Summary
Objective
Hippocampal sclerosis is the most common neuropathological finding in medically intractable cases of mesial temporal lobe epilepsy. In this study, we analyzed the gene expression profiles of dentate granule cells of mesial temporal lobe epilepsy patients with and without hippocampal sclerosis to show that next-generation sequencing methods can produce interpretable genomic data from RNA collected from small homogenous cell populations and to shed light on the transcriptional changes associated with hippocampal sclerosis.
Methods
RNA was extracted, and complementary DNA (cDNA) was prepared and amplified from dentate granule cells that had been harvested by laser capture microdissection from surgically resected hippocampi from mesial temporal lobe epilepsy patients with and without hippocampal sclerosis. Sequencing libraries were sequenced, and the resulting sequencing reads were aligned to the reference genome. Differential expression analysis was used to ascertain expression differences between patients with and without hippocampal sclerosis.
Results
Greater than 90% of the RNA-Seq reads aligned to the reference. There was high concordance between transcriptional profiles obtained for duplicate samples. Principal component analysis revealed that the presence or absence of hippocampal sclerosis was the main determinant of the variance within the data. Among the genes upregulated in the hippocampal sclerosis samples, there was significant enrichment for genes involved in oxidative phosphorylation.
Significance
By analyzing the gene expression profiles of dentate granule cells from surgically resected hippocampal specimens from mesial temporal lobe epilepsy patients with and without hippocampal sclerosis, we have demonstrated the utility of next-generation sequencing methods for producing biologically relevant results from small populations of homogeneous cells, and have provided insight on the transcriptional changes associated with this pathology.
Keywords: temporal lobe epilepsy, hippocampus, mesial temporal sclerosis, RNA-Seq
Introduction
Hippocampal sclerosis (HS), also referred to as mesial temporal sclerosis (MTS), is the most common neuropathological finding in mesial temporal lobe epilepsy (MTLE). Within the dentate gyrus, hippocampal sclerosis may be associated with granule cell dispersion and aberrant mossy fiber sprouting1, and these pathological changes are accompanied by a range of molecular alterations. HS occurs in the majority of patients referred to tertiary treatment centers for surgical treatment of drug-resistant temporal lobe epilepsy2. Neuronal loss in the CA1, CA3, and CA4 regions of the hippocampus and reactive gliosis commonly characterize HS3. Recently, there has been increasing investigation into the HS-associated changes that occur within the dentate gyrus, the primary site of hippocampal neurogenesis in the adult brain.
To date, the majority of studies have examined the molecular changes associated with MTLE in the hippocampus, as opposed to specific populations of cells. These studies have shown that miR-218 and miR-204 are downregulated in patients with MTLE with HS (MTLE-HS)4; the gene targets of these microRNAs participate in axonal guidance and synaptic plasticity. There are also reports of clear changes in epigenetic profiles. Miller-Delaney et al. recently demonstrated hypermethylation of genes involved in synaptic function and ion channels in hippocampi from TLE patients compared to control brains5; they also noted distinct methylation profiles for different subtypes of HS, which were reflected in differential expression of a select set of genes. Previous studies have noted changes in the expression of genes associated with several pathways and processes in HS; for example, HS has been associated with increased expression of pro-apoptotic genes, such as BCL-26;7 and simultaneous activation of cell survival pathways8. Johnson et al. demonstrated that in hippocampi from MTLE patients with HS compared to those from healthy controls, there is activation of inflammatory cytokines and the toll-like receptor (TLR)-signaling pathway9; these gene networks have been previously implicated in inflammation and epilepsy.
In MTLE-HS, there are pronounced changes in the organization, behavior, and growth patterns of dentate granule (DG) cells. One such marker is granule cell dispersion, which is characterized by losses in cell density10. Another is aberrant mossy fiber (axonal projection) sprouting, which leads to the reorganization of synapses and contributes to excitatory discharges within the dentate gyrus 11. Researchers have begun to examine the molecular changes in DG cells in MTLE-HS. In one narrowly focused study looking at microRNA profiles only in DG cells from patients with MTLE-HS, the authors showed that miR-1234, miR-1281, miR-30c-1, miR-92b, miR-191, and miR-1238 were more highly expressed in DG cells from patients with granule cell pathology than in those from patients without12. We aimed to expand upon the findings of these previous studies by evaluating the mRNA expression profiles associated with the presence and absence of HS at the level of the DG cells.
By examining changes in gene expression associated with HS in DG cells, this study sought to shed light on the molecular changes associated with HS in a cell type with specific HS-associated pathological changes. We hypothesize that MTLE-HS may be molecularly, and possibly pathophysiologically, distinct from MTLE without HS. Given the distinct changes in organization and behavior of dentate granule cells in HS, it is likely that the specific pathological changes seen in DG cells can be explained by molecular changes that occur only within these cells and that these changes may be overlooked in analyses studying whole hippocampal specimens. Thus, RNA-Seq was used to assess the transcriptome in DG cells from surgically resected hippocampi from five MTLE patients with HS and seven without. In addition to providing insights into the changes in gene expression associated with HS, this study demonstrates the ability of next-generation sequencing methods to produce interpretable, reproducible, and likely biologically relevant results from a small population of homogenous cells.
Methods
Patients and Sample Collection
Surgically resected hippocampal specimens from 12 patients who had undergone surgery for seizure treatment at Duke University Hospitals were included in the study under approval from the Institutional Review Board of Duke University. Written, informed consent was obtained from each patient and/or his/her guardian. En bloc surgery was performed by the same neurosurgeon in all cases without cauterization from the posterior margin to the uncus. A cortical specimen and a portion of the hippocampal tissue were submitted for routine pathological analysis. An adjacent hippocampal section containing the dentate gyrus was collected immediately after surgical resection and flash frozen in liquid nitrogen, stored frozen at −80°C, and subsequently embedded in Optimal Cutting Temperature compound (OCT) with the desired orientation on dry ice. Frozen serial sections were cut at a thickness of 8 micrometers on a cryostat at −18°C and mounted on sterile superfrost charged glass slides. The sections were immediately fixed for 30 seconds with 95% ethanol, followed by 75% ethanol for 30 seconds to remove the OCT. Sections were stained with 1% cresyl violet in 75% ethanol for 30 seconds and dehydrated in 75% and 95% ethanol for 30 seconds each, 2x in 100% ethanol for 30 seconds, and xylene for 5 minutes. Stained DG cells were visualized and identified on sections under an inverted light microscope before laser capture microdissection (LCM). Figure 1 illustrates the location of the DG cells within a representative hippocampal tissue specimen used in this study and displays these cells after LCM. The microdissection of pure DG cells was performed in duplicate for each sample with a Veritas LCM instrument (Molecular Devices), and approximately 800 to 1,000 DG cells were harvested on CapSure Macro LCM caps using an infrared laser system within 30 minutes for each duplicate. The LCM macro caps containing captured pure DG cells were immediately placed in 0.5 ml microcentrifuge tubes with 50 μl lysis buffer. The microcentrifuge tubes were inverted in a 42°C degree incubator for 30 minutes to extract RNA and stored at −80°C until RNA isolation. For each patient, the presence or absence of HS was confirmed through examination of pre-operative MRI and post-operative histopathological examination of resected tissue adjacent to the specimens used for microdissection.
Figure 1.
Laser capture microdissection (LCM) of dentate granule cells from surgically resected hippocampal tissue from a patient with mesial temporal lobe epilepsy. A) Section of hippocampal tissue showing the dentate granule cells stained with cresyl violet, 10X magnification. B) The same tissue section after LCM, 10X magnification. C) The captured dentate granule cells, 10X magnification
RNA Sequencing
Total RNA was extracted from the harvested DG cells using the ARCTURUS PicoPure RNA Isolation Kit, and the RNA integrity numbers (RIN) were determined using the 2100 Agilent Bioanalyzer 6000 Pico assay. Total RNA was amplified using the Clontech SMARTer Ultra Low RNA Kit according to the manufacturer’s instructions. Sequencing libraries were then prepared using the TruSeq RNA Sample Preparation Kit. The RNA sequencing libraries for two samples were prepared together and then multiplexed across one lane of an Illumina HiSeq 2000. For sequencing alignment, the sequencing adapters were removed, the paired-end sequencing reads were aligned to the reference genome with TopHat2 13, and the transcripts were assembled with Cufflinks14.
Principal Component Analysis (PCA)
PCA was performed in the R software environment using the DESeq2 package, version 3.1.215; 16 after accounting for batch effects using the comBat() function in the sva package, version 3.1.117. Specifically, this function was used to produce a set of transformed reads from the raw read counts for the 500 genes with the greatest variance by accounting for batch effects as a covariate; this transformed data set was then used to create a count matrix within DESeq2. These data were then log-transformed with the rld() function and passed to the plotPCA() function to produce a PCA plot of the first two principal components.
Differential Expression Analysis
Differential expression analysis was performed in edgeR, version 3.1.2, in R using the generalized linear model workflow described in section 1.4 of the edgeR manual18; 19. First, the sequencing reads for duplicate sequencing libraries were combined to produce a single set of sequencing reads for each sample, and the raw read counts for each gene were calculated with the Subread software, version 1.4.6, and used to produce a DGEList object in edgeR. Genes were only included if they were represented by at least one read in all of the samples, resulting in an expression data set of 12,605 genes. The calcNormFactors() function was used to account for differences in the library size for each sample, and an experimental design model consisting of the batch and HS status was established. The common, trended, and tagwise dispersions of the expression data were then calculated using the estimateGLMCommonDisp(), estimateGLMTrendedDisp(), and estimateGLMTagwiseDisp() functions, respectively; these dispersions are calculated with the Cox-Reid profile-adjusted likelihood model, taking into account the sources of variation provided in the experimental design model as a generalized linear model. Given the dispersions and the generalized linear model, a negative binomial model was fit for each gene with the glmFit() function. Finally, the significance of the differential expression for the model was calculated with the glmLRT() function, which compares the fit of the experimental model to the null model with a likelihood ratio test. Genes with fold changes that were > |1.5| between the two phenotypes and a false discovery rate (FDR)-adjusted p-value <0.05 were considered differentially expressed as these cut-offs have been shown to balance specificity and sensitivity for the edgeR algorithm 20. The 50 genes with the lowest p-values were used to produce a heat map in R; the data for the heat map were produced with the cpm() function in edgeR after accounting for batch effects with the sva package as described above. Taking into account the variation in library sizes for each sample, the cpm() function calculates the log2-transformed value of the number of sequencing reads for each gene that occur per one million reads.
GO Analysis
The list of differentially expressed transcripts was analyzed for enrichment for GO terms using the PANTHER Overrepresentation Test (release 20150430), which accessed the GO Ontology database that was released 2015-08-06 21. The molecular process, cellular component, and biological process annotation databases were used.
Pathway Analysis
Clusters of co-expressed genes were defined using the hopach package, version 3.1.1, in R separately in the samples with and without HS, by analyzing the 500 transcripts with the greatest variance 22. The gene lists for the largest clusters of co-expressed genes present in the HS samples that had less than 50% identity with the clusters generated for the non-HS samples was compared to the list of all Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and the significance of enrichment for a particular pathway was calculated with the hypergeometric probability test, adjusted for multiple testing with a Bonferroni correction. The sample size of the hypergeometric probability test was adjusted to account for the number of genes in the expression data set.
Quantitative Real-Time PCR (qRT-PCR) confirmation of RNA-Seq expression
RNA from DG cells was extracted as described above, and cDNA was generated with the Arcturus® RiboAmp® HS PLUS RNA Amplification Kit for random priming 1st-Strand cDNA Synthesis according to the manufacturer’s instructions. qRT-PCR was performed with TaqMan primers in an ABI 7900, and Taqman primers for β-actin were used as the endogenous control. Primers for the following transcripts were used: ARHGAP36, CDKN1C, CTGF, ELAVL2, GAD1, IL8, KCNIP2, KIT, QPCT, and SV2C. All assays were performed in duplicate. We used the ΔΔCt method to quantify the relative expression of the transcript in individuals with and without HS23. Transcript expression levels, standardized to β-actin, were normalized to the average expression of the transcript in patients without HS.
Statistical Analysis
P-values reported for the differential expression analysis were calculated with a negative binomial model in edgeR and corrected for multiple testing in R using the p.adjust() function, ‘fdr’ method. P-values reported for the pathway analysis were calculated with the hypergeometric probability distribution and corrected for multiple testing using a Bonferroni correction. P-values reported for the qRT-PCR confirmation were calculated with a one-sided Student’s t-test.
Results
The clinical characteristics of the 12 MTLE patients in this study are summarized in Table 1. All patients were of European descent. The mean ± SD age at surgery was 38+/−12 years for all patients, 39+/−9 years for the patients with HS, and 38+/−14 years for the patients without pathologic confirmation of HS. Of the five patients with HS, two were male, and three were female; of the seven patients without HS, four were male, and three were female.
Table 1.
Summary of clinical characteristics of mesial temporal lobe epilepsy patients with and without HS.
| ID | Sex | Age of Onset | Age at Surgery | Medical History | AED (at time of surgery) | Sz Types | Side | MRI summary | Pathology summary |
|---|---|---|---|---|---|---|---|---|---|
| MTLE-1-HS | M | 3–4 yrs | 46 yrs | FS @3–4 yrs. No head injury, no infections. Fam Hx: maternal GF and sister | VPA, LTG, LEV | CPSz | L | Consistent with left MTS. | Consistent with left MTS. ILAE HS type 1. Granule cell dispersion. |
| MTLE-2 | M | 36 yrs | 53 yrs | No FS, no head injury, no infections. Fam Hx: unknown/adopted | Pheno, PHT, LEV | CPSz, GTCSz (usually when non-med compliant) | L | No evidence for MTS. | No evidence for MTS. FCD type IIa. |
| MTLE-3-HS | F | 5 yrs | 27 yrs | Head injury @5 yrs (uncertain if LOC) with seizure onset (SE) 2 wks later. No FS. Fam Hx: none | CBZ, LEV, LCM | Intractable Sz disorder | R | Consistent with right MTS. | Consistent with right MTS. ILAE HS type 2. Meningitis w/secondary vasculitis - uncertain if acute (due to grid placement) or chronic. |
| MTLE-4 | M | 10 yrs | 25 yrs | Risk factor: head injury w/LOC @4yrs old. No FS, no infections. Fam Hx: none | LEV, LCM | CPSz | L | No evidence for MTS. | No evidence for MTS. FCD type IIb. |
| MTLE-5-HS | F | 16 yrs | 36 yrs | Head injury w/LOC @16yrs. No FS, no infections. Fam Hx: daughter w/FS | LEV, ZON | CPSz | L | No evidence for MTS. | Consistent with left MTS. ILAE HS type 2. Granule cell dispersion. FCD type IIIa. |
| MTLE-6-HS | F | 2 yrs | 36 yrs | No FS, no head injury, no infections. Fam Hx: none | OXC | CPSz, intractable | L | Consistent with left MTS. | Consistent with left MTS. ILAE HS type 2. |
| MTLE-7 | F | 18 mos | 45 yrs | Born “2 months” premature, twin pregnancy. No FS, no head injury, no infections. Fam Hx: none | LTG | CPSz, GTCSz, Intractable Sz | L | No evidence for MTS. | No evidence for MTS. |
| MTLE-8-HS | M | 20 yrs | 49 yrs | Head injury w/LOC at 2yrs of age, 1wk prior to sz onset. No FS, no infections. Fam Hx: paternal GF | LTG | Episodic alteration of consciousness | R | Unclear findings. | Consistent with right MTS. ILAE HS type 2. FCD type IIIa. |
| MTLE-9 | M | 17 yrs | 33 yrs | Born at 36 wks, birth complicated by collapsed lung. No FS, no head injury, no infections. Fam Hx: maternal aunt and maternal 1st cousin | CBZ monotherapy | CPSz, with rare GTCSz | R | No evidence for MTS. | No evidence for MTS. |
| MTLE-10 | F | 12 yrs | 33 yrs | No FS, no head injury, no infections. Fam Hx: maternal cousin | OXC | CPSz, intractable | L | No evidence for MTS. | No evidence for MTS. FCD type Ib. |
| MTLE-11 | M | 5 yrs | 19 yrs | No FS, no head injury, no infections. Fam Hx: none | LTG, PGB | Focal seizures | R | No evidence for MTS. | No evidence for MTS. Vascular Malformation. |
| MTLE-12 | F | 9 yrs | 56 yrs | No FS, no head injury, no infections. Fam Hx: none | LEV, LCM | CPSz, GTC (rare) | R | Faint T2 hyperintensity in R temporal lobe. | No evidence for MTS. FCD type Ib. |
CBZ=carbamazepine; CPSz = complex partial seizures; FCD= focal cortical dysplasia. FS= febrile seizures; GTCSz= generalized tonic-clonic seizures; LCM= lacosamide; LEV=levetiracetam; LOC= loss of consciousness; LTG=lamotrigine; MTS=mesial temporal sclerosis; OXC=oxcarbazepine; PGB=pregabalin; Pheno=phenobarbital; ZON=zonisamide
Six individuals had cortical dysplasia, including four without HS (MTLE-2, MTLE-4, MTLE-10 and MTLE- 12) and two with HS (MTLE-5-HS and MTLE-8-HS). Another HS case, MTLE-3-HS, had mixed meningitis with vasculitis in the lateral temporal cortex thought to be caused by a subacute infarction. Granule cell dispersion was seen in MTLE-1-HS and MTLE-5-HS and was not evident in the brain tissue of any of the patients without HS. The specimens used for neuropathology were stained with hematoxylin and eosin. Immunohistochemistry for neuron-specific nuclear protein (NeuN) and glial fibrillary acidic protein (GFAP) was used if cortical dysplasia was suspected after the preliminary review. Mossy fiber sprouting was not assessed. Supplementary Figure S1 contains neuropathological images for all samples.
There was a high rate of concordance between the duplicate sequenced transcriptional profiles for each sample, with an average correlation coefficient of >95% of transcript expression levels between the ten hippocampal specimens analyzed in duplicate. Sufficient amounts of cDNA for sequencing were not obtained for two of the 12 patients from the second capture. Approximately eight million high quality sequencing reads were obtained per sequencing run, and greater than 90% of these reads aligned to the reference genome. Transcripts were only included in the differential expression analysis if there was at least one sequencing read present in all 12 samples, leading to a final set of 12,605 genes for differential expression analysis.
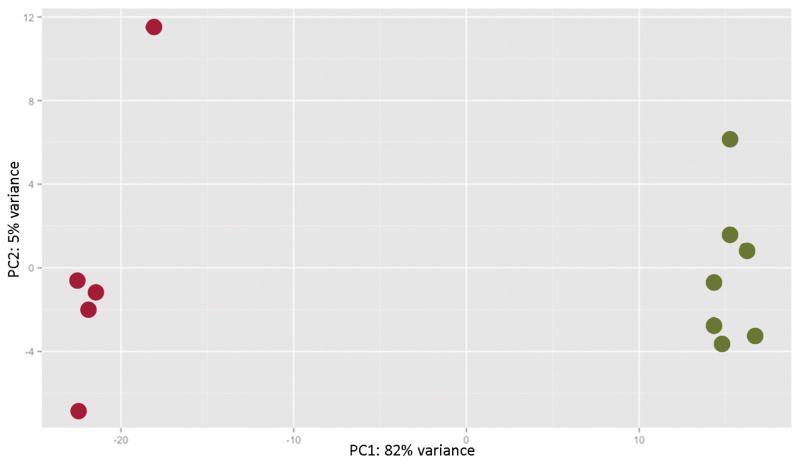
Before performing the differential expression analysis, a PCA was conducted with the 500 transcripts with the greatest variance to determine the largest source of variance in the data after correcting for batch effects. The first PC accounted for 82% of the variance in the expression data (Figure 2), with the samples segregating according to the presence or absence of HS, indicating that the variance in the expression data was predominantly explained by the differences in hippocampal pathology.
Figure 2.
Plot of the first two principal components for the 500 genes with the greatest variance in expression. The red dots represent the samples with hippocampal sclerosis (HS), and the green dots represent the samples without HS. The presence or absence of HS is the major determinant of the variance within the expression data.
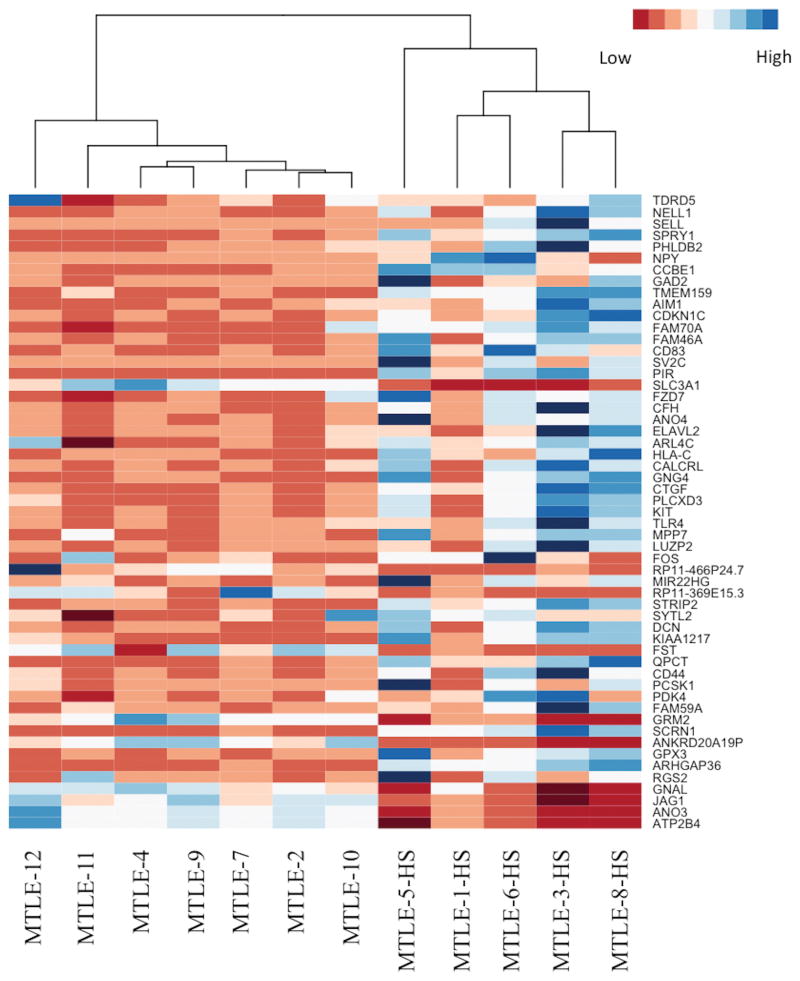
Next, the differential expression analysis was conducted in edgeR by constructing a generalized linear model that accounted for batch effects, which comprised the main source of technical variation in the data, and the presence of HS, which was the main source of biological variation in the data. The edgeR package is designed to fit a negative binomial model of the raw read counts that accounts for differences in library size and over-dispersion; as genes become more highly expressed on average, there tends to be greater variation in the expression data. By calculating the dispersions for each gene, edgeR mitigates the possibility that a gene will be considered differentially expressed as a result of outliers in the data. The fit of the negative binomial model for the generalized linear model is then compared to the fit of the negative binomial model for the null model using a likelihood-ratio test to determine whether genes are differentially expressed. Thus, the edgeR package is suited to differential expression analysis for datasets with limited biological replicates because it lessens the impact of outliers and accounts for over-dispersion within the data. In our data, 55 transcripts were identified as being significantly upregulated (>1.5 fold-change increase in expression, FDR-adjusted p-value <0.05), and 11 transcripts were identified as being significantly downregulated (<−1.5 fold-change decrease in expression, FDR-adjusted p-value<0.05) in samples with HS compared to samples without HS. The 50 most significant genes were used to create a heat map, in which the samples with HS once again clustered together separately from the samples without HS (Figure 3). The 10 most significantly differentially upregulated genes in the HS samples were CDKN1C, SV2C, PCSK1, SCRN1, KIAA1217, ELAVL2, GNG4, QPCT, KIT, and STRIP2 (Table 2a). The most significantly downregulated genes in the HS samples were ANKRD20A19P, FST, ANO3, GRM2, ATP2B4, RP11-369E15.3, SLC3A1, JAG1, GNAL, PCP4, and SLC5A12 (Table 2b). To confirm the expression differences detected by RNA sequencing, qRT-PCR was performed for ten select transcripts (ARHGAP36, CDKN1C, CTGF, ELAVL2, GAD1, IL8, KCNIP2, KIT, QPCT, and SV2C) for all samples in duplicate. The expression differences between the samples with HS and without HS were confirmed for all ten transcripts (Supplemental Figure 2). Among the genes that were significantly upregulated in HS, there were genes associated with processes such as apoptosis (BCL2), the PI3K-Akt pathway (TLR4, KIT, GNG4, and LAMA2), GABAergic synapses (GNG4 and GAD2), and the Hippo signaling pathway (CTGF and FZD7). GO ontology analysis indicated significant enrichment for genes expressed at the plasma membrane (p=0.02) and involved in the positive regulation of multicellular organismal processes (p=0.04).
Figure 3.
Heat map of the differentially expressed genes in hippocampal sclerosis (HS). The samples cluster according to the presence or absence of HS. Red indicates low levels of expression, and blue indicates high levels of expression.
Table 2.
Top differentially expressed genes
| a. Top significantly upregulated genes in samples with HS
| ||||
|---|---|---|---|---|
| Log Fold Change | Log Counts Per Million | P-Value | Adjusted | |
| CDKN1C | 3.53 | 4.35 | 1.69E-08 | 2.13E-04 |
| RP11-466P24.7 | 3.19 | 5.87 | 1.09E-07 | 4.98E-04 |
| SV2C | 4.16 | 4.66 | 1.18E-07 | 4.98E-04 |
| PCSK1 | 4.31 | 6.92 | 1.69E-07 | 5.24E-04 |
| SCRN1 | 1.93 | 6.19 | 2.08E-07 | 5.24E-04 |
| KIAA1217 | 1.85 | 5.21 | 2.80E-07 | 5.26E-04 |
| ELAVL2 | 4.00 | 4.90 | 2.92E-07 | 5.26E-04 |
| GNG4 | 2.90 | 4.97 | 3.83E-07 | 6.04E-04 |
| QPCT | 2.31 | 5.68 | 6.34E-07 | 8.88E-04 |
| KIT | 2.49 | 4.68 | 7.38E-07 | 9.31E-04 |
| STRIP2 | 1.85 | 4.93 | 1.51E-06 | 1.73E-03 |
| ARHGAP36 | 3.34 | 8.10 | 8.06E-06 | 6.77E-03 |
| CCBE1 | 2.92 | 1.80 | 1.00E-05 | 7.91E-03 |
| MIR22HG | 1.99 | 4.57 | 1.14E-05 | 8.46E-03 |
| SPRY1 | 3.05 | 2.21 | 1.29E-05 | 9.04E-03 |
| b. Significantly downregulated genes in samples with HS
| ||||
|---|---|---|---|---|
| Log Fold Change | Log Counts Per Million | P-Value | Adjusted | |
| ANKRD20A19P | −1.75 | 5.15 | 5.70E-06 | 5.57E-03 |
| FST | −3.53 | 2.91 | 5.74E-06 | 5.57E-03 |
| ANO3 | −1.68 | 8.04 | 6.24E-06 | 5.62E-03 |
| GRM2 | −1.68 | 4.66 | 5.62E-05 | 2.44E-02 |
| ATP2B4 | −1.39 | 8.32 | 6.82E-05 | 2.48E-02 |
| RP11-369E15.3 | −2.08 | 3.12 | 9.29E-05 | 2.72E-02 |
| SLC3A1 | −1.77 | 1.83 | 9.82E-05 | 2.81E-02 |
| JAG1 | −1.23 | 6.54 | 1.28E-04 | 3.36E-02 |
| GNAL | −1.60 | 6.44 | 1.91E-04 | 4.43E-02 |
| PCP4 | −1.51 | 7.83 | 2.30E-04 | 4.69E-02 |
| SLC5A12 | −2.53 | 2.16 | 2.62E-04 | 5.00E-02 |
To more systematically determine which pathways might play a role in HS, a clustering algorithm was used to identify groups of co-expressed genes. By looking for large clusters of genes that were co-expressed in the HS samples, but not in the non-HS samples, we were able to identify a network of genes that distinguished the transcriptomic profiles of the samples with HS from those without. The raw read counts for the 500 genes with the greatest variances were normalized and log-transformed in edgeR to reduce over-dispersion. From the expression data for these genes for the samples without HS, three distinct clusters of co-expressed genes emerged consisting of 252, 141, and 61 genes (henceforth clusters 1–3), respectively. For the samples with HS, there were five large clusters (henceforth clusters A–E) of co-expressed genes identified consisting of 143, 95, 68, 29, and 24 genes, respectively. Cluster A shared high identity with Cluster 1 and was therefore excluded from further analysis because it was common to both the HS and non-HS samples. The gene lists for the remaining clusters were compared to the gene lists comprising the KEGG pathways to determine if there was significant enrichment for any particular pathways. Cluster C was the only group for which there were significant findings; the significantly enriched pathways for this cluster are displayed in Table 3. Among the 68 genes in cluster C, seven participated in oxidative phosphorylation (hsa00190; p-value=5.12×10−5): NDUFA5, ATP5C1, ATP6V0D1, ATP5A1, ATP6V1A, ATP6V1B2, and ATP5B. Additionally, five genes participated in another energy-producing pathway, the glycolysis/gluconeogenesis pathway (hsa00010; p-value=5.41×10−4): ENO1, TPI1, PKM, GAPDH, and LDHA. With the exception of ENO1, these 12 genes were all upregulated in the HS samples compared to the non-HS samples, though not at a statistically significant level. There was also significant enrichment for two KEGG pathways associated with neurodegeneration: Alzheimer’s disease (hsa05010; p-value=7.12×10−3) and Parkinson’s disease (hsa05012; p-value=0.0258), which accounted for six and five genes in cluster C, respectively. There was significant overlap among the gene lists associated with these two pathways and the two energy-producing pathways described above. The genes in the cluster in the Alzheimer’s disease pathway were NDUFA5, ATP5C1, ATP5A1, GAPDH, SNCA, and ATP5B, and the genes in the Parkinson’s disease pathway were NDUFA5, ATP5C1, ATP5B, SNCA, and ATP5A1. Cluster C was also enriched for genes participating in the synaptic vesicle cycle (hsa04721; p-value=1.24×10−3); these genes were CTLC, AP2M1, ATP6V1B2, ATP6V0D1, and ATP6V1A. Two of these genes were also associated with the oxidative phosphorylation pathway.
Table 3.
Significantly enriched pathways in cluster C in the samples with hippocampal sclerosis
| Pathway Name | KEGG ID | # of Genes | P-Value(adjusted) |
|---|---|---|---|
| Oxidative phosphorylation | hsa00190 | 7 | 5.12E-05 |
| Glycolysis / Gluconeogenesis | hsa00010 | 5 | 5.41E-04 |
| Synaptic vesicle cycle | hsa04721 | 5 | 1.24E-03 |
| Alzheimer’s disease | hsa05010 | 6 | 7.12E-03 |
| Collecting duct acid secretion | hsa04966 | 3 | 1.15E-02 |
| Metabolic pathways | hsa01100 | 13 | 1.30E-02 |
| Vibrio cholerae infection | hsa05110 | 4 | 1.67E-02 |
| Huntington’s disease | hsa05016 | 6 | 1.95E-02 |
| Lysosome | hsa04142 | 5 | 2.06E-02 |
| Epithelial cell signaling in Helicobacter pylori infection | hsa05120 | 4 | 2.43E-02 |
| Parkinson’s disease | hsa05012 | 5 | 2.58E-02 |
| Biosynthesis of amino acids | hsa01230 | 4 | 2.89E-02 |
Discussion
In this study, we performed RNA sequencing of DG cells from MTLE patients with and without HS to characterize the molecular changes associated with this pathology. Differential expression analysis identified 55 transcripts that were significantly upregulated and 11 transcripts that were significantly downregulated in the samples with HS compared to the samples without HS.
By focusing specifically on DG cells, HS-associated gene expression changes specific to this key cell type involved in HS could be evaluated with higher resolution than possible with previous analyses of whole hippocampal tissue in TLE6; 8; 9; 24. In some cases, similar gene expression changes were found in DG cells as were observed in hippocampal tissue biopises, and in other cases novel associations were identified. SV2C, shown to be upregulated in DG cells of TLE patients with HS, has been shown previously to be highly expressed in HS when it is typically present at low or undetectable levels in the hippocampus25. Likewise, genes associated with inflammation, such as TLR4 and IL8, were found to be upregulated in HS in the current study, confirming findings by Johnson et al., who previously observed co-expression of genes belonging to inflammatory KEGG pathways, such as ‘cytokine-cytokine receptor interaction’ and ‘TLR-signaling’, in hippocampal tissue from TLE patients with HS 9. The set of 55 genes that were significantly upregulated in HS also included genes that had not been previously implicated in studies of whole hippocampal tissue and may represent a DG cell-specific expression change. The expression of ELAVL2, for example, which encodes a neuronal expressed RNA binding protein, was significantly upregulated in the DG cells of the samples with HS in the current study; this protein binds the 3′ end of FOS26, which was also found to be upregulated in HS, although the corrected p-value for this gene was slightly above the threshold for significance. FOS participates in the PI3K-Akt-mTOR pathway, and we identified four other genes in this pathway as being significantly upregulated in HS: KIT, GNG4, TLR4, and LAMA2. The role of this pathway has been well established in malformations of cortical development, and it has more recently been implicated in the pathogenesis of MTLE27. Xiao et al. induced an inflammatory response with IL-1 in primary cultured hippocampal neurons, which is activated by seizures and highly expressed in the hippocampi of epilepsy patients, in primary cultured hippocampal neurons and then suppressed this inflammation by introducing inhibitors of the PI3K-Akt-mTOR pathway. These researchers also observed increased levels of p-Akt in hippocampal tissue samples from MTLE patients. The upregulation of FOS, KIT, GNG4, TLR4, and LAMA2 in DG cells from MTLE patients with HS in the current study provides additional evidence for the role of the PI3K-Akt-mTOR pathway in this disease. Our study also provides support for the involvement of processes in HS that were previously implicated in animal models of epilepsy. Elliott et al. identified a list of 25 genes that were differentially regulated in both development and epileptogenesis in a rat model of epilepsy. While we would not necessarily expect individual genes to be differentially regulated in our data set because of inherent differences in animals and humans and differences in the type of epilepsy, we did observe significant upregulation of one gene identified by these authors, NPY, in the DG cells of patients with MTLE-HS. Elliot et al. noted that the expression of NPY was significantly upregulated in DG cells after seizures in several studies and is a regulator of neuronal excitability.
This study also identified a role for increased oxidative phosphorylation and glycolysis/gluconeogenesis in HS. While oxidative phosphorylation produces ATP, this process also results in the production of reactive oxygen species (ROS). If oxidative phosphorylation increases to meet the excessive energy demands of neurons involved in seizure activity, this would also increase the levels of ROS within neurons, leading to oxidative stress. While it is difficult to assess levels of ROS in human tissue due to the instability of these molecules, there is emerging evidence for the presence of oxidative stress in hippocampal tissue from patients with HS29. Ristic et al. demonstrated oxidative stress in sclerotic hippocampi from epilepsy patients by showing that the activities of antioxidative enzymes that remove ROS were significantly higher in HS samples compared to controls. The results of our study complement these findings by identifying a network of upregulated genes that could be contributing to the production of ROS in HS, specifically in DG cells.
The cluster-based analysis also implicates the process of synaptic vesicle cycling in HS. The genes in cluster C that participate in the synaptic vesicle cycle (hsa04721; p-value=1.24×10−3) included CTLC, AP2M1, ATP6V1B2, ATP6V0D1, and ATP6V1A. Winden et al. examined epileptogenic and adjacent non-epileptogenic regions of the dentate gyrus in a pilocarpine model of epilepsy and identified a network of co-expressed genes that was also enriched for synaptic processes30. Two of the genes in this network, SDCBP and NBEA, were also found in cluster C.
However, it is also crucial to understand what causes these changes, and additional research is needed to determine whether these changes are the cause or result of seizure activity. miRNAs regulate gene expression in a post-transcriptional manner, and a previous study identified miR-218 and miR-204 as being downregulated in hippocampal tissue from patients with HS4. Consistent with these findings, our study identified several genes that have been classified as targets of miR-204 as being significantly upregulated in HS in DG cells including BCL2, SOX4, and ARHGAP29.
Among the genes that were downregulated in the HS samples in this study, ANO3 and GNAL are known to be co-expressed. ANO3 is highly expressed in the hippocampus, and variants in this gene have been associated with febrile seizures31. Furthermore, hippocampal CA1 pyramidal neurons from Ano3-null rats exhibit hyperexcitability31. The downregulation of ANO3 in HS could contribute to hyperexcitability and seizure activity in patients who develop HS. Studies looking at the effects of altering the expression of particular genes identified in this study in vitro may help answer the long-standing debate of which changes in pathology contribute to or are the result of seizure activity in MTLE patients with HS11; 32; 33.
In summary, the current study demonstrates that next-generation sequencing methods can yield interpretable, reproducible, and likely biologically relevant gene expression data from a small population of cells. This approach has begun to illuminate the changes in gene expression associated with HS in DG cells and provides the foundation for studies in a larger population of MTLE patients incorporating in patient specific factors like age of onset, seizure types and severity, and antiepileptic drug treatments, to provide a better understanding of the changes that specifically underlie pathophysiology of HS. Additional studies will be needed to study the effects of differential expression of particular transcripts and networks of linked transcripts identified in this study to understand their effects on neuronal excitability, changes in granule cell pathology, and epileptogenesis.
Supplementary Material
Key Points Box.
Gene expression analysis of dentate granule cells from MTLE patients with and without HS revealed clear, likely biologically relevant differences in the transcriptome.
In dentate granule cells from patients with HS, there was co-expression of genes associated with oxidative phosphorylation, neurodegeneration, and synaptic transmission.
This study demonstrates the utility of NGS methods for characterizing expression changes in a small population of homogeneous cells.
Acknowledgments
We thank the patients and their families for participating in this study. We would also like to thank Jessica Maia and William Weir for providing bioinformatics advice. This study was supported by the NINDS (5R21NS078657 to ELH).
Footnotes
Disclosure of Conflicts of Interest
None of the authors has any conflicts of interest to disclose.
Ethical Publication Statement
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
References
- 1.Blumcke I, Kistner I, Clusmann H, et al. Towards a clinico-pathological classification of granule cell dispersion in human mesial temporal lobe epilepsies. Acta Neuropathol. 2009;117:535–544. doi: 10.1007/s00401-009-0512-5. [DOI] [PubMed] [Google Scholar]
- 2.Bernasconi A. Magnetic resonance imaging in intractable epilepsy: focus on structural image analysis. Adv Neurol. 2006;97:273–278. [PubMed] [Google Scholar]
- 3.Blumcke I, Thom M, Aronica E, et al. International consensus classification of hippocampal sclerosis in temporal lobe epilepsy: a Task Force report from the ILAE Commission on Diagnostic Methods. Epilepsia. 2013;54:1315–1329. doi: 10.1111/epi.12220. [DOI] [PubMed] [Google Scholar]
- 4.Kaalund SS, Veno MT, Bak M, et al. Aberrant expression of miR-218 and miR-204 in human mesial temporal lobe epilepsy and hippocampal sclerosis-convergence on axonal guidance. Epilepsia. 2014;55:2017–2027. doi: 10.1111/epi.12839. [DOI] [PubMed] [Google Scholar]
- 5.Miller-Delaney SF, Bryan K, Das S, et al. Differential DNA methylation profiles of coding and non-coding genes define hippocampal sclerosis in human temporal lobe epilepsy. Brain. 2015;138:616–631. doi: 10.1093/brain/awu373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engel T, Henshall DC. Apoptosis, Bcl-2 family proteins and caspases: the ABCs of seizure-damage and epileptogenesis? International Journal of Physiology, Pathophysiology and Pharmacology. 2009;1:97–115. [PMC free article] [PubMed] [Google Scholar]
- 7.Xu S, Pang Q, Liu Y, et al. Neuronal apoptosis in the resected sclerotic hippocampus in patients with mesial temporal lobe epilepsy. J Clin Neurosci. 2007;14:835–840. doi: 10.1016/j.jocn.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Dericioglu N, Soylemezoglu F, Gursoy-Ozdemir Y, et al. Cell death and survival mechanisms are concomitantly active in the hippocampus of patients with mesial temporal sclerosis. Neuroscience. 2013;237:56–65. doi: 10.1016/j.neuroscience.2013.01.050. [DOI] [PubMed] [Google Scholar]
- 9.Johnson MR, Behmoaras J, Bottolo L, et al. Systems genetics identifies Sestrin 3 as a regulator of a proconvulsant gene network in human epileptic hippocampus. Nat Commun. 2015;6:6031. doi: 10.1038/ncomms7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houser CR. Granule cell dispersion in the dentate gyrus of humans with temporal lobe epilepsy. Brain Research. 1990;535:195–204. doi: 10.1016/0006-8993(90)91601-c. [DOI] [PubMed] [Google Scholar]
- 11.Scharfman HE, Sollas AL, Berger RE, et al. Electrophysiological evidence of monosynaptic excitatory transmission between granule cells after seizure-induced mossy fiber sprouting. J Neurophysiol. 2003;90:2536–2547. doi: 10.1152/jn.00251.2003. [DOI] [PubMed] [Google Scholar]
- 12.Zucchini S, Marucci G, Paradiso B, et al. Identification of miRNAs differentially expressed in human epilepsy with or without granule cell pathology. PLoS One. 2014;9:e105521. doi: 10.1371/journal.pone.0105521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leek JT, Storey JD. Capturing Heterogeneity in Gene Expression Studies by “Surrogate Variable Analysis”. PLoS Genetics. 2005;3(9):1724–35. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YMD, Robinson M, Smyth GK. User Guide. Bioconductor. 2015. [Google Scholar]
- 20.Rapaport F, Khanin R, Liang Y, et al. Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol. 2013;14:R95. doi: 10.1186/gb-2013-14-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: A Library of Protein Families and Subfamilies Indexed by Function. Genome Research. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laan MP, KS Hybrid clustering of gene expression data with visualization and the bootstrap. Journal of Statistical Planning and Inference. 2003;117:275–303. [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Xu S, Pang Q, Liu Y, Shang W, et al. Neuronal apoptosis in the resected sclerotic hippocampus in patients with mesial temporal lobe epilepsy. J Clin Neurosci. 2007;14:835–40. doi: 10.1016/j.jocn.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Crevecoeur J, Kaminski RM, Rogister B, et al. Expression pattern of synaptic vesicle protein 2 (SV2) isoforms in patients with temporal lobe epilepsy and hippocampal sclerosis. Neuropathol Appl Neurobiol. 2014;40:191–204. doi: 10.1111/nan.12054. [DOI] [PubMed] [Google Scholar]
- 26.Abe R, Yamamoto K, Sakamoto H. Target specificity of neuronal RNA-binding protein, Mel-N1: direct binding to the 3′ untranslated region of its own mRNA. Nucleic Acids Research. 1996;24:2011–2016. doi: 10.1093/nar/24.11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao Z, Peng J, Yang L, et al. Interleukin-1beta plays a role in the pathogenesis of mesial temporal lobe epilepsy through the PI3K/Akt/mTOR signaling pathway in hippocampal neurons. J Neuroimmunol. 2015;282:110–117. doi: 10.1016/j.jneuroim.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Elliott RC, Miles MF, Lowenstein DH. Overlapping Microarray Profiles of Dentate Gyrus Gene Expression during Development- and Epilepsy-Associated Neurogenesis and Axon Outgrowth. The Journal of Neuroscience. 2003;23:2218–2227. doi: 10.1523/JNEUROSCI.23-06-02218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ristic AJ, Savic D, Sokic D, et al. Hippocampal antioxidative system in mesial temporal lobe epilepsy. Epilepsia. 2015;56:789–99. doi: 10.1111/epi.12981. [DOI] [PubMed] [Google Scholar]
- 30.Winden KD, Bragin A, Engel J, et al. Molecular alterations in areas generating fast ripples in an animal model of temporal lobe epilepsy. Neurobiol Dis. 2015;78:35–44. doi: 10.1016/j.nbd.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feenstra B, Pasternak B, Geller F, et al. Common variants associated with general and MMR vaccine-related febrile seizures. Nat Genet. 2014;46:1274–1282. doi: 10.1038/ng.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cavazos JE, Das I, Sutula TP. Neuronal loss induced in limbic pathways by kindling: evidence for induction of hippocampal sclerosis by repeated brief seizures. J Neurosci. 1994;14:3106–3121. doi: 10.1523/JNEUROSCI.14-05-03106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotloski R, Lynch M, Lauersdorf S, et al. Repeated brief seizures induce progressive hippocampal neuron loss and memory deficits. Prog Brain Res. 2002;135:95–110. doi: 10.1016/S0079-6123(02)35010-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.