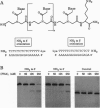
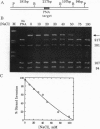
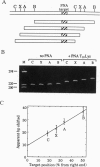
Abstract
Polyamide oligomers, termed peptide nucleic acids (PNAs), bind with high affinity to both DNA and RNA and offer both antisense and antigene approaches for regulating gene expression. When a PNA binds to a complementary sequence in a double-stranded DNA, one strand of the duplex is displaced, and a stable D-loop is formed. Unlike oligodeoxynucleotides for which binding polarity is determined by the deoxyribose sugar, the unrestrained polyamide backbone of the PNA could permit binding to a DNA target in an orientation-independent manner. We now provide evidence that PNAs can, in fact, bind to their complementary sequence in DNA independent of the DNA-strand polarity--that is, a PNA binds to DNA in both "parallel" and "antiparallel" fashion. With a mixed-sequence 15-mer PNA, kinetic studies of PNA.DNA interactions revealed that D-loop formation was rapid and the complex was stable for several hours. However, when measured either by gel-mobility-shift analysis or RNA polymerase II-elongation termination, D-loop formation was salt dependent, but PNA-strand dissociation was not salt dependent. We observed that D-loop-containing DNA fragments had anomalous gel mobilities that varied as a function of the position of the D-loop relative to the DNA termini. On the basis of permutation analysis, the decreased mobility of the PNA.DNA complex was attributed to a bend in the DNA at or near the D-loop.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhattacharyya A., Lilley D. M. The contrasting structures of mismatched DNA sequences containing looped-out bases (bulges) and multiple mismatches (bubbles). Nucleic Acids Res. 1989 Sep 12;17(17):6821–6840. doi: 10.1093/nar/17.17.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D. M., Drak J. Global features of DNA structure by comparative gel electrophoresis. Methods Enzymol. 1992;212:46–71. doi: 10.1016/0076-6879(92)12005-b. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Gartenberg M. R., Shrader T. E. DNA bending in protein-DNA complexes. Methods Enzymol. 1991;208:118–146. doi: 10.1016/0076-6879(91)08011-6. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Haran T. E., Nadeau J. G. Intrinsically bent DNA. J Biol Chem. 1990 May 5;265(13):7093–7096. [PubMed] [Google Scholar]
- Feuerstein B. G., Pattabiraman N., Marton L. J. Molecular mechanics of the interactions of spermine with DNA: DNA bending as a result of ligand binding. Nucleic Acids Res. 1990 Mar 11;18(5):1271–1282. doi: 10.1093/nar/18.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François J. C., Saison-Behmoaras T., Thuong N. T., Hélène C. Inhibition of restriction endonuclease cleavage via triple helix formation by homopyrimidine oligonucleotides. Biochemistry. 1989 Dec 12;28(25):9617–9619. doi: 10.1021/bi00451a011. [DOI] [PubMed] [Google Scholar]
- Germann M. W., Kalisch B. W., Pon R. T., van de Sande J. H. Length-dependent formation of parallel-stranded DNA in alternating AT segments. Biochemistry. 1990 Oct 9;29(40):9426–9432. doi: 10.1021/bi00492a017. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Hanvey J. C., Peffer N. J., Bisi J. E., Thomson S. A., Cadilla R., Josey J. A., Ricca D. J., Hassman C. F., Bonham M. A., Au K. G. Antisense and antigene properties of peptide nucleic acids. Science. 1992 Nov 27;258(5087):1481–1485. doi: 10.1126/science.1279811. [DOI] [PubMed] [Google Scholar]
- Hanvey J. C., Shimizu M., Wells R. D. Site-specific inhibition of EcoRI restriction/modification enzymes by a DNA triple helix. Nucleic Acids Res. 1990 Jan 11;18(1):157–161. doi: 10.1093/nar/18.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanvey J. C., Williams E. M., Besterman J. M. DNA triple-helix formation at physiologic pH and temperature. Antisense Res Dev. 1991 Winter;1(4):307–317. doi: 10.1089/ard.1991.1.307. [DOI] [PubMed] [Google Scholar]
- Hsieh C. H., Griffith J. D. Deletions of bases in one strand of duplex DNA, in contrast to single-base mismatches, produce highly kinked molecules: possible relevance to the folding of single-stranded nucleic acids. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4833–4837. doi: 10.1073/pnas.86.13.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain I., Griffith J., Sancar A. Thymine dimers bend DNA. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2558–2562. doi: 10.1073/pnas.85.8.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klysik J., Rippe K., Jovin T. M. Reactivity of parallel-stranded DNA to chemical modification reagents. Biochemistry. 1990 Oct 23;29(42):9831–9839. doi: 10.1021/bi00494a012. [DOI] [PubMed] [Google Scholar]
- Lane D., Prentki P., Chandler M. Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol Rev. 1992 Dec;56(4):509–528. doi: 10.1128/mr.56.4.509-528.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher L. J., 3rd, Dervan P. B., Wold B. J. Kinetic analysis of oligodeoxyribonucleotide-directed triple-helix formation on DNA. Biochemistry. 1990 Sep 18;29(37):8820–8826. doi: 10.1021/bi00489a045. [DOI] [PubMed] [Google Scholar]
- Maher L. J., 3rd, Wold B., Dervan P. B. Inhibition of DNA binding proteins by oligonucleotide-directed triple helix formation. Science. 1989 Aug 18;245(4919):725–730. doi: 10.1126/science.2549631. [DOI] [PubMed] [Google Scholar]
- Nielsen P. E., Egholm M., Berg R. H., Buchardt O. Sequence specific inhibition of DNA restriction enzyme cleavage by PNA. Nucleic Acids Res. 1993 Jan 25;21(2):197–200. doi: 10.1093/nar/21.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P. E., Egholm M., Berg R. H., Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991 Dec 6;254(5037):1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- Rice J. A., Crothers D. M. DNA bending by the bulge defect. Biochemistry. 1989 May 16;28(10):4512–4516. doi: 10.1021/bi00436a058. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S. L., Krawczyk S. H., Matteucci M. D., Toole J. J. Triple helix formation inhibits transcription elongation in vitro. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10023–10026. doi: 10.1073/pnas.88.22.10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sande J. H., Ramsing N. B., Germann M. W., Elhorst W., Kalisch B. W., von Kitzing E., Pon R. T., Clegg R. C., Jovin T. M. Parallel stranded DNA. Science. 1988 Jul 29;241(4865):551–557. doi: 10.1126/science.3399890. [DOI] [PubMed] [Google Scholar]