Highlight
By studying the mode of evolution of Medicago truncatula PISTILLATA-like paralogs after duplication, we provide evidence of the complex dynamics underlying the evolution by gene duplication of the B-class MADS-box subfamily.
Key words: Duplicated B-function genes, functional analyses, MADS-box transcription factors, Medicago truncatula, molecular evolution, PISTILLATA-like.
Abstract
PISTILLATA (PI) is a member of the B-function MADS-box gene family, which controls the identity of both petals and stamens in Arabidopsis thaliana. In Medicago truncatula (Mt), there are two PI-like paralogs, known as MtPI and MtNGL9. These genes differ in their expression patterns, but it is not known whether their functions have also diverged. Describing the evolution of certain duplicated genes, such as transcription factors, remains a challenge owing to the complex expression patterns and functional divergence between the gene copies. Here, we report a number of functional studies, including analyses of gene expression, protein–protein interactions, and reverse genetic approaches designed to demonstrate the respective contributions of each M. truncatula PI-like paralog to the B-function in this species. Also, we have integrated molecular evolution approaches to determine the mode of evolution of Mt PI-like genes after duplication. Our results demonstrate that MtPI functions as a master regulator of B-function in M. truncatula, maintaining the overall ancestral function, while MtNGL9 does not seem to have a role in this regard, suggesting that the pseudogenization could be the functional evolutionary fate for this gene. However, we provide evidence that purifying selection is the primary evolutionary force acting on this paralog, pinpointing the conservation of its biochemical function and, alternatively, the acquisition of a new role for this gene.
Introduction
Transcription factors (TFs) involved in developmental innovations are a paradigmatic case of evolution by gene duplication, with most TF duplications being concomitant with the emergence of major morphological innovations in angiosperms (Purugganan et al., 1995; Kramer et al., 1998; Aoki et al., 2004; Kim et al., 2004). A particularly attractive model system suited to address the mechanisms of gene diversification in plant evolution is provided by the large family of MADS-box TFs (Theissen et al., 2000; Airoldi and Davies, 2012).
Flower development is controlled by complex network interactions between TFs, most of them belonging to the MADS-box gene family (Schwarz-Sommer et al., 1990; Wellmer et al., 2014). These proteins form complexes that often interact with their own promoters and those of their targets to regulate their own and each other’s expression. They contain an N-terminal DNA-binding MADS domain, followed by the intervening (I) and keratin-like (K) regions, the latter being essential for dimerization and formation of high-order complexes (Yang et al., 2003). Moreover, the highly variable C-terminal domain may have a role in protein complex formation and transcriptional regulation (reviewed by Kaufmann et al., 2005).
B-function MADS-box genes are involved in the specification of petal and stamen identity in probably all angiosperms (Litt and Kramer, 2010). In Arabidopsis thaliana, this function is encoded by the combined activities of PISTILLATA (PI) and APETALA3 (AP3) (Bowman et al., 1989, 1991; Jack et al., 1992). PI and AP3 represent lineages that arose from a duplication event after the split between extant gymnosperms and angiosperms. Although several duplication events of the PI lineage have been documented throughout angiosperm evolution (Kramer et al., 1998, 2003; Kramer and Irish, 2000; Kim et al., 2004; Stellari et al., 2004; Viaene et al., 2009), few studies are available that combine functional and molecular analyses to understand the evolutionary forces that act on these duplicate gene lineages and the outcomes that ensure (Des Marais and Rausher, 2008).
Classic theory predicts that the genetic redundancy emerging from the duplication of genes relaxes the selection constraints on one gene copy allowing it to explore the genotype space and probe a wide range of phenotypes (Payne and Wagner, 2014). The evolutionary instability of genetic redundancy guarantees, however, the return of most duplicated genes to single-copy genes (Wolfe and Shields, 1997), which undermines the importance of the role of gene duplication in biological innovation. In contrast to this prediction, up to 30% of the genes in some organisms are duplicates (Blanc and Wolfe, 2004; Cui et al., 2006), sparking the idea that a number of factors allow duplicated genes to persist, and thus innovate, within the genomes. Determining these factors remains a major goal of evolutionary biology. Expression and functional diversification of TFs are essential to allow the persistence in duplicate (Singh and Hannenhalli, 2008). The link between gene expression and the rate of evolution (Drummond et al., 2005; Gout et al., 2010), however plausible in TFs, remains a major unknown in the evolution of TFs after duplication.
Models have been proposed to explain how selection operates on duplicated genes (Innan and Kondrashov, 2010). Studies have identified many factors influencing the functional fates of duplicates, including selection for increased dosage (Conant and Wolfe, 2008), stoichiometric and gene balance (Birchler et al., 2005; Freeling and Thomas, 2006), the position of genes in the protein interaction network (Alvarez-Ponce and Fares, 2012), and whether duplicates are the result of whole-genome, small-scale duplications or retropositions (Carretero-Paulet and Fares, 2012; Fares et al., 2013; Keane et al., 2014).
Arabidopsis homeotic floral genes are conserved in model legumes (Hecht et al., 2005). Medicago truncatula contains four B-function MADS-box genes: two AP3-like (MtNMH7 and MtTM6) (Roque et al., 2013) and two PI-like paralogs (MtPI and MtNGL9) (Benlloch et al., 2009), thus representing a good model system to study the effects of gene duplication and functional divergence within the B-function MADS-box lineages. Our previous functional characterization of the Mt AP3-like genes revealed that these paralogs have undergone a qualitative subfunctionalization process, concomitant with a complete partitioning of the expression pattern of the ancestral gene lineage (Roque et al., 2013). Previous functional characterization of MtPI, one member of the duplicated PI-like genes, suggested that MtPI plays a major role in the specification of petal and stamen identity in M. truncatula (Benlloch et al., 2009). However, the contribution of MtNGL9 to floral development was unclear and its functional fate after gene duplication remains unknown. Here, we have integrated both functional analyses and molecular evolution studies to assess comprehensively the divergence in the B-function of duplicated M. truncatula PI-like genes and determine their mode of evolution after duplication.
Materials and methods
Plant material and growth conditions
Medicago truncatula cv. Jemalong lines R108 and A17, Medicago sativa cv. Regen SY-27, Pisum sativum cv. Alaska plants, and Arabidopsis thaliana cv. Landsber erecta (Ler) plants were used in this study. Leguminous plants were grown in the greenhouse at 22 °C (day) and 18 °C (night) with a 16h light/8h dark photoperiod, in a mixture of soil/sand (3:1). Arabidopsis plants were grown in growth chambers at 21 °C under long-day (16h light) conditions, in a mixture of 1:1 perlite:vermiculite. Plants were irrigated with Hoagland No. 1 solution supplemented with oligoelements (Hewitt, 1966). The mtpi-2 mutant allele was isolated in a previous screening (Cheng et al., 2014).
Isolation and sequence analysis of PsNGL9 and MsPI
PsNGL9 and MsPI coding regions were isolated to use their sequences in the molecular evolution approaches. PsNGL9 (KJ470632) was obtained by reverse transcription–PCR (RT–PCR) from Pisum sativum floral cDNA samples using the primers MtNGL9-ATG/MtNGL9-638 (see Supplementary Table S4 at JXB online). MsPI (KJ470631) was obtained by RT–PCR from M. sativa RSY-27 floral cDNA samples using primers MtPI-ATG/MtPI-548 (see Supplementary Table S4). The MsPI 3'-untranslated region (UTR) and C-terminal region were obtained using the 3' rapid amplification of cDNA ends (RACE) system (Invitrogen) with nested gene-specific primer MsPI-363 (Supplementary Table S4). Sequence alignment and similarity comparisons of the coding region and inferred proteins were performed using CLUSTALW. Sequence alignments and similarity comparisons of several PI-like proteins were performed using the UNIPROT website (http://www.uniprot.org). MADS-domains (positions 1–61) were defined by the Prosite database (http://prosite.expasy.org/) using the AtPI protein sequence as the query.
Phylogenetic tree
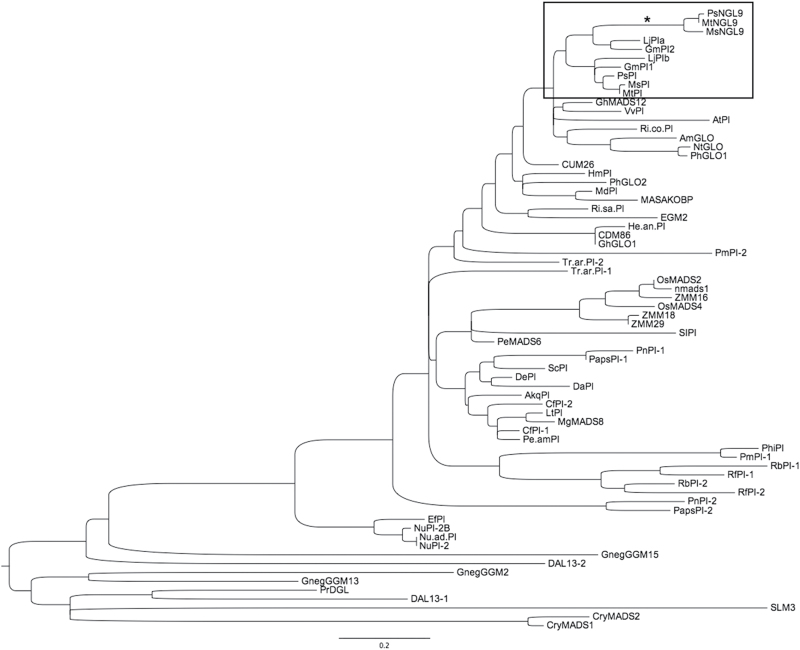
The evolutionary history was inferred by using the maximum likelihood method based on the JTT matrix-based model (Jones et al., 1992). The tree with the highest log-likelihood value (–5459.065) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Joining and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model, and then selecting the topology with the superior log-likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories +G, parameter=1.447). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 0.895% sites). The tree is drawn to scale, with branch lengths measured as the number of substitutions per site. The analysis involved 70 amino acid sequences obtained from GenBank, and two PI-like sequences that we have isolated from M. sativa and P. sativum (Supplementary Table S1). All positions containing gaps and missing data were eliminated. There were a total of 91 positions in the final data set. The substitution rates of the PI-like genes compared among different paralogs were inferred from the phylogenetic tree using the relative rate test implemented in MEGA 5 (Tamura et al., 2011).
RNA in situ hybridization
RNA in situ hybridization with digoxigenin-labeled probes was performed on 8 µm longitudinal paraffin sections of M. truncatula inflorescences as described previously (Ferrandiz et al., 2000). A 298bp fragment of MtPI (504–801 from ATG) and a 243bp fragment of MtNGL9 (511–753 from ATG) were introduced into the pGEM-T Easy vector. Digoxigenin-labeled RNA antisense and sense probes were synthesized by in vitro transcription using T7 and SP6 RNA polymerases, respectively. Signal was detected as a purple precipitate when viewed under the light microscope.
Southern blot hybridization
Plant genomic DNA was extracted from leaves of M. truncatula cv. Jemalong, line A17 using standard procedures. A 10 μg aliquot of DNA was digested with EcoRI, BamHI, and HindIII, and separated on a 0.7% agarose gel overnight. Southern blot hybridization was performed by the standard method using two different conditions (52 ºC and 65 ºC). cDNA probes were isolated by PCR using the primer pairs MtNGL9-511/MtNGL9-753 and MtPI-504/MtPI-801 for the MtNGL9 and MtPI genes, respectively (see Supplementary Table S4).
Yeast two-hybrid analysis
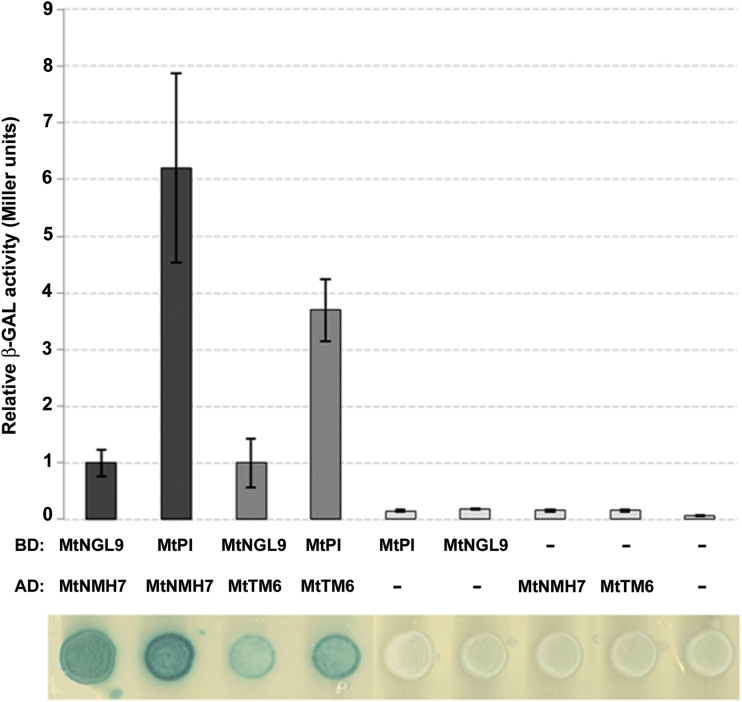
For construction of the two-hybrid plasmids, the cDNAs of the entire coding region of the MtPI and MtNGL9 genes were subcloned into the pBTM116 two-hybrid vector (Vojtek et al., 1993) to generate in-frame fusions with the LexA DNA-binding domain (BD), and the IKC fragments of MtNMH7 and MtTM6 genes were subcloned in pACT2 (Clontech) to generate in-frame fusions with the Gal4 transcriptional activation domain (AD). BamHI/SalI sites in the case of the pBTM116 constructs and BamHI/EcoRI or BamHI/XhoI sites in the case of the pACT2 constructs were added to primers used in the PCRs (see Supplementary Table S4). The yeast two-hybrid strain CTY10-5d was co-transformed with the appropriate constructs and transformants were selected on minimal medium (SD-Leu-Trp). Selection for interaction was performed on minimal medium (SD-Leu-Trp) containing X-Gal (80mg l–1) and 1× BU salts [Na2HPO4·7H2O (7g l–1); NaH2PO4 (3g l–1)]. Growth of yeast for blue staining was scored after 60h of incubation at 30 ºC (Fig. 4, bottom). Equivalent expression levels of MtPI and MtNGL9 LexA fusions were confirmed by western blot. In order to be able to quantify protein–protein interactions, we used a β-gal liquid assay with orthonitrophenyl-β-d-galactopyranoside (ONPG) as the substrate, measuring absorbance at 405nm. β-Gal activity was calculated essentially as described (Ludin et al., 1998).
Fig. 4.
Quantification of the interaction between the M. truncatula B-function MADS-box proteins. BD, LexA-binding domain; AD, GAL4 activation domain. The β-GAL activity value of MtNGL9/MtNMH7 and MtNGL9/MtTM6 interactions was set to 1.00, and the MtPI/MtNMH7 and MtPI/MtTM6 interactions were plotted relative to these values, respectively. The values were calculated using 30 samples which correspond to 10 biological replicates. (This figure is available in colour at JXB online.)
Molecular characterization of the Tnt1 insertion mutants mtngl9-1 and mtngl9-2
The M. truncatula population used for the screening of mutants has been described in detail (Roque et al., 2013; Cheng et al., 2014; Serwatowska et al., 2014). The mtngl9 alleles were identified by PCR screening of a segregating population of ~10 000 independent lines, using primers annealing to the MtNGL9 sequence (NGL9-F, Supplementary Table S4) in combination with primers annealing to the LTR borders of the Tnt1 retroelement (Tnt1-R; Supplementary Table S4). PCR products (see Supplementary Fig. S3B) were obtained and cloned into the pGEM-T easy vector for sequencing. The Tnt1 insertion in mtngl9-1 is located at 360bp from the start codon, in the second exon (Supplementary Fig. S2A) and the Tnt1 insertion in mtngl9-2 is located at 183bp from the start codon, at the end of the first exon (Supplementary Fig. S2A). The R1 plants were genotyped using the following primer pairs: NGL9-F/Tnt1-R which amplified the T-DNA insertion, and NGL9-F/NGL9-590G or NGL9-F/NGL9-612G for the wild-type fragment in mtngl9-1 and mtngl9-2, respectively (Supplementary Table S4; Supplementary Fig. S2B, C). Heterozygous lines NF14948.1 and NF14948.2 were self-pollinated. Approximately a quarter of the resultant progeny co-segregated with the Tnt1 insertion. Gene expression of MtNGL9 in the homozygous Tnt1 mutants was performed by RT–PCR (Supplementary Table S4; Supplementary Fig. S2D).
Arabidopsis thaliana transformation and genotyping
MtPI and MtNGL9 cDNAs were cloned into the SalI/BamHI sites of the pBINJIT60 vector (Guerineau and Mullineaux, 1993), a pBIN19 derivate (Clontech Laboratories). We used the primers MtPI-SalI/MtPI-BamHI for MtPI cloning, and MtNGL9-SalI/MtNGL9-BamHI for MtNGL9 cloning (Supplementary Table S4). The transcription of MtPI and MtNGL9 is under the control of a tandem repeat of the 35S promoter of Cauliflower mosaic virus. This vector was inserted into the Agrobacterium tumefaciens strain GV3101::pMP90(RK) and Arabidopsis plants were transformed according to standard procedures (Bechtold and Pelletier, 1998). For each construct, kanamycin-resistant lines were used for phenotypic and molecular characterization. Heterozygous transgenic lines 35S::MtPI (4) and 35S::MtNGL9 (22) were used as the pollen donor for crosses to the homozygous mutant pi-1 (Bowman et al., 1989). The resulting progeny were allowed to self-fertilize, and plants containing the transgenes and homozygous for the pi-1 allele (pi-1/pi-1;35S::MtPI or pi-1/pi-1;35S::MtNGL9) were identified in the next generation. To genotype the pi-1 mutation, we designed a CAPS (cleaved amplified polymorphic sequences) marker using the primers CAPS-FOR/CAPS-REV (Supplementary Table S4) that amplify 729bp and the BseGI enzyme thats cut only the wild-type PI sequence (Supplementary Fig. S6).
RT–PCR analysis
Total RNA was isolated from floral apices from wild-type M. truncatula R108, M. sativa cv. RSY-27, and P. sativum cv. Alaska, and from mtngl9 and mtpi-2 mutant flowers. Also, we isolated total RNA from leaves or floral buds of 35S::MtPI, 35S::MtNGL9, and pi-1/pi-1;35S::MtPI/MtNGL9, and Arabidopsis (Ler) plants. We used the RNeasy Plant mini Kit (Qiagen) according to the manufacturer’s instructions. Total RNA was treated with rDNaseI of the DNase Treatment and Removal Kit (Ambion). For first-strand synthesis, total RNA (1 µg) was reverse transcribed in a 20 µl reaction mixture using the PrimerScript 1st strand cDNA Synthesis Kit (Takara). Aliquots of each cDNA were used as a template for real-time PCR or semi-quantitative RT–PCR with gene-specific primers (Supplementary Table S4). For real-time RT–PCR analysis, 1 μl of the reverse transcription reaction was used with 300nM of each primer mixed with the Power SYBR® Green PCR Master Mix (Applied Biosystems) according to the manufacturer’s instructions. The reaction was carried out in 96-well optical reaction plates using an ABI PRISM 7500 Sequence Detection System and appropriate software (Applied Biosystems). The relative levels were determined by the 2–ΔΔCt method. In all cases, the efficiency of the primers was analyzed. Each experiment was done with two biological replicates, each one with three technical replicates. To normalize the variance among samples, we used Secret Agent (O-linked N-acetyl glucosamine transferase: TC77416) (Hartweck et al., 2002) for M. truncatula, PsActin-11 (Zhang et al., 2013) for P. sativum samples, elongation factor MsEF-1 for M. sativa, and Ubiquitin10 (UBQ10, At4g05320) (Czechowski et al., 2005) for A. thaliana. All used primers are listed in Supplementary Table S4.
Light microscopy and cryo-SEM
Images of M. truncatula R108, the mtpi-2 and mtngl9 mutant, Arabidopsis (Ler) wild type, 35S::MtPI, 35S::MtNGL9, and the pi-1 complementation lines flowers were obtained as described previously (Roque et al., 2007). For cryo-scanning electron microscopy (SEM), samples were frozen in slush nitrogen and attached to the specimen holder of a CT-1000C cryo-transfer system interfaced with a JEOL JSM-5410 scanning electron microscope. The samples were then transferred from the cryostage to the microscope sample stage, where the condensed surface water was sublimed by controlled warming to –85 ºC. Afterwards, the sample was transferred back to the cryostage for gold coating by sputtering. Finally, the sample was returned to the microscope sample stage and viewed at an accelerating voltage of 15 keV.
Analysis of adaptive evolution
We tested whether MtPI and MtNGL9 gene copies diverged through adaptive changes in their protein-coding gene sequences after the duplication of their ancestral gene. To this end, we ran four maximum likelihood-based models, all of which are implemented in the program CODEML from the package PAML version 4.7 (Yang, 2007). These models are based on the calculation of the log-likelihood value of a particular model that supports a given hypothesis and that allows determination of the strength of selection on the gene by estimating the non-synonymous to synonymous rates ratio (ω=dN/dS). Values of ω=1, ω>1, and ω<1 indicate neutral evolution, positive selection, and purifying selection, respectively. For each of the models, we calculate a likelihood value and then compare the log-likelihood values among models by the likelihood ratio test (LRT), such that twice the difference in the likelihood values between two models under comparison can be approached to a χ2 distribution, with the degrees of freedom (df) being the number of parameters estimated by the most complex model and which are not estimated in the simplest model. We calculated the likelihood value for four models. The Goldman and Young model (Goldman and Young, 1994) assumes a single ω value for the entire tree and alignment. The free-ratio model (FRM) assumes a single ω for the alignment, but allows this value to be inferred for each branch of the tree, so that branches with adaptive evolution (ω>1) can be identified using this model. The branch site model (BSM) allows for ω values to change among tree branches and codon sites, and this model is compared with its null model in which ω is forced to be ω=1. The model that better fits the data and phylogeny is identified comparing the Goldman and Young model with the FRM, and the BSM with its null model.
Analysis of functional divergence
We used two methods to identify functional divergence, one based on a maximum likelihood framework (Gu, 1999) and another based on a non-parametric approach (Caffrey et al., 2012). The maximum likelihood approach tests for significant changes in the rates of evolution after gene duplication (Gu, 1999; Wang and Gu, 2001). To do so, this approach calculates the log-likelihood value under two hypotheses, one in which the parameter of functional divergence is larger than 0 (H1: θ > 0) and one with θ = 0 (H0). The log-likelihood values for both hypotheses are compared through an LRT (LRT = 2Δl), with twice the difference in the log-likelihood values being approximated to a χ2 distribution with 1 df. If the null hypothesis is rejected, and thus we demonstrated the existence of functional divergence signatures at the sequence level, the posterior Bayesian probabilities at each of the amino acid sites in the alignment is calculated and those sites with probabilities >0.75 are assumed to have undergone changes responsible of the functional divergence between the two clades of paralogs. We tested two types of functional divergence, Type I and Type II. Type I functional divergence is given when at a particular amino acid site, the rate of evolution of one paralog is very slow while that of its sister clade is fast, indicating stronger functional constraints at that site in one paralog compared with the other, or a shift in the rates of evolution between the two paralogs (Gu, 1999). Alternatively, type II functional divergence measures a significant change in the amino acid properties at a site in the alignment between the two paralogous clades but with that site evolving at low rates in both of the paralogs (Gu, 2006). We used the software Diverge version 3.0 (Gu et al., 2013) to test for functional divergence Types I and II. The non-parametric approach of Caffrey et al. (2012) uses a similar method to that of Gu but without specifying the phylogenetic clades under test, and thus allows identification of very strong signatures of functional divergence in particular branches of the phylogenetic tree. For each of the branches, the method estimates the parameter of functional divergence (FD) and tests whether this parameter is larger than that estimated from 1000 alignments that are simulated using the sequence and phylogenetic parameters of the real alignment.
Results
Expression pattern of Mt PI-like paralogs during floral development
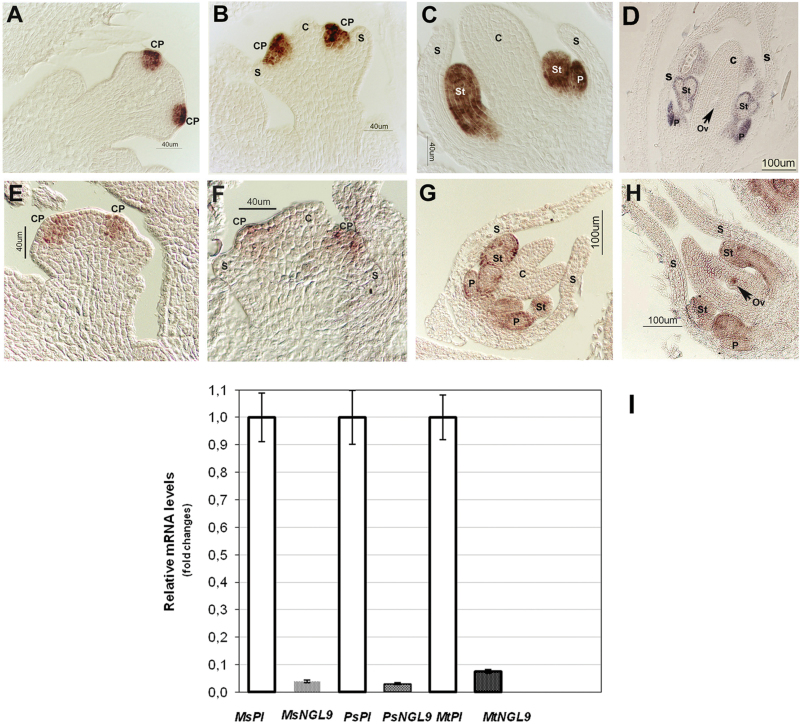
We previously isolated two M. truncatula B-function MADS-box genes (Benlloch et al., 2009) belonging to the PI/GLO subfamily (MtPI and MtNGL9; Fig. 1), which are present in the Medicago genome as single copies (Supplementary Fig. S1). MtPI expression was detected at high levels in floral buds, whereas MtNGL9 expression was very low (Fig. 2I). Similar differences in expression levels between the two PI paralogs were also observed in P. sativum and M. sativa floral buds (Fig. 2I). To determine if the Mt PI-like paralogs underwent spatial expression divergence during floral development, we compared the expression pattern of the two MtPI paralogs using in situ hybridization (Fig. 2A). MtNGL9 mRNA was first detected in floral meristems at about stage 2, in the cells that give rise to common petal–stamen primordia (Benlloch et al., 2003) (Fig. 2E). At stage 4, MtNGL9 expression was observed only in the developing common primordia (Fig. 2F). MtNGL9 expression was maintained during the entire process of common primordia compartmentalization and during the development of petals and stamens. (Fig. 2G, H). At late stages, MtNGL9 mRNA expression was also detected in ovules (Fig. 2H). MtPI and MtNGL9 had similar temporal and spatial expression patterns in petals and stamen during floral development (Fig. 2). While MtPI showed a higher uniformly distributed expression in petals and stamens during development, the MtNGL9 signal seemed to be mainly confined to their epidermal cells in late developmental stages (Fig. 2G, H). Moreover, MtNGL9 has a differential expression in ovules.
Fig. 1.
Phylogenetic tree of PI-like MADS-box genes. Sequences used in this analysis are listed in Supplementary Table S1. An accelerated branch incident on one of the Medicago truncatula PI-like gene paralogs is identified with an asterisk.
Fig. 2.
Expression patterns of the M. truncatula PI-like genes. In situ hybridization of the MtPI and MtNGL9 mRNAs in M. truncatula wild-type flower buds. (A, B) MtPI expression was detected in cells of the common primordia (CP). (C, D) At late stages, MtPI was strongly expressed in the differentiated petals (P) and stamens (St). (E, F) MtNGL9 was weakly expressed at stages 2 and 4 in cells of the common primordia. (G) At stage 5, MtNGL9 mRNA was weakly detected in the cells of differentiated petals and stamens. (H) At stage 7, MtNGL9 was also detected in ovules (Ov). (I) qRT–PCR analysis of MtPI and MtNGL9 mRNA levels in floral buds of three leguminous species. The expression value of MsPI, PsPI, and MtPI genes was set to 1.00 and the expression levels of MsNGL9, PsNGL9, and MtNGL9 were plotted relative to their respective PI values. (This figure is available in colour at JXB online.)
Loss-of-function analyses of MtNGL9 and MtPI
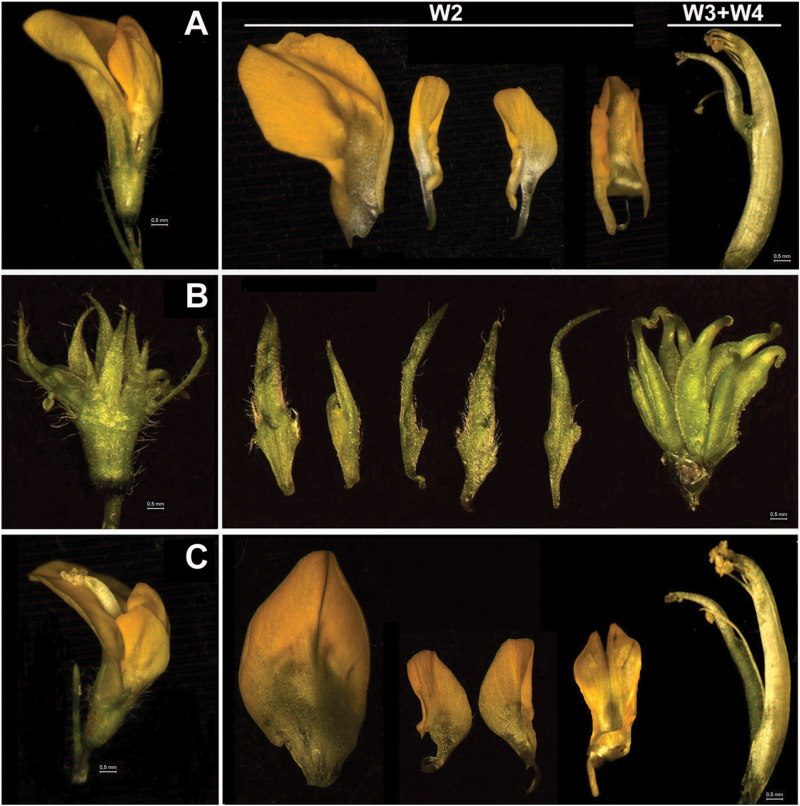
To investigate the specific contribution of the MtNGL9 gene in M. truncatula floral development, we looked for retrotransposon insertion mutants (Cheng et al., 2014). mtngl9 mutants were isolated in a reverse genetics approach (see the Materials and methods). mtngl9-1 contains a Tnt1 insertion in the coding sequence 360bp from the start codon, in the second exon (Supplementary Fig. S2A) and mtngl9-2 contains a Tnt1 insertion in the coding sequence 183bp from the start codon, at the end of the first exon (Supplementary Fig. S2A). Homozygous plants were genotyped using specific primers for both insertion lines (Supplementary Fig. S2B, C). Gene expression analysis indicated that no MtNGL9 transcript was detected in these plants (Supplementary Fig. S2D). We did not observe homeotic changes or any obvious mutant phenotype in floral organs (Fig. 3C). In contrast, mtpi-2 plants (Cheng et al., 2014) exhibited a complete conversion of petals to sepals and stamens to carpels (Fig. 3B). This phenotype resembles those of MtPI-RNAi flowers (Benlloch et al., 2009). Knockdown of MtPI resulted in a marked reduction in the expression of all B-function MADS box genes (Supplementary Fig. S3A), while the loss of MtNGL9 caused a slight decrease in their expression (Supplementary Fig. S3B). The full loss-of-B-function phenotype in mtpi-2 and the pronounced effects of MtPI absence on the expression of the other B-function genes confirms MtPI as a master regulator in establishing the regulatory pathways for petal and stamen identity, while MtNGL9 does not seem to have a role in this regard. Taken together, these results suggest that pseudogenization might be the functional evolutionary fate for this gene.
Fig. 3.
Phenotypes of mtpi-2 and mtngl9 mutants. (A) Wild-type Medicago truncatula flower. Petals present in the second whorl (W2), and stamens and carpel in W3+W4, respectively. (B) mtpi-2 flower showing full conversion of petals into sepals and of stamens into carpels. (C) mtngl9 flowers do not show homeotic floral alterations.
Evolutionary analyses
To characterize the evolutionary forces acting on MtPI-like duplicated genes, we performed molecular evolution studies. Such studies rest on the assumption that functional genes are constrained by natural selection, while pseudogenes evolve under relaxed selective constraints (Petrov and Hartl, 2000; Zheng et al., 2007; Tutar, 2012).
The phylogeny of PI-like genes reveals different rates of evolution between MtPI and MtNGL9 from different species (Fig. 1), with the lineage leading to the MtNGL9 paralog (Fig.1, asterisk) being significantly longer (i.e. has a greater rate of amino acid evolution) than that leading to MtPI (Tajima’s relative rate test: χ2=5.45, df=1, P=0.019). This accelerated evolution in the branch leading to MtNGL9 could be the result of relaxed selective constraints or be indicative of the positive selection of adaptive amino acid substitutions in that lineage.
To examine these two possibilities, we tested the action of positive selection in the MtNGL9 lineage using the BSM as implemented in the program Codeml from the PAML package version 4.7 (Yang, 2007). Briefly, we compared the log-likelihood values for two hypotheses. The simplest hypothesis (i.e. null model or null hypothesis) assumes neutral evolution, hence relaxed constraints, in the branch of interest (the non-synonymous to synonymous rates ratio; ω=dN/dS=1). The alternative hypothesis, on the other hand, allows variable strengths of selection (ω) among codons in the lineage of interest (see the Material and methods). The value of ω is a good indicator of the strength of selection, with ω=1, ω<1, and ω>1 indicating neutral evolution, purifying selection, and positive selection, respectively. The model testing for adaptive evolution (BSM) detected several amino acid sites in the MADS-box domain under positive selection (ω 3.71; Supplementary Table S2), although this model did not improve the log-likelihood value of the null model [likelihood ratio test: simple model (BSMN) 0= –10837.05, BSM 1= –10836.37; χ2=1.36, 0.10<P<0.25). We also applied a model (the FRM) in which each branch of the tree was assumed to have a different ω value. This model showed, however, no evidence of positive selection in the branch leading to MtNGL9. The mean ω value for the entire tree and alignment, as estimated using the Goldman and Yang model (Goldman and Yang, 1994), was 0.14, indicative of strong purifying selection in this gene along the phylogenetic tree. The MtNGL9 lineage showed an ω=0.56, supporting relaxed selective constraints, but no neutral evolution, in this branch. While these relaxed constraints could be indicative of a process of non-functionalization, the strong conserved amino acid nature of important functional sites in this gene and the fact that this value is lower than 1 indicate that this gene copy has evolved under strong purifying selection. Overall, these analyses discard pseudogenization (i.e. non-functionalization) as a plausible fate for the gene copy exhibiting an accelerated rate of evolution.
We also tested for amino acid substitutions that may have led to functional changes between the two genes generated by gene duplication. To this end, we examined the substitution patterns in each amino acid site of the multiple sequence alignment containing these genes. Sites that are functionally important but that have diverged after gene duplication should be highly conserved within each gene clade that includes ortholog genes from other species but variable between the paralogs. Inspection of these patterns identifies a number of sites that present a conserved amino acid in one gene copy which is different from its sister gene copy.
No signatures of functional divergence between MtNGL9 and MtPI
To test whether functional diversification events have followed the duplication giving rise to these two Mt PI-like gene copies, we searched for signatures of functional divergence in the amino acid sequences encoded by MtNGL9 and MtPI. We used two tests of functional divergence: (i) a maximum-likelihood approach (see the Materials and methods) (Gu, 1999); and (ii) a non-parametric probabilistic approach (Caffrey et al., 2012) that tests for functional divergence across the phylogenetic tree.
The maximum-likelihood test showed no evidence for either functional divergence Type I (parameter of functional divergence θ=0.087, P>0.1) or Type II (θ=0.016, P>0.1). Likewise, the non-parametric approach found no evidence of functional divergence within these groups (FD=0.01, P>0.25). Therefore, although some amino acid sites tended to be more conserved within paralogs than between them, formal statistical tests (Gu, 2003; Caffrey et al., 2012; Gu et al., 2013) could not find significant evidence supporting this observation.
Interaction strengths between Medicago truncatula PI-like proteins
It has been shown that Arabidopsis PI and AP3 proteins act jointly as heterodimers to perform the B-function (Goto and Meyerowitz, 1994; Jack et al., 1994; Riechmann et al., 1996; Krizek and Meyerowitz, 1996; McGonigle et al., 1996; Yang et al., 2003). The heterodimer formed by PI- and AP3-like proteins is the only functional DNA-binding dimer in the vast majority of angiosperms (Melzer et al., 2014).
To evaluate the effect of relaxed functional constraints on the MtNGL9 function, we carried out yeast two-hybrid assays on pairwise combinations of B-class MADS-box proteins to determine if the dimerization capabilities were conserved in Mt PI-like gene products. Our results showed that both MtPI and MtNGL9 were able to interact with the Mt AP3-like proteins MtNMH7 and MtTM6 (Fig. 4). Also, we analyzed whether differences exist in the strengths with which PI-type MADS-box proteins interact. We have quantified the interaction of Mt PI-like proteins with MtNMH7 and MtTM6 by β-galactosidase activity assays. The MtPI/MtNMH7 and MtPI/MtTM6 pairs produced stronger interactions as compared with the MtNGL9/MtNMH7 and MtNGL9/MtTM6 pairs (Fig. 4).
Constitutive expression of MtPI and MtNGL9 in Arabidopsis
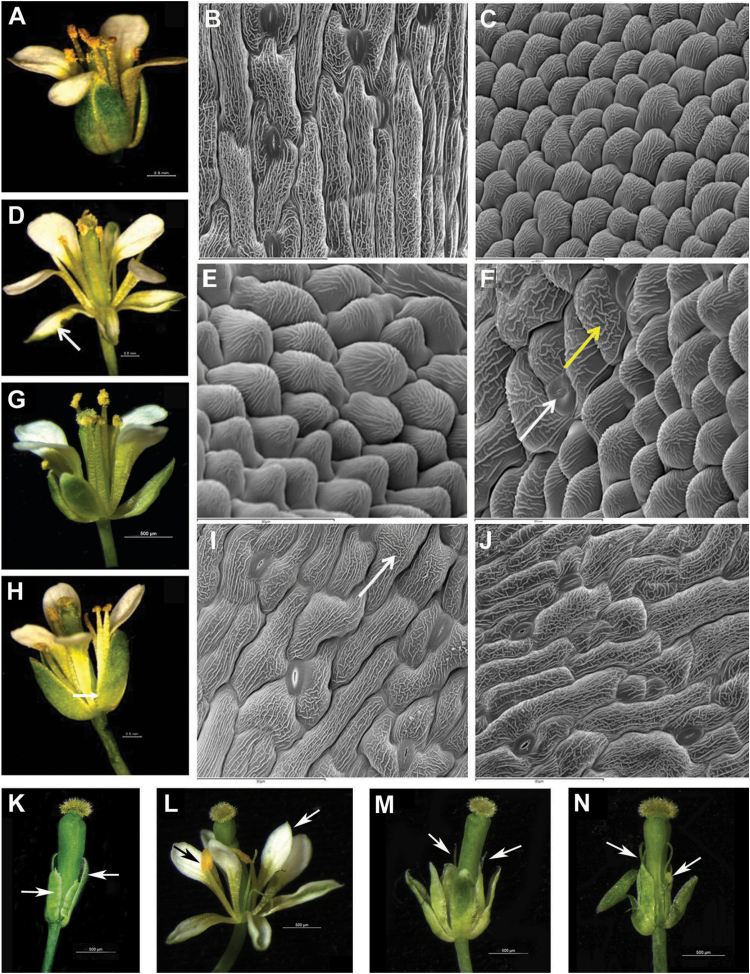
To investigate the ability of MtPI and MtNGL9 proteins to induce B-function identity, we overexpressed these two genes in Arabidopsis plants. Constitutive expression of MtPI in Arabidopsis leads to homeotic conversion of sepals into petals (Fig. 5D). This phenotype is characteristic of the constitutive expression of most PI-like genes in Arabidopsis (Krizek and Meyerowitz, 1996; Berbel et al., 2005). The majority of 35S::MtPI (64.7%) plants displayed very open sepals transformed into petaloid organs (strong phenotype) (Fig. 5D). At the cellular level, these organs contained petal-like cells rounded and cobblestone-like in appearance with prominent cuticular thickenings (Fig. 5E). Occasionally, petal-like cells co-existed with cells retaining the typical pattern of the sepal cells, showing stomata (Fig. 5F, white arrow) and typical cuticular striation (Fig. 5F, yellow arrow). The remaining 35S::MtPI plants displayed flowers with partially opened sepals which occasionally showed a slight white color (medium phenotype) (Fig. 5G). In fact, the abaxial epidermal cells of the first whorl showed some cells with a small protrusion, which is reminiscent of cells typically found in petals (Fig. 5I; arrow). Most of the plants that overexpressed MtNGL9 exhibited the medium phenotype (81.8%) described above (Supplementary Fig. S5C). The remaining 35S::MtNGL9 plants showed flowers with slightly opened sepals (weak phenotype; Fig. 5H) containing typical sepal-like cells (Fig. 5I).
Fig. 5.
Constitutive expression of MtPI and MtNGL9 in Arabidopsis and rescue of the pi-1 mutant floral phenotype. (A) Wild-type flower (Ler). (B) Abaxial epidermal cells of a wild-type sepal. (C) Abaxial epidermal cells of a wild-type petal. (D). Flower representative of the strong phenotype showing homeotic conversions of sepals into petals (arrow). (E, F) Abaxial epidermal cells of the first whorl organs of the 35S::MtPI line displaying the strong phenotype. (G) Flower representative of the medium phenotype. (H) Flower representative of the weak phenotype. Arrow: slightly opened sepals. (I) Abaxial epidermal cells of the first whorl organs of a medium 35S::MtNGL9 transgenic line. (J) Abaxial epidermal cells of the first whorl organs of a weak 35S::MtNGL9 plant. (K) pi-1 mutant flower. (L) Flowers from 35S::MtPI;pi-1 lines showing a near complete rescue of petal (white arrow) and stamen organs (black arrow). (M, N) 35S::MtNGL9;pi-1 flowers showing a weak rescue of petal and stamens (arrows).
We examined the expression levels of the MtPI and MtNGL9 transgenes in their respective overexpression lines by qRT–PCR (Supplementary Fig. S5A, B). Despite the high expression levels of MtNGL9 detected in 35S::MtNGL9 lines, neither of these lines displayed the strong phenotype (Fig. 5G–J). We were able to observe differences in the relative effects of both constructs, therefore indicating that the MtPI protein was more active than MtNGL9.
To determine if Mt PI-like genes could rescue petal and stamen organ identity in the absence of endogenous PI activity, we crossed heterozygous 35S::MtPI and 35S::MtNGL9 transgenic lines into a pi-1 mutant background. In the F2 population of kanamycin-resistant plants, a number of independent transformants homozygous for pi-1 were identified (Supplementary Fig. S6), showing a range of phenotypes (Fig. 5L–N; Supplementary Table S3).
Only pi-1/pi-1;35S::MtPI plants showed near complete rescue of petal and stamen organs. Petals were indistinguishable from wild-type petals, and, in the third whorl, mosaic organs generally appeared, possessing both stamen and carpel characteristics. The restored stamens were not fully extended and did not produce fertile pollen (Fig. 5L; Supplementary Table S3). Meanwhile, pi-1/pi-1;35S::MtNGL9 lines (Supplementary Table S3) showed a weak pi-1 mutant rescue. The second whorl organs were larger than the second organs displayed in the pi-1 mutant flowers with a whitish color, but they never reached the size of the wild-type petals (Fig. 5M, N). The third whorl organs were generally filament-like structures and occasionally the stamens showed anther-like organs (Fig. 5M, N). Similar levels of MtPI or MtNGL9 transgenes were detected in floral buds of pi-1/pi-1;35S::MtPI or pi-1/pi-1;35S::MtNGL9 lines, respectively (Supplementary Fig. S5), suggesting that the degree of rescue in the complementation lines is not correlated with the transgene expression level, but with the ability of MtPI or MtNGL9 to replace PI in A. thaliana.
Discussion
MtPI controls B-function in Medicago truncatula
Paralogous Mt PI-like genes arose from a duplication event that occurred prior to the speciation of legumes (Fig. 1). They may have originated during the whole-genome duplication (WGD) event that pre-dated speciation of Mt and other legumes ~50–60 Mya (Cannon et al., 2006). Two copies of both PI-like genes are also found in the legume subfamilies P. sativum, L. japonicus, M. sativa, and G. max. Our phylogenetic analyses consistently placed one copy from each species in separate gene clades (Fig. 1). Our results showed that following this duplication event, only the MtPI duplicated copy has retained the B-function, based on its robust expression levels and the conserved expression pattern of the B-class genes, protein interaction capabilities, and the strong homeotic phenotype observed for the mtpi-2 mutant. The co-ordinated expression of MtPI with MtNMH7 in the inner cell layers of the petal and stamen organ primordia and with MtTM6 in the outer primordia cell layers is required to specify petal and stamen identity in M. truncatula (Roque et al., 2013). However, MtNGL9 does not appear to have a role in this regard, so could potentially be on the way to becoming a pseudogene.
MtNGL9 does not show evidence of pseudogenization
Duplicate genes that are stably preserved in genomes usually have divergent functions (He and Zhang, 2005). Indeed, duplicated genes destined to ‘die’ usually do so within a few million years after duplication, having acquired mutations that result in a non-functional gene (Lynch and Conery, 2000). Pseudogenes are expected to evolve neutrally; therefore, after a sufficient amount of time, the signature of purifying selection at the amino acid level will ultimately be erased (Zou et al., 2009).
mtngl9 mutants do not present any homeotic change or any obvious floral mutant phenotype (Fig. 3C), suggesting that MtNGL9 does not have a role in the specification of petal and stamen fate. Our analyses discard pseudogenization as a plausible scenario for MtNGL9 evolution. First, the rate of evolution of MtNGL9 indicates strong purifying selection (i.e. dN/dS <1). Secondly, MtNGL9 has persisted for >60 million years, as the duplication of the gene pre-dates the speciation of legumes (Cannon et al., 2006). Since the half-life for a duplicated gene is in the order of 20 million years (Lynch, 2007), then we can assume that MtNGL9 has not lost its function.
In the basal eudicot Papaver somniferum, one of the Paps PI-like proteins has lost dimerization capabilities with the Paps AP3-like proteins, thus indicating that this gene could be on the way to becoming a pseudogene in this species (Drea et al., 2007). In contrast, in support of a functional role for MtNGL9, this gene encodes a protein that conserves this competence to interact with the Mt AP3-like proteins, which it is an essential feature for the B-class MADS box proteins.
Mode of Medicago truncatula PISTILLATA-like gene evolution after duplication
Studies of MIKCC-type MADS-box TFs in an array of species are contributing to a better understanding of how these genes may have changed their functions after duplication (Vandenbussche et al., 2004; Drea et al., 2007; Geuten and Irish, 2010; Pan et al., 2010; Fourquin et al., 2013; Roque et al., 2013; Serwatowska et al., 2014).
A number of theoretical models have been proposed to explain the persistence of duplicated genes in genomes. Paralogs may be selected for increased dosage or as a repository for gene conversion against deleterious changes in either copy and result in functional redundancy (Nadeau and Sankoff, 1997; Nowak et al., 1997; Gu, 2003; Gu et al., 2003). Alternatively, the paralogs may diverge either to generate new gene functions (neofunctionalization) (Taylor and Raes, 2004) or to partition multiple functions (subfunctionalization) through complementary degeneration (Force et al., 1999; Stoltzfus, 1999; Lynch and Force, 2000). These theoretical expectations are, however, only partially consistent with data (Lynch and Conery, 2000).
To describe the evolution of these duplicated genes, we have to invoke factors from different models of molecular evolution. The differential evolution of the two clades examined here is in agreement with the quantitative subfunctionalization model of Force and colleagues (Force et al., 1999): the unduplicated ancestor possessed elevated expression levels that were retained in only an Mt PI-like gene copy after the WGD pre-dating legumes speciation, the MtPI gene. Similar differences in expression levels between the two PI paralogs were also observed in P. sativum and M. sativa floral apices (Fig. 2I), suggesting ancestral changes in regulatory sequences. How can this differential expression between the two gene copies explain their evolutionary and functional patterns?
Whether gene expression can evolve independently from gene function remains an open question in evolutionary biology (Castillo-Davis et al., 2004). Many recent studies have explored molecular and population-genetic constraints on the rate of protein evolution and they have hypothesized that the best predictor of the evolutionary rates of proteins is the gene expression level (Pal et al., 2001; Herbeck et al., 2003; Rocha and Danchin, 2004; Subramanian and Kumar, 2004; Gout et al., 2010). Our results showed that the expression levels may have imposed additional constraints to the highly expressed MtPI protein. The lower expression of the MtNGL9 gene is likely to have relaxed the selective constraints on this gene, in agreement with the mistranslation model (Drummond et al., 2005), allowing it to evolve at a faster rate than MtPI. MtNGL9 has lost its ancestral expression level, and, as a consequence, its ancestral function seems to have decreased. In line with this, the constitutive expression of MtPI and MtNGL9 in Arabidopsis and the rescue of the pi-1 mutant by these genes revealed that MtPI had a greater ability than MtNGL9 to be a component of the transcriptional complex that specifies the second and the third whorl in a heterologous system.
TFs that function in the context of molecular networks are likely to be under stronger selective pressures (Luisi et al., 2015). Thus, the most parsimonious explanation for the preservation of both Mt PI-like duplicated gene copies is that both will be selected to achieve an optimal total dosage balance of their functional complex. In the case of ohnologs (duplicated genes generated by WGD), expression dosage is thought frequently to drive functional persistence, suggesting that the stoichiometry of expression among duplicated loci is important (Papp et al., 2003; Birchler and Veitia, 2007; Edger and Pires, 2009; Gout et al., 2010; Makino and McLysaght, 2012). MtNGL9 would have a far lower contribution to the total activity required to specify the B-function. The slight decrease of MtPI and the other B-class gene expression levels in mtngl9 floral buds suggests that MtNGL9 may be required to maintain the critical dosage for the B-function in M. truncatula.
On the other hand, we have detected MtNGL9 expression in ovules (Fig. 2H). The B-class gene MtNMH7 (Roque et al., 2013) and the euAP3 gene GmNMH7 in soybean (Wu et al., 2006) have also been detected in this floral tissue. Moreover, low levels of MtNGL9 transcript have been reported in vegetative tissues (Benlloch et al., 2009). The presence of transcripts in a given tissue does not necessarily mean that the gene activity is required for its proper development. However, it might be considered that changes in MtNGL9 regulation enabled its expression in different tissues and that the preservation of MtNGL9 and its partial functional relaxation subsequently provide the opportunity for the fixation of advantageous mutations, which can lead to new functions (He and Zhang, 2005). This makes neofunctionalization plausible as an alternative evolutionary fate for this gene, through the ability to form part of new complexes that control other development processes. The extent to which such divergences have led to a neofunctionalization of one of the duplicated MtPI-like copies and possible acquisition of other roles in different developmental processes requires further investigation.
Our results provide evidence of the complex dynamics underlying the evolution by gene duplication of the B-class MADS-box subfamily. Notwithstanding this complexity, the B-function remains conserved in all eudicots, providing robustness to the specification of petal and stamen identities. Finally, our findings confirm MADS-box TFs as promising targets for novel studies aimed at a more complete understanding of the complexity of evolution by gene duplication.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Southern blot analysis of the Medicago truncatula PI-like genes.
Figure S2. Molecular characterization of the Tnt1 insertions in the MtNGL9 locus.
Figure S3. qRT–PCR expression analyses of loss-of-function plants.
Figure S4. Analyses of 35S:MtPI and 35S:MtNGL9 plants.
Figure S5. RT–PCR expression analyses in pi-1/pi-1;35S:MtPI and pi-1/pi-1;35S:MtNGL9 complementation lines.
Figure S6. Analyses of the pi-1 complementation by MtPI and MtNGL9.
Table S1. Sequences from different plant species used in the elaboration of the phylogenetic tree with their respective GenBank accession numbers.
Table S2. Maximum-likelihood test of adaptive evolution in the duplicated M. truncatula PISTILATA-like transcription factors.
Table S3. Summary of rescue phenotypes.
Table S4. Primers used in this work.
Acknowledgements
This work was supported by grants from the Spanish Ministry of Economy and Competitiveness (MINECO; BIO2009-08134 and BIO2012-39849-CO2-01). MAF was supported by a grant from the MINECO (BFU2012-36346). We wish to thank Drs Santiago F. Elena Fito (IBMCP, CSIC-UPV), José V. Gimeno Alcáñiz (IATA, CSIC), Javier Paz-Ares (CNB, CSIC), and Carlos Alonso-Blanco (CNB, CSIC) for valuable suggestions and comments in the initial stages of this work. The technical assistance of Rafael Martínez-Pardo in the greenhouse is gratefully acknowledged.
References
- Airoldi CA, Davies B. 2012. Gene duplication and the evolution of plant MADS-box transcription factors. Journal of Genetics and Genomics 39, 157–165. [DOI] [PubMed] [Google Scholar]
- Alvarez-Ponce D, Fares MA. 2012. Evolutionary rate and duplicability in the Arabidopsis thaliana protein–protein interaction network. Genome Biology and Evolution 4, 1263–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki S, Uehara K, Imafuku M, Hasebe M, Ito M. 2004. Phylogeny and divergence of basal angiosperms inferred from APETALA3- and PISTILLATA-like MADS-box genes. Journal of Plant Research 117, 229–244. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. 1998. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods in Molecular Biology 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Benlloch R, Navarro C, Beltrán J, Cañas LA. 2003. Floral development of the model legume Medicago truncatula: ontogeny studies as a tool to better characterize homeotic mutations. Sexual Plant Reproduction 15, 231–241. [Google Scholar]
- Benlloch R, Roque E, Ferrandiz C, Cosson V, Caballero T, Penmetsa RV, Beltran JP, Canas LA, Ratet P, Madueno F. 2009. Analysis of B function in legumes: PISTILLATA proteins do not require the PI motif for floral organ development in Medicago truncatula . The Plant Journal 60, 102–111. [DOI] [PubMed] [Google Scholar]
- Berbel A, Navarro C, Ferrandiz C, Canas LA, Beltran JP, Madueno F. 2005. Functional conservation of PISTILLATA activity in a pea homolog lacking the PI motif. Plant Physiology 139, 174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA. 2005. Dosage balance in gene regulation: biological implications. Trends in Genetics 21, 219–226. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. 2007. The gene balance hypothesis: from classical genetics to modern genomics. The Plant Cell 19, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. 2004. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. The Plant Cell 16, 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. 1989. Genes directing flower development in Arabidopsis . The Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. 1991. Genetic interactions among floral homeotic genes of Arabidopsis . Development 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Caffrey BE, Williams TA, Jiang X, Toft C, Hokamp K, Fares MA. 2012. Proteome-wide analysis of functional divergence in bacteria: exploring a host of ecological adaptations. PLoS One 7, e35659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Sterck L, Rombauts S, et al. 2006. Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proceedings of the National Academy of Sciences, USA 103, 14959–14964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L, Fares MA. 2012. Evolutionary dynamics and functional specialization of plant paralogs formed by whole and small-scale genome duplications. Molecular Biology and Evolution 29, 3541–3551. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis CI, Hartl DL, Achaz G. 2004. Cis-Regulatory and protein evolution in orthologous and duplicate genes. Genome Research 14, 1530–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Wang M, Lee HK, Tadege M, Ratet P, Udvardi M, Mysore KS, Wen J. 2014. An efficient reverse genetics platform in the model legume Medicago truncatula . New Phytologist 201, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nature Reviews Genetics 9, 938–950. [DOI] [PubMed] [Google Scholar]
- Cui L, Wall PK, Leebens-Mack JH, et al. 2006. Widespread genome duplications throughout the history of flowering plants. Genome Research 16, 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis . Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais DL, Rausher MD. 2008. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454, 762–765. [DOI] [PubMed] [Google Scholar]
- Drea S, Hileman LC, de Martino G, Irish VF. 2007. Functional analyses of genetic pathways controlling petal specification in poppy. Development 134, 4157–4166. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. 2005. Why highly expressed proteins evolve slowly. Proceedings of the National Academy of Sciences, USA 102, 14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Pires JC. 2009. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Research 17, 699–717. [DOI] [PubMed] [Google Scholar]
- Fares MA, Keane OM, Toft C, Carretero-Paulet L, Jones GW. 2013. The roles of whole-genome and small-scale duplications in the functional specialization of Saccharomyces cerevisiae genes. PLoS Genetics 9, e1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF. 2000. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER . Development 127, 725–734. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourquin C, del Cerro C, Victoria FC, Vialette-Guiraud A, de Oliveira AC, Ferrandiz C. 2013. A change in SHATTERPROOF protein lies at the origin of a fruit morphological novelty and a new strategy for seed dispersal in Medicago genus. Plant Physiology 162, 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Thomas BC. 2006. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Research 16, 805–814. [DOI] [PubMed] [Google Scholar]
- Geuten K, Irish V. 2010. Hidden variability of floral homeotic B genes in Solanaceae provides a molecular basis for the evolution of novel functions. The Plant Cell 22, 2562–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N, Yang Z. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Molecular Biology and Evolution 11, 725–736. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. 1994. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA . Genes and Development 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Gout JF, Kahn D, Duret L, Paramecium Post-Genomics Consortium 2010. The relationship among gene expression, the evolution of gene dosage, and the rate of protein evolution. PLoS Genetics 6, e1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Zou Y, Su Z, Huang W, Zhou Z, Arendsee Z, Zeng Y. 2013. An update of DIVERGE software for functional divergence analysis of protein family. Molecular Biology and Evolution 30, 1713–1719. [DOI] [PubMed] [Google Scholar]
- Gu X. 1999. Statistical methods for testing functional divergence after gene duplication. Molecular Biology and Evolution 16, 1664–1674. [DOI] [PubMed] [Google Scholar]
- Gu X. 2003. Evolution of duplicate genes versus genetic robustness against null mutations. Trends in Genetics 19, 354–356. [DOI] [PubMed] [Google Scholar]
- Gu X. 2006. A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Molecular Biology and Evolution 23, 1937–1945. [DOI] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421, 63–66. [DOI] [PubMed] [Google Scholar]
- Guerineau F, Mulleneaux P. 1993. Plant transformation and expression vectors. In: Croy RRD, ed. Plant molecular biology Labfax. Oxford: BIOS Scientific Publishers, 121–148. [Google Scholar]
- Hartweck LM, Scott CL, Olszewski NE. 2002. Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 161, 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J. 2005. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169, 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Foucher F, Ferrandiz C, et al. 2005. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiology 137, 1420–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbeck JT, Wall DP, Wernegreen JJ. 2003. Gene expression level influences amino acid usage, but not codon usage, in the tsetse fly endosymbiont Wigglesworthia . Microbiology 149, 2585–2596. [DOI] [PubMed] [Google Scholar]
- Hewitt EJ. 1966. Sand and water culture methods used in the study of plant nutrition . Farnham Royal, UK: Commonwealth Agricultural Bureau. [Google Scholar]
- Innan H, Kondrashov F. 2010. The evolution of gene duplications: classifying and distinguishing between models. Natural Reviews Genetics 11, 97–108. [DOI] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. 1992. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS-box and is expressed in petals and stamens. Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- Jack T, Fox GL, Meyerowitz EM. 1994. Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determines floral organ identity. Cell 76, 703–716. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences 8, 275–282. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Melzer R, Theissen G. 2005. MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347, 183–198. [DOI] [PubMed] [Google Scholar]
- Keane OM, Toft C, Carretero-Paulet L, Jones GW, Fares MA. 2014. Preservation of genetic and regulatory robustness in ancient gene duplicates of Saccharomyces cerevisiae . Genome Research 24, 1830–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yoo MJ, Albert VA, Farris JS, Soltis PS, Soltis DE. 2004. Phylogeny and diversification of B-function MADS-box genes in angiosperms: evolutionary and functional implications of a 260-million-year-old duplication. American Journal of Botany 91, 2102–2118. [DOI] [PubMed] [Google Scholar]
- Kramer E, Di Stilio VS, Schlulter PM. 2003. Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. International Journal of Plant Sciences 164, 1–11. [Google Scholar]
- Kramer EM, Dorit RL, Irish VF. 1998. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149, 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Irish VF. 2000. Evolution of the petal and stamen developmental programs: evidence from comparative studies of the lower eudicots and basal angiosperms. International Journal of Plant Sciences 161, S29–S40. [Google Scholar]
- Krizek BA, Meyerowitz EM. 1996. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122, 11–22. [DOI] [PubMed] [Google Scholar]
- Litt A, Kramer EM. 2010. The ABC model and the diversification of floral organ identity. Seminars in Cell and Developmental Biology 21, 129–137. [DOI] [PubMed] [Google Scholar]
- Ludin K, Jiang R, Carlson M. 1998. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae . Proceedings of the National Academy of Sciences, USA 95, 6245–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi P, Alvarez-Ponce D, Pybus M, Fares MA, Bertranpetit J, Laayouni H. 2015. Recent positive selection has acted on genes encoding proteins with more interactions within the whole human interactome. Genome Biology and Evolution 7, 1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2007. The origins of genomic architecture . Sinauer Associates: Sunderland, MA. [Google Scholar]
- Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A. 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino T, McLysaght A. 2012. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proceedings of the National Academy of Sciences, USA 107, 9270–9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle B, Bouhidel K, Irish VF. 1996. Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes and Development 10, 1812–1821. [DOI] [PubMed] [Google Scholar]
- Melzer R, Harter A, Rumpler F, Kim S, Soltis PS, Soltis DE, Theissen G. 2014. DEF- and GLO-like proteins may have lost most of their interaction partners during angiosperm evolution. Annals of Botany 114, 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH, Sankoff D. 1997. Comparable rates of gene loss and functional divergence after genome duplications early in vertebrate evolution. Genetics 147, 1259–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, Boerlijst MC, Cooke J, Smith JM. 1997. Evolution of genetic redundancy. Nature 388, 167–171. [DOI] [PubMed] [Google Scholar]
- Pal C, Papp B, Hurst LD. 2001. Highly expressed genes in yeast evolve slowly. Genetics 158, 927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan IL, McQuinn R, Giovannoni JJ, Irish VF. 2010. Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. Journal of Experimental Botany 61, 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424, 194–197. [DOI] [PubMed] [Google Scholar]
- Payne JL, Wagner A. 2014. The robustness and evolvability of transcription factor binding sites. Science 343, 875–877. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Hartl DL. 2000. Pseudogene evolution and natural selection for a compact genome. Journal of Heredity 91, 221–227. [DOI] [PubMed] [Google Scholar]
- Purugganan MD, Rounsley SD, Schmidt RJ, Yanofsky MF. 1995. Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics 140, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Krizek BA, Meyerowitz EM. 1996. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proceedings of the National Academy of Sciences, USA 93, 4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP, Danchin A. 2004. An analysis of determinants of amino acids substitution rates in bacterial proteins. Molecular Biology and Evolution 21, 108–116. [DOI] [PubMed] [Google Scholar]
- Roque E, Gomez MD, Ellul P, Wallbraun M, Madueno F, Beltran JP, Canas LA. 2007. The PsEND1 promoter: a novel tool to produce genetically engineered male-sterile plants by early anther ablation. Plant Cell Reports 26, 313–325. [DOI] [PubMed] [Google Scholar]
- Roque E, Serwatowska J, Cruz Rochina M, Wen J, Mysore KS, Yenush L, Beltran JP, Canas LA. 2013. Functional specialization of duplicated AP3-like genes in Medicago truncatula . The Plant Journal 73, 663–675. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Huijser P, Nacken W, Saedler H, Sommer H. 1990. Genetic control of flower development by homeotic genes in Antirrhinum majus . Science 250, 931–936. [DOI] [PubMed] [Google Scholar]
- Serwatowska J, Roque E, Gomez-Mena C, Constantin GD, Wen J, Mysore KS, Lund OS, Johansen E, Beltran JP, Canas LA. 2014. Two euAGAMOUS genes control C-function in Medicago truncatula . PLoS One 9, e103770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh LN, Hannenhalli S. 2008. Functional diversification of paralogous transcription factors via divergence in DNA binding site motif and in expression. PLoS One 3, e2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellari GM, Jaramillo MA, Kramer EM. 2004. Evolution of the APETALA3 and PISTILLATA lineages of MADS-box-containing genes in the basal angiosperms. Molecular Biology and Evolution 21, 506–519. [DOI] [PubMed] [Google Scholar]
- Stoltzfus A. 1999. On the possibility of constructive neutral evolution. Journal of Molecular Evolution 49, 169–181. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. 2004. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics 168, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Raes J. 2004. Duplication and divergence: the evolution of new genes and old ideas. Annual Review of Genetics 38, 615–643. [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Munster T, Winter KU, Saedler H. 2000. A short history of MADS-box genes in plants. Plant Molecular Biology 42, 115–149. [PubMed] [Google Scholar]
- Tutar Y. 2012. Pseudogenes. Comparative and Functional Genomics 2012, 424526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M, Zethof J, Royaert S, Weterings K, Gerats T. 2004. The duplicated B-class heterodimer model: whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. The Plant Cell 16, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaene T, Vekemans D, Irish VF, Geeraerts A, Huysmans S, Janssens S, Smets E, Geuten K. 2009. Pistillata-duplications as a mode for floral diversification in (Basal) asterids. Molecular Biology and Evolution 26, 2627–2645. [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74, 205–214. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gu X. 2001. Functional divergence in the caspase gene family and altered functional constraints: statistical analysis and prediction. Genetics 158, 1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F, Bowman JL, Davies B, et al. 2014. Flower development: open questions and future directions. Methods in Molecular Biology 1110, 103–124. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708–713. [DOI] [PubMed] [Google Scholar]
- Wu C, Ma Q, Yam KM, Cheung MY, Xu Y, Han T, Lam HM, Chong K. 2006. In situ expression of the GmNMH7 gene is photoperiod-dependent in a unique soybean (Glycine max [L.] Merr.) flowering reversion system. Planta 223, 725–735. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fanning L, Jack T. 2003. The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. The Plant Journal 33, 47–59. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24, 1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang S, Cheng F, Wang C, Zhang L, An Y. 2013. Cloning and tissue-specific expression of predicted Pisum sativum actin isoform PEAc14-1. Biochemical Genetics 51, 722–727. [DOI] [PubMed] [Google Scholar]
- Zheng D, Frankish A, Baertsch R, et al. 2007. Pseudogenes in the ENCODE regions: consensus annotation, analysis of transcription, and evolution. Genome Research 17, 839–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C, Lehti-Shiu MD, Thibaud-Nissen F, Prakash T, Buell CR, Shiu SH. 2009. Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant Physiology 151, 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.