Abstract
Cleavage of APP (amyloid precursor protein) by BACE-1 (β-site APP cleaving enzyme-1) is the rate-limiting step in amyloid-beta (Aβ) production and a neuropathologic hallmark of Alzheimer's disease (AD); thus physical approximation of this substrate-enzyme pair is a critical event with broad biological and therapeutic implications. Despite much research, neuronal locales of APP/BACE-1 convergence and APP-cleavage remain unclear. Here we report an optical assay – based on fluorescence complementation – to visualize in-cellulo APP/BACE-1 interactions as a simple on/off signal. Combined with other assays tracking the fate of internalized APP in hippocampal neurons, we found that APP/BACE-1 interact in both biosynthetic and endocytic compartments; particularly along recycling-microdomains such as dendritic spines and presynaptic boutons. In axons, APP and BACE-1 are co-transported, and also interact during transit. Finally, our assay reveals that the AD-protective “Icelandic” mutation greatly attenuates APP/BACE-1 interactions, suggesting a mechanistic basis for protection. Collectively, the data challenge canonical models and provide concrete insights into long-standing controversies in the field.
Keywords: amyloid precursor protein (APP), β-site APP cleaving enzyme (BACE-1), Bimolecular fluorescence complementation, vesicle trafficking, recycling endosomes, Alzheimer's disease
Introduction
Extracellular Aβ deposition is a pathologic hallmark of AD and a central tenet of the decades old ‘amyloid cascade hypothesis’, positing that neuronal dysfunction, synapse loss, neurofibrillary degeneration, and the full manifestation of Alzheimer neuropathology is initiated by Aβ. Aβ is generated by sequential cleavage of APP by the enzymes β- and γ- secretases, with BACE-1-cleavage being the rate-limiting step in this pathway 1. Thus physical approximation of APP and BACE-1 is a required cell biological event in Aβ generation, and precise understanding of APP/BACE-1 interaction-sites as well as pathways leading up to this approximation is of high significance.
Where do wild-type (WT) APP and BACE-1 meet; and what is the site of β-cleavage in neurons? Despite much research, the answer remains unclear 1. A model currently favored is that after biogenesis in the Golgi, both APP and BACE-1 recycle with the plasma membrane and converge in early endosomes, and that β-cleavage of APP occurs in early endosomal compartments 2-4. However, several observations are inconsistent with this view, and it's unclear how the model applies to neurons – the relevant cell-type in this case. First, at steady state, much of the WT APP and BACE-1 in cell lines is located at the Golgi 5, and it's unclear why β-cleavage would be precluded there. Furthermore, APP and BACE-1 colocalize in multiple endocytic compartments in these cells, including early and recycling endosomes 6. Importantly, existing models of the amyloidogenic pathway are almost entirely based on experiments in non-neuronal cells that lack the anatomy of neurons – with elongated processes orders of magnitude longer than the microscopic cell bodies that resemble their non-neuronal counterparts. Anatomically and functionally distinct neuronal microdomains (such as dendritic spines and presynaptic specializations) are not considered in canonical models, and it is difficult to imagine that trafficking concepts that have emerged from studies in non-neuronal cells can be simply swapped to neurons.
In previous studies we visualized the trafficking of WT APP and BACE-1 in cultured hippocampal neurons using minimal and transient expression of fluorescent-tagged proteins 7. We found that in dendrites, BACE-1 is largely localized to recycling endosomes, where APP and BACE-1 can colocalize. However neuronal sites of APP/BACE-1 interaction and β-cleavage have remained unknown. Moreover, putative APP/BACE-1 interactions in axons and presynaptic sites were also not addressed. Here we report a tool we call the Optical Convergence of APP and BACE-1 (or OptiCAB) assay to directly visualize APP/BACE-1 interactions in-cellulo. Combining this with other assays that track the fate of internalized APP from the plasma-membrane, as well as two-color high resolution live imaging, our experiments offer concrete insights into long-standing questions and controversies in the field.
Results
Characterization of the OptiCAB assay
The OptiCAB assay is based on fluorescence complementation of Venus protein fragments (called Bimolecular Fluorescence Complementation or BiFC) - an established tool to visualize protein-protein interactions 8. In this assay, one partner of an interacting-pair is tagged to the N-terminus fragment of the Venus fluorescent protein (VN), while the other partner to the complimentary C-terminus (VC). Though individual VN/VC fragments are non-fluorescent, if and when the two interacting partners bind, Venus fragments are reconstituted and become fluorescent. We tagged the C-terminus of WT APP and BACE-1 to VN and VC respectively, following previously established tagging strategies (Fig 1a; see methods and Das et al., 2013 for details). A major advantage of this assay is that complementation is irreversible – ‘capturing’ transient interactions – and indeed this assay has been validated for several enzyme-substrate pairs (for instance, see 9, 10).
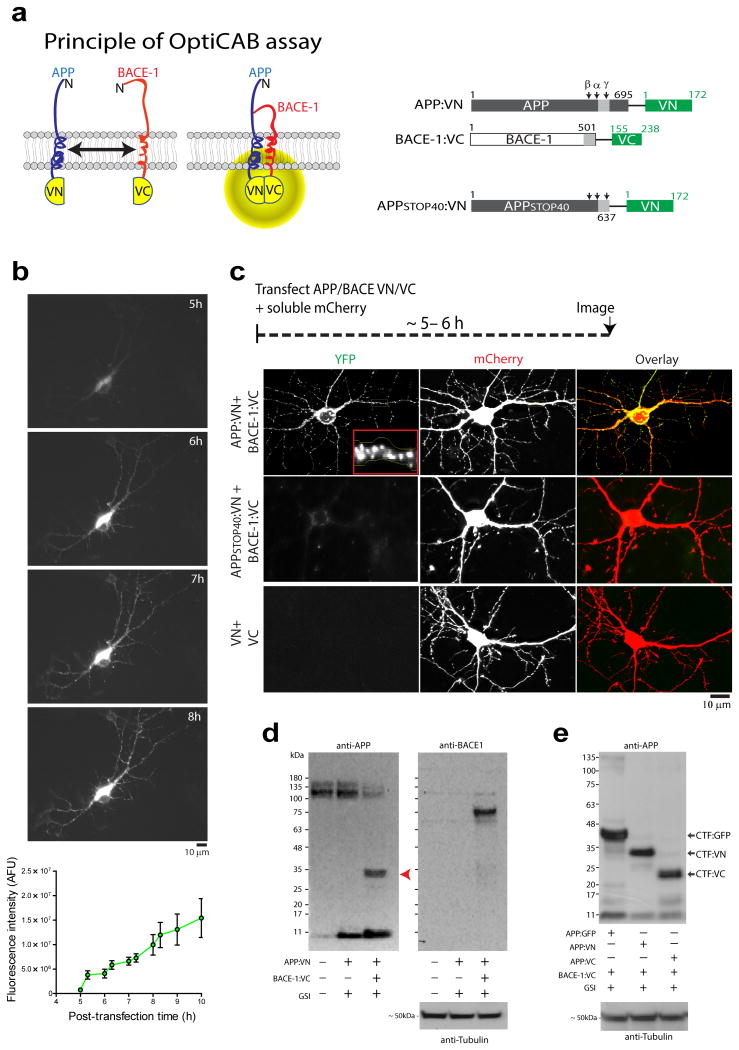
Figure 1. OptiCAB: An assay to detect APP/BACE-1 interactions in-situ.
(a) The principle: APP and BACE-1 were tagged to complementary (VN- or VC-) fragments of VFP. Note that interaction of APP/BACE-1 leads to reconstitution of VFP fluorescence. Cloning strategies on right, numbers denote amino-acid residues and light-grey box represents the transmembrane domain. (b) Hippocampal neurons were cotransfected with APP:VN and BACE-1:VC, and time course of complementation was evaluated. Post-transfection times (in hours) shown on upper right, intensities quantified below (N=7 neurons). Note the gradual increase in somatic and neuritic fluorescence over time. (c) Neurons transfected with APP:VN/BACE-1:VC and soluble mCherry (volume filler); note punctate fluorescence in soma and dendrites (top panels, zoomed in inset). Attenuated complementation was seen with an APP C-terminus mutant (APPSTOP40, middle panels, also see ‘results’). Individually transfected VN/VC fragments were non-fluorescent (bottom panels). (d) Cleavage of APP:VN by BACE-1:VC. BACE-1 knock out fibroblasts were transfected with APP:VN alone or APP:VN + BACE-1:VC (with a γ-secretase inhibitor, “GSI”) and APP-cleavage products were analyzed by Western blotting. Note that in presence of BACE-1:VC, APP:VN was cleaved to generate a fragment of ∼30 kDa (red arrowhead) – the expected β-cleavage product of APP:VN. Corresponding BACE-1 blot confirms knockdown. (e) Neuro2A cells were co-transfected with APP (APP:GFP, APP:VN or APP:VC) and BACE-1:VC in presence of a γ-secretase inhibitor and APP-cleavage products were analyzed by Western blotting. Note that the major bands are consistent with CTF:GFP (∼40 kDa), CTF:VN (∼30 kDa), and CTF:VC (∼22 kDa) respectively, indicating that β-cleavage is not influenced by APP/BACE-1 complementation.
First we transfected mature cultured hippocampal neurons with well-developed spines (days in vitro – DIV 10-14, isolated from post-natal mouse pups) with APP:VN and BACE-1:VC (APP/BACE-1-VN/VC) and soluble mCherry (volume filler); and examined the cells thereafter. The time-course of fluorescence evolution is shown in figure 1b. Note that fluorescence is first visible at ∼ 5 hours after expression, appearing in the soma and then along processes. Based on this, we decided to examine our neurons ∼ 5-6 hours after APP/BACE-1-VN/VC transfection (schematic on fig. 1c, top). It is important to note that at these early times the fluorescent proteins are just starting to fold and mature, and protein expression levels are at a minimum, thus it is unlikely that conclusions based on these experiments are confounded by over-expression artifacts (also see data with ‘crippled promoter’ below). APP/BACE-1-VN/VC fluorescence was punctate in dendrites resembling vesicles (fig. 1c – top panels).
Several observations argue that the fluorescence read-out of the OptiCAB assay is a consequence of APP/BACE-1 interactions, and not non-specific association of the Venus-fragments. The best way to determine specificity is to mutate one interacting-partner in a BiFC pair and ask if the fluorescence complementation is attenuated. Towards this we engineered an APP-fragment containing the β-cleavage site and membrane-spanning domain but lacking the C-terminus – suspected to mediate APP/BACE-1 associations [called APP-STOP40, see 11]. Neurons transfected with APP-STOP40:VN and BACE-1:VC were essentially non-fluorescent (fig. 1c, middle panels). Individually-transfected VN or VC fragments were also non-fluorescent (fig. 1c, bottom panel and data not shown). Note that these results are not due to variable expression of the transfected constructs as all tested constructs express adequately in neurons (Supp. fig. 1a). Further data shown below (APP:VN-cleavage by BACE-1:VC and effects of AD mutants) also strongly argue for the specificity of APP/BACE-1 interactions in our assay.
Does fluorescence complementation of APP:VN and BACE-1:VC lead to the cleavage of APP:VN? To address this, we co-transfected APP:VN and BACE-1:VC into cell-lines and analyzed APP-cleavage products by Western blotting (γ-secretase activity was blocked by the inhibitor BMS-299897). If the APP:VN protein was cleaved by BACE-1:VC in these experiments, one would expect to see a cleaved APP β–C-terminal fragment (β-CTF) linked to the ∼ 17-19 kDa VN protein (β-CTF:VN; note that the γ-secretase inhibitor would block further processing of this fragment). To eliminate potentially confounding effects of endogenous BACE-1, we first did these experiments in a fibroblast cell-line lacking BACE-1, generated from BACE -/- mice. As shown in figure 1d, cleavage of APP:VN by BACE-1:VC generates an expected ∼30 kD fragment consistent with β-CTF:VN (fig. 1d, red arrowhead). Similar results were obtained from neuro 2A cells (fig. 1e). Furthermore, note that cleavage patterns of various APP constructs also suggest that complementation does not influence β-cleavage (fig. 1e); and omission of the γ-secretase inhibitor leads to expected shifts in APP-cleavage (Supp. fig. 1b). Also, APP mutants linked to familial AD (“Arctic” and “London” mutations, known to enhance APP β-cleavage, see 12, 13) expectedly led to increases in β-CTF levels in the OptiCAB assay, though total fluorescence was not significantly enhanced in the short timeframes examined (4-6h, Supp. fig. 2a-b).
Figure 2. APP/BACE-1 interactions at the trans-Golgi network.
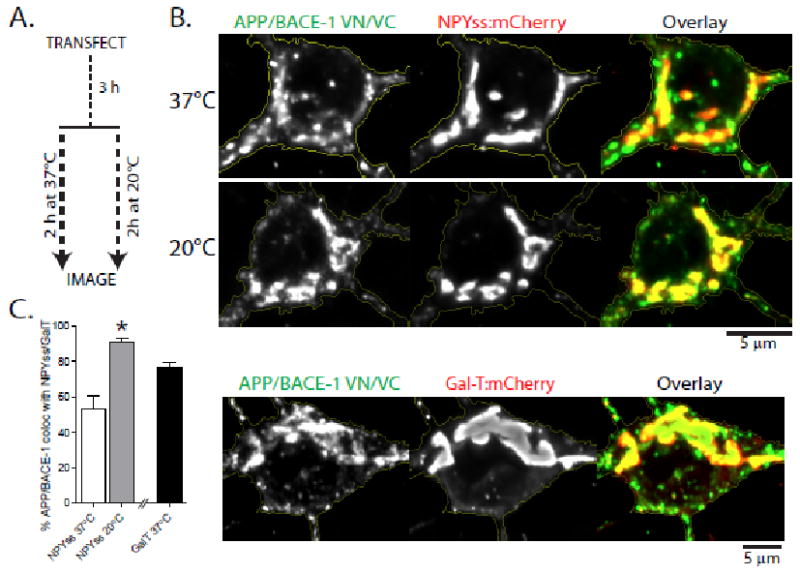
(a) Schematic: Hippocampal neurons were co-transfected with APP:VN/BACE-1:VC (to mark APP/BACE-1 interaction sites) and NPYss-mCherry (to label Golgi-derived organelles, see ‘results’); and a subset of neurons were incubated at 20 °C, known to ‘trap’ vesicles at the TGN. (b) Note colocalization of complemented APP/BACE-1 puncta with NPYss – enhanced upon temperature block. Complemented APP/BACE-1 also extensively colocalized with the TGN marker GalT. Fluorescence intensities are quantified in (C); * p = 0.0002
APP/BACE-1 interaction sites in somatodendritic domains
Precise sites where APP and BACE-1 meet within neurons are unknown. Previous studies in non-neuronal cells or neuronal cell-lines implicate a variety of locales including the ER, Golgi, endosomes (early, late, recycling) and lysosomes 14; and the OptiCAB assay offers an opportunity to pinpoint APP/BACE-1 interaction sites in hippocampal neurons. Our general strategy was to localize WT APP/BACE-1 interaction sites unambiguously by fluorescence complementation and determine its colocalization with established organelle markers. Somatic APP/BACE-1 VN/VC fluorescence was perinuclear, consistent with a TGN (trans-Golgi network)-type distribution. To confirm this, we cotransfected neurons with APP:VN + BACE-1:VC – to label interaction sites – and neuropeptide-Y signal-sequence (NPYss), a marker that labels the interior of Golgi and Golgi-derived organelles 15, 16. A subset of neurons was incubated at 20°C for 2h to block the export of proteins from the TGN (see strategy in fig. 2a). The colocalization of complemented APP/BACE-1 VN/VC with NPYss was significantly augmented after the temperature block (fig. 2b, top). Furthermore, complemented APP/BACE-1 also colocalized with GalT – a marker of TGN – confirming APP/BACE-1 interactions at this locale (fig. 2b, bottom – quantified in 2c).
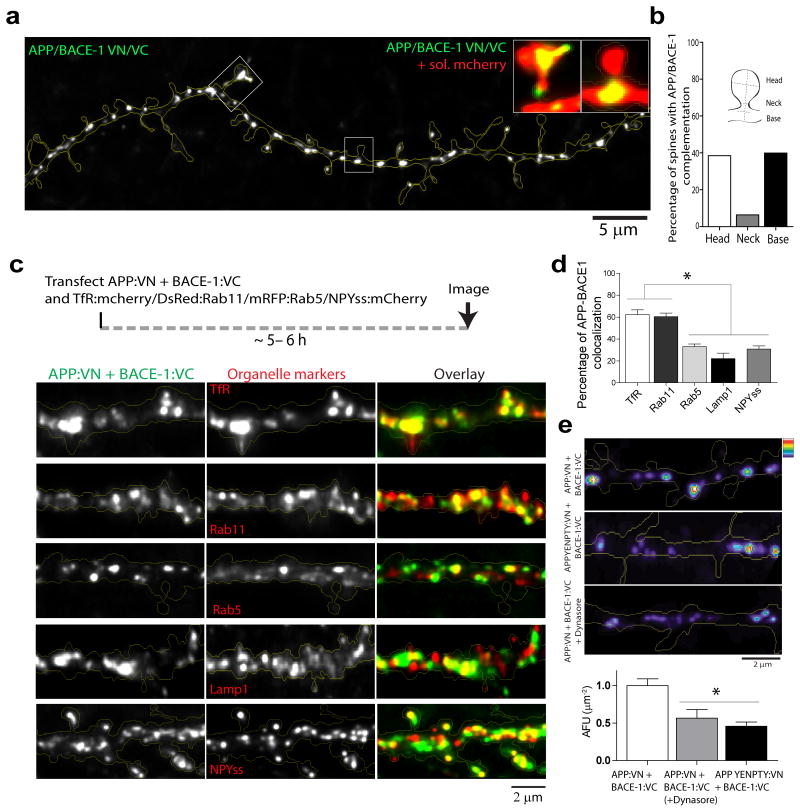
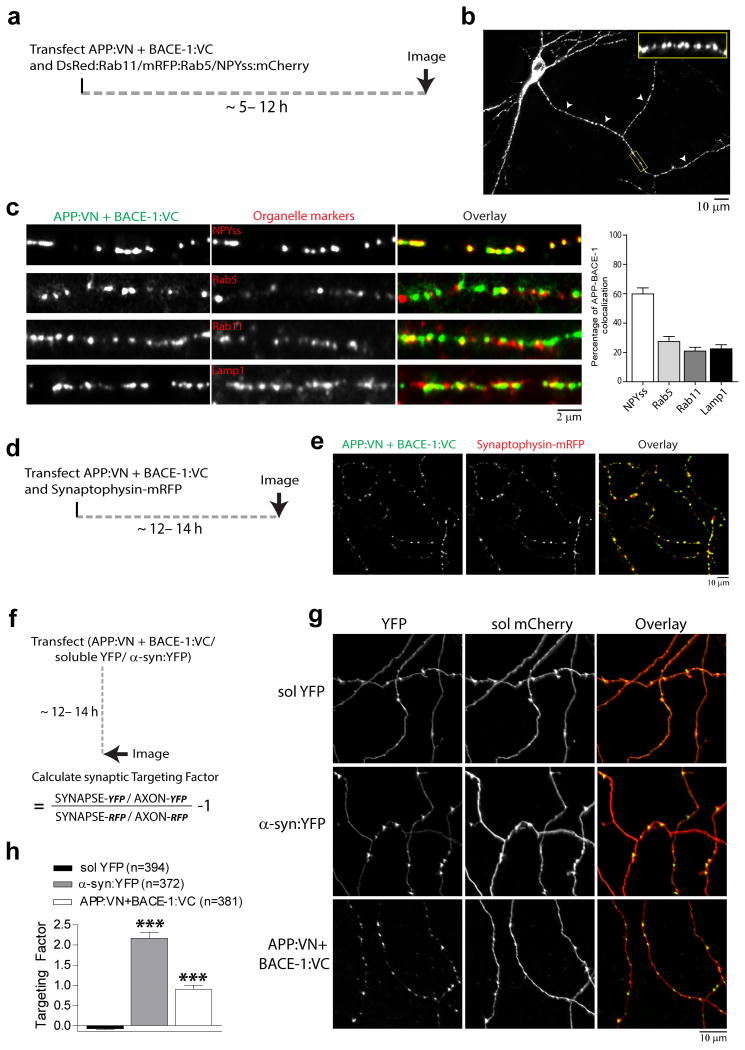
Next we focused on dendrites. As shown in figure 3a, punctate structures representing APP/BACE-1 interactions were seen throughout the dendritic profiles, including shafts and spines. Interestingly, ∼ 80% of the APP/BACE-1 complementation was seen in and around dendritic spines (fig. 3b), suggesting a role for these specialized recycling-microdomains in APP β-cleavage. To determine specific organelles where APP and BACE-1 interact, we co-transfected neurons with APP:VN + BACE-1:VC and various established organelle markers (Transferrin receptor – TfR and Rab11 for recycling endosomes, Rab5 for early endosomes, LAMP-1 for late-endosomes/lysosomes, and NPYss for Golgi-derived vesicles; also see Das et al., 2013) and analyzed the neurons after ∼ 5-6 hours (schematic, fig. 3c – top). As shown in figure 3c, complemented APP/BACE-1 most conspicuously colocalized with recycling endosome markers (∼ 60% colocalization with TfR and Rab11 as opposed to ∼ 20-30% with other markers – data quantified in fig. 3d). In previous studies, we and others found that at steady state, neuronal BACE-1 is largely localized to recycling endosomes 7, 17. A role for endocytosis in APP processing is well-established 18, and based on experiments evaluating the trafficking of APP and BACE-1 by live imaging, we proposed a model where APP undergoes endocytosis and meets BACE-1 in recycling endosomes 7. Consistent with this model, we found that APP/BACE-1 complementation was attenuated upon inhibition of APP endocytosis – either directly by mutation of a C-terminus APP endocytosis motif (YENPTY, see 19), or by a global inhibition of endocytosis by the dynamin inhibiting agent Dynasore (fig. 3e, quantified at bottom). These data are unlikely to be a consequence of over-expression, as APP/BACE-1 cDNA driven by weakened promoters lacking enhancer elements (‘broken CMV promoter’, see 20) also gave similar results (Supp. fig. 2c-d).
Figure 3. Subcellular sites of APP/BACE-1 interaction in dendrites of hippocampal neurons.
(a) Fluorescence puncta representing APP/BACE-1 interaction sites were seen throughout the dendritic shafts and spines (insets show complementation in spine head, neck and base). Note that many puncta are within spine heads or bases. (b) Quantification showing that ∼ 40% of APP/BACE-1 complementation occurs in close proximity to spines (283 spines from 12 neurons were analyzed). (c, d) To determine colocalization of APP/BACE-1 interaction sites with dendritic organelles, neurons were co-transfected with APP:VN/BACE-1:VC and the indicated organelle markers, and colocalization was determined quantitatively (see ‘methods’). Note that complemented APP/BACE-1 particles most conspicuously colocalized with the recycling endosome markers TfR and Rab11 (∼ 60%); and there was lesser (∼ 30%) colocalization with early endosomal (Rab5), lysosomal (LAMP-1), and Golgi-vesicle markers (NPYss). 20-24 dendrites from 12-18 neurons (two separate cultures) were analyzed; * p < 0.0001 (e) APP/BACE-1 complementation is also attenuated upon inhibiting clathrin-dependent endocytosis (by Dynasore) or by mutating an endocytosis motif in APP (APP-YENPTY, see ‘results’). 18-25 dendrites from 10-14 neurons (two separate cultures) were analyzed; * p = . 0.0148
APP/BACE-1 interaction sites in axons and presynapses
Next we examined WT APP/BACE-1 interactions in axons, examining colocalization of APP/BACE-1 BifC with organelle markers as above (schematic in fig. 4a). As shown in figure 4b, fluorescent puncta representing complemented APP/BACE-1 were seen throughout the length of the axon. However unlike dendrites, axonal APP/BACE-1 BifC puncta largely colocalized with NPYss – a marker of Golgi-derived vesicles – and colocalization with endosomes was lower (fig. 4c). Since dendritic APP/BACE-1 complementation was seen mostly in and around post-synaptic specializations where recycling occurs (fig. 3a), we asked if complementation was also seen in presynaptic en-passant boutons. The latter are major recycling sites in neurons, and are mainly localized along distal axons in our hippocampal cultures, where they can be easily identified by presynaptic markers 21. Towards this we first co-transfected neurons with APP/BACE-1 BifC and synaptophysin-mRFP (a marker for presynaptic boutons 21). Indeed almost all complemented APP/BACE-1 puncta colocalized with presynaptic boutons (fig. 4d-e). To determine the relative enrichment of APP/BACE-1 BifC at boutons, we used a quantitative algorithm (“targeting factor”) that compares levels of transfected proteins at boutons to the flanking axon, normalizing for potential variations in expression levels (fig 4f; also see 21, 22). As shown in figure 4g-h, presynaptic targeting of APP/BACE-1 BifC was significantly higher than a soluble marker, though expectedly lower than a classic presynaptic protein (α-synuclein). These data suggest that APP and BACE-1 interact at presynaptic boutons.
Figure 4. APP/BACE-1 interaction in axons and presynaptic boutons of hippocampal neurons.
(a, b) To determine colocalization of APP/BACE-1 interaction sites with axonal organelles, neurons were co-transfected with APP:VN/BACE-1:VC and the indicated organelle markers, and colocalization was determined quantitatively (see ‘methods’). Low power view of APP/BACE-1 BifC (arrowheads mark axon and section magnified in inset). (c) Complemented APP/BACE-1 particles most conspicuously colocalized with NPYss – a marker for Golgi-derived vesicles – while colocalization with endosomal markers was limited. 20-25 axons from two separate cultures were analyzed; * p = 0.0001 (d, e) To determine if presynapses are sites of APP/BACE-1 interaction, neurons were co-transfected with APP:VN/BACE-1:VC and synaptophysin:mRFP (to mark boutons). ∼ 96 % contained APP/BACE-1 BifC. 257 boutons from 15 neurons from two separate cultures were analyzed. (f-h) Quantitative analyses of APP/BACE-1 BifC presynaptic targeting, compared to a known presynaptic protein α-synuclein. Neurons were co-transfected with the respective BifC/YFP and soluble mCherry constructs, fluorescence at boutons was compared to flanking axons, normalizing for variations in expression (see “results” for details). Reprsentative images shown in (G); all quantified data shown in (H). Note presynaptic targeting of APP/BACE-1 BifC at boutons is significantly higher than soluble YFP, though expectedly not as high as α-synuclein (∼ 370-395 boutons from 20 neurons and two separate cultures were analyzed); p = 0.0001
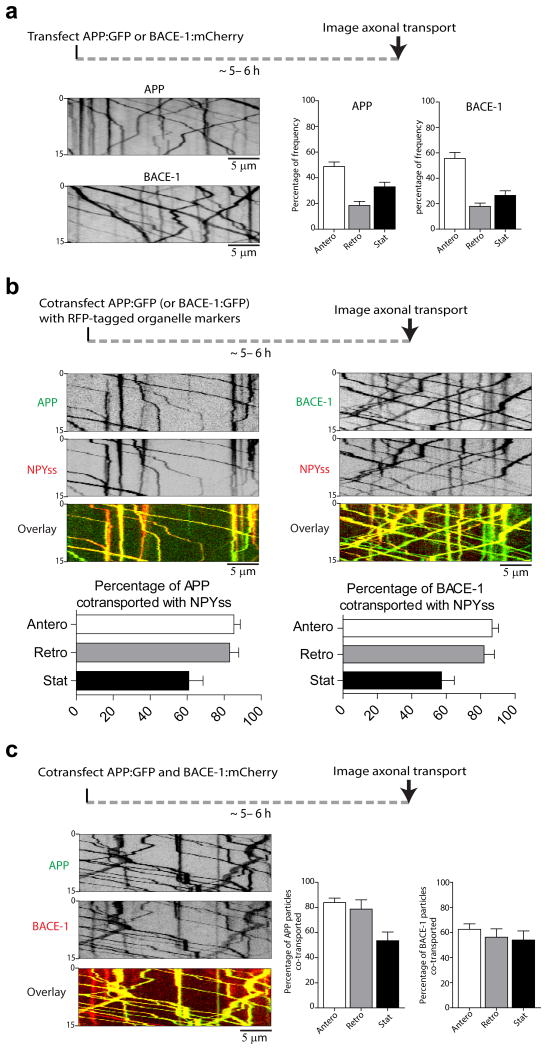
The physical interaction of APP and BACE-1 in axons suggests that they might also be transported in a common carrier. Towards this we first examined the axonal transport of fluorescent-tagged APP and BACE-1 individually, and also determined colocalization of mobile APP or BACE-1 vesicles with organelle markers (see schematic in fig. 5a-b). Transport kinetics of APP and BACE-1 containing vesicles were similar but not identical (fig. 5a), and motile particles were largely colocalized with NPYss, a marker for Golgi-derived vesicles (fig. 5b). However, colocalization of mobile axonal APP and BACE-1 vesicles with endosomal organelles was much lower (Supp. fig. 3, also see kinetic data in Supplementary Table 1). Finally we asked if APP and BACE-1 are co-transported in axons. Towards this we co-transfected neurons with APP:GFP and BACE-1:mCherry and examined their trafficking in axons by simultaneous dual-color imaging. The majority of moving APP vesicles (>80%) colocalized with BACE-1; but some BACE-1 vesicles were also transported independently (fig. 5c).
Figure 5. Axonal transport of APP and BACE-1.
(a) Neurons were transfected with either APP:GFP (left panels) or BACE-1:mCherry (right panels), and axonal transport was analyzed at a high temporal resolution (see “methods”). Schematic on top; representative kymographs with quantification shown below. Note similar kinetics of APP and BACE-1 transport.. Estimated velocities of APP vesicles: anterograde – 1.74 ± 0.03 μ/s, retrograde – 1.51 ± 0.06 μ/s and of BACE-1 vesicles: anterograde – 2.1 ± 0.06 μ/s, retrograde – 1.76 ± 0.07 μ/s. (∼ 360 APP and 300 BACE-1 vesicles from 20-22 neurons and two separate cultures were analyzed). (b) Neurons were co-transfected with APP:GFP (or BACE-1:GFP) and various RFP-tagged organelle markers as noted, and imaged live by simultaneous dual-cam imaging. Schematic on top; representative kymographs with quantification shown below. Note co-transport with NPYss, a marker for Golgi-derived vesicles. For APP/NPYss pair ∼ 320 APP and 390 NPYss particles (16-18 neurons from two separate cultures). For BACE-1/NPYss pair ∼ 410 BACE-1 and 500 NPYss (18-20 neurons from two separate cultures) were analyzed. (c) Neurons were co-transfected with APP:GFP and BACE-1:mCherry and imaged live by simultaneous dual-cam imaging. Note significant co-transport
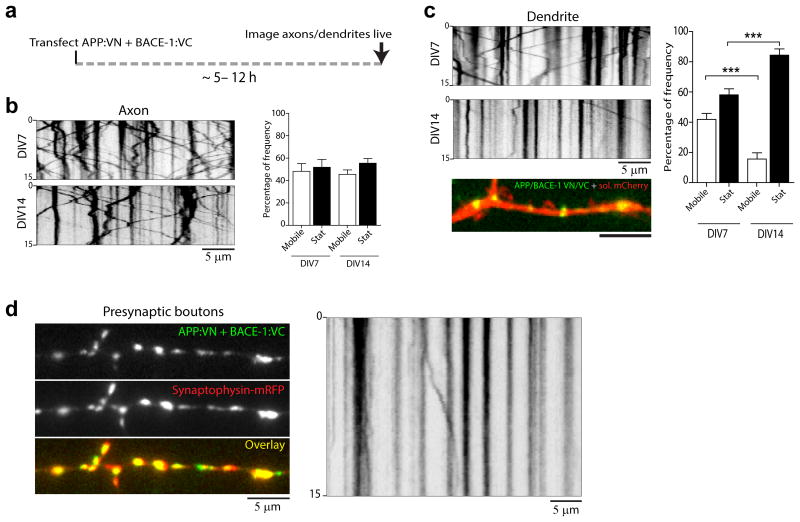
Collectively, the above data indicate that the majority of APP and BACE-1 are co-transported in hippocampal axons, and that the enzyme-substrate pair can interact during transit. In further support of the latter, we found that complemented APP/BACE-1 BifC particles in axons were also mobile (fig. 6a-b). Unlike axons, complemented APP/BACE-1 BifC particles in dendrites were largely immobile in mature (DIV 14) neurons, localizing in and around spines as noted previously (fig. 6c, kymograph and image at the bottom). Interestingly, APP/BACE-1 BifC particles were mobile in younger (DIV 7) neurons lacking mature spines, suggesting that trafficking APP/BACE-1 can be ‘captured’ around dendritic spines, as shown previously for other proteins 23. Complemented APP/BACE-1 BifC particles at presynaptic boutons were also largely immobile, though moving particles were seen in flanking axons as expected (fig. 6d).
Figure 6. Kinetics of APP/BACE-1 BifC particles.
(a) Neurons were co-transfected with APP:VN and BACE-1:VC, and kinetics of complemented YFP particles were imaged live. (b) Representative kymographs (left) and quantification (right) of APP/BACE-1 BifC movements in axons. ∼200 (DIV7) and 160 (DIV14) vesicles from 12-14 axons from two separate cultures were analyzed, p= 0.7488 (between the mobile groups) and p = 0.6634 (between the stationary groups). (c) Representative kymographs (left) and quantification (right) of APP/BACE-1 BifC movements in dendrites. Note restricted movement of APP/BACE-1 BifC in regions around dendritic spines (also see text). ∼200 (DIV7) and 170 (DIV14) vesicles from 12-14 dendrites (two separate cultures) were analyzed. Image at bottom shows APP/BACE-1 BifC in a dendrite. p= 0.0002 (between the mobile groups) and p = 0.0002 (between the stationary groups). (d) APP/BACE-1 BifC particles in presynaptic boutons were largely stationary (85.19 ± 2.53 %) compared to the mobile particles (14.79 ± 2.53 %) as shown in the representative kymograph (right). ∼150 YFP vesicles from 10-12 neurons (two separate cultures) were analyzed, * p =0.0001)
Fate of endocytosed APP in neurons
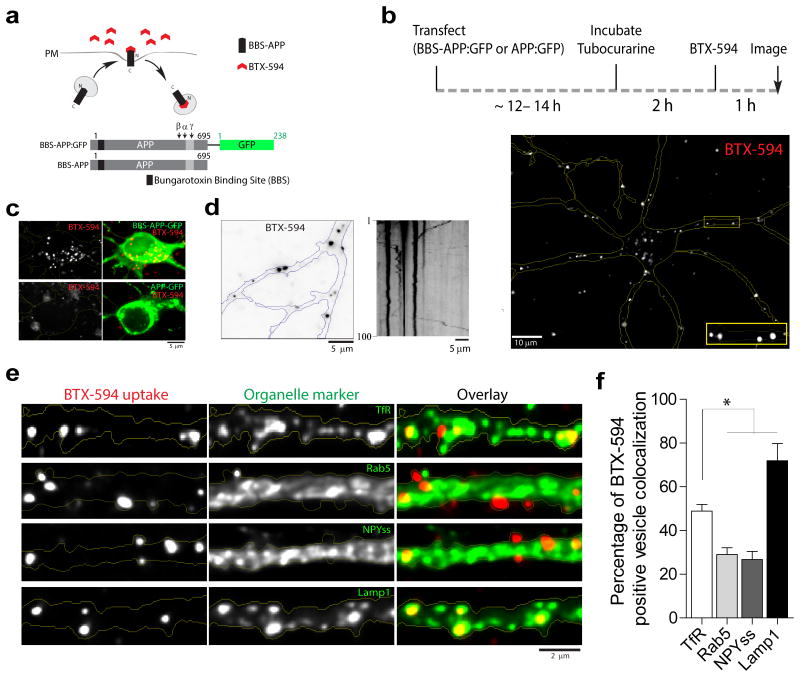
A previous study tracked the fate of endocytosed BACE-1 in neurons, showing a remarkable retrograde bias of BACE-1 containing vesicles in dendrites 17; however the fate of endocytosed WT APP in neurons is less clear. To address this, we adopted a strategy where membrane-spanning proteins are surface-labeled using fluorescent-tagged α-bungarotoxin (BTX) 24. Specifically, we inserted a 13-aa BTX-binding site to the N-terminus of APP (BBS-APP, inserted after the APP signal-sequence); transfected neurons with BBS-APP; and then incubated the cultures with BTX-Alexa-594/488 to label BBS-APP epitopes exposed to the dendritic surface (see general strategy in fig. 7a and schematic in fig. 7b, top). Internalized BBS-APP was seen throughout the somatodendritic compartment (fig. 7b, bottom). As shown in figure 7c, BTX-Alexa-594 internalization was specific for BBS-APP and was not seen with an APP:GFP construct that lacks the BBS domain. Internalized APP particles were largely stationary, and mobile particles showed no directional bias (fig. 7d), very different from the persistent retrograde kinetics reported for internalized BACE-1 17.
Figure 7. Fate of internalized APP in neurons.
(a) Strategy of APP internalization assay. Neurons were transfected with an APP construct containing a bungarotoxin (BTX)-binding site (BBS) at the luminal N-terminus (BBS-APP), and cells were incubated with Alexa-594 labeled BTX (BTX-594). Upon recycling, BTX-594 binds to BBS-APP and internalized Alexa-594 dye indicates APP endocytosis. (b) Schematic shows experimental design; a representative neuron with internalized APP shown below. Note APP internalization throughout somatodendritic compartments, magnified in inset. (c) Specificity of the BTX-594 uptake assay. BTX-594 internalization was seen in BBS-APP:GFP transfected neurons but not in neurons transfected with APP:GFP (lacking BBS domain). Representative of 5-7 neurons in each group. (d) Internalized dendritic BBS-APP particles were largely stationary (54.8 %) along with bi-directionally moving particles comprising anterograde (24.7 %) and retrograde (20.2 %). N = 128 particles from 12 neurons were analyzed; * p = 0.0001. (e, f) Colocalization of internalized BTX-594 with organelle markers. Neurons were co-transfected with BBS-APP and the stated organelle marker; and colocalization was analyzed in dendrites. Internalized BBS-APP predominantly colocalized with recycling endosomes and lysosomes (48.9% colocalization with TfR, 29% with Rab5, 26.7% with NPYss, and 71.9% with LAMP-1; 18-22 dendrites from 15-18 neurons – two separate cultures – were analyzed; * p = 0.0001).
What is the fate of endocytosed WT APP in neurons? To address this, we determined the colocalization of internalized APP with organelle markers. Neurons were transfected with BBS-APP and GFP-tagged organelle markers; incubated with BTX-Alexa-594; and colocalization of GFP/Alexa-594 was determined. In dendrites, a significant fraction (∼ 50%) of internalized APP vesicles were colocalized with TfR +ve recycling endosomes, while a smaller fraction (∼ 25-30%) colocalized with Rab5 and NPY-ss (fig. 7e, F) – reminiscent of the data with APP/BACE-1 BifC above. Surprisingly however, a large fraction (∼ 70%) of internalized APP colocalized with the late-endosomal/lysosomal marker LAMP-1 (fig. 7f). As lysosomes are not a major compartment where APP and BACE-1 interact (see fig. 3c-d), these data suggest an alternate route for internalized APP that might limit APP/BACE-1 interactions and APP cleavage (also see discussion). Internalized APP in neuronal cell bodies was distributed as puncta throughout the soma, colocalizing with both endosomal and TGN markers (40-50%); and significant colocalization with LAMP-1 (∼ 80%) was again noted (Supp. fig. 4).
Reduced APP/BACE-1 interactions in protective AD mutation
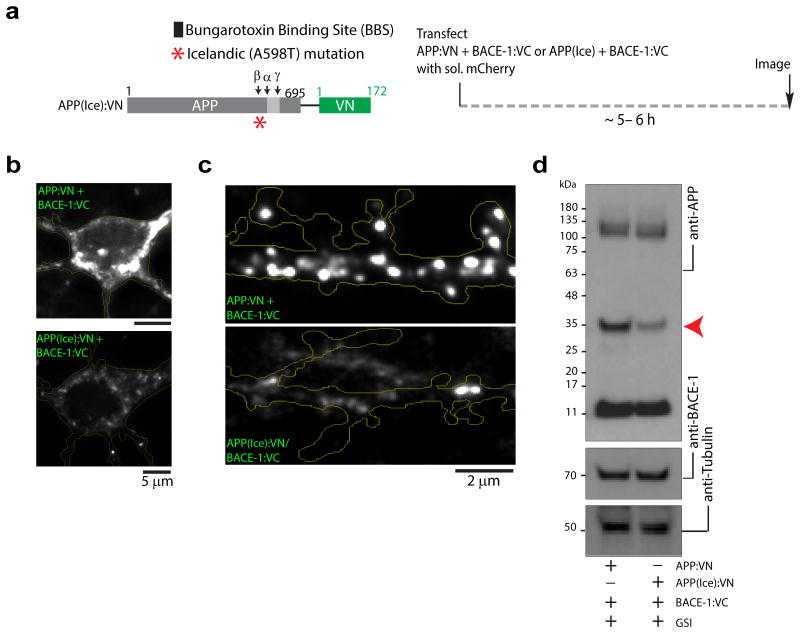
Recently, a search for low-frequency APP variants in whole genome sequencing data from Icelanders revealed a mutation that confers protection against AD [A673T or “Icelandic” mutation, henceforth called APP(Ice), 25]. Although studies suggest that the APP(Ice) mutation makes APP less susceptible to BACE-1 cleavage 26, 27, the underlying mechanism is unclear. One might imagine that this mutation alters APP trafficking pathways, perhaps rerouting the protein into aberrant organelles, diminishing its affinity for BACE-1. Alternatively, physical approximation of the mutant protein with BACE-1 may be attenuated – perhaps due to conformational changes in the mutant protein. Towards this we synthesized an APP:VN construct containing the Icelandic mutation [APP(Ice):VN], and co-transfected it with BACE-1:VC in neurons (schematic in fig. 8a). Fluorescence complementation was greatly attenuated (fig. 8b-c), despite appropriate expression of the construct (Supp. fig. 1). Consequent β-cleavage of APP(Ice):VN was greatly diminished as well (fig. 8e). To examine trafficking parameters of the APP(Ice) mutation, we determined internalization of BBS-APP(Ice) using the BBS/BTX assay described above (see schematic in Supp. fig. 5a). Internalization of BBS-APP(Ice) was similar to its WT counterpart (Supp. fig. 5b). Similarly, trafficking of APP(Ice):GFP was largely unaffected (Supp. fig. 5c). These data suggest that the protective effect of the APP(Ice) mutation may be a consequence of defective approximation of the enzyme-substrate pair prior to cleavage, and not altered vesicle-trafficking. Finally, the OptiCAB assay also gives expected readouts in other cell-types that can be easily propagated (Supp. fig. 6).
Figure 8. Attenuated APP/BACE-1 interactions with an AD protective mutation.
(a) Schematic showing location of AD protective mutation [APP(Ice)] and experimental strategy. (b, c) Neurons were transfected with APP:VN [WT or APP(Ice)] and BACE-1:VC, and fluorescence was analyzed. Complementation of APP(Ice) with BACE-1 was significantly lower in soma (normalized fluorescence intensity of APP(Ice):VN + BACE-1:VC was 0.22 ± 0.06 compared to APP:VN + BACE-1:VC – 1.0 ± 0.17; p = 0.0001) and dendrites (normalized fluorescence intensity of APP(Ice):VN + BACE-1:VC was 0.37 ± 0.07 compared to APP:VN + BACE-1:VC – 1.0 ± 0.18; p = 0.0042). 22-24 dendrites from 14-18 neurons – three separate cultures – were analyzed. (d) BACE-1 knock out fibroblasts were co-transfected with APP:VN [WT or APP(Ice)] and BACE-1:VC, and APP fragments were analyzed by Western blotting (in presence of γ-secretase inhibitor). Note that the ∼30 kDa band corresponding to the expected β-cleavage product of APP:VN (red arrowhead) is greatly attenuated in the APP(Ice) lane (performed four independent experiments); p = 0.0272.
Discussion
Subcellular locale of APP cleavage has been a topic of exceptional interest for decades, and yet, precise sites where WT APP and BACE-1 meet in neurons are essentially unknown. In this study we report optical assays to visualize the convergence and interaction of APP and BACE-1; and also to track internalized APP in hippocampal neurons. Our experiments provide the following insights (all in neurons): 1) Indicate that APP and BACE-1 interact in both biosynthetic and endocytic compartments. 2) Implicate recycling microdomains such as dendritic spines and presynapses as important sites of APP/BACE-1 convergence (and thus β-cleavage). 3) Indicate that recycling endosomes distributed throughout neuronal processes are a major locale of APP/BACE-1 convergence. 4) Indicate that axonal APP and BACE-1 are co-transported in Golgi-derived vesicles, and that they interact within these organelles. 5) Show that a significant population of APP endocytosed from the plasma membrane enters late endosomes/lysosomes – perhaps avoiding excessive cleavage; and finally, 6) suggest attenuated APP/BACE-1 approximation as a mechanistic basis for the AD-protective Icelandic mutation. To our knowledge, these are the first data to directly show interaction of this enzyme-substrate pair in hippocampal neurons – a cell-type relevant to AD – challenging long-standing models of APP/BACE-1 trafficking based on studies in non-neuronal cells and providing concrete answers to controversial topics.
Somatodendritic organelles where APP is cleaved by BACE-1
Previous studies in HEK293T and other cell-lines show that surface internalized APP and BACE-1 containing vesicles colocalize with early endosomes, suggesting that these organelles are the site of β-cleavage 4, 28. These data have led to the assumption that APP β-cleavage occurs mainly in early endosomal, and not biosynthetic compartments 2. However, other studies in cell lines suggest convergence of APP and BACE-1 at the TGN and multiple endosomal compartments including early and recycling endosomes 5, 6, 29. Regardless, the location of APP β-cleavage in neurons - the relevant cell-type in this context – is unknown. Our experiments in hippocampal neurons suggest that in the soma, WT APP and BACE-1 interact at the TGN. In dendrites, WT APP and BACE-1 largely interact in recycling endosomes (∼ 60% colocalization of APP/BACE-1:VN/VC with TfR and Rab11); with lower levels of colocalization (∼ 20-30%) with other endosomal markers (fig. 3) – thus implicating both biosynthetic and endosomal compartments in APP β-cleavage. Interestingly, about half of all APP/BACE-1 interactions were in and around dendritic spines (fig. 3a, b). Indeed these microdomains are rich in recycling endosomes that recycle with the plasma membrane of the spines 30, further supporting the concept that vesicle recycling is an important event in APP cleavage.
The collective data suggest that the majority of steady-state WT APP/BACE-1 interactions in dendrites occur in neuronal recycling endosomes, though a transient association of APP and BACE-1 in early endosomes prior to their engagement in recycling compartments cannot be excluded. The potential discrepancies between data from non-neuronal cells and neurons are difficult to pinpoint, but it is recognized that principles governing APP trafficking in non-neuronal cells are not always applicable to neurons 1. Importantly, neurons have an extensive network of highly dynamic and functional recycling endosomes extending throughout dendrites shafts and spines (7, 30, 31, and this study) – an anatomic feature that is obviously lacking in non-neuronal cells where recycling endosomes are clustered around the nuclei 6, 28. Indeed dynamics of early and recycling endosomes in neurons is quite distinct, as we have reported previously 7. Unlike early endosomes, recycling endosomes are much more dynamic and move with an anterograde bias in dendrites (Supp. Table 1 of reference 7). One might speculate that evolutionary demands of timely delivery into elongated processes have led to sophisticated endosomal networks in neurons that are very different from non-neuronal cells 32. While studies in the latter have offered fundamental insights into the basics of APP processing, we reason that critical aspects where neuronal pathways differ need to be appreciated as well. Finally, it is worth noting that neuronal recycling endosomes are acidic (31 and also see Supp. fig. 7); thus these organelles provide a suitable milieu for BACE-1 enzymatic activity, optimal at low pH 33.
APP/BACE-1 convergence and trafficking in axons
Our live imaging data unequivocally show that WT APP and BACE-1 are co-transported in axons, and that they interact in these transport-vesicles (figs. 4-5). Several studies have shown that APP is likely processed in axons and presynaptic sites 34-38. It has also been suggested that APP and BACE-1 are co-transported in a common transport-carrier 39, but this has been controversial 40-42. Importantly, arguments for or against co-transport of APP and BACE-1 have been largely based on biochemical or histological evidence, rather than direct, simultaneous visualization of the two proteins in living axons. Of note, one study did visualize axonal transport of APP or BACE-1 (separately) in axons of cultured retinal ganglion neurons, reporting much faster velocities for APP 43. However, axonal transport of WT APP and BACE-1 was not simultaneously visualized, though qualitative data from mutant APP and BACE-1 suggested a lack of co-transport 43.
In our studies, simultaneous visualization of WT APP and BACE-1 trafficking in hippocampal axons clearly show that APP and BACE-1 are co-transported (fig. 5c). Since our BifC experiments show that the two proteins also interact during transit, it is unlikely that they move in distinct (but tandem) vesicles that cannot be resolved by light microscopy. It also seems unlikely that these data are confounded by over-expression, as – besides the short time-intervals post-transfection (∼ 5-6h) – overall kinetics of APP and BACE-1 were similar whether they were transfected individually, or co-transfected in these neurons (fig. 5). Our data also indicate that APP and BACE-1 interact in presynaptic boutons (fig. 4d-h). The dichotomy of APP/BACE-1 trafficking in dendrites and axons is intriguing. While dendritic APP and BACE-1 largely move in distinct vesicles 7, they are co-transported in axons (this study). The reasons are unclear at this time, but may be related to transcytosis 32.
Fate of internalized APP in dendrites
Though previous studies have examined internalization of APP in non-neuronal cells, the fate of internalized APP in neurons has been less clear. We used a technique where the intra-luminal/extracellular domain of a transmembrane protein is fluorescent-tagged in live cells by surface-labeling, and the internalized tagged protein is then detected 24. As predicted by our working model 7, internalized APP colocalized with recycling endosomes in dendritic shafts; interestingly however, a large fraction (∼ 70-80%) colocalized with late-endosomes/lysosomes (fig. 7f). APP and BACE-1 do not interact much within lysosomes (fig. 3c-d), and BACE-1 is also not normally localized to these compartments 7; suggesting that routing of surface APP into late-endosomes/lysosomes may be a mechanism by which APP enters the degradative pathway and ‘escapes’ cleavage. This behavior is also consistent with the known short (∼ 2h) half-life of APP 44. Accumulation of lysosomes within neurites, and disruption of lysosomal pathways is an established pathology in AD, and it is possible that such disruptions cause mis-localization of APP/BACE-1 in lysosomes, leading to enhanced cleavage in the acidic milieu. Alternatively, an intriguing possibility is that lysosomal accumulation in AD is not a pathologic phenotype, but an attempt by neurons to protect excess APP from cleavage. About half of the internalized APP in neuronal soma also colocalized with the TGN (Supp. fig. 4), consistent with the idea that retromers and ESCRT pathways recycle endocytic APP with the Golgi 14, 29.
APP/BACE approximation in a protective AD mutant
A recent study discovered a novel point mutation of APP(Ice) in Icelandic populations that is protective against AD 25. Understanding the mechanistic underpinnings of this mutation might lead to novel ways of targeting AD pathology, and at least two scenarios can be imagined. One, the mutation might alter trafficking of the mutant APP – perhaps partitioning it into an aberrant organelle – and diminishing interaction of the enzyme-substrate pair. Alternatively, trafficking routes might be unaffected, but approximation of the two proteins prior to cleavage might be attenuated. Our experiments favor the latter scenario, suggesting that the APP(Ice) mutation attenuates its association with BACE-1 (and APP β-cleavage), without much effect on internalization and other trafficking parameters (fig. 8 and Supp. fig. 5). Recent studies also indicate that these mutations have additional effects on the aggregation properties of misfolded APP 26, 27, suggesting multiple pathways by which this mutation protects against AD. However precise mechanisms by which the APP(Ice) mutations confers protection still remain unknown and may have to await a refined structural understanding of the underlying APP/BACE-1 interacting interface.
Materials and Methods
Constructs, Antibodies and Reagents
Venus N-terminus (VN, amino acid residues 1-172) and Venus C-terminus (VC, amino acid residues 155-238) fragments were PCR amplified using an alpha-synuclein:Venus template 25, and cloned into pEGFP vector at AgeI and NotI sites replacing the GFP to generate pVN and pVC plasmids. Human WT APP695 isoform and APPYENPTY (described in 25) were PCR amplified and cloned at HindIII and SacII sites of pVN to generate pAPP:VN and pAPPYENPTY:VN. APP(1-636) was PCR amplified and cloned into pVN to generate pAPPSTOP40:VN. Mouse BACE-1 was cloned at XhoI and SalI sites of pVC to generate pBACE-1:VC. The “Icelandic”, “Arctic” and “London” APP mutations were generated by overlap PCR. Note that “APP(Ice)” “APP(Arc)” and “APP(Lon)” refer to the APP(A598T), APP(E618G) and APP(V642I) mutations in the APP-695 isoform used here and in other studies 26, and these correspond to the APP(A673T), APP(E693G) and APP(V717I) mutations respectively in the longest APP isoform [APP-770, 25]. All spilt Venus constructs have a 10 aa (PRARDPPVAT) linker between the gene of interest and the Venus fragments. To incorporate the BBS at the N-terminus of APP, a SpeI site was introduced by mutating APP at A30T/E31S. Annealed oligos for BBS sequence with SpeI site at both ends were ligated to obtain pBBS-APP:GFP, pBBS-APP(Ice):GFP, and untagged pBBS-APP. All constructs were cloned in-frame and fidelity was confirmed by sequencing.
The following constructs were obtained from Addgene: DsRed:Rab11a (Cat # 12679, Richard Pagano, Mayo Clinic); mRFP:Rab5 (Cat # 14437, Art Helenius, ETH, Zurich – the GFP:Rab5 was made in-house by swapping the mRFP with GFP); LAMP-1:mCherry (Amy Palmer, University of Colorado); and LAMP-1:GFP (Cat #34831, Esteban Dell'Angelica, UCLA). Antibodies and reagents used were: APP (ab32136; Abcam), BACE-1 (MAB931; R&D), Tubulin (clone DM1A; Sigma), GFP (polyclonal, ab290; Abcam), α-bungarotoxin Alexa-594 conjugate (Life Technologies), Alexa-Transferrin-594 (Life Technologies) and Tubocurarine chloride (Tocris). Dynasore was used at a final concentration of 40μm. All other reagents/culture media were obtained either from Sigma or Life Technologies.
Cell cultures and transfections
All animal studies were performed in accordance with University of California guidelines. Primary hippocampal neurons were acquired from postnatal (P0-P1) CD1 mice and transiently transfected using Lipofectamine 2000 as described previously 45. Briefly, dissociated neurons were plated at a density of 50,000 cells/cm2 on poly-D-lysine coated glass-bottom culture dishes (Mattek) and maintained in Neurobasal/B27 media with 5 % CO2. Neurons were cultured for ∼ 5-6 h or ∼ 12-15 h post-transfection as described in results. For generating BACE-1 -/- cells, fibroblasts were cultured from the lungs of P1 BACE-1 -/- mice (obtained from Jackson Labs, from mice deposited by Dr. Philip Wong, Johns Hopkins) followed by immortalization with SV40 t antigen. Both BACE-1 KO and HEK293T cells (ATCC) were maintained in DMEM with 10% FBS and transiently transfected with the respective constructs using Amaxa 4D system with SF Cell Line 4D-Nucleofector X kit (V4XC-2024), program CM137 and CM130 respectively using the manufacturer's protocol. The cell density ranged from 1×107 to 3×107 cells/ml in a 100μl transfection reaction. Cells were cultured for 24 h post-transfection before analyses.
Microscopy/analyses, and BTX-uptake experiments
All images were acquired using an Olympus IX81 inverted epifluorescence microscope with a Z-controller (IX81, Olympus), a motorized X-Y stage controller (Prior Scientific), a fast electronic shutter (SmartShutter), an ultra-stable light source (Exfo exacte) and CCD cameras (Coolsnap HQ2, Photometrics). Colocalization experiments were done as described previously 7. Briefly, cells were co-transfected with desired constructs, fixed, and distal regions of the primary dendrites, or first-order branches of secondary dendrites and axonal regions ∼ 150 μm from the soma were selected for imaging. Z-stack images were captured using a 100× objective with consistent imaging-parameters (0.2 μm z-step, 500 ms exposure, 1×1 binning), and de-convolved (Huygens, Scientific Volume Imaging B.V.). De-convolved z-stacks were subjected to a maximum intensity projection, resulting in 2-D images for processing. Images were thresholded and colocalization was calculated as the fraction of area overlapping between the above-threshold structures in each image using custom algorithms written in Matlab (Mathworks). For live imaging, cells were transferred to Hibernate-E-Low Fluorescence media (HELF, BrianBits) at 35-37°C 7.
For transport studies in dendrites, imaging parameters were set at 1 frame/sec, total 200 frames and 500 millisecond exposure with 2 × 2 camera binning. Cargo dynamics in axons was imaged at 5 frames/second and 200-millisecond exposure (total of 75 frames, with no interval between frames) using the ‘stream acquisition’ mode in Metamorph. We found that these imaging parameters are critical to capture all moving APP/BACE-1 cargoes, and lower time intervals lead to an underestimation of particle-kinetics. Axons were selected based on criteria described in our previous studies 45, 46. For simultaneous two-color imaging, two spectrally distinct fluorophores were imaged using a “dual cam” imaging device (Photometrics) with 70 % attenuation of the incident light to reduce photobleaching. Kymographs were generated in MetaMorph; segmental tracks were traced on the kymographs using a line tool; individual lines were saved as “.rgn” file; and the resultant velocity data (distance/time) was obtained for each track as described previously 45. Analyses of all transport studies were done in a blinded fashion. For the BTX-uptake experiments, Neurons were pretreated with 150 μM Tubocurarine chloride for 2 h prior to BTX-594 addition. After incubating with 7 μg/ml BTX-594 in Neurobasal/B27 media for 1 h at 37°C, cells were washed with ice cold HBSS, pH 7.6 and fixed or imaged live in HELF imaging media.
Quantitative imaging of presynaptic targeting
Cultured DIV13 hippocampal neurons (plating density of 60,000 cells/cm2) were co-transfected with YFP (or VN/VC) tagged proteins (as described), and soluble mCherry using Lipofectamine 2000, and imaged live 12-14 hours after transfection. Z-stack images were obtained and Targeting Factor was calculated as described previously 21, 22. Briefly, fluorescence intensities for each channel were measured along lines drawn perpendicularly to the axons and across the boutons. Peak fluorescence along boutons and the adjacent axon were determined by fitting Gaussian functions to the resultant intensity profiles and corrected for background. Targeting factor was expressed as [(BoutonYFP/AxonYFP)/(BoutonRFP/AxonRFP)] – 1 22. For co-localization analysis, boutons were identified and marked on synaptophysin:mRFP images and then the regions were transferred to YFP images to calculate percentage of RFP boutons occupied by YFP.
Biochemistry
Neuro2A, HEK293T cells (ATCC) or BACE-1-KO fibroblast cells were cultured for 24 hours post transfection with DNA constructs using Amaxa 4D system and lysates were prepared in 50 mM NaCl, 50 mM Tri-Cl, 5 mM EDTA and protease inhibitor cocktail (Thermo Scientific), pH 8.0. After centrifuging at 1000 ×g for 20 min at 4°C, S1 fractions were quantified, resolved on SDS-PAGE for Western blot analyses.
Statistical Analysis
Statistical analyses were performed using Prism software (Graphpad). Student's t-test (unpaired) or one-way ANOVA with Dunnet's post-hoc test was used to compare two or more groups respectively. A p-value of <0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Gary Banker (Oregon Health and Science University) for the NPYss:mCherry and TfR:mCherry/GFP constructs; George Patterson and Jennifer Lipincott-Schwartz (NIH) for the GalT:mCherry/GFP constructs; Zu-Hang Sheng (NIH) for the Synaptophysin-mRFP construct; Wolfhard Almers (Oregon Health and Science University) for the ‘crippled CMV promoter’; and Darren Boehning (The University of Texas Health Science Center) for the APP(Lon):GFP construct. Constructs from Addgene and investigators are acknowledged in ‘methods’. UD was partly supported by a pilot award from the UCSD Alzheimer's Center (P50 AG005131). This work was supported by grants from the NIH/NIA (R01AG048218 and NIH/NINDS (R01NS075233) to SR.
Footnotes
Author Contributions: UD and SR designed the assays and wrote the paper. UD designed and performed most of the experiments/data-analysis. LW designed and performed the synaptic targeting and some of the axonal transport experiments. AG and LW helped with neuronal cultures, and JS performed and analyzed some of the axonal transport experiments. SW and EK provided key reagents and technical advice.
Competing financial interests The authors declare no competing financial interests
References
- 1.O'Brien RJ, Wong PC. Amyloid precursor protein processing and Alzheimer's disease. Annu Rev Neurosci. 2011;34:185–204. doi: 10.1146/annurev-neuro-061010-113613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajendran L, Annaert W. Membrane trafficking pathways in Alzheimer's disease. Traffic. 2012;13:759–770. doi: 10.1111/j.1600-0854.2012.01332.x. [DOI] [PubMed] [Google Scholar]
- 3.Kinoshita A, et al. Demonstration by FRET of BACE interaction with the amyloid precursor protein at the cell surface and in early endosomes. J Cell Sci. 2003;116:3339–3346. doi: 10.1242/jcs.00643. [DOI] [PubMed] [Google Scholar]
- 4.Sannerud R, et al. ADP ribosylation factor 6 (ARF6) controls amyloid precursor protein (APP) processing by mediating the endosomal sorting of BACE1. Proc Natl Acad Sci U S A. 2011;108:E559–568. doi: 10.1073/pnas.1100745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenfield JP, et al. Endoplasmic reticulum and trans-Golgi network generate distinct populations of Alzheimer beta-amyloid peptides. Proc Natl Acad Sci U S A. 1999;96:742–747. doi: 10.1073/pnas.96.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prasad H, Rao R. The Na+/H+ Exchanger NHE6 Modulates Endosomal pH to Control Processing of Amyloid Precursor Protein in a Cell Culture Model of Alzheimer Disease. J Biol Chem. 2015;290:5311–5327. doi: 10.1074/jbc.M114.602219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das U, et al. Activity-induced convergence of APP and BACE-1 in acidic microdomains via an endocytosis-dependent pathway. Neuron. 2013;79:447–460. doi: 10.1016/j.neuron.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc. 2006;1:1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Virgilio M, Kiosses WB, Shattil SJ. Proximal, selective, and dynamic interactions between integrin alphaIIbbeta3 and protein tyrosine kinases in living cells. J Cell Biol. 2004;165:305–311. doi: 10.1083/jcb.200402064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Remy I, Montmarquette A, Michnick SW. PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nat Cell Biol. 2004;6:358–365. doi: 10.1038/ncb1113. [DOI] [PubMed] [Google Scholar]
- 11.Citron M, Teplow DB, Selkoe DJ. Generation of amyloid beta protein from its precursor is sequence specific. Neuron. 1995;14:661–670. doi: 10.1016/0896-6273(95)90323-2. [DOI] [PubMed] [Google Scholar]
- 12.Sahlin C, et al. The Arctic Alzheimer mutation favors intracellular amyloid-beta production by making amyloid precursor protein less available to alpha-secretase. J Neurochem. 2007;101:854–862. doi: 10.1111/j.1471-4159.2006.04443.x. [DOI] [PubMed] [Google Scholar]
- 13.Muratore CR, et al. The familial Alzheimer's disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum Mol Genet. 2014;23:3523–3536. doi: 10.1093/hmg/ddu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Small SA, Gandy S. Sorting through the cell biology of Alzheimer's disease: intracellular pathways to pathogenesis. Neuron. 2006;52:15–31. doi: 10.1016/j.neuron.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaech S, Huang CF, Banker G. Short-term high-resolution imaging of developing hippocampal neurons in culture. Cold Spring Harb Protoc. 2012;2012:340–343. doi: 10.1101/pdb.prot068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Meskini R, Cline LB, Eipper BA, Ronnett GV. The developmentally regulated expression of Menkes protein ATP7A suggests a role in axon extension and synaptogenesis. Dev Neurosci. 2005;27:333–348. doi: 10.1159/000086713. [DOI] [PubMed] [Google Scholar]
- 17.Buggia-Prevot V, et al. A function for EHD family proteins in unidirectional retrograde dendritic transport of BACE1 and Alzheimer's disease Abeta production. Cell Rep. 2013;5:1552–1563. doi: 10.1016/j.celrep.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koo EH, Squazzo SL. Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J Biol Chem. 1994;269:17386–17389. [PubMed] [Google Scholar]
- 19.Perez RG, et al. Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J Biol Chem. 1999;274:18851–18856. doi: 10.1074/jbc.274.27.18851. [DOI] [PubMed] [Google Scholar]
- 20.Knowles MK, et al. Single secretory granules of live cells recruit syntaxin-1 and synaptosomal associated protein 25 (SNAP-25) in large copy numbers. Proc Natl Acad Sci U S A. 2010;107:20810–20815. doi: 10.1073/pnas.1014840107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, et al. alpha-synuclein multimers cluster synaptic vesicles and attenuate recycling. Curr Biol. 2014;24:2319–2326. doi: 10.1016/j.cub.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gitler D, et al. Molecular determinants of synapsin targeting to presynaptic terminals. J Neurosci. 2004;24:3711–3720. doi: 10.1523/JNEUROSCI.5225-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Washbourne P, Bennett JE, McAllister AK. Rapid recruitment of NMDA receptor transport packets to nascent synapses. Nat Neurosci. 2002;5:751–759. doi: 10.1038/nn883. [DOI] [PubMed] [Google Scholar]
- 24.Sekine-Aizawa Y, Huganir RL. Imaging of receptor trafficking by using alpha-bungarotoxin-binding-site-tagged receptors. Proc Natl Acad Sci U S A. 2004;101:17114–17119. doi: 10.1073/pnas.0407563101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jonsson T, et al. A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 26.Benilova I, et al. The Alzheimer disease protective mutation A2T modulates kinetic and thermodynamic properties of amyloid-beta (Abeta) aggregation. J Biol Chem. 2014;289:30977–30989. doi: 10.1074/jbc.M114.599027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maloney JA, et al. Molecular mechanisms of Alzheimer disease protection by the A673T allele of amyloid precursor protein. J Biol Chem. 2014;289:30990–31000. doi: 10.1074/jbc.M114.589069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajendran L, et al. Alzheimer's disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci U S A. 2006;103:11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choy RW, Cheng Z, Schekman R. Amyloid precursor protein (APP) traffics from the cell surface via endosomes for amyloid beta (Abeta) production in the trans-Golgi network. Proc Natl Acad Sci U S A. 2012;109:E2077–2082. doi: 10.1073/pnas.1208635109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park M, et al. Plasticity-induced growth of dendritic spines by exocytic trafficking from recycling endosomes. Neuron. 2006;52:817–830. doi: 10.1016/j.neuron.2006.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, et al. Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell. 2008;135:535–548. doi: 10.1016/j.cell.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yap CC, Winckler B. Harnessing the power of the endosome to regulate neural development. Neuron. 2008;74:440–451. doi: 10.1016/j.neuron.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vassar R, Kovacs DM, Yan R, Wong PC. The beta-secretase enzyme BACE in health and Alzheimer's disease: regulation, cell biology, function, and therapeutic potential. J Neurosci. 2009;29:12787–12794. doi: 10.1523/JNEUROSCI.3657-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buxbaum JD, et al. Alzheimer amyloid protein precursor in the rat hippocampus: transport and processing through the perforant path. J Neurosci. 1998;18:9629–9637. doi: 10.1523/JNEUROSCI.18-23-09629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazarov O, Lee M, Peterson DA, Sisodia SS. Evidence that synaptically released beta-amyloid accumulates as extracellular deposits in the hippocampus of transgenic mice. J Neurosci. 2002;22:9785–9793. doi: 10.1523/JNEUROSCI.22-22-09785.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cirrito JR, et al. Endocytosis is required for synaptic activity-dependent release of amyloid-beta in vivo. Neuron. 2008;58:42–51. doi: 10.1016/j.neuron.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeBoer SR, Dolios G, Wang R, Sisodia SS. Differential release of beta-amyloid from dendrite- versus axon-targeted APP. J Neurosci. 2014;34:12313–12327. doi: 10.1523/JNEUROSCI.2255-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kandalepas PC, et al. The Alzheimer's beta-secretase BACE1 localizes to normal presynaptic terminals and to dystrophic presynaptic terminals surrounding amyloid plaques. Acta Neuropathol. 2013;126:329–352. doi: 10.1007/s00401-013-1152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamal A, Almenar-Queralt A, LeBlanc JF, Roberts EA, Goldstein LS. Kinesin-mediated axonal transport of a membrane compartment containing beta-secretase and presenilin-1 requires APP. Nature. 2001;414:643–648. doi: 10.1038/414643a. [DOI] [PubMed] [Google Scholar]
- 40.Lazarov O, et al. Axonal transport, amyloid precursor protein, kinesin-1, and the processing apparatus: revisited. J Neurosci. 2005;25:2386–2395. doi: 10.1523/JNEUROSCI.3089-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szodorai A, et al. APP anterograde transport requires Rab3A GTPase activity for assembly of the transport vesicle. J Neurosci. 2009;29:14534–14544. doi: 10.1523/JNEUROSCI.1546-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sisodia SS. Biomedicine. A cargo receptor mystery APParently solved? Science. 2002;295:805–807. doi: 10.1126/science.1069661. [DOI] [PubMed] [Google Scholar]
- 43.Goldsbury C, et al. Inhibition of APP trafficking by tau protein does not increase the generation of amyloid-beta peptides. Traffic. 2006;7:873–888. doi: 10.1111/j.1600-0854.2006.00434.x. [DOI] [PubMed] [Google Scholar]
- 44.Ring S, et al. The secreted beta-amyloid precursor protein ectodomain APPs alpha is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J Neurosci. 2007;27:7817–7826. doi: 10.1523/JNEUROSCI.1026-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang Y, et al. Early and selective impairments in axonal transport kinetics of synaptic cargoes induced by soluble amyloid beta-protein oligomers. Traffic. 2012;13:681–693. doi: 10.1111/j.1600-0854.2012.01340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganguly A, Roy S. Using photoactivatable GFP to track axonal transport kinetics. Methods Mol Biol. 2014;1148:203–215. doi: 10.1007/978-1-4939-0470-9_13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.