Highlight
Gene duplication has led to the existence of a large HAP gene family. In this study, three HAP genes were identified that regulate flowering in rice in addition to the previously reported Ghd8/OsHAP3H.
Keywords: Association analysis, expression profiling, gene duplication, heading date, nucleotide diversity, OsHAP.
Abstract
Heterotrimeric Heme Activator Protein (HAP) family genes are involved in the regulation of flowering in plants. It is not clear how many HAP genes regulate heading date in rice. In this study, we identified 35 HAP genes, including seven newly identified genes, and performed gene duplication and candidate gene-based association analyses. Analyses showed that segmental duplication and tandem duplication are the main mechanisms of HAP gene duplication. Expression profiling and functional identification indicated that duplication probably diversifies the functions of HAP genes. A nucleotide diversity analysis revealed that 13 HAP genes underwent selection. A candidate gene-based association analysis detected four HAP genes related to heading date. An investigation of transgenic plants or mutants of 23 HAP genes confirmed that overexpression of at least four genes delayed heading date under long-day conditions, including the previously cloned Ghd8/OsHAP3H. Our results indicate that the large number of HAP genes in rice was mainly produced by gene duplication, and a few HAP genes function to regulate heading date. Selection of HAP genes is probably caused by their diverse functions rather than regulation of heading.
Introduction
The heterotrimeric Heme Activator Protein (HAP) complex is also known as the CCAAT box factor (CBF) or nuclear factor Y (NF-Y) (Mantovani, 1999). The HAP complex consists of three subunits: HAP2 (NF-YA; CBF-B), HAP3 (NF-YB; CBF-A), and HAP5 (NF-YC; CBF-C). This complex binds to CCAAT sequences in a promoter to control the expression of target genes (Gusmaroli et al., 2001; Kusnetsov et al., 1999). In animals and yeast, there is a single gene for each HAP subunit, while in plants, there are gene families encoding each subunit. For example, in rice there are 10 genes (OsHAP2A–OsHAP2J) encoding the OsHAP2 unit, 11 genes (OsHAP3A–OsHAP3K) encoding the OsHAP3 unit, and seven genes (OsHAP5A–OsHAP5G) encoding the OsHAP5 unit (Thirumurugan et al., 2008).
Recent studies have revealed the function of members of the HAP family in multiple plant developmental processes. LEAFY COTYLEDON 1 (LEC1) and LEC1-LIKE (LIL), which share similar sequences, regulate embryogenesis in Arabidopsis (Kwong et al., 2003; Lotan et al., 1998). Additionally, NF-YA3 and NF-YA8 are functionally redundant genes that are required in the early embryogenesis of Arabidopsis (Fornari et al., 2013). The expression of NF-YA5 is strongly induced by drought stress and ABA treatments in Arabidopsis (Li et al., 2008). Overexpression of the AtNF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis (Han et al., 2013). Overexpression of AtNF-YB1 and ZmNF-YB2 has been shown to increase seed grain yield and improve performance under drought conditions (Nelson et al., 2007). The CCT (CONSTANS, CO-like, and TOC1) domain of CO in Arabidopsis and the functional domain conserved within HAP2 share important residues, and CO might replace AtHAP2 to form a trimetric CO/AtHAP3/AtHAP5 complex in the HAP complex. Overexpression of AtHAP2 or AtHAP3 delays flowering in Arabidopsis (Wenkel et al., 2006). AtHAP3b is required to regulate flowering time in Arabidopsis under osmotic stress conditions (Chen et al., 2007).
In rice, several CCT domain family genes have been confirmed to regulate heading date, such as major flowering regulators of Ghd7 (Grain number, plant height and heading date 7), Ghd7.1, and Hd1 (Xue et al., 2008; Yan et al., 2013; Yano et al., 2000). Hd1 regulates the day-length oscillations of Hd3a mRNA and promotes flowering under short-day conditions while inhibiting flowering under long-day conditions (Hayama et al., 2003). Both Ghd7 and Ghd7.1 delay flowering by suppressing expression of Ehd1 under long-day conditions (Xue et al., 2008; Yan et al., 2013). The functions of the HAP genes have been also characterized in rice. OsHAP3A, OsHAP3B, and OsHAP3C control chloroplast biogenesis (Miyoshi et al., 2003). OsLEC1/OsHAP3E participates in the determination of meristem identity in both vegetative and reproductive development (Zhang and Xue, 2013). OsHAP2E confers resistance to pathogens, salinity, and drought, and increases photosynthesis and the tiller number (Alam et al., 2015). OsNF-YB1, a rice endosperm-specific gene, encodes a cell cycle regulator and plays a role in maintaining endosperm cell proliferation (Sun et al., 2014). The evidence from these various studies shows that the OsHAP family genes might have important and diverse functions in plant development and abiotic stress tolerance. However, only one HAP gene, Ghd8/OsHAP3H, acts upstream of Ehd1, Hd3a, and RFT1 to regulate heading in rice (Yan et al., 2011b). It is not clear whether other HAP family genes are functional in rice heading.
Heading date has been well studied in rice during the past two decades. Hundreds of quantitative trait loci (QTLs) for heading date have been collected on the Gramene website (http://www.gramene.org/, last accessed: 14 September 2013). Association mapping is an efficient approach to establish the relationship between molecular markers and traits in a given population based on linkage disequilibrium (LD) (Flint-Garcia et al., 2003). Genome-wide association studies have gained prominence in recent years, especially in species with a rapid LD decay. For example, maize has a LD decay of 1–5kb, which can map QTL to a single gene (Remington et al., 2001). The extent of LD variation is greater in japonica (150–167kb) than indica (75–123kb) rice, and LD patterns vary among genomic regions (Huang et al., 2010; Mather et al., 2007). Association mapping can be adversely affected by many factors, including population structure and small sample size, which may increase the detection of false-positive associations (Yan et al., 2011a). Huang et al. (2010, 2012) made a genome-wide association mapping study for heading date in rice, but only a small number of cloned flowering genes were identified, even when using different populations and analysis methods. However, candidate gene association studies remain a key approach to gene mapping due to their high efficiency (Ehrenreich et al., 2009; Zhang et al., 2015).
The aim of the present study was to determine how many HAP family genes potentially have a function in the regulation of heading date. A total of 529 rice accessions were used to test the association between the HAP family genes and heading date. Then, we overexpressed or suppressed 18 HAP family genes in japonica rice Zhonghua 11 and Hejiang 19 (containing Ghd8/OsHAP3H) and tested the mutants of nine HAP genes under long-day and short-day conditions. Additionally, to elucidate how the HAP family genes have evolved, we analyzed the nucleotide diversity and the fixation index (Fst) of population differentiation. Our results showed that 13 HAP genes underwent selection and that at least four HAP genes regulate heading date in rice, including the previously cloned gene Ghd8/OsHAP3H.
Materials and methods
Plant materials, field experiments, and heading date
A total of 529 rice (Oryza sativa) cultivars, comprising a Chinese core collection consisting of 203 varieties except C126 and a world core collection of 330 accessions except W190, W196, and W232, and 107 common wild rice (Oryza rufipogon) accessions were sown in the field of an experimental farm of Huazhong Agricultural University, Wuhan, on 18 May 2011 and 19 May 2012. Basic information for the 529 cultivars is available on the RiceVarMap website (http://ricevarmap.ncpgr.cn/, last accessed: 10 August 2014) and information for the 107 wild rice accessions was previously reported by Zhang et al. (2015). Seven plants of each genotype were planted in a one-row plot at distances of 16.5cm within a row and 26.4cm between rows. Field management was in accordance with normal agricultural practices. Five plants in the middle of a row were used to score heading date; and the average date across the five plants was used as the heading date of the genotype for association mapping (see below). The heading dates for the 529 cultivars in 2011 and 2012 are presented in Supplementary Table S1 at JXB online.
Search for HAP family genes
To obtain sequencing data for all of the HAP genes in rice, BLASTP searches were performed in the predicted protein databases of the rice genome TIGR (http://rice.plantbiology.msu.edu/, last accessed: 10 August 2014) and NCBI (http://www.ncbi.nlm.nih.gov/, last accessed: 10 August 2014) with partial HAP proteins as queries (Thirumurugan et al., 2008). If a protein sequence satisfied E<10−10, it was selected as a candidate HAP protein. The SMART (http://smart.embl-heidelberg.de/, last accessed: 10 August 2014) and Pfam (http://pfam.sanger.ac.uk/, last accessed: 10 August 2014) databases were then used to predict the domain of all the candidate proteins. The deduced nucleotide and protein sequences of the new HAP genes in rice were downloaded from the TIGR database. In addition to the 28 HAP family genes that were reported by Thirumurugan et al. (2008), seven further HAP genes were identified; these were named sequentially according to their genome positions (Supplementary Table S2). To construct the phylogenetic tree of the HAP proteins, multiple protein sequences were executed using MEGA version 4.0 software to generate a maximum parsimony tree with bootstrapping analysis (Tamura et al., 2007).
Analysis of the expansion patterns of HAP genes
The method developed by Maher et al. (2006) was used to identify segmental duplications between HAP genes. First, we constructed a phylogenetic tree and identified all of the paralogous HAP genes at the terminal nodes. Next, 10 protein-coding genes that were upstream and downstream of each pair of paralogs were obtained from Gramene (Ware et al., 2002). Finally, the genes flanking one HAP gene were matched to the genes flanking the other HAP gene in the same pair. If these sequences resided within a region of conserved protein-coding genes, the paralogous HAP gene pair was regarded as the result of a duplication event. Tandem duplications were arbitrarily defined as ones that occur within a sequence distance of 50kb (Riechmann et al., 2000). The homology among the duplicated genes was calculated using MEGA version 4.0 software.
Nucleotide diversity and evolution analysis
The whole genomic DNA sequences of the 529 cultivar accessions and 107 O. rufipogon accessions were genotyped with approximate two-fold coverage genome sequencing using a bar-coded multiplex sequencing approach on an Illumina Genome Analyzer II (Chen et al., 2014). We extracted genomic sequences, defined as the DNA sequence of the gene from the transcription initiation site to the transcription stop site, of all HAP family genes from the TIGR database (http://rice.plantbiology.msu.edu/, last accessed: 10 August 2014). A sequence approximately 2kb in length, upstream of the genomic sequence, was extracted and considered to encompass the promoter region.
Two parameters of nucleotide diversity (the expected heterozygosity per nucleotide site, π, and theta per site from Eta, θ) were calculated. Tajima’s D statistic (Tajima, 1989) was used to search for evidence of selection. We calculated the ratio of genetic diversity in wild rice to that in cultivated rice (πw/πc) across genes to screen for selection signals. Fst, a standardized measure of the genetic variance between populations, was analyzed. All sequence analyses were conducted using DnaSP version 5.00 software (Librado and Rozas, 2009).
Expression profile analysis
The expression profile data of some duplicated OsHAP genes in indica rice Zhenshan 97 were extracted from the CREP database (http://crep.ncpgr.cn/, last accessed: 4 January 2015), including 25 RNA samples from several tissues at different developmental stages.
Candidate gene-based association mapping
For association analysis, the genome sequences of the 529 O. sativa accessions were downloaded from RiceVarMap. Using the population structure and relative kinship as covariates, single nucleotide polymorphism (SNP)–trait associations were analyzed with TASSEL software, using a mixed linear model. Analyses were conducted with population structure estimates, using the Q-matrix obtained from RiceVarMap. The parameter of the number of ancient clusters, K, was set from two to seven to obtain different inferences. The Bonferroni-adjusted significance threshold (P<0.05/n) was used to identify significant associations (n polymorphic sites) (Xu et al., 2011).
Transformation of HAP family genes
We amplified cDNAs of 18 OsHAP family genes (including Ghd8/OsHAP3H) from Nipponbare seedlings with gene-specific primers (Supplementary Table S3) using the high-fidelity LA Taq polymerase (Takara, Otsu, Japan). The cDNA of OsHAP genes was cloned into the binary vector pCAMBIA1301S-XBZ with the 35S promoter. Meanwhile, we used a 150–250bp sequence of part of the 3′ section of the target gene with the application of the double-stranded RNAi vector ds1301 to reduce expression of the gene. Additionally, the fragments of OsHAP3H, OsHAP2A, OsHAP2B, OsHAP2E, OsHAP2K, OsHAP5A and OsHAP5B for RNAi were identified by sequencing with gene-specific primers (Supplementary Table S3) and inserted into the binary vector ds1301, which inhibited the expression of RNA. All the recombinant constructs were delivered to wild type Zhonghua 11 (ZH11) or Hejiang 19 (HJ19) callus according to the rice genetic transformation method (Lin and Zhang 2005). The rice plants were grown in a paddy field under natural environmental conditions in Wuhan. Three mutants, Oshap2c, Oshap2j, and Oshap2e, were obtained from the Rice Mutant Database (http://rmd.ncpgr.cn/, last accessed: 14 October 2013), and six mutants, Oshap2h, Osha2d, Oshap3l, Oshap5b, Oshap5d, and Oshap5h, were obtained from the Rice T-DNA Insertion Sequence Database (http://an6.postech.ac.kr/pfg/index.php, last accessed: 14 October 2013).
Identification of positive transgenic plants
We extracted the total DNA from fresh leaves by using the CTAB method (Kidwell and Osborn, 1992). To identify positive transgenic plants, we first amplified the GUS fragment from the transgenic plants using the primers GUSF+GUSR. Positive transgenic plants were identified by the presence of a bright band 1.5kb in size on gel electrophoresis. Then, we randomly selected positive and negative transgenic plants checked by GUS amplification to detect the level of expression of the target genes. These two methods in combination enabled us to identify positive and negative plants.
RNA extraction and quantitative real-time reverse transcription PCR
Total RNA was extracted from leaves using an RNA extraction kit (TRIzol reagent, Invitrogen) following the manufacturer’s instructions. First-strand cDNA was reverse transcribed from DNase I-treated RNA with oligo(dT) as the primer. The expression of OsHAP5A, OsHAP5B, OsHAP3D and OsHAP3E was measured by quantitative real-time reverse transcription PCR (qRT-PCR). Approximately 3mg total RNA was reverse transcribed using M-MLV reverse transcriptase (Invitrogen) in a volume of 180 µl to obtain cDNA. The Ubiquitin gene was used as an internal control for the qRT-PCR. The primers used for qRT-PCR are listed in Supplementary Table S3. qRT-PCR was run in a total volume of 15 µl containing 3.6 µl of the reverse-transcribed product obtained as described above, 0.25mM gene-specific primers and 7.8 µl FastStart Universal SYBR Green Master (Rox) superMIX (Roche, Mannheim, Germany) on an Applied Biosystems ViiA 7 RT-PCR system, according to the manufacturer’s instructions. The relative quantification method was used to measure the quantity of qRT-PCR product. The qRT-PCR was performed using the following program: 95 °C for10min, then 40 cycles of 95 °C for 5s and 60 °C for 34s.
Results
Identification of HAP family genes
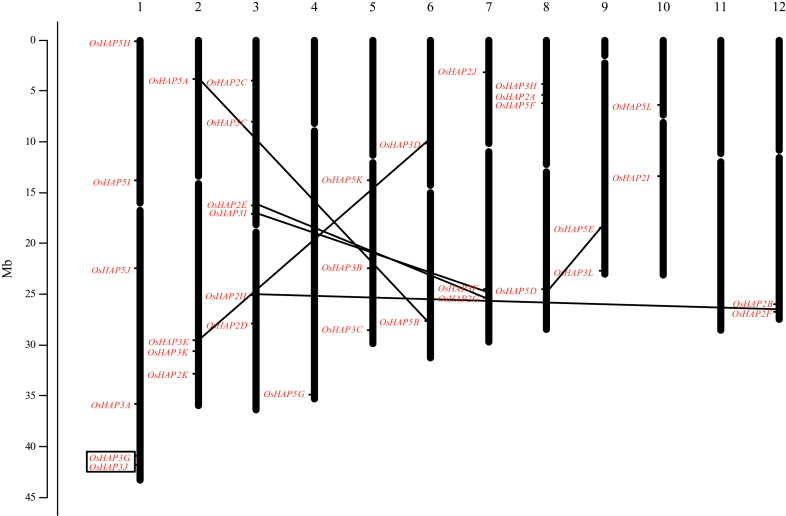
In addition to the 28 genes that were previously reported by Thirumurugan et al. (2008) in rice, we identified one additional HAP2 gene, one additional HAP3 gene, and five additional HAP5 genes (Supplementary Table S2). The newly identified genes were termed OsHAP2K, OsHAP3L, and OsHAP5H–OsHAP5L. The 35 HAP family genes included 11 HAP2, 12 HAP3, and 12 HAP5 genes. HAP genes were distributed on every chromosome except chromosome 11 (Fig.1). Chromosomes 1 and 3 each harbored the largest number (six) of HAP genes; there were four HAP genes on chromosomes 2 and 8; chromosomes 5 and 7 contained three HAP genes; chromosomes 6, 9, 10, and 12 each contained two HAP genes; and chromosome 4 carried a single HAP gene. A phylogenetic tree was constructed with a maximum parsimony method using protein sequences that were deduced from Nipponbare (Supplementary Fig. S1A). The newly identified HAP genes were grouped into their corresponding subfamilies, indicating that they are real HAP family genes.
Fig. 1.
Genetic linkage map indicating the positions of HAP family genes. The HAP genes are indicated on the chromosomes in red. The segmental duplication events corresponding to HAP genes are indicated by black lines between the chromosomes. The black-outlined box indicates tandem duplication events.
Structure of HAP genes and proteins
All the members of the HAP family identified in rice each had a complete HAP domain (Supplementary Fig. S1B). All the other OsHAP proteins contained only a CBFB_NFYA or CBFD_NFYB domain, except OsHAP5F, which included the pollen_allerg_1 and transmembrane domains. Analysis of the gene structure showed that 17 (48.6%) of the 35 OsHAP genes were intronless. OsHAP2 genes each had four to six exons, the OsHAP3 genes had one to three exons, and the OsHAP5 genes each had one exon, except OsHAP5F, which had six exons (Supplementary Fig. S1C).
Duplication of HAP family genes
A total of 10 pairs of rice paralogous genes were identified on the terminal node of the phylogenetic tree (Supplementary Fig. S1A). No highly conserved genes were observed between the flanking regions of the paralogous pairs of OsHAP2B/2D, OsHAP3B/3C, and OsHAP5I/5J, indicating that these pairs of paralogous genes were formed through random translocation and insertion events. However, the conserved genes were detected between the flanking regions of other paralogous pairs of OsHAP2E/2G, OsHAP2F/2H, OsHAP3D/3E, OsHAP3F/3I, OsHAP5A/5B, and OsHAP5D/5E (Table 1). Therefore, these paralogous genes might have arisen from segmental duplications. OsHAP3G and OsHAP3J, with 49.6% homology (Fig.1, Supplementary Table S4), were located in a tandem repeat on chromosome 1, indicating that tandem duplication events contributed to producing these two genes. There are therefore seven pairs of paralogs in duplicate (Fig.1, Supplementary Table S4). We compared the expression patterns of three pairs of genes, OsHAP2E/2G and OsHAP3E/3D, which represented segmental duplication patterns, and OsHAP3G/3J, which represented a tandem duplication pattern. The segmentally duplicated pair of OsHAP3D and OsHAP3E showed similar expression patterns in 25 tested stages/tissues (Supplementary Fig. S2A). In contrast, the expression patterns of the segmentally duplicated pair of OsHAP2E and OsHAP2G and the tandem duplication pair of OsHAP3G and OsHAP3J were different in most of these tissues (Supplementary Fig. S2B, C). qRT-PCR analysis for eight tissues at different developmental stages showed consistent results (Supplementary Fig. S3).
Table 1.
Conserved protein-coding genes in their flanking regions of segmentally duplicated HAP genes
| Duplicated gene 1 | Conserved protein-coding flanking gene 1 | Duplicated gene 2 | Conserved protein-coding flanking gene 2 | Annotation for flanking genes |
|---|---|---|---|---|
| OsHAP2E | Os03g29830 | OsHAP2G | Os07g41660 | Expressed protein |
| OsHAP2F | Os12g42420 | OsHAP2H | Os03g44580 | DNA binding protein |
| OsHAP3D | Os06g17410 | OsHAP3E | Os02g49440 | dof zinc finger domain |
| OsHAP3F | Os07g41600 | OsHAP3I | Os03g29920 | proline-rich protein |
| OsHAP5A | Os02g07430 | OsHAP5B | Os06g45650 | MADS-box with MIKCc type-box |
| OsHAP5D | Os08g38810 | OsHAP5E | Os09g30320 | BURP domain |
Nucleotide diversity of HAP genes and evolution analysis
The genomic DNA length of the 35 OsHAP genes ranged from 377–9357bp. We analyzed the DNA polymorphisms of the HAP family genes within 107 wild rice accessions and 529 cultivars (Supplementary Tables S5–7). There were 998, 496, and 856 SNPs within the HAP2, HAP3, and HAP5 subfamilies, respectively, across the entire sequence (containing the promoter and genomic sequences) among the 529 cultivar accessions (Table 2). The average nucleotide diversity of whole HAP family genes in both O. sativa and wild rice (π=3.3×10−3) was higher than that of the whole genome in O. sativa (π=2.4×10−3) (Huang et al., 2012). In the promoter region, the nucleotide diversity was not significantly different among the HAP2, HAP3, and HAP5 subfamilies. However, compared with the HAP2 and HAP5 subfamilies, the HAP3 subfamily genes, on average, demonstrated low nucleotide diversity in both O. sativa and O. rufipogon (π= 1.4×10−3 and 1.6×10−3, respectively) in the genomic sequence. The average nucleotide diversity (π=4.2×10−3) was thus apparently higher in the promoter regions of the 35 HAP family genes than in the genomic sequence (Table 2). No difference was observed between O. sativa and O. rufipogon for each subfamily.
Table 2.
Nucleotide diversity of HAP family genes in cultivar and wild rice
| HAP family | Cultivars | Wild rice | |||||
|---|---|---|---|---|---|---|---|
| SNPs | π (×10 −3) | θ (×10 −3) | SNPs | π (×10 −3) | θ (×10 −3) | ||
| Promoter | HAP2 | 398 | 4.7 | 2.2 | 470 | 4.6 | 3.2 |
| HAP3 | 370 | 3.9 | 2.1 | 533 | 4.2 | 3.4 | |
| HAP5 | 376 | 4.0 | 2.3 | 525 | 4.7 | 3.3 | |
| Whole | 1144 | 4.2 | 2.2 | 1528 | 4.5 | 3.3 | |
| Genomic sequence | HAP2 | 600 | 2.6 | 1.3 | 703 | 2.5 | 1.9 |
| HAP3 | 126 | 1.4 | 0.8 | 184 | 1.6 | 1.5 | |
| HAP5 | 480 | 3.1 | 1.9 | 505 | 3.2 | 2.7 | |
| Whole | 1206 | 2.4 | 1.3 | 1392 | 2.4 | 2.0 | |
| Entire sequence | HAP2 | 998 | 3.7 | 1.8 | 1173 | 3.6 | 2.6 |
| HAP3 | 496 | 2.7 | 1.5 | 717 | 2.9 | 2.5 | |
| HAP5 | 856 | 3.6 | 2.1 | 1030 | 4.0 | 3.0 | |
| Whole | 2350 | 3.3 | 1.8 | 2920 | 3.5 | 2.7 | |
Estimates of nucleotide diversity were calculated based on average pairwise diversity (π) and the number of segregating sites (θ). The promoter region has a length of 2kb upstream of the gene.
The pairwise nucleotide diversity in the genomic sequence ranged from 0–11 SNPs per kb (Supplementary Tables S5–7). OsHAP2A and OsHAP5J had the highest diversity, with approximately 8–11 SNPs per kb in both O. sativa and wild rice. OsHAP2C, OsHAP2J, OsHAP3B, OsHAP3E, OsHAP5D, and OsHAP5E had the lowest diversity, with less than 1 SNP per kb in the genomic sequence. In the promoter region, the pairwise nucleotide diversity ranged from 1–14 SNPs per kb, higher than in the genomic sequence. OsHAP2E and OsHAP5J had the highest diversity in the promoter region, with approximately 9–14 SNPs per kb in both O. sativa and wild rice. In cultivated rice, the Tajima’s D values in the genomic sequence of OsHAP2A, OsHAP2B, OsHAP2E, OsHAP3C, OsHAP3L, and OsHAP5J reached a significantly positive level; in contrast, no gene in wild rice reached a significantly positive level. The πw/πc ratio in the promoter region and in the genomic sequences of OsHAP2J and OsHAP5B was greater than 4 (Supplementary Tables S5–7), indicating that these genes underwent selection during domestication and genetic improvement. For most of the 35 OsHAP family genes, the population-differentiation statistic (Fst) between the populations of indica (295 varieties) and japonica (156 varieties) was greater than 0.25 (Supplementary Table S8), indicating a very strong population differentiation. Only OsHAP2J, OsHAP3D, OsHAP3F, OsHAP5B, OsHAP5E, and OsHAP5F had a lower Fst between the indica and japonica groups.
Association between heading date and HAP family genes
Four genes, including three OsHAP3 and one OsHAP5 subfamily genes, were significantly associated with heading date at P<10−3 under long-day conditions in both 2011 and 2012 (Table 3). No genes in the OsHAP2 subfamily were associated with heading date. The associated gene contained the previously cloned flowering QTL Ghd8/OsHAP3H (Yan et al., 2011b). The associated SNP site of OsHAP5A fell into an intron region. OsHAP3E has the same identity as OsLEC1, a key regulator of meristem identity determination in both vegetative and reproductive development (Kwong et al., 2003; Lotan et al., 1998). OsHAP3G exhibited the highest significant association in 2011, at P=3.2×10−11. The associated SNP sites of two, one, and one genes were located in untranslated region, intron, and exon regions, respectively.
Table 3.
HAP family genes associated with heading date
| Gene | 2012 | 2011 | ||
|---|---|---|---|---|
| Association site | P | Association site | P | |
| OsHAP3G | 3′UTR | 5.6×10–3 | 3′UTR | 3.2×10–11 |
| OsHAP3E | Exon | 2.1×10–3 | Exon | 2.4×10–3 |
| OsHAP3H (Ghd8) | 5′UTR | 2.5×10–5 | 5′UTR | 1.3×10–3 |
| OsHAP5A | Intron | 2.1E×10–3 | Intron | 7.9×10–3 |
P values of the association signals were calculated from the mixed linear model.
3′UTR, 3′untranslated region; 5′UTR, 5′ untranslated region.
Phenotype of the mutants of nine HAP family genes
To rapidly confirm the function of the HAP family genes in relation to heading date, we searched the database in Rice GE (http://signal.salk.edu/cgi-bin/RiceGE/, last accessed: 14 October 2013) and found 11 distinct mutants that targeted 11 HAP family genes. However, only nine mutants of OsHAP genes were germinated and planted, due to poor seed viability. We performed two sets of PCR to genotype each mutation, one using a pair of gene-specific primers (F and R) and the other using a gene-specific primer (F) and a corresponding T-DNA border primer (TRB2 or PGAR2) (Supplementary Fig. S4, Supplementary Table S3). However, none of the mutants had a change in heading date compared with that of wild-type plants under either long-day or short-day conditions (Supplementary Table S9).
Flowering function identification of 18 HAP family genes by transformation
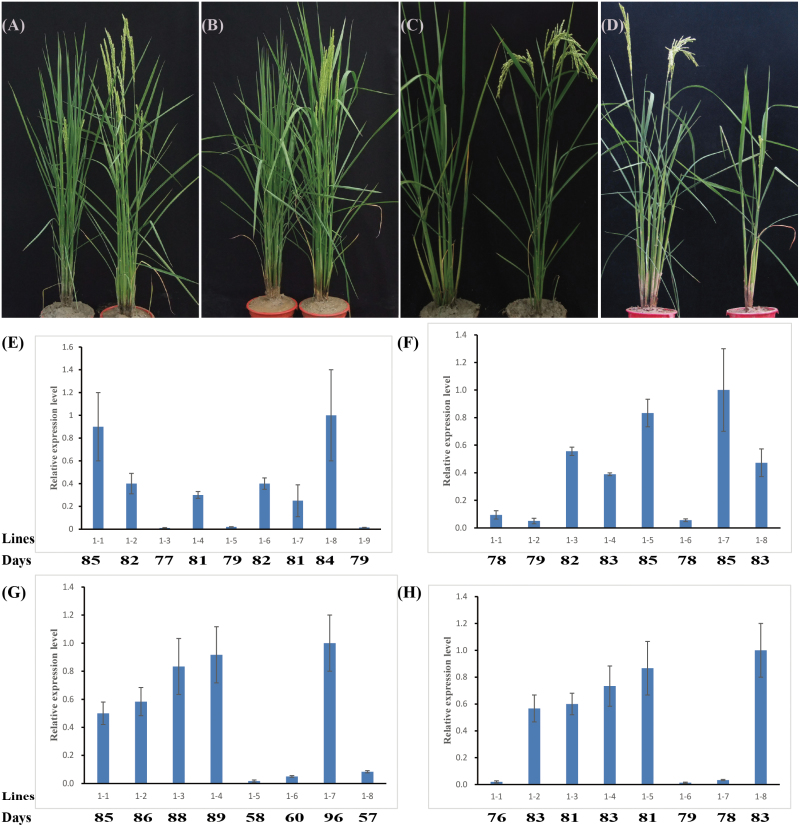
Among the four associated genes, OsHAP3H/Ghd8 has previously been confirmed to regulate heading date (Yan et al., 2011). Thus, we tested the function of the additional three genes by overexpression. In addition, to determine whether the non-associated HAP family genes affect heading date, we randomly overexpressed 14 genes—OsHAP2C, OsHAP2G, OsHAP2I, OsHAP5B, OsHAP5E, OsHAP2B, OsHAP2E, OsHAP2H, OsHAP2K, OsHAP3D, OsHAP3B, OsHAP3C, OsHAP5H, and OsHAP5K—in japonica rice ZH11 or HJ19 (Fig. 2). The positive OsHAP5A (Fig. 2A) and OsHAP5B (Fig. 2B) plants had significant differences in heading date compared with negative (wild-type) plants in the T1 generation under long-day conditions (Table 4). Moreover, significant differences in heading date were observed between negative and positive OsHAP3E (Fig. 2C) and OsHAP3D (Fig. 2D) plants in the T0 generation under long-day conditions (Table 4). qRT-PCR analyses indicated that in the overexpression lines of OsHAP5A (Fig. 2E), OsHAP5B (Fig. 2F), OsHAP3E (Fig. 2G), and OsHAP3D (Fig. 2H), the mRNA levels of these four genes were indeed higher than in wild-type plants. Co-segregation analysis in the T1 generation showed that overexpression of OsHAP5A (Chi-square=0.04, P=0.8) and OsHAP5B (Chi-square=0.4, P=0.5) resulted in delayed heading (Table 4). OsHAP3E and OsHAP3D overexpression lines were sterile, but all positive plants in the T0 generation were always heading later than wild-type and negative plants (Table 4). OsHAP5A and OsHAP3D delayed heading by 3–4 days under long-day conditions. OsHAP5B delayed heading by approximately 7 days. OsHAP3E greatly delayed the heading date of HJ19, by 30 days.
Fig. 2.
Performance in heading date of transgenic plants of four HAP genes and real-time PCR analyses. (A) Overexpression plants of OsHAP5A (left) and wild-type Zhonghua 11 (right) in the T1 generation. (B) Overexpression plants of OsHAP5B (left) and wild-type Zhonghua 11 (right) in the T1 generation. (C) Overexpression plants of OsHAP3E (left) and wild-type Hejiang 19 (right) in the T0 generation. (D) Overexpression plants of OsHAP3D (right) and wild-type Zhonghua 11 (left) in the T0 generation. (E–G) qRT-PCR analyses of (E) OsHAP5A and (F) OsHAP5B in the T1 generation, and of (G) OsHAP3E and (H) OsHAP3D in the T0 generation. The numbers beneath each graph show the number of days to heading for the plants. The relative expression levels were normalized with that of rice Ubiquitin. The highest level was set at 1.
Table 4.
Heading date of transgenic plants for the functional HAP genes under long-day conditions
| Gene | Overexpression lines | Positive plants | Negative plants | P1 | Chi-square | P2 | ||
|---|---|---|---|---|---|---|---|---|
| No | HD (d) | No | HD (d) | |||||
| OsHAP3D | OX (ZH11)-T0 | 5 | 82.2±1.1 | 3 | 77.7±1.5 | 1.2×10–3 | ||
| OsHAP3E | OX (HJ19)-T0 | 5 | 88.8±4.3 | 3 | 58.3±1.5 | 2.65×10–5 | ||
| OsHAP5A | OX (ZH11)-T1 | 23 | 81.8±4.3 | 7 | 78.3±1.0 | 5.3×10–6 | 0.04 | 0.8 |
| OsHAP5B | OX (ZH11)-T1 | 24 | 83.6±2.8 | 6 | 77.8±1.6 | 4.0×10–11 | 0.4 | 0.5 |
OX (HJ19) and OX (ZH11) represent the overexpressed plants of the Hejiang 19 and Zhonghua 11 genotype, respectively. No, number of plants investigated; HD, heading date. P1 values were obtained by t tests for significant difference in heading date between the positive and negative plants. P2 values were obtained by chi-squared test for segregation of 3:1. T0 plants came from different resistant callus. T1 plants came from one T0 plant.
In addition, we constructed OsHAP2A, OsHAP2B, OsHAP2E, OsHAP2K, OsHAP3H, OsHAP5A, and OsHAP5B for double-stranded RNA lines in the ZH11 background. However, the RNAi plants of OsHAP2A, OsHAP2B, OsHAP2E, OsHAP2K, OsHAP3H, OsHAP5A, and OsHAP5B did not show any change in heading date from the wild type.
Except for OsHAP3E, OsHAP3D, OsHAP5A and OsHAP5B, the other 13 overexpressed genes did not change heading date (Supplementary Table S10). In addition, these nine mutants, Oshap2c, Oshap2d, Oshap2e, Oshap2h, Oshap2j, Oshap3l, Oshap5b, Oshap5d, and Oshap5h, did not exhibit any change in heading date.
Some heading genes in rice were previously reported to function under short-day conditions (Yano et al., 2000; Yan et al. 2011b). Thus, we investigated the heading dates of all transgenic plants, mutants, and their controls covering 20 HAP genes under short-day conditions (winter Hainan). None of the tested HAP genes functioned under short-day conditions except Ghd8/OsHAP3H (Supplementary Table S10).
Discussion
At least four HAP genes including Ghd8/OsHAP3H control heading date in rice
In this study, we identified four HAP family genes that were associated with heading date, based on a diverse germplasm collection. We then tested the function of 18 HAP genes on heading date by overexpressing or silencing cDNAs from Nipponbare seedlings and nine mutants that were targeted at HAP genes. The overexpression of OsHAP5A and OsHAP5B genes delayed heading date under long-day conditions in the T1 generation. OsHAP3D and OsHAP3E delayed heading date under long-day conditions in the T0 generation; for these genes, data was not obtained in the T1 generation because of the transgenic were sterile plants. All five of the positive OsHAP3E plants headed 30 days later than the negative plants (Table 4), indicating with certainty that OsHAP3E regulates heading date. For OsHAP3D, although the difference in heading between positive and negative plants in T0 was only four days, all five of the positive plants consistently headed later than the negative plants, suggesting that OsHAP3D might regulate heading date. These genes belonged to the subfamilies encoding HAP5 and HAP3 subunits, in addition to our previously cloned Ghd8/OsHAP3H. No HAP2 subfamily genes regulated heading date. It should be noted that only Nipponbare alleles were used to test the function of HAP genes in this study. We cannot be sure that all the Nipponbare alleles are functional, although no premature termination mutation was observed in the Nipponbare alleles (Supplementary Table S11). Alternatively, overexpressing the alleles from other genotypes for these HAP genes, which were presumed non-functional in regulating heading by overexpressing Nipponbare alleles, may identify more HAP genes that regulate heading. In addition, we tested only 23 of the 35 identified HAP genes, and the remaining 13 genes need to be tested in future. From these findings we can conclude that at least four HAP genes, including Ghd8/OsHAP3H, regulate heading date in rice.
It is surprising that none of the nine tested HAP2 subfamily genes regulated heading date. In Arabidopsis, the HAP proteins have been suggested to work in a complex composed of HAP2/HAP3/HAP5 (Ben-Naim et al., 2006). Even though the additional two untested HAP2 genes, OsHAP2D and OsHAP2F, do not regulate heading date, this result is also understandable because the CCT domain of CO can replace HAP2 and interact with HAP3/HAP5 to regulate flowering in Arabidopsis (Wenkel et al., 2006). There are dozens of CCT family genes that are involved in the flowering pathway in rice (Zhang et al., 2015), providing flexibility to form a heterotrimeric complex.
Type I and type II errors in association mapping were identified by transformation
Four genes were associated with heading date, but one of them was non-functional in regulating heading date. Additionally, no association was detected between OsHAP5B or OsHAP3D and heading date. However, OsHAP5B- and OsHAP3D-overexpressing plants showed a significantly delayed heading date. These results indicated that both type I and type II errors occurred when performing the association analysis. In statistics, a type I error is the incorrect rejection of a true null hypothesis. In general, a type I error is frequently caused by low threshold values; before running an association analysis, the exclusion of rare causal mutations (i.e. SNPs) due to low frequency will also lead to a type I error. Similar to OsHAP3H (Ghd8), the causal mutation is a SNP that is located +322bp from the initiation codon ATG, resulting in a premature stop in protein translation (Yan et al., 2011b). Because this SNP is detected in only a few accessions, it was not included in the association analysis. Therefore, the SNP was not identified as being associated with heading date. A type II error is the failure to reject a false null hypothesis. In general, a type II error is frequently caused by high threshold values, but a close linkage between the tested HAP family genes and a QTL/gene influencing heading date is a possible reason for the type II error here because LD decay is slow in rice, extending to an approximately 200kb region (Huang et al., 2012). For example, the association between OsHAP3G and heading date is probably due to linkage to the heading date gene OsHY2 (Saito et al., 2010), because OsHAP3G itself was verified to be non-functional in regulating heading. In cases of type I and type II errors, general linear models and linear mixed models are recommended for association analysis, besides specifying a reasonable threshold value for claiming an association.
Overexpression enhanced the power of functional identification of genes in a large family
Overexpression and knockout/knockdown mutations have been confirmed to be efficient ways to identify gene function. In general, mutants are primarily recommended for testing gene function. However, the members of a large gene family frequently have a similar function in plants, and loss of function of one gene can be rescued by other members of the gene family (Hughes and Liberles, 2007). Consequently, gene function frequently cannot be identified for a single gene mutant in a large gene family. For example, in this study, for OsHAP5A and OsHAP5B, the overexpression lines significantly delayed heading date, but the RNAi lines showed no change under long-day conditions. Similar to OsHAP5A and OsHAP5B, Ghd8/OsHAP3H was confirmed to suppress flowering in rice, increasing plant height and yield potential simultaneously (Yan et al., 2011b), but Ghd8-RNAi lines did not alter heading date, plant height or yield (Supplementary Table S10). It is obvious that some redundant genes exist in the rice HAP gene family. This was the case in a previous study identifying the function of CCT family genes (Zhang et al., 2015). Although RNAi plants cannot adequately unveil the function of HAP family genes, they can be used to identify redundant genes by crossing them to produce double or triple mutants. Considering the redundancy among a large number of genes within a family, overexpression easily validates gene function and is largely encouraged to use for testing the gene function of a large gene family.
Duplication diversified the function of HAP family genes
Three mechanisms contribute to the expansion of gene families—tandem duplication, large-scale block duplication (segmental duplication), and transposition events, such as retro-transposition and replicative transposition (Kong et al., 2007). The duplication of protein-coding genes can result in loss of function (pseudogenes), maintenance of the original function (redundant function), or acquisition of a new function (neofunctionalization) (Hughes and Liberles, 2007). There are 18 pairs of duplicated segments that together cover 65.7% of the rice genome (Yu et al., 2005). In this study, among 10 pairs of paralogous HAP family genes (Supplementary Fig. S1A), one pair (OsHAP3G and OsHAP3J) was identified as having formed under tandem duplication. Six pairs of HAP genes were segmentally duplicated. Among these genes, four pairs (OsHAP3F/3I, OsHAP2E/2G, OsHAP5D/5E, and OsHAP5A/5B) were located on the corresponding duplication blocks in the rice genome; the two pairs of OsHAP3D/3E and OsHAP2F/2H were not. Both of these latter pairs probably arose from duplication events and lost their counterparts over a long period of evolution, because the rice genome underwent ancient polyploidization through two rounds of polyploidy events followed by massive gene losses and numerous chromosome rearrangements (Guyot and Keller, 2004).
Part of the short arm of chromosome 2 harboring OsHAP5A and part of the long arm of chromosome 6 harboring OsHAP5B (Fig. 1) include pairs of duplicated segments of the rice genome (Yu et al., 2005). After this duplication, OsHAP5A and OsHAP5B maintained the same function in regulating heading date, which can be explained by the mechanisms of concerted evolution and purifying selection (Li, 2006; Nei et al., 2000). Similarly, the duplicated genes OsHAP3D and OsHAP3E play critical roles in embryogenesis (Kwong et al., 2003; Lotan et al., 1998) and flowering regulation. Similar expression profiles of OsHAP3D and OsHAP3E were also observed (Supplementary Fig. S2A). These findings indicate that some segmentally duplicated genes in the OsHAP family maintained conserved functions during evolution. The expression patterns of the OsHAP2E/2G pair were different in most of the tissues that were tested (Supplementary Fig. S2B), indicating that one member of the duplicate might have gained a new function. The tandem-duplicated genes OsHAP3G and OsHAP3J demonstrated a different expression pattern in most tissues, indicating a functional differentiation. Therefore, the HAP genes have evolved to have diverse functions such as drought stress tolerance (Han et al., 2013) and pathogen resistance (Alam et al., 2015), and only a few HAP genes regulate heading in rice.
Diverse function contributed to the selection of HAP family genes
The selective signatures from domestication have been applied most commonly to data from genes (Doebley et al., 2006). In this study, we found that HAP genes had a higher Fst between indica and japonica subspecies, indicating that the genes in both populations are significantly differentiated. A positive Tajima’s D signifies that population stratification or balancing selection occurred in this locus during the evolution and breeding of rice. A negative Tajima’s D signifies an excess of low-frequency polymorphisms relative to expectations, indicating a selective sweep or purifying selection. Significant positive Tajima’s D values indicated that at least 13 genes, including OsHAP2A, OsHAP2B, OsHAP3L, and OsHAP5J, were subject to balancing selection or population stratification during the natural evolutionary process. Moreover, OsHAP2B was located in a selective sweep region (Huang et al., 2012). However, we did not observe any function of OsHAP2B in relation to heading date. These results indicate that a strong selection pressure was imposed on the region of OsHAP2B during domestication of rice. The selection of OsHAP2B was probably the result of genetic ‘hitch-hiking’.
When gene mutation affects the fitness and survival of an organism, selection naturally occurs during evolution. In particular, when a mutation leads to a trait that is desirable by humans, this mutation is artificially selected after the species has been domesticated (Sang, 2009). Therefore, it is expected that some HAP family genes that underwent selection would be responsible for important biological and agronomic traits. In this study, only four genes regulated heading date, which is an important factor when determining ecological adaptation. In addition, OsHAP2E confers resistance to pathogens, salt, and drought, and increases photosynthesis and tiller number (Alam et al., 2015). It is likely that some HAP genes may have a potential function in abiotic stress tolerance and have consequently been selected during domestication and genetic improvement. It would be worthwhile to test the stress tolerance of HAP genes using these transgenic lines in future.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Heading date data for 529 cultivars in 2011 and 2012.
Table S2. List of 35 HAP family genes identified in rice.
Table S3. Primers used for amplifying gene fragments and transcriptional expression analysis.
Table S4. Homology among duplicated HAP genes.
Table S5. DNA polymorphic sites of HAP2 genes.
Table S6. DNA polymorphic sites of HAP3 genes.
Table S7. DNA polymorphic sites of HAP5 genes.
Table S8. F statistics of HAP family genes between japonica and indica cultivars.
Table S9. Heading dates of mutants targeted nine HAP family genes.
Table S10. Functional identification on heading date of 23 HAP family genes by transformation or mutant in long-day condition (Wuhan) and short-day condition (Hainan).
Table S11. Variation in the position of the stop codon in Nipponbare allele in 529 rice cultivars.
Fig. S1. Phylogenetic tree for rice HAP family and protein structure of OsHAP proteins.
Fig. S2. Comparison of expression profiles between duplicated OsHAP genes.
Fig. S3. qRT-PCR analysis of the expression of six OsHAP genes.
Fig. S4. PCR tests for mutants Oshap2j (A) and Oshap5d (B) as an example.
Acknowledgements
This work was supported by grants from the National Special Program for Research of Transgenic Plants of China (2011ZX08009-001-002), the 863 program on functional genomics of stress resistance and nutrient utility in rice (2012 AA10A303), the National Natural Science Foundation of China (91335201), and the Bill & Melinda Gates Foundation.
References
- Alam MM, Tanaka T, Nakamura H, Ichikawa H, Kobayashi K, Yaeno T, Yamaoka N, Shimomoto K, Nishina H, Nishiguchi M. 2015. Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant Biotechnology Journal 13, 85–96. [DOI] [PubMed] [Google Scholar]
- Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, Samach A, Lifschitz E. 2006. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. The Plant Journal 46, 462–476. [DOI] [PubMed] [Google Scholar]
- Chen NZ, Zhang XQ, Wei PC, Chen QJ, Ren F, Chen J, Wang XC. 2007. AtHAP3b plays a crucial role in the regulation of flowering time in Arabidopsis during osmotic stress. Journal of Biochemistry and Molecular Biology 40, 1083–1089. [DOI] [PubMed] [Google Scholar]
- Chen W, Gao Y, Xie W, et al. 2014. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nature Genetics 46, 714–721. [DOI] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD. 2006. The molecular genetics of crop domestication. Cell 7, 1309–1321. [DOI] [PubMed] [Google Scholar]
- Ehrenreich IM, Hanzawa Y, Chou L, Roe JL, Kover PX, Purugganan MD. 2009. Candidate gene association mapping of Arabidopsis flowering time. Genetics 183, 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint-Garcia SA, Thornsberry JM, d Buckler ES. 2003. Structure of linkage disequilibrium in plants. Annual Review of Plant Biology 54, 357–374. [DOI] [PubMed] [Google Scholar]
- Fornari M, Calvenzani V, Masiero S, Tonelli C, Petroni K. 2013. The Arabidopsis NF-YA3 and NF-YA8 genes are functionally redundant and are required in early embryogenesis. PLOS ONE 8, e82043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusmaroli G, Tonelli C, Mantovani R. 2001. Regulation of the CCAAT-binding NF-Y subunits in Arabidopsis thaliana . Gene 264, 173–185. [DOI] [PubMed] [Google Scholar]
- Guyot R, Keller B. 2004. Ancestral genome duplication in rice. Genome 47, 610–614. [DOI] [PubMed] [Google Scholar]
- Han X, Tang S, An Y, Zheng DC, Xia XL, Yin WH. 2013. Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis . Journal of Experimental Botany 64, 4589–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. 2003. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. [DOI] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, et al. 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wei X, Sang T, et al. 2010. Genome-wide association studies of 14 agronomic traits in rice landraces. Nature Genetics 42, 961–967. [DOI] [PubMed] [Google Scholar]
- Hughes T, Liberles DA. 2007. The pattern of evolution of smaller-scale gene duplicates in mammalian genomes is more consistent with neo-than subfunctionalisation. Journal of Molecular Evolution 65, 574–588. [DOI] [PubMed] [Google Scholar]
- Kidwell KK, Osborn TC. 1992. Simple plant DNA isolation procedures. In: Beckmann JS, Osborn TC, eds. Plant genomes: methods for genetic and physical mapping. Dordrecht: Springer, 1–13. [Google Scholar]
- Kong HZ, Landherr LL, Frohlich MW, Leebens-Mack J, Ma H, DePamphilis CW. 2007. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. The Plant Journal 50, 873−885. [DOI] [PubMed] [Google Scholar]
- Kusnetsov V, Landsberger M, Meurer J, Oelmuller R. 1999. The assembly of the CAAT-box binding complex at a photosynthesis gene promoter is regulated by light, cytokinin, and the stage of the plastids. Journal of Biological Chemistry 274, 36009–36014. [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ. 2003. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryogenesis. The Plant Cell 15, 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. [DOI] [PubMed] [Google Scholar]
- Li WH. 2006. Molecular evolution . Sunderland: Sinauer Associates. [Google Scholar]
- Li WX, Oono Y, Zhu JH, He XJ, Wu JM, Lida K, Lu XY, Cui XP, Jin HL, Zhu JK. 2008. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. The Plant Cell 20, 2238–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YJ, Zhang QQ. 2005. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Reports 23, 540–547. [DOI] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. 1998. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. [DOI] [PubMed] [Google Scholar]
- Maher C, Stein L, Ware D. 2006. Evolution of Arabidopsis microRNA families through duplication events. Genome Research 16, 510−519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 239, 15–27. [DOI] [PubMed] [Google Scholar]
- Mather KA, Caicedo AL, Polato NR, Olsen KM, Couch SM, Purugganan MD. 2007. The extent of linkage disequilibrium in rice (Oryza sativa L.). Genetics 177, 2223–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Ito Y, Serizawa A, Kurata N. 2003. OsHAP3 genes regulate chloroplast biogenesis in rice. The Plant Journal 36, 532–540. [DOI] [PubMed] [Google Scholar]
- Nei M, Rogozin IB, Piontkivska H. 2000. Purifying selection and birth-and-death evolution in the ubiquitin gene family. Proceedings of the National Academy of Sciences of the United States of America 20, 10866–10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, et al. 2007. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proceedings of the National Academy of Sciences of the United States of America 104, 16450–16455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES. 2001. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proceedings of the National Academy of Sciences of the United States of America 98, 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Sherman BK, Yu GL. 2000. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. [DOI] [PubMed] [Google Scholar]
- Saito H, Okumoto Y, Yoshitake Y, Inoue H, Yuan QB, Teraishi M, Tsukiyama T, Nishida H, Tanisaka T. 2010. Complete loss of photoperiodic response in the rice mutant line X61 is caused by deficiency of phytochrome chromophore biosynthesis gene. Theoretical and Applied Genetics 122, 109–118. [DOI] [PubMed] [Google Scholar]
- Sang T. 2009. Genes and mutations underlying domestication transitions in grasses. Plant Physiology 149, 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XC, Ling S, Lu ZH, Ouyang YD, Liu SS, Yao JL. 2014. OsNF-YB1, a rice endosperm-specific gene, is essential for cell proliferation in endosperm development. Gene 551, 214–221. [DOI] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thirumurugan T, Ito Y, Kubo T, Serizawa A, Kurata N. 2008. Identification, characterization and interaction of HAP family genes in rice. Molecular Genetics and Genomics 279, 279–289. [DOI] [PubMed] [Google Scholar]
- Ware DH, Jaiswal P, Ni J, Yap IV, Pan X, Clark KY, Teytelman L, Schmidt SC, Stein LD, McCouch SR. 2002. Gramene, a tool for grass genomics. Plant Physiology 130, 1606−1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Gourrierec JL, Coupland G. 2006. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis . The Plant Cell 18, 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HP, Zeng H, Luo CL, Zhang DX, Wang Q, Sun L, Yang LS, Zhou M, Nie QH, Zhang XQ. 2011. Genetic effects of polymorphisms in candidate genes and the QTL region on chicken age at first egg. BMC Genetics 12, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue WY, Xing YZ, Weng XY, et al. 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics 40, 761–767. [DOI] [PubMed] [Google Scholar]
- Yan JB, Warburton M, Crouch J. 2011a. Association mapping for enhancing maize (Zea mays L.) genetic improvement. Crop Science 51, 433–449. [Google Scholar]
- Yan WH, Wang P, Chen HX, Zhou HJ, Li QP, Wang CR, Ding ZH, Yu SB, Xing YZ, Zhang QF. 2011b. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Molecular Plant 4, 319–330 [DOI] [PubMed] [Google Scholar]
- Yan WH, Liu HY, Zhou XC, et al. 2013. Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Research 23, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, et al. 2000. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS . The Plant Cell 12, 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, et al. 2005. The genomes of Oryza sativa: a history of duplications. PLOS Biology 3, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JJ, Xue HW. 2013. OsLEC1/OsHAP3E participates in the determination of meristem identity in both vegetative and reproductive developments of rice. Journal of Integrated Plant Biology 55, 232–249. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li QP, Dong HJ, et al. 2015. Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice. Scientific Reports 5, 7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.