Abstract
Threonyl-tRNA synthetase (ThrRS) participates in protein synthesis quality control by selectively editing the misacylated species Ser-tRNAThr. In bacteria and eukaryotes the editing function of ThrRS resides in a highly conserved N-terminal domain distant from the active site. Most archaeal ThrRS proteins are devoid of this editing domain, suggesting evolutionary divergence of quality-control mechanisms. Here we show that archaeal editing of Ser-tRNAThr is catalyzed by a domain unrelated to, and absent from, bacterial and eukaryotic ThrRSs. Despite the lack of sequence homology, the archaeal and bacterial editing domains are both reliant on a pair of essential histidine residues suggestive of a common catalytic mechanism. Whereas the archaeal editing module is most commonly part of full-length ThrRS, several crenarchaeal species contain individual genes encoding the catalytic (ThrRS-cat) and editing domains (ThrRS-ed). Sulfolobus solfataricus ThrRS-cat was shown to synthesize both Thr-tRNAThr and Ser-tRNAThr and to lack editing activity against Ser-tRNAThr. In contrast, ThrRS-ed lacks aminoacylation activity but can act as an autonomous protein in trans to hydrolyze specifically Ser-tRNAThr, or it can be fused to ThrRS-cat to provide the same function in cis. Deletion analyses indicate that ThrRS-ed is dispensable for growth of S. solfataricus under standard conditions but is required for normal growth in media with elevated serine levels. The growth phenotype of the ThrRS-ed deletion strain suggests that retention of the discontinuous ThrRS quaternary structure relates to specific physiological requirements still evident in certain Archaea.
To ensure accurate translation of the genetic code, aminoacyl-tRNA synthetases (aaRS) display a remarkable precision in pairing tRNAs to their cognate amino acids. They catalyze a two-step reaction, first by activating the amino acid in the form of adenylate and subsequently by transferring it to the 3′-end of the tRNA molecule (1). These enzymes reject most noncognate amino acid substrates at the level of activation, based on steric or chemical incompatibilities with their active-site architecture. However, because discrimination of isosteric and chemically related amino acids is difficult and may consequently result in the formation of misactivated and mischarged products, some aaRSs have evolved editing mechanisms. The incorrect products are usually translocated and hydrolyzed at specific sites (editing sites), either as adenylates (pretransfer) or aminoacylated tRNAs (posttransfer editing) (2). Perhaps the best-studied editing mechanisms are those of the class I enzymes isoleucyl- (3–5), leucyl- (6), and valyl-tRNA synthetase (7), all of which are supported by extensive functional and structural investigations. Among class II synthetases, prolyl-tRNA synthetase (8), alanyl-tRNA synthetase (9, 10) and, most recently, threonyl-tRNA synthetase (ThrRS) (11) have been reported to possess posttransfer editing activities.
The crystal structure of ThrRS from Escherichia coli allowed delineation of the mechanisms this enzyme employs to ensure the fidelity of Thr-tRNAThr synthesis (11, 12). The active site efficiently discerns between threonine and the isosteric valine by using a zinc ion as a cofactor; the zinc coordinates both the amino group and the side-chain hydroxyl group of threonine, whereas it rejects valine because of steric hindrance with one of its methyl groups. However, serine cannot be discriminated at the activation step; instead, incorrectly formed Ser-tRNAThr is hydrolyzed at the editing site, which is located in the N-terminal domain of the enzyme (11). Mutational studies located the editing site as a cleft in the N-terminal domain; in particular, two histidines (His-73 and His-77) and an aspartate (Asp-180) proved critical for the editing activity (11).
Although the editing domain described for E. coli ThrRS is always found in ThrRS proteins from all bacteria and eukaryotes, a homologous domain could not be identified in most archaeal ThrRSs (12). However, a recent report indicated that a distinct N-terminal domain present in the majority of archaeal ThrRSs performs the analogous function (13). In addition, it was suggested that homologous domains encoded in some Crenarchaea such as Sulfolobus solfataricus might perform the same function. Here we analyze biochemical properties of the Methanocaldococcus jannaschii and S. solfataricus ThrRSs, and demonstrate that the archaeal ThrRS editing domains specifically and efficiently deacylate Ser-tRNAThr, both in cis and trans. Furthermore, using a S. solfataricus genetic system, we provide direct evidence that the hydrolytic activity furnished by the ThrRS editing domain is critical for protein synthesis quality control in vivo.
Materials and Methods
Cloning of thrS Genes and Preparation of ThrRS Enzymes. DNA and predicted amino acid sequences were obtained from publicly available databases. S. solfataricus DNA was purchased from American Type Culture Collection bioproducts (ATCC 35092D), and M. jannaschii DNA was prepared by standard methods from cells (a gift from K. O. Stetter and M. Thomm, Regensburg University, Regensburg, Germany). The thrS gene from M. jannaschii (MJthrS) (with the primer pair 5′-GAATCTTCATATGAGGGATAATATGAAGATGC-3′ and 5′-GTAATCACTCGAGATGGAACTTTGGCTG-3′) and the two S. solfataricus genes, the ThrRS-ed gene (thrS-ed) (ORF SSO0384) (5′-GAATCTTCATATGATAATACTATTTATTCACG-3′ and 5′-GTAATCACTCGAGCAATAAAAATACGG-3′) and the ThrRS-cat gene (thrS-cat) (ORF SSO2486) (5′-GAATCTTCATATGGAAAGTTATAAACCAG-3′ and 5′-GTAATCACTCGAGTTTTAACGCTTTTACTGTAG-3′), were PCR amplified and cloned into pET20b (Novagen). A chimeric S. solfataricus thrS gene (encoding SSThrRS-ed/ThrRS-cat) consisting of the 5′ end of thrS-ed (nucleotides 1–456) attached to the 5′ end of thrS-cat was made by a two-step PCR reaction with two pairs of oligonucleotides (5′-GAATCTTCATATGATAATACTATTTATTCACG-3′ and 5′-CTGGT T TATA ACT T TCCAT TCCA A ATTTT TCACAATAC-3′, and 5′-GTATTGTGAAAAATTTGGAATGGA A AGT TATA A ACCAG-3′ and 5′-GTA ATCACTCGAGTTTTAACGCTTTTACTGTAG-3′) and cloned into pET20b. The E. coli thrS gene (ECthrS) was cloned into pET15b. The QuikChange system (Stratagene) was used for site-directed mutagenesis of MJthrS.
Expression of these genes was induced in E. coli BL21-Codon Plus (DE3)-RIL at A590 = 0.6 with 0.4 mM isopropyl-1-thio-β-d-galactopyranoside, or 0.1 mM of thrS-ed, for 2 h at 37°C. N- and C-terminally His-6-tagged proteins were purified on a nickel-column as described in ref. 14. Archaeal ThrRS preparations were flocculated at 65°C for 45 min to minimize the amount of contaminating E. coli proteins.
Aminoacylation and ATP-PPi Exchange Assay. The effective enzyme concentration was determined by active-site titration as described in ref. 14. Unfractionated mature tRNA from M. jannaschii was prepared according to the published procedure (15). M. jannaschii tRNAThr was prepared by using MJThrRS and threonine to generate Thr-tRNA, which was subsequently purified by using an EF-Tu affinity column (16). All buffer pH values were determined at 60°C.
The ATP-PPi exchange assay was performed as described in ref. 14 at 60°C in 50 mM Hepes-NaOH (pH 7.2), 15 mM MgCl2, 50 mM KCl, 5 mM DTT, 1 mM potassium fluoride, 2 mM ATP, 15–500 μM l-threonine or 5–300 mM l-serine (Sigma), and 2 mM [32P]pyrophosphate (2 cpm/pmol) (PerkinElmer). ThrRS concentrations ranged from 10 to 40 nM.
Aminoacylation was performed at 60°C in 50 mM Hepes-KOH (pH 7.2), 50 mM KCl, 15 mM MgCl2, 5 mM DTT, 10 mM ATP, 50 μM [3H]threonine (200 cpm/pmol) or 300 μM [3H]serine (200 cpm/pmol) (Amersham Biosciences), and 2 mg/ml M. jannaschii unfractionated tRNA in 120-μl reactions. Twenty-microliter aliquots were spotted on 3 mm filter disks (Whatman) and washed in 10% trichloroacetic acid, and the radioactivity was determined by liquid scintillation counting.
Posttransfer Editing Assay. Thr-tRNAThr and Ser-tRNAThr were generated by charging unfractionated M. jannaschii tRNA or purified M. jannaschii tRNAThr with MJThrRS or ThrRS-cat, respectively. Additionally, M. jannaschii unfractionated tRNA was charged with seryl-tRNA synthetase from Methanosarcina barkeri (D.K., unpublished work) to produce Ser-tRNASer. After aminoacylation, tRNA was extracted by phenol and chloroform, ethanol-precipitated, dried, and subsequently used in deacylation assays as described in ref. 14.
Cultivation and Construction of S. solfataricus Strains. S. solfataricus strains were grown at 80°C in batch culture as described in refs. 17 and 18. Strain PBL205 [del(SSO3004-3050)] is described in ref. 19. Defined minimal medium consisted of the basal salts of Allen (20), as modified by Brock (21), supplemented either with 0.2% (wt/vol) sucrose or lactose as the sole carbon and energy source. Recovery of transformed cells used a rich medium supplemented with 0.2% (wt/vol) tryptone. Growth was monitored at 540 nm by using a Cary 50 Bio spectrophotometer (Varian). Solid medium was prepared by using 0.6% (wt/vol) gelrite gellan gum (Kelco, San Diego) and 8 mM magnesium chloride. Plates were incubated at 80°C in plastic containers with sufficient hydration to prevent desiccation. Strains treated with amino acids were grown in sucrose to a cell density of 108 per milliliter (≈0.1 A540) in glass crew-cap Erlenmeyer flasks (250 ml). Filter-sterilized solutions of l-serine and/or l-threonine were then added to final concentrations of 0.5 mM. Cultures were incubated at 80°C, and growth was monitored spectrophotometrically.
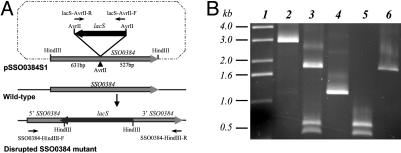
A disruption of the ThrRS-ed encoding gene SSO0384 was made (Fig. 1A) in strain PBL2025 by inserting the lacS gene as described in ref. 18. Plasmid pSSO03841 was constructed by insertion of a HindIII-digested PCR SSO0384 amplicon produced with primers SSO0384-HindIII-F (5′-ATGCAAGCTTGTGATAATACTATTTATTCACGCAT-3′) and SSO0384-HindIII-R (5′-ATGCAAGCTTAGTTATTTTTATGCCTATAAAGAGT-3′) and cloned into the HindIII site of pUC19. The complementary region of SSO0384-HindIII-F starts at the SSO0384 start codon, and the complementary region of SSO03842-R starts 1,159 nt downstream relative to the SSO0384 start codon, beginning with the SSO0384 stop codon. Plasmid pSSO0384S1 was constructed by insertion of an AvrII-digested PCR lacS amplicon into the AvrII site of pSSO03841 in the reverse orientation relative to SSO0384. To achieve this result, lacS was placed into pSSO03841 at nucleotide 527 relative to the SSO0384 stop codon. PCR primers for lacS were forward primer LacS-AvrII-F (5′-AGTCCGCCCTAGGAATACTAGGAGGAGTAGCATATAATTACGT-3′) and reverse primer LacS-AvrII-R (5′-AGTCCGCCCTAGGAGTAT TA A ATCTAAATGACTTTCCAATTAG-3′), and both encode added AvrII sites. LacS-AvrII-F starts 170 nt 5′ to the lacS start codon, whereas LacS-AvrII-R starts 165 nt 3′ to the lacS stop codon. After transformation of pSSO0384S1 into PBL2025 by electroporation, lactose-using recombinants were recovered by enrichment in lactose, and individual colonies were isolated by serial dilution onto rich medium plates. Chromosomal recombinants resulting from lacS insertion were detected by spraying the plates with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. Lac+ isolates were analyzed by PCR and restriction analysis.
Fig. 1.
Disruption of the SSO0384 locus. (A) Outline of the SSO0384 disruption strategy. The locations of PCR primers are indicated by arrows. The AvrII site of SSO0384 is located at nucleotide 524. LacS is positioned in a reverse orientation relative to SSO0384. (B) PCR analysis of the disrupted and WT SSO0384 locus: lane 1, 1-kb DNA ladder; lane 2, amplification of SSO0384 from disruption mutant by using primers SSO0384-HindIII-F and SSO0384-HindIII-R; lane 3, same as lane 2 but digested with HindIII; lane 4, SSO0384 amplified from WT; lane 5, same as lane 4 but digested with HindIII; lane 6, lacS amplified from WT by using primers lacS-AvrII-F and lacS-AvrII-R.
PCR amplification (Fig. 1B) of the WT undisrupted SSO0384 locus in strain PBL2025 by using primers SSO0384-HindIII-F and SSO0384-HindIII-R produced a single fragment of 1.16 kb and two fragments of 0.53 and 0.63 kb after digestion with AvrII. PCR of the disrupted SSO0384 locus in strain PBL2034 produced a single fragment of 3.09 kb. Three fragments were produced after digestion with AvrII that represented the 5′ and 3′ ends of SSO0384 and the 1.93-kb lacS insert.
Results
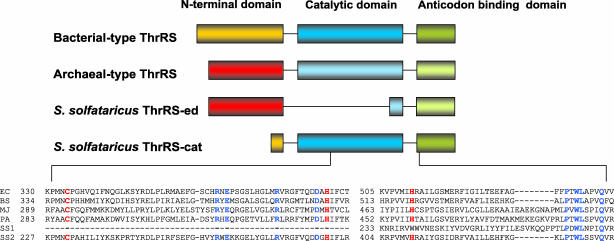
Divergence of Archaeal ThrRSs. The majority of characterized Archaea possess a distinctive archaeal-type ThrRS, whereas bacterial-type ThrRS-encoding genes are restricted to a few archaeal species (22). A survey of available archaeal genome sequences revealed bacterial-type ThrRSs encoded only in the groups Thermoplasmata and Halobacteria. Three crenarchaeal species (S. solfataricus, Sulfolobus tokodaii, and Aeropyrum pernix) were all found to possess two thrS-related genes. Initial alignments of the encoded ThrRS pairs showed that they diverge into two groups, one comprising the archaeal-type sequences and the other resembling the bacterial-type enzymes. Closer inspection revealed peculiarities in both groups: the archaeal genus Sulfolobus ThrRS shows high conservation in the archaeal-specific N-terminal domain (≈150 aa) and in the characteristic class IIa anticodon-binding domain at its C terminus; however, it lacks the putative catalytic domain in the middle (Fig. 2). Thus, we name it ThrRS-ed. Conversely, the Sulfolobus bacterial-type ThrRS possesses highly conserved catalytic and anticodon-binding domains but lacks almost the entire N-terminal editing domain, shown to be responsible for editing in bacterial-type ThrRSs (Fig. 2). We therefore name it ThrRS-cat. Because the presence of the known ThrRS editing domain appeared restricted to Thermoplasmata and Halobacteria in Archaea, we were prompted to analyze mechanisms of serine discrimination pertinent to archaeal-type ThrRS enzymes in general and the atypical crenarchaeal systems in particular.
Fig. 2.
Schematic diagram of the domain structure of various ThrRSs and detailed alignment around the region that interacts with the zinc atom. Highly conserved residues are highlighted in blue, and residues involved in binding of zinc are in red. Numbering corresponds to the residue immediately to the right. Abbreviations are as follows: EC, E. coli; BS, Bacillus subtilis; MJ, M. jannaschii; PA, P. aerophilum; SS1, S. solfataricus archaeal-type ThrRS-ed; SS2, S. solfataricus bacterial-type ThrRS-cat.
Elements in M. jannaschii ThrRS and S. solfataricus ThrRS-cat That Are Responsible for the Specificity of Threonylation. Archaeal ThrRSs were tested for their ability to activate threonine and noncognate serine. As anticipated, amino acid activation was observed only in the case of ThrRS-cat and MJThrRS, whereas ThrRS-ed was catalytically inactive. Kinetic parameters were then determined for the activation of threonine and serine by both ThrRS-cat and MJThrRS (Table 1). The bacterial-type ThrRS-cat, archaeal-type MJThrRS, and E. coli ThrRS (11) all show similar specificities for threonine over serine, attesting to similarities in their amino acid binding sites. The comparable patterns of serine misactivation among these ThrRSs suggest the need for error correction mechanisms in S. solfataricus and M. jannaschii similar to that described in E. coli (12). The archaeal ThrRS enzymes were therefore first tested for their ability to mischarge serine to tRNA in standard aminoacylation assays. As seen in Fig. 3A, MJThrRS was unable to charge serine significantly above background, whereas ThrRS-cat mischarged serine to a conspicuously high level. Strikingly, an equimolar mixture of ThrRS-ed and ThrRS-cat suppressed production of the misacylated product, suggesting that ThrRS-ed contributes to the amino acid specificity of the reaction. Furthermore, the rate of cognate threonine aminoacylation was enhanced in the presence of ThrRS-ed and ThrRS-cat together, compared with ThrRS-cat alone (Fig. 3 A and A Inset). Although this effect was not high, it consistently appeared in our assays.
Table 1. Kinetic parameters for threonine and serine activation by various ThrRSs.
| ThrRS species | Amino acid | kcat, s-1 | KM, mM | kcat/KM, s-1/mM | kcat/KM, relative* |
|---|---|---|---|---|---|
| S. solfataricus† | Threonine | 13.5 ± 0.2 | 0.11 ± 0.01 | 124 ± 11 | 1 |
| Serine | 12.3 ± 0.2 | 55 ± 5 | 0.22 ± 0.02 | 1.8 × 10-3 | |
| M. jannaschii† | Threonine | 1.9 ± 0.02 | 0.10 ± 0.01 | 19 ± 2 | 1 |
| Serine | 1.3 ± 0.01 | 25 ± 2 | 0.05 ± 0.01 | 2.6 × 10-3 | |
| E. coli‡ | Threonine | 36 | 0.11 | 320 | 1 |
| Serine | 26 | 81.5 | 0.32 | 1 × 10-3 |
kcat/KM is relative to threonine in each set, which was set at 1. Individual kinetic parameters are based on the average of three determinations.
Reactions performed at 60°C.
As reported in ref. 12, reactions performed at 37°C.
Fig. 3.
Mischarging and proofreading of tRNAThr. (A) Aminoacylation of M. jannaschii tRNAThr (2 μM) with 300 μM [3H]serine by ThrRSs: ○, 1 μM ThrRS-cat; •, 1 μM ThrRS-cat plus 1 μM ThrRS-ed; □, 0.2 μM MJThrRS. (Inset) Aminoacylation of M. jannaschii tRNAThr (2 μM) with 50 μM[3H]threonine by ThrRSs: ○, 1 μM ThrRS-cat; •, 1 μM ThrRS-cat plus 1 μM ThrRS-ed; □, 0.2 μM MJThrRS. (B) Deacylation of Ser-tRNAThr by various ThrRSs: □, 30 nM M. jannaschii ThrRS; •, 30 nM S. solfataricus ThrRS-ed; ▴, 200 nM E. coli ThrRS; ○, 200 nM S. solfataricus ThrRS-cat; ▪, no enzyme.
ThrRS-ed and MJThrRS Hydrolyze Ser-tRNAThr Specifically. The archaeal ThrRSs were analyzed for their ability to edit mischarged Ser-tRNAThr by the posttransfer route, analogous to the mechanism described for E. coli ThrRS in ref. 11. The mischarging ability of ThrRS-cat was used to produce the substrate for this reaction. ThrRS-ed and MJThrRS both hydrolyzed Ser-tRNAThr, as did ECThrRS, but not ThrRS-cat (Fig. 3B). In addition, none of the enzymes showed detectable deacylation of either Thr-tRNAThr or Ser-tRNASer (data not shown), confirming the specificity of the editing reaction for both tRNA and amino acid.
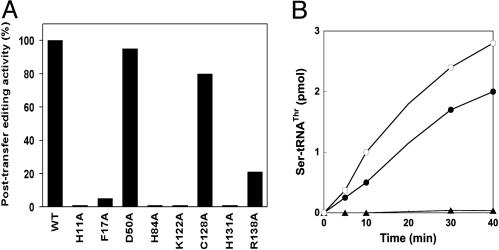
The N-Terminal Domain of the Archaeal-Type ThrRS Is Responsible for Editing. The ability of ThrRS-ed and MJThrRS to efficiently hydrolyze Ser-tRNAThr suggested that the editing function resides in their highly conserved N-terminal domains. Because these domains share no sequence similarity with the editing domain of E. coli ThrRS, we investigated their involvement in Ser-tRNAThr editing. Site-directed mutagenesis of the MJThrRS-encoding gene was used to introduce eight alanine replacements of conserved residues in the N-terminal domain of the protein (Fig. 4). All of the mutant enzymes were successfully purified, and none were significantly impaired in threonine charging (data not shown). However, when tested for their ability to deacylate Ser-tRNAThr, some of the variants (H11A, H84A, K122A, and H131A) failed to show detectable activity (Fig. 5A). These latter variants were further analyzed for serine mischarging; two of them (H84A and H131A) demonstrated notable mischarging activities (Fig. 5B), thus confirming the involvement of the N-terminal domain in editing.
Fig. 4.
Multiple sequence alignments of the editing domains in archaeal-type ThrRSs inferred by clustalx (30). Residues analyzed by alanine-scanning mutagenesis of M. jannaschii ThrRS are indicated with asterisks. Abbreviations are as follows: A.per, A. pernix (NP_146977); S.sol, S. solfataricus (NP_341922); S.tok, S. tokodaii (NP_378184); M.jan, M. jannaschii (NP_248192); M.the, Methanothermobacter thermautotrophicus (NP_276569); P.hor, Pyrococcus horikoshii (NP_142646); A.ful, Archaeoglobus fulgidus (NP_069384); P.aer, P. aerophilum (NP_559516) (GenBank accession numbers are in parentheses).
Fig. 5.
Site-directed mutagenesis of the archaeal-type editing domain. (A) Editing activity in M. jannaschii ThrRS mutants. Shown are bar graphs comparing the posttransfer editing activity of Ser-tRNAThr by M. jannaschii ThrRS mutants. Results are based on the initial rate of deacylation with WT ThrRS activity set at 100%. (B) Aminoacylation of M. jannaschii tRNAThr (2 μM) with [3H]serine (300 μM) in the presence of ThrRSs: •, 0.4 μM H84A mutant; ○, 0.4 μM H131A; ▴, 0.4 μM C128A.
Chimeric S. solfataricus ThrRS-ed/ThrRS-cat Possesses both Aminoacylation and Editing Activity. Complementing aminoacylation and editing activities from two distinct ThrRS enzymes in Sulfolobus may be suggestive of the evolutionary processes implicated in the development of the contemporary multidomain aaRS architecture. To probe this issue further, we investigated the structural compatibility of the archaeal-type editing domain with the bacterial-type ThrRS core. The editing domain of S. solfataricus ThrRS-ed was fused to the body of the ThrRS-cat enzyme from the same organism to generate a chimeric ThrRS-ed/ThrRS-cat protein. The enzyme chimera was successfully produced in E. coli and analyzed in the standard aminoacylation and posttransfer editing assays. This protein not only retained threonylation activity equivalent to ThrRS-cat, but also demonstrated the ability to efficiently and specifically hydrolyze Ser-tRNAThr with an efficiency comparable with ThrRS-ed (Figs. 6 A and B). In addition, hydrolysis of Ser-tRNASer and Thr-tRNAThr by the chimeric enzyme could not be detected (data not shown).
Fig. 6.
Aminoacylation and editing activities of the chimeric S. solfataricus ThrRS-ed/ThrRS-cat. (A) Aminoacylation of M. jannaschii tRNAThr (2 μM) with 50 μM [3H]threonine: •, 1 μM SSThrRS-cat; ○, 1 μM ThrRS-ed/ThrRS-cat. (B) Deacylation of Ser-tRNAThr (≈0.5 μM): •, 10 nM ThrRS-ed; ○, 10 nM ThrRS-ed/ThrRS-cat; ▪, no enzyme.
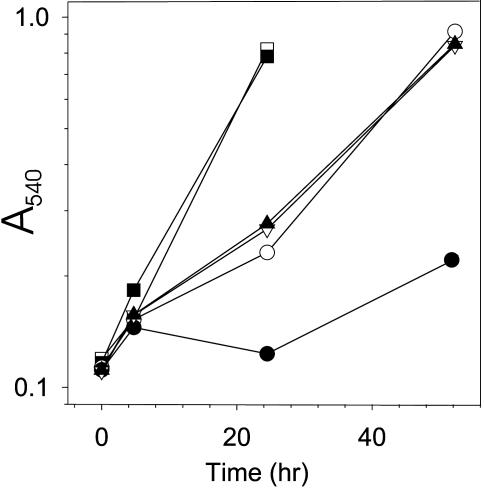
A S. solfataricus ThrRS-ed Deletion Strain Shows Reduced Growth in Serine-Rich Medium. Separation of aminoacylation and editing functions between the S. solfataricus ThrRS-cat and ThrRS-ed enzymes, respectively, provides an ideal system in which to directly test the effects of mischarged tRNA formation in vivo. To pursue this issue further, we inactivated the gene encoding S. solfataricus ThrRS-ed by introducing an insertion cassette into ORF SSO0384. The resulting S. solfataricus strain PBL2034 was viable. The growth of this mutant strain was compared with the WT strain PBL2025; in minimal medium both strains exhibited similar growth rates. Raising the serine concentration in the medium to 0.5 mM significantly reduced the growth rate of the mutant strain, whereas growth of the WT strain was much less affected (Fig. 7). Further addition of threonine (0.5 mM in equimolar concentration to serine) restored the growth rate of the deletion strain to that of the WT (Fig. 7). Thus, the editing activity of ThrRS-ed was not essential under normal growth conditions. However, elevated serine concentrations leading to unfavorable intracellular serine:threonine levels (probably causing increased generation of Ser-tRNAThr) produced a pronounced growth defect of the S. solfataricus ThrRS-ed deletion strain.
Fig. 7.
Effect of serine and threonine on growth of the S. solfataricus WT (PBL2025) and the ThrRS-ed deletion (PLB2034) strain in minimal medium. □, PBL2025, no amino acid addition; ○, PBL2025 plus 0.5 mM Ser; ▿, PBL2025 plus 0.5 mM Ser+Thr; ▪, PBL2034, no amino acid addition; •, PBL2034 plus 0.5 mM Ser; ▴, PBL2034 plus 0.5 mM Ser+Thr.
Discussion
Functional Convergence of Unrelated Domains Assures Specificity of Thr-tRNAThr Formation. Our analysis of uncharacterized N-terminal regions present in the majority of archaeal ThrRSs identified an editing domain in these aaRSs, as also recently proposed for the methanogenic archaeon M. mazei (13). Although amino acid sequence comparisons of the archaeal-type ThrRS editing domains and the known bacterial-type editing domains (found in ThrRSs of both bacteria and eukaryotes) failed to identify any significant homology (10, 23), functional studies indicate that these two types of modules are entirely analogous in their function, both possessing efficient and specific Ser-tRNAThr editing capacity (ref. 13 and this work). Consequently, ThrRS appears to be the first aaRS found to use two evolutionarily unrelated editing domains. The analogous function of the two ThrRS editing domains is likely correlated with the existence of homologous catalytic domains in archaeal and bacterial ThrRSs and, thus, with the persistence of comparable needs to avoid serine mischarging in phylogenetically distant organisms. The functional convergence between the two ThrRS editing domains is highlighted by the observation that both depend on an absolutely conserved set of histidine residues for their function. Data available on E. coli ThrRS suggest that these histidines form a zinc-binding editing site (11), and structural studies are now required to elucidate whether the essential histidines fulfill a similar role in archaeal-type ThrRS enzymes.
ThrRS Editing Domains Are Ubiquitously Distributed. Our results, together with another recent study of editing by archaeal ThrRS (13), indicate that all living systems have efficient mechanisms for Ser-tRNAThr recognition and hydrolysis. Whereas bacteria and eukaryotes all possess the bacterial-type ThrRS, Archaea hold a greater diversity of editing patterns; they use the archaeal-type editing domain both in cis and trans and, less frequently, the bacterial-type ThrRS enzyme. This diversity in archaeal threonylation may have resulted from the exchange of archaeal-type ThrRS enzymes with their bacterial-type counterparts, the results of which are still evident in Thermoplasmata, Halobacteria, and Crenarchaea. Although the potential advantages of such replacements are not apparent, our phylogenetic analyses suggest that there are at least three independent horizontal gene-transfer events from bacteria to these archaeal groups (data not shown).
The distribution of ThrRS editing domain homologues was also analyzed outside of the ThrRS system. Searches of publicly available databases failed to identify other ORFs with significant similarities to the archaeal-type ThrRS editing domain, confirming its restricted emergence in archaeal threonylation. In contrast, the bacterial-type ThrRS editing domain apparently shares an evolutionary origin with the editing domain of alanyl-tRNA synthetase, the N-terminal domain of some tryptophanyl-tRNA synthetases, and a distinct family of small autonomous proteins (10, 24).
Origin of Trans-Acting Editing Proteins in Crenarchaea. The strictly delineated role in editing of the ThrRS-ed enzyme indicates that the ThrRS-related proteins of Crenarchaea represent a previously uncharacterized class of trans-editing factor. However, unlike single-domain transediting proteins homologous to the editing domains of prolyl-tRNA synthetase and alanyl-tRNA synthetase (24, 25), ThrRS-ed should not be regarded as a freestanding progenitor of the editing module, because it already contains an anticodon-binding domain; a more likely scenario proposes that some Crenarchaea acquired a bacterial-type ThrRS by horizontal gene transfer, which then rendered aminoacylation by the archaeal ThrRS redundant. Consequently, the catalytic domain of the archaeal-type ThrRS gradually degenerated, but its editing domain retained hydrolytic function. Likewise, the editing domain of the acquired bacterial-type ThrRS may have lost its deacylating capability and become rudimentary in the presence of the archaeal-type editing module. Some support for this proposal is provided from N-terminal truncations of ThrRS-cat enzymes, which show recognizable homology to bacterial-type editing domains (Fig. 2). Such a series of events may pertain to threonylation in the two Sulfolobus species and A. pernix but not to another crenarchaeon, Pyrobaculum aerophilum, which possesses only a standard archaeal-type ThrRS and, perhaps, never acquired a bacterial enzyme.
In contrast to the various combinations of archaeal-type ThrRSs described here and in ref. 13, Thermoplasmata and Halobacteria possess only the bacterial-type enzyme, which may have completely replaced the archaeal-type ThrRS in these organisms. The rationale as to why these particular organisms found the editing activity of the bacterial-type ThrRSs of greater utility could be related to their mesophilic or moderately thermophilic character. Because all of the crenarchaeal species possessing the ThrRS-ed enzyme belong to the hyperthermophilic group of organisms, it is possible that the bacterial-type editing domain was unstable or lacking in function at high temperatures. Conversely, if archaeal-type ThrRS maintained its activity at elevated temperatures, it may have been retained in hyperthermophiles to maintain editing function, while the bacterial enzyme assumed the aminoacylation function.
We analyzed available databases in search of analogous scenarios in other editing systems and identified two organisms with duplicated prolyl-tRNA synthetase genes of different origin [Chloroflexus aurantiacus (GenBank accession nos. ZP_00018001 and ZP_00020290) and Clostridium botulinum (www.sanger.ac.uk/Projects/C_botulinum)]. One of the prolyl-tRNA synthetase genes (bacterial-type) is truncated with a putative editing function, whereas the other (archaeal-type, lacking the editing domain) is predicted to perform aminoacylation activity. Thus, the apparent complementarity of autonomous editing and catalytic proteins may not be restricted to the ThrRS system; it could accordingly be presumed as an alternative mechanism used by aaRSs to increase both their efficiency and substrate specificity by selecting a combination of independent proteins that is functionally superior to either single ancestral partner.
Modularity in Protein Synthesis Quality Control. The aminoacylation and editing capacity of the chimeric ThrRS enzyme constructed here illustrates the flexibility of aaRS domains. The fact that most modern aaRSs possess a multidomain structure comprising tRNA-binding and editing domains (26) advocates advantages for such associations. In this respect, the separation of essential ThrRS functionalities between two enzymes in Crenarchaea may represent a transitional stage before fusion into one polypeptide. This finding is consistent with our suggestion that S. solfataricus ThrRS-ed and ThrRS-cat interact in vivo because the presence of both enzymes significantly improves threonylation activity in vitro compared with ThrRS-cat alone. Moreover, association between the two enzymes would certainly increase the efficiency of editing by avoiding dissociation of the mischarged product. The other possibility is that the continued separation of archaeal type ThrRS from bacterial-type ThrRS reflects instead a particular physiological adaptation. This alternative is supported by our observation that archaeal ThrRS imparts a distinct advantage under certain growth conditions, which may be even more pronounced in nature. Thus, if S. solfataricus is naturally subjected to elevated levels of serine and, therefore, to the presence of Ser-tRNAThr, the genetic separation of ThrRS editing activity would offer a means to directly regulate a cellular response to the accumulation of misacylated tRNA independent of aminoacylation. The documentation of other naturally existing aaRSs possessing separated domains, such as Aquifex aeolicus leucyl-tRNA synthetase (27), Nanoarchaeum equitans alanyl-tRNA synthetase (28), or accessory tRNA-binding domains known to be involved in formation of aaRS complexes in eukaryotes (29), supports the notion that more flexible noncovalent complex formation is beneficial in certain contexts.
Acknowledgments
We thank K. O. Stetter and M. Thomm for the gift of M. jannaschii cells. This work was supported by grants from the National Institute of General Medical Sciences (to D.S.), the Department of Energy (to D.S.), and the National Science Foundation (to P.B. and M.I.).
Abbreviations: aaRS, aminoacyl-tRNA synthetase; ThrRS, threonyl-tRNA synthetase; ThrRS-cat, threonyl-tRNA synthetase-catalytic domain; ThrRS-ed, threonyl-tRNA synthetase-editing domain.
References
- 1.Ibba, M. & Söll, D. (2000) Annu. Rev. Biochem. 69, 617–650. [DOI] [PubMed] [Google Scholar]
- 2.Jakubowski, H. & Goldman, E. (1992) Microbiol. Rev. 56, 412–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fersht, A. R. & Dingwall, C. (1979) Biochemistry 18, 2627–2631. [DOI] [PubMed] [Google Scholar]
- 4.Nureki, O., Vassylyev, D. G., Tateno, M., Shimada, A., Nakama, T., Fukai, S., Konno, M., Hendrickson, T. L., Schimmel, P. & Yokoyama, S. (1998) Science 280, 578–582. [DOI] [PubMed] [Google Scholar]
- 5.Silvian, L. F., Wang, J. & Steitz, T. A. (1999) Science 285, 1074–1077. [PubMed] [Google Scholar]
- 6.Lincecum, T. L., Tukalo, M., Yaremchuk, A., Mursinna, R. S., Williams, A. M., Sproat, B. S., Van Den, E. W., Link, A., Van Calenbergh, S., Grotli, M., et al. (2003) Mol. Cell 11, 951–963. [DOI] [PubMed] [Google Scholar]
- 7.Fukai, S., Nureki, O., Sekine, S., Shimada, A., Tao, J., Vassylyev, D. G. & Yokoyama, S. (2000) Cell 103, 793–803. [DOI] [PubMed] [Google Scholar]
- 8.Beuning, P. J. & Musier-Forsyth, K. (2000) Proc. Natl. Acad. Sci. USA 97, 8916–8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsui, W. C. & Fersht, A. R. (1981) Nucleic Acids Res. 9, 4627–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beebe, K., Ribas De Pouplana, L. & Schimmel, P. (2003) EMBO J. 22, 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dock-Bregeon, A., Sankaranarayanan, R., Romby, P., Caillet, J., Springer, M., Rees, B., Francklyn, C. S., Ehresmann, C. & Moras, D. (2000) Cell 103, 877–884. [DOI] [PubMed] [Google Scholar]
- 12.Sankaranarayanan, R., Dock-Bregeon, A. C., Rees, B., Bovee, M., Caillet, J., Romby, P., Francklyn, C. S. & Moras, D. (2000) Nat. Struct. Biol. 7, 461–465. [DOI] [PubMed] [Google Scholar]
- 13.Beebe, K., Merriman, E., Ribas de Pouplana, L. & Schimmel, P. (2004) Proc. Natl. Acad. Sci. USA 101, 5958–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahel, I., Stathopoulos, C., Ambrogelly, A., Sauerwald, A., Toogood, H., Hartsch, T. & Söll, D. (2002) J. Biol. Chem. 277, 34743–34748. [DOI] [PubMed] [Google Scholar]
- 15.Curnow, A. W., Tumbula, D. L., Pelaschier, J. T., Min, B. & Söll, D. (1998) Proc. Natl. Acad. Sci. USA 95, 12838–12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ribeiro, S., Nock, S. & Sprinzl, M. (1995) Anal. Biochem. 228, 330–335. [DOI] [PubMed] [Google Scholar]
- 17.Bini, E., Dixit, V., Dirksen, K., Drozda, M. & Blum, P. (2002) RNA 8, 1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Worthington, P., Blum, P., Perez-Pomares, F. & Elthon, T. (2003) Appl. Environ. Microbiol. 69, 252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schelert, J., Dixit, V., Hoang, V., Simbahan, J., Drozda, M. & Blum, P. (2004) J. Bacteriol. 186, 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen, M. B. (1959) Arch. Mikrobiol. 32, 270–277. [DOI] [PubMed] [Google Scholar]
- 21.Brock, T. D., Brock, K. M., Belly, R. T. & Weiss, R. L. (1972) Arch. Mikrobiol. 84, 54–68. [DOI] [PubMed] [Google Scholar]
- 22.Woese, C. R., Olsen, G. J., Ibba, M. & Söll, D. (2000) Microbiol. Mol. Biol. Rev. 64, 202–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sankaranarayanan, R., Dock-Bregeon, A. C., Romby, P., Caillet, J., Springer, M., Rees, B., Ehresmann, C., Ehresmann, B. & Moras, D. (1999) Cell 97, 371–381. [DOI] [PubMed] [Google Scholar]
- 24.Ahel, I., Korencic, D., Ibba, M. & Söll, D. (2003) Proc. Natl. Acad. Sci. USA 100, 15422–15427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong, F. C., Beuning, P. J., Silvers, C. & Musier-Forsyth, K. (2003) J. Biol. Chem. 278, 52857–52864. [DOI] [PubMed] [Google Scholar]
- 26.Alexander, R. W. & Schimmel, P. (2001) Prog. Nucleic Acid Res. Mol. Biol. 69, 317–349. [DOI] [PubMed] [Google Scholar]
- 27.Xu, M. G., Chen, J. F., Martin, F., Zhao, M. W., Eriani, G. & Wang, E. D. (2002) J. Biol. Chem. 277, 41590–41596. [DOI] [PubMed] [Google Scholar]
- 28.Waters, E., Hohn, M. J., Ahel, I., Graham, D. E., Adams, M. D., Barnstead, M., Beeson, K. Y., Bibbs, L., Bolanos, R., Keller, M., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 12984–12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simos, G., Sauer, A., Fasiolo, F. & Hurt, E. C. (1998) Mol. Cell 1, 235–242. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]