Abstract
Bdellovibrio bacteriovorus is a Gram-negative bacterium that belongs to the delta subgroup of proteobacteria and is characterized by a predatory life cycle. In recent years, work has highlighted the potential use of this predator to control bacteria and biofilms. Traditionally, the reduction in prey cells was used to monitor predation dynamics. In this study, we introduced pMQ414, a plasmid that expresses the tdTomato fluorescent reporter protein, into a host-independent strain and a host-dependent strain of B. bacteriovorus 109J. The new construct was used to conveniently monitor predator proliferation in real time, in different growth conditions, in the presence of lytic enzymes, and on several prey bacteria, replicating previous studies that used plaque analysis to quantify B. bacteriovorus. The new fluorescent plasmid also enabled us to visualize the predator in liquid cultures, in the context of a biofilm, and in association with human epithelial cells.
INTRODUCTION
Bdellovibrio bacteriovorus is a Gram-negative motile bacterium that exhibits a predatory life cycle. Bdellovibrios are known to attack other Gram-negative bacteria by attaching to the prey/host cell, entering into the host cell periplasm, forming a structure known as a bdelloplast, growing in the periplasmic space, and, finally, releasing numerous progeny into the environment to start a new predation cycle (1).
In the past few years, the introduction of new genetic tools and a better understanding of the genetic makeup of predatory bacteria have helped to move the field forward (2, 3). Furthermore, increasing antibiotic resistance among Gram-negative pathogens has evoked new interest in the potential use of predatory bacteria for therapeutics (4, 5). Throughout the years, several methods were used to measure predation and predator activity. Predation, or the change in the prey bacteria population, is frequently evaluated indirectly by the reduction in prey cell culture turbidity or directly by standard dilution plating and viability counts of the prey (6). Luminescent prey was also used to monitor changes in the prey population and study the impact of altering specific predator genes in the predation process (7, 8). Double-layered agar plating and PFU enumeration are widely used to detect and quantify predator biomass and to investigate predator host specificity (6). Additional methods utilize PCR and real-time quantitative PCR to determine the presence and bioload of predatory bacteria (9–11). Labeled oligonucleotide probes that target Bdellovibrio 16S rRNA were also used to visualize the predator by use of in situ fluorescence hybridization (12).
In our study, tdTomato fluorescent protein was expressed in a host-independent strain and a host-dependent strain of B. bacteriovorus 109J. The reporter protein was used to monitor predator population growth in real time and to better track the predator. Here, the red fluorescent tdTomato protein was used because it is the brightest and most photostable red fluorescent protein (13). This fluorescent marker was validated by recapitulating key findings from previous studies on B. bacteriovorus predation that used traditional methods, and the marker was further used to evaluate the interaction of B. bacteriovorus with biofilms and human epithelial cells in vitro.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The predatory bacteria used in the study were Bdellovibrio bacteriovorus 109J (ATCC 43826) and a facultative host-independent (HI) variant of B. bacteriovorus 109J, i.e., HI-A (14). As prey, Bdellovibrio was routinely cultured on Escherichia coli S17-1. Additional prey microorganisms included E. coli ZK2686 (15) and WM3064, a diaminopimelic acid auxotroph (16), Acinetobacter baumannii ATCC 19606, Pseudomonas aeruginosa UCBPP-PA14 (PA14), Klebsiella pneumoniae ATCC 13883, Enterococcus faecalis ATCC 51229, Staphylococcus epidermidis ATCC 12228, Staphylococcus aureus SH1000, and the yeast Candida albicans ATCC 90029. Prey cells were grown and maintained in lysogeny broth (LB) or yeast mold broth for C. albicans. B. bacteriovorus was cultured as described previously (17). Predator stock lysates were prepared by coculturing prey cells with the predators in dilute nutrient broth (DNB), a 1:10 dilution of nutrient broth amended with 3 mM MgCl2 and 2 mM CaCl2. The cultures were incubated at 30°C. To harvest the predators, cocultures were prepared by adding 2 ml of washed host cells (∼1 × 109 CFU/ml) to 2 ml of predatory bacteria stock lysate in 20 ml of DNB. Cultures were incubated overnight until the predator reached a final concentration of ∼1 × 108 PFU/ml. Thereafter, the lysates were filtered through a 0.45-μm-pore-size Millex filter (Millipore, Billerica, MA) (harvested predator). HI-A was cultured on peptone-yeast extract (PYE) medium (10 g/liter peptone, 3 g/liter yeast extract, amended with 3 mM MgCl2 and 2 mM CaCl2) at 30°C (14). Additional experiments were conducted by placing the cultures at elevated temperatures.
Predation experiments.
Predation experiments were conducted as described before (5, 17). Cocultures were prepared by addition of 1 ml of harvested predators (∼1 × 108 PFU/ml) to 1 ml of DNB-washed prey cells (∼1 × 109 CFU/ml) and 10 ml of DNB medium. The cultures were placed on a rotary shaker at 30°C. The change in the prey population was measured by dilution plating and enumeration of CFUs. Predatory bacteria were enumerated as PFUs (6). For predation experiments, cocultures were prepared in 96-well plates as described above, and 160-μl aliquots were inserted in each well. The plate was placed in a Synergy H1 hybrid multimode microplate reader (BioTek, Winooski, VT). For growth curves, the microplate reader was set to 30°C with shaking. The change in prey population was measured by culture turbidity (optical density at 600 nm [OD600]). The change in predator population was measured by fluorescence at 548-nm excitation and 586-nm emission. To measure predation dynamics on inactivated cells, prey cells were heated to 65°C for 20 and 40 min. Plating was used to confirm the loss of cell viability, and light microscopy confirmed that the host cells maintained structural integrity. Predation on surface-attached cells was done as previously described (18). Briefly, green fluorescent protein (GFP)-labeled E. coli strain ZK2686 bacteria were grown on glass-bottomed 6-well plates for 24 h, after which the biofilm was washed with phosphate-buffered saline (PBS) to remove unattached cells. One hundred microliters of harvested predator was added to the well and incubated for 30 min to 3 h. Bacteria were observed by confocal microscopy by use of an Olympus FluoView FV-1000 laser scanner confocal microscope with a 60× oil immersion objective equipped with FluoView image viewing software version 3.1.
Construction of a B. bacteriovorus expressing tdTomato.
To express tdtomato in B. bacteriovorus, we chose to use the IncQ family RFS1010 replicon, as it has been reported to support plasmid autonomous replication in that species (19, 20). While the copy number for this replicon has not been reported for B. bacteriovorus, it has been documented to be maintained at 10 to 12 copies per chromosome in at least two different Gram-negative bacteria (21). The RSF1010 replicon was amplified from pMMB66EH (22) using primers 2955 (TACGAAGTTATATTAAGGGTTGTCGAGCCGCTGGTGCCGCGCAATACTGTGTTTACATAC) and 2958 (CCCAACAGTTGCGCAGCCTGAAAGGCAGGCCGGGCCCCTTTTCTGAGCATGGTATTTTTC).
Recombination with Saccharomyces cerevisiae was used to replace the pRO1600 replicon on plasmid pMQ125 with the RSF1010 replicon (23, 24) to make pMQ397. The PBAD expression system on pMQ397 was replaced by an nptII promoter-tdtomato fusion-containing amplicon from pMQ361 (25) using in vivo yeast cloning (23, 24), resulting in pMQ414. The PnptII-tdtomato region was amplified with primers 3056 (CCCGCGCGTTGGCCGATTCATTAATGCAGCTGGCCCGGGACGCTGCCGCAAGCACTCAGG) and 3057 (CAGACCGCTTCTGCGTTCTGATTTAATCTGTATCAGGATCCTTACTTGTACAGCTCGTCC). The pMQ414 construct has the tdtomato gene under transcriptional regulation of the PnptII promoter, p15a, and broad-host-range RSF1010 bacterial replicons, an aacC-1 gene for selection with gentamicin, an RP4 origin of conjugal transfer, and yeast replication machinery with the S. cerevisiae URA3 gene for selection. The pMQ414 plasmid was verified by PCR and the ability to confer red fluorescence to E. coli. pMQ414 was introduced into wild-type and HI-A variants of B. bacteriovorus by conjugation (14, 24). A diagram of pMQ414 is shown in Fig. S1 in the supplemental material.
Recipient B. bacteriovorus HI-A was grown for 2 days in PYE medium. Wild-type B. bacteriovorus was grown as described above (harvested predator). Donor E. coli strain WM3064 bearing pMQ414 was grown to log phase (OD600 = 0.6 to 0.8) in medium containing 0.3 mM diaminopimelic acid (DAP) and 10 μg/ml gentamicin (Gm). Recipient predatory bacteria were added at a 1:1 (vol/vol) ratio to the donor. Fifty microliters of the sample was spotted on PYE or DNB plates supplemented with DAP for HI-A and wild-type B. bacteriovorus, respectively. Plates were incubated at 30°C for 24 h. To select for HI-A plasmid recipients, cells were scraped from the PYE plate and resuspended in 1 ml of PYE broth, and 100-μl aliquots were plated on PYE agar plates containing Gm, to select for plasmid recipients, and no DAP, to select against E. coli. Plates were incubated for 3 days at 30°C until HI-A Gm-resistant colonies developed. A single colony was isolated and recultured on PYE Gm (referred to herein as BbHIpMQ). To select for wild-type plasmid recipients, cells were scraped from the DNB plate and cocultured with Gm-resistant E. coli S17-1 carrying plasmid pMQ414 (24). Cocultures were prepared in DAP-free DNB medium and supplemented with Gm (10 μg/ml). After 48 h, the culture was plated on prey cells in the presence of Gm, and a single plaque was isolated and recultured in broth (referred to herein as BbpMQ for simplicity).
Microscopy of B. bacteriovorus-prey interactions.
Cocultures were prepared as described above by using BbpMQ and E. coli strain ZK2686 bearing the pGFPmut2 plasmid as prey (26). The coculture was incubated for 3 to 4 h at 30°C and then used directly for microscopy. Alternatively, E. coli with pGFPMut2 was grown overnight in a 6-well plate with a glass bottom (MatTek Corp., Ashland, MA) at an angle to form a biofilm at the air-liquid interface across the center of the bottom of the well (27). Unattached bacteria were removed by aspiration of the LB medium and gentle rinsing with PBS (3 ml), and 3 ml of LB with 500 μl of the BbpMQ-purified lysate was added. After 3 to 4 h, the LB medium was removed and replaced with PBS, and the biofilms were imaged.
Bacteria were imaged with 60× or 100× oil immersion objectives using an Olympus IX81 inverted microscope with an FV1000 laser scanning confocal system (Olympus) and FluoView FV10-ASW 3.1 imaging software. An Olympus IX73 inverted fluorescence microscope was used for biofilm images, and the images were examined by cellSens Dimension software (Olympus). The imaging experiments were repeated three times with similar results.
Attachment of B. bacteriovorus to human corneal limbal epithelial cells.
Human corneal limbal epithelial cells (HCLE) cells were grown to confluence on poly-l-lysine-treated 12-well glass-bottomed dishes (MatTek no. P12G-1.5-14-F) to facilitate microscopy, as previously described (25). HCLE layers were coincubated with 3.67 × 1011 PFU/ml of BbpMQ. After 2 h of incubation at 37°C with 5% CO2, the wells were rinsed twice with 1 ml PBS to remove unattached bacteria, stained with calcein AM viability stain (0.5 μM) for 15 min, and resuspended in tissue culture medium. The cells and bacteria were imaged as noted for the biofilm studies above. The experiment was performed twice.
RESULTS
Expression of tdTomato fluorescent protein in a host-independent variant of B. bacteriovorus 109J.
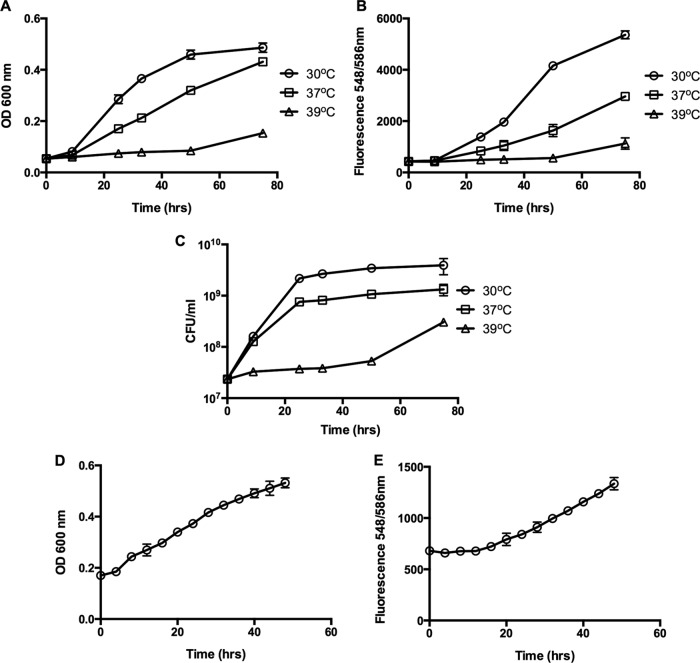
In order to determine if the tdTomato protein can be expressed in B. bacteriovorus, plasmid pMQ414 was moved into a B. bacteriovorus 109J HI variant by conjugation to generate strain BbHIpMQ. When the strain was cultured in broth and examined by fluorescence microscopy, a clear expression of the red fluorescent protein was seen (Fig. 1). BbHIpMQ was also able to form plaques when spotted on a lawn of host cells, which is characteristic of HI variants of B. bacteriovorus. To examine if fluorescence can be used to monitor HI cell growth, cultures were grown in PYE medium and placed at 30°C, 37°C, and 39°C. The bacteria were cultured for 75 h. Aliquots were removed every 12 h, and HI growth was measured by the change in culture turbidity, fluorescence, and dilution plating (Fig. 2A, B, and C, respectively). As seen in Fig. 2, similar growth patterns were registered by all three growth detection methods. Higher fluorescence readings were measured at 30°C than at higher temperatures. To measure the HI growth pattern in real time, BbHIpMQ was cultured for 48 h in a 96-well plate placed at 30°C with a microplate reader. Growth was measured by culture turbidity (Fig. 2D) and fluorescence (Fig. 2E). Similar growth patterns were observed by the two methods. No fluorescent signals were measured in HI cells that were not expressing tdTomato (data not shown).
FIG 1.
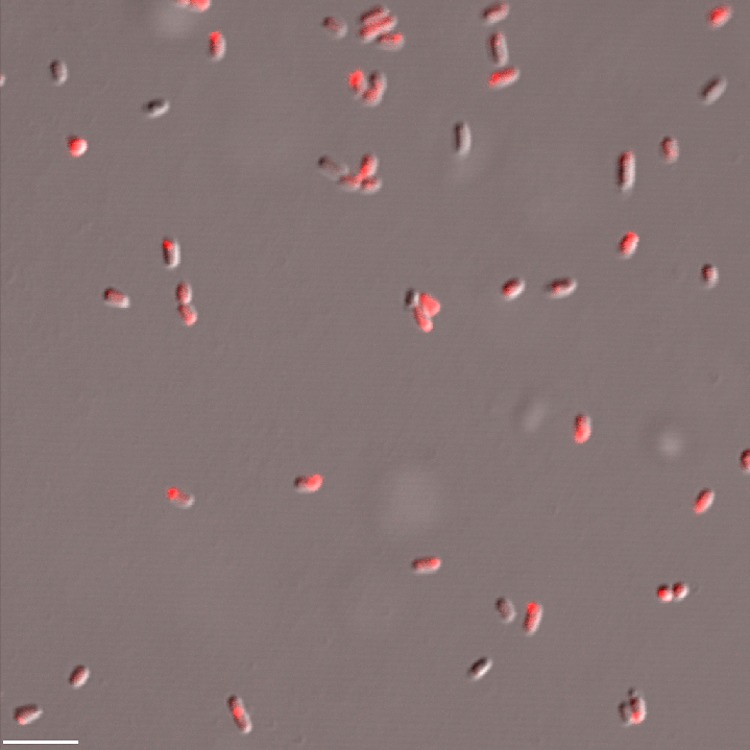
Fluorescence microscopy of HI variants expressing tdTomato. BbHIpMQ was grown in PYE medium for 24 h and examined under an Olympus IX81 inverted confocal fluorescence microscope using a 60× oil objective. Image shows an overlay of bright-field and tetramethyl rhodamine isocyanate (TRITC) filter images. Bar = 5 μm.
FIG 2.
Growth of HI variant expressing tdTomato. (A to C) BbHIpMQ was grown in PYE medium at different temperatures. Cell growth was measured by the change in culture turbidity (A), fluorescence (B), and cell enumeration (C). Each experiment was conducted in triplicate, with each value representing the mean and standard deviation. (D, E) BbHIpMQ cells were cultured in a 96-well plate and grown in a Synergy H1 microplate reader at 30°C. Growth was measured by the change in culture turbidity (D) and fluorescence (E). Each value represents the mean and standard deviation of 8 wells.
Expression of tdTomato fluorescent protein in a host-dependent B. bacteriovorus 109J.
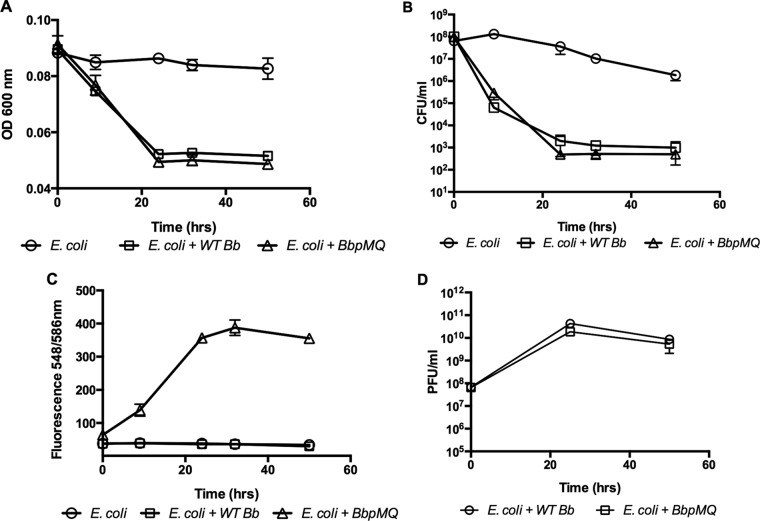
After confirming that the red fluorescent protein can be expressed in the HI-A variant, pMQ414 was mobilized into wild-type B. bacteriovorus to create strain BbpMQ. BbpMQ was cocultured with E. coli prey cells, and the changes in prey and predator quantities were evaluated. A reduction in prey cells was measured by cell turbidity and dilution plating 9 h after incubation (Fig. 3A and B, respectively). Fluorescent measurements and PFU enumeration both confirmed predator proliferation (Fig. 3C and D, respectively). The growth patterns measured by fluorescence correlated with an increase in PFU numbers and were in agreement with the reduction pattern in the prey cell population. Reductions in PFUs and fluorescence readings were detected following 32 h of incubation and may have occurred in response to predator death following the reduction in prey cell numbers. No fluorescent signal was measured in cocultures that were incubated with wild-type B. bacteriovorus (Fig. 3C, squares). These experiments demonstrated that BbpMQ has growth and predation characteristics that are similar to those of the wild-type B. bacteriovorus (Fig. 3). In a separate experiment, BbpMQ cells were concentrated by centrifugation, and PFU enumeration was conducted in concert with fluorescence readings. The relative fluorescence readings of 4,710, 2,295, 1,215, 738, 420, 214, 155, 114, 106, and 84 were correlated to 5 × 1012, 1 × 1012, 5 × 1011 1 × 1011, 5 × 1010, 1 × 1010, 5 × 109, 1 × 109, 5 × 108, and 1 × 108 PFU/ml. When cocultures were examined by fluorescence microscopy in the presence of E. coli expressing GFP, attack-phase B. bacteriovorus cells expressing the red protein were clearly seen (Fig. 4A). Over time, the appearance of spherical green bdelloplast prey cells containing red Bdellovibrio cells were detected (Fig. 4A to C, arrows). To examine predation on surface-attached cells (Fig. 4C) and biofilms (Fig. 4D), E. coli biofilms were developed on glass-bottomed multiwell dishes and incubated with BbpMQ. After 30 min of coincubation and washing, red fluorescent bacteria penetrated the GFP-labeled E. coli biofilms (Fig. 4D).
FIG 3.
Growth of host-dependent B. bacteriovorus expressing tdTomato. Cocultures of B. bacteriovorus and E. coli prey (E. coli + WT Bb), B. bacteriovorus expressing tdTomato and E. coli prey (E. coli + BbpMQ), and E. coli control were grown for 50 h at 30°C. The change in prey cell population was measured by the change in culture turbidity (A) and by dilution plating (B). The change in predator population was measured by fluorescence (C) and plaque enumeration (D). Experiments were conducted in triplicate, with each value representing the mean and standard deviation.
FIG 4.
Fluorescence microscopy of B. bacteriovorus expressing tdTomato. Cocultures of BbpMQ and E. coli prey expressing GFP were grown for 4 h. Images show attack-phase Bdellovibrio cells (A) and E. coli bdelloplasts (green) containing Bdellovibrio cells (red) (B). (C, D) Predation on surface-attached cells. E. coli GFP-labeled bacteria were developed on glass-bottomed multiwell plates and incubated for 30 min with BbpMQ. Wells were washed to remove unattached bacteria and examined using confocal laser scanning microscopy. Bars = 5 μm. Arrows indicate an E. coli bdelloplast (green) containing Bdellovibrio cells (red).
Measuring B. bacteriovorus growth and predation under different growth conditions.
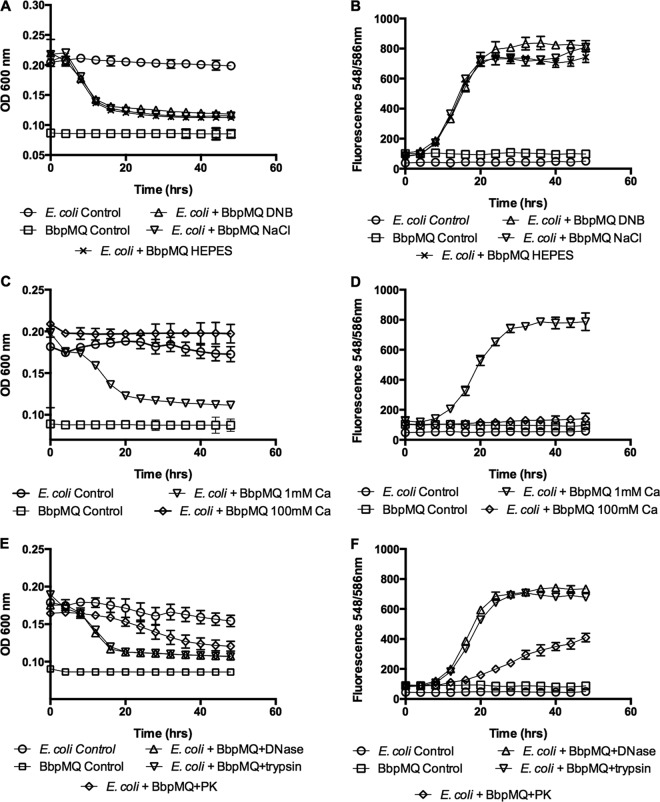
To measure predation dynamics in real time, cocultures were placed in 96-well plates for 48 h, and predation was measured by the change in culture turbidity, showing the reduction in prey and the increase in fluorescence, thus indicating proliferation of the predatory bacteria. The cocultures were prepared in DNB medium, HEPES buffer (25 mM), and saline (0.9% NaCl), with calcium (3 mM CaCl2) and magnesium (2 mM MgCl2). As shown in Fig. 5A and B, similar predation dynamics were observed in all three media; however, a slight increase in fluorescence was seen in DNB cocultures between 20 and 40 h. The effect of high salt levels on predation dynamics was also investigated. Cocultures were placed in 96-well plates containing 1 and 100 mM CaCl2 in DNB medium. The presence of 1 mM CaCl2 had no obvious effect on predation, as measured by the reduction in prey cell turbidity and predator fluorescence, whereas a high salt concentration diminished predation (Fig. 5C and D). Similarly, addition of DNase and trypsin (100 μg/ml) to the coculture did not alter predation dynamics or predator growth; however, the presence of proteinase K (100 μg/ml) reduced both prey and predator growth (Fig. 5E and F). No reduction in host population or increase in fluorescence was measured in the control cultures containing prey E. coli or predator BbpMQ alone.
FIG 5.
Measurement of predation in real time and under different growth conditions. (A, B) Predation in different media. Cocultures of E. coli prey (E. coli) and B. bacteriovorus expressing tdTomato (BbpMQ) were prepared in 96-well plates and incubated for 48 h in a Synergy H1 microplate reader. Cocultures were prepared in DNB medium, NaCl, and HEPES buffer. (C, D) Predation in high salt concentration. Cocultures of E. coli prey and BbpMQ were prepared in DNB medium containing 1 mM CaCl2 (1 mM Ca) or 100 mM CaCl2 (100 mM Ca). (E, F) Predation in the presence of enzymes. Cocultures were prepared in DNB medium containing no enzyme addition (DNB, control), DNase, trypsin, or protease K (PK). The change in prey cell population was measured by the change in culture turbidity (A, C, E). The change in predator population was measured by fluorescence (B, D, F). Each value represents the mean of results for 5 wells from one representative experiment. Error bars are shown as 1 SD. Each experiment was carried out twice, yielding similar results.
To investigate if the addition of Gm is required for maintenance of the pMQ414 plasmid, cocultures were prepared by adding Gm-resistant E. coli prey, with or without Gm, to BbpMQ prey cells. The cultures were incubated for 48 h in a Synergy H1 microplate reader, and predator growth was measured by fluorescence. No significant change (P = 0.2) in fluorescent signal was measured between the Gm-supplemented coculture and the Gm-free coculture (from a fluorescence reading of 66 ± 6 at time 0 to 596 ± 21 and 547 ± 63 at 48 h for Gm and no-Gm cocultures, respectively), supporting that the plasmid is stable for at least 48 h in the absence of antibiotic selection. Fluorescence was also maintained after cultures of BbpMQ and BbHIpMQ were initiated from frozen stocks and grown in the presence of Gm.
Predation dynamics on different host cells.
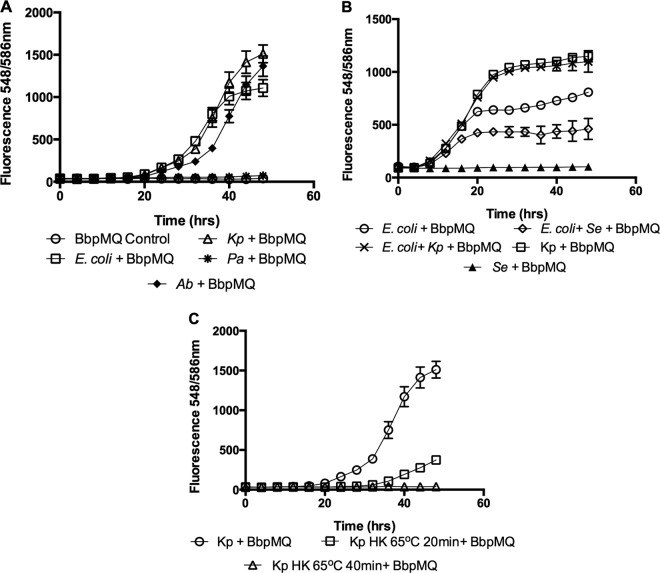
To evaluate the ability of B. bacteriovorus to proliferate on different Gram-negative prey cells in real time, cocultures were prepared by using BbpMQ as the predator and 7.4 ± 1 × 108 CFU/ml of E. coli, A. baumannii, P. aeruginosa strain PA14, and K. pneumoniae as prey. Cocultures were placed in 96-well plates in a Synergy H1 microplate reader for 48 h. The change in predator population was measured by fluorescence. Similar levels of prey proliferation were measured when E. coli and A. baumannii were used as prey. The highest predator fluorescence reading was seen with K. pneumoniae, with very little increase in fluorescence measured when P. aeruginosa was used as prey (from a fluorescence reading of 35 ± 6 at time 0 to 72 ± 5 at 48 h; P = 0.0001). No increase in fluorescence was measured in the prey-free control (from 43 ± 6 at time 0 to 46 ± 9 at 48 h; P = 0.5) (Fig. 6A). Similarly, no increase was seen in BbpMQ fluorescence measured following 48 h predation on nonhost cells, including the yeast Candida albicans ATCC 90029 (from a fluorescence reading of 83 ± 7 at time 0 to 81 ± 5.9 at 48 h; P = 0.86), the Gram-positive bacteria Enterococcus faecalis ATCC 29212 (from a fluorescence reading of 120 ± 6 at time 0 to 122 ± 13 at 48 h; P = 0.6), Staphylococcus epidermidis ATCC 12228 (from a fluorescence reading of 91 ± 7 at time 0 to 102 ± 19 at 48 h; P = 0.16), and Staphylococcus aureus SH1000 (from a fluorescence reading of 73 ± 3 at time 0 to 81 ± 8 at 48 h; P = 0.13).
FIG 6.
Measurement of predation in real time on prey cells. (A) Predation on different prey cells. K. pneumoniae (Kp), E. coli (E. coli), P. aeruginosa (Pa), and A. baumannii (Ab) prey cells were cocultured with B. bacteriovorus expressing tdTomato (BbpMQ). (B) Predation in the presence of decoy. To measure the growth dynamics in the predator population while preying on host alone or host in the presence of nonhost, cocultures were prepared using BbpMQ cocultured in the presence of E. coli (E. coli + BbpMQ), E. coli and K. pneumoniae (E. coli + Kp + BbpMQ), or E. coli and S. epidermidis (E. coli + Se + BbpMQ). (C) Predation on heat-killed prey. Cocultures were prepared using BbpMQ and viable K. pneumoniae prey cells (Kp + BbpMQ) or prey cells that were heat killed at 65°C for 20 and 40 min. Cocultures were placed in 96-well plates and incubated for 48 h in a Synergy H1 microplate reader. The change in predator population was measured by fluorescence. Each value represents the mean of 5 wells from one representative experiment. Error bars are shown as 1 SD. Each experiment was carried out twice, yielding similar results.
To measure the growth dynamics in predator populations while preying on host alone or host in the presence of nonhost (decoy) bacteria, cocultures were prepared with BbpMQ as the predator with E. coli alone, a 1:1 (vol/vol) ratio of E. coli and K. pneumoniae, or a 1:1 (vol/vol) ratio of E. coli and S. epidermidis. The addition of K. pneumoniae to E. coli prey resulted in an increase in predator growth, whereas addition of a nonprey decoy bacterium reduced proliferation of the predator compared to that with E. coli alone (Fig. 6B). The effect of heat-killed prey on the proliferation of the predator was also examined. K. pneumoniae cells were heated to 65°C for 20 and 40 min. Thereafter, BbpMQ was added, and the plates were incubated for 48 h in a microplate reader. As seen in Fig. 6C, preheating the prey cells for 20 and 40 min reduced the final predator fluorescence by 75% and 97%, respectively, compared to that in the non-heat-killed control. These data were in line with the measured change in prey cell turbidity (40%, 33%, and 3.5% reductions for treatment with 0, 20, and 40 min of heat, respectively). Similar findings were seen with E. coli (94% reduction following 40 min heat treatment) and A. baumannii (67% reduction following 40 min heat treatment), indicating that heat-killed cells are less adequate for predator proliferation.
Interaction of B. bacteriovorus 109J with human corneal epithelial cells.
The interaction of predatory bacteria with the mammalian epithelium is not well studied yet is important to understand if B. bacteriovorus is used as an alternative therapy for treatment of multidrug-resistant bacteria. BbpMQ was coincubated with HCLE cells at a multiplicity of infection (MOI) of 3 × 106. After 2 h, unattached or weakly attached cells were removed by washing with PBS, and the HCLE cells were stained with calcein AM, a green fluorescent viability dye. HCLE and B. bacteriovorus cells were observed by confocal laser scanning microscopy (Fig. 7). No red fluorescence was observed in the absence of B. bacteriovorus (Fig. 7A). Fifteen fields of HCLE cells were counted from two separate experiments representing 77 HCLE cells. Only 11.2 ± 3.4 bacterial cells per human epithelial cell were observed after 2 h, indicating a very low rate of attachment or uptake given the exceedingly high MOI. Similar to the results of a previous study on the effect of B. bacteriovorus on HCLE cell viability tested by using resazurin-based assays (28), calcein AM staining (green dye) showed that the HCLE cells remained viable despite being incubated with B. bacteriovorus (Fig. 7B).
FIG 7.
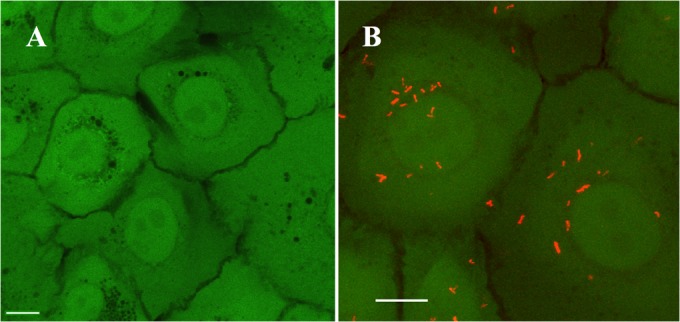
Interaction of B. bacteriovorus with a human epithelial cell line in vitro. HCLE monolayers were stained with green viability dye (calcein AM) and imaged with confocal laser microscopy. (A) Control HCLE cells without B. bacteriovorus. (B) HCLE cells were imaged after incubation with B. bacteriovorus BbpMQ at an MOI of 3 × 106 for 2 h. The red dots are B. bacteriovorus cells expressing tdTomato (BbpMQ) attached to epithelial cells. Bars = 5 μm.
DISCUSSION
From the time that predatory bacteria were first isolated in the early 1960s, researchers have used various methods to study the growth and proliferation of these unique bacteria. Given the obligate predator characteristics of these organisms, predator-prey cocultures need to be used. The use of cocultures allowed the change in prey numbers to be used as an indicator of predation. The most direct manner by which to measure the change in prey population is measurement of the reduction in culture turbidity, which occurs as the predator escapes from the bdelloplast (6). Although this method may be used in real time to measure predation, it provides only a partial indication that predation did occur. A more accurate manner of measuring the extent of predation is by the change in prey cell numbers during predation. However, as this method calls for dilution plating and viability counts of the prey, it does not allow real-time estimation of predation. In addition, the method is time-consuming, primarily when evaluating multiple cocultures. To allow real-time measurement of predation, researchers have used bioluminescent prey (7, 8). Although monitoring of the change in prey population does allow evaluation of predation and predation dynamics, it shows only one aspect of predation, with little insight into the actual proliferation of the predator.
When plated in the presence of high-density prey cells, predatory bacteria, like bacteriophages, form plaques, allowing quantification of the predator population by PFU counts (6). While an accurate measure, plaque formation usually requires several days to develop and cannot be used to monitor predator growth in real time. The time required to conduct doubled-layered agar plating is probably the main reason that the change in prey population and not the predator population is often used to monitor predation and predator biology.
In this study, we utilized a tdTomato fluorescent protein to measure the growth of B. bacteriovorus (29). The tdTomato fluorescent protein was expressed in both the host-independent (HI) and the wild-type variants of B. bacteriovorus, allowing visualization of the Bdellovibrio cells during predation. In the past few years, fluorescence microscopy was used to visualize Bdellovibrio growth within a fluorescent prey (30, 31). Expressing GFP and teal fluorescent protein in Bdellovibrio was also utilized to observe predation on the cellular level and to investigate the role of specific Bdellovibrio genes in this process (20, 32).
Patterns of HI cell growth measured by fluorescence were similar to those measured by CFU enumeration and optical density. Optimal growth was measured at 30°C, consistent with previous studies showing reduced Bdellovibrio HI cell growth at higher temperatures (33). When placed in cocultures, similar predator growth patterns were measured for both PFU enumeration and fluorescence, which correlated with predation dynamics measured by the reduction in prey cell population. By using a plate reader, we were able to monitor predation dynamics in real time. This system allowed us to conduct experiments in a 96-well plate format at various temperatures and shaking speeds; similar capabilities are available for other plate readers by several manufacturers. As noted above, predation dynamics measured by the reduction in culture turbidity was also noted by the increase in fluorescence. Data collected using this approach was in agreement with data from other studies; e.g., survival and abundance of predatory bacteria have been found to be negatively impacted by high salinity (34, 35), proteinase K but not trypsin or DNase can impair predation (36), B. bacteriovorus demonstrates dissimilar capacities to prey on different host bacteria (37), and decoy bacteria, such as Gram-positive bacteria, can competitively hinder predation (38, 39).
The data demonstrated penetration of B. bacteriovorus into biofilms. Although the biofilms were relatively immature at 24 h, the predatory bacteria penetrated into the microcolonies on the plastic surface. Bdelloplasts were clearly observed after 30 min of interaction with E. coli. The B. bacteriovorus in the bdelloplasts was visible but was relatively dim, perhaps because the fluorescence signal was reduced, as it had to travel through the bacterial cell wall and membranes or was quenched by factors within the host. It is also possible that the nptII promoter had reduced expression within the bdelloplast.
Culture of the predator in the presence of a heat-deactivated host revealed an inverse correlation between predator proliferation and the temperature by which the prey was heat killed. This finding is in agreement with earlier studies showing that B. bacteriovorus was unable to prey on E. coli cells that were killed by high heat (98°C or 120°C for 15 min), was less capable of growing on prey that was heat killed by a less severe heat treatment (70°C), and preyed “normally” on prey that was inactivated by UV irradiation (40).
In addition to validating previous studies that used fluorescence rather than PFU as an output, we investigated the interaction of B. bacteriovorus with an ocular epithelial cell line in vitro. With an MOI of >1 million, there was a very low level of attachment of B. bacteriovorus to HCLE cells at 2 h. Therefore, the pMQ414 plasmid can be of use in defining the interaction of B. bacteriovorus with cultured cells in future studies.
In conclusion, a tdTomato fluorescent protein was expressed in B. bacteriovorus. Expression of the protein was found to be a convenient and adequate tool for monitoring predator growth during predation, monitoring growth in real time, and visualizing different predator growth stages by microscopy. As the data gathered in this study support those of other studies that use the reduction in prey population to monitor predation, we believe that this fluorescent construct may be an important tool that will aid in the study of predatory bacterial biology and predator-prey interaction. For example, this plasmid can be used with a number of Bdellovibrio and potentially other Gram-negative predatory bacteria to track predation in situ and to study the spatial distribution of predators in the microenvironment, in addition to its clear utility in the analysis of predator-prey relationships with fluorescence microscopy or flow cytometry.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kira Lathrop for assistance with fluorescence microscopy with the NIH-supported (P30 EY08098) imaging module.
This work was partly supported by the Department of the Army (USAMRAA W81XWH-12-2-0067) and by unrestricted funds from the Oral Biology Department to D.E.K. R.M.Q.S. was supported by NIH grant AI085570, the Eye and Ear Foundation of Pittsburgh, and unrestricted funds from Research to Prevent Blindness. K.M.B. was supported by NIH grant EY017271.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03611-15.
REFERENCES
- 1.Stolp H, Starr MP. 1963. Bdellovibrio bacteriovorus gen. et sp. n., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie Van Leeuwenhoek 29:217–248. doi: 10.1007/BF02046064. [DOI] [PubMed] [Google Scholar]
- 2.Pasternak Z, Njagi M, Shani Y, Chanyi R, Rotem O, Lurie-Weinberger MN, Koval S, Pietrokovski S, Gophna U, Jurkevitch E. 2014. In and out: an analysis of epibiotic vs periplasmic bacterial predators. ISME J 8:625–635. doi: 10.1038/ismej.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A, Meyer F, Sockett RE, Schuster SC. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 4.Dwidar M, Monnappa AK, Mitchell RJ. 2012. The dual probiotic and antibiotic nature of Bdellovibrio bacteriovorus. BMB Rep 45:71–78. doi: 10.5483/BMBRep.2012.45.2.71. [DOI] [PubMed] [Google Scholar]
- 5.Kadouri DE, To K, Shanks RM, Doi Y. 2013. Predatory bacteria: a potential ally against multidrug-resistant Gram-negative pathogens. PLoS One 8:e63397. doi: 10.1371/journal.pone.0063397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jurkevitch E. 2006. Isolation and classification of Bdellovibrio and like organisms. Curr Protoc Microbiol Chapter 7:Unit 7B.1. doi: 10.1002/9780471729259.mc07b01s00. [DOI] [PubMed] [Google Scholar]
- 7.Lambert C, Smith MC, Sockett RE. 2003. A novel assay to monitor predator-prey interactions for Bdellovibrio bacteriovorus 109J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ Microbiol 5:127–132. doi: 10.1046/j.1462-2920.2003.00385.x. [DOI] [PubMed] [Google Scholar]
- 8.Im H, Kim D, Ghim CM, Mitchell RJ. 2014. Shedding light on microbial predator-prey population dynamics using a quantitative bioluminescence assay. Microb Ecol 67:167–176. doi: 10.1007/s00248-013-0323-z. [DOI] [PubMed] [Google Scholar]
- 9.Jurkevitch E, Ramati B. 2000. Design and uses of Bdellovibrio 16S rRNA-targeted oligonucleotides. FEMS Microbiol Lett 184:265–271. doi: 10.1111/j.1574-6968.2000.tb09025.x. [DOI] [PubMed] [Google Scholar]
- 10.Zheng G, Wang C, Williams HN, Pineiro SA. 2008. Development and evaluation of a quantitative real-time PCR assay for the detection of saltwater Bacteriovorax. Environ Microbiol 10:2515–2526. doi: 10.1111/j.1462-2920.2008.01676.x. [DOI] [PubMed] [Google Scholar]
- 11.Iebba V, Santangelo F, Totino V, Nicoletti M, Gagliardi A, De Biase RV, Cucchiara S, Nencioni L, Conte MP, Schippa S. 2013. Higher prevalence and abundance of Bdellovibrio bacteriovorus in the human gut of healthy subjects. PLoS One 8:e61608. doi: 10.1371/journal.pone.0061608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahmoud KK, McNeely D, Elwood C, Koval SF. 2007. Design and performance of a 16S rRNA-targeted oligonucleotide probe for detection of members of the genus Bdellovibrio by fluorescence in situ hybridization. Appl Environ Microbiol 73:7488–7493. doi: 10.1128/AEM.01112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaner NC, Steinbach PA, Tsien RY. 2005. A guide to choosing fluorescent proteins. Nat Methods 2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 14.Medina AA, Shanks RM, Kadouri DE. 2008. Development of a novel system for isolating genes involved in predator-prey interactions using host independent derivatives of Bdellovibrio bacteriovorus 109J. BMC Microbiol 8:33. doi: 10.1186/1471-2180-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 16.Saltikov CW, Newman DK. 2003. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci U S A 100:10983–10988. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dashiff A, Keeling TG, Kadouri DE. 2011. Inhibition of predation by Bdellovibrio bacteriovorus and Micavibrio aeruginosavorus via host cell metabolic activity in the presence of carbohydrates. Appl Environ Microbiol 77:2224–2231. doi: 10.1128/AEM.02565-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadouri D, O'Toole GA. 2005. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl Environ Microbiol 71:4044–4051. doi: 10.1128/AEM.71.7.4044-4051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotter TW, Thomashow MF. 1992. A conjugation procedure for Bdellovibrio bacteriovorus and its use to identify DNA sequences that enhance the plaque-forming ability of a spontaneous host-independent mutant. J Bacteriol 174:6011–6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flannagan RS, Valvano MA, Koval SF. 2004. Downregulation of the motA gene delays the escape of the obligate predator Bdellovibrio bacteriovorus 109J from bdelloplasts of bacterial prey cells. Microbiology 150:649–656. doi: 10.1099/mic.0.26761-0. [DOI] [PubMed] [Google Scholar]
- 21.Rawlings DE, Tietze E. 2001. Comparative biology of IncQ and IncQ-like plasmids. Microbiol Mol Biol Rev 65:481–496. doi: 10.1128/MMBR.65.4.481-496.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furste JP, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 23.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shanks RM, Kadouri DE, MacEachran DP, O'Toole GA. 2009. New yeast recombineering tools for bacteria. Plasmid 62:88–97. doi: 10.1016/j.plasmid.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shanks RM, Stella NA, Lahr RM, Wang S, Veverka TI, Kowalski RP, Liu X. 2012. Serratamolide is a hemolytic factor produced by Serratia marcescens. PLoS One 7:e36398. doi: 10.1371/journal.pone.0036398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cormack BP, Valdivia RH, Falkow S. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 27.Merritt JH, Kadouri DE, O'Toole GA. 2005. Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1:Unit 1B.1. doi: 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shanks RM, Davra VR, Romanowski EG, Brothers KM, Stella NA, Godboley D, Kadouri DE. 2013. An eye to a kill: using predatory bacteria to control Gram-negative pathogens associated with ocular infections. PLoS One 8:e66723. doi: 10.1371/journal.pone.0066723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 30.Fenton AK, Kanna M, Woods RD, Aizawa SI, Sockett RE. 2010. Shadowing the actions of a predator: backlit fluorescent microscopy reveals synchronous nonbinary septation of predatory Bdellovibrio inside prey and exit through discrete bdelloplast pores. J Bacteriol 192:6329–6335. doi: 10.1128/JB.00914-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capeness MJ, Lambert C, Lovering AL, Till R, Uchida K, Chaudhuri R, Alderwick LJ, Lee DJ, Swarbreck D, Liddell S, Aizawa S, Sockett RE. 2013. Activity of Bdellovibrio hit locus proteins, Bd0108 and Bd0109, links type IVa pilus extrusion/retraction status to prey-independent growth signalling. PLoS One 8:e79759. doi: 10.1371/journal.pone.0079759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fenton AK, Lambert C, Wagstaff PC, Sockett RE. 2010. Manipulating each MreB of Bdellovibrio bacteriovorus gives diverse morphological and predatory phenotypes. J Bacteriol 192:1299–1311. doi: 10.1128/JB.01157-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medina AA, Kadouri DE. 2009. Biofilm formation of Bdellovibrio bacteriovorus host-independent derivatives. Res Microbiol 160:224–231. doi: 10.1016/j.resmic.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Williams HN, Turng BF, Kelley JI. 2009. Survival response of Bacteriovorax in surface biofilm versus suspension when stressed by extremes in environmental conditions. Microb Ecol 58:474–484. doi: 10.1007/s00248-009-9499-7. [DOI] [PubMed] [Google Scholar]
- 35.Pineiro S, Chauhan A, Berhane TK, Athar R, Zheng G, Wang C, Dickerson T, Liang X, Lymperopoulou DS, Chen H, Christman M, Louime C, Babiker W, Stine OC, Williams HN. 2013. Niche partition of Bacteriovorax operational taxonomic units along salinity and temporal gradients in the Chesapeake Bay reveals distinct estuarine strains. Microb Ecol 65:652–660. doi: 10.1007/s00248-013-0186-3. [DOI] [PubMed] [Google Scholar]
- 36.Dashiff A, Kadouri DE. 2011. Predation of oral pathogens by Bdellovibrio bacteriovorus 109J. Mol Oral Microbiol 26:19–34. doi: 10.1111/j.2041-1014.2010.00592.x. [DOI] [PubMed] [Google Scholar]
- 37.Dashiff A, Junka RA, Libera M, Kadouri DE. 2011. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J Appl Microbiol 110:431–444. doi: 10.1111/j.1365-2672.2010.04900.x. [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson MH. 2001. Predation in the presence of decoys: an inhibitory factor on pathogen control by bacteriophages or Bdellovibrio in dense and diverse ecosystems. J Theor Biol 208:27–36. doi: 10.1006/jtbi.2000.2197. [DOI] [PubMed] [Google Scholar]
- 39.Hobley L, King JR, Sockett RE. 2006. Bdellovibrio predation in the presence of decoys: three-way bacterial interactions revealed by mathematical and experimental analyses. Appl Environ Microbiol 72:6757–6765. doi: 10.1128/AEM.00844-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varon M, Shilo M. 1969. Interaction of Bdellovibrio bacteriovorus and host bacteria. II. Intracellular growth and development of Bdellovibrio bacteriovorus in liquid cultures. J Bacteriol 99:136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.