association studies: evidence from a recent meta-analysis. Psychiatry Res 2004; 129:39–44. [DOI] [PubMed] [Google Scholar]
- 24.Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997; 315:629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hildesheim A, Wang CP. Genetic predisposition factors and nasopharyngeal carcinoma risk: a review of epidemiological association studies, 2000-2011: Rosetta Stone for NPC: genetics, viral infection, and other environmental factors. Semin Cancer Biol 2012; 22:107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canova C, Hashibe M, Simonato L, et al. Genetic associations of 115 polymorphisms with cancers of the upper aerodigestive tract across 10 European countries: the ARCAGE project. Cancer Res 2009; 69:2956–2965. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Ma K. Association between MDM2 rs769412 and rs937283 polymorphisms with alcohol drinking and laryngeal carcinoma risk. Int J Clin Exp Pathol 2015; 8:7436–7440. [PMC free article] [PubMed] [Google Scholar]
- 28.Vivenza D, Gasco M, Monteverde M, et al. MDM2 309 polymorphism predicts outcome in platinum-treated locally advanced head and neck cancer. Oral Oncol 2012; 48:602–607. [DOI] [PubMed] [Google Scholar]
- 29.Farnebo L, Tiefenbock K, Ansell A, et al. Strong expression of survivin is associated with positive response to radiotherapy and improved overall survival in head and neck squamous cell carcinoma patients. Int J Cancer 2013; 133:1994–2003. [DOI] [PubMed] [Google Scholar]
- 30.Jin L, Sturgis EM, Zhang Y, et al. Genetic variants in p53-related genes confer susceptibility to second primary malignancy in patients with index squamous cell carcinoma of head and neck. Carcinogenesis 2013; 34:1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie X, Jin H, Hu J, et al. Association between single nucleotide polymorphisms in the p53 pathway and response to radiotherapy in patients with nasopharyngeal carcinoma. Oncol Rep 2014; 31:223–231. [DOI] [PubMed] [Google Scholar]
- 32.Sam KK, Gan CP, Yee PS, et al. Novel MDM2 splice variants identified from oral squamous cell carcinoma. Oral Oncol 2012; 48:1128–1135. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Chen X, Zhai Y, et al. Combined effects of genetic variants of the PTEN, AKT1, MDM2 and p53 genes on the risk of nasopharyngeal carcinoma. PLoS ONE 2014; 9:e92135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alhopuro P, Ylisaukko-Oja SK, Koskinen WJ, et al. The MDM2 promoter polymorphism SNP309T–>G and the risk of uterine leiomyosarcoma, colorectal cancer, and squamous cell carcinoma of the head and neck. J Med Genet 2005; 42:694–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou G, Zhai Y, Cui Y, et al. MDM2 promoter SNP309 is associated with risk of occurrence and advanced lymph node metastasis of nasopharyngeal carcinoma in Chinese population. Clin Cancer Res 2007; 13:2627–2633. [DOI] [PubMed] [Google Scholar]
- 36.Tu HF, Chen HW, Kao SY, et al. MDM2 SNP 309 and p53 codon 72 polymorphisms are associated with the outcome of oral carcinoma patients receiving postoperative irradiation. Radiother Oncol 2008; 87:243–252. [DOI] [PubMed] [Google Scholar]
- 37.Huang SF, Chen IH, Liao CT, et al. Combined effects of MDM2 SNP 309 and p53 mutation on oral squamous cell carcinomas associated with areca quid chewing. Oral Oncol 2009; 45:16–22. [DOI] [PubMed] [Google Scholar]
- 38.Nakashima M, Kondo S, Shimizu Y, et al. Impact of MDM2 single nucleotide polymorphism on tumor onset in head and neck squamous cell carcinoma. Acta Otolaryngol 2008; 128:808–813. [DOI] [PubMed] [Google Scholar]
- 39.Hamid S, Yang YH, Peng KN, et al. MDM2 SNP309 does not confer an increased risk to oral squamous cell carcinoma but may modulate the age of disease onset. Oral Oncol 2009; 45:496–500. [DOI] [PubMed] [Google Scholar]
- 40.Misra C, Majumder M, Bajaj S, et al. Polymorphisms at p53, p73, and MDM2 loci modulate the risk of tobacco associated leukoplakia and oral cancer. Mol Carcinog 2009; 48:790–800. [DOI] [PubMed] [Google Scholar]
- 41.Xiao M, Zhang L, Zhu X, et al. Genetic polymorphisms of MDM2 and TP53 genes are associated with risk of nasopharyngeal carcinoma in a Chinese population. BMC Cancer 2010; 10:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X, Sturgis EM, Lei D, et al. Human papillomavirus seropositivity synergizes with MDM2 variants to increase the risk of oral squamous cell carcinoma. Cancer Res 2010; 70:7199–7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sousa H, Pando M, Breda E, et al. Role of the MDM2 SNP309 polymorphism in the initiation and early age of onset of nasopharyngeal carcinoma. Mol Carcinog 2011; 50:73–79. [DOI] [PubMed] [Google Scholar]
- 44.Yu H, Huang YJ, Liu Z, et al. Effects of MDM2 promoter polymorphisms and p53 codon 72 polymorphism on risk and age at onset of squamous cell carcinoma of the head and neck. Mol Carcinog 2011; 50:697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Hadyan KS, Al-Harbi NM, Al-Qahtani SS, et al. Involvement of single-nucleotide polymorphisms in predisposition to head and neck cancer in Saudi Arabia. Genet Test Mol Biomarkers 2012; 16:95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Sturgis EM, Zhang Y, et al. Combined p53-related genetic variants together with HPV infection increase oral cancer risk. Int J Cancer 2012; 131:E251–E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin L, Xu L, Song X, et al. Genetic variation in MDM2 and p14ARF and susceptibility to salivary gland carcinoma. PLoS ONE 2012; 7:e49361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou J, Liu F, Zhang D, et al. Significance of MDM2-309 polymorphisms and induced corresponding plasma MDM2 levels in susceptibility to laryngeal squamous cell carcinoma. DNA Cell Biol 2014; 33:88–94. [DOI] [PubMed] [Google Scholar]
- 49.Bhowmik A, Das S, Bhattacharjee A, et al. MDM2 and TP53 polymorphisms as predictive markers for head and neck cancer in northeast Indian population: effect of gene-gene and gene-environment interactions. Asian Pac J Cancer Prev 2015; 16:5767–5772. [DOI] [PubMed] [Google Scholar]
- 50.Tobias A. Assessing the influence of a single study in the meta-analysis estimate. Stata Tech Bull 1999; 8:15–17. [Google Scholar]
- 51.Yang J, Li L, Yin X, et al. The association between gene polymorphisms and risk of nasopharyngeal carcinoma. Med Oncol 2015; 32:398. [DOI] [PubMed] [Google Scholar]
- 52.Dong HJ, Fang C, Fan L, et al. MDM2 promoter SNP309 is associated with an increased susceptibility to chronic lymphocytic leukemia and correlates with MDM2 mRNA expression in Chinese patients with CLL. Int J Cancer 2012; 130:2054–2061. [DOI] [PubMed] [Google Scholar]
- 53.Sheng W, Dong M, Zhou J, et al. Cooperation among Numb, MDM2 and p53 in the development and progression of pancreatic cancer. Cell Tissue Res 2013; 354:521–532. [DOI] [PubMed] [Google Scholar]
- 54.Ware PL, Snow AN, Gvalani M, et al. MDM2 copy numbers in well-differentiated and dedifferentiated liposarcoma: characterizing progression to high-grade tumors. Am J Clin Pathol 2014; 141:334–341. [DOI] [PubMed] [Google Scholar]
- 55.Wu HC, Lu TY, Lee JJ, et al. MDM2 expression in EBV-infected nasopharyngeal carcinoma cells. Lab Invest 2004; 84:1547–1556. [DOI] [PubMed] [Google Scholar]
- 56.Kashuba E, Yurchenko M, Yenamandra SP, et al. Epstein-Barr virus-encoded EBNA-5 forms trimolecular protein complexes with MDM2 and p53 and inhibits the transactivating function of p53. Int J Cancer 2011; 128:817–825. [DOI] [PubMed] [Google Scholar]
- 57.Li L, Li Z, Zhou S, et al. Ubiquitination of MDM2 modulated by Epstein-Barr virus encoded latent membrane protein 1. Virus Res 2007; 130:275–280. [DOI] [PubMed] [Google Scholar]
- 58.Renouf B, Hollville E, Pujals A, et al. Activation of p53 by MDM2 antagonists has differential apoptotic effects on Epstein-Barr virus (EBV)-positive and EBV-negative Burkitt's lymphoma cells. Leukemia 2009; 23:1557–1563. [DOI] [PubMed] [Google Scholar]


