Abstract
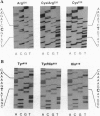
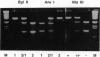
The molecular basis of the classical human phosphoglucomutase 1 (PGM1) isozyme polymorphism has been established. In 1964, when this genetic polymorphism was first described, two common allelozymes PGM1 and PGM1 2 were identified by starch gel electrophoresis. The PGM1 2 isozyme showed a greater anodal electrophoretic mobility than PGM1 1. Subsequently, it was found that each of these allelozymes could be split, by isoelectric focusing, into two subtypes; the acidic isozymes were given the suffix + and the basic isozymes were given the suffix -. Hence, four genetically distinct isozymes 1+, 1-, 2+, and 2- were identified. We have now analyzed the whole of the coding region of the human PGM1 gene by DNA sequencing in individuals of known PGM1 protein phenotype. Only two mutations have been found, both C to T transitions, at nt 723 and 1320. The mutation at position 723, which changes the amino acid sequence from Arg to Cys at residue 220, showed complete association with the PGM1 2/1 protein polymorphism: DNA from individuals showing the PGM1 1 isozyme carried the Arg codon CGT, whereas individuals showing the PGM1 2 isozyme carried the Cys codon TGT. Similarly, the mutation at position 1320, which leads to a Tyr to His substitution at residue 419, showed complete association with the PGM1+/- protein polymorphism: individuals with the + isozyme carried the Tyr codon TAT, whereas individuals with the - isozyme carried the His codon CAT. The charge changes predicted by these amino acid substitutions are entirely consistent with the charge intervals calculated from the isoelectric profiles of these four PGM1 isozymes. We therefore conclude that the mutations are solely responsible for the classical PGM1 protein polymorphism. Thus, our findings strongly support the view that only two point mutations are involved in the generation of the four common alleles and that one allele must have arisen by homologous intragenic recombination between these mutation sites.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bark J. E., Harris M. J., Firth M. Typing of the common phosphoglucomutase variants using isoelectric focusing--a new interpretation of the phosphoglucomutase system. J Forensic Sci Soc. 1976 Apr;16(2):115–120. doi: 10.1016/s0015-7368(76)71042-9. [DOI] [PubMed] [Google Scholar]
- Bowman B. H., Kurosky A. Haptoglobin: the evolutionary product of duplication, unequal crossing over, and point mutation. Adv Hum Genet. 1982;12:189-261, 453-4. doi: 10.1007/978-1-4615-8315-8_3. [DOI] [PubMed] [Google Scholar]
- Carter N. D., West C. M., Emes E., Parkin B., Marshall W. H. Phosphoglucomutase polymorphism detected by isoelectric focusing: gene frequencies, evolution and linkage. Ann Hum Biol. 1979 May-Jun;6(3):221–230. doi: 10.1080/03014467900007222. [DOI] [PubMed] [Google Scholar]
- Dai J. B., Liu Y., Ray W. J., Jr, Konno M. The crystal structure of muscle phosphoglucomutase refined at 2.7-angstrom resolution. J Biol Chem. 1992 Mar 25;267(9):6322–6337. [PubMed] [Google Scholar]
- Drago G. A., Hopkinson D. A., Westwood S. A., Whitehouse D. B. Antigenic analysis of the major human phosphoglucomutase isozymes: PGM1, PGM2, PGM3 and PGM4. Ann Hum Genet. 1991 Oct;55(Pt 4):263–271. doi: 10.1111/j.1469-1809.1991.tb00852.x. [DOI] [PubMed] [Google Scholar]
- Du R., Zhang Z., Zhao H. Distribution of PGM1 subtypes in 12 populations of China. Gene Geogr. 1992 Apr-Aug;6(1-2):21–26. [PubMed] [Google Scholar]
- Hollyoake M., Putt W., Edwards Y. H., Whitehouse D. B. Two TaqI polymorphisms at the human PGM1 locus. Hum Mol Genet. 1992 Aug;1(5):354–354. doi: 10.1093/hmg/1.5.354-a. [DOI] [PubMed] [Google Scholar]
- Lee Y. S., Marks A. R., Gureckas N., Lacro R., Nadal-Ginard B., Kim D. H. Purification, characterization, and molecular cloning of a 60-kDa phosphoprotein in rabbit skeletal sarcoplasmic reticulum which is an isoform of phosphoglucomutase. J Biol Chem. 1992 Oct 15;267(29):21080–21088. [PubMed] [Google Scholar]
- Lyons K. M., Stein J. H., Smithies O. Length polymorphisms in human proline-rich protein genes generated by intragenic unequal crossing over. Genetics. 1988 Sep;120(1):267–278. doi: 10.1093/genetics/120.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March R. E., Hollyoake M., Putt W., Hopkinson D. A., Edwards Y. H., Whitehouse D. B. Genetic polymorphism in the 3' untranslated region of human phosphoglucomutase-1. Ann Hum Genet. 1993 Jan;57(Pt 1):1–8. doi: 10.1111/j.1469-1809.1993.tb00881.x. [DOI] [PubMed] [Google Scholar]
- Marchuk D., Drumm M., Saulino A., Collins F. S. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1991 Mar 11;19(5):1154–1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J., Merbs S. L., Sung C. H., Weitz C. J., Wang Y. Molecular genetics of human visual pigments. Annu Rev Genet. 1992;26:403–424. doi: 10.1146/annurev.ge.26.120192.002155. [DOI] [PubMed] [Google Scholar]
- Neel J. V., Satoh C., Smouse P., Asakawa J., Takahashi N., Goriki K., Fujita M., Kageoka T., Hazama R. Protein variants in Hiroshima and Nagasaki: tales of two cities. Am J Hum Genet. 1988 Dec;43(6):870–893. [PMC free article] [PubMed] [Google Scholar]
- Ott J. A chi-square test to distinguish allelic association from other causes of phenotypic association between two loci. Genet Epidemiol. 1985;2(1):79–84. doi: 10.1002/gepi.1370020108. [DOI] [PubMed] [Google Scholar]
- Ray W. J., Jr, Hermodson M. A., Puvathingal J. M., Mahoney W. C. The complete amino acid sequence of rabbit muscle phosphoglucomutase. J Biol Chem. 1983 Aug 10;258(15):9166–9174. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman R. Blood group frequencies of immigrant and indigenous populations from South East England. J Forensic Sci Soc. 1985 Mar-Apr;25(2):95–134. doi: 10.1016/s0015-7368(85)72376-6. [DOI] [PubMed] [Google Scholar]
- Takahashi N., Neel J. V., Satoh C., Nishizaki J., Masunari N. A phylogeny for the principal alleles of the human phosphoglucomutase-1 locus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6636–6640. doi: 10.1073/pnas.79.21.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse D. B., Putt W., Lovegrove J. U., Morrison K., Hollyoake M., Fox M. F., Hopkinson D. A., Edwards Y. H. Phosphoglucomutase 1: complete human and rabbit mRNA sequences and direct mapping of this highly polymorphic marker on human chromosome 1. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):411–415. doi: 10.1073/pnas.89.1.411. [DOI] [PMC free article] [PubMed] [Google Scholar]