SUMMARY
La Crosse virus (LACV), a zoonotic Bunyavirus, is a major cause of pediatric viral encephalitis in the United States. A hallmark of neurological diseases caused by LACV and other encephalitic viruses is the induction of neuronal cell death. Innate immune responses have been implicated in neuronal damage, but no mechanism has been elucidated. By using in vitro studies in primary neurons and in vivo studies in mice, we have shown that LACV infection induced the RNA helicase, RIG-I, and mitochondrial antiviral signaling protein (MAVS) signaling pathway, resulting in upregulation of the sterile alpha and TIR-containing motif 1 (SARM1), an adaptor molecule that we found to be directly involved in neuronal damage. SARM1-mediated cell death was associated with induced oxidative stress response and mitochondrial damage. These studies provide an innate-immune signaling mechanism for virus-induced neuronal death and reveal potential targets for development of therapeutics to treat encephalitic viral infections.
INTRODUCTION
The innate immune response protects against virus infections by production of type I interferons that mediate antiviral host responses and cytokines that recruit inflammatory cells. This innate immune response can also induce cellular damage (Chattopadhyay et al., 2010; Fink et al., 2008; Lei et al., 2009; McAllister and Samuel, 2009). Studies have demonstrated direct innate immune signaling-mediated cell death with activation of pattern recognition receptors such as Toll-like receptors (TLRs) or RNA helicase receptors (RLRs) leading to cellular damage and, sometimes, apoptosis (Cameron et al., 2007; Lathia et al., 2008; Ma et al., 2006, 2007; Tang et al., 2008). In the brain, innate immune-induced apoptosis following virus infection may be a contributing factor to neuronal damage and neuronal dropout. Identification of specific targets that initiate apoptosis during virus infection of neurons will offer important insight into the mechanisms of neurodegeneration and will provide targets for the development of antiviral therapies.
One protein that may have a role in innate immune signaling-mediated cell death is sterile alpha and Toll/interleukin-1 (IL-1) receptor (TIR) motif-containing 1 protein (SARM1, MyD88-5). This protein is a member of the TIR-containing adaptor family and, in immune cells, acts as a negative regulator of TLR-mediated NF-κB activation (Carty et al., 2006; Peng et al., 2010) and contributes to T cell apoptosis (Panneerselvam et al., 2013). In neurons, SARM1 interacts with syndecan 2 and regulates neuronal morphogenesis (Chen et al., 2011). Studies using GFP-tagged SARM1 show that under conditions of metabolic stress in neurons, SARM1 translocates to the mitochondria, interacts with c-Jun N-terminal kinase 3 (JNK3), and mediates neuronal apoptosis (Kim et al., 2007). SARM1 has also been identified as mediating axonal death, although the mechanism is unknown (Osterloh et al., 2012). The role of SARM1 in the innate immune response and neuronal or axon death has prompted questions about the function for SARM1 in inducing neuronal damage during virus infections in the central nervous system (CNS) and whether this damage would be mediated through innate immune activation.
To investigate the role of SARM1 in virus-induced neuronal death, we utilized La Crosse virus (LACV), an enveloped trisegmented negative-sense RNA virus belonging to the family Bunyaviridae. LACV is a major cause of pediatric viral encephalitis in the USA and is an emerging pathogen due to increased vector hosts and range (Gerhardt et al., 2001; Haddow and Odoi, 2009; McJunkin et al., 2001). Neurons are the predominant cell type infected with the virus in the CNS and LACV-mediated encephalitis is associated with degenerative neuronal changes characteristic of apoptotic cells, including nuclear vacuolization and cell shrinkage (Bennett et al., 2008; Kalfayan, 1983; Pekosz et al., 1996). In this study, we have demonstrated a clear role for SARM1 in mediating LACV-induced neuronal apoptosis. We have also determined the mechanisms for SARM1 induction and SARM1-induced cell death during LACV infection. We show that both protective type I interferon (IFN) and damaging SARM1-induced responses are generated following virus stimulation of the RNA helicase, retinoic acid-inducible gene 1 protein (RIG-I), and subsequent activation of mitochondrial antiviral signaling protein (MAVS) signaling pathway in neurons.
RESULTS
SARM1 Is Induced in LACV-Infected Neurons
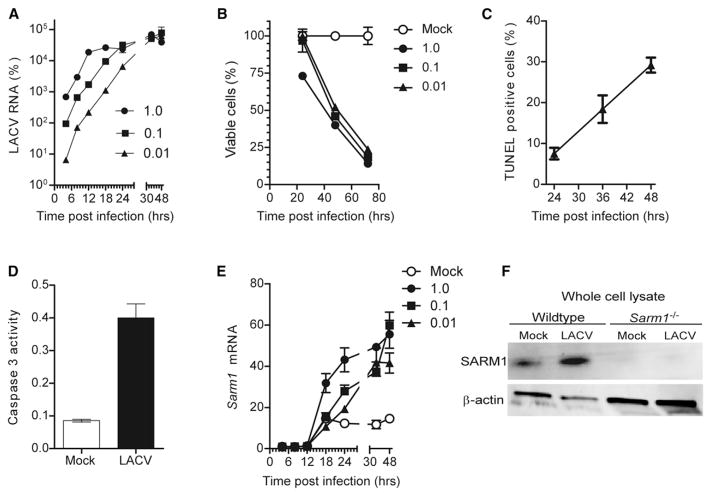
SARM1 is highly conserved from chordates to humans (Mink et al., 2001), suggesting a conserved function. Given that SARM1 influences neuronal death following oxygen and glucose deprivation (OGD) or axonal injury (Osterloh et al., 2012; Yuan et al., 2010; Kim et al., 2007), we investigated whether SARM1 also influenced virus-mediated neuronal damage. Primary cortical neurons were infected with LACV at different multiplicities of infection (MOI) and followed for gene expression and cell death. LACV RNA was detectable in infected neuronal cultures as early as 6 hr postinfection (hpi) and increased logarithmically until reaching a plateau between 18 to 36 hpi, depending on MOI (Figure 1A). LACV-induced neuronal death started at 24 hpi in cultures infected with the highest MOI of virus with widespread cell death by 72 hpi for all MOIs tested (Figure 1B). Dying neurons were TUNEL positive (Figure 1C) and associated with increased caspase-3 activity, typical of apoptotic neurons (Figure 1D). Analysis of SARM1 expression during LACV infection demonstrated increased Sarm1 messenger RNA (mRNA) as early as 18 hpi at the highest MOI and by 24 to 30 hpi for the lower MOIs (Figure 1E). A corresponding increase was observed with SARM1 protein in whole-cell lysates of LACV-infected neurons compared to mock-infected controls (Figure 1F). Thus, LACV infection resulted in the upregulation of Sarm1 mRNA and protein expression.
Figure 1. LACV Infection of Primary Cortical Neurons Induces Apoptotic Death and Increased Production of SARM1.
(A) Cortical neurons were infected with LACV at MOIs of 1, 0.1, and 0.01. Virus RNA was measured at 4, 8, 12, 18, 24, 36, and 48 hpi by real-time PCR and calculated as percent of Gapdh expression.
(B) Cell viability of neurons following LACV infection was measured by MTT assay.
(C) Neurons infected with LACV (MOI 0.01) were stained with TUNEL reagent. Cells were counted from five to six images for each group and a percentage of TUNEL-positive cells per total number of nuclei per image determined.
(D) At 36 hpi, neurons were analyzed for caspase-3 activity.
(E) RNA from cells in (A) was analyzed for Sarm1 mRNA by real-time PCR and calculated as percentage of Gapdh mRNA expression. (A–E) Data are the mean ± SEM for three or more samples per group per time point and are representative of duplicate experiments.
(F) Immunoblot analysis of SARM1 and β-actin in whole cell lysates of mock and LACV infected neurons from WT and Sarm1−/− mice at 36 hpi.
SARM1 Contributes to LACV-Mediated Neuronal Death
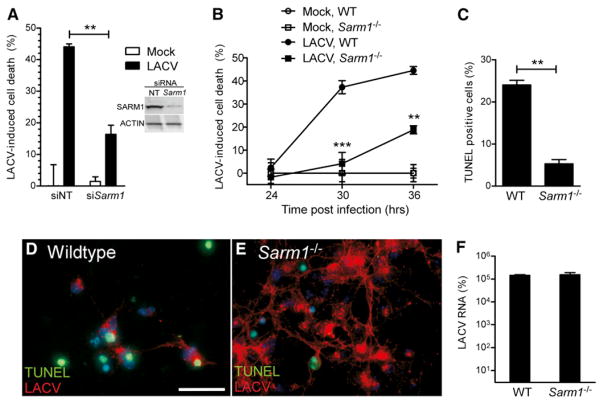
Because SARM1 was induced during LACV infection and upregulated prior to apoptosis, we examined whether SARM1 had a role in LACV-mediated cell death. Transfection of neurons with small interfering RNA (siRNA) targeted to Sarm1 prior to infection significantly reduced LACV-mediated neuronal death (Figure 2A). Similar results were observed with neurons from Sarm1−/− mice as determined by both MTT assay and TUNEL staining (Figures 2B–2D), with a significant inhibition or delay in neuronal death compared to C57BL/6 wild-type (WT) neurons. SARM1 deficiency did not inhibit virus replication with cells from Sarm1−/− cultures expressing high amounts of virus (red fluorescence) (Figure 2E), but not undergoing the same amount of cell death (green fluorescence) as observed in infected cells from WT cultures (Figures 2C–2E). Analysis of viral RNA expression in neuronal cultures, controlled for the number of cells by Gapdh mRNA expression, showed similar amounts of virus RNA (Figure 2F). Thus, SARM1 mediates neuronal death during LACV infection, by a mechanism unrelated to suppression of virus replication in neurons.
Figure 2. SARM1 Deficiency Limits LACV-Mediated Neuronal Death but Not Virus Infection or Replication in Neurons.
(A) Neurons were treated with Sarm1 siRNA or nontargeting (NT) siRNA starting at 3 days postculture. Twenty-four hours later, cells were infected with LACV and cell viability was measured by MTT assay at 36 hpi. SARM1 protein expression in neurons treated with NT or Sarm1 siRNA is shown in inset on right.
(B) Neurons from WT and Sarm1−/− mice were infected with LACV, and cell viability was measured at 24, 30, and 36 hr by MTT assay.
(C–E) Cells were stained for DNA fragmentation by TUNEL analysis at 36 hpi and counted as percent positive as described in Figure 1. (D and E) Immunofluorescence analysis of neurons from (D) WT and (E) Sarm1−/− mice infected with LACV at 36 hpi with TUNEL-stained nuclei (green) and LACV (red). The relative lack of cells in (D) compared to (E) is due to cell death. Scale bar represents 25 μm.
(F) Viral RNA expression in neurons at 36 hpi as determined by real-time PCR. All data are shown as mean ± SEM of 3–6 samples per group per time point. Data are representative of 2 or 3 replicate experiments per panel. See also Figure S3.
SARM1 Deficiency Suppresses Viral Pathogenesis of LACV Infection In Vivo
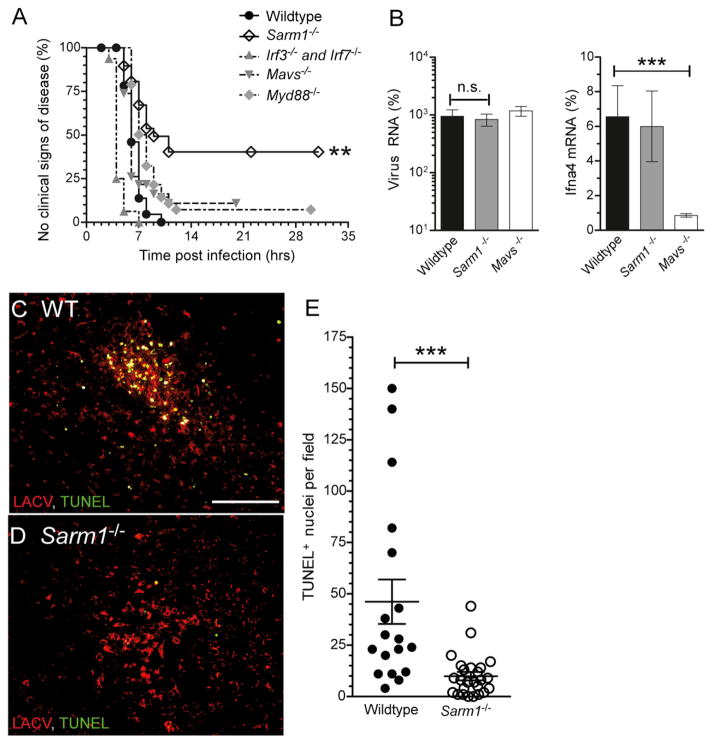
To examine whether SARM1 deficiency would alter LACV pathogenesis in vivo, we infected C57BL/6 WT and Sarm1−/− mice with 103 plaque forming units (PFU) of LACV intraperitoneally, a dose that induces 100% incidence of neurological disease in 3-week-old WT mice (Figure 3A). Sarm1−/− mice developed neurological disease at a significantly lower incidence than WT mice, indicating a detrimental role for SARM1 during LACV infection. In comparison, deficiency in MyD88, a family member of SARM1, did not significantly affect neurological disease development. Analysis of viral RNA from brain tissue of wild-type and Sarm1−/− mice at 5 dpi, just prior to the onset of disease in wild-type mice, revealed similar amounts of viral RNA in Sarm1−/− mice compared to wild-type controls (Figure 3B). In uninfected wild-type mice, SARM1 was found almost exclusively in neuronal cell bodies (see Figure S1A available online). However, in focal areas in the cortex of LACV infected mice, SARM1 was observed localized to the axons of neurons (Figure S1B). Analysis of 5 dpi preclinical WT mice demonstrated numerous TUNEL-positive cells (green) in areas of virus infection (red) (Figure 3C; Figure S1G). In contrast, Sarm1−/− mice at 5 dpi had significantly fewer TUNEL-positive cells, despite large areas of virus infection (Figures 3D and 3E; Figure S1H). Thus, SARM1 deficiency inhibits LACV-induced damage and death in vivo, through a mechanism independent of virus replication, similar to the results from neuronal cultures in vitro.
Figure 3. SARM1 Deficiency Protects Mice from LACV-Induced Neuronal Damage.
(A) Mice at 20 to 21 days of age were infected with 103 PFU of LACV by intraperitoneal infection and followed for the development of clinical disease. Survival curve for each strain was determined using Kaplan-Meier analysis of 32 WT, 16 Irf3−/− and Irf7−/−, 38 Myd88−/−, 19 Mavs−/−, and 29 Sarm1−/− mice. P value for Sarm1−/− versus WT was < 0.01.
(B) Viral RNA and Ifna4 mRNA was quantified from brains of WT and Sarm1−/− mice at 5 dpi. No difference was observed in viral RNA between WT and Sarm1−/− mice. A decrease in Ifna4 mRNA was observed in Mavs−/− mice. Data are shown as mean ± SEM of 5–8 samples per group.
(C–E) Immunohistochemical analysis of TUNEL (green) and LACV (red) in the cortex of (C) WT and (D) Sarm1−/− mice at 5 dpi. Scale bar represents 100 μm. (E) Number of TUNEL-positive cells per field of view (200×) from focal area of virus infection in brain tissue from WT and Sarm1−/− mice. Data are shown as individual points with mean +/− SEM plotted. See also Figure S1.
SARM1 Localizes to the Mitochondria and Is Associated with Mitochondrial Damage
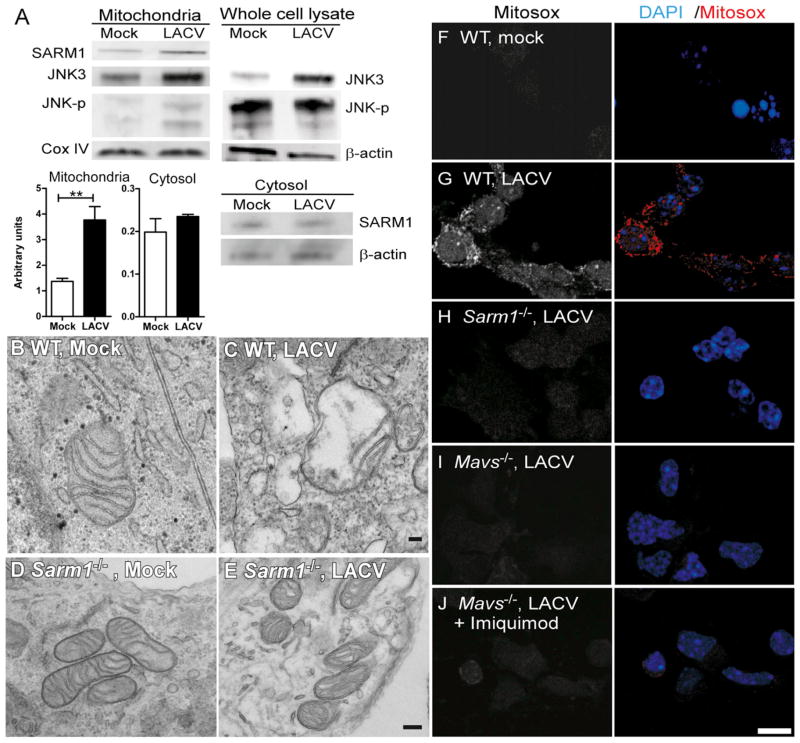
The N-terminal domain of SARM1 contains a mitochondrial localization signal sequence (Panneerselvam et al., 2012). Analysis of LACV-infected neurons demonstrated increased SARM1 in the mitochondrial fraction compared to mock-infected controls, which was not observed in the cytosol fraction (Figure 4A). Similar results were observed in the mitochondrial fraction of brain tissue from LACV-infected mice (Figure S1C). The c-Jun N-terminal kinase 3 (JNK3), which associates with SARM1 following metabolic stress (Kim et al., 2007), was also increased in mitochondrial fractions following LACV infection as was phosphorylated JNK (Figure 4A).
Figure 4. LACV Infection Induces Mitochondrial Localization of SARM1 and Mitochondrial Damage.
(A) Immunoblot analysis of SARM1, JNK3, and phosphorylated JNK in the mitochondrial fraction, whole cell lysate, or cytosol of mock and LACV infected neurons at 36 hpi. CoxIV and β-actin were used as loading controls. SARM1 whole-cell lysate is shown in Figure 1F. Data are representative of duplicate or triplicate experiments. Graphs below are the mean amounts of SARM1 protein in the mitochondrial and cytosol fractions from mock and LACV-infected neurons. Data are the mean ± SEM of densitometry readings from three experiments.
(B–E) Morphological changes in the mitochondria associated with LACV infection. TEM images were acquired at 36 hpi from (B and D) mock- and (C and E) LACV-infected neurons. Data are representative of 5–6 fields for each group. Scale bars represent 100 μm (F–J). Images of (F) mock- and (G–J) LACV-infected neurons from (F–G) WT or (H) Sarm1−/−, (I) Mavs−/−, or (J) Mavs−/− cells treated with imiquimod at 36 hpi stained with 5 uM of Mitosox red. First panel for each image is single channel for Mitosox red and the second panel is Mitosox red plus DAPI. Images are representative of cells in culture. Scale bar represents 10 μm. See also
Despite mitochondrial localization, the mechanism by which SARM1 induces neuronal apoptosis is unknown. Analysis of neurons from WT mice showed numerous neurons containing swollen and/or degenerative mitochondria in cell bodies and axons in LACV-infected cultures (Figure 4C), but not in mock-infected controls (Figure 4B). In contrast, LACV-infection of Sarm1−/− neurons did not induce mitochondrial damage (Figures 4D and 4E). Mitochondrial damage can be induced via reactive oxygen species (ROS) formation, which has been implicated in some cases of neuronal damage (Pan et al., 2009). Staining with Mitosox red, an indicator of mitochondrial super-oxide production, indicated that LACV-infected neurons had increased superoxide production following LACV infection (Figures 4F and 4G). However, superoxide production was not detected in LACV-infected neurons from Sarm1−/− mice indicating that SARM1 was necessary for LACV-induced generation of ROS (Figure 4H). Furthermore, analysis of genes that are often induced in response to oxidative stress demonstrated increased mRNA expression following LACV infection in WT neurons, but not in Sarm1−/− neurons (Figure S2A). Differences in mRNA expression of oxidative stress response genes were also observed in vivo to a lesser extent (data not shown). These data indicate that SARM1 is necessary for oxidative stress response to virus infection and contributes to LACV-induced neuronal apoptosis.
SARM1-Mediated Damage Is Independent of Interleukin 1 or Type I IFN Pathways
Mechanisms of innate immune signaling-mediated cell death have been described including caspase 1-dependent IL-1 mediated pyroptosis, IFN-mediated apoptosis, and IFN regulatory factor-3 (IRF3)-mediated apoptosis (Eitz Ferrer et al., 2011; Chattopadhyay et al., 2010; Fink et al., 2008). We therefore investigated whether SARM1-mediated neuronal death was influenced by one of these pathways. SARM1-mediated neuronal death was not related to pyroptosis because caspase-1 inhibitors did not suppress neuronal death in vitro, SARM1 deficiency did not affect Il1a or Il1b mRNA expression, and LACV infection did not induce pro-IL-1β cleavage (Figures S3A–S3C, data not shown). Stimulation of neurons with type I IFNs did not significantly alter Sarm1 mRNA expression either in the presence or absence of LACV infection (Figure S3D). Additionally, deficiency in Irf3 and Irf7 or IFN-α receptor (Ifnar1) did not influence LACV-induced cell death or SARM1 expression, suggesting that SARM1-induced neuronal death was not dependent on IRF3 or type I IFN responses (Figure S3E; data not shown). SARM1 deficiency did not suppress type I IFN responses of neurons either in vitro or in vivo (Figure S2B; data not shown), indicating that SARM1 does not mediate antiviral type I IFN responses.
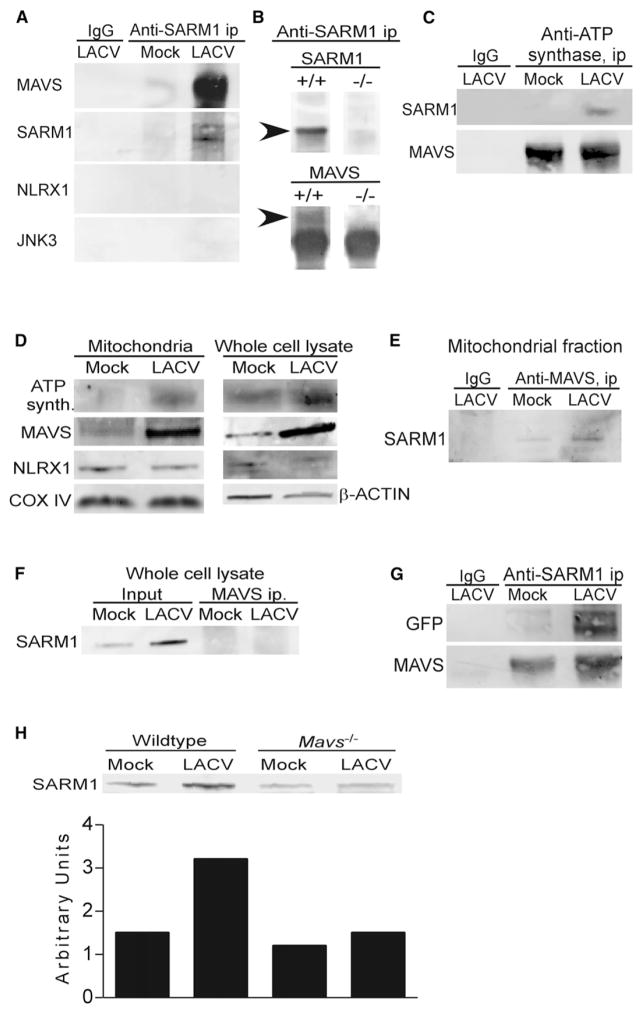
Protein Interactions with SARM1 at the Mitochondria following LACV Infection
In studies using transfected SARM1 in different cells, SARM1 was shown to interact with Syndecan 2 and JNK3 (Chen et al., 2011; Kim et al., 2007). Additionally, SARM1 was predicted to interact with the NOD-like receptor protein, NLRX1 based on computational studies (Li et al., 2011). Surprisingly, immunoprecipitation (IP) of endogenous SARM1 from the mitochondrial fraction of uninfected or infected cortical neurons did not readily precipitate detectable amounts of these proteins (Figure 5A, data not shown). Instead, IP studies with SARM1 analyzed either by tandem mass-spectroscopy (MS/MS) or immunoblot indicated two readily detectable protein interactions with SARM1; ATP synthase and MAVS (Figure 5A; Figure S5). Specificity of the IP with anti-SARM1 in precipitating SARM1 was confirmed using MS analysis (Figure S4) as well as the lack of precipitation of SARM1 in cells from Sarm1−/− mice (Figure 5B). Coimmunoprecipitation (coIP) of these proteins was also detectable by IP of mitochondrial fractions from LACV-infected brain tissue (Figure 5C). Immunoblot analysis on mitochondrial fractions from LACV-infected neurons showed increased amounts of MAVS and ATP synthase at the mitochondria during LACV infection (Figure 5D).
Figure 5. SARM1 Associates with MAVS at the Mitochondria.
(A) IP with anti-SARM1 or IgG control (IgG) from the mitochondrial fractions of mock and LACV-infected WT neurons at 36 hpi, subjected to immunoblotting by using anti-MAVS, anti-SARM1, anti-NLRX1, and anti-JNK3 (left panel).
(B) SARM1 IP readily pulled down SARM1 and MAVS (arrows) in WT, but not Sarm1−/− mice. Arrow indicates correct band size. Lower band for MAVS immunoblot is IgG, which is not shown in (A), (C), or (G).
(C) IP with anti-ATP synthase or IgG control from mitochondrial fractions of brain tissue from mock or LACV-infected mice at 5 dpi. IPs were run on a SDS-PAGE, transferred to a blot, and probed for SARM1 or MAVS.
(D) Immunoblot analysis of mitochondria fraction or whole-cell lysate from mock or LACV-infected WT neurons at 36 hpi probed for ATP synthase, MAVS, NLXR1, Cox IV, and/or β-actin.
(E and F) MAVS was IP from the (E) mitochondria or (F) whole-cell lyaste of neurons at 36 hpi. The blots were probed with anti-SARM1 to confirm coIP of SARM1.
(G) HEK293T cells were transfected with GFP-tagged full-length SARM1 and then infected with LACV. Mitochondria were isolated at 36 hpi for IP analysis using anti-SARM1 antibody. The membrane was probed with anti-GFP or anti- MAVS. (H) Mitochondrial fraction of neurons from WT and Mavs−/− mice at 36 hpi, probed with anti- SARM1. Densitometry quantitation of bands is displayed below in arbitrary units. All data are representative of at least two replicate experiments. See also Figure S4.
MAVS Localizes with SARM1 during LACV Infection
MAVS is located on mitochondria, mitochondrial associated membranes (MAMs), and peroxisomes, and forms a signaling complex following activation of RIG-I and other RLRs (Horner et al., 2011; Seth et al., 2005). LACV infection induces the activation of RIG-I (Verbruggen et al., 2011) and RIG-I protein expression was elevated in brain tissue from LACV-infected mice compared to WT controls (Figure S3F). To confirm MAVS interaction with SARM1, mitochondrial fractions from mock and LACV-infected neurons were immunoprecipitated using anti-MAVS, which resulted in the precipitation of SARM1 from mitochondria (Figure 5E), although it was undetectable in an IP of the whole cell lysate (Figure 5F). CoIP of MAVS with SARM1 was confirmed in the mitochondrial fraction of HEK cells transfected with green-fluorescent protein (GFP)-tagged SARM1 (Figure 5G).
Confocal microscopy analysis also demonstrated colocalization of SARM1 and MAVS in LACV-infected neurons compared to controls (Figure S6). Pearson’s correlation coefficient analysis, which measures degree of colocalization, was higher in LACV-infected neurons (range of 0.067 to 0.624) than in mock-infected cultures (range of −0.312 to −0.078) indicating an increase in colocalization of SARM1 with MAVS following infection. This was particularly true in axons, where SARM1 and MAVS aggregated in specific regions in LACV-infected neurons (Figures S5B and S5D), but not in mock-infected neurons (Figures S5A and S5C). SARM1 also localized with the mitochondrial marker TOMM20 in infected neurons (Figures S5C and S5E), correlating with the IP of SARM1 with MAVS in mitochondrial fractions (Figure 5E). Thus, both IP and colocalization studies demonstrate interactions of SARM1 with MAVS are increased by LACV-infection.
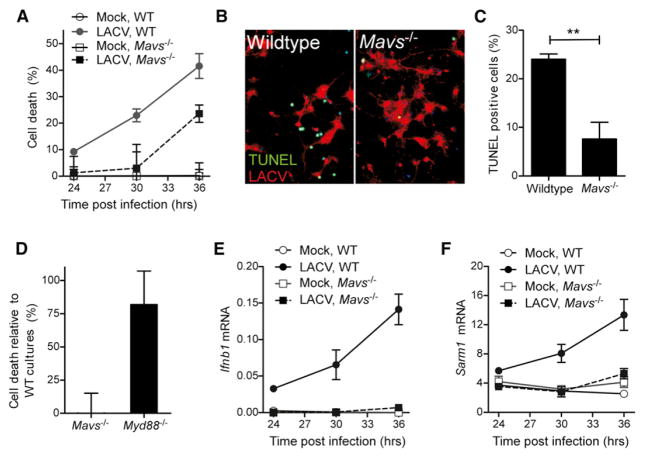
MAVS Deficiency Inhibits SARM1 Upregulation and SARM1-Induced Neuronal Apoptosis
The interaction of SARM1 with MAVS suggested that MAVS may also influence LACV-induced neuronal death. To examine the role of MAVS in SARM-mediated neuronal death, we utilized neurons from Mavs−/− mice. LACV-infection of Mavs−/− neurons did not result in increased SARM1 at the mitochondria as observed in neurons from WT mice (Figure 5H) and did not produce detectable ROS (Figure 4I). Additionally, MAVS-deficient neurons had a delay in the onset of LACV-induced neuronal death (Figures 6A–6C). In contrast, deficiency in MyD88, which is an adaptor molecule for TLR signaling and has been shown to interact with amphioxus SARM1 (Yuan et al., 2010), had no such effect (Figure 6D). This indicated that MAVS, but not MyD88, was required for SARM1-mediated neuronal death during LACV infection. Furthermore, siRNA targeted against RIG-I, which induces MAVS activation, also inhibited LACV-induced cell death (Figure S3H). As expected, deficiency in MAVS inhibited the Ifnb1 mRNA response to virus infection (Figure 6E). However, Sarm1 mRNA expression was also inhibited in Mavs−/− neurons (Figure 6F) indicating that activation through MAVS was required for LACV-induced upregulation of SARM1. Thus, in addition to interacting with SARM1 at the mitochondria, MAVS also appears to be important for the induction of SARM1 during LACV infection.
Figure 6. MAVS Is Required for SARM1 Induced Neuronal Death during LACV Infection.
(A) Cell death in neurons from WT and Mavs−/− mice infected with LACV. MTT assay was performed as described in Figure 2. P value for two-way ANOVA for strain difference was < 0.01.
(B) Neurons from WT (left panel) or Mavs−/− mice stained with TUNEL (green) and LACV (red).
(C) Quantification of the number of TUNEL-positive cells in WT and Mavs−/− cultures as described in Figure 1.
(D) Neurons from WT and either Mavs−/− or Myd88−/− mice were infected with LACV and analyzed for survival using MTT assay. Percent cell death was compared to WT control cultures generated the same day for each deficient strain and each experiment.
(E) Ifnb1 and (F) Sarm1 mRNA in the mock and LACV-infected neurons from WT and Mavs−/− mice were determined quantitatively by real time PCR at 24, 30, and 36 hpi. P value for two-way ANOVA for strain difference between LACV-infected groups was < 0.001 All data are the mean ± SEM of 3 to 6 samples per point and are representative of 2 to 4 replicate experiments. See also Figure S5.
The role for MAVS in SARM1-mediated neuronal death suggested that MAVS may contribute to neuronal pathogenesis in vivo. However, the MAVS pathway may also have an important role in the induction of type I IFN responses produced in response to LACV infection, which are protective in vivo as shown by Ifnar1−/− mice (Blakqori et al., 2007; Hefti et al., 1999) and Irf3−/−, Irf7−/− mice (Figure 3A). Therefore, in vivo, MAVS may have both protective as well as pathogenic roles during LACV infection. Mavs−/− mice infected with 103 PFU of virus had similar incidence and kinetics of neurological disease development comparable to WT mice (Figure 3A), despite reduced type I IFN responses (Figure 3B; data not shown). Thus, MAVS deficiency does not provide the same protection in vivo as SARM1 deficiency, despite the similar effect observed in reducing LACV-induced neuronal death, in vitro.
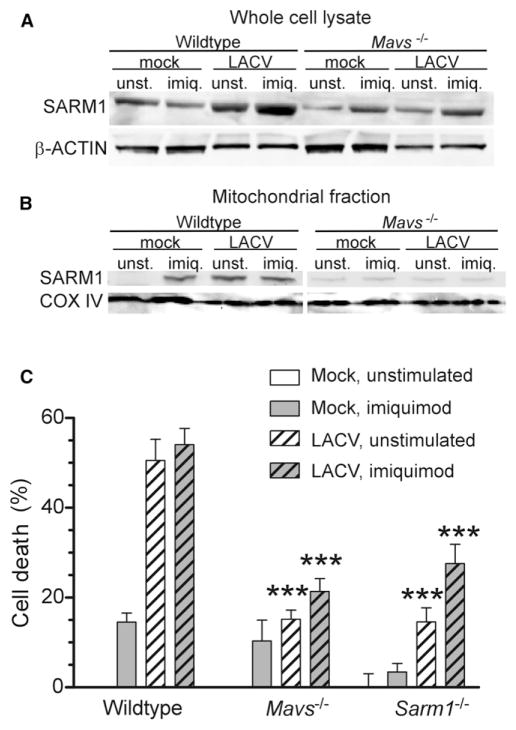
MAVS Influences SARM1 Localization to the Mitochondria during Virus Infection
The above study indicated that MAVS could influence SARM1-mediated cell death at two separate steps during LACV infection, first in the upregulation of SARM1 (Figure 6F) and second at the mitochondria where MAVS directly interacts with SARM1 (Figure 5). Because SARM1 also contributes to neuronal death in instances where MAVS would not be activated, such as OGD, we examined whether MAVS was required for OGD induced neuronal death. Mavs−/− neurons underwent OGD at a similar rate as WT controls (Figure S6), whereas Sarm1−/− neurons did not, indicating that MAVS was not required for SARM1-mediated neuronal death induced by OGD.
To determine whether we could circumvent the MAVS requirement for SARM1 upregulation during LACV infection, we stimulated Mavs−/− neurons with the TLR7 agonist, imiquimod. Stimulation with imiquimod did increase SARM1 mRNA and protein expression in Mavs−/− neurons (Figure 7A; data not shown) and induced a low amount of cell death (Figure 7C; data not shown). However, neurons from Mavs−/− mice had significantly lower amounts of cell death compared to wild-type controls even in the presence of imiquimod-induced SARM1 expression (Figure 7C). Furthermore, even though SARM1 was induced by imiquimod stimulation in Mavs−/− neurons, SARM1 did not localize to the mitochondria in the absence of MAVS (Figure 7B). Thus, MAVS appeared to influence SARM1-induced cell death at two time points during LACV infection, first in the induction of Sarm1 mRNA expression and then in translocation of SARM1 protein to the mitochondria resulting in oxidative damage and neuronal death.
Figure 7. MAVS Is Necessary for SARM1 Localization to the Mitochondria during LACV Infection.
Neurons from Mavs−/− mice were treated with 5 uM imiquimod at the time of infection, and mitochondria and whole cell lysate were generated at 36 hpi.
(A and B) Immunoblot of SARM1 in the (A) whole cell lysate or (B) mitochondrial fraction of cells. Blots from wild-type and MAVS deficient neurons were processed under identical conditions.
(C) Cell viability at 36 hpi from above experiment. Data are shown as the mean ± SEM of 3–6 samples per group per time point. A low amount of imiquimod-induced cell death was observed in most, but not all, experiments. See also Figure S6.
DISCUSSION
RLR-induced MAVS activation has an important role in the generation of type I IFN response and inhibition of virus replication (Scott, 2010). The current study provides evidence that the RLR pathway is also involved in virus-mediated neuronal death and that this pathway is mediated by SARM1. On the basis of our studies, MAVS activation during LACV-infection of neurons results in increased expression of SARM1. SARM1 then localizes to the mitochondria where it interacts with MAVS and induces oxidative stress, mitochondrial damage, and ultimately neuronal death. These studies provide mechanistic details for both SARM1 induction in neurons as well as innate immune signaling-mediated neuronal death.
Our current results indicate that SARM1-mediated cell damage, not virus replication per se, is a key mediator of bunyavirus-induced neuronal death. Cell death was significantly lower in Sarm1−/− neurons compared to WT neurons despite similar amounts of viral RNA and widespread infection of cultures. Similar results were observed in vivo, where virus RNA expression was similar between WT and Sarm1−/− mice, despite the difference in clinical outcome. The ability to inhibit or delay the onset of neuronal damage in the presence of an insult is important not only for therapeutic potential in virus-mediated disease but could also be important for other neurode-generative diseases where neuronal cell death is a hallmark of disease progression.
This study also demonstrates a direct role for the innate immune response in mediating neuronal damage during virus infection. The requirement for MAVS in the induction of SARM1 expression and the interaction of MAVS with SARM1 at the mitochondria demonstrates that the MAVS activation in the neuron can be harmful. Studies with TLR ligands also demonstrated innate immune signaling-mediated neuronal damage (Butchi et al., 2010; Cameron et al., 2007; Lathia et al., 2008; Ma et al., 2006, 2007; Tang et al., 2008), although the mechanism behind TLR-induced cell death is still unknown. The induction of neuronal cell death by stimulation of pattern recognition receptors (PRRs) may offset the benefits of the type I IFN response induced by PRRs in neurons. The role of MAVS in both protection and pathogenesis may explain why MAVS deficiency did not significantly alter the incidence of LACV-induced neurological disease in vivo. Possibly, the balance between the SARM1 pathway and the type I IFN response may determine whether the innate immune response is pathogenic or protective in neurovirulent viral diseases.
In contrast to our results with LACV infection, SARM1 did not alter viral pathogenesis during West Nile Virus (WNV) infection in mice (Szretter et al., 2009). Although both LACV and WNV are RNA viruses that activate the RLR pathway (Verbruggen et al., 2011; Fredericksen and Gale, 2006), they belong to separate virus families and replicate via different mechanisms. Both viruses are also known to express virus proteins that subvert the host immune response (Verbruggen et al., 2011; Fredericksen and Gale, 2006). The different role of SARM1 in the pathogenesis of these viruses may be partially due to different viral proteins affecting neuronal cell death. It will be important to determine whether other encephalitic RNA viruses induce SARM1 expression in neurons or perhaps have mechanisms to limit the SARM1 induction of cell death.
SARM1 has been associated with cell death in other systems, including OGD-induced neuronal death, axonal injury, and most recently T cell death (Kim et al., 2007; Osterloh et al., 2012; Panneerselvam et al., 2013). SARM1-mediated damage appears mechanistically to be similar among different cell types, with SARM1 leading to ROS generation and mitochondrial damage as demonstrated in our study with neurons and as recently shown for T cells (Panneerselvam et al., 2013). However, the mechanism of SARM1 upregulation appears to be through distinct pathways, with innate immune responses leading to SARM1 expression in neurons, whereas SARM1 upregulation in T cells was associated with activation-induced cell death and neglect-induced cell death pathways (Panneerselvam et al., 2013).
MAVS also contributes to apoptosis of other cell types (Lei et al., 2009; McAllister and Samuel, 2009; Yu et al., 2010). MAVS-mediated cell death in these systems is independent of the type I IFN response, similar to our current study where deficiency in IFNAR1 or IRF3 and IRF7 did not affect cell death. However, MAVS induces NF-κB activation (Sun et al., 2006), which is also observed following LACV infection (data not shown), and this pathway may be responsible for the increase in SARM1. It is possible that increased SARM1 expression is a common response to MAVS activation and that other factors, such as the expression of antiapoptotic molecules, determine whether MAVS activation leads to cell death. Overexpression of SARM1 through MAVS stimulation may have therapeutic potential in diseases where targeted cell death is the goal as in the case of tumors.
Other proteins may also influence MAVS and SARM1 activation during LACV infection. MAVS forms a signaling complex at the mitochondria or MAMs where it interacts with the ubiquitin protein ligase TRAF6 to induce signaling (Yoshida et al., 2008). NLRX1 can also interact with MAVS at the complex to negatively regulate MAVS (Allen et al., 2011). Although NLRX1 was predicted to interact with SARM1 based on computational studies (Li et al., 2011), we did not observe an interaction between SARM1 and NLRX1 in neurons. Furthermore, SARM1 deficiency did not affect type I IFN or other cytokine production suggesting that SARM1 does not influence the MAVS signaling pathway.
ATP synthase interacted with SARM1 both in vitro and in vivo. Disrupted ATP synthase expression and/or function has been correlated with the generation of ROS in multiple studies, including ROS generation in neurons (Natera-Naranjo et al., 2012; Comelli et al., 1998). Although our current studies have not identified suppression of ATP synthase activity by SARM1, SARM1 interactions with ATP synthase may affect ATP synthase function and influence the generation of ROS.
In the current study, SARM1 interacted with MAVS at the mitochondria. Deficiency in MAVS resulted in a decrease in SARM1 localization to the mitochondria, even when SARM1 was independently induced by imiquimod stimulation. However, SARM1 also contributes to neuronal death in instances where MAVS may not be activated, such as OGD or axonal damage (Kim et al., 2007; Osterloh et al., 2012). Indeed, MAVS was not necessary for OGD-induced cell death, suggesting that other mechanisms can induce mitochondrial localization of SARM1. SARM1 contains a mitochondrial signaling sequence (Panneerselvam et al., 2012) and overexpression of SARM1 constructs in COS-cells or HEK cells is sufficient to induce mitochondrial SARM1 localization (Kim et al., 2007; Panneerselvam et al., 2013). However, SARM1 also interacts with the cytoplasmic domain of syndecan 2 in neurons (Chen et al., 2011). Perhaps in neurons, SARM1 needs a trigger for mitochondrial localization by interacting with a complex containing MAVS or other mitochondrial proteins. Although the amount of SARM1 induced by imiquimod stimulation was similar to that induced by LACV infection alone, it may be insufficient to induce mitochondrial localization in the absence of critical cofactor. Further investigation into the mechanisms by which SARM1 localizes to the mitochondria in other disease models will be essential in understanding how SARM1 contributes to cell death.
Our studies demonstrate a mechanism of virus-induced neuronal cell death that involves activation of the innate immune response and SARM1 induction. Because SARM1 is normally expressed in neurons and has a clear role in dendritic arborization (Chen et al., 2011), the overexpression of SARM1 and resulting oxidative damage may simply be an unintended consequence of stimulation of neurons through pathways that are primarily required for neuronal development. Developing therapeutics that target SARM1, but not MAVS, may be a viable strategy for inhibiting neuronal damage during virus infection while still allowing antiviral responses induced by RIG-I and MAVS activation.
EXPERIMENTAL PROCEDURES
Infection of Mice with LACV
Sarm1−/− and Irf3−/−, Irf7−/− mice were kindly provided by Michael Diamond, Washington University (Szretter et al., 2009; Daffis et al., 2009). These mice as well as Myd88−/− mice (Adachi et al., 1998) and Ifnar1−/− mice (Auerbuch et al., 2004) were maintained on a C57BL/6 background. Mavs−/− mice were purchased from Jackson Laboratories. These strains, as well as Inbred Rocky Mountain White (IRW) mice, were maintained at Rocky Mountain Laboratories (RML). WT refers to the C57BL/6 strain unless otherwise noted. All of the animal procedures were conducted in accordance with the RML Animal Care and Use Committee guidelines under protocol RML2011-65. LACV human 1978 stock was a kind gift from Richard Bennett (NIAID, NIH) and has been previously described (Bennett et al., 2007). At 3 weeks of age, mice were inoculated intraperitoneally with 103 PFU of LACV. Mice were observed daily for signs of encephalitis. Mice at 5 dpi or at the time of clinical signs were euthanized and brains were removed for histology and RNA analysis.
Primary Cultures of Cortical Neurons and LACV Infection
Primary cultures of cortical neurons were prepared from 14 to 16 day gestation mouse embryos from indicated mouse strains or from IRW mice. Mouse cortices were digested in CMF-HBSS containing 0.125% Trypsin followed by dissociation by repeated pipetting. Cells were plated in amine-coated plates (BD Biosciences) at 8×105 cells/ml. Following attachment, the medium was replaced with neurobasal medium containing 2% B-27 and 0.5 mM glutamine. After overnight culture, primary neurons were infected with LACV at a MOI of 0.01 unless otherwise indicated. Mock-infected cultures were treated with equivalent amounts of supernatants from uninfected Vero cells. Samples were collected for RNA analysis or MTT assay at 36 hpi unless otherwise indicated. For caspase inhibition studies, cells were treated with 200 μM of Z-VAD-FMK or 10 μM of Z-WEHD-FMK (R&D Systems) at the time of LACV infection. For imiquimod studies, 5 μM of imiquimod was added to cultures at the time of LACV infection. For OGD studies, neurons were cultured in OGD buffer as described (Kim et al., 2007) and placed in an incubator containing 95% N2 and 5% CO2 for 1–3 hr.
Histology
Mouse brains were fixed in 10% neutral-buffered formalin, embedded in paraffin, and cut in 4 μm sections. Immunohistochemical analysis was completed using an antigen-retrieval protocol (Du et al., 2010). SARM was detected using an anti-SARM1 (Proscience) and detected using Alexa Fluor 555 conjugated goat anti-rabbit. SARM1 specificity was confirmed by staining tissue sections from Sarm1−/− mice. Cell body staining of SARM1 was confirmed as specific (Figures S1D–S1F, yellow arrow), however, nonspecific staining of nuclei was observed in both WT and Sarm1−/− mice as shown in spinal cord sections (Figures S1D–S1F, white arrow). Neurons were detected by using mouse anti-MAP2 (Millipore) with an Alexa Fluor 488 conjugated anti-mouse. LACV infection was detected using anti-G2 monoclonal (QED Biosciences, 18572) and Alexa Fluor 555 goat anti-mouse. TUNEL was detected by using the in situ cell death detection kit (Roche Applied Science). Slides were mounted with ProLong Gold antifade reagent (Invitrogen) with or without DAPI. Slides incubated without primary antibodies or with isotype controls were used to confirm specificity.
Real-Time PCR
RNA was isolated from primary cortical neurons using the RNA isolation kit (Zymo Research). cDNA was prepared from RNA samples as described (Du et al., 2010). Primers were designed using primer3 website (Rozen and Skaletsky, 2000) with a Tm of 60°C. SYBR green dye with ROX (Bio-Rad) was used for measurement of real-time PCR amplification. Data for each sample was calculated as the percent difference in CT value (ΔCT = CT Gapdh - CT gene of interest). The data was plotted as mean percent Gapdh values for each gene of interest for each sample. Superarray analysis was conducted as described (Du et al., 2010). Data were calculated as fold differences between mock and LACV infected neurons and brain homogenates for both WT and Sarm1−/− mice.
MTT Assay to Determine Cell Viability
Neurons were cultured in 96 well plates and inoculated with mock supernatants or LACV. At specific time points, triplicate or quadruplicate wells per treatment were incubated with MTT reagent (Invitrogen) at a concentration of 0.5 mg/ml for 3 hr. The MTT solution was aspirated and the cells were lysed in DMSO. The formazan concentration in each well was measured by absorbance at 540 nm using a cell plate reader (Synergy 4, BioTek). Data were compared with mock-infected cultures to determine percent cell death. Cultures from deficient mice were directly compared to WT control cultures generated the same day and treated with the same virus stock.
Measurement of DNA Fragmentation by Using TUNEL Assay
Apoptotic cell death following LACV infection was analyzed in the primary cortical neurons by TUNEL reaction using the in situ cell death detection kit (Roche). Cells were further stained with a mouse antibody against the G2 envelope protein of LACV (QED Bioscience, Inc.) and anti-mouse Alexa Fluor 555 (Invitrogen).
Caspase-3 Detection
Caspase-3 activity was determined using a caspase-3 colorimetric assay kit (GenScript). Cells were lysed, centrifuged at 10,000 rpm, and the supernatant was collected. We incubated 300 μg of protein with the caspase-3 substrate. The samples were incubated for 6 hr and the extinction values obtained using Synergy 4, Biotek spectrophotometer at 405 nm.
Transfection of Primary Neurons with SARM1, Mda5, or Rig-I siRNA
Primary cortical neurons were cultured for 3 days and transfected with 50 nM of Sarm1 (Dharmacon, Thermo Scientific), Mda5, or Rig-I siRNA (Santa Cruz Biotechnology) using 0.5 μl of HiPerFect (QIAGEN) for 24 hr. For confirmation of Sarm1 inhibition, siRNA for Sarm1 from Santa Cruz Biotechnology was also used and yielded similar results. All siRNAs were a pool of three target-specific siRNAs. Cells also were transfected with a nontargeting siRNA (Dharmacon, Thermo Scientific). Neurons were then infected with LACV at a MOI of 0.01 and cultured for 36 hpi. Transfection efficiency was ~35% as confirmed with cGFP siRNA transfection.
Transfection of HEK Cells with GFP-Tagged SARM1
HEK293T cells were seeded at a density of 4 × 106 cells/ml. Cells were transfected with mouse GFP-SARM1 (OriGene) and LTX reagent (Invitrogen). Following transfection, cells were infected with LACV (MOI 0.01). At 36 hpi, mitochondria were isolated from SARM1-transfected mock and LACV infected cells and immunoprecipitated using SARM1 antibody (Proscience).
Immunoprecipitation and Immunoblot
Mitochondria and cytosolic fractions from mock and LACV-infected primary neurons were isolated by using a mitochondria and cytosol fractionation kit (Millipore). The IP matrix from ImmunoCruz IP/WB Optima F system (Santa Cruz) was used for all IP. For each reaction, the IP-matrix was incubated overnight with anti-SARM1 (Proscience). Intact mitochondrial fractions were lysed in 0.1% NP-40, 50 mM Tris, pH 7.4, and 25 mM NaCl, precleared with nonspecific IgG and added to IP-matrix beads and incubated overnight prior to processing for immunoblot analysis. Immunoblotting was done using anti-rabbit SARM1 (Genetex), anti-MAVS (Cell Signaling), anti-NLRX1 (Millipore), and anti-mouse JNK3 (Novus Biologicals). Cox IV (Abcam) was used as a mitochondrial loading control. Protein was detected using a Typhoon scanner and analyzed with ImageQuant TL software (GE Healthcare). Specificity of the SARM1 antibody was confirmed by comparison to cells from Sarm1−/− mice (Figure 1F).
Tandem Mass Spectrometry Analysis
Samples isolated by IP were separated by SDS-PAGE and stained with Coomassie blue. Those bands selected for analysis by HPLC-based nanospray LC-MS/MS were processed and analyzed as described previously (Moore et al., 2010). Peak lists were searched using MASCOT Daemon (Perkins et al., 1999). The proteins identified by MASCOT were visualized using the proteomics software ProteoIQ (NuSep, Inc Athens, GA, USA). Proteins were considered present in the sample only if they were identified by at least two peptides having distinct sequences and Mascot ion scores above 35.
Transmission Electron Microscopy
Primary cortical neurons were cultured on Aclar coverslips precoated with poly-D-Lysine (0.1 mg/ml) (Sigma). Cells were infected with LACV at a MOI of 0.01 and following 36 hpi, cells were fixed with 2.5% glutaraldehyde. Samples were prepared for TEM following standard protocols and analyzed using a H7500 microscope (Hitachi high Technologies, Tokyo, Japan). Images were acquired in single-blind experiments.
Confocal Microscopy
For confocal microscopy, primary cortical neurons were grown on poly-Dlysine coated chamber slides and infected with LACV at a MOI of 0.01. For colocalization studies, cells were fixed and permeabilized at 36 hpi and costained with anti-SARM1 (a gift from Aihao Ding, Cornell University) and anti-MAVS (Cell Signaling). The slides were analyzed using a Zeiss 510 Meta confocal microscope.
Detection of Mitochondrial Superoxide Generation
To detect relative superoxide generation, MitoSox Red (Invitrogen) was used. Primary cortical neurons were cultured in 8-well chamber slides. At 36 hpi with LACV, cells were incubated with 5 uM of MitoSox Red for 10 min. The relative intensity of mitochondrial superoxide generation was analyzed using a Zeiss 510 Meta confocal microscope. Images were acquired in single-blinded experiments.
Statistical Analysis
All statistical analysis was completed using Graphpad Prism. *p < 0.05, **p < 0.01, and ***p < 0.001. For a comparison of two values, a two-tailed Mann-Whitney analysis was performed. For comparison of more than two values with one variable, a one-way ANOVA was used with a Bonferroni’s post-test. For comparison of two or more values with more than one variable, a two-way ANOVA was used with a Bonferroni’s post-test.
Supplementary Material
Acknowledgments
The authors thank Sonja Best and Vinod Nair for confocal microscopy assistance and David Dorward for transmission electron microscopy assistance. The authors also thank Austin Athman for graphics assistance and Sonja Best, Sue Priola, Byron Caughey, and Kim Hasenkrug for critical reading of the manuscript. The project was supported by the Division of Intramural Research, National Institutes of Health, National Institute of Allergy and Infectious Diseases.
Footnotes
Supplemental Information includes six figures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2013.02.013.
References
- Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, Gris D, Roney KE, Zimmermann AG, Bowzard JB, et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-κB signaling pathways. Immunity. 2011;34:854–865. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbuch V, Brockstedt DG, Meyer-Morse N, O’Riordan M, Portnoy DA. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J Exp Med. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RS, Ton DR, Hanson CT, Murphy BR, Whitehead SS. Genome sequence analysis of La Crosse virus and in vitro and in vivo phenotypes. Virol J. 2007;4:41. doi: 10.1186/1743-422X-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RS, Cress CM, Ward JM, Firestone CY, Murphy BR, Whitehead SS. La Crosse virus infectivity, pathogenesis, and immunogenicity in mice and monkeys. Virol J. 2008;5:25. doi: 10.1186/1743-422X-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakqori G, Delhaye S, Habjan M, Blair CD, Sánchez-Vargas I, Olson KE, Attarzadeh-Yazdi G, Fragkoudis R, Kohl A, Kalinke U, et al. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J Virol. 2007;81:4991–4999. doi: 10.1128/JVI.01933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchi NB, Du M, Peterson KE. Interactions between TLR7 and TLR9 agonists and receptors regulate innate immune responses by astrocytes and microglia. Glia. 2010;58:650–664. doi: 10.1002/glia.20952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JS, Alexopoulou L, Sloane JA, DiBernardo AB, Ma Y, Kosaras B, Flavell R, Strittmatter SM, Volpe J, Sidman R, Vartanian T. Toll-like receptor 3 is a potent negative regulator of axonal growth in mammals. J Neurosci. 2007;27:13033–13041. doi: 10.1523/JNEUROSCI.4290-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty M, Goodbody R, Schröder M, Stack J, Moynagh PN, Bowie AG. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol. 2006;7:1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Marques JT, Yamashita M, Peters KL, Smith K, Desai A, Williams BR, Sen GC. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J. 2010;29:1762–1773. doi: 10.1038/emboj.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Lin CW, Chang CY, Jiang ST, Hsueh YP. Sarm1, a negative regulator of innate immunity, interacts with syndecan-2 and regulates neuronal morphology. J Cell Biol. 2011;193:769–784. doi: 10.1083/jcb.201008050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comelli M, Londero D, Mavelli I. Severe energy impairment consequent to inactivation of mitochondrial ATP synthase as an early event in cell death: a mechanism for the selective sensitivity to H2O2 of differentiating erythroleukemia cells. Free Radic Biol Med. 1998;24:924–932. doi: 10.1016/s0891-5849(97)00373-0. [DOI] [PubMed] [Google Scholar]
- Daffis S, Suthar MS, Szretter KJ, Gale M, Jr, Diamond MS. Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent signal that does not require IRF-3 and IRF-7. PLoS Pathog. 2009;5:e1000607. doi: 10.1371/journal.ppat.1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Butchi NB, Woods T, Morgan TW, Peterson KE. Neuropeptide Y has a protective role during murine retrovirus-induced neurological disease. J Virol. 2010;84:11076–11088. doi: 10.1128/JVI.01022-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitz Ferrer P, Potthoff S, Kirschnek S, Gasteiger G, Kastenmüller W, Ludwig H, Paschen SA, Villunger A, Sutter G, Drexler I, Häcker G. Induction of Noxa-mediated apoptosis by modified vaccinia virus Ankara depends on viral recognition by cytosolic helicases, leading to IRF-3/IFN-β-dependent induction of pro-apoptotic Noxa. PLoS Pathog. 2011;7:e1002083. doi: 10.1371/journal.ppat.1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink SL, Bergsbaken T, Cookson BT. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc Natl Acad Sci USA. 2008;105:4312–4317. doi: 10.1073/pnas.0707370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen BL, Gale M., Jr West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J Virol. 2006;80:2913–2923. doi: 10.1128/JVI.80.6.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt RR, Gottfried KL, Apperson CS, Davis BS, Erwin PC, Smith AB, Panella NA, Powell EE, Nasci RS. First isolation of La Crosse virus from naturally infected Aedes albopictus. Emerg Infect Dis. 2001;7:807–811. doi: 10.3201/eid0705.017506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddow AD, Odoi A. The incidence risk, clustering, and clinical presentation of La Crosse virus infections in the eastern United States, 2003–2007. PLoS ONE. 2009;4:e6145. doi: 10.1371/journal.pone.0006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefti HP, Frese M, Landis H, Di Paolo C, Aguzzi A, Haller O, Pavlovic J. Human MxA protein protects mice lacking a functional alpha/beta interferon system against La crosse virus and other lethal viral infections. J Virol. 1999;73:6984–6991. doi: 10.1128/jvi.73.8.6984-6991.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner SM, Liu HM, Park HS, Briley J, Gale M., Jr Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci USA. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalfayan B. Pathology of La Crosse virus infection in humans. Prog Clin Biol Res. 1983;123:179–186. [PubMed] [Google Scholar]
- Kim Y, Zhou P, Qian L, Chuang JZ, Lee J, Li C, Iadecola C, Nathan C, Ding A. MyD88-5 links mitochondria, microtubules, and JNK3 in neurons and regulates neuronal survival. J Exp Med. 2007;204:2063–2074. doi: 10.1084/jem.20070868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Okun E, Tang SC, Griffioen K, Cheng A, Mughal MR, Laryea G, Selvaraj PK, ffrench-Constant C, Magnus T, et al. Toll-like receptor 3 is a negative regulator of embryonic neural progenitor cell proliferation. J Neurosci. 2008;28:13978–13984. doi: 10.1523/JNEUROSCI.2140-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Moore CB, Liesman RM, O’Connor BP, Bergstralh DT, Chen ZJ, Pickles RJ, Ting JP. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS ONE. 2009;4:e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang L, Berman M, Kong YY, Dorf ME. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35:426–440. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Li J, Chiu I, Wang Y, Sloane JA, Lü J, Kosaras B, Sidman RL, Volpe JJ, Vartanian T. Toll-like receptor 8 functions as a negative regulator of neurite outgrowth and inducer of neuronal apoptosis. J Cell Biol. 2006;175:209–215. doi: 10.1083/jcb.200606016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Haynes RL, Sidman RL, Vartanian T. TLR8: an innate immune receptor in brain, neurons and axons. Cell Cycle. 2007;6:2859–2868. doi: 10.4161/cc.6.23.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister CS, Samuel CE. The RNA-activated protein kinase enhances the induction of interferon-beta and apoptosis mediated by cytoplasmic RNA sensors. J Biol Chem. 2009;284:1644–1651. doi: 10.1074/jbc.M807888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin JE, de los Reyes EC, Irazuzta JE, Caceres MJ, Khan RR, Minnich LL, Fu KD, Lovett GD, Tsai T, Thompson A. La Crosse encephalitis in children. N Engl J Med. 2001;344:801–807. doi: 10.1056/NEJM200103153441103. [DOI] [PubMed] [Google Scholar]
- Mink M, Fogelgren B, Olszewski K, Maroy P, Csiszar K. A novel human gene (SARM) at chromosome 17q11 encodes a protein with a SAM motif and structural similarity to Armadillo/beta-catenin that is conserved in mouse, Drosophila, and Caenorhabditis elegans. Genomics. 2001;74:234–244. doi: 10.1006/geno.2001.6548. [DOI] [PubMed] [Google Scholar]
- Moore RA, Timmes A, Wilmarth PA, Priola SA. Comparative profiling of highly enriched 22L and Chandler mouse scrapie prion protein preparations. Proteomics. 2010;10:2858–2869. doi: 10.1002/pmic.201000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-Naranjo O, Kar AN, Aschrafi A, Gervasi NM, Macgibeny MA, Gioio AE, Kaplan BB. Local translation of ATP synthase subunit 9 mRNA alters ATP levels and the production of ROS in the axon. Mol Cell Neurosci. 2012;49:263–270. doi: 10.1016/j.mcn.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Osterloh JM, Yang J, Rooney TM, Fox AN, Adalbert R, Powell EH, Sheehan AE, Avery MA, Hackett R, Logan MA, et al. dSarm/Sarm1 Is Required for Activation of an Injury-Induced Axon Death Pathway. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Xiao Q, Sheng CY, Hong Z, Yang HQ, Wang G, Ding JQ, Chen SD. Blockade of the translocation and activation of c-Jun N-terminal kinase 3 (JNK3) attenuates dopaminergic neuronal damage in mouse model of Parkinson’s disease. Neurochem Int. 2009;54:418–425. doi: 10.1016/j.neuint.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Panneerselvam P, Singh LP, Ho B, Chen J, Ding JL. Targeting of pro-apoptotic TLR adaptor SARM to mitochondria: definition of the critical region and residues in the signal sequence. Biochem J. 2012;442:263–271. doi: 10.1042/BJ20111653. [DOI] [PubMed] [Google Scholar]
- Panneerselvam P, Singh LP, Selvarajan V, Chng WJ, Ng SB, Tan NS, Ho B, Chen J, Ding JL. T-cell death following immune activation is mediated by mitochondria-localized SARM. Cell Death Differ. 2013;20:478–489. doi: 10.1038/cdd.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekosz A, Phillips J, Pleasure D, Merry D, Gonzalez-Scarano F. Induction of apoptosis by La Crosse virus infection and role of neuronal differentiation and human bcl-2 expression in its prevention. J Virol. 1996;70:5329–5335. doi: 10.1128/jvi.70.8.5329-5335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Yuan Q, Lin B, Panneerselvam P, Wang X, Luan XL, Lim SK, Leung BP, Ho B, Ding JL. SARM inhibits both TRIF- and MyD88-mediated AP-1 activation. Eur J Immunol. 2010;40:1738–1747. doi: 10.1002/eji.200940034. [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. In: Primer3 on the WWW for general users and for biologist programmers. Krawetz S, Misener S, editors. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Scott I. The role of mitochondria in the mammalian antiviral defense system. Mitochondrion. 2010;10:316–320. doi: 10.1016/j.mito.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Szretter KJ, Samuel MA, Gilfillan S, Fuchs A, Colonna M, Diamond MS. The immune adaptor molecule SARM modulates tumor necrosis factor alpha production and microglia activation in the brainstem and restricts West Nile Virus pathogenesis. J Virol. 2009;83:9329–9338. doi: 10.1128/JVI.00836-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SC, Lathia JD, Selvaraj PK, Jo DG, Mughal MR, Cheng A, Siler DA, Markesbery WR, Arumugam TV, Mattson MP. Toll-like receptor-4 mediates neuronal apoptosis induced by amyloid beta-peptide and the membrane lipid peroxidation product 4-hydroxynonenal. Exp Neurol. 2008;213:114–121. doi: 10.1016/j.expneurol.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen P, Ruf M, Blakqori G, Överby AK, Heidemann M, Eick D, Weber F. Interferon antagonist NSs of La Crosse virus triggers a DNA damage response-like degradation of transcribing RNA polymerase II. J Biol Chem. 2011;286:3681–3692. doi: 10.1074/jbc.M110.154799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Takaesu G, Yoshida H, Okamoto F, Yoshioka T, Choi Y, Akira S, Kawai T, Yoshimura A, Kobayashi T. TRAF6 and MEKK1 play a pivotal role in the RIG-I-like helicase antiviral pathway. J Biol Chem. 2008;283:36211–36220. doi: 10.1074/jbc.M806576200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Chiang RL, Chang TH, Liao CL, Lin YL. The interferon stimulator mitochondrial antiviral signaling protein facilitates cell death by disrupting the mitochondrial membrane potential and by activating caspases. J Virol. 2010;84:2421–2431. doi: 10.1128/JVI.02174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Wu K, Yang M, Xu L, Huang L, Liu H, Tao X, Huang S, Xu A. Amphioxus SARM involved in neural development may function as a suppressor of TLR signaling. J Immunol. 2010;184:6874–6881. doi: 10.4049/jimmunol.0903675. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.