Abstract
The functioning of recirculation aquaculture systems (RAS) is essential to maintain water quality for fish health, and one crucial process here is nitrification. The investigated RAS was connected to a rainbow trout production system and operated at an average temperature of 13°C and pH 6.8. Community analyses of the nitrifying biofilm revealed a coexistence of Nitrospira and Nitrotoga, and it is hypothesized that a slightly acidic pH in combination with lower temperatures favors the growth of the latter. Modification of the standard cultivation approach toward lower pH values of 5.7 to 6.0 resulted in the successful enrichment (99% purity) of Nitrotoga sp. strain HW29, which had a 16S rRNA sequence similarity of 99.0% to Nitrotoga arctica. Reference cultures of Nitrospira defluvii and the novel Nitrotoga sp. HW29 were used to confirm differentiation of these nitrite oxidizers in distinct ecological niches. Nitrotoga sp. HW29 revealed pH and temperature optima of 6.8 and 22°C, respectively, whereas Nitrospira defluvii displayed the highest nitrite oxidation rate at pH 7.3 and 32°C. We report here the occurrence of Nitrotoga as one of the main nitrite-oxidizing bacteria in freshwater aquaculture systems and indicate that a slightly acidic pH, in addition to temperatures below 20°C, can be applied as a selective isolation criterion for this microorganism.
INTRODUCTION
Nitrogen is one of the most important elements of life and nitrification is a key process in the global N cycle, since the biological conversion of ammonia to nitrate is required wherever organic matter accumulates. This oxidation of ammonia to nitrate via nitrite is catalyzed by chemolithoautotrophic ammonia-oxidizing bacteria (AOB) and archaea (AOA) and nitrite-oxidizing bacteria (NOB). The biotechnological application of nitrification in recirculating aquaculture systems (RAS) has gained increasing attention for the production of aquatic organisms, since it allows an efficient and ecological way of fish breeding. To remove toxic ammonia released by fish gills or mineralization of feces and feed offcut, biological filtration is required. Nitrification is promoted on the surfaces of carrier elements in well-aerated moving-bed reactors. The carrier elements (plastic beads) are colonized by nitrifying bacteria, which form a dense biofilm together with heterotrophic bacteria.
In biofilters of most aquaculture systems (marine and freshwater), Nitrosomonas and Nitrospira were identified as the main ammonia and nitrite oxidizers, respectively (1–4), but members of the genera Nitrobacter and Nitrotoga are also potential candidates for nitrite removal (5, 6). Cold-adapted Nitrotoga spp. were initially found in permafrost-affected soils in Siberia (7), and the preference for low temperatures was confirmed by the detection of Nitrotoga-like 16S rRNA gene sequences in other cold soils such as recently deglaciated soils (8) or periglacial soils (9). Nitrotoga was also abundant in oligotrophic aquatic environments such as the Movile cave (10), a neutral groundwater seep (11) and inside a drinking water filter, where it constituted 4.8% in a 16S rRNA clone library (12). Nitrotoga was even more abundant in a subglacial Antarctic lake, representing 13 and 7.8% of all 16S rRNA sequences in the water column and sediment, respectively (13). Although all of these ecosystems have a pristine character, Nitrotoga seems to be also relevant for nitrite oxidation in eutrophic ecosystems, provided that the operation temperature is low. For instance, these NOB were abundant in bioreactors treating industrial wastewaters (14) and on a reverse osmosis membrane of a wastewater treatment plant (WWTP) operating at 5°C to 10°C (15). In a screen of 20 full-scale WWTPs in Germany and Switzerland, Nitrotoga was detected in about half of the sludges, together with Nitrospira, and was the only NOB found at two of the plants (6). In accordance with this, in several Danish plants Nitrotoga was even more abundant than Nitrospira, which has been to date considered the dominant nitrite oxidizer in activated sludge (16).
In addition to temperature, the pH is a key driver of niche differentiation of NOB. With the exception of Nitrobacter alkalicus (17), pH optima of 7.6 to 8.0 were determined for terrestrial species of Nitrobacter and Nitrospira in physiological experiments (18–20) and a neutral range (7.4 to 7.6) is mainly used for the cultivation of NOB in general (21). In the aquaculture plant described here, the pH was manually lowered to 6.8 to shift the equilibrium of ammonia toward ammonium, since this reduces toxicity effects to fish (<0.1 mg of NH3-N liter−1) (22). The approach of intensive fish culture at high ammonia and low pH was supposed to circumvent the necessity of nitrifying biofilters by reducing the fraction of toxic NH3 (23). In contrast to the described flowthrough system, the plant investigated here could not be operated safely without a nitrifying unit. Hence, a first biofilter (BF1) and later on an additional biofilter (BF2) inside the same module were constructed in 2008 and 2010, respectively. The goal of the present study was to monitor the development of nitrification potentials in BF1 and BF2 and to identify the dominant nitrifying microorganisms.
MATERIALS AND METHODS
Sampling sites.
The investigated freshwater recirculating aquaculture system (RAS) is located in northeast Germany and supplied with groundwater. In a RAS, biofiltration takes place in separated basins (biofilters) containing biocarrier elements. Two distinct biofilters have been used in this study. The moving-bed biofilter 1 (BF1) was initially put into operation in 2008. To improve the biofiltration power, a second biofilter (BF2) was added to the system in November 2010. Both biofilters were filled with two types of high-density polyethylene biocarriers (BCN009, surface of 497 m2 m−3; KLL012, surface of 704 m2 m−3; Stöhr, Marktrodach, Germany). To simplify matters, only carrier elements of type BCN009 were chosen for all experiments for this study. During investigation period, the respective ponds were filled with rainbow trout (Oncorhynchus mykiss) until September 2011, when stock has been changed to alsatian char (Salvelinus alpinus × fontinalis). The average temperature of the process water ranged from 10 to 14°C, and the pH was at 6.8. The oxygen concentration was kept at 9 to 10 mg liter−1.
Cultivation and physiological experiments.
Mineral salts medium (24) with a pH value of 7.4 and 0.3 mM nitrite was used as the basic medium for all cultivation procedures and physiological experiments in the present study, unless noted elsewise. Enrichments and physiological tests were done in 300-ml Erlenmeyer flasks. Enrichment cultures of biofilm material were started with two to three densely covered biocarrier elements in 150 ml of medium. Incubation was performed either at 10 or 17°C in darkness without agitation. The presence of nitrite was regularly checked by a Griess-Ilosvay spot test (25) and replaced in case of consumption. For the enrichment of Nitrotoga sp. HW29, a mineral salts medium at either pH 7.4 or 6.0 was used. Subsequent serial dilutions (10−1 up to 10−8) were performed in acidic medium at pH 5.7 according to the method of Schmidt and Belser (26) with slight modifications (21). The enrichment procedure was checked regularly for the presence of Nitrospira and Nitrotoga via PCR with genus-specific primer pairs as described below. The potential activity of nitrifying biofilms was determined by the separation of AOB and NOB approaches. Five colonized biocarrier elements were counted into 50 ml of either AOB or NOB mineral salts medium (24, 27) with a substrate concentration of 1 mM ammonia or nitrite, respectively. Incubation was performed at 17°C, and flasks were agitated (150 rpm) in the dark. Then, 1-ml samples were taken hourly until substrate depletion and analyzed via high-pressure liquid chromatography (HPLC) to control nitrification performance. Physiological tests of isolated or enriched NOB were conducted with 1 mM nitrite and inoculated with 1% of a well-grown, active preculture. To test pH optima, sterile-filtered mineral salts media of definite pH values were used (pH 5.5 to 7.5), and subsequent tests were performed in duplicates. For the determination of temperature optima, cultures were incubated at 4 to 37°C without agitation. Samples (1 ml) were taken regularly, and nitrite and nitrate were analyzed using HPLC. A substrate inhibition test was performed with nitrite concentrations from 0.3 to 10 mM, followed by incubation at 22°C.
Chemical analyses.
Nitrite and nitrate were quantified by HPLC via ion pair chromatography with a LiChrospher RP-18 column (125 by 4 mm; Merck KGaA, Germany) and UV detection in an automated system (Hitachi LaChrom Elite; VWR International GmbH, Darmstadt, Germany). Data acquisition and processing was performed with the integrated software EZChrom Elite 3.3.2.
Cell harvesting and DNA extraction.
To obtain cell material from biocarriers for DNA extraction, ten biofilm covered biocarriers were shaken vigorously in 10 ml of 0.9% NaCl by the addition of 2.5 g of sterile glass beads (2.0 and 0.5 mm in diameter) for 4 h. The liquid phase was centrifuged, and DNA was extracted from the cell pellet using the UltraClean microbial DNA isolation kit (MoBio Laboratories, Carlsbad, CA) according to the manufacturer's instructions. In the case of NOB cultures, centrifugation (8,000 rpm, 30 min) of 50- to 100-ml culture volumes was done prior to DNA extraction, as described above.
Molecular and phylogenetic analyses.
To demonstrate the presence of three different NOB genera, PCRs with genus-specific primer pairs targeting Nitrotoga (NTG 200f/NTG 840r) (28), Nitrospira (Nsp 60-kf/Nsp662r) (28, 29) and Nitrobacter (Nb 1000gf/degr; modified as described by Degrange and Bardin, Mobarry et al., and Alawi [28, 30, 31]) were performed. A Nitrotoga-specific PCR was carried out using a nested PCR approach with the eubacterial primers 27f and 1492r (32) for the first round of amplification, followed by a second round of amplification with Nitrotoga-specific primers as described above. PCR products were purified with Geneaid Gel/PCR DNA fragment extraction kit according to the manufacturer's instructions (DF100/DF300; Geneaid Biotech, Ltd, Taiwan) and subsequently sequenced using the Sanger method. For separation, the respective PCR product was ligated into a TOPO-TA cloning vector system according to the manufacturer's instructions (Invitrogen/Life Technologies, Carlsbad, CA). Prior to Sanger sequencing, plasmid primers T7 and T3 were used to reamplify the cloned inserts. The obtained nucleotide sequences were analyzed using BLASTn (33). Next, 10 ng of DNA of Nitrotoga sp. HW29 enrichment culture was used as the template for preparative PCR with the primers 515f and 806r (34) to generate 16S rRNA amplicons for 454 pyrosequencing. Preparative PCR, as well as subsequent 454 amplicon sequencing, with a sequencing depth of 3,000 reads was performed at a sequencing service facility. Denoised sequences were generated into operational taxonomic units (OTU), and chimeras were removed. The taxonomic classification of OTU was accomplished by using BLASTn compared to information from the GreenGenes, RDPII, and NCBI databases.
FISH.
Biofilm material was harvested as described above. The pellet was fixed with paraformaldehyde as described previously (35). Fluorescence in situ hybridization (FISH) was performed according to the protocol detailed by Manz et al. (36) with the Cy5-labeled universal bacterial probes EUB338, EUB338II, and EUB338III (35, 37), as well as the Fluos-labeled probes Ntspa-662 and Ntspa-772 (38) and the Cy-3-labeled probe Ntoga-122 (6), targeting the genera Nitrospira and Nitrotoga, respectively. After hybridization, the cells were stained with DAPI (4′,6′-diamidino-2-phenylindole) and embedded in Citifluor AF1 (Citifluor, Ltd., London, United Kingdom) prior to microscopic observation. Probe-conferred fluorescence was recorded on a Leica LSM SP8 with a white-light laser (Leica Microsystems GmbH, Wetzlar, Germany). Digital image analysis was carried out using the software package LAS AF Lite 2.6 (Leica Microsystems).
Transmission electron microscopy (TEM).
For transmission electron microscopic observations, cells were mechanically removed from the biocarriers (see above) and fixed in 2.5% (vol/vol) glutaraldehyde for 1.5 h, followed by an overnight incubation in 2% (wt/vol) osmium tetroxide. Fixed cells were embedded in Spurr resin (39) as described previously (21). Ultrathin sections were stained with uranyl acetate and lead citrate (40, 41). Micrographs were obtained using a Zeiss model Leo 906E charge-coupled device camera (model 794; Carl Zeiss, Jena, Germany).
Nucleotide sequence accession numbers.
All 16S rRNA gene sequences obtained in this study were deposited at GenBank (National Center for Biotechnology Information) after quality filtering and removing chimeras, with the following accession numbers: KT778545 and KT805947 to KT805950.
RESULTS AND DISCUSSION
What controls nitrification in RAS?
Biofilter performance, measured as potential nitrification rates in batch incubations, is affected by several parameters, like ammonia concentration, and is therefore strongly dependent on stocking density and feeding intensity. Further important factors influencing nitrification kinetics in biofilters are the temperature and the pH value besides the dissolved oxygen concentration, organic matter content, alkalinity, salinity, and turbulence (42, 43). Three months after the addition of new carrier elements into BF2, a well-established biofilm, consisting of AOB and NOB, as well as heterotrophic bacteria, became visible inside the carriers (see Fig. S1 in the supplemental material). Determination of potential nitrification rates revealed complete nitrification in both biofilters despite the low average temperature of 13°C and the slightly acidic pH of 6.8 (see Fig. S2 in the supplemental material). After an initial phase with low nitrifying activities following the startup in November 2010, the nitrification potential in BF2 increased compared to BF1 to maximum turnover rates of 27 g h−1 m−3 total ammonia nitrogen (TAN), mediated via AOB, and 66 g h−1 m−3 total nitrite nitrogen (NO2-N), mediated via NOB. The activity levels of BF2 remained nearly constant from February to June 2011 but decreased by 55% (AOB) and 60% (NOB) in September when the fish population was changed from rainbow trout to alsatian char with lower stocking and feeding rates.
RAS efficiency at low temperature and slightly acidic pH.
According to Strauss et al. (44), pH is one of the most important variables for regulating nitrification, and a pH of 6.5 can completely inhibit NOB activity (45). To elucidate the metabolic activity of biofilm inhabiting NOB in dependency on pH, we performed laboratory scale short-term nitrite consumption experiments at different pH values with biofilm material from BF2 (September 2011). Nitrite was oxidized over the whole spectrum investigated, with the highest nitrite oxidation rates of 160 μM nitrite/h at pH 7.1 (see Fig. S3 in the supplemental material). Only minor differences in oxidation rates were found between pH 6.0 and 7.5, with still 50% of the maximal activity at pH 6.0. Growth at low pH might be enhanced by a dense biofilm (46), since NOB are protected from adverse conditions (47).
NOB community analyses.
Although it was found earlier that nitrification proceeds at temperatures of 12°C (48), no community analyses were performed in combination with process measurements thus far (49). In this study, the NOB community composition of a freshwater RAS was identified by means of specific PCR primers, typical ultrastructural criteria, and genus-specific FISH probes.
PCR using genus-specific primers for Nitrotoga, Nitrospira, or Nitrobacter revealed the continuous presence of Nitrotoga and Nitrospira in both biofilters, with the exception of June and July 2011, when Nitrotoga could not be detected in BF2 (Table 1). Instead, Nitrobacter appeared in BF2 from July to September 2011. Sequence analyses of the 16S rRNA gene amplicons of Nitrotoga revealed a similarity of 99.7% to Nitrotoga HAM-1 from activated sludge (5). Two representatives of Nitrospira could be detected, which were 99.1 and 99.7% similar to N. defluvii, respectively. The obtained Nitrobacter 16S rRNA sequence was 98.9% identical to N. hamburgensis.
TABLE 1.
Identification of NOB present in BF1 and BF2 by genus-specific PCR and TEM
| Biofilter and sampling date (mo/yr) | PCRa |
TEMb |
||||
|---|---|---|---|---|---|---|
| Nitrotoga | Nitrospira | Nitrobacter | Nitrotoga | Nitrospira | Nitrobacter | |
| BF1 | ||||||
| 07/2010 | + | + | – | +++ | ++ | – |
| 10/2010 | + | + | – | |||
| BF2 | ||||||
| 11/2010 | + | + | – | + | ++ | – |
| 12/2010 | + | + | – | |||
| 03/2011 | + | + | – | |||
| 06/2011 | – | + | – | + | ++ | – |
| 07/2011 | – | + | + | |||
| 08/2011 | + | + | + | |||
| 09/2011 | + | + | + | |||
+, positive reaction; −, no reaction.
+, present; −, not present. The number of plus signs reflects the approximate abundance of the respective NOB visible via TEM.
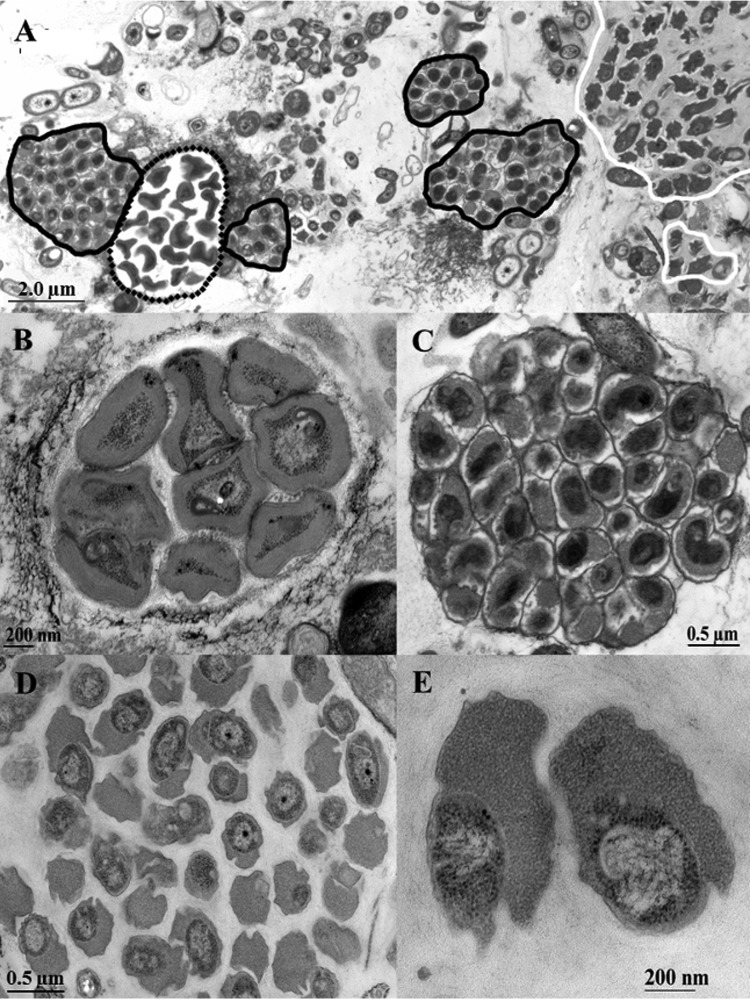
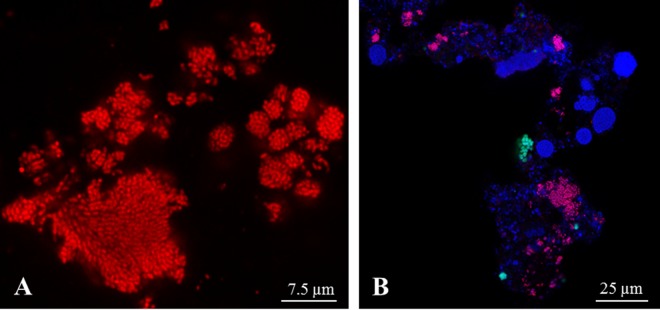
TEM of the July 2010 sample of BF1 (Fig. 1A) showed a high abundance of irregular shaped Nitrotoga cells and of Nitrospira aggregates. Both NOB grew in dense microcolonies in close vicinity to Nitrosomonas-like ammonia oxidizers (Fig. 1B). Nitrospira colonies consisted of short, slightly curved cells, which were tightly packed in extracellular polymeric substances (EPS) (Fig. 1C). Nitrotoga cells were situated in loosely grouped aggregates surrounded by thin layers of EPS (Fig. 1D) and had an extremely wide particular periplasmic space (Fig. 1E). Directly after the start-up of BF2 in November 2010, dense clusters of Nitrospira were detected by TEM, but only single microcolonies of Nitrotoga-like cells (not shown). After an operating time of 6 months, Nitrotoga formed large colonies again, which were scarcely distributed within the biofilm matrix. In accordance with previous investigations (1, 2, 4, 50), a higher density of NOB compared to Nitrosomonas-like AOB was observed in most samples. In addition to TEM, specific molecular identification via FISH was performed with BF2 samples taken in February 2011. FISH visualization revealed a high level of cells in huge colonies, which hybridized with the Nitrotoga genus-specific probe Ntoga-122 (Fig. 2A). Use of the Nitrospira-specific probes Ntspa-712 and Ntspa-662 resulted in a lower abundance of this NOB, arranged in small colonies (Fig. 2B). In accordance with the electron microscopic investigations, coccoid cells of Nitrotoga were clearly different from the densely aggregated cells of Nitrospira.
FIG 1.
Ultrathin section of the nitrifying biofilm grown on a carrier element of BF1 sampled in July 2010 and visualized via TEM. (A) Overview of the nitrifier arrangement inside the biofilm. Microcolonies of AOB Nitrosomonas are marked with dotted lines; for the second step of nitrification, Nitrospira (black outline) and Nitrotoga (white outline) were present. (B to D) Microcolonies of Nitrosomonas (B), Nitrospira (C), and Nitrotoga (D) in detail. (E) Typical ultrastructure of Nitrotoga cells, revealing the extremely wide periplasmic space with a particulate appearance.
FIG 2.
FISH visualization of 3-month-old biofilm material of BF2 sampled in February 2011, stained with genus-specific probes. (A) Section of a huge Nitrotoga colony stained with probe Ntoga-122. (B) Dense biofilm reveals a large number of Ntoga-122-stained Nitrotoga clusters (pink) and fewer Ntspa-712- and Ntspa-662-stained microcolonies of Nitrospira (green). EUB338-, EUB338II-, and EUB338III-stained cells are colored blue.
Previous studies on the RAS bioreactor community composition of nitrite oxidizers identified Nitrospira as the most abundant and active NOB inside these systems (1, 2, 51). In the cold-water system investigated here, the simultaneous presence of Nitrotoga and Nitrospira was detected by microscopic and molecular analyses. In the beginning of our survey, Nitrotoga even occurred as the most abundant NOB in BF1, but Nitrospira dominated the nitrifying biofilm when the new biofilter BF2 was connected to the running system and therefore appears as the primary resident of the new plastic beads. Nevertheless, Nitrotoga reemerged to high cell densities in February 2011, as demonstrated by FISH, but was not detectable even by PCR during the summer months June and July. However, the negative PCR results might be due to patchy distribution of the microcolonies causing bias in DNA isolation. Overall, Nitrotoga was a permanent and main NOB of the reactor biofilm in this RAS, since it was continuously detected until 2014 (data not shown). Therefore, Nitrotoga is considered a relevant nitrite oxidizer inside biofilms of cold-water-operated RAS. At temperatures of ≤17°C, where the activity of Nitrospira is drastically reduced, Nitrotoga is able to compete with other NOB (52). Nitrobacter was only occasionally detectable via PCR due to the higher sensitivity compared to TEM and FISH. The rare appearance of Nitrobacter inside RAS biofilters had already been observed in earlier studies and leads to the general assumption that this NOB seems to play a minor role in these systems (1, 2, 50, 53).
Separation of Nitrotoga from Nitrospira at low pH and low temperature.
The initial enrichment of Nitrotoga arctica was performed at 10°C as the optimal incubation temperature for the inhibition of other mesophilic NOB (7), but the pH value has not been considered as an additional criterion thus far. The results of NOB community analyses in the present study indicated that a slightly acidic pH in combination with the low temperature might have a selective growth effect on Nitrotoga. To confirm this hypothesis, a three-step modified enrichment approach with decreased pH values (5.7 and 6.0), temperatures of 10 and 17°C, and a low substrate concentration (0.3 mM nitrite) was utilized (Fig. 3). In step 1, to enrich NOB, the cultures were inoculated with densely covered biocarrier elements of BF1 (sampled in October 2010) in mineral salts medium (pH 7.4) and then incubated at 17°C. In step 2, after 7 months, the incubation was shifted to a slightly acidic pH of 6.0 and to lowered incubation temperature of 10°C to selectively advance the growth of Nitrotoga and inhibit Nitrospira, respectively. In step 3, since both NOB were still present after 6 months, a dilution series was prepared in acidic media at pH 5.7 and incubated at 17°C to accelerate growth. After incubation for 9 months, the dilution 10−6 was the last dilution step that tested positive for nitrite oxidation and contained only one Nitrotoga sp. with a 16S rRNA sequence similarity of 99.0% to Nitrotoga arctica. The absence of Nitrospira was confirmed by genus-specific PCR. Final estimation of the enrichment purity degree via 454 amplicon pyrosequencing indicated the persistence of a few heterotrophic bacteria, but the vast majority (99%) of the obtained reads were affiliated with the genus Nitrotoga. The highly enriched culture of this NOB was from now on termed Nitrotoga sp. HW29.
FIG 3.
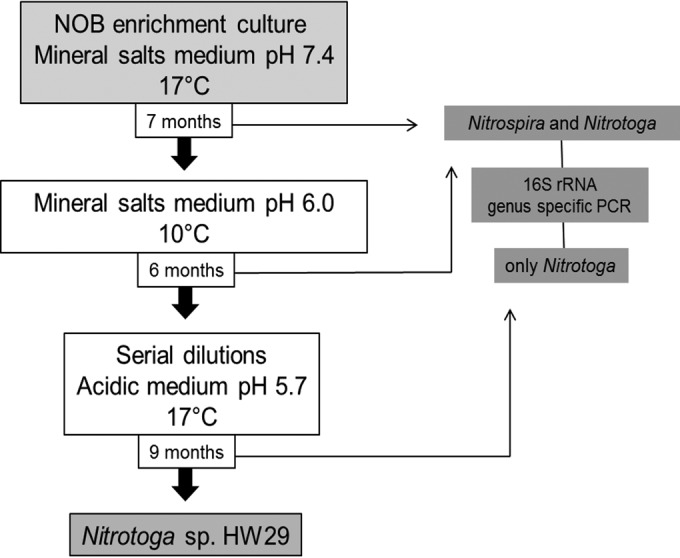
Schematic representation of the selective enrichment procedure for Nitrotoga sp. HW29.
Our low-pH strategy led to the selective enrichment of Nitrotoga from the investigated RAS biofilter and the loss of Nitrospira after a few passages and one dilution row. Nevertheless, this strategy might only be successful by the combination of further factors, such as a low incubation temperature and a low substrate concentration. Nitrospira can overgrow Nitrotoga during extensive feeding because this NOB possesses the highest affinity for nitrite (52). However, the beneficial growth of Nitrotoga versus Nitrospira in acidic medium was confirmed with other cultures, and further attempts to enrich Nitrospira in acidic medium failed (unpublished results). This might be due to the fact that, in contrast to Nitrobacter, cells of Nitrospira are easily inhibited by free nitrous acid (FNA) (54), although the FNA concentration at pH > 5 is very low (55).
Physiological characterization of Nitrotoga sp. HW29 compared to Nitrospira defluvii.
To demonstrate whether a slightly acidic pH might be one of the reasons for the predominance of Nitrotoga over Nitrospira, pH optima for Nitrospira defluvii A17 (56, 57), a member of sublineage I that is closely related to Nitrospira in the present RAS, and for highly enriched Nitrotoga sp. HW29 were evaluated. Both NOB developed distinct temperature optima and showed a drastic loss of activity when exposed to slightly higher temperatures. The highest nitrite oxidation rates were observed at 32°C (151 μM nitrite consumption/day) for Nitrospira defluvii A17, as previously shown (21), and at 22°C (121 μM nitrite consumption/day) for Nitrotoga sp. strain HW29 (Fig. 4A). This value is clearly higher than the average temperature of the pristine RAS (13°C) and the temperature optimum of Nitrotoga arctica 6680 (13°C) (data not shown). At 10°C Nitrotoga still retained about 40% of its maximal activity, whereas Nitrospira activity was reduced to 15%. Although the enrichment of HW29 revealed a sequence similarity of 99.0% on the 16S rRNA gene level to Nitrotoga arctica, these two NOB feature distinct physiological characteristics, such as the temperature optimum shown here, which makes the highly enriched HW29 a potential candidate for a new species of Nitrotoga.
FIG 4.
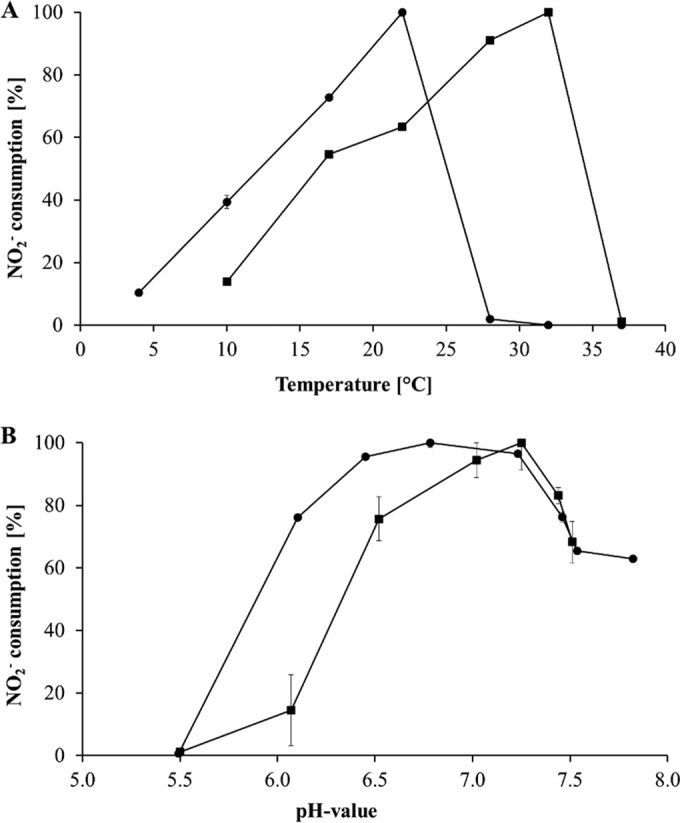
(A) Temperature optima of highly enriched Nitrotoga sp. HW29 (●) and Nitrospira defluvii A17 (■) grown at pH 7.4. (B) Influence of the pH on growth of Nitrotoga sp. HW29 (●) incubated at 17°C and Nitrospira defluvii A17 (■) incubated at 28°C. NOB growth corresponds to the capability for nitrite consumption per day.
Evaluation of pH effects on nitrite oxidation was performed close to the respective temperature optima of 17°C for HW29 and 28°C for A17 (Fig. 4B). The latter showed optimal growth at pH 7.0 to 7.3 (440 μM nitrite consumption/day). In contrast, Nitrotoga sp. HW29 achieved nearly identical nitrite oxidation rates between pH 6.5 and 7.2, revealing a broad pH spectrum with a slight maximum at pH 6.8 (261 μM nitrite consumption/day). Even at pH 6.1 a relative nitrite turnover rate of 75% was detected, whereas the activity levels of N. defluvii were reduced to ca. 14%. The results showed different pH optima for these two NOB, indicating the separation of Nitrospira and Nitrotoga into distinct ecological niches and further confirming our assumption regarding the relevance of Nitrotoga in the investigated RAS running at pH 6.8. In contrast to distinct pH optima, activity studies of enrichment cultures of Nitrospira and Nitrotoga via FISH-microautoradiography (MAR) have shown that both NOB were active when incubated at 14°C in media at pH 6.4 and 7.2 (data not shown). Nevertheless, it should be taken into account that FISH-MAR analyses were conducted under short-term incubation (6 h) and that only long-term cultivation of both NOB led to the loss of Nitrospira and a high enrichment of Nitrotoga under cultivation conditions with slightly acidic pH.
Nitrotoga is well suited for the treatment of wastewaters.
Low substrate concentrations of 0.3 mM nitrite were regarded as optimal for the growth of Nitrotoga arctica, and early enrichment cultures were already inhibited at 1.2 mM (7). In contrast, in the present study a substrate inhibition test revealed activity of Nitrotoga sp. HW29 up to a nitrite concentration of 8 mM, with total nitrite consumption within 2 weeks. Using FISH-MAR, Lücker et al. detected Nitrotoga activity from wastewater at up to 10 mM nitrite (6), but it has to be considered that the substrate conversion appears to be strain and fitness dependent. The ability of Nitrotoga to oxidize nitrite to relative high concentrations could be a reason for its appearance in WWTPs (5, 6) and aquaculture systems (the present study) with rising nitrite concentrations, although the tolerance level of 30 mM Nitrospira defluvii is much higher (57).
Ecological aspects of nitrite oxidation at low pH.
There are only a few studies examining nitrite oxidation at low pH (58, 59), and the only described acidophilic NOB culture belongs to Nitrobacter (Io acid), which is able to grow optimally at pH 5.5 (60, 61). Interestingly, Nitrotoga sp. HW29 is the first known NOB with a pH optimum in the slightly acidic range. The closest relative of the Nitrotoga sp. HW29 strain investigated here is Nitrotoga arctica (99.0% sequence similarity of the 16S rRNA gene), which originates from Siberian permafrost soil (7). Considering the sampling site pH of 5.6 to 6.5 (62), it is not surprising to find the pH optimum of this NOB in culture at lower values. With the adaptation to a slightly acidic pH, Nitrotoga might be able to occupy distinct habitats, such as acid-impacted lakes (63). This fact makes this NOB even more interesting regarding niche differentiation among NOB. Nitrotoga seems to be not only cold adapted but also moderately acid tolerant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Elke Woelken for excellent technical assistance with the electron microscopy. We also thank our project partners, Heiko Roepke and Gregor Schmidt of the Institute of Fishery Sciences (Hohen Wangelin, Germany), for providing biofilm material and chemical data. We thank Tina Sanders for physiological tests on N. arctica.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03163-15.
REFERENCES
- 1.Foesel BU, Gieseke A, Schwermer C, Stief P, Koch L, Cytryn E, de la Torré JR, van Rijn J, Minz D, Drake HL, Schramm A. 2008. Nitrosomonas Nm143-like ammonia oxidizers and Nitrospira marina-like nitrite oxidizers dominate the nitrifier community in a marine aquaculture biofilm. FEMS Microbiol Ecol 63:192–204. doi: 10.1111/j.1574-6941.2007.00418.x. [DOI] [PubMed] [Google Scholar]
- 2.Keuter S, Kruse M, Lipski A, Spieck E. 2011. Relevance of Nitrospira for nitrite oxidation in a marine recirculation aquaculture system and physiological features of a Nitrospira marina-like isolate. Environ Microbiol 13:2536–2547. doi: 10.1111/j.1462-2920.2011.02525.x. [DOI] [PubMed] [Google Scholar]
- 3.Brown MN, Briones A, Diana J, Raskin L. 2013. Ammonia-oxidizing archaea and nitrite-oxidizing nitrospiras in the biofilter of a shrimp recirculating aquaculture system. FEMS Microbiol Ecol 83:17–25. doi: 10.1111/j.1574-6941.2012.01448.x. [DOI] [PubMed] [Google Scholar]
- 4.Kruse M, Keuter S, Bakker E, Spieck E, Eggers T, Lipski A. 2013. Relevance and diversity of Nitrospira populations in biofilters of brackish RAS. PLoS One 8:e64737. doi: 10.1371/journal.pone.0064737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alawi M, Off S, Kaya M, Spieck E. 2009. Temperature influences the population structure of nitrite-oxidizing bacteria in activated sludge. Environ Microbiol Rep 1:184–190. doi: 10.1111/j.1758-2229.2009.00029.x. [DOI] [PubMed] [Google Scholar]
- 6.Lücker S, Schwarz J, Gruber-Dorninger C, Spieck E, Wagner M, Daims H. 2015. Nitrotoga-like bacteria are previously unrecognized key nitrite oxidizers in full-scale wastewater treatment plants. ISME J 9:708–720. doi: 10.1038/ismej.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alawi M, Lipski A, Sanders T, Pfeiffer EM, Spieck E. 2007. Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J 1:256–264. doi: 10.1038/ismej.2007.34. [DOI] [PubMed] [Google Scholar]
- 8.Sattin SR, Cleveland CC, Hood E, Reed SC, King AJ, Schmidt SK, Robeson MS, Ascarrunz N, Nemergut DR. 2009. Functional shifts in unvegetated, perhumid, recently-deglaciated soils do not correlate with shifts in soil bacterial community composition. J Microbiol 47:673–681. doi: 10.1007/s12275-009-0194-7. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt SK, Nemergut DR, Miller AE, Freeman KR, King AJ, Seimon A. 2009. Microbial activity and diversity during extreme freeze-thaw cycles in periglacial soils, 5400 m elevation, Cordillera Vilcanota, Perú. Extremophiles 13:807–816. doi: 10.1007/s00792-009-0268-9. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Wu L, Boden R, Hillebrand A, Kumaresan D, Moussard H, Baciu M, Lu Y, Murrell JC. 2009. Life without light: microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. ISME J 3:1093–1104. doi: 10.1038/ismej.2009.57. [DOI] [PubMed] [Google Scholar]
- 11.Roden EE, McBeth JM, Blöthe M, Percak-Dennett EM, Fleming EJ, Holyoke RR, Luther GW, Emerson D, Schieber J. 2012. The microbial ferrous wheel in a neutral pH groundwater seep. Front Microbiol 3:1–18. doi: 10.3389/fmicb.2012.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White CP, Debry RW, Lytle DA. 2012. Microbial survey of a full-scale, biologically active filter for treatment of drinking water. Appl Environ Microbiol 78:6390–6394. doi: 10.1128/AEM.00308-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christner BC, Priscu JC, Achberger AM, Barbante C, Carter SP, Christianson K, Michaud AB, Mikucki JA, Mitchell AC, Skidmore ML, Vick-Majors TJ, Adkins WP, Anandakrishnan S, Barcheck G, Beem L, Behar A, Beitch M, Bolsey R, Branecky C, Edwards R, Fisher A, Fricker HA, Foley N, Guthrie B, Hodson T, Jacobel R, Kelley S, Mankoff KD, McBryan E, Powell R, Purcell A, Sampson D, Scherer R, Sherve J, Siegfried M, Tulaczyk S. 2014. A microbial ecosystem beneath the West Antarctic ice sheet. Nature 512:310–313. doi: 10.1038/nature13667. [DOI] [PubMed] [Google Scholar]
- 14.Karkman A, Mattila K, Tamminen M, Virta M. 2011. Cold temperature decreases bacterial species richness in nitrogen-removing bioreactors treating inorganic mine waters. Biotechnol Bioeng 108:2876–2883. doi: 10.1002/bit.23267. [DOI] [PubMed] [Google Scholar]
- 15.Bereschenko L, Stams JM, Euverink GJW, van Loosdrecht MCM. 2010. Biofilm formation on reverse osmosis membranes is initiated and dominated by Sphingomonas spp. Appl Environ Microbiol 76:2623–2632. doi: 10.1128/AEM.01998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saunders AM, Albertsen M, Vollertsen J, Nielsen PH. 2015. The activated sludge ecosystem contains a core community of abundant organisms. ISME J 10:11–20. doi: 10.1038/ismej.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorokin DY, Muyzer G, Brinkhoff T, Kuenen JG, Jetten MSM. 1998. Isolation and characterization of a novel facultatively alkaliphilic Nitrobacter species, N. alkalicus sp. nov. Arch Microbiol 170:345–352. doi: 10.1007/s002030050652. [DOI] [PubMed] [Google Scholar]
- 18.Bock E, Koops H-P, Harms H, Ahlers B. 1991. The biochemistry of nitrifying organisms, p 171–199. In Barton L, Shively S (ed), Variations in autotrophic life. Academic Press Ltd, London, United Kingdom. [Google Scholar]
- 19.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. 1995. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov., and its phylogenetic relationship. Arch Microbiol 164:16–23. [DOI] [PubMed] [Google Scholar]
- 20.Lebedeva E, Alawi M, Maixner F, Jozsa P-G, Daims H, Spieck E. 2008. Physiological and phylogenetic characterization of a novel lithoautotrophic nitrite-oxidizing bacterium, “Candidatus Nitrospira bockiana.” Int J Syst Evol Microbiol 58:242–250. doi: 10.1099/ijs.0.65379. [DOI] [PubMed] [Google Scholar]
- 21.Spieck E, Lipski A. 2011. Cultivation, growth physiology, and chemotaxonomy of nitrite-oxidizing bacteria, p 109–130. In Klotz MG. (ed), Methods in enzymology. Academic Press/Elsevier, Oxford, United Kingdom. [DOI] [PubMed] [Google Scholar]
- 22.Camargo JA, Alonso Á. 2006. Ecological and toxicological effects of inorganic nitrogen pollution in aquatic ecosystems: a global assessment. Environ Int 32:831–849. doi: 10.1016/j.envint.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Eshchar M, Lahav O, Mozes N, Peduel A, Ron B. 2006. Intensive fish culture at high ammonium and low pH. Aquaculture 255:301–313. doi: 10.1016/j.aquaculture.2005.11.034. [DOI] [Google Scholar]
- 24.Bock E, Sundermeyer-Klinger H, Stackebrandt E. 1983. New facultative lithoautotrophic nitrite-oxidizing bacteria. Arch Microbiol 136:281–284. doi: 10.1007/BF00425217. [DOI] [Google Scholar]
- 25.Keeney DR, Nelson DW. 1982. Nitrogen: inorganic forms, p 643–698. In Page AL. (ed), Methods of soil analysis. 2. Chemical and microbiological properties. Soil Science Society of America, Madison, WI. [Google Scholar]
- 26.Schmidt EL, Belser LW. 1994. Autotrophic nitrifying bacteria, p 159–177. In Weaver RW, Angle JS, Bottomley PJ (ed), Methods of soil analysis. 2. Microbiological and biochemical properties. Soil Science Society of America, Madison, WI. [Google Scholar]
- 27.Krümmel A, Harms H. 1982. Effect of organic matter on growth and cell yield of ammonia-oxidizing bacteria. Arch Microbiol 133:50–54. doi: 10.1007/BF00943769. [DOI] [Google Scholar]
- 28.Alawi M. 2007. Diversität Nitrit oxidierender Bakterien in Böden des nordsibirischen Permafrostes und Sedimenten der Laptev-See. PhD thesis University of Hamburg, Hamburg, Germany. [Google Scholar]
- 29.Hovanec T, DeLong E. 1996. Comparative analysis of nitrifying bacteria associated with freshwater and marine aquaria. Appl Environ Microbiol 62:2888–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Degrange V, Bardin R. 1995. Detection and counting of Nitrobacter populations in soil by PCR. Appl Environ Microbiol 61:2093–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mobarry B, Wagner M, Urbain V, Rittmann B, Stahl D. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol 62:2156–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane DJ. 1991. 16/23S rRNA sequencing, p 115–171. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amann R, Ludwig W, Schleifer K-H. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol 15:593–600. doi: 10.1016/S0723-2020(11)80121-9. [DOI] [Google Scholar]
- 37.Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 38.Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol 67:5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spurr AR. 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26:31–43. doi: 10.1016/S0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- 40.Watson ML. 1958. Staining of tissue sections for electron microscopy with heavy metals. J Cell Biol 4:475–478. doi: 10.1083/jcb.4.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds ES. 1963. The use of lead citrate at high pH as an electron-opaque stain electron microscopy. J Cell Biol 17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S, Lee CS, Ling J, Blancheton JP. 2006. Nitrification kinetics of biofilm as affected by water quality factors. Aquac Eng 34:179–197. doi: 10.1016/j.aquaeng.2005.09.004. [DOI] [Google Scholar]
- 43.Crab R, Avnimelech Y, Defoirdt T, Bossier P, Verstraete W. 2007. Nitrogen removal techniques in aquaculture for a sustainable production. Aquaculture 270:1–14. doi: 10.1016/j.aquaculture.2007.05.006. [DOI] [Google Scholar]
- 44.Strauss EA, Mitchell NL, Lamberti GA. 2002. Factors regulating nitrification in aquatic sediments: effects of organic carbon, nitrogen availability, and pH. Can J Fish Aquat Sci 59:554–563. doi: 10.1139/f02-032. [DOI] [Google Scholar]
- 45.Jiménez E, Giménez JB, Ruano MV, Ferrer J, Serralta J. 2011. Effect of pH and nitrite concentration on nitrite oxidation rate. Bioresour Technol 102:8741–8747. doi: 10.1016/j.biortech.2011.07.092. [DOI] [PubMed] [Google Scholar]
- 46.Keen GA, Prosser JI. 1987. Steady state and transient growth of autotrophic nitrifying bacteria. Arch Microbiol 147:73–79. doi: 10.1007/BF00492908. [DOI] [Google Scholar]
- 47.Schroeder JP, Klatt SF, Schlachter M, Zablotski Y, Keuter S, Spieck E, Schulz C. 2015. Impact of ozonation and residual ozone-produced oxidants on the nitrification performance of moving-bed biofilters from marine recirculating aquaculture systems. Aquac Eng 65:27–36. doi: 10.1016/j.aquaeng.2014.10.008. [DOI] [Google Scholar]
- 48.Shammas NK. 1986. Interactions of pH, temperature and biomass on the nitrification process. Water Pollut Control Fed J 58:52–59. [Google Scholar]
- 49.Kir M. 2009. Nitrification performance of a submerged biofilter in a laboratory scale size of the recirculating shrimp system. Turkish J Fish Aquat Sci 9:209–214. [Google Scholar]
- 50.van Kessel MAHJ, Harhangi HR, van de Pas-Schoonen K, van de Vossenberg J, Flik G, Jetten MSM, Klaren PHM, Op den Camp HJM. 2010. Biodiversity of N-cycle bacteria in nitrogen removing moving-bed biofilters for freshwater recirculating aquaculture systems. Aquaculture 306:177–184. doi: 10.1016/j.aquaculture.2010.05.019. [DOI] [Google Scholar]
- 51.Itoi S, Niki A, Sugita H. 2006. Changes in microbial communities associated with the conditioning of filter material in recirculating aquaculture systems of the pufferfish Takifugu rubripes. Aquaculture 256:287–295. doi: 10.1016/j.aquaculture.2006.02.037. [DOI] [Google Scholar]
- 52.Nowka B, Daims H, Spieck E. 2015. Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation. Appl Environ Microbiol 81:745–753. doi: 10.1128/AEM.02734-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar VJR, Joseph V, Philip R, Singh ISB. 2010. Nitrification in brackish water recirculating aquaculture system integrated with activated packed bed bioreactor. Water Sci Technol 61:797. doi: 10.2166/wst.2010.849. [DOI] [PubMed] [Google Scholar]
- 54.Blackburne R, Vadivelu VM, Yuan Z, Keller J. 2007. Kinetic characterisation of an enriched Nitrospira culture with comparison to Nitrobacter. Water Res 41:3033–3042. doi: 10.1016/j.watres.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 55.Udert KM, Larsen TA, Gujer W. 2005. Chemical nitrite oxidation in acid solutions as a consequence of microbial ammonium oxidation. Environ Sci Technol 39:4066–4075. doi: 10.1021/es048422m. [DOI] [PubMed] [Google Scholar]
- 56.Spieck E, Hartwig C, McCormack I, Maixner F, Wagner M, Lipski A, Daims H. 2006. Selective enrichment and molecular characterization of a previously uncultured Nitrospira-like bacterium from activated sludge. Environ Microbiol 8:405–415. doi: 10.1111/j.1462-2920.2005.00905.x. [DOI] [PubMed] [Google Scholar]
- 57.Nowka B, Off S, Daims H, Spieck E. 2015. Improved isolation strategies allowed the phenotypic differentiation of two Nitrospira strains from widespread phylogenetic lineages. FEMS Microbiol Ecol 91:fiu031. doi: 10.1093/femsec/fiu031. [DOI] [PubMed] [Google Scholar]
- 58.Tarre S, Green M. 2004. High-rate nitrification at low pH in suspended- and attached-biomass reactors. Appl Environ Microbiol 70:6481–6487. doi: 10.1128/AEM.70.11.6481-6487.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gieseke A, Tarre S, Green M, De Beer D. 2006. Nitrification in a biofilm at low pH values: role of in situ microenvironments and acid tolerance. Appl Environ Microbiol 72:4283–4292. doi: 10.1128/AEM.00241-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hankinson TR, Schmidt EL. 1984. Examination of an acid forest soil for ammonia- and nitrite-oxidizing autotrophic bacteria. Can J Microbiol 30:1125–1132. doi: 10.1139/m84-176. [DOI] [Google Scholar]
- 61.Hankinson TR, Schmidt EL. 1988. An acidophilic and a neutrophilic Nitrobacter strain isolated from the numerically predominant nitrite-oxidizing population of an acid forest soil. Appl Environ Microbiol 54:1536–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanders T, Fiencke C, Pfeiffer EM. 2010. Small-scale variability of dissolved inorganic nitrogen (DIN), C/N ratios, and ammonia-oxidizing capacities in various permafrost affected soils of Samoylov Island, Lena River Delta, Northeast Siberia. Polarforschung 80:23–35. [Google Scholar]
- 63.Percent SF, Frischer ME, Vescio P, Duffy EB, Milano V, McLellan M, Stevens BM, Boylen CW, Nierzwicki-Bauer S. 2008. Bacterial community structure of acid-impacted lakes: what controls diversity? Appl Environ Microbiol 74:1856–1868. doi: 10.1128/AEM.01719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.