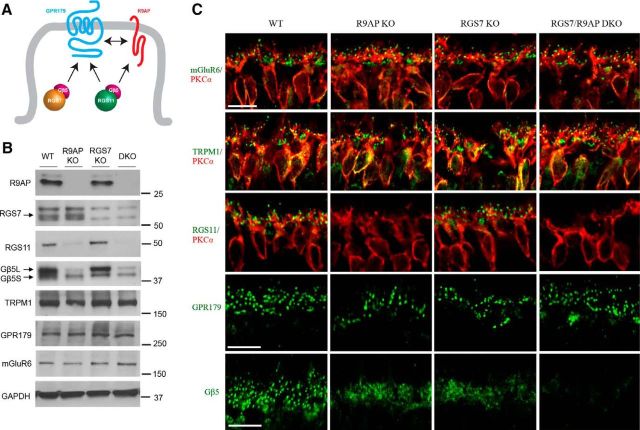
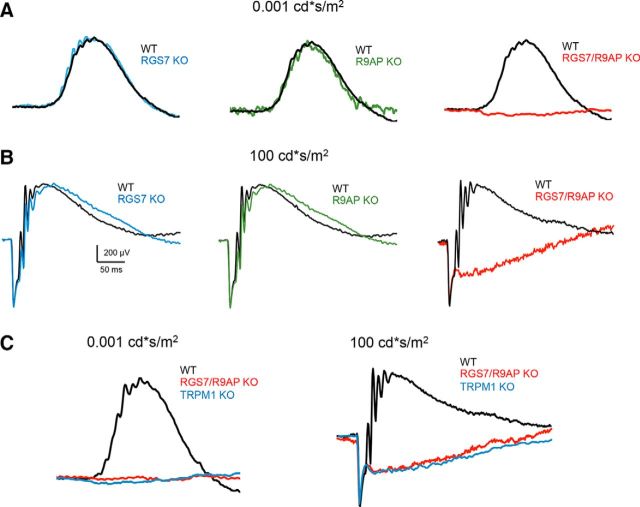
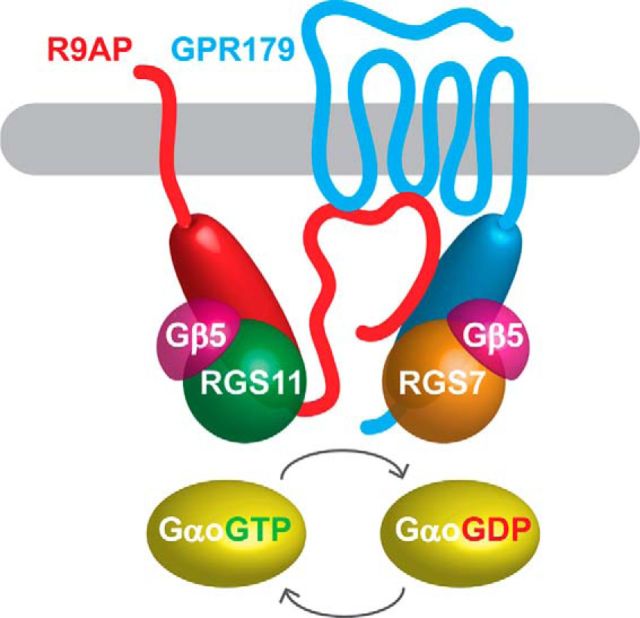
Abstract
In vertebrate retina, light responses generated by the rod photoreceptors are transmitted to the second-order neurons, the ON-bipolar cells (ON-BC), and this communication is indispensible for vision in dim light. In ON-BCs, synaptic transmission is initiated by the metabotropic glutamate receptor, mGluR6, that signals via the G-protein Go to control opening of the effector ion channel, TRPM1. A key role in this process belongs to the GTPase Activating Protein (GAP) complex that catalyzes Go inactivation upon light-induced suppression of glutamate release in rod photoreceptors, thereby driving ON-BC depolarization to changes in synaptic input. The GAP complex has a striking molecular complexity. It contains two Regulator of G-protein Signaling (RGS) proteins RGS7 and RGS11 that directly act on Go and two adaptor subunits: RGS Anchor Protein (R9AP) and the orphan receptor, GPR179. Here we examined the organizational principles of the GAP complex in ON-BCs. Biochemical experiments revealed that RGS7 binds to a conserved site in GPR179 and that RGS11 in vivo forms a complex only with R9AP. R9AP and GPR179 are further integrated via direct protein–protein interactions involving their cytoplasmic domains. Elimination of GPR179 prevents postsynaptic accumulation of R9AP. Furthermore, concurrent knock-out of both R9AP and RGS7 does not reconfigure the GAP complex and completely abolishes synaptic transmission, resulting in a novel mouse model of night blindness. Based on these results, we propose a model of hierarchical assembly and function of the GAP complex that supports ON-BCs visual signaling.
SIGNIFICANCE STATEMENT The ability of photoreceptors to transmit signals to the downstream ON-bipolar neurons in the retina is indispensible for vision. In this study, we delineate the molecular organization of the central regulatory complex, the GTPase Activating Protein (GAP) complex, that drives postsynaptic responses in ON-bipolar cells. Here, we identify an unexpected complexity and interdependence between multiple subunits of the GAP complex. We propose a model for its supramolecular assembly, where individual components hierarchically control expression and intracellular targeting of the GAP complex. Broad interest results from the crucial role of similarly organized GAP complexes throughout the nervous system, where they control a wide range of fundamental neuronal processes, including learning and memory, reward, and movement coordination.
Keywords: G-protein signaling, retina, RGS proteins, synaptic transmission, vision
Introduction
In vertebrates, vision begins when rod and cone photoreceptors signal the detection of photons by a hyperpolarization of their membrane potential. This results in light-induced suppression of the release of the neurotransmitter glutamate from the photoreceptor axon terminals. The change in glutamate concentration is detected by the postsynaptic neurons, the rod, and cone bipolar cells (BCs) (Taylor and Smith, 2004; Pahlberg and Sampath, 2011). Rod photoreceptors, which are exquisitely sensitive to low light, connect predominantly to a single type of BC, the rod ON-BC (ON-BC) (Hoon et al., 2014). Disruption of synaptic communication between rods and ON-BC results in forms of congenital stationary night blindness in humans and a loss in behavioral sensitivity of mice to low levels of light stimulation (Pardue and Peachey, 2014; Zeitz et al., 2015).
The ON-BCs use a metabotropic G-protein signaling cascade for the synaptic communication with photoreceptors. At the dendritic tips of the ON-BCs, glutamate released by the photoreceptors is sensed by the G-protein-coupled receptor, mGluR6, which activates the heterotrimeric G-protein, Go. In its activated state, Go keeps the effector ion channel TRPM1 closed. Upon light stimulation, synaptic glutamate declines, reducing the activity of mGluR6 and leading to the opening of TRPM1 channels that depolarize the ON-BCs (Snellman et al., 2008; Morgans et al., 2010). The opening of TRPM1 channels requires Go deactivation, and blockade of this step abolishes ON-BC light responses (Dhingra et al., 2000; Sampath and Rieke, 2004). Go deactivates when bound GTP hydrolyzes. Although this occurs spontaneously, the rate is too slow (τ ∼ 30 s) to account for the fast speed of the ON-BC light response (τ ∼ 60 ms). To achieve physiologically relevant speed, deactivation of Go is catalyzed by members of the Regulators of G-protein Signaling (RGS) family that accelerate the rate of the GTP hydrolysis on the Gα subunits (Hollinger and Hepler, 2002). Recent studies revealed that two RGS proteins RGS7 and RGS11 are crucial for Go deactivation in ON-BCs and generation of their depolarizing response to light. Concurrent knock-out of both RGS7 and RGS11 completely abolishes ON-BC sensitivity to light flashes (Cao et al., 2012; Shim et al., 2012). A similar phenotype is observed also in mice lacking Gβ5, an obligatory subunit of these RGS proteins (Rao et al., 2007). Furthermore, levels of RGS proteins determine key parameters of ON-BC light responses: kinetics, amplitude, and light sensitivity. Thus, it is important to understand how expression, localization, and activity of RGS7 and RGS11 are regulated (Sarria et al., 2015). A clue to this may come from analyzing the interaction of RGS proteins with their membrane anchoring subunits: RGS9 Anchor Protein (R9AP) and orphan receptor GPR179. For example, R9AP forms complexes only with RGS11, and its knock-out in mice dramatically destabilizes RGS11 (Cao et al., 2009; Jeffrey et al., 2010). Further, an orphan receptor, GPR179, binds to both RGS7 and RGS11 (Orlandi et al., 2012), and its knock-out in mice prevents postsynaptic targeting of both RGS proteins and abolishes ON-BC light responses (Peachey et al., 2012). Thus, there is a complexity of molecular interactions involving RGS proteins that regulate their membrane anchoring and proteolytic stability at the dendrites of ON-BCs. In this study, we examine the relationship between R9AP and GPR179 and provide a model for the integration of RGS proteins into the signaling complex that enables ON-BC to respond to light.
Materials and Methods
Mice, antibodies, and genetic constructs.
The generation of R9AP KO (Keresztes et al., 2004), RGS7 KO (Cao et al., 2012), RGS7/11 double knock-out (DKO) (Cao et al., 2012), and GPR179nob5 (Peachey et al., 2012) mice have been described. RGS7/R9AP DKO mice were generated by crossing R9AP KO with RGS7 heterozygous mice. Mice of either sex, 2–4 months old, were used in this study. Mice were housed in groups on a 12 h light-dark cycle with food and water available ad libitum. All studies were performed in accordance with the National Institute of Health guidelines and were granted formal approval by the Institutional Animal Care and Use Committee of the Scripps Research Institute.
Generation of the following antibodies have been described previously: sheep anti-GPR179 (Orlandi et al., 2013), rabbit anti-R9AP (Keresztes et al., 2003), and sheep anti-RGS11 (Cao et al., 2008), sheep anti-TRPM1 (Cao et al., 2011). Rabbit anti-RGS7 (7RC1) and anti-Gβ5 (ATDG) were generous gifts from William Simonds (National Institute of Diabetes and Digestive and Kidney Diseases/ National Institutes of Health). Guinea pig anti-mGluR6 antibody was a gift from Dr. Takahisa Furukawa (Osaka University). Mouse anti-PKCα (ab11723; Abcam), mouse anti-CtBP2 (612044; BD Biosciences), mouse anti-HA (Millipore), mouse anti-myc (Roche Life Science), and rabbit anti-myc (Genscript) are commercially available.
Full-length human myc-tagged-GPR179 was purchased from OriGene. Plasmids encoding human GPR179 with an internal deletion of amino acids 629-974 (GPR179Δ-CT1-myc), a C terminus deletion of amino acids 974-2367 (GPR179-ΔCT2-myc), and a plasmid encoding the amino acids 629-974 (GPR179-CT1-myc) were generated by PCR, using the plasmid encoding the full-length GPR179 as template. Cloning of full-length mouse R9AP, R9AP-ΔTM, Gβ5, RGS11, and RGS7-HA were previously described (Martemyanov et al., 2005; Baker et al., 2006; Song et al., 2006; Anderson et al., 2007; Orlandi et al., 2015).
Western blotting.
Whole retinas were removed from mice and lysed by sonication in ice-cold PBS supplemented with 150 mm NaCl, 1% Triton X-100, and Complete protease inhibitor tablets (Roche). Lysates were cleared by centrifugation at 14,000 × g for 15 min at 4°C. Total protein concentration in the supernatant was measured by using BCA Protein Assay Kit (Pierce). Supernatants were added with SDS sample buffer, pH 6.8, containing 8 m urea and were subjected to 4%–20% gradient SDS-PAGE. Protein bands were transferred onto PVDF membranes, subjected to Western blot analysis by using HRP-conjugated secondary antibodies, and detected by using enhanced chemiluminescence ECL West Pico system (Pierce). Signals were captured on film and scanned by densitometer.
Cell culture, transfection, and immunoprecipitation.
HEK293T/17 cells were cultured at 37°C and 5% CO2 in DMEM supplemented with 10% FBS, MEM nonessential amino acids, 1 mm sodium pyruvate, and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin). Cells were transfected using Lipofectamine LTX (Invitrogen) and Plus reagent (Invitrogen) and used 24 h later. For immunoprecipitation, cells were harvested and lysed in ice-cold immunoprecipitation buffer (300 mm NaCl, 50 mm Tris-HCl, pH 7.4, 1% Triton X-100 and complete protease inhibitor mixture) by sonication. Lysates were cleared by centrifugation at 14,000 × g for 15 min, and the supernatants were incubated with 20 μl Dynabeads (Invitrogen) and 2 μg antibodies on a rocker at 4°C for 1 h. After three washes with immunoprecipitation buffer, proteins were eluted with 40 μl of 2× SDS sample buffer and analyzed by SDS-PAGE. The same protocol was followed for immunoprecipitation from retina samples. Two retinas from either wild-type (WT) or R9AP KO mice were sonicated in ice-cold immunoprecipitation buffer, centrifuged at 14,000 × g for 15 min, the supernatant incubated at 4°C for 1 h with 20 μl Dynabeads and 2 μg antibodies and then eluted with 40 μl of 2× SDS sample buffer.
Electroretinography.
Electroretinograms were recorded by using the UTAS system and a BigShot Ganzfeld (LKC Technologies). Mice (4–8 weeks old) were dark-adapted (≥6 h) and prepared for recordings by using dim red light or light adapted (50 cd · s/m2, 5 min). Mice were anesthetized with an intraperitoneal injection of ketamine and xylazine mixture containing 100 and 10 mg/kg, respectively. Recordings were obtained from the right eye only, and the pupil was dilated with 2.5% phenylephrine hydrochloride (Bausch & Lomb), followed by the application of 0.5% methylcellulose. Recordings were performed with a gold loop electrode (LKC Technologies) embedded within transparent contact lenses to keep the eyes immersed in solution. The reference electrode was a stainless steel needle electrode placed subcutaneously in the neck area. The mouse body temperature was maintained at 37°C by using a heating pad controlled by ATC 1000 temperature controller (World Precision Instruments). Electroretinogram (ERG) signals were sampled at 1 kHz and recorded with 0.3 Hz low-frequency and 300 Hz high-frequency cutoffs.
Full field white flashes were produced by a set of LEDs (duration <5 ms) for flash strengths ≤2.5 cd · s/m2 or by a Xenon light source for flashes >2.5 cd · s/m2 (flash duration <5 ms). ERG responses were elicited by a series of flashes ranging from 2.5 × 10−4 to 2.5 × 101 cd · s/m2. Three trials were averaged for responses evoked by 0.001 cd · s/m2 flashes, and single flash responses were recorded for 100 cd · s/m2 flashes. To allow for recovery, interval times between single flashes were as follows. For WT and RGS7 KO mice: 5 s for 2.5 × 10−4 to 2.5 × 10−1 cd · s/m2 flashes, 30 s for 2.5 × 100 cd · s/m2 flashes, and 180 s for 2.5 × 101 cd · s/m2 flashes. For R9AP KO mice, interval times between single flashes were 180 s to allow for full recovery. If response rundown occurred between flashes of the same intensity, these trials were discarded.
Immunohistochemistry.
Dissected eyecups were fixed for 15 min in 4% PFA, cryoprotected with 30% sucrose in PBS for 2 h at room temperature, and embedded in optimal cutting temperature medium; 12-μm-thick frozen sections were cut, mounted onto slides, and blocked in PBS with 0.1% Triton X-100 and 10% donkey serum for 1 h. They then were incubated with primary antibody in PBS with 0.1% Triton X-100 and 2% donkey serum for at least 1 h. After four washes with PBS with 0.1% Triton, sections were incubated with fluorophore-conjugated secondary antibodies in PT2 for 1 h. After four washes, sections were mounted in Fluoromount (Sigma). Images were taken with a Leica SP800 confocal microscope. Quantitative analysis of GPR179 and R9AP immunofluorescence from confocal images was performed using Leica software. Sections were double stained with a combination of antibodies to GPR179, R9AP, and mGluR6. mGluR6 staining was used as a mask to define synaptic puncta, encircling the area. The fluorescence intensity within this mGluR6-positive area was then determined and defined as synaptic. Fluorescence intensity was also determined for the entire outer plexiform layer (OPL) region (defined as a DAPI negative area between outer nuclear layer and inner nuclear layer) that contained synaptic puncta to be analyzed. For the quantitative analysis of synaptic enrichment, mean fluorescence intensity across regions of interest was divided by the area they occupy for both synaptic puncta (IS) and the outlying OPL (IOPL). To determine relative synaptic enrichment (%), the fraction above the equally distributed value was calculated by the following transformation: (IS/IOPL) × 100 − 100 and reported on the graphs for each genotype. Alternatively, synaptic enrichment was analyzed by a line-scan analysis. A line of 1–1.5 μm (white bar) was drawn through the center of the distinct mGluR6-positive synapses, and the distribution of the fluorescence intensity along this line was scanned to generate the traces. For both methods of imaging, mean fluorescent intensity (measured in pixels) was averaged from ∼10 to 20 individual and randomly selected mGluR6-positive puncta per imaged section, with two or three sections per retina, and using two or three retinas per genotype. Imaging parameters were the same for all sections and retinas.
Results
R9AP and GPR179 are colocalized in all dendritic tips of ON-BCs
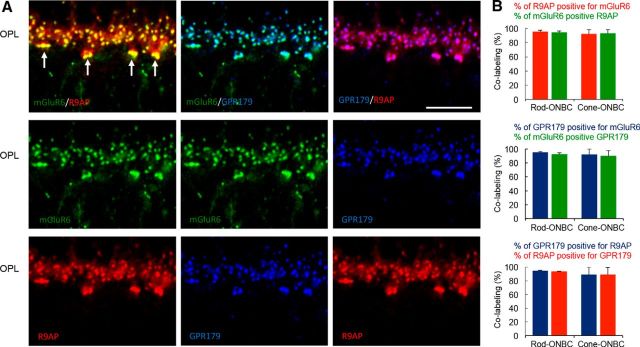
Previous studies have reported that both R9AP and GPR179 are selectively targeted to the tips of the ON-BC dendrites, where they colocalize with components of the mGluR6 cascade, including mGluR6 (Cao et al., 2009; Jeffrey et al., 2010; Peachey et al., 2012; Orlandi et al., 2013). To examine whether R9AP and GPR179 are present at the same dendritic tips and to determine the extent of possible colocalization, we performed immunohistochemical studies analyzing quantitatively the presence of each membrane anchor in rod and cone ON-BCs (Fig. 1A). We first referenced each to mGluR6 puncta and then to each other. Quantification revealed that 92 ± 3% of mGluR6-positive rod and 90 ± 8% cone synapses contained GPR179. Similarly, 95 ± 2% of rod and 92 ± 8% cone synapses with GPR179 also contained mGluR6 (Fig. 1B). The localization of R9AP similarly coincided with mGluR6 wherein 94 ± 2% of mGluR6 in rod and 93 ± 5% in cone synapses were positive for R9AP, whereas 95 ± 2% R9AP-containing rod and 92 ± 6% cone synapses had mGluR6. R9AP and GPR179 colocalization was nearly complete (>90%) at both rod and cone ON-BC synapses (Fig. 1B). The results show that both membrane anchors are present at the same synapses where they can bind to RGS proteins.
Figure 1.
R9AP and GPR179 are colocalized in the majority of ON-BC synapses. A, Immunohistochemical analysis of R9AP and GPR179 colocalization with each other and with mGluR6 in the ON BC dendrites in OPL of mouse retinas. Triple staining with indicated antibodies was performed, and each channel is individually pseudo-colored. Arrows indicate the position of the cone synapses. Scale bar, 10 μm. B, Quantitative analysis of protein distribution across ON-BC synapses with rod and cone photoreceptors.
GPR179 is required for postsynaptic accumulation of R9AP in ON-BC dendrites
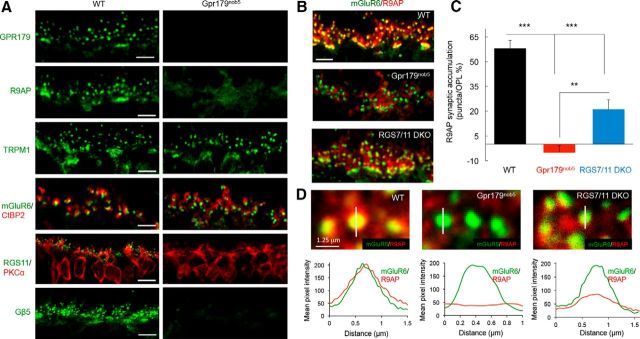
To begin analyzing the relationship between different membrane anchors of RGS proteins, we first determined the impact of disrupting expression of GPR179 or RGS on R9AP. We confirmed previous reports (Orlandi et al., 2012; Peachey et al., 2012; Ray et al., 2014) that Gpr179nob5 retinas that lack GPR179 had normal synaptic accumulation of mGluR6 and TRPM1 but not the RGS proteins (Fig. 2A). Synaptic architecture in Gpr179nob5 also appeared to be intact as evidenced by normal apposition of presynaptic and postsynaptic markers (CtBP2 and mGluR6, respectively). Consistent with these earlier findings, we also found a small but statistically significant reduction of TRPM1 immunofluorescence in the OPL of Gpr179nob5 retinas (84 ± 5% of levels present in WT controls, p < 0.05, n = 7 retinas). There was no significant change in the average levels of mGluR6 fluorescence intensity (92 ± 9%, p > 0.05, n = 9 retinas).
Figure 2.
GPR179 is necessary for the correct localization of R9AP in retinal ON-BC synapses and the expression of RGS11 in the OPL. A, Immunohistochemical analysis of ON-BC synaptic components in the OPL of WT and Gpr179nob5 mouse retinas. Retina sections were immunostained with the indicated antibodies. Synaptic architecture in Gpr179nob5 retinas is intact as evidenced by normal apposition of presynaptic and postsynaptic markers (CtBP2, mGluR6, and TRPM1, respectively). Note the lack of accumulation of R9AP and complete absence of RGS11 in Gpr179nob5 ON-BC dendritic tips. Scale bar, 5 μm. B, Double immunolabeling of WT, Gpr179nob5, and RGS7/RGS11 DKO retinas with mGluR6 and R9AP antibodies. Scale bar, 5 μm. Quantification of the accumulation of R9AP within mGluR6-positive puncta of ON-BC dendrites in OPL (described in Materials and Methods). C, Corresponding values. Error bars indicate SEM. *p < 0.05 (one-way ANOVA with Bonferroni's post hoc test). **p < 0.01 (one-way ANOVA with Bonferroni's post hoc test). ***p < 0.001 (one-way ANOVA with Bonferroni's post hoc test). n = 25–30 puncta for each group. D, Top, High-magnification images of retina sections with scan-line analyses of R9AP fluorescence intensity across the midline of mGluR6-positive synaptic puncta in OPL. Bottom, Averaged traces of R9AP fluorescent intensity across the midline of mGluR6-positive synaptic puncta reveal a complete lack of after synaptic R9AP accumulation in Gpr179nob5 retinas, but only a partial loss in RGS7/11 DKO. n = 25–30 for all groups. Scale bar, 0.125 μm.
Strikingly, R9AP accumulation at the ON-BC dendritic tips was abolished in Gpr179nob5 retinas (Fig. 2A). To confirm this observation, we performed double immunolabeling and quantitative analysis of R9AP distribution (Fig. 2B). In WT retina, R9AP is present on the ON-BC dendrites in the OPL where it is specifically clustered within mGluR6-positive puncta. Quantification of R9AP content within these synaptic puncta compared with dendrites outside of puncta reveals a 58 ± 5% higher concentration at the synapses (Fig. 2C). In the absence of GPR179 expression, the enrichment of R9AP at the dendritic tips was eliminated and R9AP fluorescence was equally distributed between the ON-BC dendrites and synapses in the OPL (−5 ± 4% puncta enrichment; Fig. 2B,C). Because GPR179 plays an essential role in localization of RGS proteins and RGS proteins also bind to R9AP, we next analyzed the impact of RGS7 and RGS11 on localization of R9AP (Fig. 2B,C). Knock-out of both RGS7 and RGS11 (RGS7/11 DKO) diminished accumulation of R9AP in mGluR6-positive puncta only partially, to ∼21 ± 6% enrichment level. Similar observations were made when R9AP accumulation at the dendritic tips was quantified separately for synapses with cone photoreceptors, where R9AP showed 75 ± 5% synaptic enrichment in WT controls but only 29 ± 6% in RGS7/11 DKO mice and 2 ± 1% in Gpr179nob5 (one-way ANOVA with Bonferroni's post hoc test; p < 0.05 for all genotypes; n = 25–30 puncta in each).
The lack of R9AP postsynaptic accumulation in Gpr179nob5 ON-BC dendrites also was evident when we scanned the distribution of its fluorescence intensity across mGluR6-positive puncta (Fig. 2D). In WT retinas, the intensity of R9AP staining inside mGluR6 puncta increased ∼4-fold. The localization of R9AP was only partially affected by the deletion of RGS7 and RGS11, reducing synaptic accumulation of R9AP ∼2-fold (Fig. 2D). In contrast, R9AP is completely absent from mGluR6-puncta in Gpr179nob5 retinas (Fig. 2D). Overall, these results establish that GPR179 plays an essential role in R9AP expression and postsynaptic targeting in the ON-BC.
Targeting of GPR179 to ON-BC dendritic tips is independent of R9AP or RGS proteins
We next examined the reciprocal influence of RGS and R9AP on GPR179 expression and localization (Fig. 3). We found GPR179-positive puncta to be present in the OPL of retinas from WT, R9AP KO, and RGS7/11 DKO retina (Fig. 3A). Elimination of either R9AP or RGS proteins did not alter the colocalization of GPR179 with mGluR6 (Fig. 3B). Quantification of GPR179 concentration within mGluR6-positive puncta revealed no difference between the genotypes with ∼170% synaptic enrichment (Fig. 3C). Similarly, when cone synapses were analyzed separately, we found no differences in GPR179 synaptic accumulation between genotypes (WT: 189 ± 6%, R9AP KO = 184 ± 6% and RGS7/11 DKO 178 ± 5%; one-way ANOVA, p > 0.05, n = 30–35 puncta for each genotype). These observations were further supported by examining the distribution of GPR179 across individual mGluR6 puncta (Fig. 3D). We found an ∼4-fold increase in the levels of GPR179 inside the postsynaptic cluster, relative to its dendritic content in the OPL and coinciding with the distribution profile of mGluR6. These data indicate that neither R9AP nor RGS7/RGS11 proteins play a role in determining clustering of GPR179 at the dendritic tips of ON-BCs.
Figure 3.
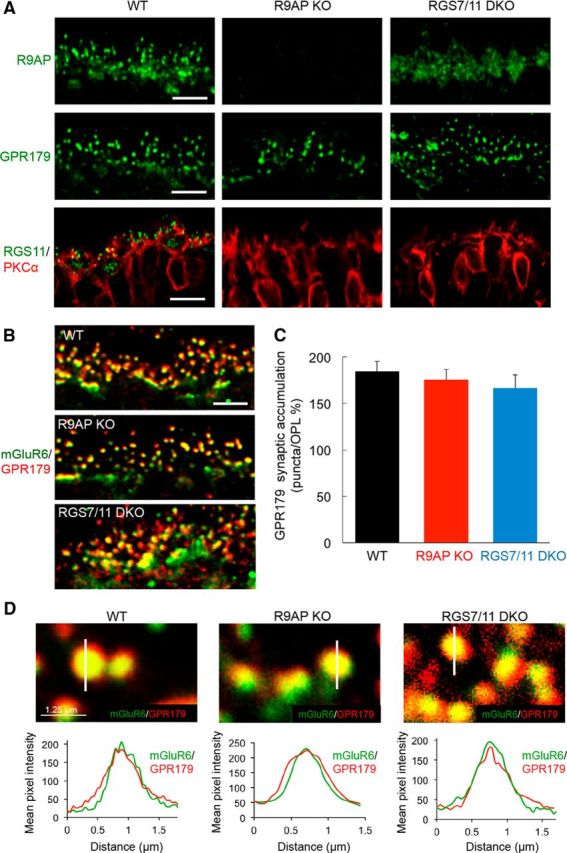
RGS and R9AP proteins are not required for the expression or correct localization of GPR179 in the retinal OPL. A, Immunohistochemical analysis of ON-BC synaptic components in the OPL of WT, R9AP, and RGS7/RGS11 DKO mouse retinas. Retinal sections were stained with the indicated antibodies. Scale bar, 5 μm. B, Double immunolabeling with mGluR6 and GPR179 antibodies. Scale bar, 5 μm. Quantification of the accumulation of GPR179 within mGluR6-positive synaptic puncta relative to the OPL (described in Materials and Methods). C, Corresponding values. Error bars indicate SEM; n = 30–35 puncta for each group. One-way ANOVA shows no significant differences. D, Top, High-magnification images of retinal sections with line-scan analyses of GPR179 fluorescence intensity across the midline of mGluR6-positive synaptic puncta in OPL. Bottom, Averaged traces of GPR179 fluorescent intensity across the midline of mGluR6-positive synaptic puncta reveal no changes to postsynaptic GPR179 accumulation in either R9AP KO or RGS7/RGS11 DKO retinas; n = 25–30 for all groups.
GPR179 and R9AP form a macromolecular complex
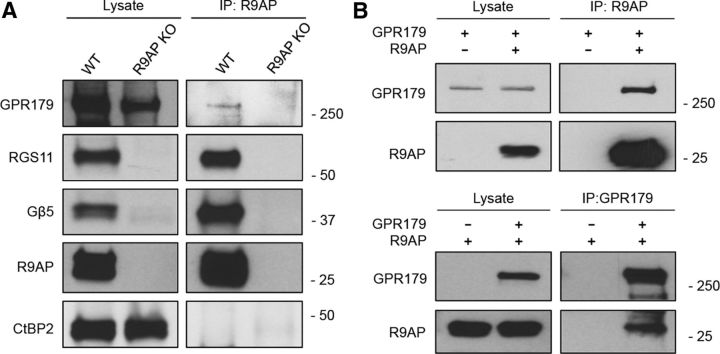
The dependence of R9AP targeting on the GPR179/RGS complex suggests that the proteins may physically bind to each other in vivo. To examine this possibility, we immunoprecipitated R9AP from retina and tested whether RGS11 and GPR179 coimmunoprecipitated (Fig. 4A). We found that both RGS11 and GPR179 were present in the eluates together with R9AP, suggesting that they form a complex. When retinas from R9AP knock-out mice were used, neither RGS11 nor GPR179 was pulled down, indicating that the interaction is specific. Furthermore, an unrelated component of the synaptic ribbon, CtBP2, also was not present in the immunoprecipitation eluates confirming the specificity of binding (Fig. 4A). We next analyzed the same binding upon reconstitution in transfected HEK293 cells. We found that both R9AP and GPR179 coimmunoprecipitated when pulled down by either R9AP or GPR179 antibodies (Fig. 4B).
Figure 4.
GPR179 and R9AP physically interact in vivo and in transfected cells. A, Immunoprecipitation of R9AP from retinas following detection of coeluting proteins by Western blotting. R9AP KO retinas were used as a negative control in a parallel experiment. GPR179 and RGS11 coimmunoprecipitate with R9AP from WT, but not from the R9AP KO retinas. B, Analysis of R9AP and GPR179 coimmunoprecipitation in transfected HEK293 cells. Top, Antibodies against R9AP are able to specifically coimmunoprecipitate GPR179. Bottom, Antibodies against GPR179 are able to coimmunoprecipitate R9AP.
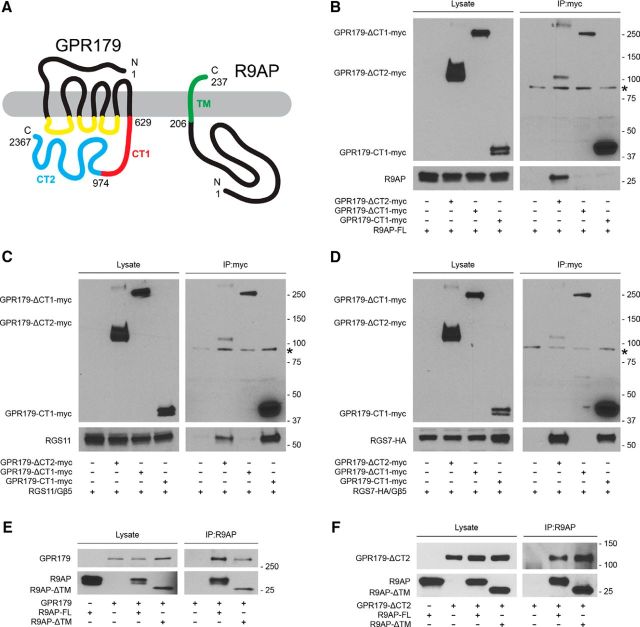
To further characterize their interaction, we mapped sites in both GPR179 and R9AP required for complex formation. GPR179 contains a heptahelical transmembrane region found in all GPCRs, flanked by extracellular N-terminal and intracellular C-terminal domains (Fig. 5A). R9AP is anchored to the membrane through a single pass C-terminal helix localizing all of its amino acid sequence to the cytoplasm (Fig. 5A). Given this spatial organization, the extracellular N terminus of the GPR179 can be excluded from the consideration as possible binding epitope to R9AP. Thus, we focused our analysis on the intracellular C-terminal portion of the molecule, which contains almost 75% of GPR179 sequence. The C terminus of GPR179 shows considerable sequence conservation with the corresponding region of its close homolog GPR158. GPR158 has two functionally and structurally distinct regions (Orlandi et al., 2015). The CT1 portion, which corresponds to the CD1/2/3 domains in GPR158 containing R9AP/R7BP homology, and the distal CT2 region, which corresponds to the CD4 domain in GPR158 containing G-protein-binding repeats (Orlandi et al., 2015). To examine the role of these regions in binding R9AP and RGS proteins, we generated myc-tagged deletion mutants of either the CT1 (GPR179-ΔCT1-myc) or CT2 (GPR179-ΔCT2-myc) region. These constructs were then tested for their binding to RGS7, RGS11, and R9AP using coimmunoprecipitation assays following coexpression in HEK293 cells (Fig. 5B–D). We found that deletion of the CT2 region of GPR179 did not affect its ability to pull down R9AP, RGS7, or RGS11. In contrast, the deletion of the CT1 region of GPR179 resulted in its failure to coimmunoprecipitate any of the three proteins (Fig. 5B–D). Interestingly, the CT1 region alone was able to immunoprecipitate RGS7 and RGS11, but not R9AP (Fig. 5B–D). These findings indicate that CT1 is both necessary and sufficient for binding to RGS proteins and that CT1 serves as a critical determinant that mediates GPR179 interaction with R9AP, although additional elements in GPR179 also are required for this interaction to occur.
Figure 5.
Mapping of the binding determinants involved in the interactions between GPR179, R9AP, and RGS proteins. HEK293 cells were cotransfected with the indicated constructs, and coimmunoprecipitation assays were conducted. A, Schematic representation of GPR179 and R9AP organization with the annotation of the protein fragments deleted in the various constructs used in the binding experiments. B, Deletional mutagenesis of R9AP binding determinants in GPR179. An antibody against the myc-tagged deletion mutants of GPR179 was used for immunoprecipitation. R9AP was detected only after coimmunoprecipitation with GPR179-ΔCT2. C, D, Deletional mutagenesis of RGS11 and RGS7 binding determinants in GPR179, respectively. An antibody against the myc-tagged deletion mutants of GPR179 was used for immunoprecipitation. RGS11 (C) and RGS7 (D) were coimmunoprecipitated with GPR179-ΔCT2 and GPR179-ΔCT1 constructs. E, Deletional mutagenesis of GPR179 binding determinants in R9AP. An antibody against R9AP was used in the experiment. Deletion of the transmembrane region of R9AP does not affect its interaction with GPR179. F, Coimmunoprecipitation of R9AP and GPR179 deletional mutants containing minimal binding determinants. Asterisk indicates the position of nonspecific band detected in the immunoprecipitation eluates, which likely corresponds to IgGs used for affinity precipitation.
Because both GPR179 and R9AP are transmembrane proteins, we next tested the contribution of membrane association to their interaction. The R9AP mutant lacking the transmembrane segment (R9AP-ΔTM) coimmunoprecipitated with full-length GPR179 in transfected HEK293 cells (Fig. 5E). Furthermore, this R9AP-ΔTM mutant interaction with GPR179 was unaffected by the removal of the CT2 domain of GPR179 (Fig. 5F). Together, these results suggest GPR179 and R9AP directly interact with each other via their cytoplasmic domains.
Elimination of RGS7 does not rescue RGS11 destabilization in R9AP knock-out
Previous studies indicate that R9AP interacts only with RGS11 (Martemyanov et al., 2005), whereas GPR179 binds both RGS7 and RGS11 (Orlandi et al., 2012). The RGS binding site in GPR179 (contained within CT1 region; see Fig. 5) shows sequence homology to the corresponding binding sites in other RGS anchors: R9AP, GPR158, and R7BP (Orlandi et al., 2015). As a result, membrane anchors compete for RGS binding (Anderson et al., 2007; Orlandi et al., 2012); and even though both RGS7 and RGS11 colocalize with GPR179, only one of them would be expected to bind directly to GPR179 via its RGS binding domain. These considerations suggest that RGS11 may be found in two distinct complexes: anchored either to GPR179 or to R9AP. To examine whether this occurs in vivo, we generated an RGS7/R9AP DKO mouse line and compared expression and localization of ON-BC signal transduction components to single R9AP or RGS7 KOs (Fig. 6B,C). Western blot analyses of retina tissues indicate that elimination of R9AP and RGS7 alone or together does not alter expression of two key components of the ON-BC cascade: mGluR6 and TRPM1 (Fig. 6B). However, the expression of GPR179 in R9AP/RGS7 DKO was reduced to 80 ± 6% of WT, consistent with earlier observations in RGS7/11 DKO mice (Orlandi et al., 2013). In contrast, we found no detectable RGS11 expression in either R9AP KO or R9AP/RGS7 DKO retinas.
Figure 6.
Elimination of RGS7 does not rescue loss of RGS11 expression and synaptic localization in R9AP knock-out retinas. A, Diagram model representing possible arrangement of RGS proteins and their membrane anchors GPR179 and R9AP at the dendritic tips of the ON-BC neurons. RGS7 is capable of binding only to GPR179, whereas RGS11 can bind to both GPR179 and R9AP. B, Western blot analysis of protein expression in retinas of RGS7/R9AP DKO mice in comparison with retinas lacking R9AP or RGS7 alone. There is no detectable RGS11 expression in either R9AP KO or R9AP/RGS7 DKO retinas. C, Analysis of synaptic targeting of proteins of interest in retinas lacking RGS7 and R9AP by immunohistochemistry. ON-BCs of R9AP/RGS7 DKO mice have normal cytoarchitecture and correct targeting of cascade components. There was no detectable accumulation of RGS11 at the synapses of ON-BCs in the R9AP/RGS7 DKO mice. Scale bar, 10 μm.
Immunohistochemical analyses of retinal cross-sections revealed normal targeting of mGluR6, TRPM1, and GPR179 to the dendritic tips of ON-BC of RGS7/R9AP DKO mice (Fig. 6C). Immunohistochemical data further confirmed the absence of RGS11 in ON-BC dendrites of both R9AP KO and RGS7/R9AP DKO retinas. Together, these observations indicate that elimination of RGS7 fails to rescue deficits in expression and localization of RGS11 in ON-BCs brought about by loss of R9AP. This indicates that, even though GPR179 contains a RGS7 binding site, in vivo this site is unable to bind RGS11, even in the absence of RGS7.
Concurrent knock-out of R9AP and RGS7 abolishes light responses of ON-BCs
Elimination of either R9AP, RGS7, or RGS11 alone causes only minor kinetic alterations in depolarizing activity of ON-BCs as measured by the ERG (Mojumder et al., 2009; Jeffrey et al., 2010; Zhang et al., 2010). In contrast, the DKO of both RGS7 and RGS11 eliminates the depolarizing response of ON-BCs to light (Cao et al., 2012; Shim et al., 2012). To test whether the lack of compensation from GPR179 in R9AP/RGS7 DKO at the cellular and biochemical level translates into functional changes, we measured responses of ON-BC to light flashes by ERG. In contrast to relatively normal ERG b-wave responses in single R9AP or RGS7 knock-outs, the b-wave component of the ERG was eliminated at all flash intensities tested in R9AP/RGS7 DKO mice (Fig. 7A,B). Direct comparison revealed that these responses were nearly identical to the ones seen in TRPM1 KO mice (Fig. 7C) consistent with complete elimination of the depolarizing activity of ON-BCs under both photopic and scotopic conditions. The a-wave component of the ERG in all the mutant lines tested was unchanged compared with WT controls, indicating that photoreceptor function is intact in these models. Thus, these data indicate that concurrent elimination of R9AP and RGS7 prevents ON-BC signal transduction resulting in a novel no b-wave (nob) mouse model of night blindness.
Figure 7.
Concurrent knock-out of R9AP and RGS7 causes no b-wave (nob) ERG phenotype. A, Representative ERG traces from dark-adapted WT, RGS7 KO, R9AP KO, and RGS7/R9AP DKO mice retinas elicited with scotopic flashes. B, Representative ERG traces from dark-adapted WT, RGS7 KO, R9AP KO, and RGS7/R9AP DKO mice retinas elicited with photopic flashes. RGS7/R9AP DKO mice have a nob ERG phenotype in both scotopic and photopic ranges and a normal a-wave. C, Comparison of ERG light responses of RGS7/R9AP DKO mice with those recorded in TRPM1 KO mice under scotopic (left) and photopic (right) conditions. Overlay of the representative traces reveals no apparent differences.
Discussion
In this study, we determine the molecular organization of the GTPase Activating Protein (GAP) complex at the dendritic tips of retinal ON-BCs. Previous studies identified several components of this complex: RGS7, RGS11 (Morgans et al., 2007; Cao et al., 2012; Shim et al., 2012), and their binding proteins Gβ5 (Rao et al., 2007), R9AP (Cao et al., 2009; Jeffrey et al., 2010), and GPR179 (Orlandi et al., 2012; Ray et al., 2014). In this complex, the main role in catalyzing GTP hydrolysis on Gαo belongs to RGS proteins that directly bind to Gαo and stabilize it in a transition state, thereby accelerating the reaction (Snow et al., 1998; Levay et al., 1999; Hooks et al., 2003). Both RGS7 and RGS11 belong to the same R7 class of RGS proteins and show substrate selectivity for the members of the Gαi/o family of G-proteins (Anderson et al., 2009). Interestingly, RGS7 and RGS11 have considerable redundancy in the retina because elimination of either does not eliminate mGluR6-mediated signaling in ON-BCs (Mojumder et al., 2009; Chen et al., 2010; Zhang et al., 2010; Sarria et al., 2015). The significance of this arrangement is not well understood. It has been proposed that excess of RGS proteins ensures that the mGluR6-mediated opening of TRPM1 channels is not rate-limited by Gαo GTP hydrolysis, and this contributes to the reliability of the information transfer at the photoreceptor to ON-BC synapse (Sarria et al., 2015).
In addition to RGS proteins, the GAP complex in ON-BCs includes a number of auxiliary subunits. Both RGS7 and RGS11 form obligatory complexes with Gβ5, which is essential for their folding (Cabrera et al., 1998; Snow et al., 1998; Chen et al., 2003; Howlett et al., 2009). The RGS/Gβ5 dimers are in turn associated with four membrane-anchored proteins: R7BP, R9AP, GPR158, and GPR179. A palmitoylated protein R7BP (Song et al., 2007) and the seven-pass transmembrane orphan receptor GPR179 (Orlandi et al., 2012; Gregg et al., 2014) both bind to RGS7 and RGS11 (Martemyanov et al., 2005; Orlandi et al., 2012). In addition, the transmembrane protein R9AP (Hu and Wensel, 2002; Cao et al., 2009; Masuho et al., 2010) binds to RGS11 but not RGS7 (Martemyanov et al., 2005; Anderson et al., 2007). We have previously shown that R7BP has no influence on the function of the GAP complex and thus is unlikely to be an essential component of ON-BC signaling cascade (Cao et al., 2008). In contrast, R9AP is essential for expression of RGS11 (Cao et al., 2009), whereas GPR179 is crucial for localization of both RGS11 and RGS7 to the tips of ON-BC dendrites (Orlandi et al., 2012). We have recently reported that all of these membrane anchors share homologous RGS binding sequences featuring coiled-coil segments (Orlandi et al., 2015). Biochemical studies in reconstituted systems show that membrane anchors (R7BP, R9AP, GPR158) compete for binding to RGS proteins that are competent to interact with more than one anchor (Anderson et al., 2007; Orlandi et al., 2012). Thus, RGS protein complexes in ON-BC can theoretically adopt different configurations and exist in multiple distinct complexes (e.g., RGS7/Gβ5/GPR179, RGS11/Gβ5/R9AP, RGS11/Gβ5/GPR179) where RGS7, RGS11 and R9AP, GPR179 are interchangeable. Contrary to this expectation, the results of this study, together with a wealth of previously accumulated evidence (Gregg et al., 2014), led us to propose a different model where the GAP complex in ON-BC adopts a distinct unitary configuration (Fig. 8). In this model, each RGS protein is bound directly to only one cognate anchor: GPR179 binds with the RGS7/Gβ5 complex via its conserved RGS binding sequence, and the RGS11/Gβ5 complex binds to R9AP. Our genetic experiments show that, even when both RGS7 and R9AP subunits are eliminated, RGS11 does not complex with a Gβ5/GPR179, showing that interchangeability and competition between individual RGS subunits do not occur in vivo. Even though RGS11 can directly associate with GPR179 in vitro, this binding does not provide proteolytic protection to the complex; thus, it is likely eliminated in native ON-BCs. In contrast, RGS11 complexes with R9AP and Gβ5 are stable and therefore are selectively expressed. Strikingly, the components of the distinct complexes are further integrated by the direct interaction between R9AP and GPR179 that involves cytoplasmic domains of the proteins, thereby leading to a distinct GPR179-RGS7-R9AP-RGS11 supramolecular assembly.
Figure 8.
Proposed model of the GAP complex configuration in ON-bipolar neurons. Cytosolic domains of GPR179 that comprise intracellular loops and coiled-coil containing RGS binding motif directly interact with SNARE-like intracellular domain of R9AP. R9AP binds exclusively to RGS11/Gβ5, and GPR179 binds exclusively to RGS7/Gβ5. Both protein complexes facilitate GTP hydrolysis on Gαo promoting its deactivation.
This model raises the question as to the significance of this particular complex organization. We know that RGS7 and RGS11 are redundant, and elimination of either protein alone is not sufficient to cause substantial alteration of signaling. Yet, the entire complex ultimately depends on GPR179 for its function. We think that this arrangement may reflect the need for the GAP complex to be targeted to the correct subcellular compartment. Indeed, the mGluR6 cascade is highly restricted to the dendritic tips of ON-BCs (Gregg et al., 2014), and its regulation likely requires that the GAP complex is present at the same domain. Consistent with this idea, GPR179 appears to contain elements that target it to the tips of ON-BC dendrites, whereas other elements of the GAP complex do not (Gregg et al., 2014; Ray et al., 2014). Furthermore, GPR179 may additionally integrate into the cascade by forming physical complexes with mGluR6 and TRPM1 (Orlandi et al., 2013; Ray et al., 2014). By making R9AP targeting dependent on GPR179, the cascade ensures that all of the elements involved in Gαo deactivation are present in the same place at the same time and with the correct stoichiometry. This organization appears to be distinct from rod photoreceptors that contain a similar GAP complex composed of RGS9-1, Gβ5, and R9AP. This GAP complex is targeted to the relevant photoreceptor compartment by R9AP, which contains intrinsic elements that guide its trafficking in this cell type (Pearring et al., 2014). Thus, the integrated arrangement of the GAP complex components in the ON-BCs may facilitate spatiotemporal regulation of the mGluR6 cascade to ensure timely and reliable generation of the light response.
Footnotes
This work was supported by National Institutes of Health Grants EY018139, MH105482, and DA026405 to K.A.M., EY 140701 to M.A.M., and EY12354 to R.G.G and through an unrestricted grant from the Research to Prevent Blindness to the Department of Ophthalmology & Visual Sciences, University of Louisville. We thank Natalia Martemyanova for performing genetic crosses needed to obtain mice used in these studies.
The authors declare no competing financial interests.
References
- Anderson GR, Semenov A, Song JH, Martemyanov KA. The membrane anchor R7BP controls the proteolytic stability of the striatal specific RGS protein, RGS9-2. J Biol Chem. 2007;282:4772–4781. doi: 10.1074/jbc.M610518200. [DOI] [PubMed] [Google Scholar]
- Anderson GR, Posokhova E, Martemyanov KA. The R7 RGS protein family: multi-subunit regulators of neuronal G protein signaling. Cell Biochem Biophys. 2009;54:33–46. doi: 10.1007/s12013-009-9052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SA, Martemyanov KA, Shavkunov AS, Arshavsky VY. Kinetic mechanism of RGS9-1 potentiation by R9AP. Biochemistry. 2006;45:10690–10697. doi: 10.1021/bi060376a. [DOI] [PubMed] [Google Scholar]
- Cabrera JL, de Freitas F, Satpaev DK, Slepak VZ. Identification of the G*5-RGS7 protein complex in the retina. Biochem Biophys Res Commun. 1998;249:898–902. doi: 10.1006/bbrc.1998.9218. [DOI] [PubMed] [Google Scholar]
- Cao Y, Song H, Okawa H, Sampath AP, Sokolov M, Martemyanov KA. Targeting of RGS7/Gbeta5 to the dendritic tips of ON-bipolar cells is independent of its association with membrane anchor R7BP. J Neurosci. 2008;28:10443–10449. doi: 10.1523/JNEUROSCI.3282-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Masuho I, Okawa H, Xie K, Asami J, Kammermeier PJ, Maddox DM, Furukawa T, Inoue T, Sampath AP, Martemyanov KA. Retina-specific GTPase accelerator RGS11/G beta 5S/R9AP is a constitutive heterotrimer selectively targeted to mGluR6 in ON-bipolar neurons. J Neurosci. 2009;29:9301–9313. doi: 10.1523/JNEUROSCI.1367-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Posokhova E, Martemyanov KA. TRPM1 forms complexes with nyctalopin in vivo and accumulates in postsynaptic compartment of ON-bipolar neurons in mGluR6-dependent manner. J Neurosci. 2011;31:11521–11526. doi: 10.1523/JNEUROSCI.1682-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Pahlberg J, Sarria I, Kamasawa N, Sampath AP, Martemyanov KA. Regulators of G protein signaling RGS7 and RGS11 determine the onset of the light response in ON bipolar neurons. Proc Natl Acad Sci U S A. 2012;109:7905–7910. doi: 10.1073/pnas.1202332109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Eversole-Cire P, Zhang H, Mancino V, Chen YJ, He W, Wensel TG, Simon MI. Instability of GGL domain-containing RGS proteins in mice lacking the G protein*-subunit G*5. Proc Natl Acad Sci U S A. 2003;100:6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FS, Shim H, Morhardt D, Dallman R, Krahn E, McWhinney L, Rao A, Gold SJ, Chen CK. Functional redundancy of R7 RGS proteins in ON-bipolar cell dendrites. Invest Ophthalmol Vis Sci. 2010;51:686–693. doi: 10.1167/iovs.09-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra A, Lyubarsky A, Jiang M, Pugh EN, Jr, Birnbaumer L, Sterling P, Vardi N. The light response of ON bipolar neurons requires G*o. J Neurosci. 2000;20:9053–9058. doi: 10.1523/JNEUROSCI.20-24-09053.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg RG, Ray TA, Hasan N, McCall MA, Peachey NS. Interdependence among members of the mGluR6 G-protein mediated Signalplex of retinal depolarizing bipolar cells. In: Martemyanov KA, editor. G protein signaling mechanisms in the retina. New York: Springer; 2014. pp. 67–79. [Google Scholar]
- Hollinger S, Hepler JR. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- Hooks SB, Waldo GL, Corbitt J, Bodor ET, Krumins AM, Harden TK. RGS6, RGS7, RGS9, and RGS11 stimulate GTPase activity of Gi family G-proteins with differential selectivity and maximal activity. J Biol Chem. 2003;278:10087–10093. doi: 10.1074/jbc.M211382200. [DOI] [PubMed] [Google Scholar]
- Hoon M, Okawa H, Della Santina L, Wong RO. Functional architecture of the retina: development and disease. Prog Retin Eye Res. 2014;42:44–84. doi: 10.1016/j.preteyeres.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett AC, Gray AJ, Hunter JM, Willardson BM. Role of molecular chaperones in G protein beta5/regulator of G protein signaling dimer assembly and G protein betagamma dimer specificity. J Biol Chem. 2009;284:16386–16399. doi: 10.1074/jbc.M900800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Wensel TG. R9AP, a membrane anchor for the photoreceptor GTPase accelerating protein, RGS9-1. Proc Natl Acad Sci U S A. 2002;99:9755–9760. doi: 10.1073/pnas.152094799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey BG, Morgans CW, Puthussery T, Wensel TG, Burke NS, Brown RL, Duvoisin RM. R9AP stabilizes RGS11-G beta5 and accelerates the early light response of ON-bipolar cells. Vis Neurosci. 2010;27:9–17. doi: 10.1017/S0952523809990319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keresztes G, Mutai H, Hibino H, Hudspeth AJ, Heller S. Expression patterns of the RGS9-1 anchoring protein R9AP in the chicken and mouse suggest multiple roles in the nervous system. Mol Cell Neurosci. 2003;24:687–695. doi: 10.1016/S1044-7431(03)00231-8. [DOI] [PubMed] [Google Scholar]
- Keresztes G, Martemyanov KA, Krispel CM, Mutai H, Yoo PJ, Maison SF, Burns ME, Arshavsky VY, Heller S. Absence of the RGS9 · Gβ5 GTPase-activating complex in photoreceptors of the R9AP knockout mouse. J Biol Chem. 2004;279:1581–1584. doi: 10.1074/jbc.C300456200. [DOI] [PubMed] [Google Scholar]
- Levay K, Cabrera JL, Satpaev DK, Slepak VZ. G*5 prevents the RGS7-G*o interaction through binding to a distinct Ggamma-like domain found in RGS7 and other RGS proteins. Proc Natl Acad Sci U S A. 1999;96:2503–2507. doi: 10.1073/pnas.96.5.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martemyanov KA, Yoo PJ, Skiba NP, Arshavsky VY. R7BP, a novel neuronal protein interacting with RGS proteins of the R7 family. J Biol Chem. 2005;280:5133–5136. doi: 10.1074/jbc.C400596200. [DOI] [PubMed] [Google Scholar]
- Masuho I, Celver J, Kovoor A, Martemyanov KA. Membrane anchor R9AP potentiates GTPase-accelerating protein activity of RGS11 · Gβ5 complex and accelerates inactivation of the mGluR6-Go signaling. J Biol Chem. 2010;285:4781–4787. doi: 10.1074/jbc.M109.058511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojumder DK, Qian Y, Wensel TG. Two R7 regulator of G-protein signaling proteins shape retinal bipolar cell signaling. J Neurosci. 2009;29:7753–7765. doi: 10.1523/JNEUROSCI.1794-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgans CW, Wensel TG, Brown RL, Perez-Leon JA, Bearnot B, Duvoisin RM. Gbeta5-RGS complexes co-localize with mGluR6 in retinal ON-bipolar cells. Eur J Neurosci. 2007;26:2899–2905. doi: 10.1111/j.1460-9568.2007.05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgans CW, Brown RL, Duvoisin RM. TRPM1: the endpoint of the mGluR6 signal transduction cascade in retinal ON-bipolar cells. Bioessays. 2010;32:609–614. doi: 10.1002/bies.200900198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi C, Posokhova E, Masuho I, Ray TA, Hasan N, Gregg RG, Martemyanov KA. GPR158/179 regulate G protein signaling by controlling localization and activity of the RGS7 complexes. J Cell Biol. 2012;197:711–719. doi: 10.1083/jcb.201202123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi C, Cao Y, Martemyanov KA. Orphan receptor GPR179 forms macromolecular complexes with components of metabotropic signaling cascade in retina ON-bipolar neurons. Invest Ophthalmol Vis Sci. 2013;54:7153–7161. doi: 10.1167/iovs.13-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi C, Xie K, Masuho I, Fajardo-Serrano A, Lujan R, Martemyanov KA. Orphan receptor GPR158 is an allosteric modulator of RGS7 catalytic activity with an essential role in dictating its expression and localization in the brain. J Biol Chem. 2015;290:13622–13639. doi: 10.1074/jbc.M115.645374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlberg J, Sampath AP. Visual threshold is set by linear and nonlinear mechanisms in the retina that mitigate noise: how neural circuits in the retina improve the signal-to-noise ratio of the single-photon response. Bioessays. 2011;33:438–447. doi: 10.1002/bies.201100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue MT, Peachey NS. Mouse b-wave mutants. Doc Ophthalmol. 2014;128:77–89. doi: 10.1007/s10633-013-9424-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peachey NS, Ray TA, Florijn R, Rowe LB, Sjoerdsma T, Contreras-Alcantara S, Baba K, Tosini G, Pozdeyev N, Iuvone PM, Bojang P, Jr, Pearring JN, Simonsz HJ, van Genderen M, Birch DG, Traboulsi EI, Dorfman A, Lopez I, Ren H, Goldberg AF, et al. GPR179 is required for depolarizing bipolar cell function and is mutated in autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet. 2012;90:331–339. doi: 10.1016/j.ajhg.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearring JN, Lieu EC, Winter JR, Baker SA, Arshavsky VY. R9AP targeting to rod outer segments is independent of rhodopsin and is guided by the SNARE homology domain. Mol Biol Cell. 2014;25:2644–2649. doi: 10.1091/mbc.E14-02-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Dallman R, Henderson S, Chen CK. Gbeta5 is required for normal light responses and morphology of retinal ON-bipolar cells. J Neurosci. 2007;27:14199–14204. doi: 10.1523/JNEUROSCI.4934-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray TA, Heath KM, Hasan N, Noel JM, Samuels IS, Martemyanov KA, Peachey NS, McCall MA, Gregg RG. GPR179 is required for high sensitivity of the mGluR6 signaling cascade in depolarizing bipolar cells. J Neurosci. 2014;34:6334–6343. doi: 10.1523/JNEUROSCI.4044-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath AP, Rieke F. Selective transmission of single photon responses by saturation at the rod-to-rod bipolar synapse. Neuron. 2004;41:431–443. doi: 10.1016/S0896-6273(04)00005-4. [DOI] [PubMed] [Google Scholar]
- Sarria I, Pahlberg J, Cao Y, Kolesnikov AV, Kefalov VJ, Sampath AP, Martemyanov KA. Sensitivity and kinetics of signal transmission at the first visual synapse differentially impact visually-guided behavior. eLife. 2015;4:e06358. doi: 10.7554/eLife.06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H, Wang CT, Chen YL, Chau VQ, Fu KG, Yang J, McQuiston AR, Fisher RA, Chen CK. Defective retinal depolarizing bipolar cells (DBCs) in regulators of G-protein signaling (RGS) 7 and 11 double null mice. J Biol Chem. 2012;287:14873–14879. doi: 10.1074/jbc.M112.345751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snellman J, Kaur T, Shen Y, Nawy S. Regulation of ON bipolar cell activity. Prog Retin Eye Res. 2008;27:450–463. doi: 10.1016/j.preteyeres.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow BE, Krumins AM, Brothers GM, Lee SF, Wall MA, Chung S, Mangion J, Arya S, Gilman AG, Siderovski DP. A G protein gamma subunit-like domain shared between RGS11 and other RGS proteins specifies binding to G*5 subunits. Proc Natl Acad Sci U S A. 1998;95:13307–13312. doi: 10.1073/pnas.95.22.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JH, Waataja JJ, Martemyanov KA. Subcellular targeting of RGS9-2 is controlled by multiple molecular determinants on its membrane anchor, R7BP. J Biol Chem. 2006;281:15361–15369. doi: 10.1074/jbc.M600749200. [DOI] [PubMed] [Google Scholar]
- Song JH, Song H, Wensel TG, Sokolov M, Martemyanov KA. Localization and differential interaction of R7 RGS proteins with their membrane anchors R7BP and R9AP in neurons of vertebrate retina. Mol Cell Neurosci. 2007;35:311–319. doi: 10.1016/j.mcn.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Taylor WR, Smith RG. Transmission of scotopic signals from the rod to rod-bipolar cell in the mammalian retina. Vision Res. 2004;44:3269–3276. doi: 10.1016/j.visres.2004.07.043. [DOI] [PubMed] [Google Scholar]
- Zeitz C, Robson AG, Audo I. Congenital stationary night blindness: an analysis and update of genotype-phenotype correlations and pathogenic mechanisms. Prog Retin Eye Res. 2015;45:58–110. doi: 10.1016/j.preteyeres.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang J, Jeffrey BG, Morgans CW, Burke NS, Haley TL, Duvoisin RM, Brown RL. RGS7 and −11 complexes accelerate the ON-bipolar cell light response. Invest Ophthalmol Vis Sci. 2010;51:1121–1129. doi: 10.1167/iovs.09-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]