Abstract
Staphylococcus aureus is an important human pathogen that causes healthcare-associated and community-acquired infections. Moreover, the growing prevalence of multiresistant strains requires the development of alternative methods to antibiotic therapy. One effective therapeutic option may be antimicrobial photodynamic inactivation (aPDI). Recently, S. aureus strain-dependent response to PDI was demonstrated, although the mechanism underlying this phenomenon remains unexplained. The aim of the current study was to investigate statistically relevant correlations between the functionality and polymorphisms of agr gene determined for 750 methicillin-susceptible and methicillin-resistant S. aureus strains and their responses to photodynamic inactivation using protoporphyrin IX. An AluI and RsaI digestion of the agr gene PCR product revealed existing correlations between the determined digestion profiles (designations used for the first time) and the PDI response. Moreover, the functionality of the agr system affected S. aureus susceptibility to PDI. Based on our results, we conclude that the agr gene may be a genetic factor affecting the strain dependent response to PDI.
Keywords: accessory gene regulator, gene polymorphism, photoinactivation, Staphylococcus aureus
1. Introduction
Staphylococcus aureus is one of the most important known human pathogens. It is the etiological agent in many infections, including local infections associated with skin and soft tissue damage such as wound infections, deep-seated infections (e.g., myositis or osteomyelitis), and device-related infections, as well as toxin-mediated diseases such as toxic shock syndrome (TSS) and staphylococcal foodborne diseases (SFD) [1;2]. Healthcare-associated infections, especially those caused by methicillin-resistant S. aureus strains (MRSA), are a great danger to both hospitalized and immunocompromised patients in whom the organism causes high morbidity and mortality [3]. S. aureus possesses a wide spectrum of virulence factors, including exoproteins (i.e., hemolysins, nucleases, and proteases) that facilitate host cell lysis and cell wall-associated adhesins (i.e., fibronectin-binding protein and protein A) required for the colonization of host tissues. In general, the expression of staphylococcal virulence factors is regulated through the quorum sensing mechanism by the accessory gene regulator (agr). Although the agr locus is conserved among staphylococcal species, it consists of a polymorphic, hypervariable fragment used to cluster S. aureus strains into one of four agr groups using polymerase chain reaction (PCR) methods [4;5].
Increasing antibiotic resistance among pathogenic bacteria has forced researchers to find alternative therapeutic options against which the bacteria will not be easily able to develop resistance. Photodynamic therapy could be one such alternative. Studies of the photoinactivation (PDI) of multiresistant pathogenic bacteria have shown that they are as susceptible to PDI as their naïve counterparts [6;7]. Photodynamic therapy (PDT), which is generally recognized as a cancer treatment, utilizes photosensitizers (PS, usually non-toxic dyes) that selectively accumulate in the target cells (i.e., malignant tissues or microorganisms; if the therapy involves microorganisms, then this therapy is termed photodynamic inactivation, or PDI) [8]. The appropriate wavelength of visible light is then used to excite the PS molecules to the singlet state, and excited sensitizers undergo triplet state reactions by either Type I or Type II pathways [9]. The Type I mechanism involves electron-transfer from the triplet state PS to the substrate, producing cytotoxic reactive species such as superoxide or hydroxyl radicals [10]. The Type II mechanism is based on energy transfer from the triplet state PS to molecular oxygen (ground triplet state) to produce highly cytotoxic singlet oxygen [11].
We have recently described the effect of PDI against different strains of S. aureus and demonstrated a strain-dependent effectiveness for PDI [12;13]. The mechanism underlying this phenomenon is still poorly understood. The current study is part of a wider project that aims to investigate genetic correlations with the bactericidal effect of PDI on S. aureus. Specifically, the aim of this study was to analyze the effect of PDI on 750 strains of both MSSA and MRSA S. aureus and to determine if the effect is related to different genetic profiles involving the agr gene. This genetic element is widely used for S. aureus typing and influences staphylococcal virulence. It is claimed that agr-group-mediated differences in the expression of various virulence factors influence strain pathogenicity and disease progression, indicating that the activity of the agr system affects strain virulence [5]. Thus, the search for genetic polymorphisms in this element and the determination of the genetic background of particular strains can have an important diagnostic value. Moreover, it was shown that the agr system has a built-in oxidation-sensing mechanism through the DNA-binding domain of the response regulator AgrA [14]. Mutagenesis studies further established that S. aureus strain expressing AgrA varying in aminoacid sequence is more susceptible to H2O2.
Moreover, microarray analysis revealed that agr function is upregulated by photodynamic treatment and is related to resistance against PDI [15]. These results show that oxidation sensing is a component of the quorum-sensing agr signaling system and it is justified to analyse the influence of the agr locus, its functionality and polymorphism on S. aureus susceptibility to PDI which acts due to oxidative mechanisms. We hypothesise that both the functionality and polymorphism of agr could influence S. aureus susceptibility to PDI-induced oxidative mechanisms.
2. Materials and Methods
2.1. Bacterial isolates
In total, 750 clinical S. aureus strains isolated from 2002 to 2012 at the Provincial Hospital in Gdansk and Hospital in Koszalin, Poland, were used; of these, 307 (41.0%) were MRSA and 443 (59.0%) were MSSA. The isolates were characterized by Gram staining and the ability to produce coagulase and the clumping factor using Slidex Staph Plus (bioMèrieux, France). Species were identified using the MALDI-TOF MS (Bruker Daltonics, Germany). Methicillin resistance was determined using a disc-diffusion method as well as a latex test detecting PBP2a protein (Staphytect Plus, OXOID, US) [16]. Of the 750 strains collected, 27.5% (206 isolates) were isolated from surveillance cultures, 52.5% (394 isolates) from patients with local infections, 16.3% (122 isolates) from patients with bacteremia and generalized infections and 3.7% (28 isolates) from infections connected with endoprostheses. Additionally, the isogenic S. aureus agr-positive/negative pairs were used (JE2, NE95, NE1532, NE873; obtained through the Network of Antimicrobial Resistance in Staphylococcus aureus (NARSA) and agr isogenic pair of S. aureus LAC strain was used (LAC wild type, and LAC agr-negative mutant), and control strains of S. aureus listed in Table 1. Moreover, for assessment of hemolysin activity the reference S. aureus RN4220 was used.
Table 1.
Primers and reference strains used in the study
| Strain used as standard | Primer | Oligonucleotide sequence (5’ – 3’) | Size (bp) | Reference |
|---|---|---|---|---|
| RN6390 | Agr | Forward: ACC AGT TTG CCA CGT ATC | 1,884 | [19] |
| Reverse: TAA ACC ACG ACC TTC ACC | ||||
| RN6390 | Agr 1 | Forward: ATG CAC ATG GTG CAC ATG C | 441 | [18] |
| Reverse: GTC ACA AGT ACT ATA AGC TGC GAT | ||||
| RN6607 | Agr 2 | Forward: ATG CAC ATG GTG CAC ATG C | 575 | [18] |
| Reverse: TAT TAC TAA TTG AAA AGT GGC CAT AGC | ||||
| RN8465 | Agr 3 | Forward: ATG CAC ATG GTG CAC ATG C | 323 | [18] |
| Reverse: GTA ATG TAA TAG CTT GTA TAA TAA TAC CCA G | ||||
| RN4850 | Agr 4 | Forward: ATG CAC ATG GTG CAC ATG C | 659 | [18] |
| Reverse: CGA TAA TGC CGT AAT CG |
2.2. MALDI-TOF MS
All strains were examined by MALDI-TOF MS using a Microflex LT instrument (Bruker Daltonics, Germany), Flexcontrol 3.0 software and the Biotyper 2.0 database (Bruker Daltonics, Germany). The data analysis was performed according to the manufacturer’s instructions. The samples were covered with 1 ml matrix solution (a saturated solution of a-cyano-4-hydroxycinnamic acid in 50% acetonitrile, 2.5% trifluoroacetic acid). The analysed mass range of spectra was 2000–20 000 m/z. Each spectrum was obtained after 240 shots in an automatic acquisition mode. For the identification approach, a mass-to-charge range of 3000–15 000 Da was used. Identification was performed in duplicate and the higher score was retained. The identification score cut-off values were applied on each measurement according to the manufacturer’s instructions. According to this score system, a score of < 2 is recommended for a probable species and a score of greater than 2.3 is recommended for a secure species identification [17].
2.3. Nucleic acid isolation
For DNA isolation, strains were grown overnight in nutrient trypticase soy broth (TSB) (bioMèrieux, France) at 37°C for 24 h. Then, 1.5 ml of the bacterial culture was centrifuged at 5000 x g for 10 min. The pellet was suspended in 180 μl of lysis buffer (20 mM Tris-HCl, pH 8.0, 2 mM EDTA, 1.2% Triton X-100, lysozyme 20 mg/ml) and the GeneJET™ Genomic DNA Purification Kit (Thermo Scientific, Lithuania) was used for DNA isolation according to the manufacturer’s procedure. The DNA concentrations were measured using NanoDrop ND-1000 (Thermo Scientific, Lithuania) and were in the range from 10 to 100 ng per microliter. In addition, quality of the isolated DNA was confirmed by gel electrophoresis.
2.4. Random Amplification of Polymorphic DNA Fragments
To assess the clonal relatedness of studied MRSA S. aureus strains, the RAPD technique was applied. The isolates were typed using primer qfaseq4 – 5’ CCC ACT GTG GTG TTC ATA 3’. The amplifications were performed with approximately 30–40 ng of S. aureus DNA in a 50 μl reaction mixture containing 5 μl of 10xTaqNova reaction buffer (750 mM Tris-HCl pH 8.8, 200 mM (NH4)2SO4, 0.1% (v/v) Tween 20, 2 μl of 50 mM MgCl2, 5 μl of dNTP mixture (2 mM of each) and 1.5 μl of primer (10 μM). This mixture was supplemented with 1 unit of TaqNova DNA polymerase (DNA-Gdansk, Poland). The PCRs were performed in a Veriti® Thermal Cycler as follows: initial denaturation at 94 °C for 2 min; ten cycles of denaturation at 94 °C for 30 s, annealing at 30 °C for 3 min and extension at 72 °C for 2 min; and 30 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 2 min and extension at 72 °C for 2 min. After the last cycle, a final extension step was performed at 72 °C for 5 min. The amplification products were analyzed by electrophoresis of 10 μl samples on 5% polyacrylamide gels. The above RAPD protocol was established after preliminary trials of various reaction parameters (DNA, polymerase and magnesium salt concentration, PCR thermal profile optimisation) and control experiments to test the reproducibility of the method. The electrophoresis conditions were standardized and applied as follows: 10 μl of sample per well, 5% polyacrylamide gel, TBE buffer, 4.4 V/cm, 14h. The DNA bands obtained by the RAPD method described above, were visualized by UV transillumination after ethidium bromide staining. All RAPD electrophoregrams were analysed with FPQuest TM II program. The RAPD profiles were compared according to the DNA Molecular Weight Marker M10kpz (DNA-Gdansk, Poland). DNA relatedness was calculated by the band-based Dice coefficient with a setting of 2% band tolerance using the un-weighed pair group method with mathematical averaging (UPGMA). Inter-gel similarity coefficient was 90%.
2.5. Agr group-specific multiplex PCR
The agr sequences were amplified from 4 μl of the purified nucleic acid solutions in a 25 μl reaction mixture containing 1 U of Taq DNA Polymerase (Thermo Scientific, Lithuania), Taq Buffer with (NH4)2SO4 (Thermo Scientific, Lithuania), 200 μM dNTPs (MetaBion, Germany), 5 mM MgCl2, and a 1.2 μM concentration of each of the primers Agr1-4 listed in Table 1. Amplification was performed in a BIOMETRA thermocycler (UNO II) using the following temperature program: 1 cycle of 5 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 55°C, 60 s at 72°C; and a final cycle of 7 min at 72°C. Amplification products were electrophoresed in a 1.5% agarose gel (BIORON, Germany) containing ethidium bromide and visualized by transillumination (Gel Doc 2000, Bio-Rad) under UV irradiation [18].
2.6. PCR amplification of the agr gene
The 1,884-bp agr gene fragment was amplified using primers Agr listed in Table 1. The agr gene was amplified from 4 μl of the purified nucleic acid solution in a 25 μl reaction mixture containing 1 U of DyNAzyme™ EXT Taq DNA Polymerase (Thermo Scientific, Lithuania), 10X EXT Buffer (Thermo Scientific, Lithuania), 200 μM dNTPs (MetaBion, Germany), 1.6 μM sense primer, and 1.6 μM antisense primer (Table 1). Amplification was performed in a BIOMETRA thermocycler (UNO II) using the following temperature program: 1 cycle of 4 min at 94°C; 30 cycles of 1 min at 94°C, 2 min at 55°C, 2 min at 72°C; and a final cycle of 4 min at 72°C. All samples were stored at −20°C prior to restriction enzyme digestion [19].
2.7. Restriction enzyme digestion of the PCR products
The agr PCR amplicons were digested with RsaI (Thermo Scientific, Lithuania) and AluI (Thermo Scientific, Lithuania) in a 31 μl reaction mixture containing 1 U of restriction enzyme and 10X Buffer Tango (Thermo Scientific, Lithuania). Digestions of the agr amplicons were performed in a BIOMETRA thermocycler (UNO II) with an incubation time of 1 h at 37°C followed by thermal inactivation of the restriction enzymes by incubation at 65°C for 20 min, according to the manufacturer’s instructions. The restriction fragments were then separated by electrophoresis on a 2% agarose gel (BIORON, Germany) containing ethidium bromide and visualized by transillumination (Gel Doc 2000, Bio-Rad) under UV irradiation.
2.8. Assessment of hemolysin activity on Sheep Blood Agar plates
Cultures to be tested were grown overnight on trypticase-soy agar plates (bioMèrieux, France), cross-streaked on sheep blood agar (Biocorp, Poland) against a culture of RN4220, incubated overnight at 37°C, and then incubated for 6 h at 4°C. The patterns could be interpreted according to previously described method [20]: beta-hemolysin appears as a partially turbid zone, as seen with RN4220, which produces only beta-hemolysin. Delta-hemolysin is synergistic with beta-hemolysin and is seen as a clearing where the two hemolysins intersect. Coproduction of beta-hemolysin and delta-hemolysin is seen as a clearing next to the streak within a wider beta-hemolysin zone [20].
2.9. Photosensitizer
Protoporphyrin IX (PPIX) (MP Biomedicals, LLC, USA) was used as a sensitizer. PPIX was dissolved in dimethyl sulfoxide (DMSO; Sigma, Germany) to make a 1 mM stock solution and was stored in the dark at −20°C until use. The concentration of the photosensitizer was determined spectrophotometrically (extinction coefficient 1.6 x 105 M−1 cm−1, wavelength 408 nm).
2.10. Light source
Illumination was performed using a Q.Light® PDT Lamp (b & p® Shweiz AG, Switzerland) (ISO 9001 & EN 46001 - CE 1275) (power of lamp 80 mW; fluence rate 102 mW/cm2; fluence 6.12 J/cm2 per minute). The delivered light energy was determined with the use of light power meter (model LM1, CARL ZEISS, Germany). The Q.Light® PDT Lamp emits polarized light (polarization level: 98%) over a wavelength range of 620 to 780 nm.
2.11. PDI studies
All bacterial isolates were subjected to photodynamic inactivation (PDI). Bacterial cultures were grown for 24 h at 37°C in nutrient trypticase soy broth (bioMèrieux, France) and then diluted with fresh broth to an appropriate density (107/ml bacterial cells) based on densitometry (Densi Meter II, EMO). Diluted S. aureus cultures were incubated with 25 μM protoporphyrin IX in the dark at 37°C for 30 min (final DMSO concentration in the sample was 2.5%). The cells were then transferred to a 96-well microtiter plate (100 μl per well) and illuminated with appropriate light (50 J/cm2) at room temperature for 8 min and 10 s. Following illumination, 10 μl aliquots were used to determine the colony forming units (CFU). The contents of the wells were mixed before sampling, and the aliquots were serially diluted 10-fold in PBS (0.13 mM NaCl, 8.1 mM Na2HPO4, 2.68 mM KCl, 1.47 mM KH2PO4) to achieve final dilutions of 10−1 to 10−4, which were then streaked horizontally onto square trypticase soy agar plates [21]. The plates were incubated at 37°C overnight. Controls consisted of untreated bacteria (no photosensitizers or light) kept in 96-well plates for the duration of the illumination. After 18 h of incubation at 37°C in the dark, CFUs were counted and the results were statistically analyzed. Survival fractions were expressed as ratios of CFUs of treated bacteria (with light and photosensitizer) to CFUs of untreated bacteria. The experiment was performed three times.
2.12. Statistical analysis
The statistical analyses have been performed using the statistical suite StatSoft. Inc. (2011). STATISTICA (data analysis software system). version 10.0. www.statsoft.com and Excel. The quantitive variables were characterized by the arithmetic mean of standard deviation or median or max/min (range) and 95% confidence interval. The qualitative variables were presented with the use of count and percentage. In order to check if a quantitive variable derives from a population of normal distribution, the W Shapiro-Wilk test has been used. Whereas to prove the hypotheses on homogeneity of variances, Leven (Brown-Forsythe) test has been utilized. Statistical significance of differences between two groups (unpaired variables model) was processed with the t-Student test (or Welch test in the case of lack of homogeneity) or U Mann-Withney test (in cases where conditions of performing the t-Student test were not satisfied or for variables measured by ordinal scale). The significance of difference between more than two groups were assessed with F test (ANOVA) or Kruskal-Wallis (if ANOVA conditions were not fulfilled). In the case of statistically significant differences between two groups post hoc tests were utilized (Tukey test for F or Dunn for Kruskal-Wallis). Chi-squared tests for independence were used for qualitative variables (with the use of Yates correction for cell counts below 10, with check of Cochrane’s conditions or with Fisher's exact test respectively). In all the calculations the statistical significance level of p < 0.05 has been used.
3. Results
3.1. Clinical isolates of S. aureus heterogeneously respond to PDI
All 750 strains, including both methicillin-susceptible and methicillin-resistant S. aureus, were subjected to photodynamic inactivation. The different strains expressed a heterogeneous response to photoinactivation, ranging from 0 to 5.1 log10-unit reductions in viable counts under the same experimental conditions. Among the MRSA strains, the viable count reduction ranged from 0 to 4.5 log10-units, while for the MSSA strains, the range was from 0 to 5.1 log10-units. Using the observed differences in strain responses to PDI to simplify the analysis, the strains were classified into three groups of PDI responses according to the extent of the log10-unit reduction in viable counts.
Terms and the ranges used for classification are shown in Table 2. The cut-offs for the categorization of PDI response was made according to lethal or sub-lethal damage caused for S. aureus strains. Lethal and sub-lethal conditions are defined as conditions where the direct damage is sufficient or insufficient to destroy the majority of the bacterial population [22]. Thus, lethal and sub-lethal damage were defined as resulted in > 2 log10 and 0–0.99 log10 unit reduction in viable count, respectively. Thus, strains reaching lethal or sub-lethal damage were assigned as sensitive or resistant to PDI, respectively. Strains revealing reduction in viable counts ranged between lethal and sub- lethal damage were assigned as intermediate sensitive.
Table 2.
Range of effectiveness of photosensitization
| log10 -unit reduction | Classificationa | No. of strains (%) |
|---|---|---|
| 0 – 0.99 | Resistant | 391 (52.1) |
| 1 – 1.99 | Intermediate sensitive | 210 (28.0) |
| 2 – 5.1 | Sensitive | 149 (19.9) |
|
| ||
| Total | 750 (100) | |
Categorization made due to lethal and sub-lethal damage caused by photoinactivation
The PDI effect on the 750 analyzed strains of methicillin-susceptible and methicillin-resistant S. aureus showed different distribution of PDI-responders across S. aureus population (Table 2).
3.2. Functionality of agr locus influences response to PDI
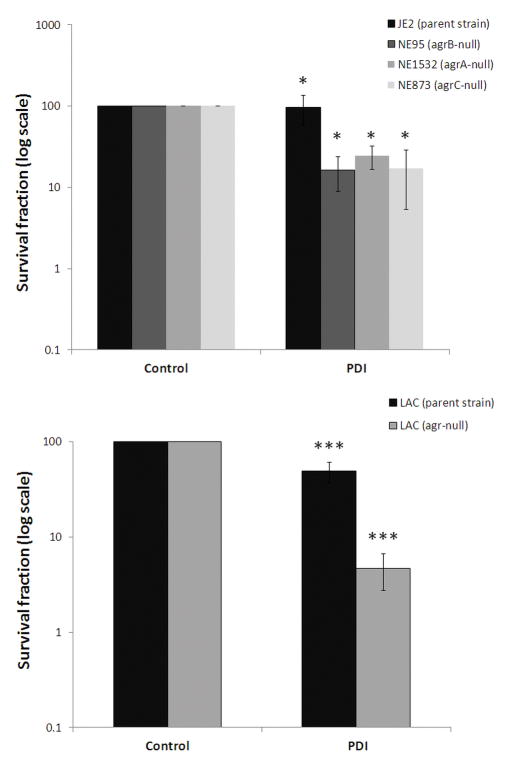
To establish whether agr gene status could correlate with PDI phenotype, the agr functionality test (delta-toxin hemolytic activity) was applied for all tested S. aureus strains. For 633 strains agr gene was determined to be active, as delta-toxin hemolytic activity was reported. Next, S. aureus groups differentiated due to the functionality of agr locus were compared according to their response to photodynamic treatment. Strains lacking functional agr system revealed increased susceptibility to PDI (p < 0.01) (Fig. 1). Moreover, to indicate the correlation between agr function and the response to PDI, the photoinactivation was applied to isogenic pairs of S. aureus that differ in agr status (agr-positive JE2 and agrA-, agrB- and agrC-negative strains, NE1532, NE95, NE873, respectively). The significant difference (p < 0.05) in response to PDI was observed for reference strains according to agr status indicating its influence on strains susceptibility to photoinactivation (Fig. 2, upper panel). When entire gene was deleted (in the case of S. aureus LAC isogenic strains) the difference in response to PDI was even more pronounced (p < 0.001) indicating the significant role of agr locus in S. aureus response to photoinactivation (Fig. 2, lower panel).
Fig. 1. Response of S. aureus strains differing in agr functionality to PPIX-mediated PDI.
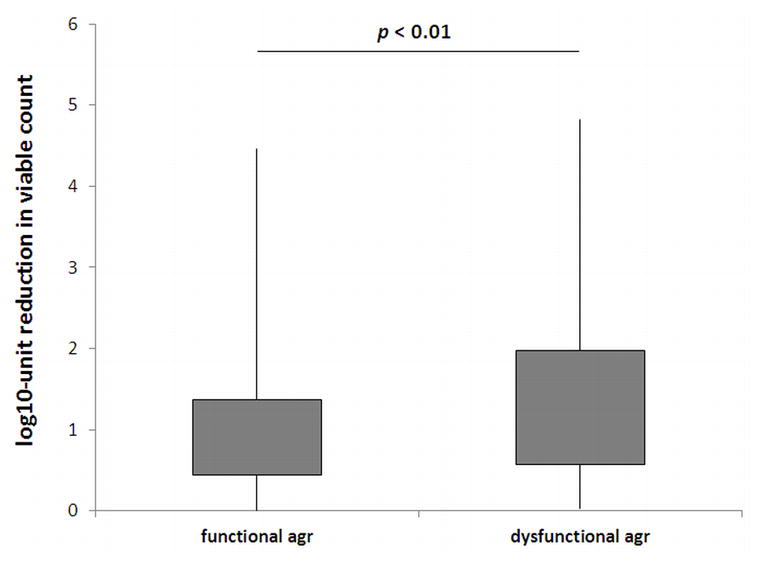
Each box plot represents the spread of bacterial response across the different clinical isolates. The error bars represent minimum and maximum value of log10-unit reduction in viable counts.
Fig. 2. Photoeffect of PPIX against isogenic S. aureus strains.
The cells were illuminated after dark incubation for 30 min at 37°C with 25 μM PPIX with the light dose of 50 J/cm2. The survival rate was calculated from the number of CFU in the PDI-treated sample divided by the number of CFU in the sample irradiated without photosensitizer. Each experiment was done three times, and error bars show S.D. * p < 0.05; *** p < 0.001
3.3. The genetic polymorphism pattern of the agr gene correlates with PDI phenotype
Next, to assess whether not only the functionality of agr locus but also its polymorphism could influences the susceptibility to PDI, 633 isolates with active agr gene were used for analysis of agr polymorphism. Thus, further analysis concerning agr grouping and polymorphism were performed. An agr group-specific multiplex PCR reaction was performed for all 633 S. aureus strains to amplify the agr sequence. The banding patterns obtained were compared to the PCR products of agr group-specific reference strains to determine the agr group for all analyzed strains. The results showed that there were statistically significant differences between agr groups according to the response to PDI. Strains with agr group 3 differently responded to photoinactivation in comparison to strains agr 1, 2 and 4 (p = 0.0001; 0.0001 and 0.0481, respectively). Agr group 1 strains revealed significant difference in response to PDI when compared to agr group 2 strains (p = 0.001) (Fig. 3).
Fig. 3. Response of S. aureus strains differing in agr groups to PPIX-mediated PDI.
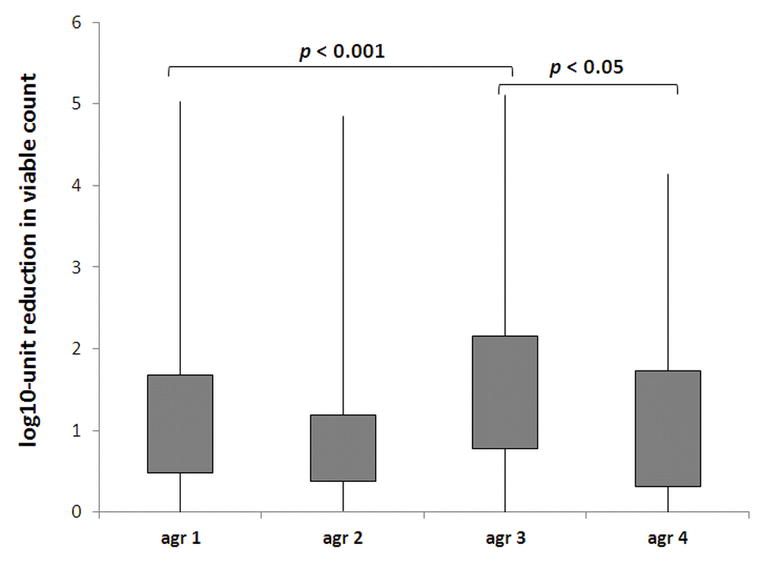
Each box plot represents the spread of bacterial response across the different clinical isolates. The error bars represent minimum and maximum value of log10-unit reduction in viable counts.
To examine the genetic elements in more detail, we used PCR to amplify the agr gene. The PCR products were then enzymatically digested with AluI, creating different digestion products that corresponded to discrete polymorphisms in the agr gene; these polymorphisms were subsequently named with roman numerals: I, II, IV, IVB, V, VI, VII, IX, and X (Supplementary Figure 1). Statistical analysis demonstrated that S. aureus strains with AluI patterns II and V differently respond to PDI in comparison to patterns I, IV and IVB (significant correlations and p values are presented in Table 3). Interestingly, the majority of strains characterized with patterns no. I, IV, IVB, IX and X were resistant to photoinactivation (Table 3).
Table 3.
Statistical analysis of AluI and RsaI determined polymorphisms over S. aureus response to PDI (N/%).
| AluI patterns | VI (n=46) | I (n=95) | IV (n=167) | IX (n=30) | V (n=81) | II (n=97) | X (n=12) | IVB (n=58) | VII (n=40) |
|---|---|---|---|---|---|---|---|---|---|
| PDI categories | |||||||||
| R | 22/47.83% | 67/70.53% | 103/61.68% | 17/56.67% | 22/27.16% | 34/35.05% | 7/58.33% | 40/68.97% | 14/35.00% |
| IS | 13/28.26% | 18/18.95% | 38/22.75% | 7/23.33% | 32/39.51% | 35/36.08% | 3/25.00% | 13/22.41% | 16/40.00% |
| S | 11/23.91% | 10/10.53% | 26/15.57% | 6/20.00% | 27/33.33% | 28/28.87% | 2/16.67% | 5/8.62% | 10/25.00% |
| Median | IS | R1,2 | R3,4 | R | IS1,3,5 | IS2,4,6 | R | R5,6 | IS |
| RsaI patterns | I (n=132) | IV (n=45) | IX (n=23) | V (n=50) | IB (n=13) | IVB (n=77) | III (n=24) | II (n=136) | IVD (n=12) | VII (n=68) | IIB (n=31) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PDI categories | |||||||||||
| R | 84/63.64% | 28/62.22% | 15/65.22% | 14/28.00% | 11/84.62% | 55/71.43% | 13/54.17% | 52/38.24% | 8/66.67% | 21/30.88% | 14/45.16% |
| IS | 28/21.21% | 9/20.00% | 6/26.09% | 16/32.00% | 2/15.38% | 16/20.78% | 7/29.17% | 41/30.15% | 3/25.00% | 31/45.59% | 13/41.94% |
| S | 20/15.15% | 8/17.78% | 2/8.70% | 20/40.00% | 0/0.00% | 6/7.79% | 4/16.67% | 43/31.62% | 1/8.33% | 16/23.53% | 4/12.90% |
| Median | R7,8 | R | R | IS7,9,10 | R9 | R10,11,12 | R | IS8,11 | R | IS12 | IS |
p < 0.001
p < 0.01
p < 0.05 (Chi-squared test (p=0.0001), Kruskal-Wallis test (p=0.0001))
PCR amplicons of the agr gene were also subjected to RsaI-based polymorphism analysis. Based on RsaI restriction enzyme digestion, different digestion patterns were obtained, allowing for the classification of the analyzed strains into classes (I, II, IIB, III, IV, IVB, IVD, V, VII, and IX). Different strains express different small polymorphisms within the agr gene, and this determines the different restriction products (Supplementary Figure 2). Statistical analysis demonstrated that the S. aureus strains revealing RsaI restriction pattern V respond significantly different to PDI than strains with patterns no. I, IB and IVB. Furthermore, the statistically relevant difference was reported between strains characterized with patterns II, IVB and VII (significant correlations and p values are presented in Table 3). Interestingly, the majority of strains revealing RsaI patterns I, IB III, IV, IVB, IVD and IX represented strains resistant to photoinactivation (Table 3).
To explore the genetic characterization of the examined strains further, we applied statistical analysis to the combined AluI/RsaI patterns with respect to the response to PDI. This combination produced 62 divergent groups and allowed for the more precise determination of genetic elements potentially involved in the observed phenomenon. The results for the 633 methicillin-susceptible and methicillin-resistant S. aureus strains show that there were statistically significant differences (p < 0.05) between AluI/RsaI digestion patterns and responses to PDI. Specifically, we can conclude that the majority of strains expressing the polymorphisms associated with AluI/RsaI patterns I/I, IV/IV, IVB/IVB, IV/IVB and IV/II were generally the PDI-resistant strains. On the contrary, the V/VII pattern was associated with intermediate sensitive strains (Table 4). These correlations involved both MRSA and MSSA strains excluding pattern IV/II which was solely associated with MRSA strains. In contrast, pattern V/VII was associated with IS PDI responders in the case of MSSA strains (Table 4). Statistical analysis demonstrated that S. aureus response to photoinactivation was significantly different between strains expressing various AluI/RsaI patterns (statistical correlations presented in Table 4).
Table 4.
Statistical analysis of AluI/RsaI patterns over S. aureus response to PDI (N/%).
| All S. aureus | VI/I (n=36) | I/I (n=82) | IV/IV (n=35) | V/V (n=43) | II/II (n=68) | IVB/IVB (n=38) | VII/VII (n=37) | IV/IVB (n=38) | IV/II (n=56) | II/IIB (n=17) | V/VII (n=30) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PDI categories | |||||||||||
| R | 16/44.44% | 58/70.73% | 25/71.43% | 11/25.58% | 24/35.29% | 30/78.95% | 13/35.14% | 24/63.16% | 26/46.43% | 4/23.53% | 8/26.67% |
| IS | 10/27.78% | 15/18.29% | 6/17.14% | 13/30.23% | 22/32.35% | 6/15.79% | 15/40.54% | 10/26.32% | 15/26.79% | 9/52.94% | 16/53.33% |
| S | 10/27.78% | 9/10.98% | 4/11.43% | 19/44.19% | 22/32.35% | 2/5.26% | 9/24.32% | 4/10.53% | 15/26.79% | 4/23.53% | 6/20.00% |
| Median | IS | R1,2 | R3 | IS1,3,4,5 | IS2,7 | R4,6,7 | IS | R5 | IS | IS | IS6 |
| MRSA | (n=0) | (n=66) | (n=25) | (n=22) | (n=24) | (n=32) | (n=3) | (n=28) | (n=13) | (n=4) | (n=15) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PDI categories | |||||||||||
| R | 0/0.00% | 49/74.24% | 19/76.00% | 7/31.82% | 9/37.50% | 27/84.38% | 0/0.00% | 19/67.86% | 11/84.62% | 3/75.00% | 7/46.67% |
| IS | 0/0.00% | 11/16.67% | 5/20.00% | 7/31.82% | 7/29.17% | 4/12.50% | 3/100.00% | 6/21.43% | 1/7.69% | 1/25.00% | 7/46.67% |
| S | 0/0.00% | 6/9.09% | 1/4.00% | 8/36.36% | 8/33.33% | 1/3.13% | 0/0.00% | 3/10.71% | 1/7.69% | 0/0.00% | 1/6.67% |
| Median | - | R8 | R | IS8,9 | IS10 | R9,10 | IS | R | R | R | IS |
| MSSA | (n=36) | (n=16) | (n=10) | (n=21) | (n=44) | (n=6) | (n=34) | (n=10) | (n=43) | (n=13) | (n=15) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PDI categories | |||||||||||
| R | 16/44.44% | 9/56.25% | 6/60.00% | 4/19.05% | 15/34.09% | 3/50.00% | 13/38.24% | 5/50.00% | 15/34.88% | 1/7.69% | 1/6.67% |
| IS | 10/27.78% | 4/25.00% | 1/10.00% | 6/28.57% | 15/34.09% | 2/33.33% | 12/35.29% | 4/40.00% | 14/32.56% | 8/61.54% | 9/60.00% |
| S | 10/27.78% | 3/18.75% | 3/30.00% | 11/52.38% | 14/31.82% | 1/16.67% | 9/26.47% | 1/10.00% | 14/32.56% | 4/30.77% | 5/33.33% |
| Median | IS | R | R | S | IS | - | IS | - | IS | IS | IS |
p < 0.001
p < 0.01
p < 0.05 (Chi-squared test (p=0.0001), Kruskal-Wallis test (p=0.0001))
It is noted that agr groups correlate with the clonal complexes [23;24]. Therefore, to examine the possibility that the correlation between agr groups and PDI response resulted from the difference in genetic background rather than agr polymorphism, the RAPD analysis was performed for all MRSA S. aureus strains. RAPD typing revealed 258 different electrophoretic patterns (with the similarity below 100%), consisting of 15–28 bands. Genetic similarity of 307 S. aureus strains was 61.6%. Statistical analysis of genetic relatedness allowed to distinguish 7 genetic groups - A to G (within min. similarity level – 70.5%). Among 307 S. aureus strains, 250 isolates of the group A proved to be clonally dependent. They were classified to 44 different genotypes (similarity level within genotype ≥90%) with the most predominant genotype “34” (represented by 46 isolates). The remaining isolates were classified as 57 different genotypes, each represented by one isolate. Interestingly, genotypically indistinguishable strains (similarity 100%) differed in their agr groups, agr polymorphisms and PDI-response indicating that the correlation between PDI susceptibility and agr did not result from genetic background.
4. Discussion
Recently we have demonstrated that a strain-dependent response to PDI by S. aureus exists, although the underlying mechanism remains unexplained [12;13]. Also Miyabe et al. tested the susceptibility of 20 Staphylococcus strains isolated from the human oral cavity to methylene blue mediated PACT. They demonstrated that various log10 units reduction could be achieved in in vitro tests [25]. Next, studies by Yow et al. suggested that the MRSA isolate might be more sensitive to cationic PS than S. aureus ATCC 25923 strains [26]. Studies by Ribeiro et al. also indicated that PDI effect is rather strain- and isolate-dependent than species-dependent [27]. One reason for the poor response to PDI could be an effective antioxidant defense system in the bacteria. S. aureus strains lacking active superoxide dismutases were significantly more susceptible to PPIX-based photo-killing than their native counterparts [28]. The same observation was true for a strain deprived of staphyloxanthin, a staphylococcal golden pigment known for its antioxidant properties (Nakonieczna J., unpublished results). PDI resistant phenotypes could also result from variations in biofilm production. The effect of extracellular slime on photodynamic inactivation of bacteria was also analyzed by another group, who reported that extracellular slime significantly influenced the sensitizer uptake by the S. aureus cells [29]. On the basis of these observations, the current study aimed to investigate statistically relevant correlations between the genetic profiles of methicillin-susceptible and methicillin-resistant S. aureus strains and their response to photodynamic inactivation.
Our investigation proceeded to a genetic element on the assumption that the effectiveness of photoinactivation may be related to some aspect of strain virulence. As the response to PDI is a multifactorial phenomenon, we conducted a genetic analysis of the agr gene, which is the main regulator of several virulence factors that either are transported outside of the bacterial cell or are bound to the cell wall. Moreover, agr system revealed to be bacterial oxidation sensing system what directly combine its action with photodynamic treatment. Additionally, it was demonstrated that clinical isolates of S. aureus with inactive agr loci were also strong biofilm producers [30], indicating the impact of agr loci to biofilm production. Recent studies by Geisinger et al. (2012) demonstrated the presence of agr allele-dependent differences in agr activation. These differences were influenced by polymorphisms within the agr regulon that affected variations in the expression of several exoproteins and surface factors involved in pathogenesis [31]. Thus, we hypothesized that agr functionality and polymorphism could influence S. aureus susceptibility to PDI-induced oxidative mechanisms.
S. aureus strains exhibit well-defined genetic polymorphisms within the agr locus. Four agr genotypes, group I to IV, have been described to date [32]. The classification of S. aureus strains into four agr groups showed that there were statistically significant differences between the agr groups according to the response to PDI. Hence, our work proceeded to a more detailed analysis of the agr gene, focusing on agr gene polymorphisms. The data obtained allowed us to determine that strains expressing specific AluI/RsaI polymorphic patterns (listed in Table 4) were connected with strain’s resistance or intermediate sensitivity to photoinactivation and that they distributed differently among MRSA and MSSA S. aureus strains. Based on these results, we can further hypothesize that the agr gene may be a genetic factor associated with strain-dependent responses to PDI, as statistically significant correlations were found between agr gene polymorphisms and strain responses to PDI. The agr locus is complex, consisting of two divergent transcription units, driven by promoters P2 and P3 (reviewed in reference [33]). The P2 operon encodes a two-component signaling module, of which AgrC is the receptor and AgrA is the response regulator. It also encodes two proteins, AgrB and -D, which combine to produce and secrete an autoinducing peptide (AIP) that is the ligand for AgrC. AgrA activates the agr P3 promoter, which drives the synthesis of RNAIII, the effector of target gene regulation [34]. As agr controls a large set of genes, including most of those encoding cell wall associated and extracellular proteins as well as biofilm formation it is hard to identify specific pathway involved in S. aureus susceptibility to photodynamic treatment [35]. The association between agr specific group, the type of infection, and also antibiotic resistance has been reported by many researchers [36]. We can assume that polymorphism within agr locus could impact RNAIII transcription, function, or stability what may result in various expression of related genes and/or biofilm production influencing S. aureus susceptibility to oxidative stress. Nevertheless, to determine underlying mechanisms further investigations must be performed i.e. RNAIII transcription profiles, or agr locus sequencing.
One could claim that because the isolates were obtained from one hospital, the agr typing could simply be clustering the isolates into particular clonal types that may be more or less susceptible to PDI and that do not influence directly the mechanism of strain-dependent responses to photoinactivation. We cannot exclude this possibility; however, if it was true, we would assume that other typing method such as RAPD analysis (current study and data previously published by our research group, Grinholc et al. 2008) would also cluster the strains according to their PDI susceptibility. As only agr typing correlates with PDI phenotypes, we can assume that this regulon influences the strain-dependent mechanism of PDI response. However, taking into consideration all of the strains, this correlation is heterogeneous and does not show a linear trend thus demonstrating the need for a broader investigation of the agr gene polymorphism, and additional agr-dependent pathways.
Applied approach to identify agr polymorphism (based on RFLP analysis using two digestion enzymes) divided the isolates into 62 subgroups based on AluI/RsaI profile. The subgroups significantly correlated with PDI response. However, 633 isolates divided into 62 subgroups suggests that the predictive power of this method may have little value in clinical use. Thus, it is required to investigate other techniques enabling detection of gene polymorphism i.e. sequencing and searching for crucial polymorphisms determining strain susceptibility to photoinactivation.
The strain dependent susceptibility towards PDI is very large, considering that there is a 5 log range in reduction in viable count after exposure. One could concern that agr null mutants are only showing an approximately 1 log reduction compared to an isogenic strain carrying a functional agr, and that means that there is a very large unexplained variation in susceptibility/resistance towards PDI between isolates that is not related to the Agr status. Thus, it is significant to remember that the mechanism underlying strain-dependent response to PDI is multifactorial and several factors could influence strain susceptibility/resistance towards photoinactivation. Nevertheless, the current work provides proof of evidence that agr polymorphism and functionality influences S. aureus response to photodynamic treatment.
Supplementary Material
Acknowledgments
Funding
Isogenic pairs of agr mutants were obtained through the Network of Antimicrobial Resistance in Staphylococcus aureus (NARSA) program supported under NIAID/NIH Contract#HHSN272200700055C. MALDI-TOF MS analysis were performed at LM Bruss Laboratory in Poland. This work was supported by the grant no. 1651/B/P01/2010/39 from the National Science Centre (NCN). M.O. was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), U.S. National Institutes of Health (NIH).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author’s contribution
Conceived and designed the experiments: MG. Performed the experiments: AN, MG, AM, AR-Z, GF, JL-C. Analyzed the data: MG, JN. Contributed reagents/materials/analysis tools: MG, JK, KPB, MO. Wrote the paper: MG, JN.
All authors read and approved the final manuscript.
Competing Interest
The authors have declared that no competing interests exist.
Reference List
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Nastaly P, Grinholc M, Bielawski KP. Molecular characteristics of community-associated methicillin-resistant Staphylococcus aureus strains for clinical medicine. Arch Microbiol. 2010;192:603–617. doi: 10.1007/s00203-010-0594-4. [DOI] [PubMed] [Google Scholar]
- 3.Kurlenda J, Grinholc M, Jasek K, Wegrzyn G. RAPD typing of methicillin-resistant Staphylococcus aureus: a 7-year experience in a Polish hospital. Med Sci Monit. 2007;13:MT13–MT18. [PubMed] [Google Scholar]
- 4.Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, Nesme X, Etienne J, Vandenesch F. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002;70:631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Traber KE, Lee E, Benson S, Corrigan R, Cantera M, Shopsin B, Novick RP. agr function in clinical Staphylococcus aureus isolates. Microbiology. 2008;154:2265–2274. doi: 10.1099/mic.0.2007/011874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wainwright M. Photodynamic antimicrobial chemotherapy (PACT) J Antimicrob Chemother. 1998;42:13–28. doi: 10.1093/jac/42.1.13. [DOI] [PubMed] [Google Scholar]
- 7.Wilson M, Pratten J. Lethal photosensitisation of Staphylococcus aureus in vitro: effect of growth phase, serum, and pre-irradiation time. Lasers Surg Med. 1995;16:272–276. doi: 10.1002/lsm.1900160309. [DOI] [PubMed] [Google Scholar]
- 8.Hamblin MR, Newman EL. On the mechanism of the tumour-localising effect in photodynamic therapy. J Photochem Photobiol B. 1994;23:3–8. doi: 10.1016/s1011-1344(94)80018-9. [DOI] [PubMed] [Google Scholar]
- 9.Ochsner M. Photophysical and photobiological processes in the photodynamic therapy of tumours. J Photochem Photobiol B. 1997;39:1–18. doi: 10.1016/s1011-1344(96)07428-3. [DOI] [PubMed] [Google Scholar]
- 10.Athar M, Mukhtar H, Bickers DR. Differential role of reactive oxygen intermediates in photofrin-I- and photofrin-II-mediated photoenhancement of lipid peroxidation in epidermal microsomal membranes. J Invest Dermatol. 1988;90:652–657. doi: 10.1111/1523-1747.ep12560814. [DOI] [PubMed] [Google Scholar]
- 11.Redmond RW, Gamlin JN. A compilation of singlet oxygen yields from biologically relevant molecules. Photochem Photobiol. 1999;70:391–475. [PubMed] [Google Scholar]
- 12.Grinholc M, Szramka B, Kurlenda J, Graczyk A, Bielawski KP. Bactericidal effect of photodynamic inactivation against methicillin-resistant and methicillin-susceptible Staphylococcus aureus is strain-dependent. J Photochem Photobiol B. 2008;90:57–63. doi: 10.1016/j.jphotobiol.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Grinholc M, Zawacka-Pankau J, Gwizdek-Wisniewska A, Bielawski KP. Evaluation of the role of the pharmacological inhibition of Staphylococcus aureus multidrug resistance pumps and the variable levels of the uptake of the sensitizer in the strain-dependent response of Staphylococcus aureus to PPArg(2)-based photodynamic inactivation. Photochem Photobiol. 2010;86:1118–1126. doi: 10.1111/j.1751-1097.2010.00772.x. [DOI] [PubMed] [Google Scholar]
- 14.Sun F, Liang H, Kong X, Xie S, Cho H, Deng X, Ji Q, Zhang H, Alvarez S, Hicks LM, Bae T, Luo C, Jiang H, He C. Quorum-sensing agr mediates bacterial oxidation response via an intramolecular disulfide redox switch in the response regulator AgrA. Proc Natl Acad Sci U S A. 2012;109:9095–9100. doi: 10.1073/pnas.1200603109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park HJ, Moon YH, Yoon HE, Park YM, Yoon JH, Bang IS. Agr function is upregulated by photodynamic therapy (PDT) for Staphylococcus aureus and is related to resistance against PDT. Microbiol Immunol. 2013;57:547–552. doi: 10.1111/1348-0421.12070. [DOI] [PubMed] [Google Scholar]
- 16.CLSI. CLSI approved standard M100-S17. Clinical and Laboratory Standards Institute; Wayne, PA: 2007. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 17.Szabados F, Woloszyn J, Richter C, Kaase M, Gatermann S. Identification of molecularly defined Staphylococcus aureus strains using matrix-assisted laser desorption/ionization time of flight mass spectrometry and the Biotyper 2.0 database. J Med Microbiol. 2010;59:787–790. doi: 10.1099/jmm.0.016733-0. [DOI] [PubMed] [Google Scholar]
- 18.Lina G, Boutite F, Tristan A, Bes M, Etienne J, Vandenesch F. Bacterial competition for human nasal cavity colonization: role of Staphylococcal agr alleles. Appl Environ Microbiol. 2003;69:18–23. doi: 10.1128/AEM.69.1.18-23.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papakyriacou H, Vaz D, Simor A, Louie M, McGavin MJ. Molecular analysis of the accessory gene regulator (agr) locus and balance of virulence factor expression in epidemic methicillin-resistant Staphylococcus aureus. J Infect Dis. 2000;181:990–1000. doi: 10.1086/315342. [DOI] [PubMed] [Google Scholar]
- 20.Ji G, Pei W, Zhang L, Qiu R, Lin J, Benito Y, Lina G, Novick RP. Staphylococcus intermedius produces a functional agr autoinducing peptide containing a cyclic lactone. J Bacteriol. 2005;187:3139–3150. doi: 10.1128/JB.187.9.3139-3150.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jett BD, Hatter KL, Huycke MM, Gilmore MS. Simplified agar plate method for quantifying viable bacteria. Biotechniques. 1997;23:648–650. doi: 10.2144/97234bm22. [DOI] [PubMed] [Google Scholar]
- 22.Dodd CER, Sharman RL, Bloomfield SF, Booth IR, Stewart GSAB. Inimical processes: Bacterial self-destruction and sub-lethal injury. Trends Food Sci Technol. 1997;8:238–241. [Google Scholar]
- 23.Monecke S, Slickers P, Ehricht R. Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol Med Microbiol. 2008;53:237–251. doi: 10.1111/j.1574-695X.2008.00426.x. [DOI] [PubMed] [Google Scholar]
- 24.Robinson DA, Monk AB, Cooper JE, Feil EJ, Enright MC. Evolutionary genetics of the accessory gene regulator (agr) locus in Staphylococcus aureus. J Bacteriol. 2005;187:8312–8321. doi: 10.1128/JB.187.24.8312-8321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyabe M, Junqueira JC, Costa AC, Jorge AO, Ribeiro MS, Feist IS. Effect of photodynamic therapy on clinical isolates of Staphylococcus spp. Braz Oral Res. 2011;25:230–234. doi: 10.1590/s1806-83242011005000006. [DOI] [PubMed] [Google Scholar]
- 26.Yow CM, Tang HM, Chu ES, Huang Z. Hypericin-mediated photodynamic antimicrobial effect on clinically isolated pathogens. Photochem Photobiol. 2012;88:626–632. doi: 10.1111/j.1751-1097.2012.01085.x. [DOI] [PubMed] [Google Scholar]
- 27.Ribeiro AP, Pavarina AC, Dovigo LN, Brunetti IL, Bagnato VS, Vergani CE, Costa CA. Phototoxic effect of curcumin on methicillin-resistant Staphylococcus aureus and L929 fibroblasts. Lasers Med Sci. 2013;28:391–398. doi: 10.1007/s10103-012-1064-9. [DOI] [PubMed] [Google Scholar]
- 28.Nakonieczna J, Michta E, Rybicka M, Grinholc M, Gwizdek-Wisniewska A, Bielawski KP. Superoxide dismutase is upregulated in Staphylococcus aureus following protoporphyrin-mediated photodynamic inactivation and does not directly influence the response to photodynamic treatment. BMC Microbiol. 2010;10:323. doi: 10.1186/1471-2180-10-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gad F, Zahra T, Hasan T, Hamblin MR. Effects of growth phase and extracellular slime on photodynamic inactivation of gram-positive pathogenic bacteria. Antimicrob Agents Chemother. 2004;48:2173–2178. doi: 10.1128/AAC.48.6.2173-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cafiso V, Bertuccio T, Santagati M, Demelio V, Spina D, Nicoletti G, Stefani S. agr–Genotyping and transcriptional analysis of biofilm-producing Staphylococcus aureus. FEMS Immunol Med Microbiol. 2007;51:220–227. doi: 10.1111/j.1574-695X.2007.00298.x. [DOI] [PubMed] [Google Scholar]
- 31.Geisinger E, Chen J, Novick RP. Allele-Dependent Differences in Quorum-Sensing Dynamics Result in Variant Expression of Virulence Genes in Staphylococcus aureus. J Bacteriol. 2012;194:2854–2864. doi: 10.1128/JB.06685-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 33.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 34.Seidl K, Chen L, Bayer AS, Hady WA, Kreiswirth BN, Xiong YQ. Relationship of agr expression and function with virulence and vancomycin treatment outcomes in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:5631–5639. doi: 10.1128/AAC.05251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 36.Wright JS, III, Traber KE, Corrigan R, Benson SA, Musser JM, Novick RP. The agr radiation: an early event in the evolution of staphylococci. J Bacteriol. 2005;187:5585–5594. doi: 10.1128/JB.187.16.5585-5594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.