SUMMARY
After many years in the family Vibrionaceae, the genus Plesiomonas, represented by a single species, P. shigelloides, currently resides in the family Enterobacteriaceae, although its most appropriate phylogenetic position may yet to be determined. Common environmental reservoirs for plesiomonads include freshwater ecosystems and estuaries and inhabitants of these aquatic environs. Long suspected as being an etiologic agent of bacterial gastroenteritis, convincing evidence supporting this conclusion has accumulated over the past 2 decades in the form of a series of foodborne outbreaks solely or partially attributable to P. shigelloides. The prevalence of P. shigelloides enteritis varies considerably, with higher rates reported from Southeast Asia and Africa and lower numbers from North America and Europe. Reasons for these differences may include hygiene conditions, dietary habits, regional occupations, or other unknown factors. Other human illnesses caused by P. shigelloides include septicemia and central nervous system disease, eye infections, and a variety of miscellaneous ailments. For years, recognizable virulence factors potentially associated with P. shigelloides pathogenicity were lacking; however, several good candidates now have been reported, including a cytotoxic hemolysin, iron acquisition systems, and lipopolysaccharide. While P. shigelloides is easy to identify biochemically, it is often overlooked in stool samples due to its smaller colony size or relatively low prevalence in gastrointestinal samples. However, one FDA-approved PCR-based culture-independent diagnostic test system to detect multiple enteropathogens (FilmArray) includes P. shigelloides on its panel. Plesiomonads produce β-lactamases but are typically susceptible to many first-line antimicrobial agents, including quinolones and carbapenems.
INTRODUCTION
One of the earliest names used in modern bacteriology to describe a group of anaerogenic Gram-negative bacilli occasionally found in association with cases of gastroenteritis was the generic term “paracolon.” The “paracolon group” of bacteria consisted of many different taxa that we now recognize today as distinct species but in the 1940s were chiefly characterized by a limited number of biochemical characteristics, including the ability to ferment lactose. From such a potpourri of enteric bacteria sprang later genera and species, including the genus Providencia, Citrobacter freundii, and the Arizona subgroup of Salmonella (Salmonella enterica subsp. arizonae) (1). Another member of this paracolon collection was an unusual bacterium with “Shigella-like” antigenic characteristics that was sometimes isolated from diarrheal stools. This bacterium, originally dubbed “C27” or the “C27 paracolon” group was years later given the name that we know it by today, Plesiomonas shigelloides (2, 3).
The existence of P. shigelloides has now been known for almost 70 years, with its original discovery and description by Ferguson and Henderson in 1947 (2). Initially, this organism received little attention due to its infrequent isolation from clinical samples and its singular association with sporadic episodes of enteritis. In fact, it wasn't until almost 3 decades later that plesiomonads were documented as human pathogens, with the first case description of P. shigelloides sepsis in a 62-year-old woman with sickle cell anemia (4). Over the next 2 decades, with an ongoing search for new causes of bacterial gastroenteritis, Plesiomonas along with other reputed pathogens, such as Aeromonas, came into the limelight and under scrutiny as potential etiologic agents of bacterial gastroenteritis, with a number of authoritative reviews published on the topic (5, 6, 7, 8). Despite these advances, many questions remained unanswered. Some of these questions included whether P. shigelloides was a bona fide enteropathogen, the lack of an association between this species and credible outbreaks of diarrheal disease in healthy persons, an inability to identify potential pathogenic factors in this group, and taxonomy and classification issues concerning the correct nomenclature and phylogenetic position of this nomenspecies.
Scientific and medical information on this pathogen has recently been published and has provided insights regarding many of these and other unanswered questions. Furthermore, with the availability of different molecular technologies, including DNA sequencing and gene amplification, many other advances have been made, including publication of the first full genome sequence of a serogroup O1 strain of P. shigelloides (9). It seems appropriate then to review where the clinical microbiology community now stands in regard to this organism in light of past viewpoints and misconceptions. This review will therefore revisit the biology and microbiology of P. shigelloides in relationship to human disease, laboratory detection, and diagnosis, primarily focusing on data generated since 2000, with previous seminal publications included for a comprehensive overview.
TAXONOMY
Nomenclature
Historical perspective.
Bacteria that originally were designated members of the C27 group have a checkered history with regard to previous taxonomic names and epithets prior to their current designation as P. shigelloides. From their initial description by Ferguson and Henderson (2), it was unclear whether they belonged within an existing genus or family or should be placed elsewhere. Bader (10) proposed the species designation “shigelloides” in 1954 to reflect their possession of Shigella sonnei phase I antigen. C27 strains were subsequently placed in a variety of families or genera over the next 2 decades, based upon selected phenotypic or serologic characteristics. These characteristics included possession of certain somatic antigens and anaerogenic fermentation of glucose (Enterobacteriaceae) or number and location of flagella and oxidase positivity (Pseudonomas). Other less frequently recognized genera proposed to include C27 strains were the genera Escherichia, Fergusonia, Scatamonas, and Vibrio (11, 12, 13).
In 1961, Ewing and associates (12) suggested that these strains possessed a number of characteristics in common with the genus Aeromonas and included it in this group as a separate species. Thus, a number of early scientific publications can be found in the literature concerning “Aeromonas shigelloides.” However, shortly after this proposal, Habs and Schubert in 1962 believed these organisms should be transferred to a new genus they named Plesiomonas by virtue of a substantially distinct G+C content (51 mol%) from that of true aeromonads (57% to 60%), among other traits (14). The Greek name “plesios,” which means neighbor, and “monas,” which means unit, implies a neighboring group to the genus Aeromonas. The genus and species name Plesiomonas shigellloides gained wide acceptance as the preferred name for these bacteria over the next decade, and only this designation was included on the Approved List of Bacterial Names in 1980 (15). There appeared a fairly substantial amount of phenotypic data indicating that plesiomonads shared some properties with members of both the Enterobacteriaceae and Vibrionaceae families. Véron proposed that both genera (Aeromonas and Plesiomonas) be included in the family Vibrionaceae, based upon a number of common properties shared by other genera (vibrios), including a cytochrome oxidase, fermentative metabolism, ecologic associations, and disease presentations (16).
Classification
Phylogenetic investigations.
Beginning in the mid-1980s, phylogenetic data began to accumulate that suggested that the placement of P. shigelloides in the family Vibrionaceae was inappropriate. 5S rRNA sequence analysis of 31 type or reference strains of members of the family Vibrionaceae indicated a closer ancestral relationship between Plesiomonas and the family Enterobacteriaceae than the Vibrionaceae (17). From strains studied, the authors found the closest relationship for plesiomonads in the enteric group to be with Proteus mirabilis (18). The authors concluded from their studies that P. shigelloides should be transferred to the tribe Proteeae (Proteus, Morganella, Providencia) within the family Enterobacteriaceae.
Subsequent phylogenetic investigations regarding the potential reclassification of plesiomonads tended to support earlier findings based upon 5S rRNA gene sequence analysis. Martinez-Murcia and others (19) found the type strain of P. shigelloides (ATCC 14029T = NCIMB 9242T = CIP 63.5T) exhibited 93% to 94.9% 16S rRNA sequence relatedness to two enteric species while being only 90.7% related to Vibrio anguillarum. Still other studies looking at the phylogenetic positions of various genera traditionally associated with the Vibrionaceae and employing different types of analysis (e.g., unrooted trees, maximum likelihood) always found P. shigelloides to genetically reside closer to enteric bacteria than to true vibrios (20). Each of these studies, however, looked at only a limited number of species within the Enterobacteriaceae in their analyses. Overall, plesiomonads display 93% to 95% 16S homology to the family Enterobacteriaceae but only 91% relatedness to either the Vibrionaceae or Aeromonadaceae (13).
Reclassification of Plesiomonas into the family Enterobacteriaceae.
In the second edition of Bergey's Manual of Systematic Bacteriology, the genus Plesiomonas was officially transferred from the family Vibrionaceae to the family Enterobacteriaceae (13). The main reasons for this transfer were the following. (i) Phylogenetic 5S, 16S, and multilocus sequence typing (MLST) data indicating that this taxon is rooted within the enterobacteria and not with the Vibrio and Photobacterium clades (17, 18, 19, 20, 21). (ii) P. shigelloides possesses a heteropolymer antigen linked to the lipopolysaccharide (LPS) termed the enterobacterial common antigen (22). This antigen is exclusively found in members of the Enterobacteriaceae and not in vibrios, including Grimontia hollisae and Photobacterium damselae (23). (iii) A number of cellular components, including polyamine composition and a lipid A structure containing a common 1,4′-bis-phosphorylated-β1,6′-linked glucosamine backbone with six amide-linked acyl-oxyacyl groups is identical to that of Shigella sonnei (24, 25).
Salient features relative to the placement of plesiomonads in the enterobacteria are listed in Table 1. Although some authors have suggested P. shigelloides should be included in the tribe Proteeae and within the genus Proteus, that recommendation has not been generally accepted. Unlike genera within this grouping, Plesiomonas lacks a number of tribe-defining traits associated with these genera and absent in most other enterobacteria. These include phenylalanine deaminase activity, production of a tyrosinase, pigmentation on d,l-tryptophan agar, urea hydrolysis, and swarming motility (1). Furthermore, neither the ecologic habitats nor the spectrum of human diseases caused by plesiomonads mirrors closely that of the Proteeae.
TABLE 1.
Common and distinguishing features between P. shigelloides and core members of the family Enterobacteriaceae
| Trait | Characteristic | Presence/absence or value for: |
|
|---|---|---|---|
| Core membersa | Plesiomonas | ||
| Biochemical | Oxidase | − | + |
| Facultatively anaerobic | + | + | |
| Acid and gas from d-glucose | +b | − | |
| Acid from d-xylose | + | − | |
| Acid from m-inositol | −b | + | |
| Nitrate reductase | + | + | |
| Cell associated | ECAc | + | + |
| Predominant polyamines (spermidine, putrescine) | + | + | |
| Genomics | G+C content (mol%) | 48–59 | 51 |
| 16S rRNA dendrograms (position) | Core or centrist | Peripheral or external | |
| Interspecies relatedness (DDH)d | 30–50% | 8% | |
Core members of the Enterobacteriaceae include the genera Citrobacter, Escherichia, Klebsiella, Enterobacter, Salmonella, and Shigella.
Infrequent exceptions occur.
ECA, enterobacterial common antigen.
Observed at 60°C.
Genomics.
Almost all present data strongly suggest that the genus and species designation P. shigelloides is composed of a collection of homogeneous bacteria at the phenotypic as well as molecular level. A population study of a diverse collection of 77 P. shigelloides strains from different geographic as well as environmental settings indicated a monophyletic clade nested within the Enterobacteriaceae (20, 21). Although extensive DNA-DNA hybridization (DDH) studies have not been formerly published, Fanning and coinvestigators (26) at the Centers for Disease Control and Prevention (CDC) studied 18 P. shigelloides strains and found them to be 87% and 81% related to the type strain at 60°C and 75°C, respectively, with only 1.5% divergence. Furthermore, the species as a whole is “phenotypically tight,” with little variability noted in carbohydrate fermentation patterns or other conventional biochemical characteristics (13, 27, 28, 29).
The complete genome of a single strain of P. shigelloides (strain 302-73, serogroup O1) was recently sequenced (9). The genome consists of a single circular chromosome of 3.9 Mbp with 3,285 coding sequences. The GenBank accession number is AQQO01000000. A population study of plesiomonads indicated high levels of nucleotide diversity (average of 1.49%) within five housekeeping genes sequenced (21). This finding suggests a very high recombination rate similar to that found in transformable taxa, such as Streptococcus pneumoniae and Neisseria meningitidis, and unlike low rates observed in other enteric bacteria, such as Escherichia coli and Yersinia. The exact ramifications of such a high recombination rate, if verified, are presently unknown.
Outstanding Issues
Presently, P. shigellodes is one of a very few of the older established genera in the family Enterobacteriaceae represented by a single species, with the recent addition of new taxa to the genera Hafnia, Morganella, and Rahnella among others. While it is apparent that Plesiomonas is not a true member of the Vibrionaceae, it is less clear whether this taxon should permanently reside within the enteric group. Most 5S and 16S rRNA phylogenetic investigations have placed plesiomonads at the periphery of the dendritic tree, with their nearest neighbors either the Proteeae (17, 18, 19, 30), Hafnia (20), or Photorhabdus (31). However, when other housekeeping genes are looked at individually or as part of concatenated sequences of five loci, P. shigelloides not only appears to be deep rooted within the Enterobacteriaceae but also aligned closer to the Hafnia-Obseumbacterium group (32) or Edwardsiella tarda (21). One 2003 study in fact found Plesiomonas branching from Vibrio cholerae rather than E. coli in an analysis of gyrB gene sequences (33). These discrepancies may arise from the limited number of enteric species analyzed in some studies, the number of loci sequenced, or analytical methodologies employed. However, it presently appears from most studies that plesiomonads assume a phylogenetic position at the periphery or margin of the family tree relative to core members (Table 1).
In addition to the controversial phylogenetic data, DDH studies have found that P. shigelloides displays low-level relatedness to the type strain of E. coli and nine other enteric strains (26). However, this same level of relatedness (8%) is also found with Aeromonas species and only slightly less with Vibrio cholerae (7%) and other vibrios (26). These data led Ruimy (20) to suggest that perhaps this taxon should be placed in its own family, the Plesiomonadaceae, a position with which others are in disagreement (21). It seems logical, however, that until the exact phylogenetic position of plesiomonads is established or confirmed by other independent groups, it is difficult to determine precisely where it resides. Even if it is found to reside on a peripheral branch of the Enterobacteriaceae, it seems untenable at present to consider placing it in its own family, given the single-species status of the genus and the lack of phylogenetic depth this situation would create.
For additional information on the taxonomy, nomenclature, and classification of plesiomonads, the reader is suggested to consult the reviews of Ewing, Hugh, and Johnson (12), Farmer, Arduino, and Hickman-Brenner (11), Janda (13), McNeely et al. (34), and Miller and Koburger (35).
ENVIRONMENTAL DISTRIBUTION AND ECOLOGIC ASSOCIATIONS
A number of Plesiomonas physiologic characteristics partially dictate the environmental distribution and ecology for this species. Plesiomonads are mesophiles with growth temperatures ranging between 8°C and 45°C (11, 13). The optimal temperature for growth for most strains occurs between 35°C and 39°C, with maximal temperatures being 40°C to 45°C. One early study described a psychrophilic strain of P. (Aeromonas) shigelloides (CDC882-69) with a minimal growth temperature of 0°C, but this seems to be an aberrant finding for the group as a whole (36). Few if any strains routinely grow below 10°C (35, 37), although a number of Plesiomonas isolates were recovered from Lake Vettasjärvi in the Arctic region of Sweden, which is north of the Polar Circle (38). The recorded water temperature there was 9°C with a pH of 6.5.
In addition to temperature characteristics, P. shigelloides primarily grows at pH ranges between 4.5 and 9 and can grow in salinities of 0% to 4% (11, 13, 35, 37). Salt concentrations exceeding 4% are problematic. Although one review (34) listed soil as a common reservoir for P. shigelloides, the major habitat for this species is water and inhabitants of such ecosystems, including fish, shellfish, and crustaceans, water fowl, marine mammals, amphibians, reptiles, and other vertebrates (28). Only soils associated with aquatic environs, such as sediments, are usually positive for P. shigelloides (35).
Aquatic Environments
Based upon the physiologic characteristics listed above, and in particular salinity, it is predicted that the primary aquatic habitats of P. shigelloides include freshwater sources (<0.5% NaCl; 0 to 5 ppt) and brackish or estuary waters (0.05% to 3%; 5 to 25 ppt), as opposed to saline/brine offshore marine environments (>4%; 35 ppt). This is exactly the case, and most studies documenting plesiomonads in these ecosystems have reported positive findings from streams, rivers, and lakes, in contrast to seawater. Unfortunately, definitive studies on the prevalence, distribution, and concentration of plesiomonads in these aquatic systems are not available, in contrast to studies for Aeromonas, for which surveys have been conducted by Hazen and others (39) of 147 natural aquatic habitats in the United States. How pH, temperature, turbidity, conductivity, and salinity affect P. shigelloides concentrations in freshwater sources is presently unknown.
Farmer et al. (11) described a limited number of studies where Plesiomonas concentrations ranged from 10 CFU/100 ml in one German river to 10,000 CFU/100 ml in a Florida estuary. Furthermore, Miller and Koburger (35) listed several early studies where plesiomonads were recovered from a variety of water sources. Over a quarter of a century ago, Arai et al. (40) analyzed 350 water samples from the Tama River using a membrane filter technique and Salmonella-Shigella and deoxycholate-hydrogen sulfide-lactose agars and recovered P. shigelloides from 8.9% to 22.4% of sampling sites as well as from 10.5% of river bed sludge samples. Plesiomonads were only recovered during the warmer months of the year in that study.
Few studies of the prevalence or concentration of Plesiomonas in freshwater samples have been published since 2000. A 2000 Brazilian study surveyed the Cambé Stream and found 7.1% of 70 sampling sites positive for P. shigelloides (41). Those authors used both MacConkey agar and a selective medium (inositol brilliant green-bile salts agar) to recover plesiomonads. Interestingly, one positive site near the stream source suggested this organism could be isolated from nonpolluted waters. A subsequent multinational study using a 23S rRNA probe recovered multiple strains of P. shigelloides from lake, river, and sewage waters in Slovakia and from river water in Sweden (42). An investigation of the Nilufer Stream in Bursa, Turkey, which employed membrane filtration and Plesiomonas isolation agar found 30 of 36 samples (83%) positive for plesiomonas, with geometric mean counts ranging from 64 to 330/100 ml (43). Higher concentrations correlated with fecal pollution, where Escherichia coli geometric means varied from 104 to 107/100 ml. Other freshwater sites have also yielded P. shigelloides, such as rearing waters for aquafarming systems for fish. One such study found 6.6% of such water samples positive for Plesiomonas (44).
Natural disasters.
Plesiomonas has not yet been associated with post-natural disaster infections, as has Aeromonas (45). However, indirect evidence suggests that plesiomonads could be involved in illnesses after major natural aquatic disasters. Kanungo and others (46) found P. shigelloides to be present in two hand pumps after a tsunami hit southern India in 2004. A decomposing cadaver was reported to harbor P. shigelloides in multiple internal organs after a powerful typhoon (47). These preliminary results suggest that this species should be entertained as a possible human pathogen subsequent to major water-related natural disasters.
Invertebrate and Vertebrate Hosts
In an excellent review in 2000, Jagger (28) provided a detailed and comprehensive overview of the isolation of P. shigelloides from mammals, marine mammals, fish, shellfish and crustaceans, reptiles and amphibian, and birds. Several points were noteworthy from this article. First, much of our current knowledge on the association of plesiomonads with animals comes from a limited number of older publications, including one study from the Antwerp Zoo (48). Thus, the isolation of plesiomonads from various species is limited in many instances to a single citation from one of four or five older studies. Second, few studies have looked specifically at the frequency and concentration of this taxon in various animal species, a finding that parallels the aquatic ecosystems mentioned above. What animal species Plesiomonas predominates in under natural conditions is for the most part unknown. Finally there is a paucity of information regarding P. shigelloides and zoonotic disease associations, with the exception of certain piscine species. How often this species causes infection in nonhuman vertebrates or other animals is again not well appreciated.
Mollusks and crustaceans.
Studies prior to 2000 occasionally reported the presence of plesiomonads in a number of different shellfish, including clams, oysters, shrimp, and crab (28, 35). However, surprisingly little additional information has accumulated over the past 15 years on this topic. One Australian study looked at the concentration of a number of bacterial species, including Plesiomonas, in wild and cultured banana prawns, Penaeus merguiensis (49). They found very low levels (<1 CFU/g [wet weight]) of plesiomonads in the gut of either group. A second California-based investigation found 4% of surf and bay mussels in the dry season and 0% in the wet season yielded P. shigelloides (50).
Fish.
Plesiomonads have been isolated from fish on multiple occasions. Jagger (28) listed over 15 different species or groups of fish from which P. shigelloides had been recovered. Virtually all of these isolations involved freshwater piscine species.
(i) Saltwater fish.
Other than for herring (sardines), there have been few reports on the prevalence or distribution of Plesiomonas in saltwater species. Using conventional as well as PCR-based methodologies, Herrera et al. (51) sampled a number of prepacked varieties of saltwater fish from two markets. They detected P. shigelloides in 23% of marine fish lots, with grouper fillets being the most common fish yielding plesiomonads; one halibut sample was also positive. Catadromous fish (such as Japanese eels) harbor Plesiomonas as well (52). P. shigelloides represented 2.2% of 183 bacterial isolates recovered from 621 farm-cultured sick eels in 26 Korean eel farms over a 7-year period. Higher proportions (5.6%) of plesiomonads were recovered from 216 healthy eels in the same study. Associated water samples from fish-rearing activities rarely yielded plesiomonads (0.8%).
(ii) Freshwater fish.
A large number of freshwater species harbor plesiomonads. In addition to those fish cited in a previous review (28), recent investigations have isolated Plesiomonas from rainbow trout (53, 54), carp (55), and tilapia (44, 56, 57). Studies of the intestinal tracts of a number of freshwater fish suggest that the genus Plesiomonas is one of the most common species composing the bacterial microbiota of these vertebrates, in addition to Fusobacterium and Aeromonas (58, 59). An Auburn University study pooled DNA samples from the intestinal contents of three commercial freshwater species and subjected these samples to 16S rRNA gene pyrosequencing (58). Of over 58,000 bacterial sequences generated and 311 operational taxonomic units identified, Plesiomonas accounted for 7.64%, 2.84%, and 0.39% (relative abundance) of the bacterial sequences present in largemouth bass (Micropterus salmoides), bluegill (Lepomis macrochirus), and channel catfish (Ictalurus punctatus), respectively (58). Aquaculture-raised fish for commercial purposes appear to be strongly associated with the presence of P. shigelloides in several studies. One survey found plesiomonad concentrations ranging from 1013 to 1016 CFU/g in the muscle or digestive contents of red hybrid tilapia (56). In another, Plesiomonas was a common pathogen in the gills and intestine of tilapia placed in earthen ponds in the Philippines as well as in rearing waters (6.6% of isolates) and pond sediment (44).
(iii) Ornamental aquaria and aquariums.
Aquaria appear to be a potentially rich source of plesiomonads, although very limited data currently exist linking this source to human infections (60). Ornamental fish from which P. shigelloides has previously been isolated include firemouths, oscars, swordtails, barbs, gouramis, and platys (60). Using next-generation high-throughput amplicon sequencing of 16S rRNA hypervariable regions, Smith and coworkers (61) identified Plesiomonas in the water of two ornamental aquarium fish, namely, goldfish and algae eaters. Other aquarium fish found to contain plesiomonads include an Asian arowana and cichlids (62, 63).
Marine mammals.
Plesiomonas is often found in association with aquatic mammals, including cetaceans, pinnipeds, and sea otters (64, 65, 66). Of the more than 35 species of dolphins known to exist, recent publications have recovered plesiomonds from bottlenose dolphins (Tursiops truncates), La Plata dolphins (Ponttopria blainvillei), and costero or Guiana dolphins (Sotalia guianensis). P. shigelloides has been identified as the most common or one of the most common microbial species recovered from bottlenose dolphins (66, 67). Common sites of isolation from this mammal include anus (29%), blowholes (21.1%), and gastric fluid (11.4%) (68). Plesiomonas has also been recovered from the feces of 5.3% of live and dead California sea otters (65) and the mouth of one South American sea lion (65). Freshwater runoff and coastal urbanization may play roles in the frequency of isolation of this organism from coastal marine mammals.
Waterfowl.
Aquatic birds that nest along shorelines, inlets, and cliffs or that prey off marine life for food or develop colonies on islands are often colonized with P. shigelloides. One of the most common groups found to harbor plesiomonads are cormorants. Studies of the pharynx, cloaca, or feces of great or black cormorants have found positivity rates ranging from 47.8% to 74.4% for these birds (69, 70). Plesiomonas has also been recovered from 3.9% to 7.4% of ring-billed and herring gull feces on bathing beaches located on southwestern Lake Michigan (71). Other avian species reported to yield P. shigelloides include whooper swan, black stork, goldeneyes, herons, and penguins (72, 73, 74).
Other animals.
A number of reports listing the sources of Plesiomonas strains have occasionally reported isolations from a number of animal species. Detailed investigations of their relative frequencies in such hosts have not been published. These taxa include reptiles (alligator, lizard), cats, dogs, hares, red wolf, foxes, and a roach (42, 72, 74, 75, 76). Table 2 lists a compilation of animal species from which Plesiomonas has been recovered since 2000.
TABLE 2.
Recent animal isolations of Plesiomonas
| Category | Specific organismsa |
|---|---|
| Invertebrates | Mussels, prawns |
| Fish | Algae eaters, arowana (Asian), bass (largemouth), bluegills, catfish (channel), carp (grass), cichlids, goldfish, grouper, halibut, eel (Japanese), roaches, tilapia, trout (rainbow) |
| Marine mammals | Dolphins, sea lions (South American), sea otters (CA) |
| Waterfowl | Cormorants, goldeneyes, gulls, heron, penguins, storks (black), swans (whooper) |
| Other | Alligators, cats, dogs, foxes, hares, lizards, wolves |
Isolations reported since 2000. Italics indicate species with multiple reports, suggesting that plesiomonads are common inhabitants. Boldface indicates species associated with sporadic or outbreak disease.
Animal Infections
Little data exist at present that can clearly separate Plesiomonas infections in animals into either enzootic or epizootic categories. Plesiomonads have been recovered from a wide array of animals that were healthy, diseased, or under autopsy conditions, yet there is little pathological, microbiological, or clinical evidence to support colonization versus infection status in these species. Rather, current data suggest that plesiomonads can cause sporadic infections in some animals and outbreak situations in others. Gál and others (75) described a case of chronic inflammation and abscess formation in a young male lizard housed at the Budapest Zoo and Botanical Gardens. The lizard died after only being in captivity for 1 month. P. shigelloides was isolated from its liver. P. shigelloides has also been found to be associated with muscle erosive disease in grass carps (55) and die-off in aquarium fish such as cichlids (63).
Plesiomonas shigelloides was previously linked to epizootic disease involving the kidneys and livers of rainbow trout in Portugal in 1984 (77) and resulted in a high mortality rate. Rapidly rising water temperature coupled with large amounts of organic matter were postulated to have contributed to this outbreak. An outbreak of septicemia in King and African penguins at the Basel Zoo has also been traced to Plesiomonas (73). P. shigelloides was recovered in pure culture from various internal organs (liver, kidney, small intestine) in two of three fatally infected chicks. We have additionally worked on another outbreak of invasive disease in a colony of rockhopper penguins located at a Midwestern zoo. It may be that outbreak disease in these instances is linked to stress brought on by one of a number of factors, such as overcrowding, oxygen levels, temperature, and climatic conditions, as well as food sources. Table 2 highlights species implicated in disease caused by plesiomonads.
EPIDEMIOLOGY OF HUMAN INFECTIONS
Our knowledge regarding the association of P. shigelloides with human infections is, at present, primarily restricted to the diarrheal disease state, as most epidemiologic data come from studies or investigations conducted on general causes of bacterial gastroenteritis or more specifically P. shigelloides. For extraintestinal manifestations, which are much less common than enteritis, epidemiologic information is limited to a smaller number of individual case reports or series of systemic infections that include a variety of maladies, such as bacteremia, peritonitis, and hepatobiliary disease.
Geographic Distribution and Seasonal Variation
P. shigelloides is a global pathogen and has a worldwide distribution, with the possible exception of the polar caps. In addition to those geographic locales previously identified as yielding plesiomonads from clinical sources in several reviews (28, 35), recent studies have documented cases or series of illnesses from Bangladesh, China, Ecuador, Germany, Hong Kong, Nigeria, Romania, Taiwan, Senegal, South Africa, Taiwan, and Thailand. Infections have a seasonal aspect to them, as most reported cases of disease occur during the warmer months of the year when freshwater temperatures rise, allowing for increased proliferation of plesiomonads via sewage contamination (78).
Some vertebrates, including domesticated pets such as healthy cats (3.8% to 10.3%) and dogs (3.8%), appear to be often colonized with Plesiomonas and excrete the bacteria in their feces (28, 40). However, this is not the case with regard to humans, as P. shigelloides is not considered to be part of the normal commensal flora of the human gastrointestinal tract. One 2010 study found a very high carriage rate of plesiomonads (11%) in healthy young children between the ages of 3 months and 5 years (79). However, this study was conducted in a remote locale of western Thailand and is probably not reflective of many other geographic locations, including urbanized centers. Cumulative data from many different surveys over the past 40 years suggest that the asymptomatic carrier rate for P. shigelloides in humans typically ranges from <0.1% to 0.3% and rarely exceeds 0.5% except under special circumstances (8, 40).
There are practically no data available by means of conventional methodologies for the continental United States regarding either the incidence or prevalence of P. shigelloides gastroenteritis in various patient populations. Most indirect data suggest that the incidence of Plesiomonas-associated enteritis in the United States is much lower than 1%, although newer molecular methods developed in the past few years could potentially alter these conclusions. In contrast, many international studies have found Plesiomonas to be a common enteric pathogen in select patient populations. The frequency of plesiomonads recovered as enteric pathogens in diarrheal stools in these settings ranges from 2% to >10% (79, 80, 81, 82, 83, 84, 85, 86), with the exception of one retrospective study which found the prevalence of P. shigelloides in inpatients with gastroenteritis from 2001 to 2012 in southeast China to be 0.009% (84). Most of these surveys have originated from tropical or subtropical regions of the world where warmer temperatures may allow for the persistence of plesiomonads in higher numbers in freshwater sources, leading to a greater infectivity rate. O'Ryan et al. (87) suggested that the higher incidence of P. shigelloides diarrheal illness in developing countries may also be related to substandard environmental sanitation conditions compared to those of industrialized nations. Support for this concept comes from army field training exercises in China (88), where the lack of personal hygiene and drinking raw water contributed to a high infectivity rate in the military (16.5%). Additionally, another Southeast Asia report described a sizeable reduction in the frequency of stool pathogens among food handlers that occurred after a continuing education program concerning food and personal hygiene was introduced (89). However, unlike Vibrio parahaemolyticus, Salmonella, and Aeromonas, plesiomonad numbers were not reduced in this exercise (17.1% before, 58.6% after) after completion of the educational program.
Case-Controlled Investigations
Table 3 summarizes four case-controlled investigations conducted on Plesiomonas-associated diarrhea published since 2000. On the surface, three of these four studies suggest that Plesiomonas is more often found in association with the diarrheal disease state rather than colonization of healthy individuals. Furthermore, in two of these studies no plesiomonads were recovered from the feces of almost 1,000 asymptomatic persons (84, 85). Still, these studies are not without issues. An Ecuadorian study found that when only single infections with P. shigelloides were considered, case prevalence rates dropped from 11.4% to <5%, which was less than the community prevalence rate (82). Those authors suggested that the pathogenicity of this organism, while apparently low, might be linked to concurrent infection with another enteric pathogen(s). Other studies have also found high isolation rates of Plesiomonas with other enteric pathogens (85). Similar to these data, a Thai study found plesiomonads in higher concentrations in controls (11%) than in cases (10%) (79). It should be pointed out that in the latter study, in which children between the ages of 3 months and 5 years were screened, a number of recognized pathogens (Campylobacter, Salmonella, and several pathogenic Escherichia coli groups) were also not found to be significantly associated with symptomatic children. The overall environmental burden of plesiomonads in this rural community may be high, and local immunity could impact such prevalence studies.
TABLE 3.
Case-controlled studies of Plesiomonas-associated gastroenteritis
| Country | Dates of survey | Population | % (no. tested) in disease group |
Referernce | |
|---|---|---|---|---|---|
| Ill | Asymptomatic | ||||
| Ecuador | 2004–2008 | All ages | 11.8 (775) | 7.2 (2,161) | 82 |
| Thailand | 2001–2002 | Children (<5 yrs) | 10 (236) | 11 (236) | 79 |
| Nigeria | 2012–2013 | All ages | 7.2 (712) | 0 (500) | 85 |
| China | 2010–2012 | All ages | 2.9 (3,536) | 0 (478) | 84 |
Outbreaks
One of the troubling aspects of Aeromonas gastroenteritis has been the inability to document a clear-cut association between outbreaks of diarrheal disease that are unquestionably epidemiologically linked to it (90). The demonstration of such species-specific outbreaks is a defining character or trait of recognized enteropathogens. In the case of Plesiomonas, much more credible evidence of outbreaks is now available.
Table 4 summarizes 11 P. shigelloides-associated outbreaks either formally published or mentioned in the scientific literature (35, 91, 92, 93, 94, 95, 96, 97, 98). Of these, 11 outbreaks, including at least 2 from Japan, had very strong microbiological as well as epidemiological evidence supporting the enteropathogenicity of this bacterium (91, 93). These data include descriptions of a predominant strain isolated from multiple ill persons epidemiologically linked by serogroup (both O17) or serotype to the same strain recovered from the implicated food or water sources causing the outbreak. In addition, the majority of P. shigelloides-associated outbreaks in Table 4 had the following characteristics: (i) plesiomonads were recovered as the predominant growth or in pure culture from the feces of ill persons, (ii) an implicated source of infection, such as water or seafood, was compatible with known environmental distributions and exposures to P. shigelloides, (iii) a high attack rate (15% to 69%) allowed for the recovery of multiple strains of Plesiomonas in 7 of the 11 outbreak-associated events, and (iv) plesiomonads were recovered as the sole or in most cases the only enteric pathogen from stools of sick individuals. These cumulative data strongly implicate P. shigelloides as an enteropathogen. Furthermore, a recent retrospective analysis of foodborne disease outbreaks in China between the years 1994 and 2005 identified four other events (outbreaks) and 227 total cases associated with P. shigelloides (99). These outbreaks included two food service units and one family event.
TABLE 4.
Outbreaks of Plesiomonas-associated gastroenteritis
| Yr | Location | Event | Attack ratea | Source | No. of strains | Serogroup | Strength of evidenceb | Reference |
|---|---|---|---|---|---|---|---|---|
| 1961 | Japan | Mill | 275/870 (31.6) | Cuttlefish salad | 88c | O:17d | A | 91 |
| 1965 | Japan | Community | 53/355 (14.9) | Salt mackerel | 10 | (−)e | B | 92 |
| 1973 | Japan | Youth center | 878/2,141 (45.7) | Tap water | 21 | O17:H2f | A | 93 |
| O22:H3 | ||||||||
| O8:H5 | ||||||||
| 1974 | Japan | Bus tour | 24/35 (68.5) | Unknown | 3 | O24:H5 | B | 93 |
| 1980 | USA (NC) | Oyster roast | 36/102 (35.2) | Oysters | 1 | C | 94 | |
| 1982 | Mexico | Vacation | Unknown | Chicken | 2f,g | O:17d | C | 95 |
| 1983 | USA (FL) | Not provided | 29/?h | Shellfish | D | 35 | ||
| 1983 | USA (FL) | Not provided | 29/? | Shellfish | D | 35 | ||
| 1990 | Netherlands | Recreational area | ? | Freshwater | 9 | D | 96 | |
| 1996 | USA (NY) | Party | 60/98 (57) | Potato salad | 11i | B | 97 | |
| Well water | ||||||||
| 2003 | Cameroon | Party | 49/78 (62.8) | Cold fish | 17j | B | 98 | |
| Ndolé |
The attack rate was based upon the number of ill people divided by the total number of people present at the event and assuming that Plesiomonas was the single infective agent. Values in parentheses are percentages.
Strength of evidence: A, multiple epidemiologically linked P. shigelloides strains isolated from outbreak and also recovered from implicated food; B, multiple epidemiologically linked P. shigelloides strains isolated from outbreak but not recovered from implicated food source; C, limited number of strains recovered from outbreak not linked to food source; D, primarily anecdotal information regarding an unpublished outbreak.
An unnamed halophilic organism was isolated from 41% of patients.
Based upon cross-reactivity with Shigella sonnei antiserum.
All strains agglutinated with Shigella dysenteriae 7.
Predominant serotype in the outbreak.
Giardia lamblia was detected in the feces of one of these two persons.
A total of 29 cases were reported from two outbreaks over a 6-day period.
Two of 11 patients from whom Plesiomonas was isolated also had Salmonella recovered from their stools.
One of 17 patients from whom Plesiomonas was isolated also had Salmonella recovered from their stools.
Risk Factors Associated with Plesiomonas Infections
Given the current prevalence of plesiomonad-associated illnesses, it is particularly difficult to determine what social, geographic, or medical factors may predispose persons to Plesiomonas infection. Much of our limited knowledge in this area comes from multiple case reports describing a common underlying risk factor that appeared associated with Plesiomonas illness. In a few other instances, medical centers or institutions retrospectively looked at a series of infections over a number of years and reported one or more risk factors associated with either P. shigelloides intestinal or extraintestinal disease. Finally, national public health investigations have occasionally linked plesiomonad disease to specific risk factors.
Gastroenteritis.
The cardinal publication concerning potential risk factors associated with Plesiomonas gastroenteritis is now 30 years old. A 1986 CDC study reported that 28 strains of P. shigelloides submitted from children and adults by 13 state health department laboratories were recovered in large numbers from stools and without any other concurrent intestinal infections (100). The isolation of Plesiomonas in this national epidemiologic survey was strongly associated with the consumption of uncooked shellfish, most notably oysters and to a much lesser extent raw shrimp (100). Thirteen of 14 patients in this study apparently ate oysters originating from the Gulf of Mexico or the Caribbean. In other reports, Plesiomonas has been linked to two outbreaks of diarrhea involving roasted oysters, including one previously cited episode (see Table 4) (94) and an unpublished Canadian outbreak involving 18 persons (101). Epidemiologic data provided from several large-scale surveys of Plesiomonas diarrhea from Southeast Asia report that between 5% and 15.4% of patients had a recent history of consuming seafood prior to their gastrointestinal disturbances (80, 84, 102). Another recent case report identified porridge containing undercooked fish as the most likely source of a woman's illness (103).
Foreign travel is a second major risk factor associated with plesiomonad gastroenteritis. In addition to seafood consumption, CDC found foreign travel linked to the isolation of Plesiomonas for 7 of 9 patients traveling to Mexico (100). A noteworthy study from Japan identified over 1,000 returning travelers through Kansai Airport with Plesiomonas-associated enteritis (104). The most common destinations travelers returned from included Thailand, Indonesia, and Vietnam. It should also be pointed out that a fairly high rate of coinfection (20.5%) was observed in this same patient population. A review of 51 published reports on travelers' diarrhea found Plesiomonas was associated with between 1.3% and 5.41% of all episodes, depending upon geographic locale (105). The lowest frequencies were seen in cases originating from Latin America and the Caribbean, while higher numbers were noted in South and Southeast Asia (105).
Persons in immunocompromised states, in particular those who are HIV positive, may be more prone to developing Plesiomonas diarrhea than healthy individuals (29). This is supported by a relatively higher prevalence rate (4.9% to 16.6%) of Plesiomonas gastroenteritis in HIV-positive or AIDS patients in comparison to frequencies of diarrhea seen in unselected patient populations (106, 107). However, in one of these two studies Plesiomonas was more prevalent in non-AIDS patients (11.7%) than in those with AIDS (4.9%) (106). Some studies have found the highest frequency (33% to 35%) of plesiomonad enteritis in infants 19 to 31 months of age (85) and in children under 2 years of age (80), suggesting a naive immunologic system as a potentially predisposing factor in young children for acquiring Plesiomonas, although a case-controlled study in a remote part of Thailand found P. shigelloides in high percentages (10% to 11%) in both cases and controls (79).
In addition to these conditions, it is apparent that the consumption of untreated water or freshwater sources in nations with low socioeconomic status or poor hygiene conditions puts anyone at risk of acquiring Plesiomonas diarrhea (Table 4) (28, 35). These facts parallel the seasonality of the disease and its association with temperate and tropical/subtropical climates, where the prevalence of the general disease appears much higher (28). Elevated aquatic temperatures in lakes and rivers may lead to the multiplication of this species (29) and higher surface concentrations.
Extraintestinal disease.
Identification of potential risk factors associated with extraintestinal P. shigelloides infections is even more difficult, given the limited number of published cases in comparison to those associated with gastrointestinal disease. The most common of these extraintestinal syndromes is bacteremia, where only 40 or so instances have been recorded in which Plesiomonas was isolated as the sole or copathogen in a case of sepsis. Woo et al. (108) compared 7 cases of Plesiomonas occurring in their institution over a 9-year period to 31 other cases reported in the literature. They found that bacteremias in their medical center were significantly associated with advancing age (>75 years), underlying biliary tract disease, acute cholangitis, and polymicrobic sepsis, compared to previous cases in the literature (108). Since many of these illnesses were polymicrobic in nature, it is difficult to ascertain how many of these risk factors can be directly associated with Plesiomonas.
Most reports involving Plesiomonas septicemia occur in persons suffering from one or more underlying medical illnesses leading to an immunocompromised state. In addition, secondary medical sequelae may result as a direct consequence of these primary medical conditions, which further increases the risk of invasive disease. Many cases of Plesiomonas sepsis are observed in individuals with multiple risk factors that may either individually or collectively be associated with plesiomonads. However, these risk factors can also be less species specific and more a reflection of Gram-negative bacteremia.
The pattern of developing Plesiomonas sepsis parallels that of gastroenteritis in the sense that most infections appear to result from ingestion of seafood or contaminated water, particularly in persons living in tropical, subtropical, or temperate regions (28, 29, 35). Conditions proposed to be associated with plesiomonad sepsis in addition to biliary disease include cancer, cirrhosis, HIV, and bloodborne dyscrasias, such as sickle cell anemia and thalassemia (4, 34, 84, 108, 109, 110, 111, 112). The latter diseases can sometimes lead to other conditions sporadically associated with P. shigelloides bacteremia, including splenectomy (108, 111) and hemochromatosis (108, 113). Approximately a dozen cases of perinatal bacteremia with central nervous system (CNS) involvement have been reported to date. In several cases the mother had diarrhea immediately prior to delivery and in a couple of cases Plesiomonas was recovered from maternal feces. This suggests vertical transmission from mother to child during the birth process (114).
A number of rare Plesiomonas infections result from unapparent injuries or traumas associated with water environs. These events include swimming in seawater or penetrating traumas to the head connected to diving or submerged projectiles (115, 116, 117).
Disease Transmission
The exact mode of disease transmission from natural reservoirs to humans, eliciting a variety of illnesses, is still speculative. Jagger (28) proposed a dual-schematic scenario for the apparently high frequency of Plesiomonas colonizing/infecting the guts of cats and for acquisition of these same bacteria by humans from environmental sources. In the former category, natural aquatic habitats and secondary reservoirs (amphibians, fish and shellfish) serve as potential sources for the transmission of plesiomonads directly to cats or indirectly through birds living near aquatic ecosystems who have fed on freshwater fish. Subsequent consumption of bird carcasses or fresh fish by cats could also lead to colonization. In the case of humans, the same two reservoirs (contaminated water, fish and shellfish) would serve as vehicles for ingestion of adulterated food or water containing plesiomonads. A cycle of reintroducing P. shigelloides into natural habitats could also occur through sewage contamination (28).
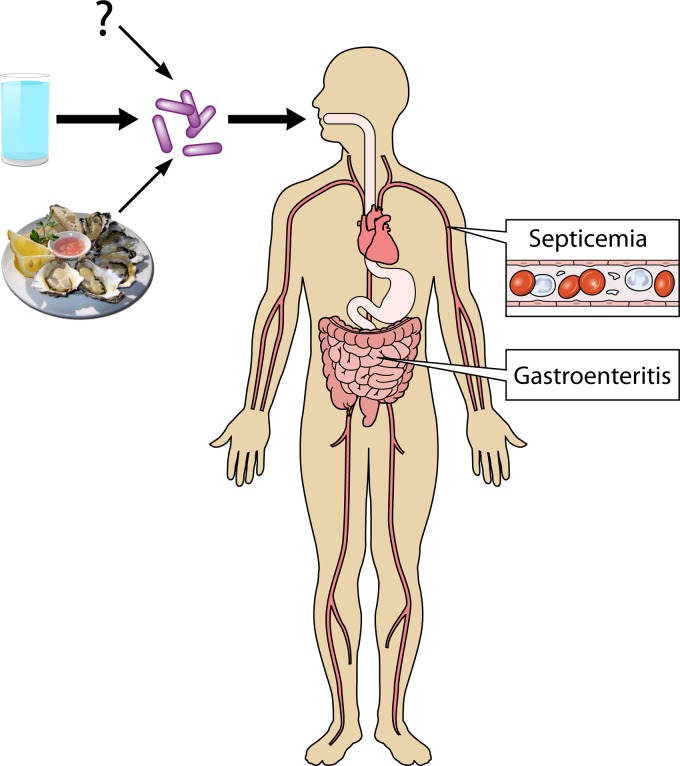
Figure 1 represents a diagrammatic flow chart concerning the potential acquisition and transmission of P. shigelloides to humans. Best available evidence suggests that the main avenue of transmission of plesiomonads from the environment to persons is via freshwater sources. Higher rates observed for cases of gastroenteritis in the Far East and other locales could be related to the greater proliferation of this species in warmer climates or socioeconomic factors or sanitary/hygiene conditions in developing countries. A second mode of acquisition involves consumption of food sources intimately linked to aquatic habitats. Ironically, these two sources together do not represent the majority of cases of illness linked to this species in epidemiologic investigations. In a CDC study by Holmberg et al. (100), 21 of 31 (68%) patients had a history of seafood consumption or foreign travel immediately preceding their diarrheal episode. More recent studies have reported even lower percentages. A Hong Kong study of 167 cases of Plesiomonas gastroenteritis found only 36% of individuals with a history of foreign travel or consumption of seafood or uncooked food (102). Similar studies from Taiwan reported cumulative values of 11% (80). The question mark in Fig. 1 suggests that there may be one or more unrecognized sources of acquisition that have not as yet been identified. Alternatively, since epidemiologic studies typically do not include questionnaires regarding water consumption (except in the case of a recognized outbreak), it may well be that the majority of unidentified sources of infection are in fact water related. Further environmental sampling studies on the frequency of plesiomonads in various aquatic environments in regions of the world where P. shigelloides disease is more common are needed.
FIG 1.
Diagrammatic representation of the main known routes of transmission and major disease manifestations associated with P. shigelloides infection.
CLINICAL INFECTIONS AND ASSOCIATED DISEASE SYNDROMES
P. shigelloides has been isolated on one or more occasions from virtually every common infectious disease syndrome or complication that has been previously reported. In practical terms, however, these infections or syndromes can be broken down into the following groups or categories, namely: (i) gastrointestinal infections or complications involving the small or large intestine, (ii) systemic disease manifested by frank septicemia with or without associated CNS complications, such as meningitis, and (iii) a myriad of miscellaneous infections, including intraabdominal disease, soft tissue and wound infections, and ocular illnesses.
Gastroenteritis and Diarrheal Disease-Related Syndromes
Although once a fairly controversial topic, Plesiomonas is nowadays considered an enteropathogen by most in the medical and scientific communities. While at one time it was rarely listed in leading publications on bacterial gastroenteritis as a pathogen, it now can be found in many authoritative reviews or references on the subject, along with more common agents such as Campylobacter, Salmonella, and Shigella (87, 118, 119, 120).
Plesiomonas enteritis can present in one of three forms: an acute secretory gastroenteritis (most common), bloody or dysenteric colitis, or chronic or persistent diarrhea of >14 days duration (121).There have been very few published case reports on this topic over the past 15 years (103). Most of our recent clinical knowledge on Plesiomonas gastroenteritis stems from prospective or retrospective studies on this syndrome from regions of the world where the prevalence of the disease is presumed to be much higher, since limited data from industrialized nations suggest that the infection rate is quite low (122, 123).
Table 5 presents cumulative data from several large retrospective or prospective investigations on P. shigelloides gastroenteritis in Southeast Asia and Africa. Plesiomonas gastroenteritis occurs in all age groups, with several studies documenting infections in infants as young as 22 days (80) to those >90 years of age (84, 102). Several reports indicate that the disease may be more common in children under 5 years of age (80, 83, 85, 102), suggesting that immunologic immaturity might be a predisposing factor to acquiring the illness. Plesiomonas has been found to be the third or fourth ranking cause of gastroenteritis in Nigeria (85) and China (84), respectively. Mixed infections are fairly frequent (16% to 28%), with common coinfecting pathogens including Salmonella, Aeromonas, and V. parahaemolyticus (80, 84, 85, 102). The association of the latter two species with Plesiomonas suggests contaminated water or seafood/fish as common vehicles of infection (80, 84).
TABLE 5.
Gastrointestinal symptomatology associated with P. shigelloides diarrhea
| Location | No. of cases | Symptoms (%)a |
Reference | ||||
|---|---|---|---|---|---|---|---|
| Diarrhea | Blood | Fever | Abdominal pain | Vomiting | |||
| Hong Kong | 167b | 99 | 25 | 51 | 72 | 38 | 102 |
| Taiwan | 111 | 96 | 45 | 51 | 45 | 41 | 80 |
| Bangladesh | 253 | 93 | 5 | 6 | 6 | 74 | 124 |
| China | 104 | 84 | 17 | 33 | 69 | 14 | 84 |
| Nigeria | 51 | 88 | 18 | 35 | 67 | 14 | 85 |
Percentage of cases or isolates.
Cumulative data (for all age groups), including single and mixed infections with P. shigelloides.
Secretory enteritis.
As previously noted, watery diarrhea is the most common clinical presentation of Plesiomonas gastroenteritis, with between 50% and 88% of all patients presenting with this type of diarrhea (8, 80, 124). Prominent symptoms associated with plesiomonad enteritis include watery diarrhea and abdominal pain (29, 85, 124) (Table 5). One study of adult patients with nonbloody P. shigelloides gastroenteritis seeking medical attention at an emergency department had a mean duration of diarrhea of 1.6 days with 10 bowel movements (bm) per day (125). Other surveys or case reports documented evacuation rates of 5 to >10 bm/day (102, 103, 124). Typical cases of untreated enteritis had a mean duration of symptoms of 11 days in one CDC study (100), while another report placed the range of symptoms from 1 day to 2 months (102). Duration of acute diarrhea in hospitalized patients in this second study was 2.2 days (102).
Watery gastroenteritis is most often observed in healthy persons (29, 80, 103). Wong et al. (102) found that almost three-fourths of the 167 persons studied with P. shigelloides diarrhea were healthy, with the remaining 25% with one or more underlying condition, including diabetes, renal disease, cirrhosis, and colon cancer. Typically, the disease is mild and self-limiting (8, 29). Klontz et al. (124) found that gastroenteritis induced by P. shigelloides was typically milder and less severe than that caused by either non-O1 V. cholerae or V. parahaemolyticus (124). Less common symptoms associated with a minority of cases include dehydration, hypokalemia, and peritonism (102).
Cholera-like diarrhea.
Although not well-documented or described, P. shigelloides has been implicated on several occasions as a rare cause of diarrhea resembling cholera, one of which occurred in a woman returning from a trip to Kenya (126, 127). Subsequently, a case from San Lazaro Hospital in the Philippines described a patient (age and sex unknown) with severe dehydration of 5-h duration and 25 bm/day (128). The stool had a “rice water” consistency with mucus, and P. shigelloides was isolated. However, the stool of this patient also contained V. cholerae and Trichuris spp. In 2002, a 53-year-old man in Djibouti was hospitalized because of severe dehydration secondary to a cholera-like illness of 1 day duration (129). He had passed 8 liquid stools of a greenish consistency each day. P. shigelloides was isolated in pure culture from his stool. Furthermore, the finding in some surveys that one or more patients experienced diarrheal episodes consisting of 30 bm/day suggests that this more severe form of secretory gastroenteritis may have been overlooked in the case of Plesiomonas (8, 102).
Dysentery.
Dysentery or infective colitis is the second most common form of Plesiomonas gastroenteritis. The syndrome is characterized by the macroscopic appearance of blood in stool accompanied by significant abdominal pain or tenderness and vomiting (Table 5). More severe disease in one study was associated with fever in 13.2% of cases (102). Mucus in stools is found in 17% to 19% of feces where P. shigelloides is the only pathogen (84, 85). Many persons with these symptoms are hospitalized with the presumptive diagnosis of bacillary dysentery.
The frequency of Plesiomonas acute dysentery in selected patient populations has been reviewed by Pfeiffer and others (123). It has ranged from a reported high of 25.1% in a combined survey of children and adults to 15.8% in children alone. Two studies conducted in the United States in the 1980s by the CDC (100) and the Texas Department of Health (130) placed the frequency of bloody diarrhea at 53% and 22%, respectively. Unfortunately, there have not been any individual case reports describing Plesiomonas dysentery over the last 15 years. One of the best-described cases of plesiomonad bloody diarrhea is that of a 42-year-old woman with persistent dysentery and histopathology indicating pseudomembranous colitis. P. shigelloides was recovered from her on two occasions in pure culture (131).
Chronic diarrhea.
P. shigelloides is an infrequent but recognized cause of chronic diarrhea (132). Wong and others (102) found 5% of their patients with chronic Plesiomonas disease had diarrhea of 2 weeks to 2 months duration. In a similar study from Taiwan, chronic diarrhea was seen in 23% of children and 12% of adults (80). Except for the lengthy duration of the illness, specific symptomatology associated with persistent gastroenteritis is poorly described.
Fatal gastrointestinal infections.
While not well appreciated, Plesiomonas gastroenteritis has been linked to a negative outcome on several instances, most of which involved invasive (bloody) diarrhea. A 19-year-old woman with bulimia nervosa who had been previously thrown into a water fountain developed unrelenting severe bloody diarrhea with dehydration of 9 days duration prior to her death (133). P. shigelloides and Aeromonas veronii biotype sobria was recovered from her stool. In 1989, Sinnott et al. (134) reported on a healthy 53-year-old woman who developed severe watery diarrhea that subsequently became bloody a day later after consuming sushi and raw oysters. After initial treatment she returned to the emergency room 24 h later hypotensive, and she subsequently expired. P. shigelloides was recovered on two separate occasions from her stool. Wong and colleagues (102) described two fatal infections associated with plesiomonad diarrhea. In one instance, an 82-year-old man with alcoholic cirrhosis presented with moderately severe diarrhea and mucus, dehydration, and hypokalemia. He died 3 days later (cause of death not mentioned). In a second case, a 68-year-old male with diabetes mellitus and end-stage renal disease had diarrhea of 1 day duration and turbid peritoneal fluid. Stool culture grew P. shigelloides, while the peritoneal fluid yielded Pseudomonas aeruginosa. Cause of death was multiorgan failure. In most of these cases, it was difficult to determine exactly to what extent Plesiomonas contributed to the person's eventual demise.
Unresolved controversies.
The conundrum concerning the evidence supporting the role of Plesiomonas in gastrointestinal disease is presented in Table 6 (5, 8, 100, 103, 131, 135). Since 2000, several case-controlled investigations have been published on the role of plesiomonads in diarrheal disease. However, of these four studies only two (84, 85) strongly supported a role for this species in gastroenteritis. This finding may not be as problematic as it first appears (Table 3) (79, 82, 84, 85). Since almost all of these studies were centered in developing nations located in tropical/subtropical climates, they all appeared to have a higher incidence of the disease. Thus, the opportunity for exposure and reexposure to plesiomonads from infancy to adulthood may be greater and thereby may be potentially protective to a certain segment of the population. This supposition is supported by the hypothesis that P. shigelloides O17 is common in surface waters in developing countries and may also have a protective effect against infection with Shigella sonnei by virtue of shared lipopolysaccharide antigens (136). Thus, there may be at least several subpopulations of residents in these locales who may be naive, partially protected, or protected from diarrheal infections produced by this enteropathogen, which has a relatively low pathogenic potential.
TABLE 6.
Outstanding issues concerning Plesiomonas gastroenteritis
| Type of study | Information from studies: |
|
|---|---|---|
| Before 2000 | Current | |
| Case reports | <10 well-described case reports or series of case reports | Only one case since 2000 (103) |
| Case-controlled investigations | One study through 2000 (5) | Four studies through 2000 (see Table 3); two with positive findings, one with negative findings, one with equivocal findings |
| Outbreaks | P. shigelloides not clearly shown to cause outbreak of gastroenteritis (5) | At least 11 outbreaks described, including 1 since 2000; several with good epidemiologic data (see Table 4); several more anecdotally described in the literature |
| Immune response | Data from a CDC study not supportive (100); several case reports more supportive (8, 131) | No new data |
| Histopathology | Good histopathology from a few selected case reports (131, 135) | No new data |
| Pathogenicity | No well-described enteropathogenic mechanisms (8) | Some now described but poorly characterized (see Pathogenicity section) |
In addition to this evidence there are a sizeable number of outbreaks described where P. shigelloides is a sole or copathogen in comparison to Aeromonas, where not a single credible outbreak has been attributed to this bacterium over the past 3 decades (90). What is needed are more case-controlled surveys and detailed case reports supported by clinical data, such as humoral immune responses (acute, convalescent) and recovery of the enteropathogen from pathological biopsy material, not simply accounts reporting isolation.
Septicemia and CNS Disease
Septicemia with or without CNS involvement is the primary clinical syndrome associated with extraintestinal P. shigelloides infection. Table 7 summarizes some retrospective reviews of cumulative data on cases of Plesiomonas sepsis reported in the English literature (29, 108, 137). Based upon the review of Woo (108) and several published case reports since 2005, there are now over 40 cases of P. shigelloides bacteremia in the literature (111, 114, 138, 139). This is almost double the number reported by Lee (137) in 1996.
TABLE 7.
Characteristics of Plesiomonas septicemia reported in the literature
| Characteristic | Value reported in study |
||
|---|---|---|---|
| Lee et al., 1996 (137) | Stock, 2004 (29) | Woo et al., 2005 (108) | |
| No. of cases reported | 21 | 24 | 38a |
| Mean age (yrs) | 22.7 | 28.8 | 38.4 |
| Age range | 1 day to 68 yrs | 2 days to 68 yrs | 1 day to 94 yrs |
| Male:female ratio | 2:1 | 2.7:1 | 1.8:1 |
| Immunocompromised (%) | 81 | 88 | 82 |
| Mortality rate (%) | 62 | 50 | 42 |
| Reference | 137 | 29 | 108 |
Includes 31 cases in the literature and 7 new cases described by the authors.
Plesiomonas sepsis is a community-acquired male-dominated disease almost invariably observed in persons with one or more underlying medical conditions leading to the immunocompromised state (Table 7). Common predisposing factors leading to sepsis in decreasing order of frequency include blood dyscrasias (thalassemia, sickle cell disease, leukemia), asplenia (functional or anatomic), and iron overload conditions, such as hemochromatosis, biliary tract disease, and cirrhosis (alcoholic, viral) (108, 111, 112). Slightly under one-third of all bacteremias occur in neonates with meningitis or meningoencephalitis (114). Almost all of these cases of infection occur within the first 5 days of life and appear to have been vertically transmitted (perinatal) rather than transplacental (114). Plesiomonas has only been recovered from maternal feces on three occasions. The mortality rate in neonates exceeds 50% (114).
About 90% of all cases of sepsis are monomicrobic. In almost half of these cases, plesiomonads are recovered from a site other than blood. These specimen sources include CSF, abscess/wound, joint fluid, pleural fluid, stool, and urine (108). The study of Woo et al. (108), which described 7 cases of Plesiomonas bacteremia, differed appreciably from the remaining body of cases in that these patients were older (median, 82 years), had biliary tract disease, and infections were more often polymicrobic. Reasons for these differences are not immediately apparent. While the source of most community-acquired infections in adults is unknown, contaminated water, raw seafood, or other foods are most often suspected (137). A case of sepsis originated in a 13-year-old girl with sickle cell who consumed many crab legs at a crab buffet 4 days prior to her admission (110). Another case of pleisomonad sepsis occurred in a 51-year-old man with liver disease that received a donor liver from a teenage boy who drowned in a freshwater lake (138). Blood cultures drawn from the donor prior to transplantation eventually yielded P. shigelloides. In another instance, a rapidly fatal case of combined Plesiomonas and Clostridium perfringens sepsis and meningitis occurred in a 71-year-old man who had just disembarked from an international flight (139). He had just returned from a hunting trip in the Swiss Alps and expired 3 h after initial presentation. Autopsy suggested death resulted from septic complications. Those authors suspected contaminated meat or water from his hunting trip as the source of infection.
Wound Infections
Unlike other aquatic pathogens, such as Aeromonas and Shewanella, there are virtually no published reports in the literature describing isolation of P. shigelloides from open or penetrating wounds, nor an association of this species with soft tissue infection (90, 140). The reason for this anomaly is presently unclear, although the lack of protease expression by plesiomonads may be a contributory factor. Two cases of cellulitis of the leg and foot, respectively, have been reported where cultures grew plesiomonads from tissue or exudate material (141, 142). On both occasions, other pathogenic bacteria were also isolated, bringing into question what contribution this microbe may have played in the infectious process. A more definitive role for Plesiomonas in bacterial cellulitis was described by Gopal and Burns (143) 20 years later in a 59-year-old man who sustained a penetrating trauma to his left hand with a knife while cleaning 20 or more freshwater fish. P. shigelloides was recovered not only from the surgical wound infection but also in pure culture from the initial blood culture. The most recent case of a wound infection caused by Plesiomonas involves a healthy 31-year-old diver who sustained a penetrating trauma to his skull when he hit submerged rocks in a pond while diving (116). He developed a frontal cutaneous abscess of the head as a result of the trauma, which required medical intervention including antimicrobial therapy.
These are currently no data available that suggest that Plesiomonas is ever an animal-related pathogen in the traditional sense of other zoonotic pathogens, including those associated with aquatic habitats. There are also no known reports of plesiomonads being associated with traumatic human wound infections acquired through penetrating injuries or bites induced by vertebrate or invertebrate species. In fact, Plesiomonas is not listed as a possible aquatic zoonotic pathogen in the broadest sense (144). Limited evidence suggests that the couple of cases of cellulitis reported are due to penetrating traumas that come in contact with material (fish, rocks) associated with freshwater environs. No aggressive wound infections, such as necrotizing fasciitis, have even been linked to the genus Plesiomonas.
Ocular Disease
There are surprisingly as many cases of Plesiomonas-associated eye infections in the literature as there are wound infections. Clinical presentations of eye disease range from endophthalmitis to keratitis (117). In three instances, the precipitating event resulted from a penetrating injury to the eye caused by a fishhook (two instances) or a rock from a creek bed (117, 145). One ocular infection in a neonate appeared to have been acquired transplacentally (146). In some cases, these devastating illnesses are solely caused by plesiomonads (117), while in other instances they are of a polymicrobic etiology involving other aquatic or marine bacteria, such as Aeromonas, Shewanella, or Vibrio (145).
Miscellaneous Infections
A number of infrequent to extremely rare complications resulting from P. shigelloides infection have been summarized in one or more reviews on the topic (27, 28, 29). These conditions include polyarthritis/osteomyelitis, cholangitis, cholecystitis, epididymoorchitis, peritonitis, pyosalpingitis, and pancreatic and splenic abscesses. Two cases of peritonitis associated with continuous ambulatory peritoneal dialysis in 62- and 73-year-old Chinese women were reported in 2004 (147). The source and portal of entry in both patients was unknown. In 2009, Schneider and associates (148) described a case of P. shigelloides pneumonia in a 76-year-old woman who had previously undergone a curative gastrectomy. X-rays and a computed tomography scan indicated a nodular lesion in the right upper lung and a cavernous lesion in the right upper lobe, respectively. Bronchial lavage yielded 105 CFU/ml; an equivalent number of E. coli was also recovered. The patient was successfully treated with multiple antimicrobial agents but suffered severe postinfectious complications, including theta coma.
Treatment
Most gastrointestinal infections caused by P. shigelloides are mild in nature and do not require antimicrobial intervention or other medical treatments. One large study of Plesiomonas diarrhea found 85% of such illnesses to be self-limiting (102). However, antimicrobial therapy or other medical interventions may be warranted for some cases of moderate to severe diarrhea or to counteract physiologic or metabolic issues arising from such illnesses. DuPont (119) recommended that for severe cases of dysentery-like diarrhea caused by P. shigelloides should be treated like shigellosis, namely, with azithromycin or ceftriaxone for children and ciprofloxacin or azithromycin for adults. Quinolone therapy for the treatment of more severe cases of plesiomonad enteritis has also been suggested by other authors (102, 103). Some earlier studies found that the duration of diarrhea (up to 10 days) could be appreciably shortened by intervention with tetracycline or trimethoprim-sulfamethoxazole (100). In cases where dehydration is clinically significant or the patient presents with cholera-like disease, the administration of oral rehydration salts or intravenous fluids may be appropriate alone or in combination with an antibiotic such as a quinolone (103, 125, 129).
Guidelines regarding the appropriate treatment of extraintestinal infections caused by Plesiomonas, of which septicemia is the most common, are difficult to determine because of a multiplicity of factors. While most illnesses are community acquired and monomicrobic, they occur so rarely that plesiomonad sepsis is almost never suspected as part of the initial diagnosis upon clinical presentation. Furthermore, most patients with Plesiomonas sepsis suffer from other serious underlying diseases, including sickle cell disease (110), thalassemia (111, 112), or HIV infection (149). The interval in time between each of these case reports is significant (four cases in the last 15 years), making comparisons in treatment regimens virtually impossible. Once P. shigelloides was identified, in each instance, antimicrobial regimens were switched to quinolones (ciprofloxacin), carbapenems (imipenem), or combinations thereof. A review of a dozen cases of neonatal plesiomonad sepsis accompanied by meningitis or menigoencephalitis indicated that all neonates, with one exception, received combination therapy (a two- or three-drug regimen) with such combinations, including a penicillin derivative, an aminoglycoside (most often gentamicin), cephalosporin (cefotaxime), or carbapenem (114). Again, because of the length in years separating publication of these single case reports, it is difficult to draw any firm conclusions on the effectiveness of directed treatment on patient outcomes. In a series of elderly persons with biliary disease and polymicrobic bacteremia, including P. shigelloides, four of five patients responded favorably to one of several drug regimens administered, including cefuroxime and cefotaxime (108).
PATHOGENICITY
While P. shigelloides has been associated with acute bacterial gastrointestinal illness and rare extraintestinal infections (112, 114, 150, 151), conflicting data on virulence factors and a lack of relevant animal models have stalled elucidation of the mechanisms involved in its pathogenicity. A variety of virulence factors have been associated with infections, including β-hemolysin (152), enterotoxins (153), cholera-like toxins (154), and a cytotoxin LPS complex (155). However, seminal work and the evolution of molecular biology have advanced our understanding of this organism's pathogenicity, particularly with respect to the structure of LPS, somatic antigenicity, cytotoxin activity, iron acquisition, and genetic diversity.
Lipopolysaccharide
The barrier properties of the outer membrane of Gram-negative bacteria are attributed to the LPS structure, which is highly immunogenic. Also referred to as an “endotoxin,” cell-bound LPS is associated with stimulation of severe pathological manifestations of immuno-activation, such as septic shock. As in other members of the Enterobacteriaceae family, the LPS in P. shigelloides is comprised of three domains which have significant biological activity and involvement in host-bacterium interactions. It includes an endotoxic glycolipid (lipid A), an O-polysaccharide (O-PS) or O-specific antigen, and an intervening core oligosaccharide (core-OS) region.
To date, LPS structures have been studied for 11 strains of P. shigelloides (Table 8) (156, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170). The majority of LPS regions studied in the Enterobacteriaceae that link the core to the lipid A moiety (KdoI) contain at least one residue of 3-deoxy-d-manno-oct-2-(ketodeoxyoctonic) acid (KdoI) (156, 159). In addition, they may possess l-glycero-d-manno-heptopyranose (L,D-Hep), the oligosaccharide L-α-d-Hep-(1→3)-l-α-d-Hep-(1→5)-[α-Kdo-(2→4)]-α-Kdo (D,D-Hep), or do not have a heptose (156, 159, 171). Among the Enterobacteriaceae, P. shigelloides is included in the Klebiella-Serratia-Proteus LPS group. This is based on the possession of the outer core disaccharide galactose A-glucosamine N and the substitution of β-glucose-HEPI, which is absent in the other major enteric LPS group, the Escherichia-Salmonella-Shigella LPS group (159). However, peculiar to P. shigelloides is the presence of β-galactose–HEPI as the substituted moiety instead of β-glucose–HEPI (157, 158, 159, 172).
TABLE 8.
Recent structural analytic studies of lipopolysaccharide in strains of Plesiomonas shigelloides
| Somatic antigen | Strain no.a | Reference(s) |
|---|---|---|
| O1 | 302-73 | 157, 158, 159 |
| O13 | CNCTC 80/89 | 160 |
| O17 | PCM 2231, 7-63 | 161, 162 |
| O24 | CNCTC 92/89 | 163 |
| O33 | CNCTC 34/89 | 156 |
| O37 | CNCTC 39/89 | 164 |
| O51 | CNTC 110/92 | 162 |
| O54 | CNTC 113/92 | 159, 165, 166, 167 |
| O74 | CNCTC 144/92, AM36565, 22074, 12254 | 168, 169, 170 |
CNCTC, Czech National Collection of Type Cultures (Prague, Czech Republic).
As is found in other Enterobacteriaceae, the chromosomal genes of P. shigelloides that are involved in LPS core biosynthesis are found in the waa gene cluster (formerly rfa) (172, 173). In a study by Aquilini et al. (159), P. shigelloides 302-73 and CNCTC 113/92 (serotypes O1 and O54, respectively) shared all the genes of the waa cluster except for one common core biosynthetic gene (wapG), which was found at a different chromosome location outside the cluster. Further, these workers were also able to assign a proteomic function to all the P. shigelloides waa genes that were identified in these two strains to encode six new glycosyltransferases (WapA, -B, -C, -D, -F, and -G). Thus, despite there being a single species of Plesiomonas, strains studied have showed a difference in LPS genetics as well as a high homologous recombination level in housekeeping genes.
Enteropathogenicity
The enteropathogenicity of P. shigelloides continues to be nebulous, but it has long been suspected that manifestation of illness in the host is multifactorial (174). For example, ingested plesiomonads do not always cause gastrointestinal disease, and these bacteria have been isolated from the stools of healthy individuals (175). Global reports of isolation as enteric pathogens also differ significantly. An assessment by Shah et al. (105) of 51 published studies between 1973 and 2004 of the etiology of diarrhea in 57 groups of travelers showed the isolation of P. shigelliodes to differ between regions. These findings contrasted with those of an earlier study by Ueda et al. (176) of overseas travelers presenting with diarrhea at Osaka Airport Quarantine Station between January 1992 and September 1994. Here, P. shigelloides was isolated in 1,127 of 1,882 (59.9%) stool samples and was the most common bacterial pathogen recovered in this period.
Such is also the diversity of virulence-associated properties in P. shigelloides. This was demonstrated by Čižnár (175) in a study of 20 isolates from environmental sources, human cases of gastroenteritis, and animal sources. Here, the isolates belonged to 13 different serovars; the majority of strains were both hydrophobic, indicating their ability to interact with a wide variety of surfaces including mammalian cells, and had a relatively high level of flagellar motility to facilitate attachment; all produced triacylglycerol lipase, which is a virulence factor found in Staphylococcus aureus (177), Staphylococcus cohnii (178), and Pseudomonas (Burkholderia) cepacia (179). However, among these strains, there was significant variation in elastase, proteinase, histidine decarboxylase, and hemolysin activities. Furthermore, these workers were unable to detect signal molecules such as C4 to C8 unsubstituted N-acylhomoserine lactones (AHLs), such as those evident in metabolic activities of many other bacterial species (180).
Using a different phenotypic methodology, Salerno et al. (151) demonstrated greater diversity of putative virulence markers (including motility, hemolysin, and four exoenzymes) in 60 biochemically diverse strains of P. shigelloides that were isolated from human (n = 25), animal (n = 18), and environmental (n = 17) specimens in different countries. All environmental and animal isolates were motile but, notably, 4 of the 25 human strains (16%) were nonmotile. Of the exoenzymes, DNase activity was prevalent, particularly among the human strains; gelatinase activity was detected in none of the human strains and in only 22% and 35% of animal and environmental strains, respectively. All isolates were negative for lipase and elastase activities (151). Most strains in this study demonstrated hemolytic activity, although it was predominantly found among the animal and environmental strains. This contrasted with the findings of a study by Čižnár et al. (175), in which hemolysin activity was demonstrable in 7 of 30 (23.3%) of the isolates tested. Such variation is expected given that detection of hemolysis is dependent on the method used, and it has been shown previously to be influenced by the concentrations of iron and calcium, medium composition, level of oxygen, and the type of erythrocytes used (151, 152, 175, 181, 182). Hemolysin activity may be a useful maker for pathogenicity, as was implicated in a study by Baratéla et al. (182), who found a correlation between the vacuolation of various cell lines with such cytotoxic activity in filtrates of P. shigelloides (Fig. 2). This correlation has also been shown for Aeromonas hydrophila (183), Serratia marcescens (184), and Vibrio cholerae (185).
FIG 2.
Effect of P. shigelloides toxin filtrates on Y1 adrenal cells.
The diversity of the prevalence of virulence factors in recent studies of P. shigelloides (above) and the technical difficulties associated with their detection continues to confound the association with gastrointestinal disease. Thus, the organism's role as an enteric pathogen is suspected only when it dominates in a stool culture of a symptomatic individual (176). Yet, the organism possesses a large plasmid (>120 mDa) that has been shown to assist in cell invasion (186). In addition, it is known to produce three forms of enterotoxin, including forms that have cholera-like (153, 154), thermo-stable (153, 187), and thermo-labile (182, 188) properties. Furthermore, it has long been suspected of being adhesive and invasive (8, 175).
Theodoropoulos et al. (189) was able to demonstrate for the first time the ability of P. shigelloides to adhere and enter mammalian cells in vitro. Using human colonic adenocarcinoma cells (Caco-2) as a model, these workers showed the organism's ability to invade the cytosol and escape from the cytoplasmic vacuoles. Following this work, Okawa et al. (155) isolated and characterized a 40-kDa cytotoxic outer membrane protein (ComP) produced by P. shigelloides P-1 strain. This virulence factor was found to be a heat-stable complex comprised of three major LPS-binding proteins and an LPS moiety. However, in this study, only the former component expressed cytotoxic or enterotoxigenic activity in suckling mouse studies. Using the same in vitro model of Theodoropoulos et al. (189), Tsugawa et al. (190) demonstrated bacterial attachment and the induction of apoptosis. Further, they hypothesized that these properties may evade an immune response conducive to metastatic infection and related to the pathology and clinical manifestations.
Seminal work on a number of other pathogenic organisms has also established the involvement of GroEL (a chaperone) or heat shock protein in cell attachment and invasion (191, 192, 193, 194, 195, 196). Tsugawa et al. (197) also showed the importance of P. shigelloides GroEL by demonstrating induction of its expression by contact with host cells (Caco-2 cells) and a subsequent increased cellular surface association of the protein. These workers speculated that the involvement of GroEL in attachment of P. shigelloides to Caco-2 cells may be due to overexpression of a receptor involved in attachment, such as they demonstrated by the upregulation of the gene for intercellular adhesion molecule-1 (ICAM-1) by this chaperone or by binding with Caco-2 cell surface protein and forming a bridge with the organism. The latter proposal was not explored by that group. However, further molecular studies by Tsugawa et al. (198) on the cytotoxin previously characterized by Okawa et al. (155) showed that not only GroEL but also ComP is an important factor in the interaction of P. shigelloides and host cells. These workers showed that ComP is the predominant virulence factor that causes apoptotic cell death in the infected host cell. However, they observed that both a wild-type strain and a mutant strain with a comP deletion were not insignificantly different in their abilities to internalize and multiply within Caco-2 cells. This observation suggests that apoptosis associated with ComP is not involved in the internalization and growth within the host cells (198).
Iron Acquisition
Iron is an essential element for most bacteria, including P. shigelloides (199, 200). However, in its oxidized form the element is insoluble, and it is highly toxic for most macromolecules in its reduced form. Consequently, bacteria have acquisition systems that involve iron and heme carrier proteins for iron sequestration and assimilation from other sources (201). Thus, iron acquisition systems have been shown to be involved in the pathogenicity of a range of bacterial pathogens as they acquire iron in host cells (199).
Siderophore-mediated iron assimilation systems are widely distributed in bacteria (202) yet have not been found in P. shigelloides (153, 200). In a study by Daskaleros et al. (200), strains of P. shigelloides did not use siderophores such as enterobactin, aerobactin, or vibriobactin for growth in medium low in iron. Similarly, Henderson et al. (203) found that P. shigelloides could not utilize siderophores of ferrichrome or schizokinen or be cross-fed by Vibrio parahaemolyticus strains that produced vibrioferrin or by an identified siderophore in Vibrio alginolyticus. Daskaleros et al. (200) further established that the growth of P. shigelloides was supported by hemin and hemoglobin in iron-deficient media, but neither transferrin nor lactoferrin served as a source of iron. Thus, the acquisition of iron from these two compounds is thought to be associated with the liberation of intracellular iron-containing compounds by the lytic activity of hemolysins or cytotoxins. Daskaleros et al. (200) investigated the relationship between acquisition of iron and the production of hemolysins or cytotoxins by using cloned DNA sequences of P. shigelloides that encoded hemolysin production and the ability to utilize heme or hemoglobin for expression in E. coli.They established that the genes for hemolysin production were not linked to those for heme iron utilization. Furthermore, they showed that production of P. shigelloides hemolysin was derepressed under conditions of iron starvation, suggesting the presence of a fur-like regulatory system for ferric uptake that could influence the expression of iron transport system genes as well as other genes, as previously described for E. coli (203). Henderson et al. (203) characterized 10 genes in P. shigelloides that encode the heme iron utilization system, including HugA, the putative heme receptor in the outer membrane, TonB and ExbBD, which facilitate HugA movement of heme into the periplasm, HugB, the putative periplasmic binding protein which moves heme across the periplasm, HugC and HugD, the putative inner membrane permeases that allow movement of heme into the cytoplasm, HugW and HugX, which may be required for metabolism of iron and are not involved in transport, and HugZ, which is essential for binding and utilization of heme (203, 204, 205). Three of the genes, hugA, hugZ, and tonB, may be controlled by iron, as they contain a FUR (ferric uptake repressor) box in their putative promoters (203, 206, 207). Interestingly, a study by Villarreal et al. (205) showed that coexpression of human hemoglobin genes and P. shigelloides heme transport genes enhanced recombinant hemoglobin production in E. coli BL21(DE3) grown in medium containing heme. These findings have implications for the use of recombinant hemoglobin as an alternative to human blood for patients requiring transfusions (205).
Genetic Diversity
Salerno et al. (21) investigated the diversity of 77 P. shigelloides strains from different sources and countries via multilocus sequencing typing based on five housekeeping genes (fusA, leuS, pyrG, recG, and rpoB). In the study, they found significant diversity, implicating a high propensity for homologous recombination. This reduces resemblance between phylogenetic trees based on single genes and compromises interaction between serotypes and the genomic background (21). Furthermore, the rate of recombination observed in P. shigelloides strains studied by these workers exceeded that of other species of the Enterobacteriaceae (Escherichia coli, Klebsiella pneumoniae, Serratia marcescens, and Yersinia pseudotuberculosis) known to have a low rate homologous recombination (21, 208). The ubiquitous living environment of P. shigelloides (209) and the high density of bacteria in the infected host cell are conducive to DNA exchange with cohabitating donor and recipient strains, transducing phages, and bacteria (21). Homologous recombination is thought to mediate genetic change in bacteria, giving rise to pathogenic properties, such as evasion of the immune response and dissemination of virulence genes to other clones (208). Whole-genome sequencing has been completed on P. shigelloides strain 302-73 (serotype O1) (9). This information can advance our understanding of the pathogenicity of P. shigelloides, particularly in elucidation of the role played by homologous recombination and exploration for novel virulence factors.
LABORATORY IDENTIFICATION
Isolation and Identification
Isolation.
Stools used to test for plesiomonads should be collected and handled the same as those for any routine enteric pathogens, with feces preferable over rectal swabs (210). Preservative fluids (all are acceptable) and/or refrigeration of the specimen is necessary if testing cannot be performed within 2 to 4 h of collection (211). Plesiomonas should be easy to isolate and identify despite the fact it is an infrequently seen microorganism in the laboratory. Although specialized media have been devised for its isolation, it usually grows well on routine enteric media such as MacConkey (MAC), xylose lysine decarboxylase (XLD), or Hektoen (HE) agars. Salmonella-Shigella agar may be inhibitory for some strains. Since this organism variably ferments sucrose, lactose, or salicin, the differential sugars utilized in these media, colonies will usually be colorless with the exception of HE agar, in which colonies often appear green due to the medium (Fig. 3). If cefsulodin-irgasan-novobiocin (CIN) medium is used as part of a laboratory's protocol, P. shigelloides can also be successfully isolated from this agar. Colonies will be colorless on CIN; screening media, described below, will be needed to separate it from other oxidase-positive organisms (Pseudomonas, also colorless, and Aeromonas species, colorless with pink centers) that grow on this agar (211). Of the several specialized media developed specifically for the isolation of Plesiomonas, inositol-Brilliant green-bile salts (IBB) is generally described as the most effective for both clinical and environmental specimens (1, 13, 212). von Graevenitz and Bucher (212) described sensitivity and specificity rates of 90% and 100%, respectively, in studies with seeded stools on IBB. Plesiomonads appear as whitish or slightly pink colonies while coliforms are generally greenish in color, however, as some coliforms can also appear pink, colonies must again be further tested with screening tests (212). Plesiomonads will grow on CHROMagar orientation medium (Becton Dickinson, Sparks, MD); however, descriptions of the colony appearance are limited to stock strains (one in each of two large studies), as it has not been isolated from any specimens tested (213, 214). It was described as transparent pink in one study (214), and a photo showed it as white with a slightly pink center (213) in another. It could not be definitely identified from its appearance and required additional tests for confirmation. Wong et al. (102) reported the isolation of this organism from Monsur agar (taurocholate tellurite gelatin agar), a medium designed for vibrio detection. In this medium, P. shigelloides appears as a small colony with a clear apron and dark center, as do vibrios.
FIG 3.
P. shigelloides on Hektoen enteric agar.
If enrichment is necessary, as might be the case for stools from patients with chronic diarrhea, alkaline peptone water (APW; pH 8.6) is the one of the most commonly used, although this organism will grow in Gram-negative broth as well (1). von Graevenitz and Bucher (212) found the use of enrichment in APW to be more effective than direct plating when the ratio of coliforms to plesiomonads is high, as would be expected in chronic diarrhea. However, another enrichment broth, bile peptone broth, has been touted as twice as effective as APW (215) and may be the enrichment of choice in studies where plesiomonads are specifically sought.
Colonies selected from enteric plating media usually cannot be tested directly for oxidase because acid produced from carbohydrate fermentation causes false-negative reactions (216). Because plesiomonads ferment few sugars in these types of media, oxidase testing may be tried but, if negative, should be repeated from a source containing no carbohydrates. If a blood agar plate is included in the testing regimen, nonhemolytic, smooth, opaque colonies approximately 1 to 2 mm in size should undergo the oxidase and spot indole tests (1), for which P. shigelloides is positive for both tests. Other nonselective agars may also be used for oxidase testing but not for spot indole unless they contain a tryptophan source. In triple sugar iron agar (TSI) P. shigelloides resembles shigellae with an alkaline slant over an acid butt with no gas or H2S production (K/A−). All oxidase-positive organisms with the above TSI reaction should be selected for work-up by conventional, kit, or automated systems or molecular identification methods. Other agents from which P. shigelloides must be differentiated are Aeromonas and non-salt-requiring Vibrio species.
Identification.
Because P. shigelloides shows remarkably little strain-to-strain variation in its biochemical profile (217; this statement is also based on our unpublished data regarding over 50 strains) and the combination of key identifying reactions is fairly unique, making it is easy to identify with conventional tests. The TSI reaction coupled with positive oxidase, lysine, and ornithine decarboxylase and arginine dihyrolase reactions and myo-inositol fermentation separate this organism from other Enterobacteriaceae, Aeromonadaceae, and Vibrionaceae (13). It is also 100% positive for indole, d-glucose, trehalose, and nitrate, 95% positive for maltose and 90% positive for ONPG (o-nitrophenyl-β-d-galactopyranoside) (217). Citrate, urea, Voges-Proskauer, and potassium cyanide tests, gas production from any carbohydrate fermented, and utilization tests with most sugars are uniformly negative. Lactose and salicin reaction results are variable. P. shigelloides is susceptible to O129 (2,4-diamino-6,7-diiopropyl-pteridine), with the reaction most clearly defined on Mueller-Hinton agar (1). P. shigelloides does not require salt for growth but can grow in salt concentrations up to 4% (13). While hemolysis is usually not present on plates with 5% sheep blood agar, β-hemolysin can be detected in greater than 90% of strains in contact-dependent or agar overlay assays (152) or on Luria agar (182).
(i) Conventional and commercial systems.
Very few comprehensive studies evaluating manual or automated identification systems have included P. shigelloides, and even then usually only 1 to 3 strains are used, so it is difficult to assess their ability to accurately identify this organism. Additionally, versions of automated equipment or systems (with the exception of API strips [bioMérieux, Marcy l'Etoile, France]) that were evaluated in older publications have been replaced with newer equipment versions or next-generation platforms for which publications including plesiomonads were not found by us. Three articles published since 2000 evaluated 14 strains of P. shigelloides in various systems. In a 2001 publication, one strain was identified correctly by the Vitek 2 system (bioMérieux, Marcy l'Etoile, France) and confirmed by API (218). In 2002, O'Hara and Miller (219) tested 3 strains, 2 of which were correctly identified by the MicroScan Walk/Away 96 instrument with a Rapid Neg ID 3 card (produced at the time by DadeBehring, Inc., West Sacramento, CA), while the third isolate was incorrectly identified, giving three different responses. This was followed by a 2006 article where 10/10 strains of P. shigelloides were correctly identified by the Phoenix 100 ID/AST NID card (Becton Dickinson, Sparks, MD) (220).
(ii) PCR amplification and gene sequencing.
Molecular techniques, particularly those based on PCR, are increasingly being utilized in public health and diagnostic laboratories because of their ability to rapidly detect microorganisms and circumvent conventional culture methods. Such technologies aid in the public health imperative to detect P. shigelloides and other enteric pathogens rapidly in foodborne outbreaks, in gastroenteritis prevalence studies, and in studies of other extraintestinal infections in immunocompromised individuals.
A range of PCR assays targeting different genomic fragments have been developed for the detection of P. shigelloides (Table 9) (42, 51, 221, 222, 223, 224, 225). One of the earliest assays was configured by González-Rey et al. (42) and targeted a specific variable region of the 23S rRNA gene previously identified by Van Camp et al. (226). This assay focused on the 5′ half of this gene, for which the variability affords optimal discernment from closely related genera, including Aeromonas, Vibrio, and Proteus. As it has been shown to be highly specific, the assay has been used to detect P. shigelloides in aquatic settings, animals, and human infections (42). Other PCR assays targeting the 23S rRNA gene have been described (222) and adapted to real-time PCR (221). However, Herrera et al. (51) found the use of the primers designed by González-Rey et al. (42) to target 23S rRNA yielded a similar-sized PCR product (284 bp) when they tested a verotoxigenic E. coli stx2+ strain. When tested for the hugA gene, which encodes an outer membrane receptor required for heme iron utilization in P. shigelloides, a larger discerning product (435 bp) was amplified (51). Primers targeting the hugA gene were shown to have high specificity for P. shigelloides when tested with a range of Gram-negative strains of different genera and were used by these workers to detect the organism in saltwater fish. Meng et al. (225) developed a detection assay for P. shigelloides targeting hugA (Table 9) that is based on a loop-mediated isothermal amplification (LAMP) reaction mediated by Bst DNA polymerase (227). When used to test spiked human stool samples, the assay proved to be more accurate, sensitive, and faster (in assay time) than quantitative PCR. Furthermore, the LAMP assay may facilitate rapid and reliable detection of P. shigelloides in basic clinical and field laboratories (225).
TABLE 9.
Primers and targets used to detect Plesiomonas shigelloides in environmental and clinical specimens by PCR and LAMP assays
| Target gene | Methoda | Primer or probe sequence (5′–3′) | PCR product size (bp) | Reference(s) |
|---|---|---|---|---|
| 23S rRNA | C | Forward (PS23FW3), CTCCGAATACCGTAGAGTGCTATCC | 284 | 42 |
| Reverse (PS23RV3), CTCCCCTAGCCCAATAACACCTAAA | ||||
| 23S rRNA | RT | Forward, AGCGCCTCGGACGAACACCTA | 112 | 221 |
| Reverse, GTGTCTCCCGGATAGCAC | ||||
| LCRed640, GGTAGAGCACTGTTAAGGCTAGGGGGTCATC-P | ||||
| 23S rRNA | C | Forward (PS-F), GCAGGTTGAAGGTTGGGTAA | 628 | 222 |
| Reverse (PS-R), TTGAACAGGAACCCTTGGTC | ||||
| gyrB | SY | Forward (237-F), TTCCAGTACGAGATCCTGGCTAA | 68 | 223, 224 |
| Reverse (304-R), TGAATCGACACGCCAGAGTTC | ||||
| hugA | C | Forward, GCGAGCGGGAAGGGAAGAACC | 435 | 51 |
| Reverse, GTCGCCCCAAACGCTAACTCATCA | ||||
| hugA | LA | F3, AACACGTTGCAGCCCATC | 225 | |
| B3, ACTTTACCGCCGAAGACAAG | ||||
| FIP, CGTTACGACGAAGCGTTCCGTGAAGTGAGTACCGGTGGTGTb | ||||
| BIP, GTCAGCCAATCAGTCGCCGCAATATCGCCGGCTCCGAGc | ||||
| LF- ACCGAGCATGGAAGAGATGT | ||||
| LB, GCGACAGGTGATCTTCGCTAC | ||||
| hugA | RT | Forward, GGAATATCGGCCTGTACAT | 116 | 225 |
| Reverse, TATGGCGGCGATATTTA | ||||
| Probe, FAM-CCCCAGACTTTGCTGCGACCATCGG-BHQ-1 |
Abbreviations: C, conventional PCR; RT, real-time PCR using hybridization probes; SY, real-time PCR using SYBR green stain; LA, loop-mediated isothermal amplification; FAM, 6-carboxyfluorescein; BHQ-1, Black Hole quencher 1.
FIP, forward inner primer. The sequence for target recognition is underlined, and the sequence for loop formation is not underlined.
BIP, backward inner primer. The sequence for target recognition is underlined, and the sequence for loop formation is not underlined.
(iii) 16S rRNA and gyrB gene sequencing.
16S rRNA gene sequencing is a common tool used for reference identification of unusual or infrequent pathogenic microorganisms, as well as for defining phylogenetic lineages (228). In the clinical microbiology laboratory, it has rarely been used due to the ease of identification of this microbial species via conventional tests or commercial panels or strips. Sequencing of this gene has also been used to confirm the isolation of P. shigelloides in disease outbreaks in fish (63, 229).
However, sequences of the gyrB gene, which encodes the ATPase domain of the DNA gyrase essential for DNA replication, has been sequenced in a wide range of microorganisms and has been shown to be more useful than 16S rRNA for determining evolutionary relationships. Furthermore, phylogenetic analysis of gyrB gene sequences in P. shigelloides showed phylogenetic characteristics distinguishable from the closely related species of the Enterobacteriaceae (230) and the Aeromonadaceae (33, 230) families. This is an advance on 16S rRNA sequencing, where P. shigelloides previously showed a closer affinity with species of the Enterobacteriaceae than with the Aeromonadaceae (19). The gyrB gene has also been used as a target in a PCR assay to detect P. shigelloides in investigations of foodborne outbreaks (223, 224).
(iv) Culture-independent diagnostic testing (CIDT).
One of the more recent applications of PCR-based technology has been the development of FDA-approved commercial panels for the detection and diagnosis of syndromic diseases such as gastroenteritis, respiratory tract infections, and bacteremia. Several such panels exist for gastrointestinal illnesses that have received FDA approval for the detection of enteric pathogens in stools (231, 232). One such assay is the FilmArray gastrointestinal panel (Biofire Diagnostics, Salt Lake City, UT), which is a nested PCR multiplex real-time PCR with melting point analysis targeting 23 pathogens (14 bacterial, 5 viral, and 4 parasitic), including P. shigelloides. Preliminary studies suggest that both the sensitivity and specificity of the FilmArray assay for the detection of P. shigelloides approach 100%, although the prevalence rate for this pathogen in two of these studies was exceedingly low (233, 234, 235). However, in a multicenter study, 1,556 specimens from four geographically distinct clinical sites across the United States were examined, and P. shigelloides was detected in 18 of 1,180 (1.5%) samples, as opposed to only 3 isolates recovered by routine culture (234). Discordant results were arbitrated by hugA PCR, which confirmed the test results of FilmArray. Furthermore, 16 of the 18 (88.9%) detections were associated with coinfection (234). FilmArray appears to be a more sensitive means of detecting uncommon or infrequent enteric pathogens, compared to traditional culture methods.
(v) MALDI-TOF MS.
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is a relatively new technology for the clinical laboratory and has been traditionally used in proteomic studies, including the elucidation of the structure of the LPS of P. shigelloides (157, 159, 163, 165, 168). Its main interest to clinical microbiologists, however, is as an easy, rapid, and high-throughput system to provide accurate microbial identifications in a matter of minutes (236). Kolínská et al. (237) used this technology to identify unique biomarker ions ranging from 3 to 12 kDa in mass and species-specific spectral database constructed from 74 isolates to create an accurate identification system for P. shigelloides. Commercial MALDI-TOF MS systems with their own databases have been marketed for this purpose and include the Vitek MS system (bioMérieux) and Microflex LT Biotyper (Bruker Daltonics, GmbH, Bremen, Germany). Neville et al. (238) tested two isolates of P. shigelloides in triplicate on the MicroFlex MALDI Biotyper 2.0, all of which were correctly identified. Five of the six runs which yielded correction identifications had scores of >2.0. Other studies have generated similar results, although the number of strains/isolates tested to date on these commercial MS systems is very low (239, 240, 241). In 2014, Deng and others (242) tested 3 isolates of P. shigelloides on the Vitek MS system, which gave correct identifications for each strain. Although the numbers are small, these results suggest that both systems are highly accurate in the identification of this single-species genus.
Serology
An antigenic schema has been used to epidemiologically type strains of P. shigelloides for over 20 years; it currently is comprised of 102 somatic (O) and 51 flagellar (H) antigens (243). In addition to agglutinating in their homologous sera, many strains of P. shigelloides cross-react in various shigella antisera and, conversely, a number of Shigella spp. have been found to react in antisera made from strains of P. shigelloides (244). In the latter situation, strains of S. dysenteriae 1 and S. flexneri serotypes 1b, 2a, 3a, and 6 agglutinated in serum made from a single strain of P. shigelloides, and those authors identified the common group 1 antigen found on these serotypes to be responsible (244). Of the 102 O types of P. shigelloides, O17 is most notable because it is identical to the O-antigen of Shigella sonnei and will agglutinate in S. sonnei antisera (29, 245). Lefebvre et al. (246) noted in a large study on commercially available sonnei antisera that one strain of P. shigelloides cross-reacted in 5 of 7 products tested. An interesting hypothesis has been advanced to account for the low incidence of S. sonnei infections in developing countries, and it is based on the possible natural immunization of the population due to reoccurring exposure to Plesiomonas O17, which is prevalent in aqueous environments (29). Shepherd et al. (245) sequenced the entire O-antigen gene cluster in a strain of P. shigelloides O17 and compared it to the plasmid-borne O-gene cluster in S. sonnei and found they differed in only two genes, wzz and wbgZ. Further, in sequencing the chromosome S. sonnei O-antigen gene cluster, it was determined that only a remnant remains, indicating that it was lost by deletion following acquisition of the Plesiomonas O-genes via an invasion plasmid. Based on the high level of similarity and the fact that the O-antigen of S. sonnei resides on a plasmid, it is suggested that there has been a recent (within the last 10,000 years) transfer of the O-gene cluster from P. shigelloides to E. coli (of which S. sonnei is actually a species), allowing this organism to adapt to a new niche, that of a diarrheal pathogen.
Antimicrobial Susceptibility Testing
The methodology and breakpoints described for the Enterobacteriaceae in Clinical and Laboratory Standards Institute document M100-S24 should be used for P. shigelloides susceptibility testing (247).
There have been a number of large studies (84, 102, 248, 249) testing a combined 392 human, environmental, and animal isolates. Where the serotype was included there was no association between serotype and susceptibility pattern (249), nor was there a difference seen with the source of the isolate and susceptibility (248). It was difficult to compare results from these studies, as a variety of methods were used and the antimicrobial agents tested differed from study to study. However, Table 10 gives the percent resistance reported for a number of antimicrobial agents. Bravo et al. (249) reported that 22 of 54 strains (22%) were multiresistant, with the most common patterns being ampicillin/tetracycline/kanamycin (n = 4; 7%).
TABLE 10.
P. shigelloides susceptibility to a variety of antimicrobial agents
| Antimicrobial agent | % resistance | Reference(s) |
|---|---|---|
| Ampicillin | 72–92 | 84, 102, 248 |
| Piperacillin | 8 | 248 |
| Amoxicillin-clavulanic acid | 0–3 | 84, 248 |
| Cefoperazone-sulbactam | 0 | 84 |
| Ampicillin-sulbactam | 0 | 84, 248 |
| Piperacillin-tazobactam | 0 | 84, 248 |
| Cefazolin | 0–16 | 84, 248 |
| Cefuroxime | 0–3 | 84, 248 |
| Ceftazidime | 0–31 | 84, 248, 249 |
| Cefotaxime | 0–1 | 84, 248 |
| Cefepime | 2–8 | 84, 248 |
| Cefoxitin | 0–6 | 84, 249 |
| Ceftriaxone | 0 | 102, 248, 249 |
| Cefoperazone | 0–22 | 248, 249 |
| Aztreonam | 4–7 | 84, 248, 249 |
| Imipenem | 0 | 84, 248 |
| Meropenem | 0 | 84, 248 |
| Amikacin | 1–46 | 84, 248, 249 |
| Gentamicin | 0–37 | 84, 248 |
| Ciprofloxacin | 0–9 | 84, 248 |
| Levofloxacin | 0–3 | 84, 102 |
| Ofloxacin | 0 | 102, 248 |
| Trimethoprim-sulfamethoxazole | 2–47 | 84, 102, 248, 249 |
| Tetracycline | 0–67 | 84, 102, 248, 249 |
| Chloramphenicol | 0–5 | 102, 248 |
| Nitrofurantoin | 0 | 248, 249 |
| Erythromycin | 19–58 | 248, 249 |
Plesiomonads are reported to produce β-lactamases (248, 250, 251). Avison et al. (250) found 10 of 20 human and environmental strains of P. shigelloides, all of which were resistant to ampicillin and carbenicillin, expressed chromosomally encoded noninducible β-lactamases. The β-lactamases produced were diverse, having 5 different isoelectric focusing points and belonging to 2a, 2c, and 2d class enzymes. No correlation with the MIC and the production of β-lactamase was found, and the authors surmised strains without a β-lactamase that are resistant to penicillins produce resistance by another mechanism. Since both environmental and human strains contained β-lactamases and some strains with β-lactamases were isolated prior to the introduction of ampicillin and carbenicillin, it appears that resistance has not arisen solely from the selective pressure of treatment (228). Stock and Wiedemann (248) maintain that P. shigelloides produces two patterns of susceptibility, depending on the inoculum used. Pattern 1 contains strains susceptible to aztreonam and cephalosporins when a 1 × 104 CFU/ml inoculum is used, whereas at 1 × 107 CFU/ml there is decreased susceptibility to these drugs. They feel this phenomenon is due to quorum sensing, where high cell density mediates a change in gene expression. Data from Wiegand and Burak (251), however, showed that a strain without a detectable β-lactamase experienced the same inoculum effect as strains with a β-lactamase and that the MIC seen with higher inocula were not influenced by the presence of a β-lactamase inhibitor. These facts led the investigators to believe resistance was not due to inoculum-dependent regulation of β-lactamases but rather to the formation of filamentous forms ranging in size from 100 μm to 2 mm. Thus, it is the presence of nonviable filamentous forms in the bottom of the wells in MIC tests that account for appearance of resistance. It is not known if these filamentous forms occur in vivo during antimicrobial therapy. Also, while Wiegand and Burak's (251) data indicate that plesiomonads are actually susceptible to cephalosporins, use of these antibiotics may lead to an antibiotic-induced release of endotoxins, which may be detrimental to the patient.
CONCLUSIONS
Our knowledge of the biology, taxonomy, and pathogenicity of P. shigelloides has increased significantly over the past 2 decades, although many questions remain unanswered. Some of these questions still concern the species correct taxonomic and phylogenetic position, as well as the need for better, well-documented case reports and series of infections. Other areas in need of better data include risk factors associated with both intestinal and systemic infections and molecular investigations into the genetic makeup of several strains with different biologic characteristics. In another decade, many of these questions should be answered.
While P. shigelloides was once regarded as only a potential enteropathogen, convincing data from most case-controlled investigations and disease-associated outbreaks now support this conclusion (Tables 3 and 4). Still unresolved are issues concerning potential enteropathogenic mechanisms and why the prevalence of P. shigelloides enteritis varies dramatically in relationship to geographic location. It may well be that sanitary conditions, dietary choices, or environmental factors and stimuli play key roles in determining the global incidence of this disease. However, many more case-controlled surveys of bacterial diarrhea need to be undertaken, as well as environmental studies looking at other potential natural reservoirs for P. shigelloides, to definitively resolve these issues.
While this bacterium is easy to identify biochemically in the laboratory, it is often overlooked for a number of reasons, including a perceived low frequency and cultural characteristics, including screening reactions. Some studies have suggested that P. shigelloides is often found in association with other enteric pathogens (82, 234), and this needs to be more fully investigated. The development of CIDT systems, such as the FilmArray gastrointestinal panel that includes this enteropathogen, may help to resolve some of these issues in the near future.
ACKNOWLEDGMENTS
None of the authors has any conflict of interest to disclose or any financial interest related to the submission of this review. No author received any financial support from or has any interest in institutions or companies mentioned in the manuscript.
Biographies

J. Michael Janda received his undergraduate degree from Loyola University (Westchester), his M.S. in Microbiology from California State Univerisity—Los Angeles, and his Ph.D. in Microbiology and Immunology from UCLA. From 1979 to 1986, he was first a postdocoral fellow (ABMM) at The Mt. Sinai Medical Center and then Assistant/Associate Professor of Clinical Microbiology there, where we was Assistant Director of the Microbiology Laboratory and Codirector of the ABMM program. From 1986 to 2011, he was a Section Chief and then Branch Chief for the Microbial Diseases Laboratory, California Department of Public Health. Subsequent positions have included Public Health Laboratory Director for Los Angeles and Alameda Counties. Currently he is the Public Health Laboratory Director for Kern County. Over his career of more than 30 years, Dr. Janda has focused on the study of bacterial gastroenteritis, gram-negative bacteria, including the family Enterobacteriaceae, and aquatic pathogens, including Aeromonas, Plesiomonas, and Vibrio.

Sharon L. Abbott received her Bachelor of Arts degree from California State University at Chico. For over 35 years she worked in the Microbial Diseases Laboratory Branch (MDL) of the California Department of Public Health, primarily in enteric bacteriology. In 1990, she became the Supervisor of the Enterics Unit for almost 15 years. After retiring from the State she was employed by the University of California at Berkeley as a Training Coordinator for a Postdoctoral Fellowship program at MDL. Her research focus has centererd on the identification of enteric bacteria, particularly Aeromonas species, and the development of in vitro pathogenicity assays.

Christopher J. McIver completed a Ph.D. in 1993 at The University of Technology, Sydney (Australia). That work was a study of the physiology and epidemiology of clinically isolated cysteine-requiring enterobacteria. He has also completed postgraduate studies in public health and management. His research interests are in diverse areas of clinical microbiology, including detection, epidemiology, physiology, and antibiotic resistance mechanisms of enterobacteria. In 2007, he was awarded a Winston Churchill Memorial Fellowship to study innovative molecular detection methods for enteric pathogens. This study was undertaken at the Microbial Diseases Laboratory, California Department of Public Health, and the Centers for Disease Control and Prevention, Atlanta, GA, USA. Currently, he is employed as a Principal Hospital Scientist in the Microbiology Department, St. George Hospital, Sydney, where he is the Scientist-in-Charge of the molecular detection section. He is also a Conjoint Associate Professor in the School of Medical Science, University of New South Wales, Sydney.
REFERENCES
- 1.Janda JM, Abbott SL. 1998. The enterobacteria. Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- 2.Ferguson WW, Henderson ND. 1947. Description of strain C27: a motile organism with major antigens of Shigella sonnei. J Bacteriol 54:179–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmid EE, Velaudapilllai T, Niles GR. 1954. Study of paracolon organisms with the major antigen of Shigella sonnei form I. J Bacteriol 68:50–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellner PD, McCarthy LR. 1974. Aeromonas shigelloides bacteremia: a case report. Am J Clin Pathol 59:216–218. [DOI] [PubMed] [Google Scholar]
- 5.Holmberg SD, Farmer JJ III. 1984. Aeromonas hydrophila and Plesiomonas shigelloides as causes of intestinal infections. Rev Infect Dis 6:633–639. [DOI] [PubMed] [Google Scholar]
- 6.Sack DA, Chowdhury KA, Huq A, Kay BA, Sayeed S. 1988. Epidemiology of Aeromonas and Plesiomonas diarrhoea. J Diarrhoeal Dis Res 6:107–112. [PubMed] [Google Scholar]
- 7.Khardori N, Fainstein V. 1988. Aeromonas and Plesiomonas as etiological agents. Annu Rev Microbiol 42:395–419. [DOI] [PubMed] [Google Scholar]
- 8.Brenden RA, Miller MA, Janda JM. 1988. Clinical disease spectrum and pathogenic factors associated with Plesiomonas shigelloides infections in humans. Rev Infect Dis 10:303–316. [DOI] [PubMed] [Google Scholar]
- 9.Piqué N, Aquilini E, Alioto T, Miana-Galbis D, Tomás JM. 2013. Genome sequence of Plesiomonas shigelloides strain 302-73 (serotype O1). Genome Announc 1:e00404–13. doi: 10.1128/genomeA.00404-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bader RE. 1954. Ueber die Herstllung eines agglutineerenden Serums gegen die Rundform von Shigella sonnei mit einem Stamm der Gattung Pseudomonas. Ziet Hyg Infektionskr 140:450–456. [PubMed] [Google Scholar]
- 11.Farmer JJ III, Arduino MJ, Hickman-Brenner FW. 2006. The genera Aeromonas and Plesiomonas, p 564–596. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes, a handbook on the biology of bacteria. Proteobacteria: gamma subclass, 3rd ed, vol 6 Springer, New York, NY. [Google Scholar]
- 12.Ewing WH, Hugh R, Johnson JG. 1961. Studies on the Aeromonas group. U.S. Department of Health and Human Services, Centers for Disease Control, Atlanta, GA. [Google Scholar]
- 13.Janda JM. 2005. Genus XXVII. Plesiomonas, Habs and Schubert 1962, 324AL, p 740–744. In Brenner DJ, Krieg NR, Staley JT, Garrity GM (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2 Springer, New York, NY. [Google Scholar]
- 14.Habs H, Schubert RHW. 1962. Uber die biochemischen Merkmale und die taxonomische Stellung von Pseudomonas shigelloides (Bader). Zentralbl Bakteriol I Abt Orig 186:316–327. [Google Scholar]
- 15.Skerman VBD, McGowan V, Sneath PHA. 1980. Approved lists of bacterial names. Int J Syst Bacteriol 30:225–420. [PubMed] [Google Scholar]
- 16.Véron MM. 1965. La position taxonomiqe des Vibrio et de certaines bacteries comparables. C R Hebd Seances Acad Sci 261:5243–5246. (In French.) [PubMed] [Google Scholar]
- 17.MacDonell MT, Colwell RR. 1985. Phylogeny of the Vibrionaceae, and recommendation for two new genera, Listonella and Shewanella. System Appl Microbiol 6:171–182. [Google Scholar]
- 18.MacDonell MT, Swartz DG, Ortiz-Conde BA, Last GA, Colwell RR. 1986. Ribosomal RNA phylogenies for the vibrio-enteric group of eubacteria. Microbiol Sci 3:172–178. [PubMed] [Google Scholar]
- 19.Martinez-Murcia AJ, Benlloch S, Collins MD. 1992. Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: lack of congruence with results of DNA-DNA hybridizations. Int J Syst Bacteriol 42:412–421. [DOI] [PubMed] [Google Scholar]
- 20.Ruimy R, Breitmayer V, Elbaze P, Lafay B, Boussemart O, Gauthier M, Christen R. 1994. Phylogenetic analysis and assessment of the genera Vibrio, Photobacterium, Aeromonas, and Plesiomonas deduced from small-subunit rRNA sequences. Int J Syst Bacteriol 44:416–426. [DOI] [PubMed] [Google Scholar]
- 21.Salerno A, Delétoile A, Lefevre M, Ciznar I, Krovacek K, Grimont P, Brisse S. 2007. Recombining population structure of Plesiomonas shigelloides (Enterobacteriaceae) revealed by multilocus sequence typing. J Bacteriol 189:7808–7818. doi: 10.1128/JB.00796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whang HY, Heller ME, Neter E. 1972. Production by Aeromonas of common enterobacterial antigen and its possible taxonomic significance. J Bacteriol 110:161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramia S, Neter E, Brenner DJ. 1982. Production of enterobacterial common antigen as an aid to classification of newly identified species of the families Enterobacteriaceae and Vibrionaceae. Int J Syst Bacteriol 32:395–398. [Google Scholar]
- 24.Yamamoto S, Chowdhury MA, Kuroda M, Nakano T, Koumoto Y, Shinoda S. 1991. Further study on polyamine compositions in Vibrionaceae. Can J Microbiol 37:148–153. [DOI] [PubMed] [Google Scholar]
- 25.Basu S, Tharanathan RN, Kontrohr T, Mayer H. 1985. Chemical structure of the lipid A component of Plesiomonas shigelloides and its taxonomical significance. FEMS Microbiol Lett 28:7–10. [Google Scholar]
- 26.Fanning GR, Hickman-Brenner FW, Farmer JJ III. 1985. DNA relatedness and phenotypic analysis of the genus Aeromonas, p 319 Abstr 85th Gen Meet Am Soc Microbiol. American Society for Microbiology, Washington, DC. [Google Scholar]
- 27.Janda JM, Abbott SL. 1999. Unusual food-borne pathogens. Listeria monocytogenes, Aeromonas, Plesiomonas, and Edwardsiella species. Clin Lab Med 19:553–582. [PubMed] [Google Scholar]
- 28.Jagger TD. 2000. Plesiomonas shigelloides: a veterinary perspective. Infect Dis Rev 2:199–210. [Google Scholar]
- 29.Stock I. 2004. Plesiomonas shigelloides: an emerging pathogen with unusual properties. Rev Med Microbiol 15:129–139. [Google Scholar]
- 30.Priest FG, Barker M. 2010. Gram-negative bacteria associated with brewery yeasts: reclassification of Obesumbacterium proteus biogroup 2 as Shimwellia pseudoproteus gen. nov., sp. nov., and transfer of Escherichia blattae to Shimwellia blattae comb. nov. Int J Syst Evol Microbiol 60:828–832. doi: 10.1099/ijs.0.013458-0. [DOI] [PubMed] [Google Scholar]
- 31.Spröer C, Mendrock U, Swiderski J, Lang E, Stackebrandt E. 1999. The phylogenetic position of Serratia, Buttiauxella and some other genera of the family Enterobacteriaceae. Int J Syst Bacteriol 49:1433–1438. [DOI] [PubMed] [Google Scholar]
- 32.Paradis S, Boissinot M, Paquette N, Bélanger SD, Martel EA, Boudreau DK, Picard FJ, Ouellette M, Roy PH, Bergeron MG. 2005. Phylogeny of the Enterobacteriaceae based on genes encoding elongation factor Tu and F-ATPase β-subunit. Int J Syst Evol Microbiol 55:2013–2025. doi: 10.1099/ijs.0.63239-0. [DOI] [PubMed] [Google Scholar]
- 33.Yáñez MA, Catalán V, Apráiz D, Figueras MJ, Martínez-Murcia AJ. 2003. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int J Syst Evol Microbiol 53:875–883. doi: 10.1099/ijs.0.02443-0. [DOI] [PubMed] [Google Scholar]
- 34.McNeeley D, Ivy P, Craft JC, Cohen I. 1984. Plesiomonas: biology of the organism and diseases in children. Pediatr Infect Dis J 3:176–181. [PubMed] [Google Scholar]
- 35.Miller ML, Koburger JA. 1985. Plesiomonas shigelloides: an opportunistic food and waterborne pathogen. J Food Prot 48:449–457. [DOI] [PubMed] [Google Scholar]
- 36.Rouf MA, Rigney MM. 1971. Growth temperatures and temperature characteristics of Aeromonas. Appl Microbiol 22:503–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.European Food Safety Authority. 2012. Scientific opinion on the minimum hygiene criteria to be applied to clean seawater and on public health risks and hygiene criteria for bottled seawater intended for domestic use. EFSA J 10:1–85. [Google Scholar]
- 38.Gonzalez-Rey C, Svenson SB, Eriksson LM, Ciznar I, Krovacek K. 2003. Unexpected finding of the “tropical” bacterial pathogen Plesiomonas shigelloides from lake water north of the Polar Circle. Polar Biol 26:495–499. [Google Scholar]
- 39.Hazen TC, Fliermans CB, Hirsch RP, Esch GW. 1978. Prevalence and distribution of Aeromonas hydrophila in the United States. Appl Environ Microbiol 36:731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arai T, Ikejima N, Itoh T, Sakai S, Shimada T, Sakazaki R. 1980. A survey of Plesiomonas shigelloides from aquatic environments, domestic animals, pets and humans. J Hyg (Lond) 84:203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibotti A, Saridakis HO, Pelayo JS, Tagliari KC, Falcão DP. 2000. Prevalence and virulence properties of Vibrio cholera non-O1, Aeromonas spp. and Plesiomonas shigelloides isolated from Cambé Stream (State of Paraná, Brazil). J Appl Microbiol 89:70–75. doi: 10.1046/j.1365-2672.2000.01077.x. [DOI] [PubMed] [Google Scholar]
- 42.González-Rey C, Svenson SB, Bravo L, Rosinsky J, Ciznar I, Krovacek K. 2000. Specific detection of Plesiomonas shigelloides isolated from aquatic environments, animals and human diarrhoeal cases by PCR based on 23S rRNA gene. FEMS Immunol Med Microbiol 29:107–113. doi: 10.1111/j.1574-695X.2000.tb01512.x. [DOI] [PubMed] [Google Scholar]
- 43.Bülger B. 2004. Occurrence of Plesiomonas shigelloides and relationship with faecal pollution in Nilufer Stream, Bursa, Turkey. Turkish Electron J Biotechnol 2:22–29. [Google Scholar]
- 44.Pakingking R Jr, Palma P, Usero R. 2015. Quantitative and qualitative analyses of the bacterial microbiota of tilapia (Oreochromis niloticus) cultured in earthen ponds in the Philippines. World J Microbiol Biotechnol 31:265–275. doi: 10.1007/s11274-014-1758-1. [DOI] [PubMed] [Google Scholar]
- 45.Presley SM, Rainwater TR, Austin GP, Platt SG, Zak JC, Cobb GP, Marsland EJ, Tian K, Zhang B, Anderson TA, Cox SB, Abel MT, Leftwich BD, Huddleston JR, Jeter R, Kendall RJ. 2006. Assessment of pathogens and toxicants in New Orleans, LA following hurricane Katrina. Environ Sci Technol 40:468–474. [DOI] [PubMed] [Google Scholar]
- 46.Kanungo R, Shashikala Karunasagar I, Srinivasan S, Sheela D, Venkatesh K, Anitha P. 2007. Contamination of community water sources by potentially pathogenic vibrios following sea water inundation. Commun Dis 39:229–232. [PubMed] [Google Scholar]
- 47.Kakizaki E, Kozawa S, Matsuda H, Muraoka E, Uchiyama T, Sakai M, Yukawa N. 2010. Freshwater bacterioplankton cultured from liver, kidney and lungs of a decomposed cadaver retrieved from a sandy seashore: possibility of drowning in a river and then floating out to sea. Leg Med (Tokyo) 12:195–199. doi: 10.1016/j.legalmed.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 48.Bauwens L, De Meurichy W, Lemmens P, Vandepitte J. 1983. Isolation of Plesiomonas shigelloides and Edwardsiella species in the Antwerp zoo. Acta Pathol Antwerp 77:61–74. [Google Scholar]
- 49.Oxley APA, Shipton W, Owens L, McKay D. 2002. Bacterial flora from the gut of the wild and cultured banana prawn, Penaeus merguiensis. J Appl Microbiol 93:214–223. doi: 10.1046/j.1365-2672.2002.01673.x. [DOI] [PubMed] [Google Scholar]
- 50.Miller WA, Miller MA, Gardner IA, Atwill ER, Byrne BA, Jang S, Harris M, Ames J, Jessup D, Paradies D, Worcester K, Melli A, Conrad PA. 2006. Salmonella spp., Vibrio spp., Clostridium perfringens, and Plesiomonas shigelloides in marine and freshwater invertebrates from coastal California ecosystems. Microb Ecol 52:198–206. doi: 10.1007/s00248-006-9080-6. [DOI] [PubMed] [Google Scholar]
- 51.Herrera PC, Santos JA, Otero A, Garcia-López ML. 2006. Occurrence of Plesiomonas shigelloides in displayed portions of saltwater fish determined by a PCR assay based upon the hugA gene. Int J Food Microbiol 108:233–238. doi: 10.1016/j.ijfoodmicro.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 52.Joh S-J, Ahn E-H, Lee H-J, Shin G-W, Kwon J-H, Park C-G. 2013. Bacterial pathogens and flora isolated from farm-cultured eels (Anguilla japonica) and their environmental waters in Korean eel farms. Vet Microbiol 163:190–195. doi: 10.1016/j.vetmic.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 53.Huber I, Spanggaard B, Appel KF, Rossen L, Nielsen T, Gram L. 2004. Phylogenetic analysis and in situ identification of the intestinal microbial community of rainbow trout (Oncorhynchus mykiss, Walbaum). J Appl Microbiol 96:117–132. doi: 10.1046/j.1365-2672.2003.02109.x. [DOI] [PubMed] [Google Scholar]
- 54.Salgado-Miranda C, Palomares E, Jurado M, Marín A, Vega F, Soriano-Vargas E. 2010. Isolation and distribution of bacterial flora in farmed rainbow trout from Mexico. J Aquatic Anim Health 22:244–247. doi: 10.1577/H09-004.1. [DOI] [PubMed] [Google Scholar]
- 55.Hu Q, Lin Q, Shi C, Fu X, Li N, Liu L, Wu S. 2014. Isolation and identification of a pathogenic Plesiomonas shigelloides from diseased grass carp. Wei Sheng Wu Xue Bao 54:229–235. (In Chinese.) [PubMed] [Google Scholar]
- 56.Nadirah M, Ruhil HH, Jalal KCA, Najiah M. 2012. Occurrence of Plesiomonas shigelloides in cultured red hybrid tilapia (Oreochromis niloticus) from tropical rivers, east coast Malaysia. Pak J Biol Sci 15:600–603. doi: 10.3923/pjbs.2012.600.603. [DOI] [PubMed] [Google Scholar]
- 57.Liu Z, Ke X, Lu M, Gao F, Cao J, Zhu H, Wang M. 2015. Identification and pathological observation of a pathogenic Plesiomonas shigelloides strain isolated from cultured tiplapia (Orechromis niloticus). Wei Sheng Wu Xue Bao 55:96–105. (In Chinese.) [PubMed] [Google Scholar]
- 58.Larsen AM, Mohammed HH, Arias CR. 2014. Characterization of the gut microbiota of three commercially valuable warm water fish species. J Appl Microbiol 116:1396–1404. doi: 10.1111/jam.12475. [DOI] [PubMed] [Google Scholar]
- 59.Duarte S, e Silva FC, Zauli DA, Nicoli JR, Araújo FG. 2014. Gram-negative intestinal indigenous microbiota from two Siluriform fishes in a tropical reservoir. Braz J Microbiol 45:1283–1292. doi: 10.1590/S1517-83822014000400019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tippen PS, Meyer A, Blank EC, Donnell HD. 1989. Aquarium-associated Plesiomonas shigelloides infection—Missouri. MMWR Morb Mortal Wkly Rep 38:617–619. [PubMed] [Google Scholar]
- 61.Smith KF, Schmidt V, Rosen GE, Amaral-Zettler L. 2012. Microbial diversity and potential pathogens in ornamental fish aquarium water. PLoS One 7:e39971. doi: 10.1371/journal.pone.0039971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jun JW, Kim JH, Choresca CH Jr, Shin SP, Han JE, Jeong DS, Park SC. 2011. Isolation and molecular detection of Plesiomonas shigelloides containing tetA gene from Asian arowana (Scleropages formosus) in a Korean aquarium. Afr J Microbiol Res 5:5019–5021. [Google Scholar]
- 63.Nisha RG, Rajathi V, Manikandan R, Prabhu NM. 2014. Isolation of Plesiomonas shigelloides from infected cichlid fishes using 16S rRNA characterization and its control with probiotic Pseudomonas sp. Acta Sci Vet 42:1–7. [Google Scholar]
- 64.Buck JD, Wells RS, Rhinehart HL, Hansen LJ. 2006. Aerobic microorganisms associated with free-ranging bottlenose dolphins in coastal Gulf of Mexico and Atlantic Ocean waters. J Wildl Dis 42:536–544. doi: 10.7589/0090-3558-42.3.536. [DOI] [PubMed] [Google Scholar]
- 65.Miller MA, Byrne BA, Jang SS, Dodd EM, Dorfmeier E, Harris MD, Ames J, Paradies D, Worcester K, Jessup DA, Miller WA. 2010. Enteric bacterial pathogen detection in southern sea otters (Enhydra lutris nereis) is associated with coastal urbanization and freshwater runoff. Vet Res 41:1 doi: 10.1051/vetres/2009049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pereira CS, Amorim SD, Santos AF, Siciliano S, Moreno IB, Ott PH, Rodrigues Ddos P. 2008. Plesiomonas shigelloides and Aeromonadaceae family pathogens from marine mammals of southern and southeastern Brazilian coast. Braz J Microbiol 39:749–755. doi: 10.1590/S1517-83822008000400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schaefer AM, Goldstein JD, Reif JS, Fair PA, Bossart GD. 2009. Antibiotic-resistant organisms cultured from Atlantic bottlenose dolphins (Tursiops truncates) inhabiting estuarine waters of Charleston, SC and Indian River Lagoon, FL. Ecohealth 6:33–41. doi: 10.1007/s10393-009-0221-5. [DOI] [PubMed] [Google Scholar]
- 68.Morris PJ, Johnson WR, Pisani J, Bossart GD, Adams J, Reif JS, Fair PA. 2011. Isolation of culturable microorganisms from free-ranging bottlenose dolphins (Tursiops truncates) from the southeastern United States. Vet Microbiol 148:440–447. doi: 10.1016/j.vetmic.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 69.Stenkat J, Krautwald-Junghanns M-E, Schmitz Ornés A, Eilers A, Schmidt V. 2014. Aerobic cloacal and pharyngeal bacterial flora in six species of free-living birds. J Appl Microbiol 117:1564–1571. doi: 10.1111/jam.12636. [DOI] [PubMed] [Google Scholar]
- 70.Matsuyama R, Kuninaga N, Morimoto T, Shibano T, Sudo A, Sudo K, Asano M, Suzuki M, Asai T. 2015. Isolation and antimicrobial susceptibility of Plesiomonas shigelloides from great cormorants (Phalacrocorax carbo hanedae) in Gifu and Shiga prefectures, Japan. J Vet Med Sci 77:1179–1181. doi: 10.1292/jvms.15-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kinzelman J, McLellan SL, Amick A, Preedit J, Scopel CO, Olapade O, Gradus S, Singh A, Sedmak G. 2008. Identification of human enteric pathogens in gull feces at southwestern Lake Michigan bathing beaches. Can J Microbiol 54:1006–1015. doi: 10.1139/W08-096. [DOI] [PubMed] [Google Scholar]
- 72.Niskanen M, Salmela P. 2000. Plesiomonas shigelloides: a possible pathogen in animals. Suomen Eläinlääkärilehti 106:451–455. [Google Scholar]
- 73.Nimmervoll H, Wenker C, Robert N, Albini S. 2011. Septicaemia caused by Edwardsiella tarda and Plesiomonas shigelloides in captive penguin chicks. Schweiz Arch Tierheilk 153:117–121. doi: 10.1024/0036-7281/a000165. [DOI] [PubMed] [Google Scholar]
- 74.González-Rey C, Siitonen A, Pavlova A, Ciznar I, Svenson SB, Krovacek K. 2011. Molecular evidence of Plesiomonas shigelloides as a possible zoonotic agent. Folia Microbial (Praha) 56:178–184. doi: 10.1007/s12223-011-0032-2. [DOI] [PubMed] [Google Scholar]
- 75.Gál J, Sós E, Medösi , Radványi LS, Mándoki M, Tóth T. 2006. Abscess formation caused by Plesiomonas shigelloides in the body cavity of the lizard Ameiva ameiva (Linnnaeus, 1758). Salamandra 42:53–56. [Google Scholar]
- 76.Johnston MA, Porter DE, Scott GI, Rhodes WE, Webster LF. 2010. Isolation of faecal coliform bacteria from the American alligator (Alligator mississippiensis). J Appl Microbiol 108:965–973. doi: 10.1111/j.1365-2672.2009.04498.x. [DOI] [PubMed] [Google Scholar]
- 77.Cruz JM, Saraiva A, Eiras JC, Branco R, Sousa JC. 1986. An outbreak of Plesiomonas shigelloides in farmed rainbow trout, Salmo gairdneri Richardson, in Portugal. Bull Eur Assoc Fish Pathol 6:20–22. [Google Scholar]
- 78.Greenlees KJ, Machado J, Bell T, Sunlof SF. 1998. Food borne microbial pathogens of cultured aquatic species. Vet Clin North Am 114:101–112. [DOI] [PubMed] [Google Scholar]
- 79.Bodhidatta L, McDaniel P, Sornsakrin S, Srijan A, Serichantalergs O, Mason CJ. 2010. Case-control study of diarrheal disease etiology in a remote rural area of western Thailand. Am J Trop Med Hyg 83:1106–1109. doi: 10.4269/ajtmh.2010.10-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tseng H-K, Liu C-P, Li W-C, Lee C-M. 2002. Characteristics of Plesiomonas shigelloides infections in Taiwan. J Microbiol Immunol Infect 35:47–52. [PubMed] [Google Scholar]
- 81.Ahmed D, Hoque A, Elahi MSB, Endtz HP, Hossain MA. 2012. Bacterial aetiology of diarrhoeal diseases and antimicrobial resistance in Dhaka, Bangladesh, 2005-2008. Epidemiol Infect 140:1678–1684. doi: 10.1017/S0950268811002135. [DOI] [PubMed] [Google Scholar]
- 82.Escobar JC, Bhavani D, Trueba G, Ponce Karina, Cevallos W, Eisenberg J. 2012. Plesiomonas shigelloides infection, Ecuador, 2004-2008. Emerg Infect Dis 18:322–324. doi: 10.3201/eid1802.110562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sire J-M, Garin B, Chartier L, Fall NK, Tall A, Seck A, Weill F-X, Breurec S, Vray M. 2013. Community-acquired infectious diarrhoea in children under 5 years of age in Dakar, Senegal. Paediatr Int Child Health 33:139–144. doi: 10.1179/2046905512Y.0000000046. [DOI] [PubMed] [Google Scholar]
- 84.Chen X, Chen Y, Yang Q, Kong H, Yu F, Han D, Zheng S, Cui D, Li L. 2013. Plesiomonas shigelloides infection in southeast China. PLoS One 8:e77877. doi: 10.1371/journal.pone.0077877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nwokocha ARC, Onyemelukwe NF. 2014. Plesiomonas shigelloides diarrhea in Enugu area of the southeastern Nigeria: incidence, clinical and epidemiological features. J Dent Med Sci 13:68–73. [Google Scholar]
- 86.Qu M, Deng Y, Zhang X, Liu G, Huang Y, Lin C, Li J, Yan H, Li X, Jia L, Kan B, Huang F, Wang Q. 2012. Etiology of acute diarrhea due to enteropathogenic bacteria in Beijing, China. J Infect 65:214–222. doi: 10.1016/j.jinf.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 87.O'Ryan M, Prado V, Pickering LK. 2005. A millennium update on pediatric diarrheal illness in the developing world. Semin Pediatr Infect Dis 16:125–136. doi: 10.1053/j.spid.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 88.Bai Y, Dai Y-C, Li J-D, Nie J, Chen Q, Wang H, Rui Y-Y, Zhang Y-L, Yu S-Y. 2004. Acute diarrhea during army field exercises in southern China. World J Gastroenterol 10:127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Danchaivijitr S, Riongrungruang Y, Kachintorn U, Techasathit V, Pakaworavuthi S, Kachintorn K. 2005. Prevalence and effectiveness of an education program on intestinal pathogens of food handlers. J Med Assoc Thai 10:S31–S35. [PubMed] [Google Scholar]
- 90.Janda JM, Abbott SL. 2010. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev 23:35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ueda S, Yamazaki S, Hori M. 1963. The isolation of paracolon C27 and pathogenic halophile organisms from an outbreak of food poisoning. Jpn J Public Health 10:67–70. (In Japanese.) [Google Scholar]
- 92.Hori M, Hayashi K. Maeshima K, Kigawa M, Miyasato T, Yoneda Y, Hagihara Y. 1966. Food poisoning caused by Aeromonas shigelloides with an antigen common to Shigella dysenteriae 7. J Jpn Assoc Infect Dis 39:433–441. (In Japanese.) [Google Scholar]
- 93.Tsukamoto T, Kinoshita Y, Shimada T, Sakazaki R. 1978. Two epidemics of diarrhoeal disease possibly caused by Plesiomonas shigelloides. J Hyg (London) 80:275–280. doi: 10.1017/S0022172400053638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rutala WA, Sarubbi FA Jr, Finch CS, MacCormack JN, Steinkraus GE. 1982. Oyster-associated outbreak of diarrhoeal disease possibly caused by Plesiomonas shigelloides. Lancet i:739. [DOI] [PubMed] [Google Scholar]
- 95.Newsom R, Gallois C. 1982. Diarrheal disease caused by Plesiomonas shigelloides. Clin Microbiol Newslett 4:158–159. doi: 10.1016/S0196-4399(82)80049-4. [DOI] [Google Scholar]
- 96.Medema G, Schets C. 1993. Occurrence of Plesiomonas shigelloides in surface water: relationship with faecal pollution and trophic state. Zentralbl Hyg 194:398–404. [PubMed] [Google Scholar]
- 97.Van Houten R, Farberman D, Norton J, Ellison J, Kielbauch J, Morris T, Smith P. 1998. Plesiomonas shigelloides and Salmonella serotype Hartford infections associated with a contaminated water supply—Livingston County, New York, 1996. MMWR Morb Mortal Wkly Rep 47:394–396. [PubMed] [Google Scholar]
- 98.Wouafo M, Pouillot R, Kwetche PF, Tejiokem M-C, Kamgno J, Fonkoua M-C. 2006. An acute foodborne outbreak due to Plesiomonas shigelloides in Yamounde, Cameroon. Foodborne Pathog 3:209–211. doi: 10.1089/fpd.2006.3.209. [DOI] [PubMed] [Google Scholar]
- 99.Wang S, Duan H, Zhang W, Li J-W. 2007. Analysis of bacterial foodborne outbreaks in China between 1994 and 2005. FEMS Immunol Med Microbiol 51:8–13. doi: 10.1111/j.1574-695X.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- 100.Holmberg SD, Wachsmuth IK, Hickman-Brenner FW, Blake PA, Farmer JJ III. 1986. Plesiomonas enteric infections in the United States. Ann Intern Med 105:690–694. doi: 10.7326/0003-4819-105-5-690. [DOI] [PubMed] [Google Scholar]
- 101.Potasman I, Paz A, Odeh M. 2002. Infectious outbreaks associated with bivalve shellfish consumption: a worldwide perspective. Clin Infect Dis 35:921–928. doi: 10.1086/342330. [DOI] [PubMed] [Google Scholar]
- 102.Wong TY, Tsui HY, So MK, Lai JY, Tse CWS, Ng TK. 2000. Plesiomonas shigelloides infection in Hong Kong: retrospective study of 167 laboratory-confirmed cases. Hong Kong Med J 6:375–380. [PubMed] [Google Scholar]
- 103.Sule AA. 2009. Severe Plesiomonas shigelloides gastroenteritis in a young healthy patient. Crit Care Shock 12:120–122. [Google Scholar]
- 104.Shigematsu M, Kaufmann ME, Charlett A, Niho Y, Pitt TL. 2000. An epidemiological study of Plesiomonas shigellolides diarrhoea among Japanese travelers. Epidemiol Infect 12:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shah N, DuPont H, Ramsey DJ. 2009. Global etiology of travelers' diarrhea: systematic review from 1973 to the present. Am J Trop Med Hyg 80:609–614. [PubMed] [Google Scholar]
- 106.Suthienkul O, Aiumlaor P, Siripanichgon K, Eampokalap B, Likhanonsakul S, Utrarachkij F, Rakus Y. 2001. Bacterial causes of AIDS-associated diarrhea in Thailand. Southeast Asian J Trop Med Public Health 32:158–170. [PubMed] [Google Scholar]
- 107.Obi C, Bessong PO. 2002. Diarrhoeagenic bacterial pathogens in HIV-positive patients with diarrhoea in rural communities of Limpopo Province, South Africa. J Health Popul Nutr 20:230–234. [PubMed] [Google Scholar]
- 108.Woo PCY, Lau SKP, Yuen K-Y. 2005. Biliary tract diseases as a risk factor for Plesiomonas shigelloides bacteraemia: a nine-year experience in a Hong Kong hospital and review of the literature. New Microbiol 28:45–55. [PubMed] [Google Scholar]
- 109.Brann OS. 2001. Infectious complications of cirrhosis. Curr Gastroenterol Rep 3:285–292. doi: 10.1007/s11894-001-0051-2. [DOI] [PubMed] [Google Scholar]
- 110.Ampofo K, Graham P, Ratner A, Rajagopalan L, Della-Latta P, Saiman L. 2001. Plesiomonas shigelloides sepsis and splenic abscess in an adolescent with sickle cell disease. Pediatr Infect Dis J 20:1178–1179. doi: 10.1097/00006454-200112000-00019. [DOI] [PubMed] [Google Scholar]
- 111.Tzanetea R, Konstantopoulos K, Xanthaki A, Kalotychou V, Spiliopoulou C, Michalopoulos A, Rombos Y. 2002. Plesiomonas shigelloides sepsis in a thalassemia intermedia patient. Scand J Infect Dis 34:687–689. doi: 10.1080/00365540210147877. [DOI] [PubMed] [Google Scholar]
- 112.Auxiliadora-Martins M, Bellissimo-Rodrigues F, Viana JM, Teixeira GCA, Nicolini EA, Cordeiro KSM, Colozza G, Martinez R, Martins-Filho OS, Basile-Filho A. 2010. Septic shock caused by Plesiomonas shigelloides in a patient with sickle beta-zero thalassemia. Heart Lung 39:335–339. doi: 10.1016/j.hrtlng.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 113.Khan FA, Fisher MA, Khakoo RA. 2007. Association of hemochromatotosis with infectious diseases: expanding spectrum. Int J Infect Dis 11:482–487. doi: 10.1016/j.ijid.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 114.Xia F-Q, Liu P-N, Zhou Y-H. 2015. Meningoencephalitis caused by Plesiomonas shigelloides in a Chinese neonate: case report and literature review. Ital J Pediatr 41:3. doi: 10.1186/s13052-014-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roth T, Hentsch C, Erard P, Tschantz P. 2002. Pyosalpinx: not always a sexual transmitted disease? Pyosalpinx caused by Plesiomonas shigelloides in an immunocompetent host. Clin Microbiol Infect 8:803–805. [DOI] [PubMed] [Google Scholar]
- 116.Herve V, Bigaillon C, Duhamel P, Petit MP, Soler C. 2007. Cutaneous abscess due to Plesiomonas shigelloides consecutive to a trauma in fresh water. Med Mal Infect 37:840 (In French.) doi: 10.1016/j.medmal.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 117.Klatte JM, Daswtjerdi MH, Clark K, Harrison CJ, Grigorian F, Stahl ED. 2012. Hyperacute infectious keratitis with Plesiomonas shigelloides following traumatic lamellar corneal laceration. Pediatr Infect Dis J 31:1200–1201. doi: 10.1097/INF.0b013e318266b61f. [DOI] [PubMed] [Google Scholar]
- 118.Gadewar S, Fasano A. 2005. Current concepts in the evaluation, diagnosis and management of acute infectious diarrhea. Curr Opin Pharmacol 5:559–565. doi: 10.1016/j.coph.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 119.DuPont HL. 2009. Bacterial diarrhea. N Engl J Med 361:1560–1569. doi: 10.1056/NEJMcp0904162. [DOI] [PubMed] [Google Scholar]
- 120.Schlenker C, Surawicz CM. 2009. Emerging infections of the gastrointestinal tract. Best Pract Res Clin Gastroenterol 23:89–99. doi: 10.1016/j.bpg.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 121.Kelly P. 2011. Diarrhoeal disease. Clin Med 11:488–491. doi: 10.7861/clinmedicine.11-5-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tam CC, O'Brien SJ, Tompkins DS, Bolton FJ, Berry L, Dodds J, Choudhury D, Halstead F, Iturriza-Gómara M, Mather K, Rait G, Ridge A, Rodrigues LC, Wain J, Wood B, Gray JJ, IID2 Study Executive Committee. 2012. Changes in causes of acute gastroenteritis in the United Kingdom over 15 years: microbiologic findings from 2 prospective, population-based studies of infectious intestinal disease. Clin Infect Dis 54:1275–1286. doi: 10.1093/cid/cis028. [DOI] [PubMed] [Google Scholar]
- 123.Pfeiffer ML, DuPont HL, Ochoa TJ. 2012. The patient presenting with acute dysentery: a systematic review. J Infect 64:374–386. doi: 10.1016/j.jinf.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 124.Klontz EH, Faruque ASG, Das SK, Malek MA, Islam Z, Luby SP, Klontz KC. 2012. Clinical and epidemiologic features of diarrheal disease due to Aeromonas hydrophila and Plesiomonas shigelloides infections compared with those due to Vibrio cholera non-O1 and Vibrio parahaemolyticus in Bangladesh. ISRN Microbiol 120:654819. doi: 10.5402/2012/654819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chan SSW, Ng KC, Lyon DJ, Cheung WL, Cheng AFB, Rainer TH. 2003. Acute bacterial gastroenteritis: a study of adult patients with positive stool cultures treated in the emergency department. Emerg Med J 20:335–338. doi: 10.1136/emj.20.4.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chatterjee BD, Neogy KN. 1972. Studies on Aeromonas and Plesiomonas species isolated from cases of choleraic diarrhoera. Indian J Med Res 60:520–524. [PubMed] [Google Scholar]
- 127.Jandl G, Linke K. 1976. Report about two cases of gastroenteritis caused by Plesiomonas shigelloides. Zentralbl Baketeriol Orig A 236:136–140. (In German.) [PubMed] [Google Scholar]
- 128.Alcantara AK, Escamilla J, Ranoa CP, Cross JH. 1984. Plesiomonas shigelloides in 11 cases with diarrhea at San Lazaro Hospital. Phil J Microbiol Infect Dis 13:5–9. [Google Scholar]
- 129.Chakour M, Koeck JL, Boulland P, Trueba F, Ruttimann M, Teyssou R. 2002. Diarrhee choleriforme severe due to a Plesiomonas shigelloides chez un expatrie a Djibouti. Med Trop (Mars) 62:205 (In French.) [PubMed] [Google Scholar]
- 130.Martin DL, Gustafson TL. 1985. Plesiomonas gastroenteritis in Texas. JAMA 254:2063. doi: 10.1001/jama.1985.03360150039013. [DOI] [PubMed] [Google Scholar]
- 131.van Loon FPL, Rahim Z, Chowdhury KA, Kay BA, Rahman R. 1989. Case report of Plesiomonas shigelloides-associated persistent dysentery and pseudomembranous colitis. J Clin Microbiol 27:1913–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kaiser L, Surawicz CM. 2012. Infectious causes of chronic diarrhoea. Best Pract Res Clin Gastroenterol 26:563–571. doi: 10.1016/j.bpg.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 133.Sawle GV, Das BC, Acland PR, Heath DA. 1986. Fatal infection with Aeromonas sobria and Plesiomonas shigelloides. Br Med J 292:525–526. doi: 10.1136/bmj.292.6519.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sinnott JT, Turnquest DG, Milam MW. 1989. Plesiomonas shigelloides gastroenteritis. Clin Microbiol Newslett 11:103–104. doi: 10.1016/0196-4399(89)90051-2. [DOI] [Google Scholar]
- 135.Penn RG, Giger DK, Knoop FC, Preheim LC. 1982. Plesiomonas shigelloides overgrowth in the small intestine. J Clin Microbiol 15:869–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sack DA, Hoque ATMS, Huq A, Etheridge M. 1994. Is protection against shigellosis induced by natural infection with Plesiomonas shigelloides? Lancet 343:1413–1415. doi: 10.1016/S0140-6736(94)92531-3. [DOI] [PubMed] [Google Scholar]
- 137.Lee ACW, Yuen KY, Ha SY, Chiu DCK, Lau YL. 1996. Plesiomonas shigelloides septicemia: case report and literature review. Pediatr Hematol Oncol 13:265–269. doi: 10.3109/08880019609030826. [DOI] [PubMed] [Google Scholar]
- 138.Bonatti H, Sifri C, Sawyer RG. 2012. Successful liver transplantation from donor with Plesiomonas shigelloides sepsis after freshwater drowning: case report and review of literature on gram-negative bacterial aspiration during drowning and utilization of organs from bacteremic donors. Surg Infect 13:114–120. doi: 10.1089/sur.2010.018. [DOI] [PubMed] [Google Scholar]
- 139.Okon E, Bishburg E, Ugras S, Chan T, Wang H. 2013. Clostridium perfringens meningitis, Plesiomonas shigelloides sepsis: a lethal combination. Am J Case Rep 14:70–72. doi: 10.12659/AJCR.883830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Janda JM, Abbott SL. 2014. The genus Shewanella: from the briny depths below to human pathogen. Crit Rev Microbiol 40:293–312. doi: 10.3109/1040841X.2012.726209. [DOI] [PubMed] [Google Scholar]
- 141.von Graevenitz A, Mensch AH. 1968. The genus Aeromonas in human bacteriology. Report of 30 cases and review of the literature. N Engl J Med 278:245–249. [DOI] [PubMed] [Google Scholar]
- 142.McCracken AW, Barkley R. 1972. Isolation of Aeromonas species from clinical sources. J Clin Pathol 25:970–975. doi: 10.1136/jcp.25.11.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gopal V, Burns FE. 1991. Cellulitis and compartment syndrome due to Plesiomonas shigelloides: a case report. Mil Med 156:43. [PubMed] [Google Scholar]
- 144.Lowry T, Smith SA. 2007. Aquatic zoonoses associated with food, bait, ornamental, and tropical fish. J Am Vet Med Assoc 231:876–880. doi: 10.2460/javma.231.6.876. [DOI] [PubMed] [Google Scholar]
- 145.Butt AA, Figueroa J, Martin DH. 1997. Ocular infection caused by three unusual marine organisms. Clin Infect Dis 24:740. doi: 10.1093/clind/24.4.740. [DOI] [PubMed] [Google Scholar]
- 146.Marshman WE, Lyons CJ. 1998. Congenital endophthalmitis following maternal shellfish ingestion. Aust N Z J Ophthamol 26:161–163. doi: 10.1111/j.1442-9071.1998.tb01534.x. [DOI] [PubMed] [Google Scholar]
- 147.Woo PCY, Lau SKP, Wong SSY, Yuen K-Y. 2004. Two cases of continuous ambulatory peritoneal dialysis-associated peritonitis due to Plesiomonas shigelloides. J Clin Microbiol 42:933–935. doi: 10.1128/JCM.42.2.933-935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schneider F, Lang N, Reibke R, Michaely HJ, Hiddemann W, Ostermann H. 2009. Plesiomonas shigelloides pneumonia. Med Mal Infect 39:397–400. doi: 10.1016/j.medmal.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 149.Young AZ, Neujahar D, Estok L. 2001. Epididymo-orchitis and bacteremia caused by Plesiomonas shigelloides in an HIV-infected patient. AIDS Read 11:617–619. [PubMed] [Google Scholar]
- 150.Ozdemir O, Sari S, Terzioglu S, Zenciroglu A. 2010. Plesiomonas shigelloides sepsis and meningoencephalitis in a surviving neonate. J Microbiol Immunol Infect 43:344–346. doi: 10.1016/S1684-1182(10)60053-9. [DOI] [PubMed] [Google Scholar]
- 151.Salerno A, Čižnar I, Krovacek K, Conte M, Dumontet S, González-Rey C, Pasquale V. 2010. Phenotypic characterization and putative virulence factors of human, animal and environmental isolates of Plesiomonas shigelloides. Folia Microbiol (Praha) 55:641–647. doi: 10.1007/s12223-010-0104-8. [DOI] [PubMed] [Google Scholar]
- 152.Janda JM, Abbott SL. 1993. Expression of hemolytic activity by Plesiomonas shigelloides. J Clin Microbiol 31:1206–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Abbott SL, Kokka RP, Janda JM. 1991. Laboratory investigations on the low pathogenic potential of Plesiomonas shigelloides. J Clin Microbiol 29:148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gardner SE, Fowlston SE, George WL. 1987. In vitro production of cholera toxin-like activity by Plesiomonas shigelloides. J Infect Dis 156:720–722. doi: 10.1093/infdis/156.5.720. [DOI] [PubMed] [Google Scholar]
- 155.Okawa Y, Ohtoma Y, Tsugawa H, Matsuda Y, Kobayashi H, Tsukamoto T. 2004. Isolation and characterization of a cytotoxin produced by Plesiomonas shigelloides P-1 strain. FEMS Microbiol Lett 239:125–130. doi: 10.1016/j.femsle.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 156.Nestor G, Lukasiewicz J, Sandström C. 2014. Structural analysis of the core oligosaccharide and the O-specific polysaccharide from the Plesiomonas shigelloides O33:H3 (strain CNCTC 34/89) lipopolysaccharide. Eur J Org Chem 2014:1241–1252. doi: 10.1002/ejoc.201301399. [DOI] [Google Scholar]
- 157.Pieretti G, Carillo S, Lindner B, Lanzetta R, Parrilli M, Jimenez N, Regué M, Tomás JM, Corsaro MM. 2010. The complete structure of the core of the LPS from Plesiomonas shigelloides 302-73 and the indentification of its O-antigen biological repeating unit. Carbohydr Res 345:2523–2528. doi: 10.1016/j.carres.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 158.Aquilini E, Merino S, Tomás JM. 2013. The Plesiomonas shigelloides wbO1 gene cluster and the role of O1-antigen LPS in pathogenicity. Microb Pathog 63:1–7. doi: 10.1016/j.micpath.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 159.Aquilini E, Merino S, Regué M, Tomás JM. 2014. Genomic and proteomic studies on Plesiomonas shigelloides lipopolysaccharide core biosynthesis. J Bacteriol 196:556–567. doi: 10.1128/JB.01100-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kaszowska M, Jachymek W, Niedziela T, Koj S, Kenne L, Lugowski C. 2013. The novel structure of the core oligosaccharide backbone of the lipopolysaccharide from Plesiomonas shigelloides strain CNCTC 80/89 (serotype O13). Carbohydr Res 380:45–50. doi: 10.1016/j.carres.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 161.Kubler-Kielb J, Schneerson R, Mocca C, Vinogradov E. 2008. The elucidation of the structure of the core part of the LPS from Plesiominas shigeloides serotype O17 expressing O-polysaccharide chain identical to the Shigella sonnei O-chain. Carbohyd Res 343:3123–3127. doi: 10.1016/j.carres.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Maciejewska A, Lukasiewicz J, Niedziela T, Szewczuk Z, Lugowski C. 2009. Structural analysis of the O-specific polysaccharide isolated from Plesiomonas shigelloides O51 lipopolysaccharide. Carbohyd Res 344:894–900. doi: 10.1016/j.carres.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 163.Lundqvist L, Kaszowska CEM, Sandström C. 2015. NMR study of the O-specific polysaccharide and the core oligosaccharide from the lipopolysaccharide produced by Plesiomonas shigelloides O24:H8 (strain CNCTC 92/89). Molecules 20:5729–5739. doi: 10.3390/molecules20045729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kasowska M, Jachymek W, Lukasiewicz J, Niedziela T, Kenne L, Lugowski C. 2013. The unique structure of complete lipopolysaccharide isolated from semi-rough Plesiomonas shigelloides O37 (strain CNCTC 39/89) containing (2S)-O-(4-oxopentanoic acid)-α-d-Glcp (α-d-lenose). Carbohyd Res 378:98–107. doi: 10.1016/j.carres.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 165.Czaja J, Jachymek W, Niedziela T, Lugowski C, Aldova E, Kenne L. 2000. Structural studies of the O-specific polysaccharide from Plesiomonas shigelloides strain CNCTC 113/92. J Biol Chem 267:1672–1679. doi: 10.1046/j.1432-1327.2000.01161.x. [DOI] [PubMed] [Google Scholar]
- 166.Niedziela T, Lukasiewicz J, Jachymek W, Dzieciatkowska M, Lugowski C, Kenne L. 2002. Core oligosaccharides of Plesiomonas shigelloides O54:H2 (strain CNCTC 113/92). J Biol Chem 277:11653–11663. doi: 10.1074/jbc.M111885200. [DOI] [PubMed] [Google Scholar]
- 167.Lukasiewicz J, Niedziela T, Jachymek W, Kenne L, Lugowski C. 2006. Structure of the lipid A-inner core region and biological activity of Plesiomonas shigelloides O54 (strain CNCTC 113/92) lipopolysaccharide. Glycobiology 16:538–550. doi: 10.1093/glycob/cwj094. [DOI] [PubMed] [Google Scholar]
- 168.Säwén E, Östervall J, Landersjö C, Edblad M, Weintraub A, Ansaruzzaman M, Widmalm G. 2012. Structural studies of the O-antigenic polysaccharide from Plesiomonas shigelloides strain AM36565. Carbohydr Res 348:99–103. doi: 10.1016/j.carres.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 169.Das R, Mukhopadhyay B. 2013. Chemical synthesis of the tetrasaccharide repeating unit of the O-antigenic polysaccharide from Plesiomonas shigelloides strain AM36565. Carbohydr Res 376:1–6. doi: 10.1016/j.carres.2013.04.033. [DOI] [PubMed] [Google Scholar]
- 170.Linnerborg M, Widmalm G, Weintraub A, Albert MJ. 1995. Structural elucidation of the O-antigen lipopolysaccharide from two strains of Plesiomonas shigelloides that share a type-specific antigen with Shigella flexneri 6, and the common group 1 antigen with Shigella flexneri spp. and Shigella dysenteriae 1. Eur J Biochem 231:839–844. doi: 10.1111/j.1432-1033.1995.0839d.x. [DOI] [PubMed] [Google Scholar]
- 171.Caroff M, Karibian D. 2003. Structure of bacterial lipopolysaccharides. Carbohyd Res 338:2431–2447. doi: 10.1016/j.carres.2003.07.010. [DOI] [PubMed] [Google Scholar]
- 172.Izquierdo L, Abtiu N, Coderch N, Hita B, Merino S, Gavin R, Tomás JM, Regué M. 2002. The inner-core lipopolysaccharide biosynthetic waaE gene: function and genetic distribution among some Enterobacteriaceae. Microbiology 148:3485–3496. doi: 10.1099/00221287-148-11-3485. [DOI] [PubMed] [Google Scholar]
- 173.Raetz C, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vitovec J, Aldova E, Vladik P, Krovacek K. 2001. Enteropathogenicity of Plesiomonas shigelloides and Aeromonas spp. in experimental mono- and coinfection with Cryptosporidium parvum in the intestine of neonatal BALB/c mice. Comp Immunol Microbiol Infect Dis 24:39–35. doi: 10.1016/S0147-9571(00)00012-6. [DOI] [PubMed] [Google Scholar]
- 175.Ciznár I, Hostacka A, González-Rey C, Krovacek K. 2004. Potential virulence-associated properties of Plesiomonas shigelloides strains. Folia Microbiol (Praha) 49:543–548. doi: 10.1007/BF02931531. [DOI] [PubMed] [Google Scholar]
- 176.Ueda Y, Suzuki N, Mori H, Miyagi K, Noda K, Hirose H, Takegaki Y, Hashimoto S, Oosumi Y, Miyata Y, Taguchi M, Ishibashi M, Honda T. 1996. Bacteriological studies of traveller's diarrhoea. Analysis of enteropathogenic bacteria at Osaka Airport Quarantine Station from January 1992 through September 3rd, 1994. Kansenshogaku Zasshi 70:29–41. [DOI] [PubMed] [Google Scholar]
- 177.Rollof J, Braconier JH, Söderström C, Nilsson-Ehle P. 1988. Interference of Staphylococcus aureus lipase with human granulocyte function. Eur J Clin Microbiol Infect Dis 7:505–510. doi: 10.1007/BF01962601. [DOI] [PubMed] [Google Scholar]
- 178.Waldon E, Sobiś-Glinkowska M, Szewcyk EM. 2002. Evaluation of selected features of Staphylococcus cohnii enabling colonization of humans. Folia Microbiol (Praha) 47:565–572. doi: 10.1007/BF02818799. [DOI] [PubMed] [Google Scholar]
- 179.Straus DC, Lonon MK, Hutson JC. 1992. Inhibition of rat aveolar macrophage function by a Pseudomonas cepacia lipase. J Med Microbiol 37:335–340. doi: 10.1099/00222615-37-5-335. [DOI] [PubMed] [Google Scholar]
- 180.Fuqua C, Winans S, Greenberg EP. 1996. Census and concensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol 50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 181.Santos JA, Gonzalez CJ, Lopez TM, Otero A, Garcia LML. 1999. Hemolytic and elastolytic activities influenced by iron in Plesiomonas shigelloides. J Food Prot 62:1475–1477. [DOI] [PubMed] [Google Scholar]
- 182.Baratéla KC, Saridakis HO, Gaziri LCJ, Pelayo JS. 2001. Effects of medium composition, calcium, iron and oxygen on haemolysin production by Plesiomonas shigelloides isolated from water. J Appl Microbiol 90:482–487. doi: 10.1046/j.1365-2672.2001.01270.x. [DOI] [PubMed] [Google Scholar]
- 183.Abrami L, Fivaz M, de Groot FG. 1998. A pore-forming toxin interacts with a GPI-anchored protein and causes vacuolation of the endoplasmic reticulum. J Cell Biol 140:525–540. doi: 10.1083/jcb.140.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hertle R, Hilger M, Weingart-Kocher S, Walev I. 1999. Cytotoxic action of Serratia marscesens haemolysin on human epithelial cells. Infect Immun 67:817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Coelho A, Andrade JRC, Vincente ACP, DiRita VJ. 2000. Cytotoxic cell vacuolating activity from Vibrio cholerae haemolysin. Infect Immun 68:1700–1705. doi: 10.1128/IAI.68.3.1700-1705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Herrington DA, Tzipori S, Robins-Browne RM, Tall BD, Levine MM. 1987. In vitro and in vivo pathogenicity of Plesiomonas shigelloides. Infect Immun 55:979–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Matthews B, Douglas H, Guiney D. 1988. Production of a heat stable enterotoxin by Plesiomonas shigelloides. Microb Pathog 5:207–213. doi: 10.1016/0882-4010(88)90023-X. [DOI] [PubMed] [Google Scholar]
- 188.Levin RE. 2008. Plesiomonas shigelloides: an aquatic foodborne pathogen. A review of its characteristics, pathogenicity, ecology, and molecular detection. Food Biotechnol 22:189–202. [Google Scholar]
- 189.Theodoropoulos C, Wong TH, Obrien M, Stenzel D. 2001. Plesiomonas shigelloides enters polarized human intestinal Caco-2 cells in an in vitro model system. Infect Immun 69:2260–2269. doi: 10.1128/IAI.69.4.2260-2269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tsugawa H, Ono T, Murakami H, Okawa Y. 2005. Invasive phenotype and apoptosis induction of Plesiomonas shigelloides P-1 strain to Caco-2 cells. J Appl Microbiol 99:1435–1443. doi: 10.1111/j.1365-2672.2005.02721.x. [DOI] [PubMed] [Google Scholar]
- 191.Ensgraber M, Loos M. 1992. A 66-kilodalton heat shock protein of Salmonella typhimurium is responsible for binding of the bacterium to intestinal mucus. Infect Immun 60:3072–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Galdiero M, Delero GC, Marcatili A. 1997. Cytokine and adhesion molecule expression in human monocytes and endothelial cells stimulated with bacterial heat shock proteins. Infect Immun 65:699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Frisk A, Ison CA, Lagergård T. 1998. GroEL heat shock protein of Haemophilus ducreyi: association with cell surface and capacity to bind to eukaryotic cells. Infect Immun 66:1252–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Garduño RA, Garduño E, Hoffman PS. 1998. Surface-associated Hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect Immun 66:4602–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Goulhen F, Hafezi A, Uitto VJ, Hinode D, Nakamura R, Grenier D, Mayrand D. 1998. Subcellular localzaton and cytotoxc activity of the GroEL-lke protein solated from Actnobacllus actinomycecomtans. Infect Immun 66:5307–5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hennequin C, Collignon A, Karjalainen T. 2001. Analysis of expression of GroEL (Hsp60) of Clostridium difficile in response to stress. Microb Pathog 31:255–260. doi: 10.1006/mpat.2001.0468. [DOI] [PubMed] [Google Scholar]
- 197.Tsugawa H, Ito H, Ohshima M, Okawa Y. 2007. Cell adherence-promoted activity of Plesiomonas shigelloides GroEL. J Med Microbiol 56:23–29. doi: 10.1099/jmm.0.46766-0. [DOI] [PubMed] [Google Scholar]
- 198.Tsugawa H, Ogawa A, Takehara S, Kimura M, Okawa Y. 2008. Primary structure and function of a cytotoxic outer-membrane protein (ComP) of Plesiomonas shigelloides. FEMS Microbiol Lett 281:10–16. doi: 10.1111/j.1574-6968.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- 199.Payne SM, Neilands JB. 1988. Iron and virulence in the family Enterobacteriaceae. Crit Rev Microbiol 16:81–111. doi: 10.3109/10408418809104468. [DOI] [PubMed] [Google Scholar]
- 200.Daskaleros PA, Stoebner JA, Payne SM. 1991. Iron uptake in Plesiomonas shigelloides: cloning of the genes for the heme-iron uptake system. Infect Immun 59:2706–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wandersman C, Delepelaire P. 2004. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol 58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 202.Neilands JB. 1984. Siderophores of bacteria and fungi. Microbiol Sci 1:9–14. [PubMed] [Google Scholar]
- 203.Henderson DP, Wyckoff EE, Rashidi CE, Verlei H, Oldham AL. 2001. Characterization of the Plesiomonas shigelloides genes encoding the heme iron utilization system. J Bacteriol 183:2715–2723. doi: 10.1128/JB.183.9.2715-2723.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Oldham AL, Wood TA, Henderson DP. 2008. Plesiomonas shigelloides hugZ encodes an iron-regulated heme binding protein required for heme iron utilization. Can J Microbiol 54:97–102. doi: 10.1139/W07-122. [DOI] [PubMed] [Google Scholar]
- 205.Villarreal DM, Phillips CL, Kelley AM, Villarreal S, Villaloboz A, Hernandez P, Olson JS, Henderson DP. 2008. Enhancement of recombinant hemoglobin production in Escherichia coli BL21(DE3) containing the Plesiomonas shigelloides heme transport system. Appl Environ Microbiol 74:5854–5856. doi: 10.1128/AEM.01291-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.de Lorenzo V, Wee S, Herrero M, Neilands JB. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol 169:2624–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Baichoo N, Helmann JD. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J Bacteriol 184:5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Feil EJ, Spratt BG. 2001. Recombination and the population structure of bacterial pathogens. Annu Rev Microbiol 55:561–590. doi: 10.1146/annurev.micro.55.1.561. [DOI] [PubMed] [Google Scholar]
- 209.Krovacek K, Eriksson LM, González-Rey C, Rosinsky J, Ciznar I. 2000. Isolation, biochemical and serological characterization of Plesiomonas shigelloides from freshwater in northern Europe. Comp Immunol Microbiol Infect Dis 23:45–51. doi: 10.1016/S0147-9571(99)00058-2. [DOI] [PubMed] [Google Scholar]
- 210.Humphries RM, Linscott AJ. 2015. Laboratory diagnosis of bacterial gastroenteritis. Clin Microbiol Rev 28:3–31. doi: 10.1128/CMR.00073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Abbott SL. 2007. Klebsiella, Enterobacter, Citrobacter, Serratia, Plesiomonas, and other Enterobacteriaceae, p 698–715. In Murray PR, Baron EJ, Jorgeson JH, Landry ML, Pfaller MA (ed), Manual of clinical microbiology, 9th ed ASM Press, Washington, DC. [Google Scholar]
- 212.von Graevenitz A, Bucher C. 1983. Evaluation of differential and selective media for isolation of Aeromonas and Plesiomonas spp. from human feces. J Clin Microbiol 17:16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ohkusu K. 2000. Cost-effective and rapid presumptive identification of gram-negative bacilli in routine urine, pus, and stool cultures: evaluation of the use of CHROMagar orientation medium in conjunction with simple biochemical tests. J Clin Microbiol 38:4586–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Merlino J, Siarakas S, Robertson GJ, Funnell GR, Gottlieb T, Bradbury R. 1996. Evaluation of CHROMagar Orientation for differentiation and presumptive identification of gram-negative bacilli and Enterococcus species. J Clin Microbiol 34:1788–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Rahim Z, Kay BA. 1988. Enrichment for Plesiomonas shigelloides from stools. J Clin Microbiol 26:789–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kohlerschmidt DJ, Mingle LA, Dumas NB. 2015. Identification of aerobic gram-negative bacteria, p 77–92. In Goldman E, Green LH (ed), Practical handbook of microbiology, 3rd ed CRC Press, Boca Raton, FL. [Google Scholar]
- 217.Farmer JJ, Boatright KD, Janda JM. 2007. Enterobacteriaceae: introduction and identification, p 649–669. In Murray PR, Baron EJ, Jorgeson JH, Landry ML, Pfaller MA (ed), Manual of clinical microbiology, 9th ed ASM Press, Washington, DC. [Google Scholar]
- 218.Ling TKW, Tam PC, Liu ZK, Cheng AFB. 2001. Evaluation of VITEK 2 rapid identification and susceptibility testing system against gram-negative clinical isolates. J Clin Microbiol 39:2964–2966. doi: 10.1128/JCM.39.8.2964-2966.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.O'Hara CM, Miller JM. 2002. Ability of the MicroScan rapid gram-negative type 3 panel to identify nonenteric glucose-fermenting and nonfermenting gram-negative bacilli. J Clin Microbiol 40:3750–3752. doi: 10.1128/JCM.40.10.3750-3752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.O'Hara CM. 2006. Evaluation of the Phoenix 100 ID/AST system and NID panel for identification of Enterobacteriaceae, Vibrionaceae, and commonly isolated nonenteric gram-negative bacill. J Clin Microbiol 44:928–933. doi: 10.1128/JCM.44.3.928-933.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Loh JP, Yap EPH. 2001. Rapid and specific detection of Plesiomonas shigelloides directly from stool by LightCycler PCR, p 161–169. In Reischl U, Wittwer C, Cockerill ER (ed), Rapid cycle real-time PCR, methods and applications, microbiology and food analysis. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 222.Gu W, Levin RE. 2006. Factors affecting quantitative PCR assay of Plesiomonas shigelloides. Food Technol 20:219–230. [Google Scholar]
- 223.Fukushima H, Tsunomori Y. 2005. Study of real-time PCR assays for rapid detection of food-borne pathogens. Kansenshogaku Zasshi 9:644–655. [DOI] [PubMed] [Google Scholar]
- 224.Fukushima H, Katsube K, Tsunomori Y, Kishi R, Atsuta J, Akiba Y. 2009. Comprehensive and rapid real-time PCR analysis of 21 foodborne outbreaks. Int J Microbiol 2009:1–13. doi: 10.1155/2009/917623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Meng S, Xu J, Xiong Y, Ye C. 2012. Rapid and sensitive detection of Plesiomonas shigelloides by loop-mediated isothermal amplification of the hugA gene. PLoS One 7:e41978. doi: 10.1371/journal.pone.0041978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Van Camp G, Chapelle S, De Wachter R. 1993. Amplification and sequencing of variable regions in bacterial 23S ribosomal RNA gene with conserved primer sequences. Curr Microbiol 27:147–151. doi: 10.1007/BF01576012. [DOI] [PubMed] [Google Scholar]
- 227.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Janda JM, Abbott SL. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, pitfalls. J Clin Microbiol 45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wang X, Xu L, Cao H, Wang J, Wang S. 2013. Identification and drug sensitivity of a Plesiomonas shigelloides isolated from diseased sturgeons. Wei Sheng Wu Xue Bao 53:723–729. [PubMed] [Google Scholar]
- 230.Dauga C. 2002. Evolution of the gyrB gene and the molecular phylogeny of Enterobacteriaceae: a model molecule for molecular systematic studies. Int J Syst Evol Microbiol 52:531–547. doi: 10.1099/00207713-52-2-531. [DOI] [PubMed] [Google Scholar]
- 231.Reddington K, Tuite N, Minogue E, Barry T. 2014. A current overview of commercially available Nucleic acid diagnostics approaches to detect and identify human gastroenteritis pathogens. Biomol Detect Quantif 1:3–7. doi: 10.1016/j.bdq.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Janda JM, Abbott SL. 2014. Culture-independent diagnostic testing: have we opened Pandora's box for good? Diagn Microbiol Infect Dis 80:171–176. doi: 10.1016/j.diagmicrobio.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 233.Khare R, Espy MJ, Cebelinski E, Boxrud D, Sloan LM, Cunningham SA, Pritt BS, Patel R, Binnicker MJ. 2014. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J Clin Microbiol 52:3667–3673. doi: 10.1128/JCM.01637-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Buss SN, Leber A, Chaplin K, Fey PD, Bankowski MJ, Kones MK, Rogatcheva M, Kanack KJ, Bourzac KM. 2015. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiological diagnosis of infectious gastroenteritis. J Clin Microbiol 53:915–925. doi: 10.1128/JCM.02674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Spina A, Kerr KG, Barbut F, Eigentier A, Zerva L, Tassios P, Popescu GA, Rafila A, Eerola E, Batista J, Maass M, Aschbacher R, Olsen KEP, Allerberger F. 2015. Spectrum of enteropathogens detected by the FilmArray GI panel in a multicenter study of community-acquired gastroenteritis. Clin Microbiol Infect Dis 21:719–728. doi: 10.1016/j.cmi.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 236.Croxatto A, Prod'hom G, Greub G. 2012. Applications of MALD-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol 36:380–407. doi: 10.1111/j.1574-6976.2011.00298.x. [DOI] [PubMed] [Google Scholar]
- 237.Kolínská R, Dřevínek M, Aldová E, Žemličková H. 2010. Identification of Plesiomonas spp.: serological and MALDI-TOF MS methods. Folia Microbiol (Praha) 55:669–672. doi: 10.1007/s12223-010-0109-3. [DOI] [PubMed] [Google Scholar]
- 238.Neville SA, LeCordier A, Ziochos H, Chater MJ, Gosbell IB, Maley MW, van Hal SJ. 2011. Utility of matrix-Assisted laser desorption ionization-time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J Clin Microbiol 49:2980–2984. doi: 10.1128/JCM.00431-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Jamal W, Albert MJ, Rotimi VO. 2014. Real-time comparative evaluation of bioMerieux VITEK MS versus Bruker Microflex MS, two matrix-assisted laser desorption-ionization time-of-flight mass spectrometry systems, for identification of clinically significant bacteria. BMC Microbiol 14:289. doi: 10.1186/s12866-014-0289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Martiny D, Burson L, Wybo I, Ait El Haj R, Dediste A, Vanderberg O. 2012. Comparison of the Microflex LT and Vitek MS systems for routine identification of bacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 50:1313–1325. doi: 10.1128/JCM.05971-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ford BA, Burnham C-DD. 2013. Optimization of routine identification of clinically relevant gram-negative bacteria by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry and the Bruker Biotyper. J Clin Microbiol 51:1412–1420. doi: 10.1128/JCM.01803-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Deng J, Fu L, Wang R, Yu N, Ding X, Jiang L, Fang Y, Jiang C, Lin L, Wang Y, Che X. 2014. Comparison of MALDI-TOF MS, gene sequencing and the Vitek 2 for the identification of seventy-three clinical isolates of enteropathogens. J Thorac Dis 6:539–544. doi: 10.3978/j.issn.2072-1439.2014.02.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Aldova E. 2000. New serovars of Plesiomonas shigelloides: 1992-1998. Cent Eur J Public Health 8:150–151. [PubMed] [Google Scholar]
- 244.Albert MJ, Ansaruzzaman M, Qadri F, Hossain A, Kibriya AKM, Haider K, Nahar S, Faruque SM, Alam AN. 1993. Characterisation of Plesiomonas shigelloides strains that share type-specific antigen with Shigella flexneri 6 and common group 1 antigen with Shigella flexneri spp. and Shigella dysenteriae 1. J Med Microbiol 39:211–217. doi: 10.1099/00222615-39-3-211. [DOI] [PubMed] [Google Scholar]
- 245.Shepherd JG, Wang L, Reeves PR. 2000. Comparison of O-antigen gene clusters of Escherichia coli (Shigella) sonnei and Plesiomonas shigelloides O17: sonnei gained its current plasmid-borne O-antigen genes from P. shigelloides in a recent event. Infect Immun 68:6056–6061. doi: 10.1128/IAI.68.10.6056-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Lefebvre J, Gosselin F, Ismail J, Lorange M, Lior H, Woodward D. 1995. Evaluation of commercial antisera for Shigella serogrouping. J Clin Microbiol 33:1997–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Humphries RM, Schuetz AN. 2015. Antimicrobial susceptibility testing of bacteria that cause gastroenteritis. Clin Lab Med 35:313–331. doi: 10.1016/j.cll.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 248.Stock I, Wiedemann B. 2001. Natural antimicrobial susceptibilities of Plesiomonas shigelloides strains. J Antimicrob Chemother 48:803–811. doi: 10.1093/jac/48.6.803. [DOI] [PubMed] [Google Scholar]
- 249.Bravo LF, Correa YM, Clausell JFI, Fernandez AA, Ramirez MA, Nunez FF, Ledo YG, CruzYI. 2009. Caracterizacion de factores de virulencia y susceptibilidad antimicrobiana en cepas de Plesiomonas shigelloides aisladas de pacientes con diarrea en Cuba. Rev Chil Infect 26:233–238. [PubMed] [Google Scholar]
- 250.Avison MB, Bennett PM, Walsh TR. 2000. β-Lactamase expression in Plesiomonas shigelloides. J Antimicrob Chemother 45:877–880. doi: 10.1093/jac/45.6.877. [DOI] [PubMed] [Google Scholar]
- 251.Wiegand I, Burak S. 2004. Effect of inoculum density on susceptibility of Plesiomonas shigelloides to cephalosporins. J Antimicrob Chemother 54:418–423. doi: 10.1093/jac/dkh322. [DOI] [PubMed] [Google Scholar]