Abstract
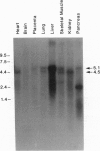
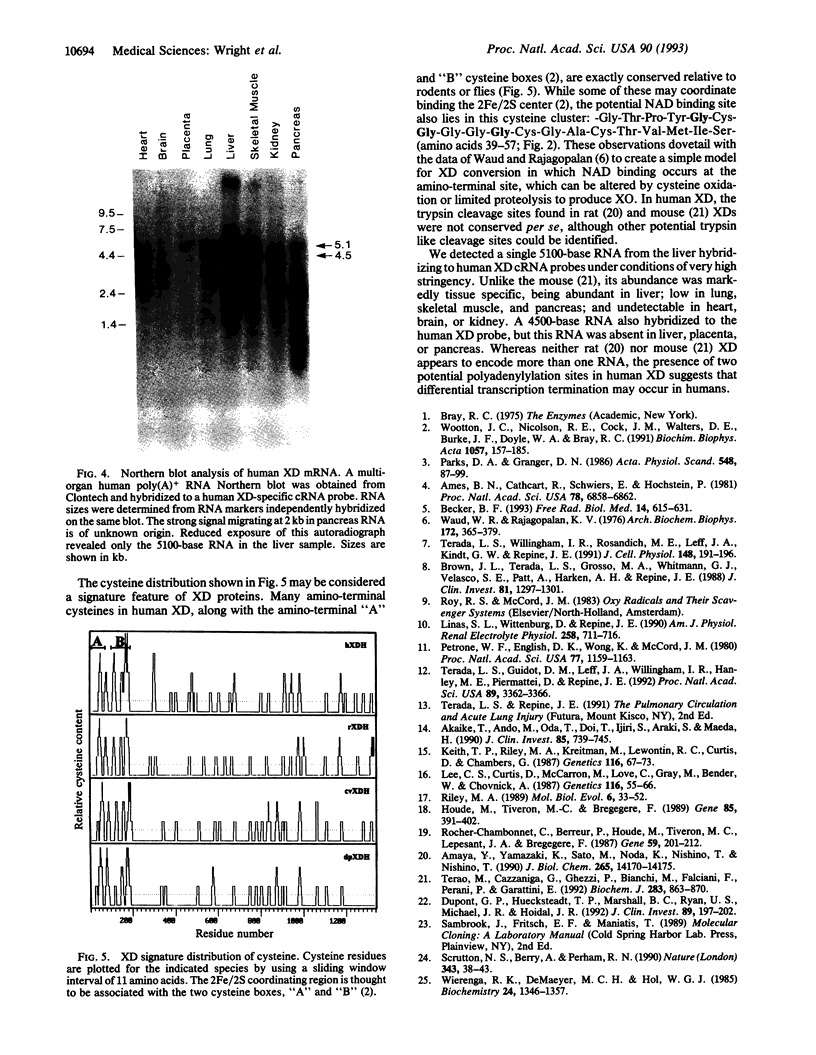
We isolated cDNAs encoding xanthine dehydrogenase (XD; xanthine:NAD+ oxidoreductase, EC 1.1.1.204) from a human liver cDNA library. The complete nucleotide sequence of human XD was determined; the deduced amino acid sequence encoded a protein of 1336 amino acid residues of M(r) 147,782. Human XD possessed many of the signature sequences typical of XDs from flies and rodents, including an unusual cysteine distribution, a potential 2Fe/2S binding site, and a putative molybdopterin cofactor binding domain. Analysis of potential NAD binding sites suggested a simple hypothesis for the conversion of human XD into the oxygen metabolite forming xanthine oxidase (XO; xanthine:oxygen oxidoreductase, EC 1.1.3.22). Using a human XD complementary RNA hybridization probe, we found a 5100-base RNA in human liver by RNA blot-hybridization analysis. This RNA exhibited tissue-specific distribution that may be pertinent to XD- and XO-mediated oxygen radical injury in ischemia/reperfusion and inflammation. A second 4500-base RNA was detected in some tissues and may arise through differential transcription termination.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akaike T., Ando M., Oda T., Doi T., Ijiri S., Araki S., Maeda H. Dependence on O2- generation by xanthine oxidase of pathogenesis of influenza virus infection in mice. J Clin Invest. 1990 Mar;85(3):739–745. doi: 10.1172/JCI114499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaya Y., Yamazaki K., Sato M., Noda K., Nishino T., Nishino T. Proteolytic conversion of xanthine dehydrogenase from the NAD-dependent type to the O2-dependent type. Amino acid sequence of rat liver xanthine dehydrogenase and identification of the cleavage sites of the enzyme protein during irreversible conversion by trypsin. J Biol Chem. 1990 Aug 25;265(24):14170–14175. [PubMed] [Google Scholar]
- Ames B. N., Cathcart R., Schwiers E., Hochstein P. Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: a hypothesis. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6858–6862. doi: 10.1073/pnas.78.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B. F. Towards the physiological function of uric acid. Free Radic Biol Med. 1993 Jun;14(6):615–631. doi: 10.1016/0891-5849(93)90143-i. [DOI] [PubMed] [Google Scholar]
- Brown J. M., Terada L. S., Grosso M. A., Whitmann G. J., Velasco S. E., Patt A., Harken A. H., Repine J. E. Xanthine oxidase produces hydrogen peroxide which contributes to reperfusion injury of ischemic, isolated, perfused rat hearts. J Clin Invest. 1988 Apr;81(4):1297–1301. doi: 10.1172/JCI113448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont G. P., Huecksteadt T. P., Marshall B. C., Ryan U. S., Michael J. R., Hoidal J. R. Regulation of xanthine dehydrogenase and xanthine oxidase activity and gene expression in cultured rat pulmonary endothelial cells. J Clin Invest. 1992 Jan;89(1):197–202. doi: 10.1172/JCI115563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde M., Tiveron M. C., Brégégère F. Divergence of the nucleotide sequences encoding xanthine dehydrogenase in Calliphora vicina and Drosophila melanogaster. Gene. 1989 Dec 28;85(2):391–402. doi: 10.1016/0378-1119(89)90432-0. [DOI] [PubMed] [Google Scholar]
- Keith T. P., Riley M. A., Kreitman M., Lewontin R. C., Curtis D., Chambers G. Sequence of the structural gene for xanthine dehydrogenase (rosy locus) in Drosophila melanogaster. Genetics. 1987 May;116(1):67–73. doi: 10.1093/genetics/116.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. S., Curtis D., McCarron M., Love C., Gray M., Bender W., Chovnick A. Mutations affecting expression of the rosy locus in Drosophila melanogaster. Genetics. 1987 May;116(1):55–66. doi: 10.1093/genetics/116.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D. A., Granger D. N. Xanthine oxidase: biochemistry, distribution and physiology. Acta Physiol Scand Suppl. 1986;548:87–99. [PubMed] [Google Scholar]
- Petrone W. F., English D. K., Wong K., McCord J. M. Free radicals and inflammation: superoxide-dependent activation of a neutrophil chemotactic factor in plasma. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1159–1163. doi: 10.1073/pnas.77.2.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M. A. Nucleotide sequence of the Xdh region in Drosophila pseudoobscura and an analysis of the evolution of synonymous codons. Mol Biol Evol. 1989 Jan;6(1):33–52. doi: 10.1093/oxfordjournals.molbev.a040529. [DOI] [PubMed] [Google Scholar]
- Rocher-Chambonnet C., Berreur P., Houde M., Tiveron M. C., Lepesant J. A., Brégégère F. Cloning and partial characterization of the xanthine dehydrogenase gene of Calliphora vicina, a distant relative of Drosophila melanogaster. Gene. 1987;59(2-3):201–212. doi: 10.1016/0378-1119(87)90328-3. [DOI] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990 Jan 4;343(6253):38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- Terada L. S., Guidot D. M., Leff J. A., Willingham I. R., Hanley M. E., Piermattei D., Repine J. E. Hypoxia injures endothelial cells by increasing endogenous xanthine oxidase activity. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3362–3366. doi: 10.1073/pnas.89.8.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada L. S., Willingham I. R., Rosandich M. E., Leff J. A., Kindt G. W., Repine J. E. Generation of superoxide anion by brain endothelial cell xanthine oxidase. J Cell Physiol. 1991 Aug;148(2):191–196. doi: 10.1002/jcp.1041480202. [DOI] [PubMed] [Google Scholar]
- Terao M., Cazzaniga G., Ghezzi P., Bianchi M., Falciani F., Perani P., Garattini E. Molecular cloning of a cDNA coding for mouse liver xanthine dehydrogenase. Regulation of its transcript by interferons in vivo. Biochem J. 1992 May 1;283(Pt 3):863–870. doi: 10.1042/bj2830863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waud W. R., Rajagopalan K. V. The mechanism of conversion of rat liver xanthine dehydrogenase from an NAD+-dependent form (type D) to an O2-dependent form (type O). Arch Biochem Biophys. 1976 Feb;172(2):365–379. doi: 10.1016/0003-9861(76)90088-6. [DOI] [PubMed] [Google Scholar]
- Wootton J. C., Nicolson R. E., Cock J. M., Walters D. E., Burke J. F., Doyle W. A., Bray R. C. Enzymes depending on the pterin molybdenum cofactor: sequence families, spectroscopic properties of molybdenum and possible cofactor-binding domains. Biochim Biophys Acta. 1991 Mar 29;1057(2):157–185. doi: 10.1016/s0005-2728(05)80100-8. [DOI] [PubMed] [Google Scholar]