Key Points
Patients with e13a2 transcripts have inferior outcomes with imatinib 400; e14a2 has favorable outcomes regardless of treatment modality.
Multivariate analysis showed that the expression of e14a2 or both e14a2 and e13a2 predicts optimal ELN responses and longer EFS and TFS.
Abstract
The most common breakpoint cluster region gene-Abelson murine leukemia viral oncogene homolog 1 (BCR-ABL) transcripts in chronic myeloid leukemia (CML) are e13a2 (b2a2) and e14a2 (b3a2). The impact of the type of transcript on response and survival after initial treatment with different tyrosine kinase inhibitors is unknown. This study involved 481 patients with chronic phase CML expressing various BCR-ABL transcripts. Two hundred patients expressed e13a2 (42%), 196 (41%) expressed e14a2, and 85 (18%) expressed both transcripts. The proportion of patients with e13a2, e14a2, and both achieving complete cytogenetic response at 3 and 6 months was 59%, 67%, and 63% and 73%, 81%, and 82%, respectively, whereas major molecular response rates were 27%, 49%, and 50% at 3 months, 42%, 67%, and 70% at 6 months, and 55%, 83%, and 76% at 12 months, respectively. Median (international scale) levels of transcripts e13a2, e14a2, and both at 3 months were 0.2004, 0.056, and 0.0612 and at 6 months were 0.091, 0.0109, and 0.0130, respectively. In multivariate analysis, e14a2 and both predicted for optimal responses at 3, 6, and 12 months. The type of transcript also predicted for improved probability of event-free (P = .043; e14a2) and transformation-free survival (P = .04 for both). Compared to e13a2 transcripts, patients with e14a2 (alone or with coexpressed e13a2) achieved earlier and deeper responses, predicted for optimal European Leukemia Net (ELN) responses (at 3, 6, and 12 months) and predicted for longer event-free and transformation-free survival.
Introduction
The Philadelphia (Ph) chromosome resulting from the balanced reciprocal translocation between chromosomes 9 and 22 t(9;22)(q34;q11.2) is the cytogenetic hallmark of chronic myeloid leukemia (CML).1-3 This balanced reciprocal translocation results in the formation of the BCR-ABL1 oncogene, which is translated into a protein with constitutive tyrosine kinase activity, possibly the most effectively therapeutically targeted oncoprotein.4,5 The breakpoints in the BCR gene on chromosome 22 most commonly occur between exons e12 (b2) and e13 (b3) or between e13 (b3) and e14 (b4), in the major breakpoint cluster region (M-BCR), generating 2 slightly different chimeric transcripts.6-8 The breakpoint in the ABL1 gene is usually located between exons a1 and a2. These breakpoints result in various BCR-ABL rearrangements, most commonly the e13a2 (b2a2) and e14a2 (b3a2), which code for a 210-kDa protein: p210. In some patients, both transcripts can be coexpressed: e13a2 (b2a2) with e14a2 (b3a2). Less frequently, the break in BCR occurs between exons 1 and 2, generating the e1a2 transcript, which codes for a 190-kDa protein, or between exons 19 and 20, generating the e19a2 transcript that codes for a 230-kDa protein.6,9-13 More rarely, other variants such as e14a3 (b3a3)14 and e8a2 transcripts15 are described.
The prognostic significance of the BCR-ABL1 transcripts16,17 has been reported from patients treated with interferon alone18 or imatinib 400 mg.19 Improved response has been reported in patients carrying the e14a2 (b3a2) transcript compared with those with the e13a2 (b2a2) transcripts after treatment with standard-dose imatinib.14,19-24 This observation correlates with higher activity of phospho CrKL (CT10 regulator of kinase-like) a surrogate marker of BCR-ABL1 tyrosine kinase activity in patients with e13a2 transcripts.25 Secondary structure elements are different in e14a2 due to the presence of extra 25 amino acids not seen in e13a2 transcripts, possibly indicating that e14a2 and e13a2 transcripts may have different roles in mediating signal transduction pathways in CML.26
Our group has previously reported higher rates of molecular response and a better trend for transformation-free survival (TFS) for patients treated with imatinib who presented with e14a2 transcripts.24 The second-generation tyrosine kinase inhibitors (2GTKIs) dasatinib and nilotinib have improved the responses in patients when used either as front-line therapy or as second-line treatment after imatinib failure.27-29 To our knowledge, none of the previously published studies have systematically analyzed the responses and survival outcomes in patients treated with imatinib 400, imatinib 800, and 2GTKI as initial therapy for CML according to the type of BCR-ABL1 transcripts.
In this analysis, we evaluated the prognostic relevance of commonly expressed BCR-ABL1 transcripts in patients with chronic phase CML treated with 4 different front-line TKI modalities. The objective of this analysis was to determine the prognostic significance of transcript types across patients with CML–chronic phase treated with different TKI modalities.
Patients and methods
Patients
All patients with chronic phase CML enrolled in consecutive or parallel clinical trials at The MD Anderson Cancer Center using TKI as front-line therapy from July 31, 2000 to September 10, 2013 were included in this analysis. Patients were treated on protocols approved by the institutional review board, and informed consent was obtained in accordance with the Declaration of Helsinki. Eligibility criteria, follow-up, and response assessment were similar for all trials: cytogenetic analysis every 3 months for the first year, then every 6 months for the next 2 to 3 years, and then every 1 to 2 years. Real-time polymerase chain reaction was generally assessed every 3 months for the first year and then every 6 months. Response criteria were as previously described.30 Only patients with BCR-ABL transcripts e13a2 (previously b2a2), e14a2 (previously b3a2), and coexpressed e13a2 (b2a2) with e14a2 (b3a2) were included in the analyses. Demographic and baseline disease characteristics were collected at baseline. The differences between variables were analyzed by the χ2 test and the Kruskal-Wallis test for categorical and continuous variables. Of note, all patients treated with imatinib 400 initiated therapy between May 2001 and June 2001 when molecular analysis was not routinely done before achievement of complete cytogenetic response (CCyR); therefore, molecular response at 3 months is not available for the imatinib 400 cohort.
Statistical analysis
Event-free survival (EFS) was measured from the start of treatment to the date of any of the following events (as defined in the International Randomized Study of Interferon and STI571)31 while on therapy: loss of complete hematologic remission, loss of major cytogenetic response (MCyR), progression to accelerated (defined as blasts ≥15%, blasts + promyelocytes ≥30%, basophils ≥20%, platelets <100 × 109/L, unrelated to therapy, or cytogenetic clonal evolution) or blast phase (defined as blasts ≥30%, or extramedullary disease), or death from any cause at any time while on study. Overall survival (OS) was measured from the time treatment was started to the date of death from any cause at any time or date of last follow-up. TFS was measured from the start of therapy to the date of transformation to accelerated or blast phase while on therapy or deaths on study (ie, deaths on initial TKI).
Survival probabilities were estimated by the Kaplan-Meier method and compared by the log-rank test. Univariate and multivariate analyses were performed to identify whether the type of transcript can predict for cytogenetic and molecular responses at different time points and/or survival outcomes. Variables with P ≤ .10 in the univariate analysis were entered into a multivariate model and analyzed using the Cox proportional hazard regression. P < .05 was considered significant. Survival end points were analyzed using the Kaplan-Meier method, and differences were calculated by the log-rank test. Statistical analyses were carried out using STATA/SE version 13.1 statistical software (Stata Corp. LP, College Station, TX).
Results
Patients
Overall, 487 patients with previously untreated chronic phase CML were treated with different TKI modalities, of which 481 patients expressing e13a2, e14a2, or coexpression of e13a2 with e14a2 transcripts (both) were included in this analysis. Six patients with variant transcripts (e1a2, n = 4; b3a3, n = 2) were excluded from this analysis. Patients were treated with 4 different frontline TKI modalities: imatinib 400 mg daily (n = 69), imatinib 800 mg daily (n = 199), dasatinib 50 mg twice daily or 100 mg daily (n = 105), or nilotinib 400 mg twice daily (n = 108). The baseline characteristics of the patients are summarized in Table 1. Two hundred patients (42%) expressed e13a2, 196 (41%) expressed e14a2, and 85 (18%) expressed both transcripts. Patients with e13a2 had significantly lower platelets (median, 288 K/µL; range, 15-1906 K/µL) compared with e14a2 or both (median, 405 K/µL; range 77-1476 K/µL; and 358 K/µL; range, 100-2928 K/µL, respectively; P < .001). Patient characteristics were also comparable for patients treated with the different TKI modalities. Median follow-up was longer for patients treated with imatinib than with 2GTKI. Disease transformation occurred in 21 patients (4%) (blast phase, n = 7; accelerated phase, n = 14), and 14 (3%) patients died on study. Of the 21 patients with transformation to accelerated or blast phase, 15 expressed e13a2 (representing 8% of all patients with e13a2), 6 patients expressed e14a2 (3% of all e14a2 patients), and none coexpressed both transcripts (P = .04). The distribution of patients who transformed to accelerated or blast phase (n = 21) by Sokal risk categories were as follows: low risk, 13 (62%); intermediate, 6 (28%); high risk, 2 (9%). A total of 52 (11%) patients died (n = 26 for e13a2, n = 20 for e14a2, and n = 6 for both), including the 14 who died while on study. Events occurred in 77 (16%) patients.
Table 1.
Patient characteristics according to the type of BCR-ABL transcript
| Characteristic | No. (%),or median [interquartile range] | P | ||
|---|---|---|---|---|
| e13a2 | e14a2 | e13a2 and e14a2 | ||
| N | 200 (42) | 196 (41) | 85 (18) | |
| Age, years [range] | 47 [37-57] | 49 [39-58] | 52 [41-61] | .11 |
| Hemoglobin (g/dL) | 12 (11-13) | 12 (11-13) | 13 (12-14) | .32 |
| WBC (K/µL) | 31 (12-71) | 28 (17-63) | 30 (13-70) | .94 |
| Platelet (K/µL) | 288 (200-418) | 405 (276-592) | 358 (268-493) | .001 |
| Sokal score | ||||
| High | 11 (5) | 11 (6) | 9 (11) | .53 |
| Intermediate | 47 (23) | 48 (24) | 21 (25) | |
| Low | 142 (71) | 137 (70) | 55 (65) | |
| Treatment type | ||||
| Imatinib 400 mg | 31 (15) | 28 (14) | 10 (12) | .32 |
| Imatinib 800 mg | 82 (41) | 89 (45) | 28 (33) | |
| Dasatinib | 41 (20) | 38 (19) | 26 (31) | |
| Nilotinib | 46 (23) | 41 (21) | 21 (25) | |
| Median follow-up (months) | 88 [43-122] | 98 [49-127] | 84 [46-115] | .18 |
Cytogenetic and molecular responses according to transcript type
Only patients with values available at the time of assessment were included in this analysis. Absolute numbers of evaluable patients according to type of transcript are shown in Figure 1. Cumulative response rates according to the transcript type were as follows: CCyR, e13a2 (89%), e14a2 (94%), and both (94%), P = not significant; MMR, e13a2 (79%), e14a2 (91%), and both (95%), P = .0001; and MR4.5, e13a2 (57%), e14a2 (79%), and both (80%), P = .00001 (supplemental Figure 1A, available on the Blood Web site). We then assessed the response rates by TKI modality for each transcript cohort (supplemental Figure 1B-D). For CCyR, patients with e13a2 who received imatinib 400 had an inferior response rate (77%) compared with other TKI modalities (90-95%); this trend was not observed in patients with e14a2 or both transcripts where CCyR rate with imatinib 400 (93%) was similar to other treatment modalities (93-96%). Similarly, for MMR and MR4.5, patients with e13a2 treated with imatinib 400 had a trend for an inferior response rate compared with those treated with other TKI modalities. Of note, MMR and MR4.5 rates were generally similar in all TKI modalities for patients with e14a2 transcripts except for patients treated with nilotinib, who showed a somewhat inferior rate of MR4.5 in both the e13a2 and e14a2 cohorts compared with patients treated with imatinib 800 or dasatinib (supplemental Figure 1D).
Figure 1.
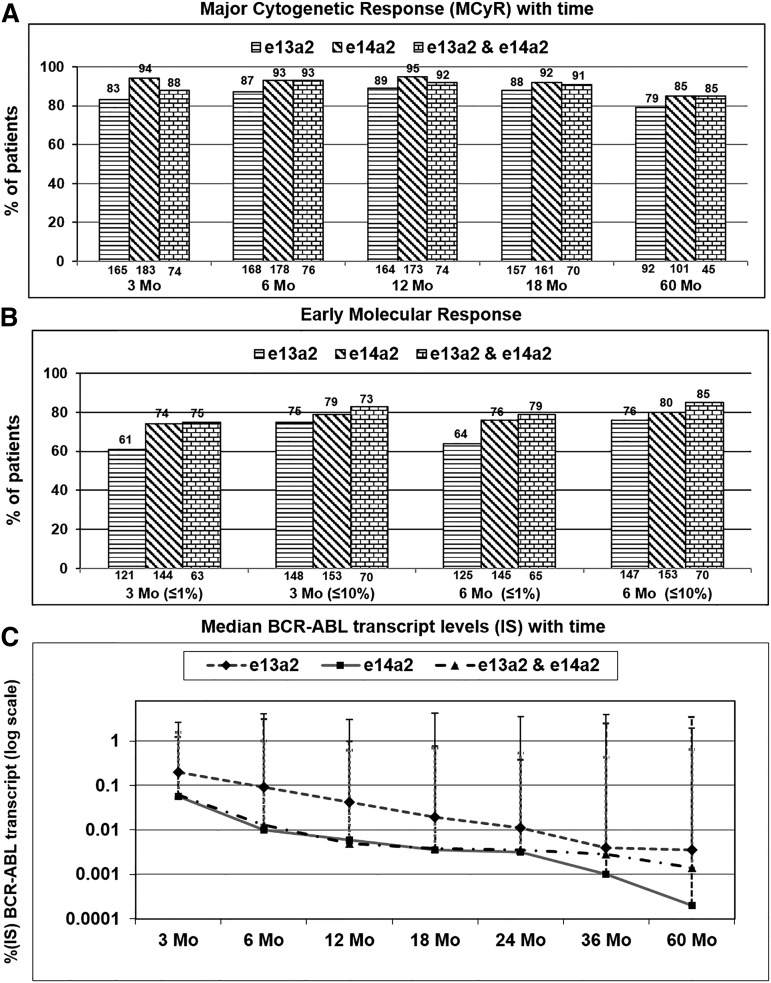
Analysis of major cytogenetic and molecular responses (≤10% or ≤1%) for BCR-ABL at 3 and 6 months according to the type of BCR-ABL transcript (e13a2, e14a2, or both). (A) Achievement of MCyR (≤35% Ph-positive metaphases). (B) Achievement of molecular response (≤1% and ≤10% BCR-ABL-IS) at 3 and 6 months. Percentages are shown at the top, and the absolute numbers of evaluable patients are shown at the bottom of each graph. (C) Pattern of changes in median BCR-ABL transcript levels (IS) over time (3, 6, 12, 18, 24, 36, and 60 months) according to the type of BCR-ABL transcript (log scale). IS, international scale.
We then analyzed response rates at different time points. For e13a2, e14a2, and coexpression cohorts, the proportion of patients achieving CCyR at 3 months was 59%, 67%, and 63%, respectively, and 73%, 81%, and 82% at 6 months, respectively (supplemental Figure 2A). Corresponding rates of MCyR were 83%, 94%, and 88% at 3 months and 87%, 93%, and 93% at 6 months, respectively (Figure 1A). The trend for lower rates of CCyR and MCyR for the e13a2 transcript cohort compared with the e14a2 cohort persisted even at 60 months.
Rates of MMR for the e13a2, e14a2, and coexpression cohorts were 27%, 49%, and 50% at 3 months, 42%, 67%, and 70% at 6 months, and 55%, 83%, and 76% at 12 months (equivalent to optimal European Leukemia Net [ELN] response32), respectively (supplemental Figure 2B). Similarly, rates of MR4.5 were lower for the e13a2 cohort over time compared with the e14a2 and coexpression cohorts (supplemental Figure 2C). Patients with e13a2 transcripts achieved lower rates of MMR or MR4.5 at all time points compared with those with e14a2.
Patterns of decline in different transcripts after treatment
We then analyzed the changes in transcript levels (in international scale) over time. The median transcripts levels (e13a2, e14a2, and both) at 3 months were 0.2004, 0.056, and 0.0612 and at 6 months were 0.091, 0.0109, and 0.0130, respectively. Patients with e13a2 transcripts demonstrated a slower rate of decline over time after start of treatment compared with the e14a2 and coexpression cohorts (Figure 1C). When analyzing the decline in transcript levels according to TKI used, generally the decline was slowest with imatinib 400 compared with imatinib 800 and dasatinib. Patients who received imatinib 400 or 800 mg or dasatinib demonstrated similar pattern of decline in all types of transcripts, with the slowest decline seen among the e13a2 cohort. This difference was less noticeable among those treated with nilotinib (supplemental Figure 3A-D). Treatment discontinuation is an important treatment goal for patients receiving TKI therapy, and the possibility to consider this end point depends on the achievement of sustained MR4.5, defined as MR4.5 that has been observed consecutively for ≥2 years with assessments at least every 6 months. We thus analyzed the cumulative incidence of achievement of sustained MR4.5 by transcript type. Patients with e14a2 and coexpressed transcripts demonstrated a significantly higher incidence of sustained MR4.5 compared with those with e13a2 (8-year probability, 43% vs 24%; P = .0021; supplemental Figure 4).
The times to achieve CCyR, MMR, and MR4.5 are shown in supplemental Figure 5A. The median time to CCyR was similar (3 months each) for the 3 transcript cohorts. However, time to MMR and MR4.5 was longer in patients with e13a2 transcripts compared with those with e14a2 and coexpressed transcripts (6, 3.5, and 3.5 months for MMR and 20, 10, and 11 months for MR4.5, respectively). When analyzing the time to CCyR, MMR, and MR4.5 according to the TKI modality used (supplemental Figure 5B-D), patients treated with imatinib 400 had the longest time to response, irrespective of type of transcripts, with patients with e13a2 generally having longer times to MMR and MR4.5 across all treatment types.
Survival outcomes according to the type of transcript
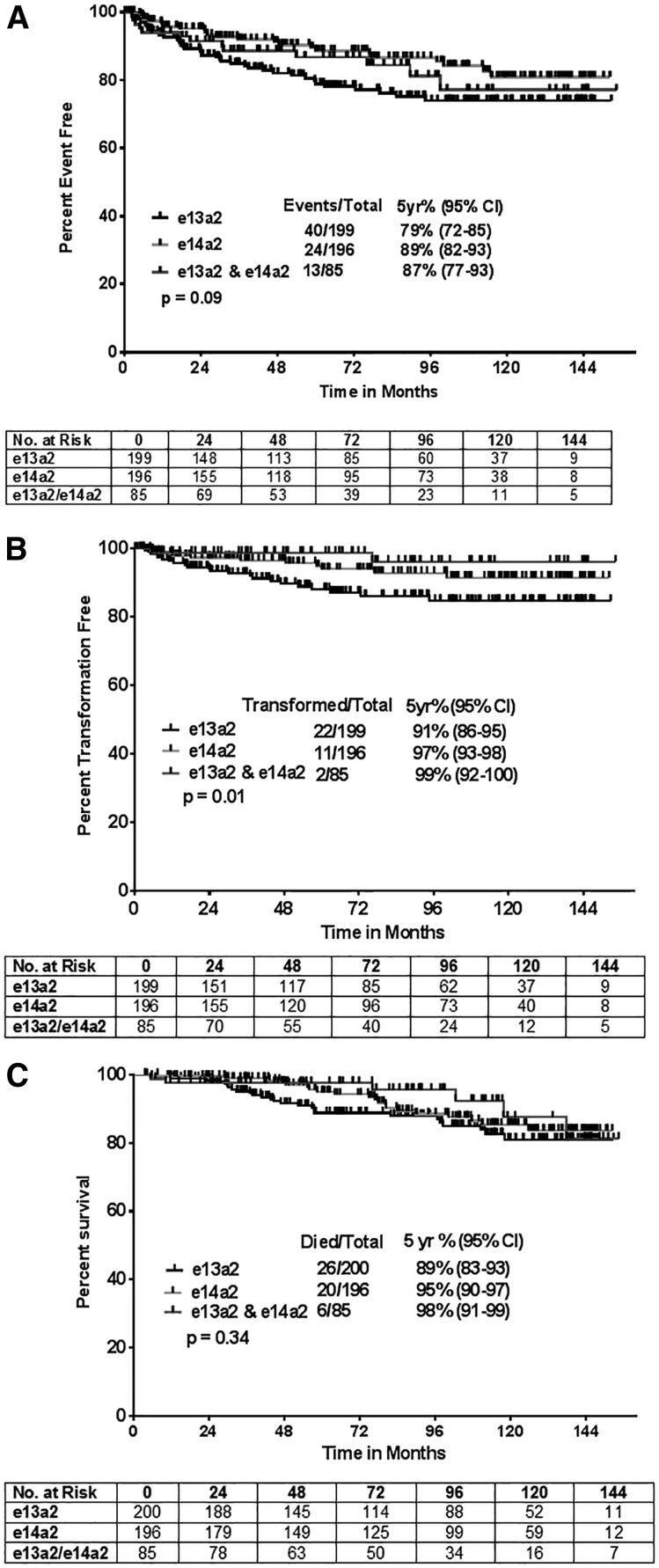
There were more events, transformations, and deaths among patients with e13a2, resulting in a general trend for inferior survival outcomes in such patients (Figure 2A-C). The 5-year probabilities for EFS for patients expressing e13a2, e14a2, and both transcripts were 79%, 89%, and 87%, respectively (P = .09). The corresponding figures for TFS were 91%, 97%, and 99% (P = .01), respectively. Five-year probabilities of survival were 88%, 95%, and 98% (P = .34) for each cohort, respectively. Because achievement of MCyR at 3 months or BCR-ABL ≤10% is an important hallmark for response to treatment, we compared survival outcomes in different transcripts based on the achievement of optimal ELN response at 3 months.32 Only TFS was significantly longer in patients who expressed e14a2 and/or both transcripts and achieved optimal response at 3 months (P = .03; supplemental Figure 6). Other survival outcomes, EFS and OS, were not significantly different in the 3 types of transcripts when categorized by 3- and 6-month optimal ELN response (Table 2). Furthermore, survival outcomes based on ELN responses categories were not significantly different in various transcript types, irrespective of the TKI modality used (supplemental Table 1). This could be due to lower number of patients in each transcript type when analyzed according to the TKI modality and type of ELN response. In general, survival outcomes were inferior in patients with e13a2 transcripts in response to different TKI modalities.
Figure 2.
Survival outcomes according to the type of BCR-ABL transcript (e13a2, e14a2, or both). (A) EFS; median survival not reached in all for all transcripts (P = .091). (B) TFS; median survival not reached for all transcripts (P = .01). (C) OS; median survival not reached in any group (P = .346).
Table 2.
Summary of time to event outcomes of patients expressing different BCR-ABL transcripts (e13a2, e14a2, and coexpressed e13a2 and e14a2) according to optimal ELN responses achieved at the 3- and 6-month landmark: EFS, TFS, and OS
| % 5-year outcome EFS/TFS/OS | ||||
|---|---|---|---|---|
| e13a2 | e14a2 | Both | ||
| Cytogenetic response* | ||||
| 3 months | MCyR | 83/90/90 | 90/94/95 | 91/99/97 |
| No MCyR | 54/78/85 | 61/90/91 | 61/100/100† | |
| 6 months | CCyR | 88/91/93 | 94/96/97 | 94/100/99 |
| No CCyR | 50/77/80 | 65/84/93 | 63/100/100† | |
| Molecular response‡ | ||||
| 3 months | ≤10% | 82/89/91 | 90/95/96 | 87/99/97 |
| >10%§ | −/−/100 | −/−/− | −/−/− | |
| 6 months | <1% | 89/92/94 | 94/96/97 | 94/100/98 |
| ≥1% | 55/71/79 | 43/91/89 | 30/100/100 | |
Optimal ELN response by cytogenetic response: major cytogenetic response at 3 months or complete cytogenetic response at 6 months.
n = 11 for coexpressed transcripts with no MCyR at 3 months and n = 14 with no MCyR at 6 months.
Optimal ELN response by molecular response: BCR-ABL1 ≤10% at 3 months or <1% at 6 months.
Less than 5 patients in this group.
Type of BCR-ABL transcript predict for survival and response: multivariate analysis
We then conducted univariate and multivariate analyses to determine whether effect of transcript on outcome is independent of other variables. Table 3 shows the results of these analyses for EFS. Covariates with P < .10 were included in the final multivariate model for EFS. Compared with e13a2, e14a2 (but not coexpressed e13a2 and e14a2 transcripts) significantly predicted for longer EFS (e14a2 hazard ratio [HR] = 0.59, 95% confidence interval [CI] = 0.36-0.98 [P = .04], coexpressed transcripts HR = 0.80, 95% CI = 0.43-1.50 [P = .48]). Treatment with imatinib 800, dasatinib, and nilotinib also predicted for longer EFS (P = .03, .004, and .02, respectively) and the presence of splenomegaly at the time of initial presentation was predictive of inferior EFS (P = .03). Similarly, e14a2 and coexpressed transcripts predicted for significantly improved probability of TFS compared with e13a2 (supplemental Table 2; e14a2, HR = 0.52, 95% CI = 0.25-1.07 [P = .07]; coexpressed transcripts, HR = 0.22, 95% CI = 0.05–0.94 [P = .04]). Treatment with imatinib 800 and dasatinib also predicted for longer TFS (P = .02 and .04, respectively). Coexpression of both transcripts was marginally significant for predicting OS (HR = 0.45, 95% CI = 0.18-1.11; P = .08).
Table 3.
Factors predictive of EFS including the type of BCR-ABL transcripts
| N | Events | Log-rank | HR | 95% CI HR | P value | |
|---|---|---|---|---|---|---|
| Univariate* | ||||||
| TKI type | ||||||
| Imatinib 400† | 69 | 21 | 0.013 | |||
| Imatinib 800 | 199 | 38 | 0.58 | (0.34-0.99) | .045 | |
| Dasatinib | 104 | 7 | 0.28 | (0.12-0.67) | .004 | |
| Nilotinib | 108 | 11 | 0.45 | (0.22-0.95) | .035 | |
| Transcript type | ||||||
| e13a2‡ | 199 | 40 | 0.091 | |||
| e14a2 | 196 | 24 | 0.57 | (0.35-0.95) | .032 | |
| Both | 85 | 13 | 0.74 | (0.40-1.39) | .355 | |
| Splenomegaly (≥10 cm) | ||||||
| No | 453 | 68 | 0.012 | |||
| Yes | 27 | 9 | 2.38 | (1.19-4.77) | .014 | |
| Multivariate | ||||||
| TKI type | ||||||
| Imatinib 400† | 69 | 21 | ||||
| Imatinib 800 | 199 | 38 | 0.55 | (0.32-0.94) | .030 | |
| Dasatinib | 104 | 7 | 0.28 | (0.12-0.67) | .004 | |
| Nilotinib | 108 | 11 | 0.44 | (0.21-0.92) | .029 | |
| Transcript type | ||||||
| e13a2‡ | 199 | 40 | ||||
| e14a2 | 196 | 24 | 0.59 | (0.36-0.98) | .043 | |
| Both | 85 | 13 | 0.80 | (0.43-1.50) | .489 | |
| Splenomegaly (≥10 cm) | ||||||
| No | 453 | 68 | ||||
| Yes | 27 | 9 | 2.18 | (1.07-4.44) | .031 |
White blood cell count, age, Sokal score, hemoglobin, platelet count, peripheral blood blasts, and serum lactate dehydrogenase are not significant (P = not significant; data not shown).
Imatinib 400 is the reference for comparison with the other 3 TKI modalities.
e13a2 is the reference for comparison with e14a2 and both (coexpressed transcripts).
We also assessed whether the type of transcript can predict for optimal ELN response at different time points (at 3, 6, and 12 months).32 The expression of e14a2 transcripts predicted for improved probability of MCyR at 3 months (supplemental Table 3). Expression of e14a2 or coexpressed transcripts predicted for MMR at 6 and 12 months (optimal ELN response; supplemental Table 4). Similarly, expression of e14a2 transcripts predicted for 6-month CCyR (P = .04; supplemental Table 5). In addition, the type of transcript (e14a2 and both) predicted for MMR at 12 months (Table 4), OR for e14a2 was 5.85, 95% CI (3.01-11.37; P < .001) and for both transcripts OR was 3.18, 95% CI (1.50-6.74; P = .003).
Table 4.
Factors predictive for MMR at 12 months (optimal response) including the type of BCR-ABL fusion transcript
| No MMR, N (%) | MMR, N (%) | OR | 95% CI OR | P | |
|---|---|---|---|---|---|
| Univariate* | |||||
| TKI type | |||||
| Imatinib 400† | 20 (23) | 24 (8) | |||
| Imatinib 800 | 33 (38) | 140 (45) | 3.54 | (1.75-7.15) | <.001 |
| Dasatinib | 22 (25) | 72 (23) | 2.73 | (1.27-5.84) | .010 |
| Nilotinib | 11 (13) | 77 (25) | 5.83 | (2.45-13.88) | <.001 |
| Transcript type | |||||
| e13a2‡ | 60 (70) | 101 (32) | |||
| e14a2 | 15 (17) | 151 (48) | 5.98 | (3.22-11.11) | <.001 |
| Both | 11 (13) | 61 (19) | 3.29 | (1.61-6.75) | .001 |
| Splenomegaly (≥10 cm) | |||||
| No | 76 (88) | 300 (96) | |||
| Yes | 10 (12) | 13 (4) | 0.33 | (0.14-0.78) | .012 |
| Platelets (>300 K/µL) | |||||
| No | 46 (53) | 123 (39) | |||
| Yes | 40 (46) | 190 (61) | 1.78 | (1.10-2.87) | .019 |
| Multivariate§ | |||||
| TKI type | |||||
| Imatinib 400† | 20 (23) | 24 (8) | |||
| Imatinib 800 | 33 (38) | 140 (45) | 4.83 | (2.20-10.62) | <.001 |
| Dasatinib | 22 (25) | 72 (23) | 3.39 | (1.45-7.89) | .005 |
| Nilotinib | 11 (13) | 77 (25) | 8.24 | (3.16-21.43) | <.001 |
| Transcript type | |||||
| e13a2‡ | 60 (70) | 101 (32) | |||
| e14a2 | 15 (17) | 151 (48) | 5.85 | (3.01-11.37) | <.001 |
| Both | 11 (13) | 61 (19) | 3.18 | (1.50-6.74) | .003 |
White blood cell count, age, Sokal score, hemoglobin, peripheral blood blasts, and serum lactate dehydrogenase are not significant (P = not significant; data not shown).
Imatinib 400 is the reference for comparison with the other 3 TKI modalities.
e13a2 is the reference for comparison with e14a2 and both (coexpressed transcripts).
Platelet count and spleen size were not significant in multivariate analysis (data not shown).
Discussion
In this study, we analyzed the influence of the transcript type on molecular and cytogenetic responses achieved in response to different TKI regimens in newly diagnosed chronic phase CML patients and also compared different transcripts with respect to survival outcomes. Patients who expressed e14a2 (and to some extent those with coexpression of e14a2 and e13a2) achieved earlier and deeper cytogenetic and molecular responses compared with those with only e13a2 transcripts and maintained these responses longer. In multivariate analysis, the expression of e14a2 or both types of transcripts independently predicted for achievement of optimal ELN response32 and an improved probability of EFS and TFS but not OS.
The question of whether the type of transcript has prognostic significance for patients with newly diagnosed CML–chronic phase has been debated for many years.13,33 More than 95% of patients with CML have e13a2 (b2a2), e14a2 (b3a2), or both transcripts coding for p210 BCR-ABL1 tyrosine kinase, whereas only a minority express rare variants such as e1a2 transcripts,34 which code for p190 BCR-ABL1, which is associated with rapid disease progression. Previous reports have suggested that patients with e14a2 achieve deeper molecular response to standard dose imatinib23 and have higher platelet counts compared with patients with e13a2 transcripts.19,21 To our knowledge, no prior analyses have suggested any influence of transcript type on survival outcomes or tested whether the type of transcript can predict for an optimal ELN response. Of further note, we showed that patients expressing the e13a2 transcript have a significantly lower incidence of sustained MR4.5 response compared with e14a2 and coexpressed transcripts (supplemental Figure 4). Our data indicate that the type of transcript may also increase the probability of reaching end points required for treatment discontinuation. Unfortunately, too few patients in this series have elected to discontinue therapy. Thus, we cannot address whether the probability of recurrence after treatment discontinuation is affected by the transcript type.
Consistent with previous reports, our analysis shows a higher platelet count in patients with e14a2 transcripts21,35; however, we did not detect any difference in white blood cell counts as had been reported by Hanfstein et al,21 nor did we identify any other significant difference in the baseline characteristics of patients with different transcripts. In our analysis, the rates of MCyR and CCyR, as well as MMR and MR4.5, were lower in patients with e13a2 transcripts at various time points. However, the median time to achieve CCyR was similar for all cohorts (3 months in each; supplemental Figure 5A). This is consistent with what was reported by Hanfstein et al in 1105 imatinib-treated patients.21
Superior molecular responses in patients with e14a2 transcripts were reported previously in patients treated with standard-dose imatinib.21,23 The median time to achieve MMR or MR4.5 was longer in patients with imatinib 400 compared with the other 3 TKI modalities, irrespective of the type of transcript. However, patients with e13a2 treated with imatinib 400 had the longest time to achieve MMR (supplemental Figure 5C-D), again suggesting that these patients have a worse outcome when treated with this treatment modality.
Because the type of TKI can influence the time and depth of cytogenetic and molecular response, we analyzed the impact of TKI type in each transcript for attaining cytogenetic and molecular responses. Interestingly, there was a suggestion that treatment with imatinib 400 mg daily was associated with a lower probability of response only among patients with e13a2. The rates of CCyR and MMR for patients with e14a2 or with coexpression of e13a2 and e14a2 treated with imatinib 400 mg daily was similar to that of patients treated with imatinib 800, dasatinib, or nilotinib. If these observations were to be confirmed in a prospective study, it would suggest that transcript type could be a tool to help select the TKI to be used for patients with newly diagnosed CML. For example, patients with e14a2 could be offered imatinib 400, whereas those with e13a2 may derive more benefit from the use of 2GTKI as initial therapy.
Although our study does not provide a mechanism with which these differences between e13a2 vs e14a2 and coexpressed transcripts could be elucidated, we could hypothesize some biological reasons to explain these observations. Based on the observed differences in attaining response, higher platelet counts, trend for better survival, and lower occurrence of disease transformation in patients with e14a2, it could be hypothesized that disease with e13a2 and e14a2 or coexpressed transcripts have different biological features that may participate differently in the pathogenesis of CML. One possible explanation for inferior outcome for patients with e13a2 is the reported higher tyrosine kinase activity reflected by higher pCrKL levels with e13a2 transcripts.19 This could result in a given TKI being able to more effectively suppress the kinase activity associated with the less active e14a2 transcripts. Because imatinib 400 has the weakest inhibitory activity, this could also explain the possible inferior outcome for patients with the e13a2 when treated with standard dose imatinib. It has also been described that the presence of an extra 25 amino acids in the e14a2 transcripts leads to changes in the structure of the binding domain of BCR-ABL kinase leading to reduced tyrosine kinase activity thereby causing better responses in e14a2 transcripts.7,19,36 Hai et al26 reported that the presence of the additional 25 amino acids encoded by the e14 exon not present in e13a2 lead to differences in SRC homology domain (SH1-SH3) and DNA binding domains, which regulate the tyrosine kinase activity in p210 BCR-ABL protein isoform. Interestingly, this difference was shown to influence difference in platelet counts in the 2 transcripts.36 Overall, this variability in the structure of the 2 isoforms has the potential to differentially influence signal transduction in CML cells.
The transcript type not only influenced response outcomes, but also long-term survival outcomes. Previous studies21 have not shown any difference in EFS or TFS according to transcript types among patients treated with imatinib. In our analysis, we showed that patients with e13a2 transcripts show a trend for inferior EFS and TFS compared with e14a2 and/or both transcripts among patients treated with various TKI modalities. By multivariate analysis, transcript type remained statistically significant and an independent variable affecting long-term EFS and TFS. One possible explanation for these results could be due to differences in treatments because our study population included a mix of 4 TKI modalities, whereas previous studies included only patients treated with imatinib. Finally, overall results were similar in patients with e14a2 and those who expressed both e13a2 and e14a2. This equivalence in outcome is expected because at the genomic level, these patients have an e14a2 rearrangement with an alternative splicing that gives rise to the coexpression of e13a2.
In summary, we showed that type of transcript may impact the probability of response to TKI among patients with CML treated with various TKI regimens. The difference in response translates in inferior probability of EFS and TFS for patients with e13a2 transcripts. The inferior outcome appears to be more evident for patients treated with imatinib 400, whereas patients with e14a2 treated with imatinib 400 may have a similarly favorable outcome as those treated with dasatinib or nilotinib. If confirmed, transcript type could be used to select TKI regimen for patients with CML. The biological mechanism behind this effect and the possible modulating effect of treatment modality in this possible differential outcome by transcript type deserves further study.
Acknowledgments
Funding for these studies was provided in part by the MD Anderson Cancer Center support grant CA016672 (principal investigator [PI]: Dr Ronald DePinho) and award P01 CA049639 (PI: Dr Richard Champlin) from the National Institutes of Health, National Cancer Institute (NCI). None of the authors are employed by the National Institutes of Health. J.C. is the recipient of a grant from the NCI (PI of project 1 of P01 CA049639).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: P.J. and J.C. designed the study; G.N.G., P.J., H.K., and J.C. analyzed results; P.J., G.N.G., K.S., C.G.R., and J.C. wrote the paper; P.J., G.N.G., R.L., K.P.P., R.K.S., H.K., and J.C. did the clinical correlation; H.K., S.O., F.R., E.J., Z.E., N.P., N.D., T.M.K., G.B., and J.C. contributed patient samples; and all authors reviewed and gave the final approval for the paper.
Conflict-of-interest disclosure: J.C. is a consultant for Pfizer, Ariad, and Teva and has received research support from Pfizer, Ariad, Chemgenex, Bristol Myers Squibb (BMS), and Novartis. F.R. has received research funding from BMS and honoraria from BMS, Novartis, and Pfizer. N.P. is a consultant for Novartis. The remaining authors declare no competing financial interests.
Correspondence: Jorge Cortes, Department of Leukemia, Unit 428, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: jcortes@mdanderson.org.
References
- 1.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243(5405):290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 2.Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36(1):93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- 3.Lugo TG, Pendergast AM, Muller AJ, Witte ON. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science. 1990;247(4946):1079–1082. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 4.Druker BJ, Tamura S, Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2(5):561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 5.Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344(14):1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189(9):1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melo JV. The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood. 1996;88(7):2375–2384. [PubMed] [Google Scholar]
- 8.Melo JV. BCR-ABL gene variants. Baillieres Clin Haematol. 1997;10(2):203–222. doi: 10.1016/s0950-3536(97)80003-0. [DOI] [PubMed] [Google Scholar]
- 9.Faderl S, Talpaz M, Estrov Z, O’Brien S, Kurzrock R, Kantarjian HM. The biology of chronic myeloid leukemia. N Engl J Med. 1999;341(3):164–172. doi: 10.1056/NEJM199907153410306. [DOI] [PubMed] [Google Scholar]
- 10.Shtivelman E, Lifshitz B, Gale RP, Roe BA, Canaani E. Alternative splicing of RNAs transcribed from the human abl gene and from the bcr-abl fused gene. Cell. 1986;47(2):277–284. doi: 10.1016/0092-8674(86)90450-2. [DOI] [PubMed] [Google Scholar]
- 11.Verschraegen CF, Kantarjian HM, Hirsch-Ginsberg C, et al. The breakpoint cluster region site in patients with Philadelphia chromosome-positive chronic myelogenous leukemia. Clinical, laboratory, and prognostic correlations. Cancer. 1995;76(6):992–997. doi: 10.1002/1097-0142(19950915)76:6<992::aid-cncr2820760612>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Neriah Y, Daley GQ, Mes-Masson AM, Witte ON, Baltimore D. The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science. 1986;233(4760):212–214. doi: 10.1126/science.3460176. [DOI] [PubMed] [Google Scholar]
- 13.Langabeer SE. Is the BCR-ABL1 transcript type in chronic myeloid leukaemia relevant? Med Oncol. 2013;30(2):508. doi: 10.1007/s12032-013-0508-9. [DOI] [PubMed] [Google Scholar]
- 14.Xiaomin G, Yong Z, Jinlan P, et al. Chronic myeloid leukemia with e14a3 BCR-ABL transcript: analysis of characteristics and prognostic significance. Leuk Lymphoma. 2015;56(12):3343–3347. doi: 10.3109/10428194.2015.1037751. [DOI] [PubMed] [Google Scholar]
- 15.Huet S, Dulucq S, Chauveau A, et al. GBMHM (Groupe des Biologistes Moléculaires des Hémopathies Malignes, French Molecular Biology Group in Hematology) Molecular characterization and follow-up of five CML patients with new BCR-ABL1 fusion transcripts. Genes Chromosomes Cancer. 2015;54(10):595–605. doi: 10.1002/gcc.22263. [DOI] [PubMed] [Google Scholar]
- 16.Prejzner W. Relationship of the BCR gene breakpoint and the type of BCR/ABL transcript to clinical course, prognostic indexes and survival in patients with chronic myeloid leukemia. Med Sci Monit. 2002;8(5):BR193–BR197. [PubMed] [Google Scholar]
- 17.Tefferi A, Bren GD, Wagner KV, Schaid DJ, Ash RC, Thibodeau SN. The location of the Philadelphia chromosomal breakpoint site and prognosis in chronic granulocytic leukemia. Leukemia. 1990;4(12):839–842. [PubMed] [Google Scholar]
- 18.Shepherd P, Suffolk R, Halsey J, Allan N. Analysis of molecular breakpoint and m-RNA transcripts in a prospective randomized trial of interferon in chronic myeloid leukaemia: no correlation with clinical features, cytogenetic response, duration of chronic phase, or survival. Br J Haematol. 1995;89(3):546–554. doi: 10.1111/j.1365-2141.1995.tb08362.x. [DOI] [PubMed] [Google Scholar]
- 19.Lucas CM, Harris RJ, Giannoudis A, et al. Chronic myeloid leukemia patients with the e13a2 BCR-ABL fusion transcript have inferior responses to imatinib compared to patients with the e14a2 transcript. Haematologica. 2009;94(10):1362–1367. doi: 10.3324/haematol.2009.009134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Lemos JA, de Oliveira CM, Scerni AC, et al. Differential molecular response of the transcripts B2A2 and B3A2 to imatinib mesylate in chronic myeloid leukemia. Genet Mol Res. 2005;4(4):803–811. [PubMed] [Google Scholar]
- 21.Hanfstein B, Lauseker M, Hehlmann R, et al. SAKK and the German CML Study Group. Distinct characteristics of e13a2 versus e14a2 BCR-ABL1 driven chronic myeloid leukemia under first-line therapy with imatinib. Haematologica. 2014;99(9):1441–1447. doi: 10.3324/haematol.2013.096537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanfstein B, Shlyakhto V, Lauseker M, et al. SAKK and the German CML Study Group. Velocity of early BCR-ABL transcript elimination as an optimized predictor of outcome in chronic myeloid leukemia (CML) patients in chronic phase on treatment with imatinib. Leukemia. 2014;28(10):1988–1992. doi: 10.1038/leu.2014.153. [DOI] [PubMed] [Google Scholar]
- 23.Lin HX, Sjaarda J, Dyck J, et al. Gender and BCR-ABL transcript type are correlated with molecular response to imatinib treatment in patients with chronic myeloid leukemia [published online ahead of print June 8, 2015]. Eur J Haematol. doi: 10.1111/ejh.12597. doi:10.1111/ejh.12597. [DOI] [PubMed] [Google Scholar]
- 24.Vega-Ruiz A, Kantarjian H, Shan J, et al. Better molecular response to imatinib for patients (pts) with chronic myeloid leukemia (CML) in chronic phase (CP) carrying the b3a2 transcript compared to b2a2 [abstract]. Blood. 2007;110(11). Abstract 1939. [Google Scholar]
- 25.White D, Saunders V, Lyons AB, et al. In vitro sensitivity to imatinib-induced inhibition of ABL kinase activity is predictive of molecular response in patients with de novo CML. Blood. 2005;106(7):2520–2526. doi: 10.1182/blood-2005-03-1103. [DOI] [PubMed] [Google Scholar]
- 26.Hai A, Kizilbash NA, Zaidi SH, Alruwaili J, Shahzad K. Differences in structural elements of Bcr-Abl oncoprotein isoforms in Chronic Myelogenous Leukemia. Bioinformation. 2014;10(3):108–114. doi: 10.6026/97320630010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jabbour E, Kantarjian HM, Saglio G, et al. Early response with dasatinib or imatinib in chronic myeloid leukemia: 3-year follow-up from a randomized phase 3 trial (DASISION). Blood. 2014;123(4):494–500. doi: 10.1182/blood-2013-06-511592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain P, Kantarjian H, Nazha A, et al. Early responses predict better outcomes in patients with newly diagnosed chronic myeloid leukemia: results with four tyrosine kinase inhibitor modalities. Blood. 2013;121(24):4867–4874. doi: 10.1182/blood-2013-03-490128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larson RA, Hochhaus A, Hughes TP, et al. Nilotinib vs imatinib in patients with newly diagnosed Philadelphia chromosome-positive chronic myeloid leukemia in chronic phase: ENESTnd 3-year follow-up. Leukemia. 2012;26(10):2197–2203. doi: 10.1038/leu.2012.134. [DOI] [PubMed] [Google Scholar]
- 30.Baccarani M, Cortes J, Pane F, et al. European LeukemiaNet. Chronic myeloid leukemia: an update of concepts and management recommendations of European LeukemiaNet. J Clin Oncol. 2009;27(35):6041–6051. doi: 10.1200/JCO.2009.25.0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Brien SG, Guilhot F, Larson RA, et al. IRIS Investigators. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348(11):994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 32.Baccarani M, Deininger MW, Rosti G, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;122(6):872–884. doi: 10.1182/blood-2013-05-501569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braekeleer M. BCR-ABL1 b3a2 and b2a2 transcripts in chronic myeloid leukemia: does it matter [published online ahead of print July 24, 2015]? doi: 10.1111/ejh.12639. Eur J Haematol., doi:10.1111/ejh.12639. [DOI] [PubMed] [Google Scholar]
- 34.Verma D, Kantarjian HM, Jones D, et al. Chronic myeloid leukemia (CML) with P190 BCR-ABL: analysis of characteristics, outcomes, and prognostic significance. Blood. 2009;114(11):2232–2235. doi: 10.1182/blood-2009-02-204693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inokuchi K, Inoue T, Tojo A, et al. A possible correlation between the type of bcr-abl hybrid messenger RNA and platelet count in Philadelphia-positive chronic myelogenous leukemia. Blood. 1991;78(12):3125–3127. [PubMed] [Google Scholar]
- 36.Perego RA, Costantini M, Cornacchini G, et al. The possible influences of B2A2 and B3A2 BCR/ABL protein structure on thrombopoiesis in chronic myeloid leukaemia. Eur J Cancer. 2000;36(11):1395–1401. doi: 10.1016/s0959-8049(00)00128-3. [DOI] [PubMed] [Google Scholar]