Abstract
Persistent mucosal inflammation and microbial infection are characteristics of chronic rhinosinusitis (CRS). Mucosal microbiota dysbiosis is found in other chronic inflammatory diseases; however, the relationship between sinus microbiota composition and CRS is unknown. Using comparative microbiome profiling of a cohort of CRS patients and healthy subjects, we demonstrate that the sinus microbiota of CRS patients exhibits significantly reduced bacterial diversity compared with that of healthy controls. In our cohort of CRS patients, multiple, phylogenetically distinct lactic acid bacteria were depleted concomitant with an increase in the relative abundance of a single species, Corynebacterium tuberculostearicum. We recapitulated the conditions observed in our human cohort in a murine model and confirmed the pathogenic potential of C. tuberculostearicum and the critical necessity for a replete mucosal microbiota to protect against this species. Moreover, Lactobacillus sakei, which was identified from our comparative microbiome analyses as a potentially protective species, defended against C. tuberculostearicum sinus infection, even in the context of a depleted sinus bacterial community. These studies demonstrate that sinus mucosal health is highly dependent on the composition of the resident microbiota as well as identify both a new sino-pathogen and a strong bacterial candidate for therapeutic intervention.
INTRODUCTION
Bacterial rhinosinusitis (sinusitis), which affects more than 15% of the U.S. population annually, is one of the most common problems presented to the primary care practitioner in the ambulatory setting and results in more than $5.8 billion in direct health care expenditures (1). Typically classified by duration of symptoms, sinusitis may be acute (less than 4 weeks in duration), sub-acute (4 to 12 weeks), or chronic (>12 weeks with or without acute exacerbations) (2). Chronic rhinosinusitis (CRS) alone represents a large portion of sinusitis cases, affecting more than 30 million Americans (3) and resulting in an annual economic health care burden in excess of $2.4 billion. Culture-based studies have demonstrated chronic bacterial colonization of CRS patient sinus cavities and implicated these species in the pathophysiology of the disease (4, 5). Despite these findings, the microbiology and immunology underlying CRS remain poorly described and controversial, and to date, no clear etiology has been described (6).
Confounding the field are recent findings demonstrating that known bacterial pathogens, such as Staphylococcus and Streptococcus species isolated from CRS sinuses and implicated in the disease (4, 5, 7, 8), have also been detected in the nasopharynx of healthy individuals with no sinus symptomatology (9). Thus, the composition of the microbiota at discrete mucosal sites may define the abundance and pathogenic behavior of specific members of the assemblage. Indeed, perturbation of the native gastrointestinal microbiota of mice, before Clostridium difficile instillation, results in increased fecal abundance of this species and enhanced murine susceptibility to infection (10). Similarly, disruption of the hindgut microbiota of the mosquito (Anopheles gambiae), the natural vector for Plasmodium falciparum, results in altered vectoral capacity characterized by increased host susceptibility to P. falciparum infection (11). Collectively, these observations suggest that microbiota composition plays a key role in the protection against pathogen overgrowth and virulence gene expression and that perturbations to these assemblages must be considered as potential contributing factors to infectious and inflammatory disease etiology.
Although the sinuses are primarily lined with respiratory epithelia, which readily supports colonization by a diverse microbiome at other upper respiratory sites such as the oropharynx (9, 12), to date little is known of the composition of the resident microbiome of the paranasal sinuses and the contribution of these assemblages to sinus mucosal health. Most microbiological studies of this niche are based on culture-based approaches, which underestimate the diversity of species present. The application of culture-independent approaches offers the opportunity to provide a broader picture of sinus microbiome composition and identify both gross community characteristics and discrete species associated with health status that contribute to or protect against disease development.
Here, we present findings from a high-resolution culture-independent comparative analysis of the sinus microbiota of CRS patients and healthy subjects without CRS, undergoing open nasal or sinus surgery. Surgical patients were preferred because they typically exhibit severe disease, and surgery provides access to affected sinus mucosal surfaces that are otherwise inaccessible. Specifically, efforts aimed to determine whether specific features of the sinus mucosal microbiota were associated with disease state and severity and to identify both pathogenic and protective species in this niche.
RESULTS
Comparative sinus microbiota analysis
Maxillary sinus samples from 20 subjects (10 CRS and 10 healthy individuals) were used for this study. Patient details are provided in Table 1. Mucin hypersecretion is a hallmark of sinus disease (13, 14). Therefore, to confirm that the CRS patients exhibited a phenotype consistent with disease, we performed quantitative polymerase chain reaction (qPCR) analysis of Muc5A gene [encoding mucin primarily secreted from surface epithelium goblet cells in humans (15, 16)] expression. This analysis confirmed significant (P < 0.007, Student's t test) up-regulation of this gene in CRS patients compared to healthy control subjects (fig. S1), thus validating the presence of sinus disease in our CRS cohort. qPCR analysis of bacterial burden by total 16S ribosomal RNA (rRNA) copy number demonstrated that perioperative CRS patients and healthy subjects exhibited no significant difference in sinus bacterial burden (2.10 × 106 ± 1.01 × 106 versus 2.92 × 106 ± 2.17 × 106 copies of 16S rRNA gene per microgram of total DNA in CRS and healthy control subjects, respectively; t = 0.93; P = 0.37, Student's t test). These data suggest that the sinus niche supports a defined bacterial load and that microbiota composition and relative taxonomic distribution, rather than the number of bacteria present, are related to disease state.
Table 1.
Patient information. TMP/SMX, trimethoprim/sulfamethoxazole; AMP/SUL, ampicillin/sulbactam; AMO/CLU, amoxicillin/clavulanate.
| Group | Study ID | Gender | Age | SNOT-20 | Antimicrobial treatment |
|
|---|---|---|---|---|---|---|
| Preoperative | Perioperative | |||||
| CRS | CRS-001* | M | 54 | 2.75 | None† | Cefazolin |
| CRS | CRS-002 | M | 54 | 2.35 | TMP/SMX | Vancomycin |
| CRS | CRS-003* | M | 48 | 2.65 | None | AMP/SUL |
| CRS | CRS-004 | M | 33 | 3.60 | None | Cefazolin |
| CRS | CRS-005* | M | 56 | 2.65 | Clarithromycin | Cefazolin |
| CRS | CRS-006 | F | 41 | 1.85 | None | Cefazolin |
| CRS | CRS-007 | M | 60 | 3.00 | Clarithromycin | Levofloxacin |
| CRS | CRS-008 | M | 53 | 1.55 | Ciprofloxacin | Cefazolin |
| CRS | CRS-009 | F | 46 | 2.35 | AMO/CLU | Cefazolin |
| Control | CRS-010 | M | 62 | 0.90 | Levofloxacin | Ceftriaxone |
| CRS | CRS-011 | M | 42 | 3.35 | AMO/CLU | Clindamycin |
| Control | CRS-012* | M | 43 | 0.15 | None | Clindamycin |
| Control | CRS-013 | F | 73 | 1.70 | None | Cefazolin |
| Control | CRS-014 | M | 41 | 2.60 | None | Clindamycin |
| Control | CRS-015* | M | 39 | 0.00 | None | Clindamycin |
| Control | CRS-016* | F | 37 | 2.25 | None | Cefazolin |
| Control | CRS-017 | F | 46 | 0.10 | None | Clindamycin |
| Control | CRS-018 | M | 46 | 2.15 | None | Clindamycin |
| Control | CRS-019 | F | 31 | 0.50 | None | Clindamycin |
| Control | CRS-020 | F | 18 | 0.30 | None | Clindamycin |
Microbiota profiling was not performed for these subject samples because of insufficient 16S rRNA amplicon.
No antibiotics administered.
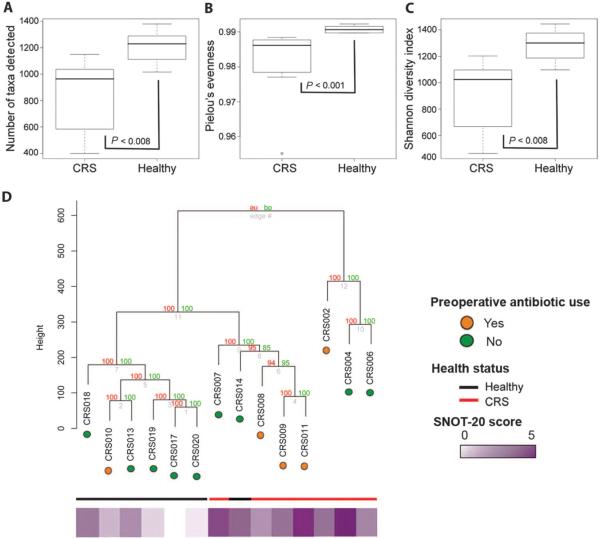
A standardized phylogenetic microarray, the 16S rRNA PhyloChip, was used to compare the presence and relative abundance of about 8500 bacterial taxa, representing broad membership of all known bacterial phyla (Hugenholtz phylogenetic classification). Because the primary aim of this study was to generate and compare a high-resolution profile of the microbiota present in healthy subjects and CRS patients, a phylogenetic microarray approach was used in favor of shotgun sequencing with limited read depth, which primarily identifies the higher-abundance species in a given community. We specifically sought to include lower-abundance microbiome members in this comparative analysis because recent evidence has demonstrated that such species can contribute considerably to microbiome function (17) and act as keystone species that shape microbial community composition (18). Using this tool, we profiled bacterial communities present in 14 subjects (7 healthy and 7 CRS) with sufficient amplified 16S rRNA product to be analyzed. Comparative analyses of gross bacterial community metrics between the CRS and the healthy groups demonstrated that compared to healthy individuals, CRS patients exhibited substantial microbiota perturbation characterized by significantly reduced bacterial richness (number of bacterial types detected; P ≤ 0.005, Welch's t test; Fig. 1A), evenness (relative distribution of bacterial types; P ≤ 0.04, Welch's t test; Fig. 1B), and Shannon's diversity (metric calculated using richness and evenness indices; P ≤ 0.01, Welch's t test; Fig. 1C). These observations were further supported by hierarchical cluster analysis (HCA), which demonstrated that most healthy subjects clustered in a single tightly knit group. The bacterial communities from healthy subjects were compositionally distinct from CRS patients, whose bacterial communities clustered into two distinct groups (Fig. 1D). Healthy subject CRS 14 clustered with CRS patients; however, it was subsequently determined that this individual had historically suffered from chronic nasal allergies. This analysis confirmed that the sinus microbiota composition of healthy subjects is distinct from that of CRS patients and that, within the CRS population, discrete subgroups with distinct microbial community profiles exist.
Fig. 1.
Comparative analyses of bacterial community. (A to C) Richness (A), evenness (B), and Shannon diversity (C) indices of array-detected sinus microbiota of CRS patients and healthy subjects. Values represent means ± SEM. (D) Hierarchal cluster analysis based on a Canberra distance matrix of microbiota detected in CRS patients and healthy subjects. Significant clusters determined with approximately unbiased (au) P values and bootstrap values (bp) are indicated. A heat plot of sinus symptom severity based on SNOT-20 scores and use of preoperative antimicrobials are also indicated (details provided in Table 1).
Although the numbers in this study were insufficient to comprehensively assess factors responsible for the observed clustering pattern, antibiotic use was examined because of its well-defined influence on microbiota composition. As is standard surgical procedure, all study subjects (CRS and healthy) received prophylactic antibiotics immediately (1 hour) before surgery and sample collection. Long-term preoperative antibiotic administration was absent for most healthy subjects and variable in the CRS patient group (Table 1). However, patients with disparate long-term antimicrobial administration histories (for example, CRS 4, 6, and 2; group II; Table 1) clustered closely together. Moreover, this group, which included two patients who had not received long-term antibiotics, exhibited significantly lower (P < 0.006, Student's t test) sinus community diversity compared to microbiota from patients in group I, all of whom had received long-term prophylactic antibiotic administration. These data suggest that although it may be a contributory factor in shaping the sinus mucosal microbiome, antimicrobial therapy is not the sole selective pressure defining bacterial community composition and loss of diversity in this niche.
Discriminatory taxa characteristic of healthy and CRS sinuses
Using the 20-question Sino-Nasal Outcome Test (SNOT-20) survey as a metric to score sinus symptomatology, we confirmed that compared to healthy controls, CRS patients reported significantly higher scores, indicating more severe sinus symptomatology (P ≤ 0.003, Student's t test), and, subsequently, that the observed microbiota clustering patterns were consistent with patient-reported sinus symptomatology (Fig. 1D). Because of this level of independent validation of disease activity, we next characterized at the taxon level (at least 97% 16S rRNA sequence identity) the specific community members that differentiated healthy subjects and CRS patients. Known pathogenic members of the Pseudomonadaceae, Lachnospiraceae, Ralstoniaceae, Mycobacteriaceae, and Helicobacteriaceae were detected in both CRS with and healthy subjects without sinonasal symptoms (table S1). Thus, the mere detection of a suspected or known pathogen in a given niche is not necessarily associated with pathogenicity; these data suggest that the microbiota composition at that site may play a large role in defining the activity of community members. After correction for false discovery (P ≤ 0.05, q ≤ 0.05), a total of 1482 taxa were detected in significantly lower relative abundance in CRS patient sinuses (table S2), which underscores the extent of sinus microbiota collapse in our CRS patient population. A large number of taxa exhibiting the most significant reductions in relative abundance in the CRS patients belonged to the order Lactobacillales; these species included known probiotic species such as Lactobacillus sakei as well as other phylogenetically distinct lactic acid bacteria such as Carnobacterium alterfunditum, Enterococcus mundtii, and Pediococcus pentosaceus. In contrast, only a single taxon with the representative species, Corynebacterium tuberculostearicum, exhibited a significant increase in abundance in CRS patients (P ≤ 0.03, q ≤ 0.003, Welch's t test).
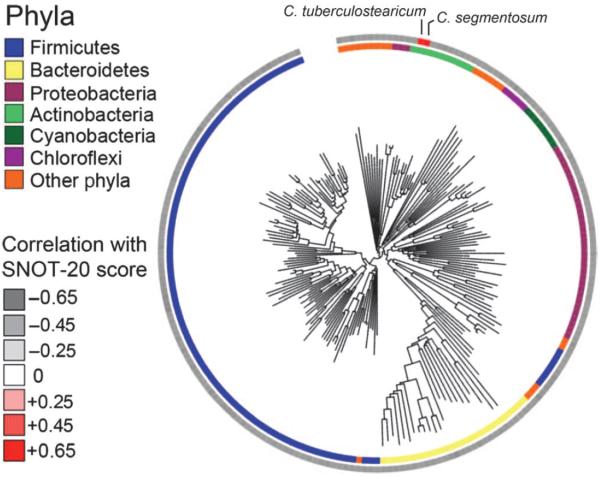
To further underscore the clinical significance of these findings, we examined the microbiota data to identify those species that correlated with SNOT-20 symptom severity scores. Two hundred twenty-eight taxa were significantly (P < 0.05, Welch's t test) correlated with lower SNOT-20 scores (indicative of healthy sinuses; Fig. 2 and table S3). Among these taxa were members of the Lactobacillaceae, Enterococcaceae, Aerococcaceae, and Streptococcaceae, which supports the hypothesis that members of these families represent key protective groups in healthy sinuses. In contrast, the relative abundance of only two taxa was positively correlated with increased symptom severity; both belonged to the Corynebacteriaceae (table S3). Of these two, the taxon most positively correlated with symptom severity was again represented by C. tuberculostearicum (r = 0.62; P ≤ 0.02). To validate these findings, we performed qPCR analysis with primers designed to specifically amplify C. tuberculostearicum. Linear regression of C. tuberculostearicum qPCR-derived copy number against both array-reported fluorescence intensity and SNOT-20 score demonstrated good concordance (r = 0.66; P ≤ 0.01 and r = 0.68; P ≤ 0.01, respectively), corroborating the array-based findings and confirming that a strong relationship between the abundance of this species and sinus symptom severity existed (fig. S2).
Fig. 2.
Phylogenetic tree demonstrating the breadth of bacterial community members exhibiting significant positive or negative correlations with sinus symptom severity (determined by SNOT-20 score).
Murine model of sinusitis
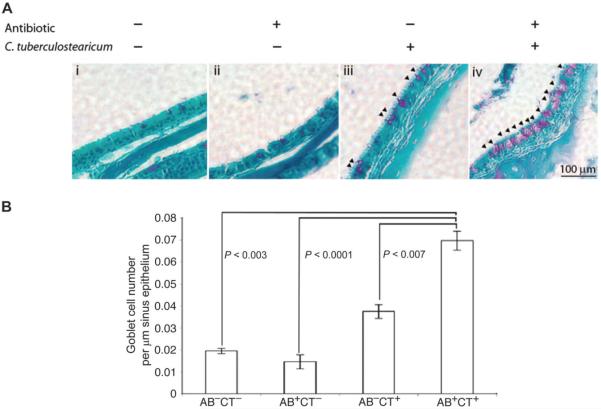
To determine whether C. tuberculostearicum, which is typically considered a skin commensal, exhibited any pathogenic potential and whether this may be influenced by resident microbiota in the sinus cavity, we developed a murine model of sinus infection and used goblet cell hyperplasia and mucin hypersecretion, characteristic indicators of pathology in human populations (13), as the outcome measures to define pathogenic activity in the sinuses. Four groups of mice (n = 5 animals per group) representing (i) untreated control, (ii) antibiotic-treated (to elicit microbiome depletion), (iii) C. tuberculostearicum [American Type Culture Collection (ATCC) 35694]–inoculated, and (iv) antibiotic-treated and C. tuberculostearicum–inoculated animals were used (fig. S3). qPCR analyses of total 16S rRNA copy number from the sinuses of these animals confirmed that the burden of bacteria in the antibiotic-treated groups was significantly lower (P < 0.03, Student's t test) than that of untreated animals, confirming acute antimicrobial depletion of bacterial burden and presumably mucosal sinus microbiota diversity in antibiotic-treated groups of animals. Histological examination of the sinus mucosa from each group of animals demonstrated that the untreated control and the antibiotic-treated animals did not exhibit aberrant epithelial physiology (Fig. 3A, i and ii), whereas instillation of large numbers of C. tuberculostearicum in the presence of a replete sinus microbiota elicited a modest increase in the number of mucin-secreting goblet cells (Fig. 3Aiii). However, animals treated with both an antimicrobial and C. tuberculostearicum exhibited profound goblet cell hyperplasia (Fig. 3, Aiv and B, and table S6) that was significantly greater than that observed in any other group (table S4). Because mucin hypersecretion is a hallmark of respiratory infection (19) and chronic sinusitis (13, 14), thesedata confirm that C. tuberculostearicum is capable of inducing a characteristic host response to pathogenic microbes and that this phenotype is significantly augmented under conditions of depleted sinus microbiota.
Fig. 3.
(A) Depletion of sinus microbiome and instillation of C. tuberculostearicum induce overt sinus pathology. PAS-stained histological sections of murine sinuses representative of animals in each treatment group treated with a combination of antibiotic and C. tuberculostearicum. Triplicate views of maxillary sinuses from two mice per treatment group were used to determine physiology (representative images are shown; panels i to iv). Mice treated with a combination of antibiotic and C. tuberculostearicum exhibit evidence of significantly increased goblet cell hyperplasia (indicated by arrows) and mucin hyper-secretion compared to other treatment groups (panel iii). (B) Enumeration of goblet cells across murine treatment groups (n = 3 animals per group). Values represent means ± SEM.
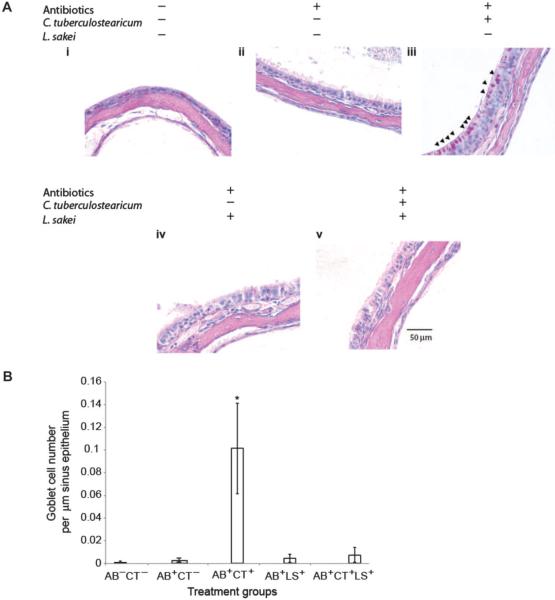
To demonstrate that goblet cell hyperplasia and mucin hypersecretion were induced specifically by C. tuberculostearicum and that this is not simply a host response to instillation of large numbers of any bacterial species into the sinus niche, we repeated the above experiment and included an additional group of animals who were treated with antibiotics before instillation of L. sakei (ATCC 15521; fig. S3). We specifically chose L. sakei because this bacterium was present in high abundance in healthy mucosal samples and was among those taxa exhibiting the most significant depletion in CRS patients. Thus, we postulated that it may represent a protective sinus mucosal colonizer and, as such, should not induce mucosal responses characteristic of infection of this niche.
qPCR was performed on all treatment groups to confirm that animals receiving bacterial inocula exhibited the presence of these species (fig. S4). Histological imaging of the maxillary sinuses (Fig. 4A) demonstrated that the antibiotic-treated and C. tuberculostearicum–inoculated group exhibited significant increases in goblet cell hyperplasia and mucin hypersecretion (Fig. 4Aiii). However, mice that received identical numbers of L. sakei demonstrated epithelial physiology comparable to that of control animals (no significant differences in goblet cell numbers; table S5; (Fig. 4Aiv), thus confirming that the observed sinus histopathology was specifically due to C. tuberculostearicum instillation and that potentially protective species such as L. sakei do not induce this host response. Enumeration of goblet cell numbers in each treatment group confirmed these observations (Fig. 4B and table S5).
Fig. 4.
(A) L. sakei protects sinus mucosa from C. tuberculostearicum–induced pathogenesis. PAS-stained histological sections of murine sinuses representative of animals in each treatment group at ×60 magnification (panels i to v). Triplicate views of maxillary sinuses from two mice per treatment group were used to determine physiology (representative images are shown). (B) Enumeration of goblet cells across murine treatment groups (n = 3 animals per group). Values represent means ± SEM.
Because the collective data generated in this study suggested that L. sakei may represent a putative protective species of sinus mucosa, we also determined whether co-instillation of L. sakei with C. tuberculostearicum could abrogate the goblet cell hyperplasia and mucin hypersecretion phenotype induced by the pathogenic species, even in the context of a depleted native microbiota. After treatment with antibiotics, equal numbers of both species were instilled into the sinuses of mice. Histological examination revealed sinus epithelia comparable to that of animals in the control groups (Fig. 4Av), with no significant differences in goblet cell numbers observed across these groups (Fig. 4B and table S5). qPCR analyses demonstrated significantly (P < 0.02, Student's t test) reduced C. tuberculostearicum abundance in the co-instilled animals when compared to animals infected with C. tuberculostearicum alone (fig. S4); however, L. sakei numbers in these animals were similar to those in animals treated with this species alone (fig. S4). This finding indicates that L. sakei protects the sinus epithelium, putatively through competitive inhibition of C. tuberculostearicum, and may represent a novel therapeutic option for amelioration or prevention of sinus pathology, even in patients with severe sinus microbiome depletion.
DISCUSSION
It is becoming increasingly apparent that human health status is highly dependent on the diverse microbial assemblages that inhabit discrete host niches, particularly mucosal-associated surfaces. Recent advances in culture-independent technologies have increased our ability to describe the diversity of microbial species that colonize specific host sites (20–25), the compositional and functional changes that occur in disease states (26), and the specific community members that are highly correlated with clinical symptom severity (21) or induction of specific immune responses (27). Because of extensive use of conventional laboratory culture approaches to detect microbial species, we have been conditioned to view chronic or acute infections as exclusively due to a single pathogenic species. However, recent studies have demonstrated that the composition of the resident microbiota in a given niche can strongly influence the behavior of specific species, particularly pathogens (10, 11, 28), and, as such, represents an important contributory factor to disease etiology.
To date, culture-based approaches to characterize the etiological agent of CRS have provided a reductionist and somewhat discordant view of the microbiology associated with this disease. In an attempt to better define this aspect, we examined whether sinus microbiota composition was altered with this chronic inflammatory disease. Despite our relatively small number of subjects, a clear signal emerged, demonstrating that the sinus microbiota of our patient cohort was characterized by both grossly depleted communities and a significant increase in relative abundance of a single organism, C. tuberculostearicum. Although phylogenetically its closest bacterial relatives include Mycobacteria and Nocardia, genera synonymous with pathogenesis, C. tuberculostearicum is customarily considered an innocuous member of the healthy skin microbiota and an unlikely etiological agent of CRS. Therefore, we developed a murine model of sinusitis to recreate the observed microbiological features of CRS and confirmed both the pathogenic potential of this species and that its impact on sinus epithelial responses, associated with pathogenesis, was significantly enhanced in the absence of a replete sinus microbiota.
The critical role of the microbiome in modulating the impact of C. tuberculostearicum on sinus mucosal responses likely explains why it (and putatively multiple other microbial species) has not previously been considered a pathogen. The observed necessary combination of microbiota depletion and C. tuberculostearicum outgrowth to elicit a pathophenotype confirms that microbiome composition plays a key role in defining the behavior of specific species in the consortium. Several lines of evidence in the recent literature support this. For example, perturbation of the gastrointestinal microbiota of mice results in proliferation of C. difficile and induces a supershedder phenotype (10), demonstrating the role a diverse microbiota plays in suppressing outgrowth of specific pathogenic species. This phenomenon may also explain why, despite detection of known pathogens in healthy sinus microbiota, these individuals exhibit no symptomatology and, more broadly, provide an explanation as to why seemingly similar patients, exhibiting comparable quantities of known pathogenic species, may exhibit markedly different clinical outcomes. These studies highlight the critical role microbiome composition plays in defining microbial behavior and underscore the need to better understand the complex microbe-microbiome-host interplay that governs microbial-host responses and undoubtedly defines disparate clinical outcomes in patient populations.
One limitation of this study is the small sample size and the inclusion of only surgical chronic sinusitis patients with severe disease. It is entirely plausible that subgroups of sinusitis patients with distinct pathophenotypes because of overgrowth of other microbial species such as fungal pathogens exist. Nonetheless, we believe that a depleted mucosal microbiome forms the universal context for proliferation of such species. These observations have obvious implications for other sinonasal pathogens such as Staphylococcus aureus as well as for inflammatory diseases in distinct niches in which gross perturbations to the microbiota have been described, such as inflammatory bowel disease (29), necrotizing enterocolitis (30), and asthma (31, 32).
One advantage to this comparative study design is the ability to identify those species associated with healthy sinuses that, presumably, provide mucosal protection. The murine model developed in this study provided an opportunity to determine whether such species may afford protection against the pathophysiology induced by a combination of depleted microbiota and C. tuberculostearicum. From these efforts, it is clear that L. sakei, a probiotic species that has demonstrated efficacy in human studies of children with atopic eczema-dermatitis syndrome (33), represents a potentially novel therapeutic for the treatment of sinusitis subtypes, including CRS. The observation that several members of the Lactobacillaceae, including L. sakei, as well as other known lactic acid–producing members of the Firmicutes, were significantly depleted suggests that characteristic physiological features of these species, such as bacteriocin or lactic acid production (34, 35), may serve to outcompete pathogenic species. This competition may plausibly shape the sinus mucosal microbiota and protect this niche from pathogen overgrowth, although these hypotheses require further study.
Because of the crucial role microbiome composition plays in CRS, it is tempting to speculate that key events that disturb the sinus microbial ecosystem such as viral infection may serve to initiate disease. Indeed, viral infection commonly precedes development of CRS (36) and is implicated in the genesis of other chronic inflammatory diseases (37–39). Our findings also have significant implications for clinical practice, particularly for the excessive use of antimicrobials, which contribute to microbiota depletion, in the treatment of viral sinusitis and other upper respiratory infections. Our data suggest that a more appropriate strategy may involve microbial supplementation during periods of acute sinusitis with one or a combination of species identified in this study. Certainly, restoration ecology has proven effective and received much media attention in other host niches such as the lower gastrointestinal tract where fecal transplant has been shown to resolve recalcitrant C. difficile infection with a high rate of success (40). Although further studies are necessary to determine the efficacy of using a multispecies sinus supplementation approach, there is precedence for this concept. A double-blind placebo-controlled trial of more than 300 children demonstrated significant decreases in fever incidence, coughing, and rhinorrhea in probiotic-supplemented children relative to the control group. This impact was even more significant in those children receiving a combination of probiotic strains (Lactobacillus and Bifidobacterium species), thus providing strong supporting evidence for the protective effect of combinations of such species against upper respiratory pathogens (41).
Demonstration that a bacterial species that commonly inhabits human skin is an etiological agent of CRS invites a change in how we perceive pathogens. Our data suggest that apparently commensal species in one setting may produce disease in another, based at least in part on the local microbial composition. The study also illustrates why the etiology of CRS, and likely other chronic inflammatory diseases, has been so difficult to define. CRS pathophysiology is multifactorial and, at least in our patient population, co-dependent on microbiota composition and increased relative abundance of C. tuberculostearicum. Simply investigating either aspect in isolation is insufficient. However, the advanced technologies we now have for profiling microbiota composition and function provide us with an unprecedented opportunity to consider microbiota composition as a contributor to disease state, to identify novel etiological agents, and to better determine the congruence of mechanisms underlying chronic inflammatory diseases.
MATERIALS AND METHODS
Patient sample collection
Patient and disease stratification was performed on the basis of recent clinical history, nasal endoscopy, computed tomography sinus review, and a validated quality-of-life instrument, the disease-specific SNOT-20 survey (42). Sinus brushings were obtained during functional endoscopic sinus surgery of CRS patients or surgery for non-CRS complaints (for example, obstructive sleep apnea or post-traumatic malocclusion) in healthy subjects. Endoscopically guided brush samples of mucosal surfaces of the lateral, central, and medial portions of the maxillary sinus were obtained and pooled together in 1 ml of RNAlater and placed at 4°C for 24 hours before storage at −80°C until processed for analysis.
DNA extraction and PhyloChip analysis
Mucosal brushings were transferred to Lysis Matrix B (MP Biomedicals) tubes containing 600 μl of RLT buffer (Qiagen). Samples were subjected to 30 s of bead beating at 5.5 m/s followed by centrifugation for 1 min at 2000 rpm. Supernatant was transferred to the AllPrep DNA spin column, and nucleic acid purification (DNA and RNA) was carried out according to the manufacturer's instructions (Qiagen) as previously described (43). Nucleic acid concentrations were determined with a NanoDrop spectrophotometer (Thermo Scientific). PhyloChip analysis was performed as previously described (44) with 250 ng of purified pooled 16S rRNA amplicon per sample generated using 27F and 1492R universal primers (45). Data sets were conservatively filtered (44), and probe set fluorescence intensity was normalized and log-transformed before analysis with packages in the R statistical environment. HCA was performed on Canberra dissimilarities calculated with the `vegdist' function in the `vegan' R package. Canberra dissimilarities have been demonstrated to perform well in cluster detection (46). HCA of the Canberra dissimilarity matrix was performed with averaging linkage, and significant clusters were determined with approximately unbiased P values and bootstrap values by a multiscale bootstrap resampling approach implemented in the R package pvclust (47). The 16S rRNA sequences of significant taxa were used to construct a neighbor-joining with nearest-neighbor interchange tree with FastTree (48), which was annotated with the Interactive Tree of Life [http://itol.embl.de/; (49)].
Murine sinus studies
Female 4- to 5-week-old C57BL6/J mice weighing 16 to 18 g were purchased from The Jackson Laboratory and housed in microisolator cages in the Parnassus Services Building at the University of California, San Francisco, Parnassus Campus. All protocols performed were approved by the Animal Care and Use Committee of the University of California, San Francisco. Animals were permitted to acclimatize for 2 weeks with food and water ad libitum before study. Mice were orally administered amoxicillin/clavulanate at a dose of 100 mg/kg once a day for 5 days before bacterial instillation. Before intranasal inoculation with C. tuberculostearicum (ATCC 35694), L. sakei (ATCC 15521), or combination of both species, mice were anesthetized by intraperitoneal injection with avertin (250 mg/kg). ATCC strains were used to confirm that the phenotypes elicited by these species were not exclusive to clinical isolates of these organisms. Inoculation was performed once a day for 3 days by applying 25 μl of either C. tuberculostearicum [1.0 × 1011 colony-forming units (CFU)/ml] or L. sakei (1.0 × 1011 CFU/ml), or combination of both microbes in equal ratios (total 2.0 × 1011 CFU/ml), in phosphate-buffered saline (PBS) onto the external nares and inhalation by the animals. This concentration was based on previously described murine sinusitis studies (50, 51). Mice were monitored during the entire process until fully recovered from anesthesia. Twenty-four hours after the final bacterial inoculation, mice were euthanized by CO2 asphyxiation followed by induced pneumothorax. The heads were decapitated, and sinuses were dissected for histological (n = 2 animals per treatment group) and molecular analysis (n = 3 animals per treatment group). Dissected sinuses used for molecular analyses were placed in RNAlater (Ambion) and stored at 4°C until processed the following day in the same manner as described above for human samples.
Sinus histology
Sinuses were fixed overnight in 4% paraformaldehyde, followed by overnight decalcification in Decalcifying Solution A (Fisher). Samples were then dehydrated, rinsed with PBS (1 hour), soaked in 30% ethanol (1 hour) and 50% ethanol (1 hour), transferred to 70% ethanol, and stored at 4°C until subsequent preparation. Samples were sectioned to 5-μm thickness and mounted onto glass microscope slides. Hematoxylin and eosin and PAS staining were performed as previously described (52). PAS-stained sections prepared from four groups of mice (CT−AB−, CT−AB+, CT+AB−, and CT+AB−) were imaged at ×20 and ×60 magnifications, and PAS-positive cells were counted for three different sections per mice (two mice in each group). The number of goblet cells was expressed as the total number of PAS-positive cells per micrometer length of epithelium.
Quantitative polymerase chain reaction
Bacterial burden was determined with extracted DNA (10 ng per sample, triplicate reactions) and universal 16S rRNA qPCR primers [338F, 5′-ACTCCTACGGGAGGCAGCAG-3′ (45); 518R, 5′-ATTACCGCGGCTGCTGG-3′; (53)]. QuantiTect SYBR Green (Qiagen) was used according to the manufacturer's instructions. Reaction mixtures (25 μl total) contained 12.5 μl of 2× QuantiTect SYBR Green (Qiagen), 2.5 μl each of 3 μM forward and reverse primers, and 6.5 μl of H2O. Reactions were amplified with the Mx3000P Real-Time PCR System (Stratagene) and the following cycling conditions: 95°C for 10 min followed by 40 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 30 s with data acquisition at 55°C. C. tuberculostearicum abundance was determined by qPCR with CT-F (5′-GAACGGAAAGGCCCTGCTTGCA-3′) and CT-R(5′-GGCTCCTATCCGGTATTAGACC-3′); L. sakei abundance was determined with the primer pair LS-F (5′-GGTAAAGGCTCACCAAGACCGTGAT-3′) and LS-R (5′-TCACGCGGCGTTGCTCCATC-3′). Reaction mixtures and PCR conditions were as described above.
For Muc5A expression analysis, confirmed DNA-free total RNA (1 μg) was reverse-transcribed at 42°C for 50 min in a 20-μl reaction mixture with 1 μl of SuperScript II (Invitrogen). Complementary DNA (cDNA) was diluted 1:5 in molecular-grade water. qPCRs were performed in a 25-μl final volume with 12.5 μl of SYBR Green PCR master mix; 10 μM each of primers Muc5AF (5′-TGTGGCGGGAAAGACAGC-3′), Muc5AR (5′-CCTTCCTATGGCTTAGCTTCAGC-3′), β-actinF (5′-CACCACACCTTCTACAATGAGCTGC-3′), and β-actinR (5′-ACACCCTGGATAGCAACGTACATGC-3′); 4 μl of diluted cDNA; and 6 μl of molecular-grade water. Reactions were amplified under the following conditions: 94°C for 10 min followed by 40 cycles of 94°C for 30 s, 58°C for 1 min, and 72°C for 30 s.
Supplementary Material
Acknowledgments
We thank the patients who provided samples for this study, H. Boushey for initiating this collaboration, and his and K. Norman's critical assessment of the manuscript. We are grateful for microbiome analyses assistance provided by E. Brodie, M. Cox, and K. Fujimura; technical assistance from D. Mandal; and sample collection by W. Ma.
Funding: This study was supported by an American Rhinological Society grant awarded to F.C.R. and S.V.L. S.V.L. is also funded by the Rainin Foundation and through a National Institute of Allergy and Infectious Disease award AI097172. N.A.N. is also funded through a National Institute of Allergy and Infectious Diseases award AI097172. N.A.A. is funded through the Minority Biomedical Research Support–Research Initiative for Scientific Enhancement grant 5R25-GM059298 and California Institute for Regenerative Medicine grant TB1-01194 at San Francisco State University, and A.N.G. and W. Ma are funded, in part, by the Rebecca Susan Buffett Foundation.
Footnotes
SUPPLEMENTARY MATERIALS www.sciencetranslationalmedicine.org/cgi/content/full/4/151/151ra124/DC1
Author contributions: N.A.A., Y.S., and N.A.N. contributed to experimental design, performed the experiments, and contributed to manuscript preparation. F.C.R., S.D.P., and A.N.G. provided clinical samples and contributed to human study design and manuscript preparation. S.V.L. designed the study, conceived the experiments, and prepared the manuscript.
Competing interests: S.V.L. is a member of the Scientific Advisory Board of Second Genome. S.V.L., A.N.G., and S.D.P. are authors on patent application no. 61/624,105, “Sinusitis diagnostics and treatments.” None of the other authors declare any competing interests.
Data and materials availability: PhyloChip data used in this study are available at the Gene Expression Omnibus Web site (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE37850.
REFERENCES AND NOTES
- 1.Ray NF, Baraniuk JN, Thamer M, Rinehart CS, Gergen PJ, Kaliner M, Josephs S, Pung YH. Healthcare expenditures for sinusitis in 1996: Contributions of asthma, rhinitis, and other airway disorders. J. Allergy Clin. Immunol. 1999;103:408–414. doi: 10.1016/s0091-6749(99)70464-1. [DOI] [PubMed] [Google Scholar]
- 2.Report of the Rhinosinusitis Task Force Committee Meeting Alexandria, Virginia, August 17, 1996. Otolaryngol. Head Neck Surg. 1997;117:S1–S68. [PubMed] [Google Scholar]
- 3.Wallace DV, Dykewicz MS, Bernstein DI, Blessing-Moore J, Cox L, Khan DA, Lang DM, Nicklas RA, Oppenheimer J, Portnoy JM, Randolph CC, Schuller D, Spector SL, Tilles SA. Joint Task Force on Practice; American Academy of Allergy; Asthma & Immunology; American College of Allergy; Asthma and Immunology; Joint Council of Allergy, Asthma and Immunology, The diagnosis and management of rhinitis: An updated practice parameter. J. Allergy Clin. Immunol. 2008;122:S1–S84. doi: 10.1016/j.jaci.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Biel MA, Brown CA, Levinson RM, Garvis GE, Paisner HM, Sigel ME, Tedford TM. Evaluation of the microbiology of chronic maxillary sinusitis. Ann. Otol. Rhinol. Laryngol. 1998;107:942–945. doi: 10.1177/000348949810701107. [DOI] [PubMed] [Google Scholar]
- 5.Brook I, Frazier EH, Gher ME., Jr. Microbiology of periapical abscesses and associated maxillary sinusitis. J. Periodontol. 1996;67:608–610. doi: 10.1902/jop.1996.67.6.608. [DOI] [PubMed] [Google Scholar]
- 6.Stephenson MF, Mfuna L, Dowd SE, Wolcott RD, Barbeau J, Poisson M, James G, Desrosiers M. Molecular characterization of the polymicrobial flora in chronic rhinosinusitis. J. Otolaryngol. Head Neck Surg. 2010;39:182–187. [PubMed] [Google Scholar]
- 7.Doyle PW, Woodham JD. Evaluation of the microbiology of chronic ethmoid sinusitis. J. Clin. Microbiol. 1991;29:2396–2400. doi: 10.1128/jcm.29.11.2396-2400.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang RS, Lin JF, Hsu CY. Correlation between bacteriology of the middle meatus and ethmoid sinus in chronic sinusitis. J. Laryngol. Otol. 2002;116:443–446. doi: 10.1258/0022215021911248. [DOI] [PubMed] [Google Scholar]
- 9.Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio. 2010;1:e00129–10. doi: 10.1128/mBio.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, Mastroeni P, Scott P, Raisen C, Mottram L, Fairweather NF, Wren BW, Parkhill J, Dougan G. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 2009;77:3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim DH, Chu HS, Lee JY, Hwang SJ, Lee SH, Lee HM. Up-regulation of MUC5AC and MUC5B mucin genes in chronic rhinosinusitis. Arch. Otolaryngol. Head Neck Surg. 2004;130:747–752. doi: 10.1001/archotol.130.6.747. [DOI] [PubMed] [Google Scholar]
- 14.Baraniuk JN, Petrie KN, Le U, Tai CF, Park YJ, Yuta A, Ali M, Vandenbussche CJ, Nelson B. Neuropathology in rhinosinusitis. Am. J. Respir. Crit. Care Med. 2005;171:5–11. doi: 10.1164/rccm.200403-357OC. [DOI] [PubMed] [Google Scholar]
- 15.Aust MR, Madsen CS, Jennings A, Kasperbauer JL, Gendler SJ. Mucin mRNA expression in normal and vasomotor inferior turbinates. Am. J. Rhinol. 1997;11:293–302. doi: 10.2500/105065897781446685. [DOI] [PubMed] [Google Scholar]
- 16.Jung HH, Lee JH, Kim YT, Lee SD, Park JH. Expression of mucin genes in chronic ethmoiditis. Am. J. Rhinol. 2000;14:163–170. doi: 10.2500/105065800782102690. [DOI] [PubMed] [Google Scholar]
- 17.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, MetaHIT Consortium. Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M'rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honda K. Porphyromonas gingivalis sinks teeth into the oral microbiota and periodontal disease. Cell Host Microbe. 2011;10:423–425. doi: 10.1016/j.chom.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Yuta A, Ali M, Sabol M, Gaumond E, Baraniuk JN. Mucoglycoprotein hypersecretion in allergic rhinitis and cystic fibrosis. Am. J. Physiol. 1997;273:L1203–L1207. doi: 10.1152/ajplung.1997.273.6.L1203. [DOI] [PubMed] [Google Scholar]
- 20.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujimura KE, Johnson CC, Ownby DR, Cox MJ, Brodie EL, Havstad SL, Zoratti EM, Woodcroft KJ, Bobbitt KR, Wegienka G, Boushey HA, Lynch SV. Man's best friend? The effect of pet ownership on house dust microbial communities. J. Allergy Clin. Immunol. 2010;126:410–412. doi: 10.1016/j.jaci.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang YJ, Kim E, Cox MJ, Brodie EL, Brown R, Wiener-Kronish JP, Lynch SV. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. OMICS. 2010;14:9–59. doi: 10.1089/omi.2009.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox MJ, Huang YJ, Fujimura KE, Liu JT, McKean M, Boushey HA, Segal MR, Brodie EL, Cabana MD, Lynch SV. Lactobacillus casei abundance is associated with profound shifts in the infant gut microbiome. PLoS One. 2010;5:e8745. doi: 10.1371/journal.pone.0008745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, Nelson KE, Gill SR, Fraser-Liggett CM, Relman DA. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R, Gordon JI. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanov II, Atarashi K, Manel N, Brodie L, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan K, Dammel C, Stein J, Rabin H, Surette MG. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 2003;50:1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 29.Nishikawa J, Kudo T, Sakata S, Benno Y, Sugiyama T. Diversity of mucosa-associated microbiota in active and inactive ulcerative colitis. Scand. J. Gastroenterol. 2009;44:180–186. doi: 10.1080/00365520802433231. [DOI] [PubMed] [Google Scholar]
- 30.Mai V, Young CM, Ukhanova M, Wang X, Sun Y, Casella G, Theriaque D, Li N, Sharma R, Hudak M, Neu J. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLoS One. 2011;6:e20647. doi: 10.1371/journal.pone.0020647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, Moffatt MF, Cookson WO. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, Woyke T, Allgaier M, Bristow J, Wiener-Kronish JP, Sutherland ER, King TS, Icitovic N, Martin RJ, Calhoun WJ, Castro M, Denlinger LC, Dimango E, Kraft M, Peters SP, Wasserman SI, Wechsler ME, Boushey HA, Lynch SV. National Heart, Lung, and Blood Institute's Asthma Clinical Research Network, Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J. Allergy Clin. Immunol. 2011;127:372–381.e3. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo SI, Kim JY, Lee YJ, Kim NS, Hahn YS. Effect of Lactobacillus sakei supplementation in children with atopic eczema–dermatitis syndrome. Ann. Allergy Asthma Immunol. 2010;104:343–348. doi: 10.1016/j.anai.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 34.Corr SC, Li Y, Riedel CU, O'Toole PW, Hill C, Gahan CG. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coffey A, Ryan M, Ross RP, Hill C, Arendt E, Schwarz G. Use of a broad-host-range bacteriocin-producing Lactococcus lactis transconjugant as an alternative starter for salami manufacture. Int. J. Food Microbiol. 1998;43:231–235. doi: 10.1016/s0168-1605(98)00115-9. [DOI] [PubMed] [Google Scholar]
- 36.Das S, Palmer OP, Leight WD, Surowitz JB, Pickles RJ, Randell SH, Buchman CA. Cytokine amplification by respiratory syncytial virus infection in human nasal epithelial cells. Laryngoscope. 2005;115:764–768. doi: 10.1097/01.MLG.0000159527.76949.93. [DOI] [PubMed] [Google Scholar]
- 37.Sankaran-Walters S, Ransibrahmanakul K, Grishina I, Hung J, Martinez E, Prindiville T, Dandekar S. Epstein–Barr virus replication linked to B cell proliferation in inflamed areas of colonic mucosa of patients with inflammatory bowel disease. J. Clin. Virol. 2011;50:31–36. doi: 10.1016/j.jcv.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou JK, Velayos F, Terrault N, Mahadevan U. Viral hepatitis and inflammatory bowel disease. Inflamm. Bowel Dis. 2010;16:925–932. doi: 10.1002/ibd.21284. [DOI] [PubMed] [Google Scholar]
- 39.Verdonk RC, Haagsma EB, Kleibeuker JH, Dijkstra G, Sudan DL. Cytomegalovirus infection increases the risk for inflammatory bowel disease. Am. J. Pathol. 2010;176:3098. doi: 10.2353/ajpath.2010.100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garborg K, Waagsbo B, Stallemo A, Matre J, Sundøy A. Results of faecal donor instillation therapy for recurrent Clostridium difficile-associated diarrhoea. Scand. J. Infect. Dis. 2010;42:857–861. doi: 10.3109/00365548.2010.499541. [DOI] [PubMed] [Google Scholar]
- 41.Leyer GJ, Li S, Mubasher ME, Reifer C, Ouwehand AC. Probiotic effects on cold and influenza-like symptom incidence and duration in children. Pediatrics. 2009;124:e172–e179. doi: 10.1542/peds.2008-2666. [DOI] [PubMed] [Google Scholar]
- 42.Piccirillo JF, Merritt MG, Jr., Richards ML. Psychometric and clinimetric validity of the 20-Item Sino-Nasal Outcome Test (SNOT-20) Otolaryngol. Head Neck Surg. 2002;126:41–47. doi: 10.1067/mhn.2002.121022. [DOI] [PubMed] [Google Scholar]
- 43.Roediger FC, Slusher NA, Allgaier S, Cox MJ, Pletcher SD, Goldberg AN, Lynch SV. Nucleic acid extraction efficiency and bacterial recovery from maxillary sinus mucosal samples obtained by brushing or biopsy. Am. J. Rhinol. Allergy. 2010;24:263–265. doi: 10.2500/ajra.2010.24.3472. [DOI] [PubMed] [Google Scholar]
- 44.Brodie EL, Desantis TZ, Joyner DC, Baek SM, Larsen JT, Andersen GL, Hazen TC, Richardson PM, Herman DJ, Tokunaga TK, Wan JM, Firestone MK. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 2006;72:6288–6298. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lane D. In: 16S/23S rRNA sequencing, in Nucleic Acid Techniques in Bacterial Systematics. Stackebrandt E, Goodfellow M, editors. John Wiley & Sons; New York: 1991. [Google Scholar]
- 46.Kuczynski J, Liu Z, Lozupone C, McDonald D, Fierer N, Knight R. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat. Methods. 2010;7:813–819. doi: 10.1038/nmeth.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki R, Shimodaira H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 48.Price MN, Dehal PS, Arkin AP. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Letunic I, Bork P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 50.Bomer K, Brichta A, Baroody F, Boonlayangoor S, Li X, Naclerio RM. A mouse model of acute bacterial rhinosinusitis. Arch. Otolaryngol. Head Neck Surg. 1998;124:1227–1232. doi: 10.1001/archotol.124.11.1227. [DOI] [PubMed] [Google Scholar]
- 51.Blair C, Naclerio RM, Yu X, Thompson K, Sperling A. Role of type 1 T helper cells in the resolution of acute Streptococcus pneumoniae sinusitis: A mouse model. J. Infect. Dis. 2005;192:1237–1244. doi: 10.1086/444544. [DOI] [PubMed] [Google Scholar]
- 52.Ahn MH, Kang CM, Park CS, Park SJ, Rhim T, Yoon PO, Chang HS, Kim SH, Kyono H, Kim KC. Titanium dioxide particle-induced goblet cell hyperplasia: Association with mast cells and IL-13. Respir. Res. 2005;6:34. doi: 10.1186/1465-9921-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.