Abstract
Tau Filaments are found in >20 neurodegenerative diseases. Yet, because of their enormous molecular weights and poor tendency to form highly ordered 3D crystal lattices, they have evaded high-resolution structure determination. Here, we studied 25 derivatized tau mutants by using electron paramagnetic resonance and fluorescence spectroscopy to report structural details of tau filaments. Based on strong spin exchange and pyrene excimer formation of core residues, we find that individual tau proteins form single molecule layers along the fiber axis that perfectly stack on top of each other by in-register, parallel alignment of β-strands. We suggest a model of filament growth wherein the existing filament serves as a template for the incoming, unfolded tau molecule, resulting in a new structured layer with maximized hydrogen-bonded contact surface and side-chain stacking.
In addition to its role in stabilizing microtubules in neuritic extensions, tau has gained prominence as the protein constituent of filamentous inclusions in numerous neurodegenerative diseases (1). These inclusions, together with extracellular fibril deposits of the amyloid-β (Aβ)-peptide, constitute the pathological hallmarks of Alzheimer's disease.
In the adult human CNS, six different tau isoforms, ranging in size from 352 to 441 aa, are produced by alternative mRNA splicing. These isoforms vary by the absence or presence of the second of four microtubule-binding repeats in the C-terminal half and two inserts in the near N-terminal half of the protein (Fig. 1). The tau inclusions in Alzheimer's disease contain all six isoforms (2) and consist of paired helical and straight filaments (3, 4). Viewed under an electron microscope, tau filaments have a fuzzy coat that can be cleaved off by pronase (5). Cleavage leaves a core that is comprised of three microtubule-binding repeats (6). The importance of these repeats in filament formation was underscored by the finding that tau fragments comprising only the repeat region aggregate in vitro (7). In contrast, recombinant full-length tau is remarkably unreactive and aggregates only when anionic cofactors such as heparin are present (8, 9).
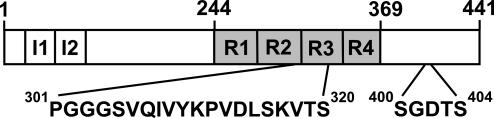
Fig. 1.
Bar diagram of the microtubule-associated protein tau. The large isoform of tau (441 aa) contains two inserts in the near N-terminal half (I1 and I2, 29 aa) and four microtubule-binding regions (R1–R4, 31–32 aa) in the C-terminal half. Sites that were investigated in this study are depicted by single-letter amino acid code.
A recent study involving x-ray and selected area electron diffraction of both straight and paired helical filaments (10) revealed a common cross-β structure, with β-strands running perpendicular to the fiber axis. These data agreed with earlier findings of diffraction patterns from filaments obtained from shorter tau fragments (11, 12). Importantly, they resolved some controversy concerning the filament structure of full-length tau (12–14). Thus, with respect to the cross-β structure and a seeded growth mechanism (15), tau filaments share similarities with a whole range of amyloidogenic protein aggregates (16). Beyond this, however, little is known about the structure of tau filaments. For example, it is not known how individual β-strands are arranged with each other and whether tau molecules align along or across the fiber axis, (i.e., whether hydrogen bonding among β-strands is intramolecular or intermolecular). To address these questions, we performed site-directed spin labeling of single cysteines in combination with electron paramagnetic resonance (EPR) spectroscopy. This approach allows us to monitor the mobility of a nitroxide spin label at any site and provides structural information at the level of the backbone fold (17–19). Furthermore, spin interactions between labels provide distance constraints, which in addition with other structural information can be used for model building. Whereas dipolar interactions yield distance information in the range of 8 to 25 Å (19), a second type of interaction, spin exchange, is observed at smaller distances and requires orbital overlap (20).
Here, we examined 25 derivatized residues within (amino acids 301–320) and outside (amino acids 400–404) the repeat region of tau (Fig. 1). Spin exchange and fluorescent excimer formation within the repeat region revealed that in the filament strands from different tau molecules are aligned in parallel resulting in a backbone geometry that allows same amino acids to stack along the fiber axis. This suggests a mechanism of filament growth in which the incoming tau molecule forms a hydrogen-bonded layer that perfectly stacks onto an outermost layer in the filament.
Materials and Methods
Plasmid Construction. pRK172, containing a DNA insert of the large isoform of human tau40 (21), served as a template for PCR, generating NheI and XhoI cleavage sites at the 5′ and 3′ ends of the tau sequence. The 3′ end also contained two stop codons. After proper cleavage, the new fragment was cloned into the NheI/XhoI restriction sites of pET-28b. The two native cysteines in tau (positions 291 and 322) were replaced by serines [previous substitutions of these sites did not change the morphology of the filaments (22, 23)], and single cysteines were introduced at positions 301–320 and 400–404 by using the QuikChange protocol (Stratagene). Sequences were always verified by DNA sequencing.
Protein Expression and Purification. The BL21 (DE3) strain of Escherichia coli was transformed with pET-28 plasmids by heat shock. Overnight cultures from single colonies were diluted 1/100 and served for large-scale amplification. Growth occurred in LB medium at 37°C under vigorous shaking. At an OD600 of 0.7–0.8, protein expression was induced with 1 mM isopropyl β-d-thiogalactoside. Bacteria were incubated for an additional 3 h and then centrifuged for 20 min at 3,500 × g. Pellets were resuspended in 500 mM NaCl, 20 mM Pipes (pH 6.5), 1 mM EDTA, and 50 mM 2-mercaptoethanol and then frozen at –80°C until further use.
Protein extraction began by heating the frozen pellets for 20 min at 80°C. After 10 min on ice, samples were sonicated 3 × 20 sec at power setting 10 in an ultrasonic cell disruptor (Microson, Misonic, Farmingdale, NY). Bacterial debris was pelleted for 20 min at 13,000 × g and soluble tau protein was precipitated from supernatant by adding ammonium sulfate (60%, m/V). After 1 h on ice, samples were centrifuged for 10 min at 13,000 × g. Pellets were resuspended in purified H2O, 2 mM DTT, passed through Acrodisc syringe filters (Pall) with a pore size of 0.45 μm, and loaded onto a Uno S cation exchange column (Bio-Rad). Protein was eluted from the column in a 0.05–1 M NaCl gradient containing 20 mM Pipes (pH 6.5), 0.5 mM EDTA, and 2 mM DTT. Fractions were analyzed by SDS/PAGE; tau protein was pooled, and 5 mM DDT was added. For further purification, samples were loaded onto a preparative-grade Superdex 200 column (Amersham Pharmacia). Elution occurred with 100 mM NaCl, 10 mM Pipes (pH 6.5), 1 mM EDTA, and 2 mM DTT. Monomeric tau was pooled and, again, 5 mM DTT was added. At that point, the protein was >95% pure, as assessed by SDS/PAGE.
For storage, tau was precipitated overnight on ice with an equal volume of MetOH, centrifuged for 20 min at 10,500 × g, washed once with MetOH and 2 mM DTT, and placed at –80°C with a layer of wash solution on top. Protein concentrations were determined at 276 nm in 6 M guanidinium chloride.
Spin Labeling and EPR. Protein pellets (1–3 mg) were solubilized in 6 M guanidinium chloride and labeled with a 10-fold molar excess of MTSL spin label [1-oxy-2,2,5,5-tetramethyl-pyrroline-3-methyl]-methanethiosulfonate (Toronto Research Chemicals, Downsview, Canada) or with a dilution of spin label and nonparamagnetic analog (24) for 1–2 h at room temperature. Excess label was removed on PD-10 columns (Amersham Pharmacia) with 100 mM NaCl and 10 mM Hepes (pH 7.4) as elution buffer. Crystals of spin label (Toronto Research Chemicals) were formed by evaporation of solvent (hexane/ethylacetate). Samples were measured in a Bruker EMX spectrometer fitted with an ER 4119HS resonator. For low-temperature scans, the spectrometer was equipped with an Oxford continuous flow cryostat and an HS cylindrical cavity. In all EPR measurements, the scan width was 150 G and the field modulation 1.5 G. The power was set at 12 mW for room temperature measurements and at 2 mW for all other measurements. If not otherwise noted, spectra were normalized to the same amount of spins. A background originating from 0.5–2% soluble tau was subtracted from all spectra.
Pyrene Labeling and Fluorescence Spectroscopy. Solubilized protein was labeled for 2 h with 5-fold molar excess of pyrene maleimide. The reaction was stopped with 10 mM DTT, and excess label was removed by using PD-10 columns. Samples were loaded onto a small Mono S column (Amersham Pharmacia) and eluted with 1 M NaCl, 20 mM Pipes (pH 6.5), 0.5 mM EDTA, and 2 mM DTT. Collected proteins were concentrated in Microcon YM-10 (Mr cut off, 10,000) (Amicon), and buffer was replaced by 50 mM NaCl, 20 mM Pipes (pH 6.5), 0.5 mM EDTA, and 2 mM DTT. Subsequently, proteins were precipitated with an equal volume of MetOH, kept on ice over night, centrifuged for 20 min at 10,500 × g, solubilized in 6 M guanidinium chloride, and desalted over PD-10 columns. Protein concentrations were determined by Bradford assay. Solutions containing 1 μM of soluble tau or tau filaments were placed into a quartz cuvette with 10-mm path length and measured in a Jasco FP-6500 spectrofluorometer. On excitation at 344 nm, emission spectra were taken from 360 to 600 nm. Filament samples were stirred during measurement.
Tau Filament Assembly. Labeled proteins were mixed with heparin (Mr 6,000, used in a tau: heparin molar ratio of 4:1) and incubated for 10 days with stirring at room temperature. Filaments were sedimented for 30 min at 160,000 × g and washed once with elution buffer.
Negative Stain Electron Microscopy (EM). One hundred fifty mesh carbon-coated copper grids were placed for 1 min onto 10-μl drops of tau sample (diluted to 2–20 μM) and for 40 s onto 10-μl drops of 2% uranyl acetate, and subsequently air-dried. EM images were taken with a JOEL JEM1200EX microscope at 80 kV.
Results
Spin Exchange Reveals Crystal-Like Order in the Core of Tau Filaments. To monitor structural changes on filament assembly of the large isoform of tau, we introduced a single cysteine into the third repeat at position 308 (Fig. 1). This site was chosen because it resides in a short hydrophobic stretch of amino acids that is part of the filament core and has high β-sheet propensity (25). As a monomer in solution, the spin-labeled mutant gave rise to an EPR spectrum with three sharp lines (Fig. 2A). This spectrum is characteristic for highly flexible sites and agrees well with previous data including CD measurements (26) showing that soluble tau is mostly unstructured.
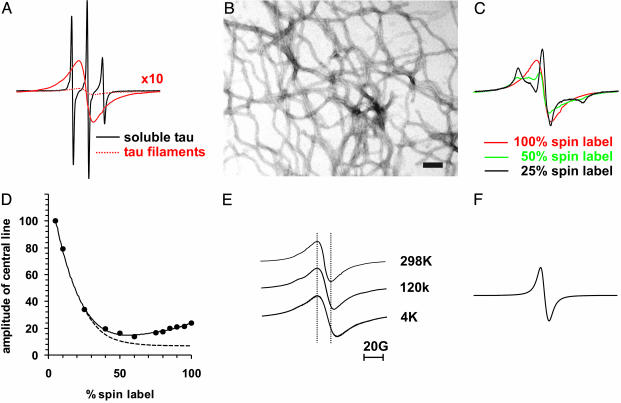
Fig. 2.
Spin exchange in a core residue of tau. Tau spectra and EM micrograph were taken from a cysteine mutant labeled at position 308. (A) Overlay of spectra from soluble tau (black), and filamentous tau (dotted red), for better comparison the amplitude of the spectrum of filamentous tau is multiplied by 10 (red line). (B) EM micrograph (×50,000) of uranyl acetate stained tau filaments. (Bar = 100 nm.) (C) Spectra of tau filaments containing 100% spin label (red as in A), 50% spin label (green), and 25% spin label (black); see also Materials and Methods. (D) Amplitudes of central lines from various dilutions are plotted vs. the percentage of spin label. Dotted line schematically illustrates decrease in amplitude as observed for dipolar coupling (see, e.g., refs. 28 and 49). (E) Spectra from filaments taken at 298, 120, and 4 K. Spectra are normalized to the same amplitude. (F) Spectrum of crystals from MTSL spin label.
Addition of heparin resulted in the formation of straight filaments with diameters of 10–15 nm (Fig. 2B), as expected for four-repeat tau isoforms (8). Thus, the spin label neither prevented the formation of filaments nor altered their morphology. The EPR spectrum had a dramatically reduced amplitude (Fig. 2 A, dotted line). To resolve its line shape, we replotted the spectrum at 10-fold magnification. Surprisingly, it contained only a single peak. Loss of the two outer peaks (hyperfine lines) is usually associated with spin exchange, and requires that, on the EPR time scale (<10–7 s), multiple spin labels are in orbital overlap (20). Because of such temporal and spatial constraints, a single peak EPR spectrum is highly unusual for a spin-labeled protein. It is important to note that two spin labels in Van der Waals contact as observed in a derivatized mutant of T4-lysozyme were not sufficient for a single-peak spectrum (27). Also, protein oligomers with three or four spin labels in very close proximity did not yield single-peak EPR spectra (24, 28). In other systems, however, where spin labels come together by frequent collision as is the case of spin-labeled lipids in membranes or free spin labels at high concentration in solution, spin exchange is a well established phenomenon (20). Furthermore, single-peak EPR spectra are observed in systems where spin labels are fixed at Van der Waals distance as for example in crystals (29).
In analogy to these latter systems, individual tau molecules in the filament must be arranged such that multiple spin labels at position 308 are in orbital overlap. Consequently, we expect that mixed filaments that contain paramagnetic as well as nonparamagnetic (EPR-inactive analog) label at position 308 should reveal a characteristic three-line spectrum. To address this point, we labeled soluble tau with two mixtures of para- and nonparamagnetic labels and subsequently initiated filament formation. Indeed, the outer peaks reappear with decreasing spin-label ratios (Fig. 2C). At 25% spin label, a typical three-line spectrum is observed. This spectrum, however, differs from that of soluble tau (Fig. 2 A) by an increased separation of the outer peaks, indicating that, in the filament, this site is buried. To further quantitate the effect of spin dilution, we collected spectra from an extended set of dilutions and plotted the amplitudes against percentage spin label (Fig. 2D). With increasing spin-label concentrations, we first observe a reduction in amplitude. This reduction is typical for dipolar interactions because it coincides with a broadening of the spectrum (19). If only dipolar interactions were present, one would expect the signal amplitude to continue to drop as the percentage of label increases (Fig. 2D, dotted line). In our case, however, we observe a rise in the signal amplitude at higher percentage of spin label. This can only be explained by exchange narrowing of the spectrum (Fig. 2C and data not shown).
Next, we asked whether spin exchange in tau filaments is modulated by motion or whether contacts between labels are fixed such as in crystals. For this purpose we monitored EPR spectra at varying temperatures. Although a decrease in temperature from 298 to 120 K and then to 4 K caused spectral broadening (13.8, 17.4, and 20.4 G) (Fig. 2E), hyperfine lines were never observed. This means that collisional effects as seen in solution systems are not required for spin exchange. The arrangement of spin labels in tau filaments is thus more similar to solid/crystal systems in which arrays of spin labels are in orbital overlap (29). As a control, we also confirmed that crystals of our spin label indeed show spin exchange (Fig. 2F).
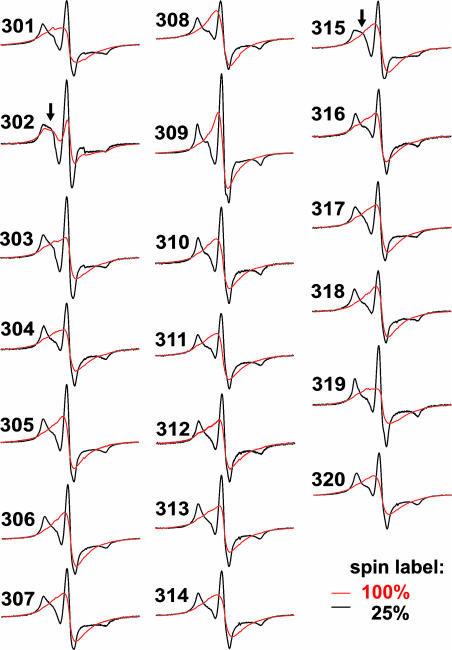
Parallel Alignment in the Filament Core. To test whether spin exchange is limited to position 308 or whether neighboring positions, too, have multiple contact partners, we introduced single spin labels at positions 301–320. All sites in soluble tau produced spectra with sharp lines similar to those seen at position 308 (data not shown), indicating that the entire region is unstructured. As for WT and the example shown in Fig. 2B, all mutant proteins formed filaments with predominantly straight appearance (data not shown), and nearly all sites gave single-peak EPR spectra (Fig. 3), indicating that spin exchange occurs in an extended region. Only for position 302 hyperfine splitting could be observed, suggesting that, in this case, spin labels were not in perfect contact. These data reveal that individual tau molecules in the filament are aligned in parallel with a backbone geometry that allows same residues to stack on top of each other. Because α-helices cannot be aligned such that consecutive positions of neighboring molecules are in direct contact they can be excluded as structural elements in this region. Parallel β-strands, however, would allow for extended stacking and thus are in full agreement with our data.
Fig. 3.
EPR spectra of spin-labeled positions 301–320 in tau filaments. Spectra of tau filaments labeled with 100% spin label (red) or 25% spin label (black). Arrows point to more mobile components in the dilution spectra of positions 302 and 315.
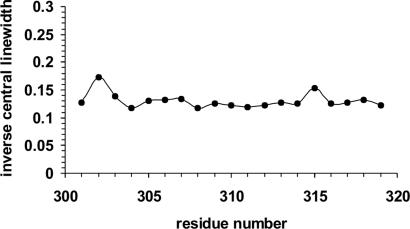
Next, we recorded the EPR spectra of filaments grown from tau derivatized with a mixture of 25% paramagnetic and 75% nonparamagnetic label. All spectra revealed hyperfine splitting and were characteristic for immobilized sites. Interestingly, two spectra (positions 302 and 315) showed an additional component closer to the central line (Fig. 3, arrows), indicating that these sites have somewhat increased mobility. This can also be seen when plotting the inverse central linewidth against residue number (Fig. 4). Residues 302 and 315 have slightly increased inverse central linewidths. Similar increases in inverse central linewidths were recently observed in a possible turn between β-strands in the Aβ-peptide of Alzheimer's disease (30). It should be emphasized, however, that spin labels at these sites are still very immobile.
Fig. 4.
Inverse linewidth plot. The inverse central linewidths from dilution spectra of tau filaments are plotted against residue number.
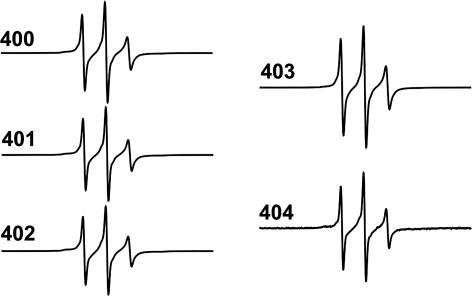
An Unstructured Region Outside the Filament Core. All measurements thus far involved residues that are within the pronase resistant core of the filament, as a control we next tested whether residues in the C-terminal region outside this core also become structured upon filament formation. Thus, we introduced single cysteines at positions 400–404. Although all five spin-labeled mutants formed filaments that were undistinguishable from those shown in Fig. 2B (data not shown), all EPR spectra revealed three sharp peaks (Fig. 5), demonstrating that this region remains unstructured even in the filament.
Fig. 5.
EPR spectra of spin-labeled positions 400–404 in the tau filament.
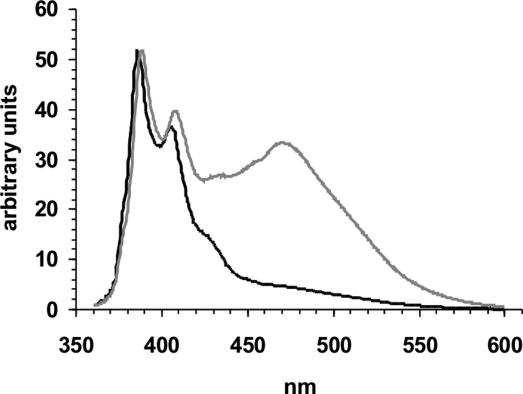
Pyrene Eximer in the Filament Core. For independent confirmation of our EPR data, we used a fluorescence-based approach. It is well established that two pyrene molecules that are separated by only a few angstroms and are correctly oriented can form excimers (excited dimers), resulting in a characteristic broad band in the emission spectrum centered ≈460–480 nm. Here, we used cysteine mutant 308 in the core region and cysteine mutant 403 outside the core and labeled them with pyrene maleimide. Filaments formed from mutant 308 result in an emission spectrum (Fig. 6) with a peak at ≈473 nm, confirming that pyrene molecules must come into contact. Filaments formed from mutant 403, however, exhibit no excimer band in their spectrum, in agreement with our EPR data that revealed this residue to be unstructured. Excimer bands were also absent when spectra were taken from the unpolymerized tau mutants (data not shown).
Fig. 6.
Excimer formation. Emission spectra (360–600 nm) from tau filaments derivatized at positions 308 (gray) and 403 (black) normalized to the 385-nm peak. Samples were excited at 344 nm.
Thus, using two independent approaches, we have shown that tau molecules within filaments are aligned in register.
Discussion
Determining the molecular structure of tau filaments poses a significant challenge in view of their insolubility and enormous size. The study presented herein introduced single cysteines and used spin labels and fluorophores to report structural changes from monomer to filament, as well as to outline higher-order structural arrangements in the filament core. Our findings have important implications for filament growth.
For our study, we chose residues 301–320 inasmuch as they are known to be located in the pronase-resistant filament core (6, 31) and because their direct involvement in filament formation has been firmly established (25, 32). As controls, we studied residues 400–404, which are located outside of the core. In soluble tau, all residues were found to be completely unstructured. Upon filament formation, residues within the core region became highly ordered, whereas those outside of the core region remained unstructured.
Our principal finding relevant to positions in the core region is that the same sites in adjacent tau molecules were in direct contact, resulting in spin exchange and pyrene excimers. Because spin exchange was observed in consecutive positions, tau molecules must form parallel β-strands that are connected by intermolecular hydrogen bonds. In combination with fiber diffraction data, which revealed a cross-β structure (10), we conclude that parallel strands from different tau molecules stack along the fiber axis. A similar structural arrangement was suggested for filaments of Aβ (30, 33, 34) and α-synuclein (a protein forming intracellular aggregates in Parkinson's disease) (35), although, particularly in the case of Aβ, the spin exchange was not as pronounced. Our data show that tau filaments must be very homogeneous with respect to the studied core region. Antiparallel alignment and mixed alignment cannot be detected. In both cases, we would expect heterogeneous spectra with hyperfine splitting but not the single peaks that we have observed (see Results). At this point, it is important to mention that tau dimers were shown to be aligned antiparallel (7). This appears to be contradictory to our data; however, in light of observations on amyloid formation of an SH3 domain from phosphatidylinositol-3′-kinase (36), antiparallel alignment might well occur in an early stage of oligomerization. It has been suggested that the unfolded Src homolog 3 domain first transitions through an antiparallel intermediate before parallel filaments are formed. Thus, filament formation appears to accommodate large conformational rearrangements. The stacking of spin labels in mature tau filaments must be very precise because the spin exchange observed here requires orbital overlap between multiple N-O moieties and does not appear to result from collisional contacts. The exchange interaction persists even at 4 K, suggesting a crystal-like order along the fiber axis.
In soluble proteins, individual strands within β-sheets often have a substantial twist (37). Because of this twist, stacking of amino acids can only occur along a single ridge (see, e.g., ref. 38). Given our observation of stacking over a large range of consecutive positions, the twist between strands must either be very small or nonexistent. Although we have a good understanding of the structural alignment along the fiber axis (quaternary structure), at this time we do not know how individual β-strands in one molecule are arranged with respect to each other (tertiary structure); i.e., what the structure is in the plane perpendicular to the fiber axis. In our EPR experiments, we observed slightly increased mobilities for positions 302 and 315, suggesting that they could be part of turn regions. Even in this case, residue 315 would have to be stacked and, to a lesser degree, residue 302 as well.
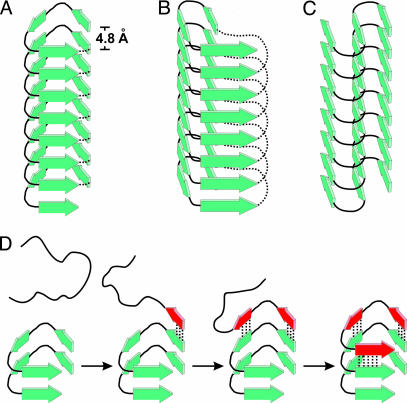
Of the known protein folds, only β-helical structures are in agreement with all structural constraints. They contain parallel strands with little twist and stacking even in turns (39). In fact, the β-helix had been suggested previously as one possible structure for tau (10). Notably, in another amyloidogenic protein, the prion protein in scrapie, β-helices have been proposed as likely structural elements for the misfolded form (40). Studies of soluble proteins revealed a number of β-helical folds (39, 41). Fig. 7 A and B schematically depicts examples of the traditional left- and right-handed β-helices. Of course, in the case of tau filaments, the β-helices would have to be discontinuous because one tau molecule represents one layer (Fig. 7A). The typical diameter of a β-helix (2–3 nm) is considerably smaller than that of the tau filament (10–15 nm). Thus, the formation of β-helical tau filaments would require not one β-helix as depicted here, but multiple β-helices arranged side-by-side. If homologous repeats formed separate β-helices, one tau molecule could extend over multiple β-helices, which would thus be interconnected within the cross-sectional plane.
Fig. 7.
Substructure of tau filament and layer extension. β-Strands in tau filaments have parallel, in-register alignment and are perfectly stacked along the fiber axis. Individual layers of tau molecules are hydrogen bonded and separated by 4.8 Å. (A–C) Models of tau filaments with different possible lateral arrangements are depicted: left-handed β-helix (A), right-handed β-helix (B), and intersheet-hairpin (C). Dotted lines represent intramolecular connections in β-helices of soluble proteins as observed in crystal structures (39). In tau filaments this continuity would be disrupted and neighboring repeat regions would project from both ends, possibly forming individual domains. (D) Filament growth is, for convenience, demonstrated on the left-handed “helix” model. The incoming unstructured tau molecule forms β-strands (red) and hydrogen bonds (dotted lines) to a filament template adding one new layer through a zipper-like mechanism.
Our data would also be consistent with intersheet hairpin models that have been suggested for Aβ and α-synuclein (30, 33, 35) (Fig. 7C). Importantly, β-helical structures and multiple intersheet hairpin structures would fit into the density map obtained from EM projectional analysis of tau filaments (42, 43).
Aside from stabilizing contributions from backbone dipole–dipole interactions, all structures allow stacking of same residues. In the case of hydrophobic and hydrogen-bonded residues, such stacking would likely be very stabilizing. Charged residues (K311, K317, and D314) pointing toward the protein interior would result in large energetic penalties unless counterbalanced by salt bridges as suggested in the case of Aβ fibrils (44), or protein dipoles as for example in soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE) complexes (45). The electrostatic repulsion due to stacking of same charges at solvent exposed sites would be considerably less because of the high dielectric environment, especially if splaying occurs at the end of longer charged side chains. In addition, salts, polyanions (heparin), or charged and polar site chains from other parts of the tau molecule could further stabilize the stack.
Regardless of the precise lateral arrangement of tau, the parallel stacking of single molecule layers along the axis has important implications for filament growth (Fig. 7D). A tau molecule in the outermost layer of the filament contains edge strands with highly exposed H-bond donors and acceptors. In natural β-sheet proteins, including β-helices, such edge strands are usually covered or otherwise modified as to avert further β-sheet interactions (46). In the case of tau filaments, these strands now serve as freely accessible templates. An incoming molecule becomes structured upon hydrogen bonding to the protruding carbonyls and amides and stacking of the perpendicularly oriented side chains.
We do not know which residues are involved in the initial contact, but subsequent layer formation would occur by hydrogen bonding in a zipper-like fashion and result in a maximized hydrogen-bonded contact surface. Filament extension would resume by stacking of individual tau molecules layer by layer. Because the contact surfaces on both ends are not the same, i.e., tau filaments contain inherent polarity, depending on conditions filament growth could preferentially occur in one direction. Indeed, when paired helical filaments were used to seed filament formation the elongation reaction was reported to be unidirectional (47).
A similar template assisted growth mechanism as for tau may also apply to Aβ and α-synuclein. In this respect, it is interesting to note that α-synuclein and tau enhance each other's filament formation and that α-synuclein is able to form joined filaments with tau by end-to-end annealing (48). In light of our findings, these results could be rationalized in terms of highly similar structures that, at least to some degree, are compatible with each other.
In summary, our study sheds light on the arrangement of tau filaments along the fiber axis and suggests a mechanism of filament growth. Future EPR analysis should allow us to obtain a more precise picture of the expansion and alignment of β-strands in the plane.
Acknowledgments
The pRK172 construct of human tau40 was generously provided by M. Goedert (Medical Research Council Laboratory, Cambridge, U.K.). Nonparamagnetic label was a kind gift of K. Hideg (University of Pécs, Pécs, Hungary). We thank A. DiBilio (California Institute of Technology, Pasadena) for help with low-temperature EPR measurements. EPR analysis software was generously provided by C. Altenbach (University of California, Los Angeles). This work was supported by the Beckman Foundation (R.L.) and a fellowship grant from the Larry L. Hillblom Foundation (to M.M.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: EPR, electron paramagnetic resonance; Aβ, amyloid-β; EM, electron microscopy.
References
- 1.Lee, V. M., Goedert, M. & Trojanowski, J. Q. (2001) Annu. Rev. Neurosci. 24, 1121–1159. [DOI] [PubMed] [Google Scholar]
- 2.Goedert, M., Spillantini, M. G., Cairns, N. J. & Crowther, R. A. (1992) Neuron 8, 159–168. [DOI] [PubMed] [Google Scholar]
- 3.Metuzals, J., Montpetit, V. & Clapin, D. F. (1981) Cell Tissue Res. 214, 455–482. [DOI] [PubMed] [Google Scholar]
- 4.Yagishita, S., Itoh, Y., Nan, W. & Amano, N. (1981) Acta Neuropathol. 54, 239–246. [DOI] [PubMed] [Google Scholar]
- 5.Wischik, C. M., Novak, M., Edwards, P. C., Klug, A., Tichelaar, W. & Crowther, R. A. (1988) Proc. Natl. Acad. Sci. USA 85, 4884–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novak, M., Kabat, J. & Wischik, C. M. (1993) EMBO J. 12, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wille, H., Drewes, G., Biernat, J., Mandelkow, E. M. & Mandelkow, E. (1992) J. Cell Biol. 118, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goedert, M., Jakes, R., Spillantini, M. G., Hasegawa, M., Smith, M. J. & Crowther, R. A. (1996) Nature 383, 550–553. [DOI] [PubMed] [Google Scholar]
- 9.Perez, M., Valpuesta, J. M., Medina, M., Montejo de Garcini, E. & Avila, J. (1996) J. Neurochem. 67, 1183–1190. [DOI] [PubMed] [Google Scholar]
- 10.Berriman, J., Serpell, L. C., Oberg, K. A., Fink, A. L., Goedert, M. & Crowther, R. A. (2003) Proc. Natl. Acad. Sci. USA 100, 9034–9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Bergen, M., Barghorn, S., Li, L., Marx, A., Biernat, J., Mandelkow, E. M. & Mandelkow, E. (2001) J. Biol. Chem. 276, 48165–48174. [DOI] [PubMed] [Google Scholar]
- 12.Giannetti, A. M., Lindwall, G., Chau, M. F., Radeke, M. J., Feinstein, S. C. & Kohlstaedt, L. A. (2000) Protein Sci. 9, 2427–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirschner, D. A., Abraham, C. & Selkoe, D. J. (1986) Proc. Natl. Acad. Sci. USA 83, 503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schweers, O., Schonbrunn-Hanebeck, E., Marx, A. & Mandelkow, E. (1994) J. Biol. Chem. 269, 24290–24297. [PubMed] [Google Scholar]
- 15.Friedhoff, P., von Bergen, M., Mandelkow, E. M., Davies, P. & Mandelkow, E. (1998) Proc. Natl. Acad. Sci. USA 95, 15712–15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stefani, M. & Dobson, C. M. (2003) J. Mol. Med. 81, 678–699. [DOI] [PubMed] [Google Scholar]
- 17.Hubbell, W. L., McHaourab, H. S., Altenbach, C. & Lietzow, M. A. (1996) Structure (London) 4, 779–783. [DOI] [PubMed] [Google Scholar]
- 18.Hubbell, W. L., Gross, A., Langen, R. & Lietzow, M. A. (1998) Curr. Opin. Struct. Biol. 8, 649–656. [DOI] [PubMed] [Google Scholar]
- 19.Hubbell, W. L., Cafiso, D. S. & Altenbach, C. (2000) Nat. Struct. Biol. 7, 735–739. [DOI] [PubMed] [Google Scholar]
- 20.Molin, Y. N., Salikhov, K. M. & Zamaraev, K. I. (1980) Spin Exchange (Springer, Berlin).
- 21.Goedert, M. & Jakes, R. (1990) EMBO J. 9, 4225–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamblin, T. C., King, M. E., Kuret, J., Berry, R. W. & Binder, L. I. (2000) Biochemistry 39, 14203–14210. [DOI] [PubMed] [Google Scholar]
- 23.Barghorn, S. & Mandelkow, E. (2002) Biochemistry 41, 14885–14896. [DOI] [PubMed] [Google Scholar]
- 24.Gross, A., Columbus, L., Hideg, K., Altenbach, C. & Hubbell, W. L. (1999) Biochemistry 38, 10324–10335. [DOI] [PubMed] [Google Scholar]
- 25.von Bergen, M., Friedhoff, P., Biernat, J., Heberle, J., Mandelkow, E. M. & Mandelkow, E. (2000) Proc. Natl. Acad. Sci. USA 97, 5129–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cleveland, D. W., Hwo, S. Y. & Kirschner, M. W. (1977) J. Mol. Biol. 116, 227–247. [DOI] [PubMed] [Google Scholar]
- 27.Langen, R., Oh, K. J., Cascio, D. & Hubbell, W. L. (2000) Biochemistry 39, 8396–8405. [DOI] [PubMed] [Google Scholar]
- 28.Langen, R., Isas, J. M., Luecke, H., Haigler, H. T. & Hubbell, W. L. (1998) J. Biol. Chem. 273, 22453–22457. [DOI] [PubMed] [Google Scholar]
- 29.Lajzerowicz-Bonneteau, J. (1976) in Spin Labeling Theory and Applications, ed. Berliner, L. J. (Academic, New York).
- 30.Torok, M., Milton, S., Kayed, R., Wu, P., McIntire, T., Glabe, C. G. & Langen, R. (2002) J. Biol. Chem. 277, 40810–40815. [DOI] [PubMed] [Google Scholar]
- 31.Wischik, C. M., Novak, M., Thogersen, H. C., Edwards, P. C., Runswick, M. J., Jakes, R., Walker, J. E., Milstein, C., Roth, M. & Klug, A. (1988) Proc. Natl. Acad. Sci. USA 85, 4506–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abraha, A., Ghoshal, N., Gamblin, T. C., Cryns, V., Berry, R. W., Kuret, J. & Binder, L. I. (2000) J. Cell. Sci. 113, 3737–3745. [DOI] [PubMed] [Google Scholar]
- 33.Tycko, R. (2003) Biochemistry 42, 3151–3159. [DOI] [PubMed] [Google Scholar]
- 34.Tycko, R. (2004) Curr. Opin. Struct. Biol. 14, 96–103. [DOI] [PubMed] [Google Scholar]
- 35.Der-Sarkissian, A., Jao, C. C., Chen, J. & Langen, R. (2003) J. Biol. Chem. 278, 37530–37535. [DOI] [PubMed] [Google Scholar]
- 36.Zurdo, J., Guijarro, J. I., Jimenez, J. L., Saibil, H. R. & Dobson, C. M. (2001) J. Mol. Biol. 311, 325–340. [DOI] [PubMed] [Google Scholar]
- 37.Chothia, C. (1973) J. Mol. Biol. 75, 295–302. [DOI] [PubMed] [Google Scholar]
- 38.Isupov, M. N., Fleming, T. M., Dalby, A. R., Crowhurst, G. S., Bourne, P. C. & Littlechild, J. A. (1999) J. Mol. Biol. 291, 651–660. [DOI] [PubMed] [Google Scholar]
- 39.Jenkins, J. & Pickersgill, R. (2001) Prog. Biophys. Mol. Biol. 77, 111–175. [DOI] [PubMed] [Google Scholar]
- 40.Wille, H., Michelitsch, M. D., Guenebaut, V., Supattapone, S., Serban, A., Cohen, F. E., Agard, D. A. & Prusiner, S. B. (2002) Proc. Natl. Acad. Sci. USA 99, 3563–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weigele, P. R., Scanlon, E. & King, J. (2003) J. Bacteriol. 185, 4022–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crowther, R. A. & Wischik, C. M. (1985) EMBO J. 4, 3661–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crowther, R. A. (1991) Proc. Natl. Acad. Sci. USA 88, 2288–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petkova, A. T., Ishii, Y., Balbach, J. J., Antzutkin, O. N., Leapman, R. D., Delaglio, F. & Tycko, R. (2002) Proc. Natl. Acad. Sci. USA 99, 16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fasshauer, D., Sutton, R. B., Brunger, A. T. & Jahn, R. (1998) Proc. Natl. Acad. Sci. USA 95, 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson, J. S. & Richardson, D. C. (2002) Proc. Natl. Acad. Sci. USA 99, 2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.King, M. E., Ahuja, V., Binder, L. I. & Kuret, J. (1999) Biochemistry 38, 14851–14859. [DOI] [PubMed] [Google Scholar]
- 48.Giasson, B. I., Forman, M. S., Higuchi, M., Golbe, L. I., Graves, C. L., Kotzbauer, P. T., Trojanowski, J. Q. & Lee, V. M. (2003) Science 300, 636–640. [DOI] [PubMed] [Google Scholar]
- 49.Berengian, A. R., Parfenova, M. & McHaourab, H. S. (1999) J. Biol. Chem. 274, 6305–6314. [DOI] [PubMed] [Google Scholar]