Dear Editor,
Cereal endosperms, which consist of an outer aleurone layer and an inner starchy endosperm, provide over 70% staple food for human [www.fao.org]. Rice endosperm development occurs in the caryopsis of a grain. During the grain filling stage, sucrose produced in photosynthetic leaves is transported to grains via the phloem1,2. It has been proposed that sucrose, upon arrival in the caryopsis through the phloem located at the dorsal vascular bundle, is partially hydrolyzed by cell wall invertases (CINs) into glucose and fructose3,4. Since endosperm is symplastically isolated from maternal tissues, it is assumed that these sugars are delivered to the endosperm actively through an apoplastic pathway by sugar transporters located in aleurone5,6. However, the exact role of aleurone in regulating sugar transport remains unknown.
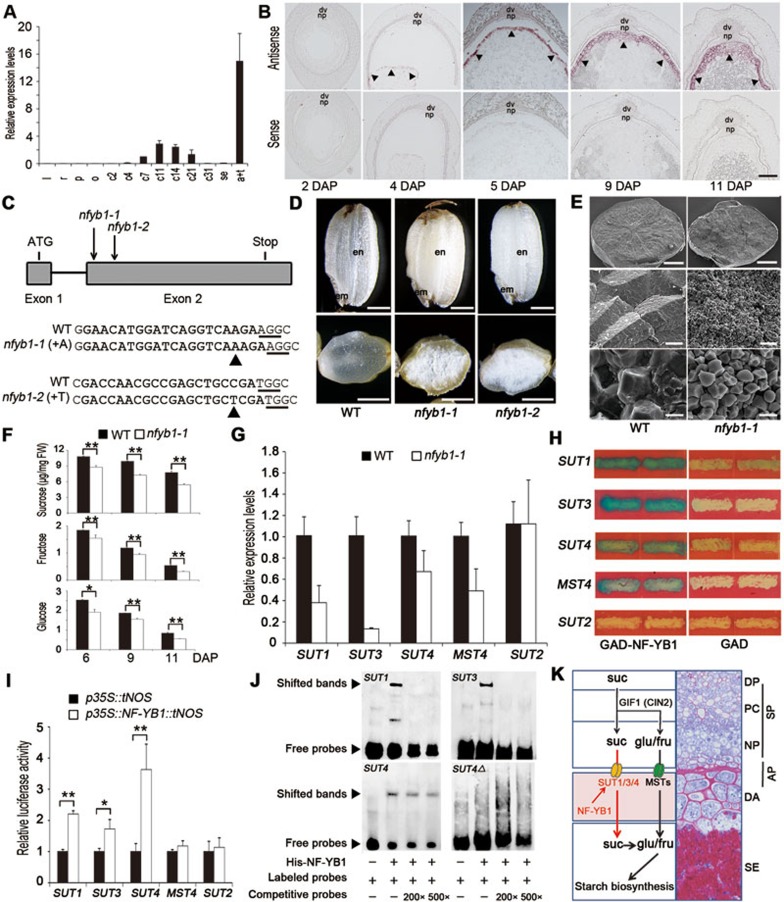
In this study, microarray analyses were performed in rice (Oryza sativa L., Zhonghua 11) to identify transcription factors (TFs) differentially expressed between starchy endosperm and aleurone in caryopses collected at 11 to 14 days after pollination (DAP), a stage when grain filling is active. Out of 949 TFs that were differentially expressed, 505 were up-regulated in the aleurone. Real-time PCR was performed to examine their expression patterns further. Os02g0725900 encoding Nuclear Factor Y B1 (NF-YB1) was shown to be expressed in caryopses from 4 to 21 DAP (c4 to c21), similar to the result published recently7, while no expression was detected in vegetative tissues examined (Figure 1A). Within caryopses, abundant NF-YB1 expression was detected in mixed aleurone-testa sample (a+t), but not in starchy endosperm (se; Figure 1A).
Figure 1.
NF-YB1-regulated expression of three sucrose transporters facilitates grain filling in rice. (A) Real-time PCR analysis showing that NF-YB1 is expressed in caryopses at 4, 7, 11, 14 and 21 DAP; and in caryopses collected at 11 DAP, NF-YB1 is highly expressed in mixed aleurone and testa tissues. l, leaves; r, roots; p, panicles; o, ovaries; c2 to c31, caryopses collected at 2 to 31 DAP; se and a+t, starchy endosperm and mixed aleurone and testa samples collected at 11 DAP, respectively. Data are shown as means ± SD (n = 3). (B) In situ hybridization in sectioned caryopses collected at 2, 4, 5, 9 and 11 DAP using antisense (upper panel) and sense probes (lower panel), showing that NF-YB1 is strongly expressed in aleurone (arrowheads), but not in maternal tissues, or in starchy endosperm. dv, dorsal vascular bundle; np, nucellar projection. Bar = 100 μm. (C) nfyb1-1 and nfyb1-2 knockout mutants generated by CRISPR-Cas9. Upper panel, a gene model of NF-YB1 to show positions of single-nucleotide insertions in nfyb1-1 and nfyb1-2. Lower panel, mutation sites in nfyb1-1 and nfyb1-2, as compared to WT sequences. Sequence in bold, target sites; underlined, protospacer-adjacent motif sequences (PAMs). Arrowheads indicate inserted single nucleotides. (D) Mature grains of nfyb1-1 and nfyb1-2, showing the chalky endosperm phenotype as compared to the semi-transparent endosperm in WT. Upper panel, side views of intact caryopses; lower panel, surface views of cracked caryopses. en, endosperm; em, embryo. Bars = 1 mm. (E) Scanning electron microscopy images of cracked mature WT and nfyb1-1 caryopses at different magnifications, showing smaller and loosely packed starch grains in nfyb1-1, as compared to large and tightly packed ones in the WT. Scale bars: 500 μm (top), 25 μm (middle), 5 μm (bottom). (F) Sucrose, fructose, and glucose contents (μg in mg fresh weight) in developing caryopses collected at 6, 9 and 11 DAP, showing significantly reduced accumulation of all these sugars in nfyb1-1, as compared to that in the WT caryopses at the same stage. Data shown as means ± SD (n = 3; *P< 0.05, **P< 0.01, based on Student's t-test). (G) Expression levels of SUT1, SUT3, SUT4 and MST4 were reduced in nfyb1-1 caryopses collected at 5 DAP, as compared to those in WT. Expression levels of these genes in the WT were normalized to 1. No significant reduction was observed for SUT2. Data shown as means ± SD (n = 3). (H) Yeast one-hybrid assay to show bindings of NF-YB1 to SUT1, SUT3, SUT4 and MST4 regulatory elements. NF-YB1 fused to GAL4 transcriptional activation domain (GAD-NF-YB1) was used as prey; 5′ regulatory sequences of SUT1, SUT3, SUT4, MST4 and SUT2 fused to LacZ as baits. Yeast cells co-transformed with GAD-NF-YB1 (left) or empty vector (GAD, right) and one of these baits were grown on a synthetic dropout medium without tryptophan and uracil. Note no activation was detected for SUT2. (I) LUC assays performed in rice protoplasts to show NF-YB1 activated SUT1, SUT3 and SUT4 promoters. LUC activities in protoplasts co-transfected with p35S::tNOS and reporter constructs (pSUT1::LUC, pSUT3::LUC, pSUT4::LUC, pMST4::LUC or pSUT2::LUC) were normalized to 1, and LUC activities in protoplasts co-transfected with p35S::NF-YB1::tNOS and these reporter constructs were relative to this value. Data shown as means ± SD (n = 3; *P< 0.05, **P< 0.01, based on Student's t-test). (J) EMSA assays to show His-tagged NF-YB1 bound directly to CCAAT box-containing 5′ upstream sequences of SUT1, SUT3 and SUT4. SUT4Δ, SUT4 with a mutated CCAAT box was used as a negative control. (K) A working model, illustrating the role of NF-YB1 in regulating sucrose loading to developing rice endosperms by activating three sucrose transporters in aleurone, while glucose and fructose are loaded to endosperms through an independently regulated route. DP, dorsal phloem; PC, parenchyma cells in the vascular bundle; NP, nucellar projection; DA, dorsal aleurone; SE, starchy endosperm; SP, symplastic; AP, apoplastic; suc, sucrose; glu, glucose; fru, fructose.
NF-YB1 belongs to NF-Y family TFs that are conserved in all eukaryotic organisms. NF-Ys usually form trimers with one NF-YA, one NF-YB and one NF-YC, and bind to CCAAT-containing regulatory elements8. Ten NF-YAs, eleven NF-YBs and seven NF-YCs are present in the rice genome, which could potentially produce over 700 heterotrimeric complexes8.
In situ hybridization showed that, during caryopsis development, expression of NF-YB1 was first detected at the periphery of coenocytic endosperms at 4 DAP, and confined to dorsal aleurone (adjacent to the primary vascular bundle and nucellar projection) at 5 DAP, and then expanded to the whole aleurone at 7, 9 and 11 DAP (stronger at the dorsal side). No expression was observed in maternal tissues such as the dorsal vascular bundle and the nucellar projection (Figure 1B and Supplementary information, Figure S1A). The expression correlated with the rapid increase of fresh caryopsis weight during this period (Supplementary information, Figure S1B). GUS staining in transgenic plants carrying 3 018 base pairs (bp) of NF-YB1 5′ upstream sequence fused to β-Glucuronidase (GUS) confirmed the expression pattern, and revealed no GUS expression in any other tissues examined (Supplementary information, Figure S1C). These results together suggest NF-YB1 is specifically expressed in aleurone.
Two CRISPR-Cas9 constructs expressing guide RNAs targeting different regions of the second exon of NF-YB1 were made and used to transform into rice (Zhonghua 11). Sequencing of PCR-amplified NF-YB1 genomic DNA from transgenic plants allowed us to identify two knockout mutants: nfyb1-1 with an adenine insertion and nfyb1-2 with a thymine insertion into the second exon (Figure 1C and Supplementary information, Figure S2A), predicted to produce truncated polypeptides with only 16 and 34 amino acids (AA) identical to NF-YB1 in nfyb1-1 and nfyb1-2, respectively (Supplementary information, Figure S2B). Although no visible phenotype was observed during vegetative growth, grains produced from homozygous nfyb1-1 and nfyb1-2 plants showed a chalky endosperm phenotype, suggesting a defect in grain filling (Figure 1D). Additionally, an RNA interference (RNAi) construct with a 350-bp fragment targeting the second exon of NF-YB1, expressed under the control of maize Ubiquitin 1 promoter, was made and transformed into rice (Zhonghua 11). Caryopses excised from 3 independent transgenic lines showed reduced NF-YB1 expression (Supplementary information, Figure S2C) and a similar chalky endosperm phenotype as nfyb1-1 and nfyb1-2 (Supplementary information, Figure S2D). These data together suggest that NF-YB1 plays an important role in rice grain filling.
Since nfyb1-1 and nfyb1-2 showed indistinguishable phenotype, nfyb1-1 was thus used in subsequent experiments. The mature nfyb1-1 caryopsis showed 18% reduction in dry weight (Supplementary information, Figure S2E). Scanning electron microscopy examinations revealed that endosperms in nfyb1-1 carried small and loosely packed starch granules, which differed markedly from the large and tightly packed starch granules in the wildtype (WT; Figure 1E). The average length of nfyb1-1 starch granules along the long axes was 4.4 ± 0.6 μm, as compared with 7.8 ± 1.0 μm in the WT (Supplementary information, Figure S2F). Gas chromatography triple quadrupole mass spectrometry (GC-QQQ-MS/MS) analyses showed that contents of sucrose, fructose and glucose in nfyb1-1 caryopses collected at 6, 9 and 11 DAP were significantly reduced as compared to those in the WT (Figure 1F). These data suggest that the chalky endosperm phenotype in nfyb1-1 might be caused by compromised sugar delivery.
We then examined expression patterns of genes involved in sugar loading in nfyb1-1. All five Sucrose Transporters (SUTs, 1 to 5) in the rice genome, CIN2 and Monosaccharide Transporter 4 (MST4) are known to be expressed at different stages of rice caryopses9,4,10. Real-time PCR performed in caryopses collected at 5 DAP showed that levels of SUT1, SUT3, SUT4 and MST4 expression were reduced in nfyb1-1, while no evident reduction was observed for SUT2 (Figure 1G), suggesting that SUT1, SUT3, SUT4 and MST4 may be regulated by NF-YB1.
To address if NF-YB1 can bind to regulatory elements of these sugar transporter genes, yeast one-hybrid experiment was performed using NF-YB1 fused with GAL4 transcriptional activation domain as prey, and 5′ upstream sequences of SUT1, SUT3, SUT4, MST4 or SUT2 fused to LacZ as baits. After co-transformation of the prey with individual baits into yeast, we observed that NF-YB1 is able to activate LacZ expression when SUT1, SUT3, SUT4 or MST4 5′ sequences were used, while no activation was detected for SUT2 (Figure 1H).
To further address if NF-YB1 is able to activate expression of these sugar transporters in plants, luciferase (LUC) assay was performed in vitro using protoplasts isolated from etiolated rice seedlings by co-transformation of p35S::NF-YB1::tNOS with either pSUT1::LUC, pSUT3::LUC, pSUT4::LUC, pMST4::LUC or pSUT2::LUC. As showed in Figure 1I, significantly elevated LUC activities were detected when pSUT1::LUC, pSUT3::LUC and pSUT4::LUC were used, while no significant elevation was observed for pMST4::LUC and pSUT2::LUC, indicating MST4 and SUT2 might not be direct targets of NF-YB1.
Several putative CCAAT boxes were identified in 5′ upstream sequences of SUT1, SUT3 and SUT4 (Supplementary information, Data S1). To elucidate if NF-YB1 is able to bind to these boxes, electrophoretic mobility shift assay (EMSA) was performed using His-NF-YB1 produced in E. coli (Supplementary information, Data S1). As shown in Figure 1J, bands with retarded migrations were observed when SUT1, SUT3 or SUT4 upstream sequences were incubated with His-NF-YB1. The bands disappeared (SUT1 and SUT3) or became significantly weakened (SUT4) when excess competitive probes were added. These results together suggest that NF-YB1 is able to bind to CCAAT boxes of SUT1, SUT3 and SUT4.
Previous studies have shown that SUT1 is expressed in aleurone, dorsal vascular bundle, nucellar projection and nucellar epidermis11, and SUT4 in aleurone and the embryo12. In situ hybridization performed in this study showed that SUT3 was expressed specifically in the dorsal aleurone. No expression was detected in maternal pericarp and testa tissues, nor in the starchy endosperm (Supplementary information, Figure S2G). Co-expression of NF-YB1 with SUT1, SUT3 and SUT4 in aleurone is consistent with the possibility that they form part of a regulatory module.
In summary, the aleurone-specific NF-YB1, with stronger expression at the dorsal side, plays an important role in regulating rice grain filling through activating the expression of SUT1, SUT3 and SUT4. It has been shown in several cereal crops that phloem in the dorsal vascular bundle, to which the dorsal aleurone and nucellar projection adjoin, is the primary route for sucrose delivery from leaves to developing endosperms13,14. Current hypothesis suggests that sucrose is partially hydrolyzed to glucose and fructose by CINs expressed in the dorsal vascular bundle region, and move first symplastically through maternal phloem parenchyma and nucellar projection through plasmadesmata, and then apoplastically to the filial endosperm by sugar transporters located in aleurone transfer cells2,5,6. Results provided in this study showed that the expression of SUT1, SUT3 and SUT4 in rice aleurone is regulated in concert by NF-YB1. Knockout or down-regulation of NF-YB1 led to defective grains with chalky endosperms. Similar grain-filling phenotype has been observed in rice when CIN2/GIF1 or SUT1 is mutated or down-regulated4,15. We showed further using yeast one-hybrid, LUC assay and EMSA experiments that NF-YB1 binds specifically to 5′ regulatory sequences of SUT1, SUT3 and SUT4. As expected, expression of these genes was down-regulated in nfyb1-1 caryopses. It is noted that SUT1 and SUT4 are expressed in a much broader pattern than NF-YB1. We reckon that the aleurone-specifically expressed NF-YB1 may form a trimeric complex with an unknown NF-FA and NF-YC to regulate the expression of these three SUTs in aleurone, while the expression of these SUTs in other tissues could be regulated by different NF-Y complexes, or other families of TFs. A model is thus proposed to illustrate the role of rice NF-YB1 in regulating sucrose loading to the developing endosperm for starch biosynthesis through transcriptional activation of SUT1, SUT3 and SUT4 in aleurone (Figure 1K). This route may act in parallel with the glucose and fructose loadings that are mediated by CINs and MSTs. Further studies on this molecular module may deepen our understanding of cereal grain filling in general, and could potentially contribute to improving crop yield.
Acknowledgments
We thank Rongcheng Lin for providing yeast one-hybrid and LUC assay vectors, Zhen Xue for performing GC-QQQ-MS/MS analyses, and Wenhao Zhang for critical reading of the manuscript. This work was funded by the CAS-CSIRO Cooperation Research Program (GJHZ1406), 863 National High-Tech R&D Program of China (2012AA02A707) and the National Transgenic Science and Technology Program (2014ZX08009-003-005).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
NF-YB1 is specifically expressed in rice aleurone during grain filling.
Genetic analyses of NF-YB1 gene in rice grain filling.
Supplementary Methods
References
- Lucas WJ, Groover A, Lichtenberger R, et al. J Integr Plant Biol 2013; 55:294–388. [DOI] [PubMed]
- Braun DM, Wang L, Ruan YL. J Exp Bot 2014; 65:1713–1735. [DOI] [PubMed]
- Lim JD, Cho J, Park Y, et al. Physiol Plant 2006; 126:572–584.
- Wang ET, Wang JJ, Zhu XD, et al. Nat Genet 2008; 40:1370–1374. [DOI] [PubMed]
- Patrick JW, Offler CE. J Exp Bot 2001; 52:551–564. [PubMed]
- Weschke W, Panitz R, Sauer N, et al. Plant J 2000; 21:455–467. [DOI] [PubMed]
- Sun XC, Ling S, Lu ZH, et al. Gene 2014; 551:214–221. [DOI] [PubMed]
- Petroni K, Kumimoto RW, Gnesutta N, et al. Plant Cell 2012; 24:4777–4792. [DOI] [PMC free article] [PubMed]
- Aoki N, Hirose T, Scofield GN, et al. Plant Cell Physiol 2003; 44:223–232. [DOI] [PubMed]
- Wang YQ, Xu HL, Wei XL, et al. Plant Mol Biol 2007; 65:439–451. [DOI] [PubMed]
- Furbank RT, Scofield GN, Hirose T, et al. Aust J Plant Physiol 2001; 28:1187–1196.
- Chung P, Hsiao HH, Chen HJ, et al. Acta Physiol Plant 2014; 36:217–229.
- Oparka KJ, Gates P. Planta 1981; 152:388–396. [DOI] [PubMed]
- Radchuk V, Borisjuk L. Front Plant Sci 2014; 5:510. [DOI] [PMC free article] [PubMed]
- Scofield GN, Hirose T, Gaudron JA, et al. Funct Plant Biol 2002; 29:815–826. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NF-YB1 is specifically expressed in rice aleurone during grain filling.
Genetic analyses of NF-YB1 gene in rice grain filling.
Supplementary Methods