SUMMARY
Molecular typing has revolutionized epidemiological studies of infectious diseases, including those of a mycobacterial etiology. With the advent of fingerprinting techniques, many traditional concepts regarding transmission, infectivity, or pathogenicity of mycobacterial bacilli have been revisited, and their conventional interpretations have been challenged. Since the mid-1990s, when the first typing methods were introduced, a plethora of other modalities have been proposed. So-called molecular epidemiology has become an essential subdiscipline of modern mycobacteriology. It serves as a resource for understanding the key issues in the epidemiology of tuberculosis and other mycobacterial diseases. Among these issues are disclosing sources of infection, quantifying recent transmission, identifying transmission links, discerning reinfection from relapse, tracking the geographic distribution and clonal expansion of specific strains, and exploring the genetic mechanisms underlying specific phenotypic traits, including virulence, organ tropism, transmissibility, or drug resistance. Since genotyping continues to unravel the biology of mycobacteria, it offers enormous promise in the fight against and prevention of the diseases caused by these pathogens. In this review, molecular typing methods for Mycobacterium tuberculosis and nontuberculous mycobacteria elaborated over the last 2 decades are summarized. The relevance of these methods to the epidemiological investigation, diagnosis, evolution, and control of mycobacterial diseases is discussed.
INTRODUCTION
Mycobacteria are speculated to have existed as early as 150 million years ago, in the Jurassic period (1), and today they are ubiquitous, occurring in every habitat and ecosystem of the world, perhaps except for the polar regions. The first known representative of this group was discovered, under the name of Bacillus leprae, by Hansen in 1875 (2), and the first scientific taxonomy of mycobacteria began in 1896, when the genus Mycobacterium was originally erected by Lehmann and Neumann. According to the most recent version of the List of Prokaryotic Names with Standing in Nomenclature (LPSN) database (http://www.bacterio.net/m/mycobacterium.html), the genus Mycobacterium now accommodates a total of 169 distinct species. They all fall into three major groups, that is, the Mycobacterium tuberculosis complex (MTBC), M. leprae, and mycobacteria other than the MTBC and M. leprae, collectively referred to as nontuberculous mycobacteria (NTM). Members of the MTBC are the causative agents of tuberculosis (TB). They are strict, intracellular pathogens of humans and animals without any defined environmental reservoirs. The MTBC comprises typical human-associated species, i.e., M. tuberculosis, M. africanum, and M. canettii, as well as several other species specifically adapted to infect domestic and wild animals: M. bovis (cattle), M. caprae (sheep and goats), M. microti (rodents), M. mungi (banded mongooses), M. orygis (members of the Bovidae family), M. pinnipedii (seals and sea lions), the dassie bacillus (Procavia capensis), and chimpanzee bacilli. Interestingly, some of those animal-adapted lineages have a documented zoonotic potential for humans. For instance, milk-borne transmission of TB by M. bovis was common in the prepasteurization era. Currently, 1 to 2% of TB cases in the United States and in Europe are attributable to M. bovis infection (3, 4). Mycobacterium caprae is another MTBC species capable of causing TB in humans as a result of transmission from livestock. This pathogen accounted for 0.3% of human TB cases in Spain and one-third of human TB cases formerly attributed to M. bovis in Germany (5, 6). There have been several reports on M. microti-induced TB in humans. However, zoonotic transmission could be established in only a few of these cases, with a raccoon, mice, and pets (dogs and cats) as the sources of infection (7, 8). Finally, M. pinnipedii was recognized as a human pathogen when its transmission from sea lions to humans was evidenced by using the tuberculin skin test and an interferon gamma release assay (9).
Mycobacterium tuberculosis, the most famous member of the MTBC and the most common cause of TB, has been one of the most devastating pathogens in the history of humankind. It is estimated that 2 billion people, or one-fourth of the world's population, are infected with M. tuberculosis. This pathogen produces nearly 9 million new infections and 1.5 million deaths every year (one-quarter of which are deaths of TB patients coinfected with human immunodeficiency virus [HIV]), ranking second, only to HIV, as the leading cause of death from an infectious agent (10). TB remains an enormous health and economic problem not only in developing regions but also in high-income countries due to TB-HIV coinfection and the emergence of multidrug-resistant (MDR) and, more recently, extensively drug-resistant (XDR) M. tuberculosis strains, currently accounting for 5% and 0.5% of all incident cases of TB, respectively (10). The increasing prevalence of drug resistance in M. tuberculosis has compounded the management of the disease and has considerably augmented TB-associated mortality among immunocompromised patients.
Mycobacterium leprae and M. lepromatosis, etiological agents of leprosy, differ from M. tuberculosis and other mycobacteria in many ways but perhaps most pointedly by their inability to be cultured in vitro. Differences are also evident at the DNA level, with leprosy bacilli having a much reduced genome (3.3 Mbp for M. leprae versus 4.4 Mbp for M. tuberculosis), due to massive gene loss, and considerably low G+C content (58% for M. leprae versus 66% for M. tuberculosis). A unique characteristic of M. leprae, not only among mycobacteria but also among all bacterial pathogens, is its capacity to invade the peripheral nervous system (11). Leprosy, like TB, is an ancient disease that has plagued humans for thousands of years. However, throughout its long history, never has TB been associated with such social stigmatization and discrimination as leprosy was in the Middle Ages. Although leprosy has already been eradicated from many parts of the world and is no longer a global health problem, it still persists in some regions of Asia, Africa, and Latin America where the disease is endemic, affecting a quarter million people annually (12). However, interest in leprosy has somewhat decreased over the past 2 decades, one reason for this being a dramatic decline in the prevalence of this disease with the advent of multidrug therapy and the other being an upsurge of TB cases in the mid-1990s, which resulted in a shift of the focus of attention toward tubercle bacilli.
Whereas the MTBC and M. leprae are obligate pathogens, NTM are for the most part environmental organisms, found predominantly in soil and waterways but also in animal species and food products. Despite being free-living saprophytes, they may, under certain conditions, usually linked with underlying immunodeficiency of the host, act as opportunistic pathogens, leading to a wide array of clinical syndromes (13). Of the more than 150 NTM species currently recognized, about 25 have consistently been associated with NTM diseases in humans and/or animals (14). The most prominent pathogenic NTM species include the M. avium complex (MAC) and M. kansasii, associated with pulmonary disease; M. ulcerans, which produces a disfiguring, ulcerative skin infection known as “Buruli ulcer,” which has emerged as the third most common mycobacterial disease worldwide after TB and leprosy; M. marinum, responsible for granulomatous skin lesions often referred to as “fish tank” or “swimming pool” granulomas; M. scrofulaceum, implicated in cervical lymphadenitis in children; or M. abscessus, M. fortuitum, and M. chelonae, which are the top causes of NTM soft tissue and skeletal infections (15–19). The known spectrum of NTM species involved in human disease has substantially expanded in recent years, and thus, the number of reported cases of NTM infections has risen (20). This can be ascribed to the development of new molecular tools for the identification of novel or previously neglected mycobacterial species. Despite the growing significance of NTM infections, only a very few comprehensive reports on NTM disease prevalence are available. To establish an accurate global picture of the epidemiology of NTM infections, a number of population-based, cross-sectional studies from different geographical locales need to be carried out. (For more information on clinical aspects of NTM disease, see reference 21.)
The modern epidemiology of TB and NTM diseases, as for virtually all infectious diseases, has become a multidisciplinary field of research, consuming most recent achievements in biology, genetics, pharmacology, medicine, and statistics. By incorporating molecular biology methods, epidemiological studies have reached the molecular level. Molecular epidemiology has emerged from a synergistic combination of genotyping techniques and conventional epidemiological approaches. This new discipline has established itself as a resource for understanding the key issues in the epidemiology of TB and other mycobacterial diseases. These issues include disclosing sources of infection, quantifying recent transmission, identifying transmission links and risk factors for transmission, discerning reinfection from relapse, tracking the geographic distribution and clonal expansion of specific strains, and determining the genetic basis behind specific phenotypic characteristics, including virulence, organ tropism, transmissibility, or resistance to antimicrobial drugs.
In this review, molecular typing methods for MTBC and NTM elaborated over the last 2 decades are summarized. The relevance of these methods to the epidemiological investigation, diagnosis, evolution, and control of mycobacterial diseases is discussed.
METHODS FOR MOLECULAR TYPING OF MYCOBACTERIA
The methods for molecular typing of mycobacteria rely on the diversity of genetic structures of these organisms. The genetic compositions of MTBC and NTM species are quite different, the former constituting an exceptionally homogeneous group genetically, with various MTBC members sharing on average >99.7% nucleotide identity (22). Polymorphisms in DNA occur, in the context of both species and strain divergence, as a consequence of imperfect DNA repair mechanisms, infidelity of DNA replication, and development of genome instability due to the malfunctioning of DNA metabolism-related proteins or effects of horizontal gene transfer (HGT). In the genomes of mycobacteria are homologs of genes involved in several DNA repair mechanisms, including base excision repair, nucleotide excision repair, homologous recombination (HR), and nonhomologous end joining (NHEJ) (23). Notably, even though M. tuberculosis does not possess homologs of genes of mismatch repair (MMR), the number of single nucleotide polymorphisms (SNPs) occurring in this species is not elevated (24–26). The reason for the limited occurrence of mutations in M. tuberculosis is a matter of dispute, but it is generally attributed to the activities of other DNA repair mechanisms, namely, base excision repair and nucleotide excision repair (27), and purifying/negative selection pressure (24). It is also important to note that pathogenic species of mycobacteria are exposed to a far more acute environment within the host organism than are nonpathogenic or opportunistic species (28). However, even though bacteria specialized to occupy host cells reside in a potentially highly mutagenic environment, a permanent switch-on of the DNA repair systems (29) and the activity of detoxifying enzymes (30) restrict the occurrence of mutations. It was recently shown that at least one double-strand break (DSB) repair system, HR or NHEJ, is required for M. tuberculosis to replicate within human macrophages, suggesting that some DSBs accumulate in DNA of tubercle bacilli in this environment (31). On the other hand, an M. tuberculosis mutant defective in both systems (HR and NHEJ) appeared not to be attenuated in various animal models (32). The amount of mutations generated in pathogenic species seems to be dependent on the stage of the disease. A comparison of mutations in M. tuberculosis isolated from cynomolgus macaques with active and latent disease revealed that the generation time-versus-mutation rate curves are similar during active disease, latency, and disease reactivation (33). This observation was contrasted with data from another study on M. tuberculosis isolates of human origin. Here, the authors observed that mutation rates were substantially lower during latency, although it is important to note that due to technical issues, those authors were unable to calculate replication and mutation rates separately (34).
Although HGT has been thought to contribute only very marginally to the variability of mycobacteria, there is increasing evidence that HGT plays an important role in shaping the diversity of this group. In M. tuberculosis, HGT is thought to be restrained, yet it was postulated to have occurred in the distant past, prior to the evolution of the M. tuberculosis complex (35, 36). Plasmids have been found in M. avium (37, 38), M. intracellulare (37), M. scrofulaceum (37), M. ulcerans (39), M. marinum (40), and M. fortuitum (41), opening the possibility of gene transfer. In fact, conjugation has been observed in M. smegmatis (42–44), in M. abscessus subsp. bolletii (45), and between M. kansasii and M. avium (46). Additionally, M. smegmatis is suspected to be capable of spontaneous plasmid transformation (47). Theoretically, genes may also be transferred by phages (48, 49), but to date, there have been no reports of naturally occurring transduction in mycobacteria.
Considering that HGT seems to be limited in mycobacteria, typing methods for this group rely largely on minor changes of DNA sequences resulting from internal mutagenesis. Comparative studies have shown that genomic variation in the MTBC has its source in deletions (22, 50, 51), duplications (52), insertions (53), mobile genetic element movements (51), and SNPs (54–58). These polymorphism-borne genetic events have been extensively explored over the last years and have been employed to develop a number of typing methods.
Phenotype-Based Methods
The first methods used for the identification and discrimination of mycobacteria relied on individual strain phenotypic characteristics, including colony morphology (59), susceptibility to antimicrobial agents (60), as well as biochemical (61–63) and serological reactivity, the latter being the basis of mycobacterial phage typing (64–66). Nowadays, some of these methods are still in use and are commonly incorporated in routine laboratory diagnostics (67). Of particular importance is drug susceptibility testing (DST), which has become a key component of global TB control programs. The World Health Organization (WHO) has approved a number of old methods, including conventional phenotypic DST on both solid and liquid media (68), microscopic observation of drug susceptibility (MODS) (69), colorimetric redox indicator (CRI) methods (70), and nitrate reductase assays (NRAs) (71). Regarding phage typing, although sometimes useful (72, 73), the limited number of mycobacteriophages identified and poor reproducibility make this method impractical for epidemiological studies (74, 75). Phenotype-based typing methods, currently under intense investigation, analyze the composition of the mycobacterial cell wall by thin-layer chromatography (76–78), high-performance liquid chromatography (79, 80), gas chromatography (81, 82), and mass spectrometry (MS) (83–85). One promising methods is matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS. It can be used to differentiate mycobacteria in two ways, that is, either by analyzing ions desorbed directly from the cell surface (83) or by producing unique spectral fingerprints from extracted proteins (86, 87). It appears that this method, regardless of the approach, has the potential for both identification and typing of Mycobacterium spp. (84, 86, 87). Note, however, that MALDI-TOF MS could not discriminate sufficiently between some mycobacterial species, including members of the MTBC and other closely related species, or the identification results often lacked reliability (86, 87). Next to the close genetic relatedness of the species, an important limitation of the MS approach is the restricted coverage of spectral entries of reference. The MS system may produce a species-specific spectral profile, yet unless it is accommodated in the database, a correct identification cannot be accomplished.
Advantageously, MS can be performed directly on clinical samples, making it an attractive alternative to genotyping methods for robust and rapid identification to the species level (85).
In comparison to DNA-based techniques, the usefulness of phenotypic modalities for typing is considered limited or doubtful. First, they usually do not provide sufficient discrimination, sometimes even at the species level. Second, the biological properties of each strain may easily change depending on the current composition of the bacterial population and environmental conditions. For example, the drug susceptibility pattern may change for the same isolate over the course of treatment due to the acquisition of drug resistance. Likewise, certain strains of the same species may exhibit different biochemical properties, leading to confusing results. Nevertheless, phenotype-based methods should not be underestimated. First, detectable phenotypic changes among closely related M. tuberculosis strains may alter the immunogenic properties and pathogenicity of the bacterial population, and hence, they may also alter the epidemiological patterns in the human population (88). Second, phenotype-based differentiation may be used in combination with other methods to determine the relationships between species and strains of mycobacteria, especially in samples where DNA is degraded and the results of genetic analysis are unclear. For example, multilocus enzyme electrophoresis (MLEE), based on the variation in enzyme electrophoretic mobility, has been successfully used with a combination of DNA-based methods to distinguish M. paraffinicum from M. scrofulaceum (89) and to differentiate among M. avium (90, 91) or M. abscessus (92) isolates. It is important to note that phenotype may significantly influence the data obtained by genotyping methods. For example, with the amplified fragment length polymorphism (AFLP) method, dendrogram analysis grouped M. avium subsp. paratuberculosis isolates into two groups according to the isolation scheme (tissue-associated versus fecal isolates). However, no genetic sequence differences were observed between the two groups. An explanation for this finding was that epigenetic modifications, such as DNA methylation, prevented restriction enzymes from recognizing their sites (93). Since DNA methylation impacts gene expression (94), it may have a tremendous impact on phenotype-derived data.
Methods Based on Nonrepetitive Sequences
Gene sequence analysis.
Sequences of several genes are considered good targets for species identification, although it is usually necessary to analyze several sequences at a time. In particular, 16S rRNA and internal transcribed spacers (ITSs) have been shown to be useful for the rapid identification of many mycobacterial species (95), including M. malmoense, M. szulgai, and M. flavescens, which are hardly identified with conventional methods (96). A list of major genes that might be used for differentiation between mycobacterial species is shown in Table 1.
TABLE 1.
Genes used for differentiation of mycobacterial species
| Gene | Product | Reference(s) |
|---|---|---|
| rrs | 16S rRNA | 142, 598 |
| ITS | Internal transcribed spacer region | 599, 600 |
| hsp65 | Heat shock protein 65 | 601–606 |
| groES | 10-kDa chaperonin | 607 |
| recA | Recombination protein | 146, 608 |
| rpoB | DNA-directed RNA polymerase beta chain | 609–612 |
| dnaJ | Chaperone protein | 613, 614 |
| oxyR | Probable hydrogen peroxide-inducible gene activator | 615, 616 |
| pncA | Pyrazinamidase/nicotinamidase | 615, 617 |
| rnpB | Catalytic subunit of RNase P | 618 |
| sodA | Superoxide dismutase | 146, 619 |
| gyrB | DNA gyrase subunit B | 620, 621 |
| secA1 | Preprotein translocase | 622 |
An important application of gene sequence analysis in mycobacterial diseases is the detection of drug resistance. It not only provides clinically relevant information but also assists in deciphering strain relatedness. Several genes of mycobacteria can be analyzed to detect drug resistance; some of them that overlap housekeeping genes are used for species identification. A list of major M. tuberculosis genes that have been linked with the acquisition of drug resistance is presented in Table 2.
TABLE 2.
Major genes of M. tuberculosis linked with acquisition of drug resistance
| Drug | Gene | Product | Reference(s) |
|---|---|---|---|
| First line | |||
| Isoniazid | katG | Catalase-peroxidase-peroxynitritase T | 623–626 |
| inhA | NADH-dependent enoyl-acyl carrier protein reductase | 626–629 | |
| ndh | NADH dehydrogenase | 626, 630, 631 | |
| ahpC | Alkyl hydroperoxide reductase C | 626, 632, 633 | |
| Rifampin | rpoB | DNA-directed RNA polymerase β chain | 634, 635 |
| Pyrazinamide | pncA | Pyrazinamidase/nicotinamidase | 636–638 |
| Ethambutol | embCAB | Membrane indolylacetylinositol arabinosyltransferase | 639–642 |
| Streptomycin | rpsL | 30S ribosomal protein S12 | 643, 644 |
| rrs | 16S rRNA | 643–645 | |
| gidB | Glucose-inhibited division protein B | 645–647 | |
| Second line | |||
| Amikacin-kanamycin | rrs | 16S rRNA | 648, 649 |
| eis | Enhanced intracellular survival protein | 650, 651 | |
| Ethionamide | ethA | Monooxygenase | 652 |
| inhA | NADH-dependent enoyl-acyl carrier protein reductase | 652 | |
| ethR | TetR family transcriptional repressor | 653 | |
| ndh | NADH dehydrogenase | 653 | |
| Fluoroquinolones | gyrAB | DNA gyrase | 654, 655 |
| para-Aminosalicylic acid | thyA | Thymidylate synthase | 656, 657 |
| folC | Folylpolyglutamate synthase C | 658, 659 |
(i) PCR-RFLP.
At first, analysis of gene sequences was addressed with PCR-restriction fragment length polymorphism (RFLP) analysis, otherwise called the PRA, or PCR-restriction enzyme analysis (REA), method. This method combines PCR amplification and restriction analysis. The pattern obtained after electrophoresis is species or strain specific. Traditional PCR-RFLP produces patterns that may be difficult to distinguish by eye. Therefore, automated versions of this method have been developed. For example, the sizes of the fragments can be assessed by fluorescence capillary electrophoresis when samples are amplified by using fluorophore-labeled primers (97). Whereas this method has proven useful for the differentiation of many mycobacterial species (98–100), novel technologies for gene sequencing provide more in-depth information about the sequences of the analyzed genes.
(ii) Hybridization.
Hybridization probes, while not used for typing at the strain level, are currently widely used for species differentiation. They have guided the development of tests that utilize nucleic acid probes to specifically identify a target sequence in the organism under investigation. In short, labeled probes are mixed with nucleic acids from the target organism. Hybrids are separated or discriminated from nonhybridized probes and detected by using labels. There are two main variants of gene probe tests. Direct nucleic acid tests (NATs) require a large amount of bacterial material. The addition of an amplification step in nucleic acid amplification tests (NAATs) allows the detection of bacterial DNA directly in clinical samples. These tests are rapid (can be performed within 2 to 8 h) and can detect a specific organism in paucibacillary clinical samples. Because of the low detection limit, the performance of these tests requires strict contamination prevention and quality control. It is also important to take into consideration that inhibitors in clinical samples can result in false-negative results. While these methods require specialized equipment, the speed of pathogen detection makes them a useful alternative to culture-based typing.
Several variants of gene probe tests for the detection of mycobacteria have been described, and either RNA or DNA can be detected. Depending on the type of nucleic acid to be targeted, several methods of amplification can be used, including standard PCR (101), real-time PCR (102), nucleic acid sequence-based amplification (103), strand displacement amplification (104), and transcription-mediated amplification (105). The target sequences may be PCR products that are either amplified in their entirety (101) or digested by restriction enzymes (106). Hybridization can be performed in solution (103), on nitrocellulose strips (107), or on microdilution plates, with products being labeled colorimetrically (108) or biotinylated and detected by an enzyme-linked immunosorbent assay (ELISA) (109). When hybridization is performed in solution, the products may be detected by TaqMan real-time PCR (110) or by molecular beacons (103).
Gene probes for detection of a large number of mycobacteria, including the MTBC and NTM, have been described (106, 108, 111). For most clinically significant mycobacterial species, a number of commercially available tests have been developed. Depending on the target sequence analyzed, they can be used to either detect a group of organisms or specifically differentiate mycobacteria. Major commercial tests used for the detection and differentiation of mycobacteria are summarized in Table 3. NAATs other than Xpert MTB/RIF, though approved by some local health departments, are not recommended by the WHO since they are not sufficiently sensitive to exclude TB and have problems in identifying M. tuberculosis isolated from extrapulmonary sites (112–116).
TABLE 3.
Major commercial NAATs used for identification and differentiation of mycobacteria
| Test (manufacturer) | Target sequence(s) | Target organisms |
|---|---|---|
| AccuProbe (Hologic) | rRNA | MTBC, M. avium, M. intracellulare, M. avium, M. avium complex, M. gordonae complex, M. kansasii complex |
| BD ProbeTec ET (Becton, Dickinson) | IS6110 and 16S rRNA | MTBC |
| Mycobacteria Identification Array kit (CapitalBio) | 16S rRNA | MTBC, M. avium, M. kansasii, M. scrofulaceum, M. terrae, M. phlei, M. marinum and M. ulcerans, M. szulgai and M. malmoense, M. smegmatis, M. intracellulare, M. gordonae, M. fortuitum, M. gilvum, M. chelonae and M. abscessus, M. nonchromogenicum, M. aurum, M. xenopi |
| GenoType MTBC (Hain Lifescience) | gyrB SNPs and RD1 presence/absence | MTBC |
| Gen-Probe Amplified Mycobacterium tuberculosis Direct test (Hologic) | rRNA | MTBC |
| Inno-LiPA Mycobacteria v2 (Fujirebio) | 16S-23S rRNA spacer region | MTBC, M. kansasii, M. xenopi, M. gordonae, M. genavense, M. simiae, M. marinum and M. ulcerans, M. celatum, M. avium, M. intracellulare, M. scrofulaceum, M. malmoense, M. haemophilum, M. chelonae complex, M. fortuitum complex, M. smegmatis |
| Inno-LiPA Rif.TB (Fujirebio) | rpoB | MTBC |
| Cobas TaqMan MTB (Roche) | rRNA | MTBC |
| Truenat MTB (Molbio Diagnostics) | Ribonucleoside diphosphate reductase gene | MTBC |
| Xpert MTB/RIF (Cepheid) | rpoB | MTBC |
(iii) SNP typing.
Two major lines of research based on SNP analysis include lineage-specific typing and determination of the occurrence of mutations leading to drug resistance. SNPs exhibit low levels of homoplasy; however, convergent evolution, especially within drug susceptibility-related genes, is considered common.
The SNP at a particular location might be detected by REA (117) or by a variety of PCRs (118, 119). Modern technology provides several efficient methods to analyze several SNP sites at a time. They can be addressed with molecular beacons, as they are able to distinguish sequences that differ by even a single nucleotide substitution (120–122). Next, they can be detected by identifying shifts in melting temperatures obtained by real-time PCR curve analysis (123–126). SNaPshot analysis (Thermo Fisher Scientific) allows the detection of up to 10 SNPs in a single experiment with a capillary electrophoresis instrument (127). This analysis consists of multiplex PCR using primers of different lengths, resulting in length-specific products for each SNP. These primers are elongated by one specifically labeled dideoxynucleotide (128, 129). Similar in principle is the iPLEX Gold technology. Here, products are amplified by using mass-modified terminators detected by MALDI-TOF (130). Furthermore, microarray technology can be used to simultaneously detect a number of mutations within different genes (131, 132). Multiplexing can also be achieved by a multiplex ligation-dependent probe amplification assay (MLPA) (133). This method combines the analysis of SNPs and large sequence polymorphisms (LSPs). Short oligonucleotides hybridize with the target sequence and are ligated with each other. When SNPs are present at the end of this oligonucleotide, adjacent fragments are unable to ligate. When a region of difference (RD) contains a deletion, the ligated product lacks the oligonucleotide that would hybridize with this sequence. Detection of ligation products is performed by PCR, and the products can be further separated by capillary electrophoresis (133) or read on a Magpix reader (Luminex, Austin, TX, USA) (134). Since this method is capable of analyzing up to 50 markers, it has shown the potential to identify several mycobacterial species, including those within the MTBC, as well as a range of drug resistance markers (134, 135). Similar in principle is analysis by ligation-dependent PCR with fluorescence signal detection and a Luminex flow cytometer, i.e., multiplexed oligonucleotide ligation PCR (MOL-PCR) (136), or padlock probes (137, 138).
The detection of SNPs in drug resistance genes has laid the basis for the development of commercially available tests for detecting drug resistance within the MTBC, including Inno-LiPA Rif.TB (Innogenetics, Belgium), GenoType MTBDR and GenoType MTBDRplus (Hain Lifescience GmbH, Germany), TB-biochip (OOO Biochip-IMB, Russia), and Xpert MTB/RIF (Cepheid). GenoType MTBDR and Xpert MTB/RIF have been approved by the WHO for drug susceptibility testing (139; for a review, see reference 140).
(iv) Gene sequencing.
Sequencing provides the ultimate level of detail for gene sequence analysis, as it analyzes the entire sequence of a given gene. It is used for species identification and SNP detection, including new SNPs possibly involved in drug resistance (141).
Gene sequence analysis restricted to small parts of the genome can be efficiently performed by using Sanger sequencing or pyrosequencing. Alternatively, this analysis may be performed with MALDI-TOF MS (142) or PCR-electrospray ionization (ESI) MS (143). However, as for all gene sequence analysis methods, better results can be achieved when multiple loci are analyzed. The sequencing of groups of alleles in order to differentiate species is a procedure called multilocus sequence typing (MLST). It has been successfully used to differentiate members of the MAC (144, 145) and fast-growing mycobacteria (146). A scheme to differentiate members of the M. abscessus complex has also been developed (147–149), but some reports suggest that MLST might not be sufficient to properly differentiate all strains (150). There is currently no efficient scheme for MLST to differentiate members of the MTBC; however, sequencing of groups of genes has been shown to disclose interstrain diversity (151–154). To avoid confusion, sequencing of groups of genes in order to disclose interstrain diversity is termed multilocus sequence analysis (155).
Genome analysis.
(i) PFGE.
The first molecular typing methods for the M. tuberculosis genome were based on RFLP analysis of bacterial DNA. Here, chromosomal DNA isolated from different mycobacterial strains is digested by using various restriction enzymes. The resulting restriction fragments are separated by gel electrophoresis and visualized with UV light. The observed fingerprint patterns are strain specific. Such a procedure has several disadvantages. First, it is arduous to provide high-resolution electrophoretic separation of fragments within a broad range of sizes. Moreover, it is difficult to analyze DNA fragments when more restriction enzymes are used for digestion (156). A more accurate separation of DNA fragments is obtained with an RFLP-related method called pulsed-field gel electrophoresis (PFGE), which was designed to simplify and improve the discriminatory capacity of the standard RFLP method. The principle of PFGE is that the electric field is periodically alternated, which forces DNA fragments to change direction and thus allows large molecules to be separated from each other. A major strength of PFGE is the ability to separate larger DNA fragments, beyond the 50-kb limit of unidirectional electrophoresis (157, 158). This is achieved by using rare-cutting restriction enzymes. An essential step in the PFGE procedure is the preparation of genomic DNA. Since large DNA molecules are prone to shearing and crushing, DNA is isolated in a gentle manner by first embedding a suspension of the bacterium in agarose plugs, lysing the cells in situ, and digesting the chromosomal DNA with restriction enzymes. The plugs are then loaded into the gel wells and sealed into place with agarose.
PFGE typing has successfully been used to differentiate between strains of M. tuberculosis (159), M. bovis (160), and M. bovis BCG (161). The reproducibility and discriminatory power of PFGE-based methods are high, although their use seems to be restricted to scientific or reference laboratories. PFGE is expensive and technically demanding, has a long turnover time (usually about 5 days), and, similarly to RFLP, requires large amounts of high-quality DNA. Moreover, discrimination between strains might not always be produced (159, 162, 163). The above-mentioned limitations discourage the use of PFGE for molecular epidemiological studies of MTBC infections. Whereas rarely applied for the MTBC, PFGE still remains the most powerful typing system for many NTM species. This method has been applied, with different degrees of success, to both slow-growing NTM, including M. kansasii (164–166), the MAC (167), M. avium subsp. paratuberculosis (168), M. gordonae (169), and M. haemophilum (170), and rapidly growing mycobacteria, such as M. fortuitum (171–173), M. chelonae (174), and M. abscessus (92, 174). PFGE has also been used as a confirmatory method for the typing of M. abscessus, M. massiliense, and M. bolletii already genotyped by repetitive-sequence-based PCR (rep-PCR) (175).
(ii) RAPD analysis.
Randomly amplified polymorphic DNA (RAPD) analysis or arbitrarily primed PCR (AP-PCR) is a typing method that has increasingly been used for estimating genetic variability among different bacterial taxons. This method requires no previous knowledge of the template DNA sequence. By using a single, arbitrarily designed primer with a length of 5 to 50 bp and low-stringency conditions, the primer anneals to template DNA at both perfectly and partially matched sites, resulting in strain-specific multiband DNA profiles (165). Although this method has high discriminatory power, it suffers from several limitations. Of these limitations, poor reproducibility is the most important. It is believed that the differences among RAPD strain profiles are due to the technical and operating parameters of the method rather than true interstrain genetic polymorphism. Variations in RAPD patterns are thus driven by variations in the priming efficiency during early rounds of amplification, and these variations in turn depend on template concentration and purity, the primer/template ratio, or thermal ramping rates of the thermocycler used. Nevertheless, RAPD analysis has been used for genotyping of both MTBC bacilli (176–179) and NTM species. Regarding the latter, RAPD analysis, when applied to M. fortuitum strains, had a lower discriminative power than PFGE (173). RAPD analysis based on several primer sets was used to analyze nonpigmented, rapidly growing mycobacteria (180). It was found that differentiation of epidemiologically unrelated strains requires at least 3 primer sets to be used, as the use of a single primer leads to false clustering. A combination of several primer sets yielded RAPD profiles that were at least comparably polymorphic with those of PFGE (180). RAPD analysis was also applied for genotyping of M. abscessus and M. chelonae, which often lyse spontaneously during gel electrophoresis and cannot be assessed by PFGE (165, 174), as well as for M. phocaicum (181), M. gordonae (182), M. szulgai (183), and M. malmoense (184). Abed et al. performed RAPD analysis on M. tuberculosis strains by using a 16-23S rRNA gene ITS as a PCR template, which gave the method higher discriminatory power (185).
(iii) AFLP.
AFLP analysis is a PCR-based method in which DNA is digested with two restriction enzymes, a rare cutter and a frequent cutter, which have 6- and 4-bp recognition sites, respectively. Such enzyme pairs were HindIII and TaqI (186), EcoRI and MseI (187, 188), or ApaI and TaqI (189). The resulting restriction fragments are ligated to double-strand adaptors (10 to 30 bp) recognized by PCR primers that are complementary to the adaptor sequence, carry the restriction site sequence, and contain selective bases at their 3′ ends. The use of radiolabeled primers allows visualization of PCR products by means of autoradiography (186).
In the fluorescent AFLP (fAFLP) method, DNA restriction fragments obtained upon the cooperative action of a frequent cutter (MseI) and a rare cutter (EcoRI) are PCR amplified with five primers, including one nonselective, unlabeled forward primer targeting the MseI adaptor site and four reverse primers targeting the EcoRI adaptor site and differing from each other by containing one selective base, A, G, C, or T, each labeled with a different fluorescent dye. An automated DNA sequencer allows visualization of PCR products, and precise estimation of their molecular weights is possible due to fluorescent internal-lane standards (190). While the traditional AFLP method has lower discriminatory potential than IS6110-based RFLP (IS6110-RFLP) typing (188, 189), the resolution of the fAFLP method is comparable to that of IS6110-RFLP typing (190, 191).
The AFLP method provides an interesting option for typing of NTM species, including members of the MAC (192) and M. haemophilum (193). Furthermore, AFLP analysis has successfully been used to differentiate M. marinum from M. ulcerans, which are otherwise difficult to distinguish (194, 195).
(iv) Deletion mapping.
Deletions or, rather, LSPs can be used as molecular markers to study genetic variability among mycobacteria. The LSP-based methodology relies on previous knowledge of the analyzed sequences, and it usually requires relatively large quantities of DNA (micrograms). However, as deletions tend to be unique and irreversible events, data obtained by analyzing their patterns provide information about strain relatedness over protracted periods of time. For example, LSP analysis revealed that M. tuberculosis does not originate from present-day M. bovis, as previously thought, but originates from an unknown ancestor (196). As a major application, these polymorphisms can be used to differentiate MTBC strains at the species level by demonstrating the presence or absence of so-called regions of difference (RDs) (197–199). For M. tuberculosis, even though this method can be used to determine genetic differences between individual clinical isolates (22), it is rather harnessed to decipher phylogeographical relationships between strains (200–202). LSP analysis also revealed significant variations between different M. bovis BCG vaccine strains (203). Furthermore, LSPs have been used to identify differences between MAC (204) and M. avium subsp. paratuberculosis (205, 206) strains. A plasmid-based analysis allowed the identification of multiple DNA deletions among M. ulcerans clinical isolates (207). However, this method turned out to be impractical for studying genetic diversities within local populations (208).
Two major ways to analyze LSPs include PCR typing, tailored for small-scale observations, and microarrays, allowing screening at the whole-genome level. An intermediate method, allowing the detection of LSPs within a few chosen loci, is deligotyping (209), which conceptually and technically simulates the spoligotyping method (described below). Here, 43 genomic regions prone to LSPs among M. tuberculosis complex strains are interrogated. Deligotyping is an efficacious approach for the rapid screening of M. tuberculosis clinical isolates and, more importantly, a useful marker for delineating phylogenetic relationships (200, 210).
While the major advantage of the microarray technology is that it may analyze the entire genome, there are some important limitations of this method. Some platforms are unable to detect deletions smaller than 350 bp; hence, the sensitivity of this method may be seriously affected (22). Furthermore, cross-hybridization of similar sequences restricts the use of this method to nonrepetitive fractions of the bacterial genome.
(v) Whole-genome sequencing.
The completion of the genomic sequence of M. tuberculosis H37Rv (23) has commenced a whole new chapter in the epidemiological study of mycobacteria. The development of second-generation sequencing (SGS) and further-generation sequencing platforms has made studies on mycobacterial epidemiology as informative as they have never been before. The first mycobacterial whole-genome studies exploited the Sanger sequencing method described above. However, it was the introduction of SGS that allowed whole-genome sequencing (WGS) at a large scale.
To date, five major SGS platforms used in mycobacterial research were or have been 454 pyrosequencing (Roche), discontinued in 2014; HiSeq/MiSeq (Illumina); SOLiD (Thermo Fisher Scientific), announced to be discontinued after 2017; and Ion Torrent/Ion Proton (Thermo Fisher Scientific), followed by a third-generation sequencing technology, PacBio (Pacific Biosystems) (for more information on high-throughput sequencing platforms, see references 211 and 212).
All SGS technologies share a similar protocol. The genome is first fragmented and amplified on a solid support in order to increase the signal emission. During actual sequencing, the signal is then generated by the incorporation of a nucleotide or oligonucleotide and read in real time. SGS sequencers read out short sequences ranging from 150 bp to 800 bp, depending on the platform. These sequences are further either mapped to a known template or assembled de novo based on the overlapping regions.
Noteworthy, data obtained by currently available high-throughput sequencing technologies suffer from sequencing errors characteristic of each individual platform. For example, the Ion Torrent and 454 pyrosequencing platforms have had difficulty in sequencing through repetitive regions (e.g., homopolymers and microsatellites). It is important to take this into consideration during epidemiological studies, where minor sequence differences may result in a false interpretation of the results (213–215). Furthermore, mistakes generated during sequencing might influence the efficiency of the de novo sequence assembly. The latter is also impeded because the readouts from SGS are short. While a fourth-generation sequencing technology, MinION (Oxford Nanopore Technologies), allows sequencing of a single molecule of a few thousand bases without the need for amplification (216), further reductions in sequencing error rates are needed before these technologies will be useful for epidemiological studies of mycobacteria.
WGS provides information about the whole genome, and this method can identify virtually all varieties of markers detected by the above-mentioned genotyping methods. It is therefore much more accurate and precise in detecting variability among strains and provides a wealth of information at every level possible, from global (population), through local (community) and individual host (single patient), to pathogen (strain) itself.
At the global level, WGS can be used to investigate genetic relationships between different species, which has already been tried for M. ulcerans and M. marinum (217) and members of the M. abscessus complex (218). It can also be helpful in exploring important questions about the biology of mycobacteria, including evolutionary mechanisms shaping mycobacterial populations (219–221), antibiotic resistance (222–225), or virulence and immunogenicity (54, 226, 227).
At the local level, compared with traditional genotyping methods such as IS6110-RFLP typing, spoligotyping, or 24-locus mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) typing, WGS provides much higher resolution and in-depth knowledge about each strain under study. Large-scale investigations have demonstrated that WGS is useful in delineating outbreaks and tracking transmission routes. When the same strains that were subjected to WGS were examined by traditional methods, they were, in part, wrongly clustered. By disclosing superspreaders, WGS allows prediction of undiagnosed cases and therefore early administration of proper treatment to newly infected individuals (55–57, 215, 228, 229). However, it was also observed that strains that were clustered by WGS were not necessarily linked by patient interview data. Therefore, it needs to be pointed out that due to the exceptionally stable genetic profile of some mycobacterial species like M. tuberculosis, even WGS resolution might not be sufficient to delineate routes of transmission among all cases (230).
At the patient level, WGS provides an excellent tool to investigate the transmission and microevolution of mycobacteria. For example, in studies performed by Merker et al., WGS enabled observation of the intrapatient evolution of serial M. tuberculosis isolates recovered from three MDR TB patients undergoing longitudinal treatment, which finally led to the development of XDR TB. The results showed that such evolution may be more complex than previously anticipated. While a mutator phenotype was not detected in any of the strains, for two patients, this analysis revealed the presence of long-term coexistent clonal subpopulations that displayed different drug-resistant allele combinations. The third patient turned out to be reinfected by an exogenous strain whose IS6110-RFLP pattern was identical to that of the resident strain. Thus, the high resolution of WGS was essential to accurately detect exogenous reinfection (231). It has to be stressed that 2 to 20% of patients are estimated to be infected with multiple strains (232–235), and mixed infections are usually difficult to detect by using conventional methods. Nevertheless, Chan et al. (236) described a mixed infection in an 18th century mummy. Those authors implemented metagenomic analysis of DNA that had been obtained directly from an infected sample. The usefulness of metagenomic analysis in detecting mixed infections was further confirmed by Doughty et al. (237). In the same paper, the potential of shotgun metagenomics, sequencing of DNA from samples without culture, or target-specific amplification was shown (237). Hence, metagenomic analysis might open a new chapter in diagnostics, at least in the case of MDR/XDR strains (238). Furthermore, metagenomic research might yet bring much new information about the evolution of mycobacteria, as it allows the reconstruction of ancient bacterial genomes from skeletal remains, even those that are 1,000 years old (239).
Methods Based on Repetitive Sequences
Insertion sequences.
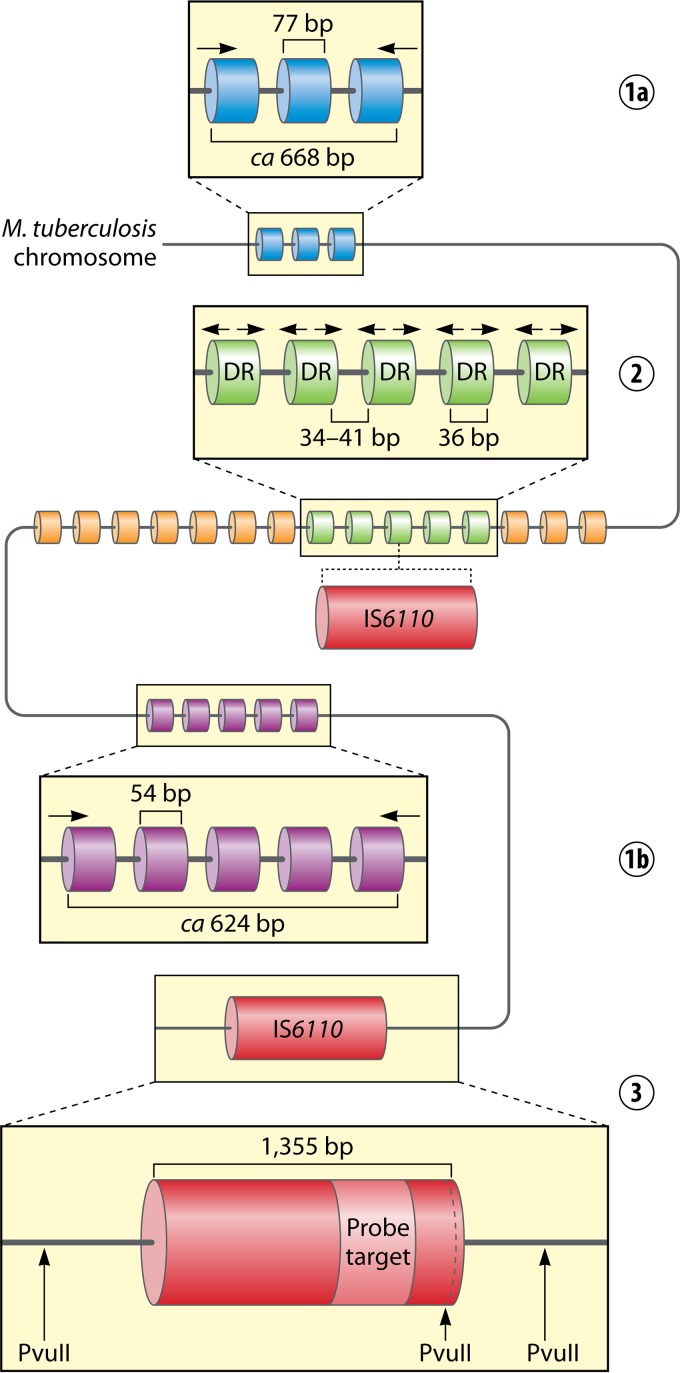
Insertion sequences (ISs) are small mobile genetic elements that encode only their capacity for transposition and regulation (240). Insertion of IS elements may cause gene disruptions and lead to the alteration of the expression of adjacent genes (241). In the MTBC, a total of 29 different IS elements belonging to 4 IS families were identified (23). Eight of these IS elements are present in M. tuberculosis H37Rv in more than a single copy, with 3 occurring in 3 copies (IS1557) or more (IS1081 and IS6110) (242). IS6110, the most extensively studied IS element, is a member of the IS3 family, which was originally described in Enterobacteriaceae (243). It is 1,355 bp long with imperfect 28-bp terminal inverted repeats (TIRs) at its ends and contains two, partially overlapping, open reading frames, orfA and orfB, coding for a transposase (243–245). IS6110 is unique to the MTBC, and in most of its members, the sequence is present in multiple copies. The exception is M. bovis, which contains only one to three IS6110 sequences (246). Generally, the copy number of IS6110 varies from 0 to 25. These elements are scattered throughout the genome, although certain hot spots for their integration into the chromosome were described. The regions where IS6110 is preferentially inserted include the direct repeat (DR) locus, the phospholipase C gene (plc) region, the Pro-Pro-Glu (PPE) family genes, the dnaA-dnaN intergenic region, or other ISs such as IS1547 (247). Due to its large number of copies and variability in location, IS6110 has become a useful genetic marker to differentiate M. tuberculosis strains (248–254). The presence of IS6110 has also been exploited to identify M. tuberculosis infection in ancient mummified samples (255–257).
In contrast to IS6110, IS1081 is present in 5 to 7 copies per genome and is rather homogeneously distributed among M. tuberculosis strains, which precludes its use as a diagnostic marker (258, 259). Likewise, IS1547 is present in low numbers of copies per genome and thus cannot be used for strain discrimination (23, 260).
A number of different ISs have been described for various NTM species. IS1245 and IS1311 are the most relevant for molecular typing of MAC strains (261–263). Besides IS1245 and IS1311, the genome of MAC bacilli contains other insertion elements. These insertion elements include IS900, present in M. avium subsp. paratuberculosis (264); IS901, present in M. avium subsp. avium (265); IS902, present in M. avium subsp. silvaticum (266); and IS666, IS1110, and IS1626, which are not yet thoroughly studied among M. avium isolates (267–269). Of the ISs described so far, IS900 (270, 271), IS901 (272), and IS902 (273) have been used for both the identification and differentiation of various M. avium strains.
Finally, there is a number of ISs used for epidemiological studies of NTM other than the MAC. These ISs include IS1395 in M. xenopi (274), IS1511/IS1512 in M. gordonae (275), IS1407 in M. celatum (276), IS6120 in M. smegmatis (277), IS2404 in M. ulcerans, IS2606 in M. ulcerans and M. lentiflavum (278), and IS1652 in M. kansasii (111).
REA typing.
A combination of traditional REA and a DNA hybridization assay gave one of the most popular typing technologies, referred to as RFLP analysis. The principle of this technology is that by detecting homologous sequences distributed across the genome, it generates strain-specific genetic patterns or fingerprints (279, 280).
Early studies with RFLP typing as a typing system were unsuccessful, producing little discrimination among M. tuberculosis strains. This lack of polymorphism was later explained by the fact that the probes initially targeted highly conserved regions and were of low specificity (281). The usefulness of the RFLP method has greatly increased with the discovery and application of ISs as hybridization probes (282, 283).
The most widely applied RFLP assay for the MTBC exploits IS6110 (284) (Fig. 1). According to a standardized protocol, genomic DNA is digested with the PvuII restriction enzyme, which cleaves IS6110 only once. The resulting DNA fragments are separated electrophoretically, transferred onto a membrane, and hybridized with a labeled probe complementary to the 3′ end of the IS6110 sequence. Hybridization band patterns are visualized on X-ray film. Each band denotes a single copy of IS6110 surrounded by flanking DNAs of different lengths, depending on the distance between the PvuII sites (284). Since the time when the IS6110-RFLP methodology was described and standardized, laboratories across the world have implemented it into routine epidemiological investigations of M. tuberculosis strains (253, 254, 285–289). The IS6110-RFLP method has a high discriminatory capacity, is easily reproducible in a laboratory, and therefore has long been considered the “gold standard” among typing methods applicable for the MTBC.
FIG 1.
Distribution of the three major types of genetic loci, i.e., mycobacterial interspersed repetitive units (MIRUs) (MIRU20 [1a] and MIRU40 [1b]), the direct-repeat (DR) locus (2), and IS6110 (3), within the chromosome of a hypothetical Mycobacterium tuberculosis strain.
However, IS6110-RFLP typing has several limitations. First, it is a labor-intensive and time-consuming method. Second, large quantities (ca. 2 to 10 μg) of high-quality DNA are required for restriction enzyme digestion, and this necessitates culturing. Furthermore, this method is technically demanding and requires both sophisticated and expensive computer software and qualified personnel with high technical expertise for its operation. A severe disadvantage of this typing method is the lack of discrimination for M. tuberculosis strains harboring fewer than 6 copies of IS6110 (290–293) as well as for M. bovis isolates (294–299). Another important limitation is that the data obtained in independent studies are difficult to compare because of the lack of reproducibility and portability of the results between different laboratories (300) in spite of attempts to standardize the procedure (284). Results of IS6110 analysis can be compared with data in an international database of IS6110 patterns (RIVM-Bionumerics), yet differentiation can be completed only upon providing cluster information obtained by spoligotyping and MIRU-VNTR typing. Finally, some NTM species have multiple copies of sequences that are homologous to IS6110 and may thus hybridize with the IS6110 probe (301). As for difficulties in performing RFLP analysis on NTM, IS1245-RFLP hybridization patterns for the MAC are difficult to interpret due to large numbers of bands with various intensities (262, 302, 303).
For all of these reasons, from 2006 on, RFLP typing was abandoned as the gold standard for typing and was replaced by MIRU-VNTR typing.
Methods based on ligation of oligonucleotide adaptors.
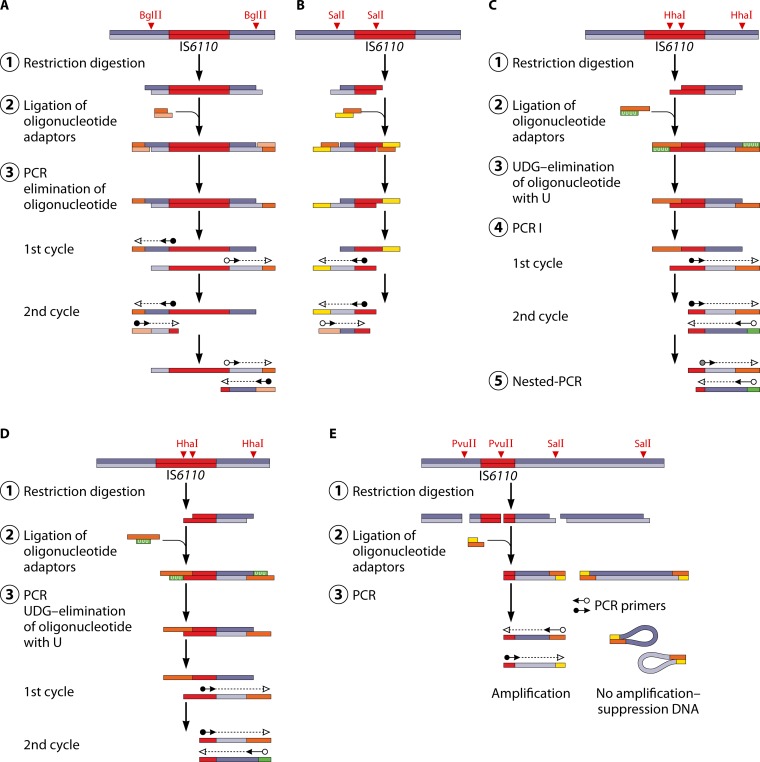
Ligation-mediated PCR (LM-PCR) is the name for a group of alternative genotyping methods exploiting the variability of IS6110. Compared with traditional IS6110 genotyping, these methods significantly reduce the time required for analysis, as the small quantity of chromosomal DNA used (1 ng) eliminates the need for bacterial culture. Several variants of these method have been successfully applied to differentiate MTBC strains (304–311). All of these variants follow a common four-stage scheme: (i) genomic DNA fragmentation using restriction enzymes that generate protruding ends, (ii) ligation of restriction fragments with synthetic oligonucleotide linkers (adaptors), (iii) amplification of the ligation products with appropriate primers, and (iv) analysis of the amplicons. A comparison of LM-PCR methods is shown in Fig. 2.
FIG 2.
Variation of LM-PCR methods used for molecular typing of M. tuberculosis strains. For details, see the text. (Adapted from reference 308 with permission of the International Union against Tuberculosis and Lung Disease [copyright The Union].)
LM-PCR, which amplifies the flanking sequences on both sides of IS6110, was first proposed by Palittapongarnpim et al. (304). Here, genomic DNA is digested with BglII, and the resulting fragments are ligated with a specifically designed linker. As the linker is not phosphorylated, it can be ligated only to the 5′ ends of each restriction fragment. Amplification is done with two primers that are complementary to both sides of IS6110, heading outwards from the sequence, with one of the primers also being identical to the shorter strand of the linker. The resulting PCR products are separated by electrophoresis, producing discriminative bands (Fig. 2A). A variation of this method was used by Prod'hom et al. to amplify only the upstream IS6110-flanking regions (305). This method exploits SalI digestion sites within the genome and uses one primer specific for IS6110 and a second one specific for a linker ligated to a SalI restriction fragment (Fig. 2B). Another variation was proposed, under the name of mixed-linker PCR (ML-PCR), where a primer specific for IS6110 and a primer complementary to a linker ligated to the HhaI restriction site are used (Fig. 2C). Instead of using a nonphosphorylated linker, which could be a potential source of unspecific amplification products, a so-called mixed linker is used, which contains uracil instead of thymidine in one of the strands. Before the amplification step, the unwanted strand of the linker is removed by treatment with uracil N-glycosylase. In order to further improve the specificity of this method, a heminested PCR is performed, using PCR products from the first amplification round as the templates. For this nested PCR, a primer complementary to the linker and a novel primer complementary to the internal sequence of the IS6110 fragment are employed (306). Butler et al. automated the ML-PCR method by using fluorescently labeled IS6110-specific oligonucleotides. The obtained fluorescently labeled PCR products are amenable to rapid and accurate analysis with a Sanger sequencer (312).
The LM-PCR method (306) was further modified to become the fast ligation-mediated PCR (FLiP) method (307). Due to the optimized workflow, FLiP is much easier and faster than the original method and allows typing within only 6.5 h, as opposed to the 2-day workflow for the original protocol. Following restriction digestion, genomic DNA is ligated with an adaptor composed of two oligonucleotides, one of which is complementary to the protruding end and the other of which contains uracil instead of thymidine. A pair of starters is used for amplification: one of them is specific for IS6110, and the other is complementary to the oligonucleotide ligated with restricted genomic DNA fragments (Fig. 2D). By omitting the steps of additional restriction digestion and heminested PCR (as in the original mixed-linker method), both the time and labor required to complete the protocol are considerably reduced (307, 313). The specificity of the FLiP products was verified by Southern hybridization with the IS6110 fragment as a probe (307) and by cloning and sequencing followed by determination of IS6110 localization in the genome (254). The discriminatory power of FLiP was reported to be close to that of the IS6110-RFLP method (307, 314) or to surpass that of 15-locus MIRU-VNTR typing (315, 316).
The recently developed fast ligation amplification polymorphism (FLAP) method is a novel variation of LM-PCR (Fig. 2E). This method involves double digestion of chromosomal DNA with the SalI and PvuII endonucleases. The PvuII enzyme recognizes a single nucleotide sequence within IS6110 and generates blunt ends. After the digestion step, oligonucleotide adaptors are ligated to SalI cohesive ends. All restriction fragments are used as the templates for PCR amplification, with one primer being complementary to the adaptor sequence and the second primer being complementary to the inner fragment of IS6110. The amplification of fragments carrying adaptors at both sites (SalI-SalI fragments) is inhibited by the suppression of substrate hybridization; the amplification of fragments carrying a single adaptor sequence (SalI-PvuII) requires the presence of the 5′ end of IS6110 as a template for the second primer. The fragment containing two PvuII blunt ends can be amplified only when two closely located IS6110 elements are in a head-to-head orientation (308). The discriminatory power of the FLAP method was shown to be very similar to that of IS6110-RFLP typing and greater than that of 15-locus MIRU-VNTR typing (308).
LM-PCR methods are highly reliable, inexpensive, and relatively fast. They do not require large quantities of purified DNA and can be valuable molecular tools for analyzing collections with limited numbers of strains. These methods exhibit significant discriminatory potential, slightly lower than that of IS6110-RFLP analysis (307, 317, 318).
LM-PCR patterns usually contain 5 to 12 bands, compared with up to 20 for IS6110-RFLP fingerprinting. The different number of bands in LM-PCR patterns may not necessarily reflect the number of IS6110 copies in the strains and should not be equated with them (315). The observed differences may be caused by visual misinterpretation of the patterns, a failure of the gels to sufficiently separate the PCR fragments, or difficulty in amplification of large products by PCR. Furthermore, highly polymorphic DNA fingerprints can be due to nonspecific amplification (307) or overlapping of the bands.
The major constraint of LM-PCR-based methods is the difficulty in establishing any reference database. The reproducibility of the results is relatively low, and the performance conditions lack standardization. Hence, comparisons of fingerprints produced in different laboratories may be problematic. Moreover, LM-PCR methods, similarly to the IS6110-RFLP method, have little applicability for isolates with low IS6110 copy numbers. Altogether, LM-PCR-based methods are better suited to assist other genotyping methods rather than act autonomously in defining epidemiological links (309, 319).
Spoligotyping.
Clustered regularly interspaced short palindromic repeats (CRISPRs) comprise a family of widely encountered repetitive DNA elements. While initially detected in Escherichia coli (320), these elements have been subsequently identified in ∼40% of bacteria and 90% of archaea (321, 322). The CRISPR loci generally consist of a noncoding, A/T-rich leader sequence and variable numbers of identical direct repeats (DRs) interspersed with unique spacer sequences or spacers. Adjacent to CRISPRs are often CRISPR-associated (Cas) genes, together forming a CRISPR-Cas genomic region. CRISPR loci are thought to represent a sort of prokaryotic adaptive immunity system that confers resistance to phages (323). The number of spacers within CRISPR loci is variable. Spacers may be acquired from a viral invader as a specific way of memorizing phage infection (323). On the other hand, some spacers may be deleted as a result of transposition and homologous recombination between neighboring or distant DRs. After the incorporation of the spacer, the mechanism of phage resistance is conferred by the expression of this sequence, hybridization, and cleavage of foreign RNA or DNA (322).
CRISPR loci have been identified in several mycobacterial species (324, 325). However, long CRISPRs have been found in M. tuberculosis, M. bovis, and M. avium. As the integrated CRISPR-Cas system can be found only in M. tuberculosis and M. bovis (324), different systems in NTM are thought to have been acquired by horizontal gene transfer from other bacteria (325). It is still unknown whether the CRISPR system is functional in mycobacteria. It seems that while it might interfere with incoming nucleic acids, it might have lost the capability of incorporating new spacers (324, 325).
Spacer oligonucleotide typing (spoligotyping) is a PCR-based technique for MTBC strain differentiation that takes advantage of the structure and polymorphism of the DR locus (Fig. 1). In spoligotyping, the entire locus is amplified by PCR by using two inversely oriented primers complementary to the sequences of DRs. A biotinylated reverse primer is used so that all the reverse strands are labeled. Next, PCR products are hybridized to a membrane with a set of 43 immobilized, covalently bound, synthetic oligonucleotides, each representing a unique spacer identified by sequencing of the DR locus in M. tuberculosis H37Rv (spacers 1 to 19, 22 to 32, and 37 to 43) and M. bovis BCG vaccine strain P3 (spacers 20, 21, and 33 to 36). After hybridization, the membrane is incubated with a streptavidin-peroxidase or streptavidin-alkaline phosphatase conjugate, and the hybridization signals are detected by chemiluminescence. Strain-specific patterns (spoligotypes) are then visualized on X-ray film. Strains are differentiated by the presence or absence of individual spacers in the complete 43-spacer set (326). Since spoligotyping results can be presented as a binary system (present/absent), they can be easily interpreted, digitized, and compared among different laboratories (327). Two major databases of spoligotyping patterns available online are SpolDB4 (http://www.pasteur-guadeloupe.fr:8081/SITVITDemo/) and SITVIT (http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE/), which are discussed in detail below.
Since the introduction of spoligotyping into laboratory practice in the late 1990s, two automatic variants of this method have been proposed. The Luminex technology uses synthetic spacer oligonucleotide probes immobilized on microspheres and detected via fluorochromes attached to the beads and hybridized PCR products. This method offers higher reproducibility due to the elimination of membrane hybridization and avoidance of manual interpretation of the spoligopatterns. This method allows analysis of 96 isolates in a single run, compared with 45 isolates with the standard spoligotyping protocol (328, 329). Another alternative to conventional spoligotyping is a multiplexed primer extension-based spoligotyping assay using automated MALDI-TOF MS. In short, spacers are amplified by PCR in a multiplex manner using primer extensions. Furthermore, amplicons are analyzed by using a mass spectrophotometer, with each product occupying separate masses in the resulting spectrum. This technique delivers greater reproducibility, higher throughput, greater ease of use, and better data analysis than the classical method, but its major limitation is the need for expensive equipment (330).
Spoligotyping is less discriminatory than IS6110-RFLP analysis for strains with high copy numbers (≥6) of IS6110 (331–337). Even with an increase in the number of spacers to 51, mostly originating from the DR region of the M. canettii genome, the resolution of this method was only marginally improved (338). Application of a 68-spacer format, with 25 out of 51 new spacers, increased the discrimination for M. africanum subspecies and for the East African-Indian (EAI) clade of M. tuberculosis (329, 339). The limited discriminatory power of spoligotyping is due to the fact that it targets a single genetic locus, covering <0.1% of the M. tuberculosis complex genome, unlike IS6110-based RFLP analysis, where IS6110 is distributed over the whole mycobacterial genome. Additionally, convergent deletions in the CRISPR locus occur frequently (340). Spoligotyping, when used alone, is not sufficient for epidemiological studies, but it is sometimes recommended as a first-line screening test, especially when a large collection of isolates is being tested (331, 341, 342).
Spoligotyping has many important advantages. It allows the detection of M. tuberculosis complex bacteria in noninfectious samples without the need for culturing of bacteria. This means that detection and genotyping can be performed directly on clinical samples. The sensitivity of spoligotyping was estimated to be 10 fg of chromosomal DNA, corresponding to DNA isolated from 2 to 3 bacterial cells (343). As such, spoligotyping has proven useful for typing on nonviable cultures, Ziehl-Neelsen smear slides, or paraffin-embedded tissue sections (344, 345). Spoligotyping is especially useful for detecting fragmented mycobacterial DNA with a minimal average, continuous size of only ∼75 bp. Such fragmented DNA is present in samples that have been treated with formalin or in samples extracted from ancient skeletal and mummified material (346–350). Second, since spoligotyping interrogates more conserved genetic information than, for instance, IS6110-RFLP typing, this method can be used for the identification of members of the MTBC to the species or subspecies level. For example, spoligotypes of M. tuberculosis are characterized by the absence of spacers 33 to 36; most M. bovis strains lack spacers 39 to 43, and all M. bovis BCG strains lack spacers 3, 9, and 16 (351). Moreover, some genotype families of tubercle bacilli are recognized based on a characteristic spoligotype pattern. For example, a key indicator of the Beijing family of strains is that they react with only the last 9 spacers (35–43) in the panel of 43 spacers (352). Third, spoligotyping is much faster than IS6110-RFLP analysis, with the laboratory turnaround time of the former being <48 h. Finally, for strains with no or few IS6110 copies, the discriminatory power of spoligotyping is higher than that of IS6110-RFLP typing (353).
Because of its simplicity, binary-result format, and high reproducibility, spoligotyping is widely used for investigations of MTBC molecular epidemiology. Indeed, the result of a PubMed search for “spoligotyping” or “spoligotype” gives over 1,000 (1,062) articles (interrogation made on 18 August 2015). Interestingly, today, spoligotyping patterns can also be predicted from WGS reads (354) to enable backwards comparisons.
Minisatellite sequences.
(i) Multilocus VNTR analysis.
VNTRs are tandem-repeat regions, scattered throughout the genome of M. tuberculosis, that resemble polymorphic minisatellites in eukaryotic genomes. The nomenclature for this type of repetitive element is somewhat confusing. This is because as new VNTR-type loci were discovered, they were recorded under different names.
The first VNTR loci detected in M. tuberculosis complex strains were described by Frothingham and Meeker-O'Connell (355). These loci comprised five major polymorphic tandem repeats (MPTRs) (MTPR-A to -E) and six exact tandem repeats (ETRs) (ETR-A to -F). The MPTRs contain 10-bp repeats separated by unique 5-bp spacer sequences. They are widely distributed among Mycobacterium species, including not only the MTBC but also M. gordonae, M. kansasii, and M. szulgai (356). The MPTR sequences are part of the 3′ ends of genes coding for the PPE proteins, named after the conserved Pro-Pro-Glu (PPE) motifs in the highly conserved N-terminal domains of the proteins. It is speculated that the polymorphism of the PPE proteins in their C-terminal domains, linked to the presence of the MPTR motifs, is related to antigen variability in M. tuberculosis (357). The MPTRs have been demonstrated to be useful in RFLP typing of M. kansasii with MPTR sequences as a probe (111, 358). The ETRs represent large tandem repeats, ranging in length from 53 to 79 bp. All six ETR loci and only one MPTR locus (MPTR-A) were found to be polymorphic when analyzed by DNA sequencing. However, since the allelic variability of MPTR-A is low (only 3 alleles) and ETR-F contains two types of tandem repeats of different lengths, only five ETR loci (ETR-A to -E) are used for genotyping of M. tuberculosis strains.
MIRUs were first described as tandem repeats of 46 to 101 bp dispersed at 41 loci in the genomes of M. tuberculosis H37Rv, CDC155, and AF2122/97 (300, 359). Among these loci, two, namely, locus 4 (VNTR0580) and locus 31 (VNTR3192), were identical to VNTR loci designated ETR-D and -E, respectively (355).
The most polymorphic VNTR/MIRU loci have been used to develop MIRU-VNTR typing, a PCR-based method which differentiates between strains by assessing the number and length of tandem repeats at each locus of each isolate (Fig. 1). The variability of particular loci often depends on the sample collection, the sample's geographical source, and natural genetic strain variability. The protocol for MIRU-VNTR typing includes PCR amplification of each locus by using specific primers complementary to the flanking regions and analysis of the resulting PCR products, which are separated by gel electrophoresis. The size of the amplified products corresponds to the number of tandem-repeat units, which is determined in reference to the known size of the repeat unit within each targeted locus. In well-equipped laboratories, MIRU-VNTR typing can be performed with the help of capillary electrophoresis and fluorescently labeled PCR primers, with an additional advantage of multiplexing (360).
Typing with a first set of 12 MIRU-VNTR loci was shown to be slightly less discriminatory than IS6110-RFLP analysis for M. tuberculosis isolates with high copy numbers of IS6110 (361, 362). Therefore, 12-locus MIRU-VNTR analysis was suggested to be used in combination with other genotyping methods (e.g., spoligotyping) to approximate the discriminatory power of IS6110 profiling (361, 363). At the same time, 12-locus MIRU-VNTR analysis can be more discriminatory than IS6110-RFLP analysis if M. tuberculosis isolates with low copy numbers of IS6110 are under study (291–293, 364, 365). Nevertheless, 12-locus MIRU-VNTR analysis cannot be used as the sole typing method, as it overestimates the number of true epidemiological links, especially in large, population-based studies (366, 367).
In 2006, an optimized 24-locus (including 12 loci previously investigated) MIRU-VNTR typing scheme was proposed as an international standard (292, 360, 368). A 24-locus format is more informative phylogenetically, and it can be used to trace transmission paths of TB (210, 369, 370). The discriminatory power of the 24-locus MIRU-VNTR typing system provides a resolution comparable to that of IS6110-RFLP profiling (252, 363, 370, 371). Therefore, 24-locus MIRU-VNTR typing has been suggested to be the new “gold standard” for molecular typing of MTBC strains. While the increased number of analyzed loci increases the discriminatory power of this method, it raises the cost of analysis and complicates the interpretation of the data gathered. Interestingly, only 15 out of the 24 loci (including 6 previously investigated loci) accounted for 96% of all detected polymorphisms in M. tuberculosis strains (372). It should be noted that due to the genome homoplasy commonly observed for Beijing family strains, the usefulness of standard MIRU-VNTR typing has been shown to be limited (373, 374). Additional loci not included in the standard 24-locus set, VNTRs 3232, 3820, and 4120, were proposed to be of use for the differentiation of Beijing genotype strains (375).
Due to possible homoplasy caused by convergent evolution and due to the various differentiation strengths of each locus depending on the MTBC lineage, it was suggested that MIRU-VNTR analysis should be performed in a lineage-dependent manner (376). Such an approach limits the amount of loci inspected and therefore decreases the costs of the analysis. Customized sets of 12 MIRU-VNTR primers have been successfully used for the differentiation of strains originating from Ghana (377) and China (378).
Generally, genotyping based on MIRU-VNTR regions is fast, easy to perform, sensitive, highly reproducible, and discriminative. It can be performed in large-scale genetic or evolutionary investigations for tracking key epidemiological events. MIRU-VNTR genotyping provides significantly higher resolution than spoligotyping and a resolution close to or even better than that obtained with IS6110-RFLP analysis. Therefore, MIRU-VNTR typing could be used to further investigate strains that were matched by both IS6110-RFLP and spoligotyping methods or that had a known epidemiological link. The results of MIRU-VNTR typing are expressed in a simple, digital format in which each digit represents the number of copies at a particular locus. Digitized data allow comparison of the results among laboratories worldwide (379, 380) (http://www.miru-vntrplus.org/). One of the largest publicly available international databases, SITVITWEB (http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE/), includes genotyping results based on MIRU-VNTR typing and spoligotyping for 62,582 MTBC clinical isolates from 153 countries all over the world (381, 382).
MIRU-VNTR analysis can also be used for typing of NTM. Thibault et al. described eight MIRU-VNTR-type loci in M. avium and M. avium subsp. paratuberculosis (271). Furthermore, newly discovered tandem repeats in M. avium were applied for genotyping of M. avium isolates and indicated significant discriminatory power (383). In M. intracellulare, 45 potential MIRU-VNTR loci were identified, 7 of which, due to high variability, could be used for differentiation (271, 384). Thirteen MIRU-VNTR loci have been used for finding links between M. ulcerans isolates recovered from clinical and environmental samples (385). Twelve MIRU-VNTR loci have been described for M. marinum and used for genetic differentiation (386).
(ii) ERIC sequences.
Enterobacterial repetitive intergenic consensus (ERIC) sequences are imperfect palindromes with a length of 126 bp that are present in the genomes of mainly Gram-negative bacteria (387). Their presence in the genome of M. tuberculosis was shown to be restricted to transcribed regions of the genome, either intergenic regions of polycistronic operons or nontranslated regions upstream or downstream of open reading frames (388). The variability of ERIC sequences allowed the proposal of a new typing method, ERIC-PCR typing, which has been applied for estimating the genetic diversity of many mycobacterial species, including M. tuberculosis (388), M. gordonae (389, 390), M. intracellulare, M. szulgai, M. fortuitum (173, 390), M. chelonae, and M. abscessus (391).
ERIC-PCR showed higher discriminatory power than IS6110-RFLP analysis for typing of M. tuberculosis (388). On the other hand, when tested on M. paratuberculosis strains, ERIC-PCR did not detect any differences in strains presenting distinct RFLP patterns and was therefore not considered a good methodological alternative for typing of this species. However, the species-specific band pattern suggested that IS900-based ERIC-PCR could be used as a suitable genetic marker for distinguishing M. paratuberculosis from other mycobacteria (392).
(iii) Trinucleotide repeat sequences.
All bacterial genomes carry various numbers of trinucleotide repeat sequences (TRSs), which can be useful for genotyping purposes. Most of the TRSs in the genomes of mycobacteria have been localized within the genes of the PE and PPE families (393). A subgroup of the PE proteins, whose N termini harbor characteristic Pro-Glu (PE) motifs, contains polymorphic GC-rich sequences (PGRSs), just as a subgroup of the above-discussed PPE proteins contains MPTRs (394). It was suggested that DNA polymorphisms associated with these repetitive elements may result in antigenic variation of mycobacteria (23). PGRSs occur at multiple loci and consist of several repeats of a 9-bp consensus sequence (5′-CGGCGGCAA-3′), tandemly arranged in up to 1.5-kb segments. PGRS-based typing, with the PGRS cloned into a recombinant plasmid, pTBN12, as a probe, was used to differentiate M. tuberculosis strains with no or low copy numbers of IS6110 (280). Further studies that employed the PGRS-RFLP method confirmed its relatively high discriminatory power (395–397). Interestingly, PGRSs were also identified in various NTM species, and consequently, a similar PGRS-RFLP assay was also used to differentiate strains of M. kansasii (398) and M. ulcerans (399).
Similarly, a (CGG)5 probe was used with success as a hybridization probe for differential typing of M. tuberculosis and M. bovis strains. However, the (CGG)5 probe and digestion of chromosomal DNA with various enzymes failed to differentiate the M. tuberculosis H37Rv and H37Ra reference strains, which are differentiated by IS6110-RFLP typing. Therefore, (CGG)5-based fingerprinting was suggested to be useful for typing of M. tuberculosis strains with no or only a few IS6110 copies or as a method complementing IS6110-RFLP analysis. The (CGG)5 repeat sequences were also identified in NTM species, including M. marinum, M. kansasii, M. szulgai, suggesting that (CGG)5-based typing could be extended beyond MTBC strains (393). Recently, single-primer PCR targeting the TRS within the M. gordonae genome was proposed. The highest discrimination was achieved when primers containing (CAC)4 and (CGG)4 sequences were used (389).
A PCR-based method where fragments located between IS6110 and the PGRS are amplified by using primers directed outwards from the ends of both of these repetitive elements was termed double-repetitive-element PCR (DRE-PCR). As strains differ in the distances between IS6110 and the PGRS, as well as in their copy numbers, this method produces different numbers of variably sized DNA fragments (400). Although the discriminatory power of DRE-PCR is even higher than that of IS6110-RFLP typing, this method has poor reproducibility, and interpretation of the data is difficult (400).
Following an approach similar to that used for DRE-PCR, another typing method, called ampliprinting, was designed (351). Here, the variability in the distances between IS6110 elements and copies of MPTRs of M. tuberculosis is measured by using unilateral nested PCR with IS6110- and MPTR-targeted primers, followed by hybridization with an IS6110-specific probe. However, this method has not been widely adopted, mainly because of the limited number and size of the amplicons (290).
A synthetic (GTG)5 oligonucleotide was used as one of the primers to amplify DNA fragments between IS6110 and (GTG)5 repeats, the other primer being an IS6110 outlooking primer. This method again demonstrated higher discriminatory power than IS6110-RFLP typing alone (401). The synthetic (GTG)5 sequences were also used to recognize specific sequences from M. tuberculosis, and these sequences, after being cloned into the pMTB484(1) and pMTB484(2K4) vectors, were themselves used as probes. They were found useful for Southern blot genotyping of M. tuberculosis strains previously shown to have identical IS6110-RFLP fingerprints (402). The (GTG)5 sequences are most informative for typing of M. tuberculosis strains with low copy numbers of IS6110 (402, 403) but also for typing of NTM species such as M. gordonae, M. scrofulaceum, members of the MAC, and M. diernhoferi (404).
Another similar method, although not amplifying fragments between TRSs and IS6110 elements but amplifying fragments between 16-bp GC-rich sequences, designated Mtb1 and Mtb2, and IS6110 elements, was proposed by Kotłowski et al. (405). The calculated discriminatory power of this method was higher than those of spoligotyping and MIRU-VNTR typing and almost equivalent to that of IS6110-RFLP typing (406–408).
(iv) Rep-PCR.
Rep-PCR (DiversiLab system; bioMérieux, France) is a commercially available, high-throughput, automated system for typing of multiple Mycobacterium species based on the variability generated by repetitive sequences interspersed within the genome (409). This procedure involves the amplification of repetitive, noncoding sequences and their separation using microfluidic electrophoresis over a chip. As the fragments migrate over the chip, their size and fluorescence intensity are measured by a laser, thereby generating a graph. Data generated with this method showed high concordance with those generated by IS6110-RFLP and IS1245-RFLP typing for M. tuberculosis and MAC strains, respectively (410). This method was also able to discriminate between M. tuberculosis Beijing strains whose IS6110-RFLP patterns were nearly identical (411). More recently, automated rep-PCR was applied for typing of M. abscessus and compared with the PFGE method. The rep-PCR patterns of the strains that were identical upon PFGE analysis showed 90% similarity, suggesting that rep-PCR may be more discriminatory than PFGE (175, 412, 413). When applied for typing of other NTM species, rep-PCR achieved improved reproducibility over RAPD analysis, and for M. avium, this method provided a higher level of discrimination than MIRU-VNTR typing (414).
Among all methods for molecular epidemiology described above, three are of particular importance for M. tuberculosis typing (IS6110-RFLP, spoligotyping, and MIRU-VNTR analyses), and one is universal for all mycobacteria (WGS). The first reliable method for molecular epidemiology of mycobacteria was IS6110-RFLP analysis, long viewed as a gold standard, which allowed initiation of the molecular typing of M. tuberculosis. Spoligotyping has been extremely useful for phylogenetic classifications and reconstructions of M. tuberculosis. However, strain lineages of the MTBC should rather be defined based on phylogenetically robust markers such as SNPs or LSPs (376). MIRU-VNTR analysis, which has replaced IS6110-RFLP typing as a gold standard for studying the molecular epidemiology of M. tuberculosis, if standardized, may also be useful in epidemiological studies of NTM. Finally, WGS, which has emerged as a new tool to investigate mycobacterial epidemiology (151), might soon be broadly implemented, especially as, once cost prohibitive, it is now becoming cheaper and more accessible. Nevertheless, all the remaining methods discussed here might still be useful to confirm or exclude particular transmission links or to preliminarily characterize strains originating from relatively small and limited communities.
EVALUATION OF THE METHODS
An ideal typing method should be highly discriminatory, easy to perform, fast, and inexpensive and should generate results that are reproducible within a laboratory and between laboratories. It should be able to analyze very small quantities of bacterial material, preferentially without the need for culturing, at the very early stages of diagnostic and epidemiological investigations. At this moment, no such method exists. All the methods currently available and briefly outlined here have their benefits and shortfalls (Table 4). Therefore, the choice of method or a combination of methods for a particular study is somewhat challenging. The answer to the question of which typing scheme would work best depends heavily on the sample under investigation, the setting where the study is to be done, and the expected outcome. Whereas some methods (e.g., spoligotyping) are better suited for defining the phylogeographic specificity of circulating clades of tubercle bacilli, others (e.g., IS6110-RFLP analysis) are tailored specifically to assess the genetic relatedness and the epidemiological links among outbreak-related cases. Moreover, different methods are chosen to study the epidemiology of mycobacterial diseases over different periods of time (415).
TABLE 4.
Various genotyping methods used for epidemiological studies of mycobacteria
| Technique(s) | Application(s) | Advantage(s) | Disadvantage(s) |
|---|---|---|---|
| Phenotype-based methods | |||
| Phage typing, drug susceptibility profiling | Species differentiation; detection of drug resistance | Relatively inexpensive and easy to perform; allow detection of drug resistance that may not be detected by molecular methods (i.e., unknown mutations, rare genetic variants within a population) | Time-consuming due to the need for bacterial culture; some methods (e.g., MS) require specialized equipment and highly trained staff; results of differentiation may not be sufficiently discriminative even at the species level |
| Methods based on nonrepetitive sequences | |||
| Gene sequence analysis | |||
| PCR-PRA | Species differentiation | Does not require specialized equipment | Time-consuming; analysis restricted to a small fraction of the genome; requires trained staff; different sequences may share identical RFLP patterns |
| Hybridization probes | Species differentiation and detection; detection of drug resistance | Quick results, as analysis may be performed directly on clinical samples | Requires specialized equipment and trained staff; only specific mutations leading to drug resistance are detected, hence the possibility of false-negative results; presence of inhibitors within a sample may lead to false-negative results |
| SNP typing | Species/lineage differentiation; detection of drug resistance | May be performed directly on clinical samples | Analysis restricted to a small fraction of the genome; for species differentiation, SNP typing needs to be coupled with other genotyping methods, e.g., deletion mapping; detection of drug resistance only by typing previously known mutations |
| Gene sequencing | Species differentiation; detection of drug resistance | Detects previously unknown mutations | Analysis restricted to a small fraction of the genome; requires costly specialized equipment |
| Genome analysis | |||
| REA | Strain differentiation | Inexpensive; does not require specialized equipment | Time-consuming; requires trained staff; difficult data analysis, which may require specialized equipment; requires a large quantity of high-quality DNA |
| PFGE | Strain differentiation | Inexpensive; data analysis easier than with REA | Time-consuming; requires trained staff; requires a large quantity of high-quality DNA |
| RAPD | Strain differentiation | Can be performed on unknown DNA sequence | Poor intra- and interlaboratory reproducibility |
| AFLP | Species/strain differentiation | Broad range of possible adjustments to improve discriminatory power of the method | Time-consuming; requires trained staff |
| Deletion mapping | Species/lineage differentiation | Provides information about strain relatedness over protracted periods of time | Requires large quantities of bacterial DNA; analysis restricted to nonrepetitive fractions of the genome; needs to be coupled with SNP typing for species differentiation |
| WGS | Species/lineage differentiation; detection of drug resistance; monitoring transmission routes and acquisition of drug resistance | May be performed directly on clinical samples (metagenomic approach); provides information on (nearly) the entire genome; allows detection of different genetic variants within the same population | Still very expensive; difficult data analysis; drug-resistant variants may be undetected if the drug-susceptible variant is in the majority; currently available sequencing platforms have problems with analysis of microsatellites |
| Methods based on repetitive sequences | |||
| Insertion sequence-based RFLP typing | Strain differentiation; monitoring of transmission routes | Standardized protocol; high discriminatory power; provides high resolution for analysis of Beijing genotype strains | Time-consuming; requires trained personnel and large quantities of bacterial DNA; cannot be used to correctly type MTBC isolates with <6 copies of IS6110; different sequences may share identical RFLP patterns |
| LM-PCR | Strain differentiation | Requires small amt of bacterial DNA; inexpensive and fast; high discriminatory power | Lack of reproducibility between laboratories; cannot be used to correctly type MTBC isolates with <6 copies of IS6110; greater possibility of misinterpretation and false results than with insertion sequence-based RFLP typing |
| Spoligotyping | Species/strain differentiation | Simple and cost-efficient; standardized protocol; international databases available; can be performed directly on cell lysates or on nonviable bacteria; requires small amt of bacterial DNA | Low discriminatory power compared with those of IS6110-RFLP and MIRU-VNTR typing; not discriminative for Beijing genotype strains |
| Analysis of minisatellite sequences | |||
| MIRU-VNTR typing | Strain differentiation | Fast, easy to perform, sensitive, highly reproducible, and discriminative; more discriminatory than IS6110-RFLP for M. tuberculosis isolates with low copy no. of IS6110; well suited for large-scale, genetic, or evolutionary investigations; digitized results; can be performed directly on cell lysates; applicable for typing of NTM | Similar patterns may be sometimes found for unlinked strains |
| ERIC-PCR typing | Species/strain differentiation | Simple and cost-effective | Lack of portability between laboratories; variable discriminative power; difficult interpretation of results |
| Trinucleotide repeat sequence typing | Species/strain differentiation | PGRS typing may be used to differentiate M. tuberculosis strains with no or a low no. of copies of IS6110; relatively high discriminatory power; PGRS typing applicable for various NTM species; (CGG)5-based typing can be used for NTM species, e.g., M. marinum, M. kansasii, M. szulgai; DRE-PCR typing has discriminatory power comparable to that of IS6110-RFLP typing | Limited to few strain collection analyses; databases are not available; for DRE-PCR typing, poor reproducibility; difficult interpretation of results |
| Rep-PCR | Species/strain differentiation | Commercially available; high-throughput automated system for typing of many Mycobacterium species; able to discriminate among M. tuberculosis Beijing genotype strains with identical IS6110-RFLP patterns; achieves higher level of discrimination than MIRU-VNTR typing for M. avium and provides better reproducibility | Expensive and requires specialized equipment as well as highly trained staff; databases not available for comparative analysis |
To assess the ability of a method to assign different types to two unrelated strains is to measure its discriminatory power. This is achieved by using the index of discriminatory power, a derivative of Simpson's diversity index (416), as the average probability that the typing system will distinguish between randomly chosen unrelated strains. This index can be used to assess the discriminatory power of either a single typing method or a combination of two or more typing schemes (417).
The discriminatory power of any molecular method is strictly related to the genetic stability of the marker that a given method exploits. This stability in turn depends on the mutability of the marker over time, which reflects the evolutionary rate, also referred to as the molecular clock. This variable can be used to determine when two organisms diverged (418). Although SNPs seem to be relatively common in mycobacteria, they are still very rarely observed in slow-growing species, with an estimated rate of occurrence of 0.3 to 0.5 SNPs per genome per year (56, 419). The half-time of change of the IS6110-RFLP pattern in M. tuberculosis was calculated to be 3 to 4 years, and this relates to the number of IS6110 elements in the genome; the more copies, the higher the concentration of transposase and, thus, the higher the rate of transposition (353, 420). Even slower is the molecular clock based on spoligotyping variability. It was shown that multiple M. tuberculosis isolates from the same patients, corresponding to reinfection episodes, even after several years, yielded identical spoligotypes (421). Therefore, spoligotyping is useful for detecting persistent infection (422), while for this particular purpose, SNP patterns may provide confusing results. On the other hand, SNP typing might be useful to detect recent transmission (423). Ideally, the molecular clock of a typing method should be fast enough to distinguish unrelated cases and, on the other hand, sufficiently slow to capture epidemiologically linked cases. Markers evolving rapidly or those evolving at low rates may lead to either an underestimation or an overestimation of the amount of recent transmission of the disease, respectively (415). The discriminatory capacity of different typing methods impacts the definition of clustering or nonrelatedness. There is currently no accepted number of SNPs to delineate a new strain, as there are few reports on mutation dynamics in mycobacteria. In M. tuberculosis, for example, it seems that its molecular evolution in vivo is characterized by periods of relative genomic stability followed by bursts of mutations (424). Generally, a relapse of disease is considered when 6 to 14 SNPs are observed, while reinfection is considered if the number of SNPs exceeds 400 (56, 132). A transmission link is suspected when more than 14 SNPs are found (423). Microevolution is not restricted to SNPs and may occur at any other mutation site, resulting in a change of the typing pattern. In M. tuberculosis, a difference of one band in the IS6110-RFLP pattern or one repeat unit in a single MIRU-VNTR locus does not necessarily mean that the individuals represent different strains (423, 425). Importantly, even identical genotyping patterns may not reflect an actual transmission link. They may be a result of convergent evolution, laboratory cross-contamination, or simply a predominance of a particular strain circulating within the population (215, 426).
For now, the method providing the most in-depth knowledge in terms of species identification and strain typing is WGS (229). Although the costs of WGS are still too high to ensure large-scale application, these costs are expected to drop soon, and the availability of this method will expand greatly. Nevertheless, what may really obstruct the usefulness of WGS is data processing. WGS produces large amounts of data, and transforming them into an informative product, even with new programs being developed for easier data analysis and cloud storage to save computer space, is effortful and demanding (225, 427, 428). WGS requires very efficient computer analysis, high data storage capacity, and/or high-speed internet access to upload results to external databases. The capacity of computer systems available in laboratories, especially in developing countries, may not be high enough to handle such tasks. Therefore, WGS might perhaps be used to develop new epidemiological tools, identifying novel hallmarks of mycobacterial variability, rather than as a genotyping method per se. To exemplify this, WGS allowed the mapping of several SNP loci in M. ulcerans isolates, some of which further served as the templates for the development of a novel epidemiological differentiation pattern for this species (429). It also allowed the typing of several SNPs in M. leprae (430). Sequencing of the genome of M. septicum allowed the identification of >700 genes absent in M. fortuitum, which could be used for setting up novel molecular tools used for discrimination between the two species (431). SGS can also be combined with traditional genotyping methods. It has already been used to determine precisely IS6110 elements in the so-called ISseq method. Such an approach combines high-throughput sequencing using the Illumina platform with combinatorial sample multiplexing. This method presents 100% specificity, a positive predictive value of 98%, and reproducibility exceeding 90% (432).
As the costs of WGS are plummeting and more sophisticated bioinformatic devices required for analysis of the large amounts of generated data are being developed, the availability of this method will increase. Now, when SGS of mycobacterial genomes becomes an everyday method, it will provide us with the answers to many pivotal questions regarding the biology, pathogenicity, diversity, and evolution of mycobacteria. However, the next step is to look for a technique that would be able to detect all genetic variants present within a single, nonhomogenous mycobacterial population. These data can then be used to discern the inconsistencies in interview-based transmission links.
MOLECULAR EPIDEMIOLOGY: A CLINICAL PERSPECTIVE
Applications of Strain Typing to M. tuberculosis Complex Isolates
Monitoring transmission of TB.
Besides being performed at the global level, where molecular typing of MTBC bacteria is employed to study the successful spread of particular genotypes and the eclipse of others, molecular typing is also performed at the local level to study the spread of strains between patients. These data inform public health authorities on patterns of spread and potential areas for action to curb the spread of TB in communities.
Patterns of TB transmission differ greatly between settings, as they reflect an interplay of various factors, including existing TB control strategies, endemicity, population density, subpopulations with lower immunity to TB, migration patterns, geography, demography, and transmissibility of locally relevant M. tuberculosis complex strains (433). Genotyping data have given important clues about risk factors for TB transmission. These risk factors, if assessed in a timely manner, can help to inform health authorities to plan adequate action to curb local TB transmission.
Population-based studies focused on risk factors for TB transmission, as evidenced by clustering of isolates by genotyping, were systematically reviewed by Fok et al. in 2008 (434). Data from the 36 studies included in their meta-analysis showed that the rate of clustering is lower in settings of low endemicity (≤25/100,000/year) than in settings of higher endemicity, at 41 versus 45%. Risk factors associated with clustering are local birth, pulmonary TB, smear-positive disease, HIV seropositivity, alcohol abuse, intravenous drug abuse, homelessness, and, only in low-endemicity settings, male sex (434). For most of these factors, however, the heterogeneity among the studies analyzed was high.
A nationwide study in England in 1998 showed that of 1,808 typeable isolates from TB cases, 372 were in 152 clusters, yielding a proportion due to recent transmission of 12% [(372 − 152)/1,808]. Risk factors for clustering included diagnosis in London, previous TB treatment, pulmonary disease, and homelessness (435). A similar nationwide study in The Netherlands yielded similar results, identifying urban residence, male sex, long-term residence in The Netherlands, and Dutch or Surinamese nationality as risk factors for recent transmission (288). In the greater Paris region, Gutiérrez et al. demonstrated that 36% of 272 patients in a 1-year period were clustered on the basis of spoligotyping and IS6110-RFLP typing and that, in multivariate analysis, the major risk factors for clustering were homelessness and male sex but not HIV status, country of birth outside France, age, and imprisonment (436). In a 9-year study of a similar nature in New York City, isolates from 48% of 546 patients were in IS6110-RFLP clusters. Here, homelessness was the only risk factor for clustering in multivariate analysis. Being foreign born and >60 years of age were both associated with nonclustered isolates (437). In Shinjuku, Tokyo, Japan, M. tuberculosis isolates from 158/388 patients (39%) were clustered upon IS6110-RFLP typing. Here too, the clustering rate was significantly higher among the homeless population; by multivariate analysis, homelessness, male sex, an age of <40 years, the presence of pulmonary TB, work as a day laborer or unemployment, and being born in Japan were all associated with clustering, as was disease caused by Beijing genotype strains (438). In two urban areas in Northern Italy (Milan and Brescia), a 7-year population-based study showed that risk factors for strain clustering included an immigrant status, particular nationalities (Somali, Senegalese, or Peruvian), and city of residence: clustering was more frequent among immigrants with TB in Milan than in Brescia (49% versus 31%) (439). In Alabama, risk factors for clustering on the basis of recent transmission were young age, homelessness, alcohol abuse, urban residency, history of imprisonment, and black race but not HIV infection (440); HIV infection was a risk factor for recent transmission in an earlier study in San Francisco, along with being born in the United States, substance abuse, and homelessness. A later nationwide follow-up study that used MIRU-VNTR typing and geospatial clustering analysis to identify recent transmission of TB confirmed these findings; homelessness, being born in the United States, male sex, substance abuse, and being a member of a minority race or ethnic group were all risk factors for recent transmission (441). A recent study from San Francisco added an interesting angle to the latter aspect by revealing differences in risk factors for recent transmission and pace of progression to active disease between members of different ethnic groups within this city (442). Transmission typically involved M. tuberculosis strains of genotypes that were characteristic of the patients' ethnic background (East Asian lineage among Chinese, Indo-Oceanic lineage among Filipinos, or Euro-American lineage among Mexicans). Among people born in China, being >50 years of age was a risk factor for being a secondary case, whereas among people born in Mexico, male sex and an age of <50 years were risk factors. Young age was also a risk factor for persons born in the United States, as were HIV infection and, again, homelessness and substance abuse (442). Studies have also used different approaches to study TB transmission patterns by applying genotyping data. In Brazil, high rates of TB transmission within households were associated with cough frequency, M. tuberculosis strains of the Latin American and Mediterranean (LAM) and T genotypes, and cavitary lung disease, although only cough frequency remained statistically significant in multivariate analysis (443). In The Netherlands, the risk of becoming a member of a large cluster (≥5 patients in 2 years) was greatest when there was a time period of <3 months between the diagnoses of the first two patients, at least one of these two patients was <35 years of age, both patients lived in an urban area, and both patients originated from sub-Saharan Africa (444).
These data show that transmission of TB in settings of low endemicity occurs primarily among people living under poor socioeconomic conditions and, in some settings, within specific immigrant groups. In general, TB in immigrant populations in low-endemicity settings is thought to result mostly from reactivation of infection acquired prior to or shortly after immigration (435, 437–439). Also, the studies described above have shown that urban settings are a risk factor for TB transmission (445). As a result, at least in Europe, much effort has recently been devoted to monitoring TB in large agglomerations, where the incidence is typically twice the national average and where specific risk groups contribute to ongoing transmission (446).
It is important to realize that almost all of the studies described above applied IS6110-RFLP typing, so the conclusions from those studies could have been different if methods with lower or higher discriminatory power had been used. The spread of M. tuberculosis strains from established high-risk groups (e.g., particular immigrant groups) to the general population is often referred to as transmission permeability. Various studies have applied molecular typing techniques to assess the extent of this permeability. For example, molecular typing revealed that spread from homeless to nonhomeless populations occurred in both Tokyo (438) and Paris (436). In England, too, clusters often consisted of patients of multiple ethnicities and both homeless and nonhomeless populations (435). The aspect of transmission from immigrant to native populations in European Union/European Economic Area (EEA) countries was recently systematically reviewed (447). This review showed that the proportions of all studied TB cases that result from recent transmission within mixed (immigrant/native) clusters varied greatly between studies, from 0% to 34%; this proportion was <15% in 11 of the 15 studies analyzed (447). However, in the European Union/EEA, foreign-born patients are less likely to belong to a cluster than are native-born patients, and those authors conclude that TB in foreign-born populations has no significant impact on TB in the native European Union/EEA population (447). One study in Denmark actually estimated that infection of a migrant by a native Dane was up to 2.5 times more frequent than vice versa (448). The percentage of all clusters that consist of both natives and immigrants has also been used as a measure of transmission permeability; this amounted to 24% of all clusters in San Francisco (449); 29% in Almeria, Spain (450); 34% in Barcelona, Spain (451); and 36% in a recent study in Israel (452). The difference in transmission permeability between settings is well illustrated by a study in the Eastern Province of Saudi Arabia, where 25/33 (76%) clusters of 532 culture-confirmed TB cases occurring in a 2-year period were mixed autochthonous/immigrant clusters (453).
The results of individual analyses and meta-analyses should be interpreted with caution; the areas of the world with the highest TB burden are underserved by genotyping and other epidemiological investigations. There is also an important bias toward areas of increased incidence in many typing efforts seeking risk factors for TB transmission. Many typing efforts have focused on small geographic areas, typically urban in nature, while these settings themselves are a risk factor for transmission (435, 440, 445). The cost of genotyping and the laboratory infrastructure required are reasons for this bias. Clustering on the basis of genotyping might not differentiate well between remote and recent transmission of M. tuberculosis in rural areas (454). Also, access to detailed patient information is best at the local level, hampering larger (inter)national efforts at seeking risk factors for transmission (455). Nonetheless, given the observed high TB incidence in major cities, the lessons on local risk factors for transmission are important for national and regional control programs (446). Observed populations at risk and risk factors for transmission may also change over time, so lessons from typing studies may have an expiration date. Political situations, health care system funding and status, changes in demography, and migrant flows can all affect TB epidemiology and control programs (456).
Molecular typing to detect cross-contamination events.
Given the hardiness and disinfectant resistance of M. tuberculosis, cross-contamination is a serious risk for diagnostic laboratories. Cross-contamination should be suspected if cultures from patients without signs and symptoms of active TB yield M. tuberculosis and if the patients recover fully without TB treatment. Laboratory features suggestive of contamination include acid-fast bacillus (AFB) smear negativity of the clinical sample, growth only in liquid medium or only on solid medium if both are used, and very low numbers of colonies on solid medium (294, 457); however, small quantity alone is not a good marker for contamination (458).
One of the first recognized uses of strain typing of M. tuberculosis was thus the identification of such laboratory cross-contamination events. Despite its low discriminatory power, phage typing was applied for this purpose (459). In the early years of molecular typing by the IS6110-RFLP method, a series of studies examined its role in the identification of cross-contamination. One of these studies describes how samples from two smear-positive patients with signs of active pulmonary TB led to two small pseudo-outbreaks involving 8 isolates from 7 patients. A thorough investigation of laboratory practices revealed the risks involved in batchwise processing of slides for staining and the now-phased-out BacTec460 automated liquid culture system, involving the use of a needle to sample the culture headspace in culture bottles (460). The first estimation of the frequency of such events was reported in a reference laboratory in Denmark. There, in 1994, a contamination event was recognized and sparked a 1-year study of possible contamination events. During 1994, processing of 30,000 to 35,000 samples yielded 1,439 positive cultures, of which 49 cultures (3.4%) from 48 patients were considered, according to typing results and clinical features, to be the result of contamination (294). Similar studies were subsequently reported in other areas of low TB incidence. Early studies were reviewed by Burman and Reves in 2000 (461). In their review, they found that 13/14 (93%) studies including isolates from >100 patients reported cross-contamination events or false-positive cultures; 12 studies applied genotyping as part of their algorithm to detect false-positive cultures (461). The median false positivity rate was 3.1% (interquartile range [IQR], 2.2 to 10.5%). Of 236 patients for whom sufficient clinical data were available, 158 (67%) had received TB treatment. After this review, several new studies in settings of low endemicity were reported. In London, a 2.5-year retrospective study found that 0.54% of all TB cases were in retrospect likely to have resulted from laboratory cross-contamination; this percentage rose to 0.93% if both possible and likely cross-contamination events were counted (462). In the same period, a reference laboratory in Leeds performed genotyping for an 18-month period after the introduction of liquid culture methods, which are more prone to contamination events owing to their high sensitivity: 34/397 positive cultures (8.6%) evaluated were retrospectively considered false-positive cultures after 9 separate contamination events, on the basis of VNTR typing results and clinical data (463). In Massachusetts, a 5-year typing effort revealed that 18/1,043 positive cultures (1.7%) were the result of two separate contamination events (464). In the same period, a prospective study specifically to detect contamination events was performed in California. For 6/296 patients (2%) identified during the 18-month study period, positive cultures were ultimately considered to be the result of contamination, on the basis of IS6110-RFLP typing, laboratory investigation, and clinical features (465).
Identification of contamination events is more difficult in settings where TB is highly endemic, where identical genotypes are more likely as a result of high transmission rates. In addition, in many settings where TB is highly endemic, financial constraints limit the submission of samples for TB diagnostics to patients with suggestive clinical signs and thus with a high likelihood of having active TB. As a result, cross-contamination may more likely be overlooked. However, genotyping has also been applied in settings where TB is highly endemic to detect cross-contamination events. In Taiwan, two studies were performed, both using VNTR typing. The first study was a 1-year, retrospective, single-center study and showed that for 8/215 (3.7%) patients, cultures were considered false-positive cultures (466). A later study of a similar type investigated 400 patients and found that for 3 patients (0.8%), clinical and genotyping data were suggestive of false positivity of cultures, arising from laboratory cross-contamination events (467). A recent study from Nigeria assessed 83 M. tuberculosis complex isolates and concluded that 3/83 isolates (3.7%), all MDR M. tuberculosis isolates, were likely the result of a single contamination event, as determined by VNTR typing (468).
Thus, the percentage of false-positive cultures varies widely between studies but typically ranges from 1 to 3%. Reported risk factors for false-positive cultures include inadequate training of technicians (294) and the use of batch processing and liquid culture systems (463). A low volume of mycobacterial cultures performed annually may also increase the risk of false-positive cultures. In a 4-year nationwide study in The Netherlands, false-positive cultures were significantly more frequent in laboratories processing <3,000 mycobacterial cultures per year (469).
Cross-contamination events and false-positive cultures can result in unwarranted hospital admissions and drug treatment, with the potential for adverse events (469, 470). In a study in the United States, the cost of three false TB diagnoses was estimated to amount to $10,873 per patient (471). Real-time genotyping can shorten the time of detection of cross-contamination and aid in the decision to halt treatment and contact investigations where applicable.
However, cross-contamination resulting in false-positive cultures can also occur outside the laboratory. Contamination of cultures through the use of improperly disinfected bronchoscopes has been reported in various settings (472, 473). Here, however, a contaminated bronchoscope can lead to false-positive cultures as well as to actual transmission and TB disease. This aspect was recently reviewed (474).
Strain typing to discern relapse from reinfection.
For patients with TB infected with pansusceptible strains who receive and take adequate treatment, recurrences occur in about 2 to 20% of all patients, depending on the setting (475, 476). After an early study showed that the stability of IS6110-RFLP patterns was such that in patients with a relapse after TB treatment, relapse isolates would have RFLP patterns identical to those of the pretreatment isolate (477), follow-up studies have assessed which proportion of recurrent TB cases is due to relapse versus reinfection. One of the early contributions to this subject assessed reinfection events in five HIV-coinfected patients in Kenya. Among them, one of the five patients was labeled reinfected on the basis of unrelated IS6110-RFLP patterns of the original and recurrence isolates (478). In Estonia, a similar approach was used to investigate recurrent TB in 11 patients. While their first episode of infection was with pansusceptible M. tuberculosis isolates, 5 out of 11 (45%) patients experienced reinfection by MDR M. tuberculosis isolates of the Beijing genotype that were unrelated to the original isolates as determined by IS6110-RFLP typing. The remaining six patients had isogenic pansusceptible follow-up isolates and thus likely suffered true relapses (479). The extent of reinfection in settings with high HIV prevalence where TB is highly endemic was assessed in a miner community in South Africa. Sixty-five (20%) of 326 patients had a recurrence of TB after treatment. Among 39 patients for whom IS6110-RFLP typing could be performed on original and recurrent isolates, 25 (64%) and 14 (36%) were considered to have relapse and reinfection, respectively. While HIV infection was a risk factor for reinfection but not for relapse, persisting cavitary lesions and continuation of work as a miner were risk factors for relapse of the disease (480). Conversely, in Cape Town, South Africa, a similar study measured a recurrence rate of TB of 18%. Here, IS6110-RFLP typing showed that 24 (77%) of 31 recurrences after successful treatment were attributable to reinfection, whereas only 4 (11%) of 37 recurrences after treatment default were reinfections (476). García de Viedma et al. performed a similar analysis, investigating recurrent TB among 43 patients in Madrid, Spain, most of whom had been poorly adherent to TB therapy (481). Even in this moderate-incidence setting, for 14 (33%) of these 43 patients, the recurrence isolates were unrelated to the original isolates, and thus, these patients were in fact reinfected. Poor treatment adherence and HIV status were not associated with either relapses or reinfections (481).
It was shown that in low-endemicity settings, recurrences are infrequent and more likely to be caused by disease relapse than by reinfection. In a nationwide study in Denmark spanning 13.5 years, 73 cases of recurrent TB (occurring in 1.3% of all TB cases) were notified, 54 (74%) of which were relapses and 19 (26%) of which proved to be reinfections (475). In New South Wales, Australia, an analysis of 3,731 culture-positive TB cases in the 1994-2006 period revealed that 15 (0.4%) patients had recurrent TB. Based on IS6110-RFLP typing, 11 (73%) cases were attributable to reactivation, and 4 (27%) were attributable to reinfection (482). Very similar data had previously been reported from The Netherlands, where 16% of all recurrent TB cases proved to result from reinfection on the basis of IS6110-RFLP typing (483).
Whole-genome sequencing has now also been applied to distinguish relapse from reinfection. Forty-seven patients enrolled in the ReMox-TB trial experienced recurrent TB and had pretreatment and recurrence isolates available for sequencing. Whole-genome sequencing identified 33 cases with little genetic distance (≤6 SNPs) between strains, which were thus considered relapses. In three cases, the distance ranged from 1,306 to 1,419 SNPs, which was considered indicative of reinfection. Six relapses defined upon whole-genome sequencing would have been considered reinfections by MIRU-VNTR analysis, based on 1- to 3-locus differences (484).
Strain typing in drug resistance studies.
Linking population-based strain typing data with data on drug resistance can be useful on two levels. The first is monitoring the spread of individual drug-resistant strains in communities to assist local public health authorities. On a more global level, typing can help in monitoring the spread of genotypes or clones that are more able to become resistant or that are better transmissible despite resistance. We say “despite” since one of the classic concepts that has been the subject of molecular typing studies is the transmissibility of drug-resistant M. tuberculosis strains. It has long been held that drug resistance in M. tuberculosis incurs a fitness cost that decreases its virulence and transmissibility; this lower virulence was observed in mouse models of isoniazid-susceptible and -resistant strains (485). In The Netherlands and San Francisco, CA, isoniazid-resistant isolates proved to be 30 and 80% less likely to be clustered than drug-susceptible isolates, respectively (288, 486), and MDR M. tuberculosis isolates in South Africa and Mexico were 80 and 70% less likely to be clustered than drug-susceptible strains, respectively (487, 488). From these animal model and epidemiological data, one might conclude that drug resistance incurs a fitness cost leading to lower transmissibility. However, large outbreaks of MDR TB and XDR TB have been recorded (489, 490); compensatory mutations that restore fitness but sustain resistance have been identified (224); and recent genotyping efforts have provided strong evidence of the international spread of an MDR M. tuberculosis clone of the Beijing genotype, particularly in East and Central Asia, Eastern Europe, and South Africa (455, 491), as well as the large-scale spread of an MDR M. tuberculosis clone of the LAM genotype family in, and beyond, South Africa (492).
The distribution of MDR M. tuberculosis strains over families of different genotypes differs by region. In Johannesburg, South Africa, a selection of 434 MDR M. tuberculosis isolates belonged mainly to the Beijing (16%) and LAM (16%) families and the various families that make up the Euro-American lineage (T, Haarlem, LAM, S, and X; 30%) (493). Further north in Africa, in Ethiopia, between 2009 and 2012, 183 MDR M. tuberculosis isolates were genotyped by spoligotyping, revealing that the T (27%), Central Asian (CAS) (25%), U (13%), Manu (11%), Haarlem (2%), and East African-Indian (1%) genotype families were most frequent (494). In Europe, too, the situation differs by country. In Portugal, MDR TB epidemiology is driven largely by the successful spread of an M. tuberculosis clone of the LAM genotype family; 55% of MDR M. tuberculosis isolates in the Lisbon area belonged to this clone (495). In Spain, 33% of 480 MDR M. tuberculosis isolates isolated between 1998 and 2008 formed 31 clusters; Euro-American family genotypes (T, Haarlem, and LAM) are the most frequently detected genotypes among MDR M. tuberculosis strains. Strains of the Beijing genotype were noted but were restricted to immigrants from Eastern Europe (496). In Poland, analysis of 46 MDR TB isolates obtained in 2004 showed that 30% (n = 14) of them formed 5 clusters by MIRU-VNTR typing, although evidence for direct transmission was found for only 4 of the clustered cases (497). By spoligotyping, the isolates belonged mostly to the T (28%), Haarlem (17%), LAM (13%), and Beijing (9%) genotype families (498). In countries of the former Soviet Union, the MDR TB epidemic is driven largely by strains of the Beijing genotype, as it is in countries in East Asia, although the two areas see distinct clonal complexes within this genotype family (491). In East Asian countries, Beijing genotype strains make up most of the MDR M. tuberculosis isolates. In a recent study in Thailand, 72% of 192 isolates collected were of the Beijing genotype. There, 49.5% of all MDR TB isolates were clustered, suggesting active transmission (499). In a small-scale study in Japan, of 55 isolates in 2002, 62% (n = 34) proved to belong to the Beijing genotype, and 13% (n = 7) belonged to the T genotype family (500). In the Shandong province of China, 95% of all MDR isolates belonged to the Beijing genotype family, versus 79% of the pansusceptible isolates, a statistically significant difference (501). The importance of the Beijing genotype in the epidemiology of TB worldwide and possible mechanisms behind the successful emergence of multidrug resistance in this genotype have been reviewed elsewhere (502).
The high rates of clustering observed among MDR TB isolates of the Beijing genotype do not necessarily indicate recent transmission. For some of the clonal complexes within this genotype, IS6110-RFLP typing and routine 24-locus MIRU-VNTR typing have low discriminatory power and overestimate the extent of recent transmission (215). This issue can be partly solved by adding a defined set of auxiliary VNTR loci specifically for this genotype (503) or by performing WGS (215, 491). The evolutionary history and global spread of the Beijing lineage were recently studied by genetic analysis of 4,987 isolates from 99 countries and WGS of 110 representative isolates linked to drug resistance (491), confirming its origin around 6,600 years ago in the Far East (geographical zone centered on northeastern China, Korea, and Japan), followed by its spread worldwide in several waves, with major successive increases in population size coinciding with the Industrial Revolution, the First World War, and HIV epidemics. This landmark study underlined that mutations identified in genes putatively under positive selection and associated with virulence probably favored the expansion of the most successful branches of this lineage; incidentally, the latest steps of this evolution include the specific epidemic expansion of two MDR clones throughout Central Asia and Russia, concomitantly with the collapse of the public health system in the former Soviet Union.
Molecular Typing of Nontuberculous Mycobacteria
Molecular typing of NTM is still in its infancy, a result of the rather recent recognition of the pathogenic potential of these bacteria in patients with local or systemic impairment of immunity. The infrequent use of molecular typing of NTM implies that comparative studies of discriminatory power and the stability of various molecular markers are still few and far between. Also, studies of the discriminatory power of a single technique across various NTM species are notoriously lacking. Most studies focus on typing of a single NTM species and use a convenience sample. Hence, in this review, we are not able to present an optimal typing algorithm for NTM but rather present an overview of potential uses. While PFGE was without doubt the most frequently used method, it is not available to most laboratories, requires adaptation to NTM species, and is now phased out, in select settings, because of the advent of whole-genome sequencing.
Given the environmental nature of these organisms, typing has been applied to investigate possible environmental sources of strains infecting individual patients or strains causing (pseudo)outbreaks. Also, typing has been applied to distinguish between reinfection and relapse in patients with recurrent disease, to investigate human-to-human transmission events, and to investigate relationships between mycobacterial genotype and clinical disease manifestations or response to therapy.
Environmental sources of human NTM disease.
The exact environmental sources of human NTM disease remain largely unknown, but several studies have applied molecular typing methods to find sources for individual patients or patient cohorts.
Recently, a research group in Australia performed extensive environmental sampling and typing by rep-PCR to find environmental sources of M. kansasii (504), M. lentiflavum (505), M. fortuitum (506), and M. abscessus (413) and related these strains to those in patients diagnosed in this region. This led to the interesting conclusion that in Queensland, Australia, clinical isolates of M. abscessus and M. lentiflavum are (413, 505) but isolates of M. kansasii and M. fortuitum are not (504, 506) related to environmental—mostly water—isolates of the same species from the same region. Hence, potable water may be the source of some but not all NTM species infecting humans in this region.
In the United States, great effort has been made to investigate environmental sources of M. avium complex (MAC) strains that cause pulmonary disease. Many of these investigations have focused on tap or shower water as a possible source of infection for humans. MAC bacteria are commonly found by culture or molecular detection in household water and showerheads in the United States (507). However, linking these MAC strains to cases of human (pulmonary) disease has been successful for only a few patients. One frequently cited case report focuses on a woman with M. avium pulmonary disease whose shower water yielded several strains of M. avium. However, the most closely related strain by IS1245/IS1311-RFLP typing was only 72% similar (508). Stronger evidence for water or plumbing as a source of NTM infection has been reported. In one study, household water sampling was performed for 37 patients, and for 7 of them, M. avium strains identical to their clinical isolates were cultured from household water samples (509). Similar results were obtained in a similar study in Japan, where household water sampling for 49 patients with MAC pulmonary disease yielded M. avium strains identical to the clinical isolates of 2 patients (510). Another similar case from Japan was presented, where a woman with M. avium pulmonary disease had five strains of M. avium isolated from her bathtub tap, one of which was identical to her respiratory isolates as determined by 15-locus VNTR typing (511). Thus, the strongest evidence for household water and plumbing as sources of NTM pulmonary disease (NTM-PD) has been gathered for M. avium in the United States and Japan. This does not pertain to M. intracellulare, which is frequently a causative agent of NTM pulmonary disease but remarkably absent from household water systems in the United States; all household water isolates in a recent study appeared to be M. chimaera by molecular identification methods (512). Thus, for M. intracellulare, the source of infection may best be sought elsewhere.
Potting soil has also been investigated as a source of NTM disease. Cultures yielded known causative agents of disease, such as M. avium, M. intracellulare, and M. chelonae. For one patient, M. avium isolates from his/her own potting soil and clinical specimens were closely related on the basis of PFGE typing, suggesting that potting soil was a possible source of infection (513). In Japan, soil samples near the households of 100 patients with MAC pulmonary disease were also sampled. While 60% of all soil samples yielded MAC isolates, strains identical to the patients' clinical samples by 16-locus VNTR typing were identified for only 6 patients (5 M. avium strains and 1 M. intracellulare strain) (514).
For the majority of the patients in the above-mentioned studies, no source of infection was found. In The Netherlands, there was even a striking discrepancy between NTM detected by both culture (515) and molecular tools (516, 517) in tap water and water systems and NTM species found in clinical samples; the absence of MAC DNA was particularly striking (516, 517).
Thus, while both soil and water in the home environment can be the source of NTM pulmonary disease in some areas, particularly for M. avium (water and soil in the United States and Japan) and M. abscessus (water in Australia), the source of infection for the majority of patients remains elusive. Typing efforts in this field should continue in order to detect environmental niches that increase our understanding of disease epidemiology and perhaps offer clues for the prevention of infection in susceptible populations.
Typing to resolve (pseudo)outbreaks of NTM disease.
Given the fact that NTM are ubiquitous in the environment, contamination of medical devices or water sources with specific NTM can lead to outbreaks or pseudo-outbreaks that can be detected by molecular typing. Here, two scenarios are apparent. In the first scenario, a susceptible host is exposed to NTM through a contaminated device, which then leads to disease because the host is immunocompromised or because the NTM are inoculated into a normally sterile site. Many such outbreaks have occurred, and their descriptions mirror the development of typing tools for NTM. An interesting example is an outbreak of M. xenopi spondylodiscitis after hernia surgery in France using a trocar washed in tap water; even though no typing was applied at the time, M. xenopi was isolated from the hospital water system (518). Crude typing methods (SDS-PAGE of whole-cell proteins or MLST using only two loci) helped track outbreaks of injection site abscesses after intramuscular beta-lactam antibiotic administration caused by M. abscessus in China and South Korea (519, 520). One of the first well-described outbreaks where PFGE typing was applied was an outbreak of M. fortuitum furunculosis related to footbath use at a nail salon in California (521). In the following years, several cases of skin and soft tissue infections by rapidly growing mycobacteria (M. abscessus, M. chelonae, and M. fortuitum) after cosmetic surgery or other therapies performed in the Caribbean and South America were recorded, including cases involving so-called lipotourists, who travel to these countries for relatively inexpensive access to these therapies (522). In Brazil, several large outbreaks of M. abscessus disease after such therapies have occurred and have sparked genotyping efforts (523, 524), including comparative studies of typing techniques (148), which are otherwise very rare for NTM. For M. abscessus, an MLST approach using 7 housekeeping genes has been designed, with the primary goal of standardizing M. abscessus typing efforts (149). By using a collection of 93 outbreak strains from Brazil, this approach was compared to PFGE: MLST identified 33 sequence types, but PFGE identified 49 unique patterns and thus had a higher discriminatory power (148).
Very recently, multiple European countries have reported cases of M. chimaera endocarditis and disseminated disease in patients who had undergone cardiac surgery, typically with valve replacement. Typing efforts are still in progress, but early results of RAPD analysis and whole-genome sequencing hint that contaminated heater-cooler units that stabilize extracorporeal blood temperature using a water-based system with a ventilator that produced M. chimaera-containing aerosols are the source (525, 526).
The second scenario is a pseudo-outbreak where contaminated materials or specific environmental exposures to NTM lead to series of positive cultures in patients who have no NTM disease. An example of nosocomial exposure to NTM in patients without signs of mycobacterial disease was reported by Conger et al., who described an outbreak of M. simiae infection in 22 patients, 19 of whom had isolates representing a single clone as determined by PFGE. This clone was also abundant in the hospital's water system but not in the patients' home water systems. Exposure to M. simiae led to disease in one patient, making this more than only a pseudo-outbreak (527).
Pseudo-outbreaks can also be a result of contamination events in the microbiology laboratory. The best-known NTM involved in such pseudo-outbreaks is M. gordonae. In Spain, M. gordonae was isolated from 19 of 21 cultures set up in 1 day for patients suspected of having TB. RAPD typing showed that these isolates were identical and different from unrelated control strains. Environmental sampling in the laboratory did not yield M. gordonae, but after sterilization or replacement of all consumables and reagents, no more related M. gordonae isolates were detected (528). Similarly, a pseudo-outbreak of M. terrae was reported in Philadelphia, PA, where a laboratory noted that 14 cultures handled in a 6-day period were positive; all were from AFB-negative clinical samples and yielded growth only with the ESP liquid medium system but not on simultaneously inoculated Lowenstein-Jensen slants. Although cultures of reagents were all negative, the introduction was considered to be related to the setting up of the liquid cultures. PFGE typing of a subset of five of the isolates showed that they were identical to each other and different from reference strains. The outbreak ended without specific action; the slow growth of the outbreak strain delayed the recognition of the pseudo-outbreak (529), a feature common to NTM pseudo-outbreaks.
NTM are particularly prone to causing true outbreaks and pseudo-outbreaks owing to their omnipresence in the environment, particularly in treated water systems; biofilm formation; and resistance to commonly used methods of disinfection such as chlorination or the use of glutaraldehyde (530). To assist in detecting and combating such outbreaks, NTM reference laboratories should have access to a typing tool.
Typing to distinguish reinfection from relapse of NTM disease.
Despite long-term multidrug treatment regimens, rates of recurrence of NTM disease are high. Particularly in patients with NTM-PD, there has been considerable interest in using molecular typing to distinguish true relapse from reinfection. Different from M. tuberculosis, the latter is probable given the ubiquitous presence of NTM in our environment. However, very few studies have addressed this issue. The landmark study in this field was reported by Wallace and colleagues in 2002, when they showed by PFGE typing that 85% of new positive cultures after prolonged (>10 months) culture conversion during treatment of nodular-bronchiectatic M. intracellulare pulmonary disease represented reinfections by different strains (531). In patients with cavitary disease, new positive cultures were related mostly to treatment failure and proved identical to the baseline isolate (531). In a large-scale follow-up study, the same group found the same result. Of 180 patients treated for nodular-bronchiectatic M. avium complex lung disease, 14% experienced a microbiological recurrence. Of these cases, 73% represented reinfections, as PFGE demonstrated patterns different from those of the baseline isolates (532).
Very recently, a recurrence of disseminated M. avium disease in a patient from Japan with anti-interferon gamma autoantibodies was also proven to be a reinfection by PFGE and 16-locus VNTR typing (533).
Treatment of reinfections is not necessarily different from treatment of relapses, but no trials have been performed. Typing for this specific purpose is performed by very few laboratories worldwide, and its merit is still a subject of debate.
What does typing tell us about human transmission of NTM disease?
For decades, transmission of NTM between humans subsequently leading to disease was thought not to occur, given the low pathogenicity of NTM and the prerequisites of an active spreader and a susceptible recipient. When human transmission of M. malmoense infection was suspected in an area of Scotland with an unexpectedly high incidence rate, this hypothesis was refuted after PFGE typing revealed strain diversity (534). However, in recent years, transmission of M. abscessus strains in cystic fibrosis (CF) centers was shown to occur. The first recognized possible transmission event involved a CF patient with M. abscessus subsp. massiliense (formerly “M. massiliense”) disease caused by a strain with mutational resistance to amikacin and macrolides. This patient transferred to a new CF center, which, over the next 8 months, noted four additional patients with new positive cultures yielding the same species and an identical susceptibility pattern. Typing by PFGE and rep-PCR revealed identical isolates for the five patients, and four of the five patients were found to have had overlapping clinic visit days; environmental sampling did not yield M. abscessus (412). A second report covered a large-scale single-center typing study using whole-genome sequencing. Using whole-genome sequences from 168 isolates of 31 patients, two clustered outbreaks of M. abscessus subsp. massiliense involving 11 patients were uncovered. The limited variance between patient isolates and the fact that all patients had overlapping clinic visits strongly suggested human transmission. Transmission was thought to occur through fomites and not through direct human transmission, even though environmental sampling has yet to reveal the exact routes (484). A second study of a pediatric CF cohort could not confirm these findings and found identical strains only in two siblings with CF (535). Possible human transmission of M. kansasii pulmonary disease in an elderly couple was also noted in a study using AFLP typing (536).
Therefore, the dogma has been challenged, but exact routes of transmission and other populations susceptible to NTM pulmonary disease and capable of transmitting it need to be further studied. Molecular typing is important for understanding routes of transmission as well as for monitoring the emergence of successfully spreading clones, particularly clones of M. abscessus, akin to the spread of the Beijing genotype of M. tuberculosis.
Is the mycobacterial genotype associated with disease manifestation or response to treatment?
Diagnosis of NTM-PD can be difficult, and if it is treated, the outcomes are relatively poor. Rates of treatment failure differ by species but may range from 5% (M. kansasii) to 50% (M. abscessus or fibrocavitary M. avium complex isolates) (537). Molecular typing of NTM to assess virulence (i.e., the chance that the isolate signifies a true NTM-PD isolate) or to predict the response to treatment has thus been of significant interest. The results from the few available studies highlight interesting differences between species and between patient cohorts in different settings.
For M. kansasii, it has long been realized that there are 5 distinct genetic groupings based on 16S-23S ITS sequencing, PFGE analysis, or RFLP analysis using the major polymorphic tandem repeat (MPTR) probe (111). Mycobacterium kansasii subtype 1 causes mostly pulmonary disease and disseminated disease in immunocompromised individuals; subtype 2 causes disseminated disease, particularly in HIV-infected patients, but few cases of pulmonary disease; and the remaining types are typically environmental strains that do not cause disease, as studied in a cohort in Switzerland (358). Despite its clinical significance, this phenomenon has not been confirmed in cohorts in other countries. The different subtypes later proved to have differences in the gene sequences of two key virulence factors (the early secreted antigenic target 6-kDa protein [ESAT-6] and culture filtrate protein [10 kDa] [CFP-10]) (538) and to differ in their ability to translocate from the phagolysosome to the cytosol of macrophages. Mycobacterium kansasii subtype 1 can do this but is less able to do so than M. tuberculosis, whereas subtype 5 strains cannot (539).
In South Korea, two studies looked into strain variation in M. intracellulare/M. chimaera and potential relationships with clinical disease manifestations and the course of disease. The first study applied a basic MLST approach using two partial hsp65 gene sequences, a full 16S-23S ITS sequence, and a partial 16S rRNA gene sequence on a set of 94 isolates from 94 patients. Five distinct genotypes were discerned, one of which was M. chimaera and one of which was a fused genotype combining traits of both M. intracellulare and M. chimaera, similar to the M. chimaera variant reported from The Netherlands (540). The genotypes were not associated with true disease according to diagnostic criteria, a particular disease manifestation, or particular patient characteristics, except for a possible trend (P = 0.051) toward correlation between genotype and the percentage of patients with cavitary disease (541). The second study assessed the potential of 16-locus VNTR typing to distinguish strains or groupings of M. intracellulare associated with particular disease manifestations and disease courses. While this typing technique proved to be highly discriminatory, no grouping was associated with any disease manifestation, patient category, or disease course (542).
A relationship between grouping on the basis of 16-locus VNTR typing and disease progression was postulated for M. avium pulmonary disease. In Japan, a study of 37 patients reported that 15 patients showed progressive disease in the first year after diagnosis. Based on genotyping, strains from 8 of these patients grouped together; no strain from the 22 patients who had stable disease fell into the same group. From this, those authors concluded that progressive M. avium pulmonary disease was associated with specific VNTR genotypes (543). In a follow-up study with 59 patients with M. avium pulmonary disease, the same research group attempted to link grouping on the basis of 16-locus VNTR typing to the response to treatment. Again, VNTR typing revealed three groups of strains, but no information on whether these groups are the same as those in the previous study was provided. Looking at these groupings, no significant relation with treatment outcome was noted. Only after principal-component analysis and multivariate analyses were statistically significant associations between genotype and response to treatment seen, but these associations drew heavily on results for just two patients (544). At the same time, a similar study was performed in South Korea; in 102 patients with M. avium pulmonary disease, VNTR typing showed the same three groups of strains, but no associations between groupings and disease manifestation, clinical course, or drug susceptibility could be established (545).
A very similar study was also performed on patients with pulmonary disease caused by M. abscessus group bacteria in South Korea. A total of 53 M. abscessus and 58 “M. massiliense” isolates were assembled into 3 clusters based on VNTR genotyping. The patients in cluster A typically had stable nodular-bronchiectatic disease and were monitored without antibiotic treatment for >2 years after diagnosis. Patients with isolates in cluster B mostly had progressive nodular-bronchiectatic disease and started antibiotic treatment within 2 years after diagnosis. All patients whose isolates fell into cluster C had fibrocavitary disease and had to start treatment directly after diagnosis. The clinical isolates that were genetically most distant from the species type strain showed the highest likelihood of disease progression and fibrocavitary disease manifestation (546).
While these results for M. abscessus are absolutely striking, as are the results of M. avium typing in Japan (543, 544), they are difficult to match with the biology of these opportunistic infections, where host, bacterial, and environmental factors ultimately lead to infection and disease. This concept is of interest, and bacteriological factors are likely to impact the chance of having progressive disease and experiencing treatment failure. It was previously observed that the M. avium strains that cause disseminated disease in HIV-infected patients are genetically distinct from the strains that cause chronic pulmonary infections in patients with preexisting pulmonary disease (547). Specific traits were associated with subgroups within the latter group. However, the observed effects are based on groups and cannot be applied to individual patients. Moreover, the bacterial factor is likely the result of specific virulence factors and not the difference in the number of tandem repeats in a noncoding region of the genome. Rather than focusing on results of VNTR typing, which are most likely independent of virulence factors and can thus be misleading, it is better to find these virulence factors and integrate them, as well as data on host genetics and immunological status, into patient diagnostics (548). Also, these observations stem from studies performed in East Asia; given the observed differences in clinical relevance and frequency of isolation of NTM species between East Asia and other regions (549, 550), these findings remain to be validated in studies in other regions.
PHYLOGEOGRAPHY AND EVOLUTION OF MYCOBACTERIA
The epidemiological expansion of TB, throughout its whole history, has been a consequence of the global human population increase and cross-border and cross-continental movements of people. The ever-increasing rate of travel/migration of populations for leisure or work in the last decades has led to a new challenge in countries where TB was declining due to changes in the socioepidemiological scenarios generated by massive immigration from countries where TB is highly endemic (551). Some key questions include comparison of the role of recent transmission with that of reactivation/importation of TB among foreign-born cases, the impact of potential importation of previously unidentified M. tuberculosis strains, and cross-transmission between cases of different nationalities. It is therefore important to understand how tubercle bacilli are transmitted and which clones are involved in drug-resistant cases and/or outbreaks, identify new clones that may be emerging in a setting versus those that may be facing extinction, identify subpopulations with the highest threat of and risk factors for contracting infection, and be able to interpret these results within the evolutionary framework of the MTBC. It is clear today that these questions would prove difficult to answer without the support of molecular epidemiology.
Until recently, all human-adapted strains of the MTBC were traditionally considered to be essentially identical; hence, the question of individual genetic variation within MTBC strains gained little attention, which led to most previous research being focused on individual organisms. The advent of molecular methods and their widespread use in population-based studies introduced both new conceptual and technological developments. Although the MTBC constitutes a remarkably homogeneous group genetically with proven evidence of clonal evolution, recent studies have shown that the genetic diversity among individual clones is much higher than previously assumed, with much of it being driven by genetic drift potentially linked to human demographic and migratory events and with a potential impact on pathobiological properties and functional consequences, such as the emergence and spread of drug-resistant TB (151). In this section, various successive global TB genotyping databases initiated since 1999 as well as currently available Web-based tools developed to offer the possibility of creating worldwide geographical distribution maps displaying the frequencies of TB genotypes worldwide at various geographical scales are summarized.
TB: Origin, Spread, and Coadaptation with Its Hosts
The isolation of M. tuberculosis complex DNA from an extinct bison dated to 17000 BC suggested its presence in America in the late Pleistocene era (552). A study thereafter looked at ancient DNA in human remains and helped trace the presence of M. tuberculosis and M. africanum DNAs in Egyptian mummies (350). Subsequently, the origin of human TB was thought to be associated with the Neolithic demographic transition (NDT) starting around 11,000 years ago, as the development of animal domestication increased the likelihood of zoonotic transfer of novel pathogens to humans, while agricultural innovations supported increased population densities that helped sustain the infectious cycle (553). Although TB displayed a pattern of chronic progression, latency, and reactivation that is characteristic of pre-NDT disease (554), it was not clear whether (i) human TB descended from a ruminant mycobacterium that recently infected humans from domestic animals or from an ancient human mycobacterium that came to infect domestic and wild ruminants and (ii) whether TB originated independently in both hemispheres or was brought to the Americas by Europeans.
The answers to these questions were potentially answered thanks to new research by Comas et al. (555), who proposed that TB is probably as old as humanity itself. By studying the diverse genetic variations in MTBC strains, those researchers were able to show that TB probably spread around the world with the first modern humans to emerge from Africa. This study analyzed the whole genomes of a collection of 259 contemporary strains of the MTBC from around the world and compared MTBC phylogenic diversity to human diversity inferred from mitochondrial genome data. These results suggested that the MTBC accompanied migrations of anatomically modern humans out of Africa and expanded as a consequence of increases in human population density during the Neolithic period (555). This early origin of TB and the fact that the genome-based phylogeny of the MTBC mirrored that of human mitochondrial genomes further showed that TB lung infection did not spread to humans from domesticated animals, since farming came much later. This study also showed striking similarities in the evolutionary paths of humans and the MTBC and suggested not only that pathogen evolution paralleled human evolution but also that bacterial diversity directly benefited from human demographic explosions. In parallel, the latency and chronicity of this intracellular pathogen possibly allowed it to adapt to lower host densities, survive, and strike back when favorable conditions allowed massive host infections.
In the context described above, a recent paper on seals as a source of tuberculosis in pre-Columbian Peru (556) provides an interesting perspective on the origin of “New World” TB. Although the strains found in the Americas today are closely related to those in Europe, earlier archaeological evidence suggested that the disease was present in the New World before contact with Europeans (557). Soon after the study by Comas et al. (555) on out-of-Africa migration and Neolithic coexpansion of M. tuberculosis with modern humans during the Pleistocene epoch, subsequent analyses of three 1,000-year-old mycobacterial genomes from human remains in Peru showed that a member of the M. tuberculosis complex caused human disease before contact with Europeans (556). The ancient DNA of the pathogen causing human disease in the precontact New World was found to be most closely related to M. tuberculosis complex strains adapted to seals and sea lions, leading to a suggestion that sea mammals probably contracted the disease from an African host species and carried it across the oceans, leading to zoonotic transfer in coastal populations of South America. Whatever the exact route, this particular M. tuberculosis complex strain was probably well adapted to humans before being replaced by European strains introduced postcontact. Nonetheless, regular contacts would be required for anthroponotic transmission; since humans did not herd or farm seals, the possibility of transmission from both “seal to human” and “human to seal” remains equally rare, and human transfer of the bacterium to marine mammals cannot be totally ruled out with these data. Finally, one cannot definitely exclude the possibility that besides ancient M. pinnipedii-related strains, other members of the M. tuberculosis complex may have caused TB in pre-Columbian Americans.
Last but not least, the dating of the origin of key MTBC lineages remains argumentative, since estimates of the dates of origin human- and animal-associated lineages varied by an order of magnitude, from 70,000 years ago (555) to <6,000 years ago (556). Indeed, the substitution rate used to calculate the date of origin as 70,000 years ago in the out-of-Africa theory is much lower than those suggested by subsequent studies (556, 558). The higher rate does not preclude a role for Neolithic expansion, but it does call into question the idea that TB has been coevolving with humans since they first emerged from Africa. Ongoing and future studies will certainly bring new information in this regard in the coming years.
Molecular Typing Methods for TB Phylogeny
For the purposes of evolutionary studies, two sets of molecular markers are essentially applicable. These markers include (i) IS6110-RFLP types, spoligotypes, and MIRU-VNTRs, which are extensively used for epidemiological investigations, and (ii) large sequence polymorphisms (LSPs)/regions of difference (RDs) and single nucleotide polymorphisms (SNPs), which are specifically useful for phylogenetic and evolutionary studies. Note that spoligotyping and MIRU-VNTR typing also provide concomitant phylogenetic information but that IS6110-RFLP typing does not, since strain lineage cannot be derived from a band pattern/position alone.
One of the earliest studies used subtractive genomic hybridization to identify three distinct genomic regions between virulent M. bovis, M. tuberculosis, and the avirulent M. bovis BCG strain, designated RD1, RD2, and RD3, respectively (197). In another study, a distinction among three genetic groups of M. tuberculosis was achieved based on two polymorphisms occurring at high frequency in the genes encoding catalase-peroxidase (katG) and the A subunit of gyrase (gyrA), which led to classification into three principal genetic groups (PGGs), with group 1 bacteria being ancestral to groups 2 and 3 (559). Almost immediately thereafter, restriction-digested bacterial artificial chromosome (BAC) arrays of the H37Rv strain were used to reveal the presence of 10 regions of difference between M. tuberculosis and M. bovis (RD1 to -10), 7 of which (RD4 to RD10) were deleted in the latter species (560). In a major contribution, Brosch et al. analyzed the distribution of 20 variable regions resulting from insertion-deletion events in the genomes of tubercle bacilli in a collection of strains belonging to all MTBC subspecies and showed that based on the presence or absence of M. tuberculosis-specific deletion 1 (TbD1) (a 2-kb sequence), M. tuberculosis strains could be divided into “ancient” TbD1-positive (TbD1+) and “modern” TbD1-negative (TbD1−) strains (53). In this new evolutionary scenario of the M. tuberculosis complex, the RD9 deletion identifies an evolutionary lineage represented by M. africanum, M. microti, and M. bovis that diverged from the progenitor of the present M. tuberculosis strains before TbD1 occurred, a finding which contradicts previous assumptions that M. tuberculosis evolved from a precursor of M. bovis (53). Since M. canettii and other ancestral M. tuberculosis complex strains lacked none of these regions, they are supposed to be direct descendants of the tubercle bacilli that existed before the “M. africanum-M. bovis” lineage separated from the M. tuberculosis lineage.
Using a global MTBC collection and 212 SNPs, Filliol et al. (561) identified six deeply branched phylogenetically distinct SNP cluster groups (SCGs) and five subgroups. The SCGs were strongly associated with the geographical origin of the M. tuberculosis samples and the birthplace of the human hosts. Those authors proposed an algorithm that was able to identify two minimal sets of either 45 or 6 SNPs out of 212 SNPs tested, which could be used for screening of global MTBC collections for studies on evolution, strain differentiation, and biological differences among strains. In another study, Gutacker et al. (562) examined MTBC genetic relationships by analyzing 36 synonymous SNPs (sSNPs) among a large collection of strains from patients enrolled in 4 population-based studies in the United States and Europe and assigned the strains in the collection to 1 of 9 major genetic clusters. A similar classification was revealed by analysis of other extended SNPs. Since the classification patterns of the SNP-based phylogenetic lineages were nonrandomly associated with IS6110 profiles, spoligotypes, and MIRU-VNTRs, those authors argued for a strongly clonal MTBC population structure.
In parallel, by using DNA microarrays to comprehensively identify LSPs, a stable association between MTBC strains and their human host populations was observed (200); phylogenetic analysis indicated not only that horizontal gene transfers were rare among MTBC strains but also that associations between host and pathogen populations were stable even in a cosmopolitan urban setting (like San Francisco) and were dictated largely by the composition of the local immigrant population. The authors of that study concluded that M. tuberculosis is organized into several large, genetically differentiated populations, which in turn are directly and stably associated with host populations delineated according to their place of origin. A subsequent report by the same group confirmed this variable host-pathogen compatibility, with the global M. tuberculosis population structure being defined by six RD/LSP-defined phylogeographic lineages, each associated with specific, sympatric human populations, i.e., the Indo-Oceanic lineage, the East Asian lineage, the East African-Indian (EAI) lineage, the Euro-American lineage, and two West African lineages (201).
The correspondence among various lineage nomenclatures is summarized in Table 5. It is important to underline that one must keep in mind the marker used when discussing a lineage, particularly for the East African-Indian lineage, which denotes 2 completely different groups of M. tuberculosis by spoligotyping versus LSP analysis. Interestingly, good congruence between spoligotyping and SNP analysis was observed (561), with the East African-Indian and Beijing spoligotypes being concordant with SCG-1 and SCG-2, respectively; X and Central Asian spoligotypes were also associated with one SCG or subgroup combination. Other clades had less consistent associations with SCGs. Furthermore, the various spoligotyping-defined lineages fit well with the previous PGGs; hence, MTBC strains can be tentatively classified as the ancestral TbD1+ group/PGG1 (subset 1; M. africanum and EAI), the modern TbD1− group/PGG1 (subset 2; Beijing and CAS), and the evolutionarily recent TbD1− group/PGG2/PGG3 (subset 3; Haarlem, X, S, T, and Latin American and Mediterranean [LAM]). Nonetheless, proper epidemiological and phylogenetic inferences are not always an easy task due to a lack of understanding of the mechanisms behind the mutations leading to the polymorphisms of these genomic targets. Recent studies have shown that phylogenetically unrelated MTBC strains were sometimes found to have the same spoligotype pattern as a result of independent mutational events (563), an observation that corroborates the fact that spoligotyping is more prone to homoplasy to a greater extent than MIRU-VNTR typing (376). Furthermore, spoligotyping has little discriminative power for families associated with the absence of large blocks of spacers, e.g., the Beijing lineage (503). For all these reasons, we recommend that a finer phylogenetic analysis of the most significant circulating MTBC clones as determined by multiple genetic markers should be made and that the data obtained should be compared to the existing data worldwide, a complicated task by itself if not for the availability of large international databases.
TABLE 5.
Comparison of spoligotype-based nomenclature for M. tuberculosis lineages versus PGGs, SNP-based grouping, SCGs, and LSP-based lineagese
| Spoligotype-based nomenclaturea | PGG | SCGb | SNP-based groupingc | LSP-based lineaged |
|---|---|---|---|---|
| East African-Indian | PGG1 | 1 | sSNP-I | Indo-Oceanic |
| Beijing | PGG1 | 2 | sSNP-II | East Asian |
| CAS | PGG1 | 3a | sSNP-IIA | East African-Indian |
| Haarlem | PGG2 | 3b | sSNP-III | Euro-American |
| X1 | PGG2 | 3c | sSNP-IV | Euro-American |
| X1, X2, X3 | PGG2 | 4 | sSNP-V | Euro-American |
| LAM | PGG2 | 5 | sSNP-VI | Euro-American |
| T (miscellaneous) | PGG2-3 | 6 | sSNP-VII sSNP-VIII | Euro-American |
| Bovis | PGG1 | 7 | MTBC | MTBC |
| M. africanum | PGG1 | NA | NA | West African 1 |
| M. africanum | PGG1 | NA | NA | West African 2 |
TB Genotyping Databases and Tools: What Is Available?
Knowledge about TB is wider today than ever before, but what is our ability to compare the data generated to all the data that have accumulated over years? Is it possible to instantaneously compare the genetic information on the circulating MTBC strains in conjunction with all demographical, clinical, bacteriological, and epidemiological information available in various registers? The necessity of databases in such a context is obvious, and the conception and design of databases for the control/surveillance of TB as well as other communicable diseases will certainly be essential for the “End TB Strategy” (to reduce TB deaths by 95% and to cut new cases by 90% between 2015 and 2035) proposed by the WHO (564). Indeed, databases allow the storage of large amounts of information in a structured way, facilitating data processing and interrogation and streamlining the decision process by knowledge-based data mining (565). In the last couple of years, various databases and Web tools have been developed in the TB field, devoted mostly to studying TB molecular epidemiology and evolution; some examples are as follows.
SpolDB4 and SITVITWEB are genotyping databases developed at the Pasteur Institute of Guadeloupe (381, 566). The later version of SITVITWEB is a multimarker database with genotyping data on 62,582 clinical isolates corresponding to 153 countries of patient origin (105 countries of isolation) (http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE).
SpolTools (http://www.emi.unsw.edu.au/spolTools/) is a collection of online browser programs and a visualization tool designed to manipulate and analyze MTBC spoligotyping data (567, 568). It also contains an online repository of data on spoligotyped isolates collected from the literature (currently 30 data sets containing 1,179 spoligotype patterns corresponding to 6,278 isolates). In particular, it allows one to draw spoligoforest trees that illustrate evolutionary relationships between spoligotypes in a given setting. GraphViz software (http://www.graphviz.org/) (569) allows one to color the spoligoforests as a function of the lineages.
WebLogo version 2.8.2 (http://weblogo.berkeley.edu/) (570, 571) allows evaluation and visualization of the allelic diversity of the spoligotyping patterns as a function of their associated lineages. This method of representation, adapted to 43-spacer spoligotyping, has been labeled “Spoligologos” (572).
MIRU-VNTRplus (http://www.miru-vntrplus.org/) is a Web-based tool dedicated to analysis of molecular typing data on TB, particularly the 12-, 15-, and 24-locus MIRU-VNTR formats (379, 380). Tools for data exploration include searches for similar strains, creation of phylogenetic and minimum spanning trees, and mapping of geographic information. It also provides detailed results (geographical origin, drug susceptibility profiles, genetic lineages, spoligotyping patterns, SNP and LSP profiles, and IS6110-RFLP fingerprints) for a collection of 186 well-characterized reference strains.
The TB Genotyping Information Management System (TB GIMS) is a secure Web-based system designed to improve access to and dissemination of genotyping information nationwide in the United States (573). It stores and manages genotyping data on TB patients in the United States; allows authorized users to submit and track MTBC isolates to and from the contracted genotyping laboratories; provides immediate notification of genotyping results and updates to TB laboratories and programs; links isolate data to patient-level surveillance data; provides reports on genotype clusters, including national genotype distribution; and provides national, state, and county maps of genotype clusters. This database is not publicly available.
Mbovis.org (http://www.mbovis.org/) is a spoligotype database with over 1,400 patterns belonging to the following RD9-deleted MTBC lineages: M. africanum, M. bovis (antelope), M. microti, M. pinnipedii, M. caprae, and M. bovis (574).
MycoDB.es (http://www.vigilanciasanitaria.es/mycodb/) is a Spanish database of animal TB (575), which was created as an epidemiological tool at the national level (Spain). It contains 401 different spoligotype patterns comprising 17,273 isolates of M. bovis, M. caprae, and M. tuberculosis as well as limited amounts of MIRU-VNTR data. It is restricted to those with authorized access, limited to the Spanish Animal Health Agency, Centro de Vigilencia Sanitaria Veterinaria (VISAVET).
TB-Lineage (http://tbinsight.cs.rpi.edu/run_tb_lineage.html) is an online tool for analysis of MTBC genotypes and classification of MTBC genotypes into major lineages using spoligotyping and, optionally, 24-locus MIRU analysis (576). It was developed and tested by using genotyping data from the Centers for Disease Control and Prevention (CDC), Atlanta, GA, on 37,066 clinical isolates corresponding to 3,198 spoligotype patterns and 5,430 MIRU-VNTR patterns. However, if 24-locus MIRU data are not available, the system utilizes predictions made by a naive Bayes classifier based on spoligotype data alone. The accuracy of automated classification using both spoligotyping and 24-locus MIRU typing is >99%, and that of spoligotyping alone is >95%. This website also provides a tool to generate spoligoforests in order to visualize the genetic diversity and relatedness of genotypes and their associated lineages.
tbvar (http://genome.igib.res.in/tbvar/) is a searchable database with a systematic computational pipeline that allows annotation of potential functional and/or drug resistance-associated variants from clinical resequencing data on MTBC strains (577). For this purpose, the authors reanalyzed resequencing data sets corresponding to >450 MTBC isolates available in the public domain to create a comprehensive variome map comprising >29,000 single nucleotide variations. This database can be accessed by browsing the location of variants (e.g., position 1417019, 3037367, or 4222628, etc.), genes (e.g., katG, pncA, or gyrA, etc.), Rv gene code number (RvID, e.g., Rv1059, Rv1069c, or Rv3693, etc.), or genome position range (positions 10000 to 15000, 30000 to 35000, or 80000 to 85000, etc.).
InTB (http://www.evocell.org/inTB) is a Web-based interface/system for integrated warehousing and analysis of clinical, sociodemographic, and molecular typing data on TB (578). It allows users to insert and download standard genotyping data in conjunction with an extensive array of clinical and sociodemographic variables that are used to characterize the disease. It also allows classification of new isolates into a well-characterized set of isolates based on internal references and multiple types of data plotting and to generate trees for filtered subsets of data combining molecular and clinical/sociodemographic information.
Although it is difficult to keep up with all databases and online resources, it is worthwhile to mention efforts to determine the drug resistance of M. tuberculosis isolates from genotyping data. One such example was one of the first comprehensive online databases on TB drug resistance mutations (TB Drug Resistance Mutation Database [TBDReaM] [https://tbdreamdb.ki.se/Info/]), which was established by Sandgren et al. (579). However, it required prior processing of nucleotide sequences for mutation detection prior to interrogation, which is no longer required in a new database of mutations associated with antibiotic resistance in M. tuberculosis (MUBII-TB-DB [http://umr5558-bibiserv.univ-lyon1.fr/mubii/mubii-select.cgi]) (580). MUBII is an analysis-and-interpretation engine that uses reconstructed mutated gene sequences, which can be searched by BLAST, aligned against the wild-type gene sequence, and compared with data in the mutation database. Results printed as graphs (alignments) and text (description of the mutation and therapeutic significance) can be generated. Moreover, this system provides interpretation in biological and therapeutic terms and can evolve by the addition of newly described mutations.
Another example of a useful online resource is PhyResSE (581), a simple and reliable viewer for mycobacterial DNA reads that allows delineation of M. tuberculosis drug resistance and lineages from WGS data (https://bioinf.fz-borstel.de/mchips/phyresse/). In parallel, a library of 1,325 mutations predictive of resistance to 15 antituberculosis drugs was recently compiled and validated for 11 of them by using genomic-phenotypic data from 792 M. tuberculosis strains by Coll et al. (225). Those authors also developed the rapid online “TB-Profiler” tool that allows detection of drug resistance mutations and strain type profiles directly from raw sequences (http://tbdr.lshtm.ac.uk/).
Finally, an ongoing new development is ReSeqTB (Relational Sequencing TB Data Platform), which seeks to increase the understanding of the genetic basis of resistance by correlating molecular data with results from drug susceptibility testing and, optimally, associated patient outcomes (582). The ReSeqTB consortium is jointly managed by the Critical Pathways to TB Regimens (CPTR), the Foundation for Innovative New Diagnostics (FIND), the WHO, the CDC, the New Diagnostics Working Group (NDWG), and the National Institute of Allergy and Infectious Diseases (NIAID), and investigators are invited to contribute anonymized genotypic, phenotypic, demographic, and/or clinical data from M. tuberculosis studies that reside in their respective repositories to the ReSeqTB database.
Publicly Available Global TB Genotyping Databases
The first public database, SpolDB1, was initiated at the Pasteur Institute of Guadeloupe more than 15 years ago by sorting available spoligotype patterns (n = 610) in an Excel spreadsheet (583); this led to the first description of 69 major spoligotype patterns, allowing distinction of predominant patterns, to trace the origin of the strains and their potential movements. This was followed by the launch of SpolDB2, containing data on 3,319 isolates (584), and SpolDB3, containing data on 13,008 isolates grouped into 813 shared types (containing 11,708 isolates) and 1,300 orphan patterns (585, 586). More recently, development of the fourth MySQL-based version, SpolDB4, in 2006 (n = 39,295 clinical isolates) (566) and SITVITWEB in 2012 (n = 62,582 clinical isolates) (381) permitted a finer phylogeographical snapshot of circulating MTBC genotypic lineages worldwide. The most recent version, designated SITVIT2 (to be publicly released in 2016 for online consultation), contains spoligotyping information on approximately twice the number of strains (n = 111,635 isolates) as that in the previous SITVITWEB version (381). A Web-based interface in SITVIT2 allows users to search for strains in the database by individual criteria or combined searches (year, country of isolation, country of origin, investigator's name, genotype, genotypic lineage, or drug resistance, etc.), making it possible to retrieve genotyping data in conjunction with data on geographical distribution, drug resistance, and demographic and epidemiological characteristics.
Here, we briefly describe pertinent information retrieved from the SITVIT2 database (Nalin Rastogi, personal communication), since it constitutes a useful tool for analyzing TB genotyping data within the global context of TB molecular population genetics, historical demography, and epidemiological monitoring and modeling.
Table 6 shows a comparison of the SITVITWEB and SITVIT2 databases and the corresponding phylogenetic lineages in the 2 versions, and Fig. 3 shows a map of the worldwide distribution of major lineages in the SITVIT2 database. The most noticeable increase in prevalence between the 2 versions is observed for M. bovis strains (10.36% versus 23.06%), underlining the increased potential of SITVIT2 for studying not only M. tuberculosis epidemiology but also bovine TB. Furthermore, some lineages were relabeled in the SITVIT2 database compared to the previous version; e.g., some spoligotypes previously classified as belonging to H3/H4 sublineages within the Haarlem family were recently relabeled “Ural” (587). This includes patterns belonging to the H4 sublineage, relabeled “Ural-2,” and some patterns previously classified as belonging to the H3 sublineage but with an additional specific signature (presence of spacer 2 and absence of spacers 29 to 31 and 33 to 36), relabeled “Ural-1.” Additionally, two LAM sublineages were recently raised to the independent-lineage level: LAM10-CAM was designated the Cameroon lineage (588), and LAM7-TUR was designated the Turkey lineage (589, 590). A major contribution of publicly available global TB genotyping databases is the ease with which one can map genotyping data to describe the TB genetic landscape. Indeed, georeferencing of genotyping data will represent an efficient bacteriological complement to the WHO's Communicable Disease Global Atlas (http://apps.who.int/globalatlas/) by bringing together analysis and interpretation of MTBC genotyping data in conjunction with information on demography, socioeconomic conditions, and environmental factors in a single electronic platform. Accompanied with suitable tools and software, a user today is better armed to describe the TB genetic landscape; potential tools include STATA and R software for statistical comparisons and bioinformatic software (list not exhaustive) such as BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium) and MLVA-Compare (Ridom GmbH and Genoscreen, Lille, France), which come with a variety of plug-ins and applications to analyze data generated by using CRISPR analysis, SNP analysis, MLPA, and MIRU-VNTR typing, etc.
TABLE 6.
Summary of data in the SITVITWEB and SITVIT2 databases and corresponding major phylogenetic lineages of the M. tuberculosis complex
| Major lineagea | SITVITWEB (n = 62,582) |
SITVIT2 (n = 111,635) |
||
|---|---|---|---|---|
| No. of isolates | % of isolates | No. of isolates | % of isolates | |
| Beijing | 6,159 | 9.84 | 10,850 | 9.72 |
| M. africanum | 695 | 1.11 | 965 | 0.86 |
| M. bovis | 6,486 | 10.36 | 25,741 | 23.06 |
| M. canettii | 12 | 0.02 | 12 | 0.01 |
| CAS | 2,480 | 3.96 | 4,362 | 3.91 |
| EAI | 4,674 | 7.47 | 6,617 | 5.93 |
| Haarlem/Ural | 7,058 | 11.28 | 10,580 | 9.48 |
| LAM | 8,042 | 12.85 | 12,245 | 10.97 |
| Cameroon | 650 | 1.04 | 1,095 | 0.98 |
| Turkey | 370 | 0.59 | 593 | 0.53 |
| Manu | 675 | 1.08 | 1,064 | 0.95 |
| M. microti | 29 | 0.05 | 29 | 0.03 |
| M. pinnipedii | 152 | 0.24 | 159 | 0.14 |
| S | 1,151 | 1.84 | 1,606 | 1.44 |
| T | 12,038 | 19.24 | 17,947 | 16.08 |
| X | 4,088 | 6.53 | 4,683 | 4.19 |
The strains are classified into major phylogenetic clades assigned according to signatures provided previously (381), which include various MTBC members as well as lineages/sublineages of M. tuberculosis sensu stricto (note that the sublineages are not shown), i.e., the Beijing clade, the Central Asian (CAS) clade, the East African-Indian (EAI) clade, the Haarlem/Ural clades, the Latin American-Mediterranean (LAM) clade, the Cameroon (previously LAM10-CAM) and Turkey (previously LAM7-TUR) lineages, the “Manu” family, the IS6110 low-banding X clade, and the ill-defined T clade.
FIG 3.
Worldwide distribution of lineages contained in the SITVIT2 database. AFRI, M. africanum; BOV, M. bovis; TUR, Turkey lineage; CAM, Cameroon lineage; Unk, unknown.
Examples of Recent Studies Using the SITVIT2 Database
Findings from recent studies showing an association between phylogenetic lineages and demographic and epidemiological parameters using the SITVIT2 database include (i) the finding of high phylogeographical specificity of M. africanum for Western Africa, with Guinea-Bissau being the epicenter (591); (ii) evidence that the TB epidemic in Sweden a century ago was caused by a closely knit pool of evolutionarily recent strains restricted to Sweden and its immediate neighbors (592); (iii) evidence for a prolonged, clonal, hospital-based outbreak of MDR TB among HIV-positive patients in Peru (593); (iv) determination of the predominance of the Euro-American family among elderly TB patients in Finland just as in Sweden, with the main difference being the presence of significant proportions of isolates of the Ural lineage among Finnish-born cases (also found in Russia, Latvia, and Estonia) but not in Sweden (594); and (v) determination of the association of the Beijing lineage with excessive drug resistance, including MDR and/or XDR TB (286, 502, 595), and the finding that even though the proportion of drug-resistant strains is significantly higher for Beijing than for non-Beijing strains globally, there are important variations in the distribution of drug resistance (596). Thus, drug resistance was significantly linked to Beijing strains in Russia, Southern Asia, Southeast Asia, and European countries but not to those in the Americas, Western Asia, China, and Japan. Furthermore, global data in the SITVIT2 database show that the SIT190/Beijing pattern, a rare but emerging spoligotype pattern, was significantly more associated with MDR TB than was the traditional SIT1/Beijing pattern (P < 0.0001) (596).
Finally, regarding information provided by bioinformatic tools and online georeferencing using Google API, a recent study was conducted in Baghdad, Iraq (597), where spoligoforest analysis showed that the bulk of TB isolates in postwar Iraq is limited to two phylogenetically related groups of MTBC strains belonging to the T and CAS lineages (SIT1144/T1 and SIT309/CAS1-Delhi), while georeferencing highlighted statistically significant differences between isolates from Baghdad and those from other cities in Iraq, regarding both demographic and drug resistance information.
Although many evolutionary and pathobiological characteristics of the prevailing TB epidemic remain to be discovered, studies of associations between MTBC phylogenetic lineages and demographic and epidemiological characteristics may significantly help to achieve an improved global overview of the worldwide TB situation. Collections of various databases and Web tools in conjunction with extended genotyping markers can make a difference in our understanding of the ongoing circulation of MTBC strains. Future developments should ideally include other markers such as RDs/LSPs and SNPs as well as future information that will be generated thanks to next-generation sequencing.
CONCLUSION
Molecular typing has revolutionized epidemiological studies of infectious diseases, including those of a mycobacterial origin. With the advent of fingerprinting techniques, many of the traditional concepts regarding the transmission, infectivity, or pathogenicity of mycobacterial bacilli have been revisited, and their conventional interpretation has been challenged. Since the mid-1990s, when the first typing methods were introduced, a plethora of other modalities have been proposed, with some designed as multipurpose tools and some tailored to address specific research questions. Common to all typing methods is that they disclose genetic differences at both the species and strain levels. These differences can either be used as a proxy for tracking strains, delineating transmission routes, and identifying sources of infection or be translated into phenotypic differences and act as surrogate markers of pathogenic characteristics such as virulence and drug resistance. Apart from epidemiological and diagnostic purposes, DNA fingerprinting methods are and have been successfully applied to resolve taxonomic ambiguities and evolutionary pathways within the Mycobacteriaceae. Currently, there is no single genotyping method that could serve as a panacea for all research problems. Each method has its pros and cons and differs from other methods in terms of discriminatory power, stability of genetic profiles, reproducibility, or amenability to computer-assisted analyses and databasing. Nevertheless, amid a myriad of typing approaches that have evolved over the last 2 decades, researchers can now configure a typing scheme that will most appropriately answer the question posed. Several methodological criteria have been established to assess the resolution of a particular typing system, and several approaches have been developed to evaluate its efficacy in different research contexts.
As whole-genome sequencing strategies are progressing at a fast pace, generating immense information on genome organization and diversity, a significant improvement of fingerprinting methods is to be expected. A key issue to consider is that the refinement of typing methodologies should be in tune with the financial capacities of the laboratories performing routine mycobacterial diagnostics, particularly in low- and middle-income countries, which account for the vast majority of the burden of TB and other mycobacterial diseases.
Molecular typing has been incorporated as a frontline strategy in most epidemiological studies of mycobacterial infections, and molecular epidemiology has become an integral component of modern mycobacteriology. Since genotyping continues to unravel the biology of mycobacteria, it offers enormous promise in the fight against and prevention of the diseases caused by these pathogens.
ACKNOWLEDGMENTS
This review was supported by a grant from the National Centre for Research and Development, Poland, under contract number LIDER/044/457/L-4/12/NCBR/2013.
Biographies

Tomasz Jagielski received his M.Sc. in biotechnology from the University of Warsaw in 2005 and five years later received a Ph.D. in medical sciences from the National Tuberculosis and Lung Diseases Research Institute, which serves as the National Tuberculosis Reference Laboratory for Poland. Dr. Jagielski works as a group leader at the Department of Applied Microbiology, University of Warsaw. His research interests are interdisciplinary and span a breadth of topics related to infectious diseases, with special emphasis on how to translate pathogens' genomic characteristics into diagnostic markers. His research explores sequence polymorphisms, allowing for inter/intraspecies identification and genetic determinants of drug resistance and virulence to be exploited in new molecular assays for rapid detection of these phenotypes. Dr. Jagielski is an author of over 120 articles in peer-reviewed journals, conference proceedings, educational periodicals, and popular science magazines. He is a member of several scientific societies, including the European Society of Clinical Microbiology and Infectious Diseases, the European Respiratory Society, and the American Society for Microbiology.

Alina Minias graduated in microbiology at the University of Lódź, Poland, and in genetics at the Autonomous University of Barcelona, Spain. She completed her Ph.D. in genetics of bacteria at the University of Lódź, in collaboration with the Institute of Medical Biology of the Polish Academy of Sciences. Her research interests focus on the maintenance of genome stability.

Jakko van Ingen, M.D., Ph.D., received a Ph.D. with honors at Radboud University, Nijmegen, The Netherlands, for his thesis on the clinical significance of isolation of nontuberculous mycobacteria (supervisors, Prof. Richard Dekhuijzen, Martin Boeree, and Dick van Soolingen). After a brief postdoctoral study at the National Jewish Health (Denver, CO; supervisors, Prof. Charles Daley and Prof. Leonid B. Heifets), he returned to Nijmegen for his residencies in clinical microbiology. He is currently a clinical microbiologist at the Radboud University Medical Center in Nijmegen, The Netherlands, where he also heads the mycobacteriology reference laboratory. He is an author of more than 100 peer-reviewed articles and 10 book chapters on mycobacterial disease. His research focus is to improve treatment outcome in nontuberculous mycobacterial disease, through therapy simulations in pharmacodynamic models and improving methods for drug susceptibility testing.

Nalin Rastogi is Director of Research at the Pasteur Institute in Paris. He earned a Ph.D. in biochemistry and a D.Sc. in microbiology from the University of Paris and diplomas in microbiology and in immunology from the Pasteur Institute in Paris. Between 1980 and 1993, he worked in Paris on the mycobacterial cell envelope and its role as a permeability barrier, intracellular drug screening models, host-parasite interactions, and pathogenicity. In 1993, he set up the Tuberculosis and Mycobacteria Unit in Guadeloupe, designated the WHO Supranational Reference Laboratory in 2009. Since 1994, he has worked on various aspects of TB, including diagnostics, drug resistance, taxonomy, molecular epidemiology, phylogeny, global TB transmission and geolocalization of emerging clones, and development of genotyping databases. He is an author of 350 peer-reviewed articles and 15 book chapters and is a contributor to various TB manuals. He served as the President of the European Society of Mycobacteriology (1993 to 1998) and as a Country Liaison of the American Society for Microbiology (2008 to 2012).

Anna Brzostek received her M.Sc. and Ph.D. at the University of Łódź. Since 1997, she has worked at the position of assistant professor in the Mycobacterium Genetics and Physiology Unit at the Institute of Medical Biology of the Polish Academy of Sciences in Łódź. Her research focuses on genetics and metabolism of the fast- and slow-growing mycobacteria, particularly the significance of cholesterol metabolism in the pathogenicity of tubercle bacilli. Her research interests also include the molecular epidemiology of tuberculosis and molecular mechanisms of drug resistance in mycobacteria, identification of targets for new antitubercular drugs, and understanding of the DNA repair processes in mycobacteria. Dr. Brzostek has published over 40 peer-reviewed articles on these issues in high-impact scientific journals. She has participated in several national and international research projects and has performed professional, academic training for 11 M.Sc. and Ph.D. students.

Anna Żaczek received her M.Sc. at the Jagiellonian University in Cracow and her Ph.D. at the University of Łódź, Poland. Since 1998, she has worked at the University of Rzeszów, currently at the position of assistant professor in the Department of Biochemistry and Cell Biology at the Faculty of Biology and Agriculture. Her research activities concentrate on developing new methods for the identification and differentiation of bacteria and pathogenic species in particular, such as Mycobacterium spp. She is specifically interested in the molecular epidemiology of tuberculosis and molecular mechanisms of drug resistance in tubercle bacilli. Dr. Żaczek has published articles on these issues and has participated in several research projects on tuberculosis. She is an experienced academic lecturer, with 10 B.Sc. and M.Sc. projects completed under her supervision. She is a member of the Polish Society of Microbiologists.

Jarosław Dziadek received most of his academic education at the University of Łódź, where he was appointed to a professorship in 2008. Since 2013, he has held the position of Director of the Institute of Medical Biology of the Polish Academy of Sciences in Łódź. There, he is also the Head of the Mycobacterium Genetics and Physiology Unit, leading a research group investigating the genetics of different mycobacterial species. The group has particular expertise in DNA repair, cell division, cholesterol metabolism, and cell wall biosynthesis with respect to the pathogenesis of tuberculosis. Prof. Dziadek has been the leader of several research projects founded by different national and international agencies, for over €10 million in total. He has published over 90 articles in peer-reviewed, top-ranked microbiology journals. Under his supervision, more than 20 M.Sc. and 11 Ph.D. projects have been completed.
REFERENCES
- 1.Hayman J. 1984. Mycobacterium ulcerans: an infection from Jurassic time? Lancet ii:1015–1016. [DOI] [PubMed] [Google Scholar]
- 2.Hansen GH. 1875. On the etiology of leprosy. Br J Foreign Med Chir Rev 55:459–489. [PMC free article] [PubMed] [Google Scholar]
- 3.Gallivan M, Shah N, Flood J. 2015. Epidemiology of human Mycobacterium bovis disease, California, USA, 2003-2011. Emerg Infect Dis 21:435–443. doi: 10.3201/eid2103.141539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majoor CJ, Magis-Escurra C, van Ingen J, Boeree MJ, van Soolingen D. 2011. Epidemiology of Mycobacterium bovis disease in humans, The Netherlands, 1993-2007. Emerg Infect Dis 17:457–463. doi: 10.3201/eid1703.101111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kubica T, Rüsch-Gerdes S, Niemann S. 2003. Mycobacterium bovis subsp. caprae caused one-third of human M. bovis-associated tuberculosis cases reported in Germany between 1999 and 2001. J Clin Microbiol 41:3070–3077. doi: 10.1128/JCM.41.7.3070-3077.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodríguez E, Sánchez LP, Pérez S, Herrera L, Jiménez MS, Samper S, Iglesias MJ. 2009. Human tuberculosis due to Mycobacterium bovis and M. caprae in Spain, 2004-2007. Int J Tuberc Lung Dis 13:1536–1541. [PubMed] [Google Scholar]
- 7.de Jong E, Rentenaar RJ, van Pelt R, de Lange W, Schreurs W, van Soolingen D, Sturm PDJ. 2009. Two cases of Mycobacterium microti-induced culture-negative tuberculosis. J Clin Microbiol 47:3038–3040. doi: 10.1128/JCM.00772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panteix G, Gutierrez MC, Boschiroli ML, Rouviere M, Plaidy A, Pressac D, Porcheret H, Chyderiotis G, Ponsada M, Van Oortegem K, Salloum S, Cabuzel S, Bañuls AL, Van de Perre P, Godreuil S. 2010. Pulmonary tuberculosis due to Mycobacterium microti: a study of six recent cases in France. J Med Microbiol 59:984–989. doi: 10.1099/jmm.0.019372-0. [DOI] [PubMed] [Google Scholar]
- 9.Kiers A, Klarenbeek A, Mendelts B, Van Soolingen D, Koëter G. 2008. Transmission of Mycobacterium pinnipedii to humans in a zoo with marine mammals. Int J Tuberc Lung Dis 12:1469–1473. [PubMed] [Google Scholar]
- 10.World Health Organization. 2014. Global tuberculosis report 2014 (WHO/HTM/TB/2014.08). World Health Organization, Geneva, Switzerland. [Google Scholar]
- 11.Rodrigues LC, Lockwood DN. 2011. Leprosy now: epidemiology, progress, challenges, and research gaps. Lancet Infect Dis 11:464–470. doi: 10.1016/S1473-3099(11)70006-8. [DOI] [PubMed] [Google Scholar]
- 12.Scollard DM, Adams LB, Gillis TP, Krahenbuhl JL, Truman RW, Williams DL. 2006. The continuing challenges of leprosy. Clin Microbiol Rev 19:338–381. doi: 10.1128/CMR.19.2.338-381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falkinham JO. 2002. Nontuberculous mycobacteria in the environment. Clin Chest Med 23:529–551. doi: 10.1016/S0272-5231(02)00014-X. [DOI] [PubMed] [Google Scholar]
- 14.Van Ingen J. 2013. Diagnosis of nontuberculous mycobacterial infections. Semin Respir Crit Care Med 34:103–109. doi: 10.1055/s-0033-1333569. [DOI] [PubMed] [Google Scholar]
- 15.Wolinsky E. 1995. Mycobacterial lymphadenitis in children: a prospective study of 105 nontuberculous cases with long-term follow-up. Clin Infect Dis 20:954–963. doi: 10.1093/clinids/20.4.954. [DOI] [PubMed] [Google Scholar]
- 16.De Groote MA, Huitt G. 2006. Infections due to rapidly growing mycobacteria. Clin Infect Dis 42:1756–1763. doi: 10.1086/504381. [DOI] [PubMed] [Google Scholar]
- 17.Petrini B. 2006. Mycobacterium marinum: ubiquitous agent of waterborne granulomatous skin infections. Eur J Clin Microbiol Infect Dis 25:609–613. doi: 10.1007/s10096-006-0201-4. [DOI] [PubMed] [Google Scholar]
- 18.Sizaire V, Nackers F, Comte E, Portaels F. 2006. Mycobacterium ulcerans infection: control, diagnosis, and treatment. Lancet Infect Dis 6:288–296. doi: 10.1016/S1473-3099(06)70464-9. [DOI] [PubMed] [Google Scholar]
- 19.Daley CL, Griffith DE. 2010. Pulmonary non-tuberculous mycobacterial infections. Int J Tuberc Lung Dis 14:665–671. [PubMed] [Google Scholar]
- 20.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Winthrop K, ATS Mycobacterial Diseases Subcommittee, American Thoracic Society, Infectious Disease Society of America. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 21.Tortoli E. 2009. Clinical manifestations of nontuberculous mycobacteria infections. Clin Microbiol Infect 15:906–910. doi: 10.1111/j.1469-0691.2009.03014.x. [DOI] [PubMed] [Google Scholar]
- 22.Kato-Maeda M, Rhee JT, Gingeras TR, Salamon H, Drenkow J, Smittipat N, Small PM. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res 11:547–554. doi: 10.1101/gr.166401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 24.Dos Vultos T, Mestre O, Rauzier J, Golec M, Rastogi N, Rasolofo V, Tonjum T, Sola C, Matic I, Gicquel B. 2008. Evolution and diversity of clonal bacteria: the paradigm of Mycobacterium tuberculosis. PLoS One 3:e1538. doi: 10.1371/journal.pone.0001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dos Vultos T, Mestre O, Tonjum T, Gicquel B. 2009. DNA repair in Mycobacterium tuberculosis revisited. FEMS Microbiol Rev 33:471–487. doi: 10.1111/j.1574-6976.2009.00170.x. [DOI] [PubMed] [Google Scholar]
- 26.Mizrahi V, Andersen SJ. 1998. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol Microbiol 29:1331–1339. [DOI] [PubMed] [Google Scholar]
- 27.Kurthkoti K, Varshney U. 2011. Base excision and nucleotide excision repair pathways in mycobacteria. Tuberculosis (Edinb) 91:533–543. doi: 10.1016/j.tube.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Gorna AE, Bowater RP, Dziadek J. 2010. DNA repair systems and the pathogenesis of Mycobacterium tuberculosis: varying activities at different stages of infection. Clin Sci (Lond) 119:187–202. doi: 10.1042/CS20100041. [DOI] [PubMed] [Google Scholar]
- 29.Fu LM, Fu-Liu CS. 2007. The gene expression data of Mycobacterium tuberculosis based on Affymetrix gene chips provide insight into regulatory and hypothetical genes. BMC Microbiol 7:37. doi: 10.1186/1471-2180-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulder MA, Nair S, Abratt VR, Zappe H, Steyn LM. 1999. Involvement of the N- and C-terminal domains of Mycobacterium tuberculosis KatG in the protection of mutant Escherichia coli against DNA-damaging agents. Microbiology 145(Part 8):2011–2021. [DOI] [PubMed] [Google Scholar]
- 31.Brzostek A, Szulc I, Klink M, Brzezinska M, Sulowska Z, Dziadek J. 2014. Either non-homologous ends joining or homologous recombination is required to repair double-strand breaks in the genome of macrophage-internalized Mycobacterium tuberculosis. PLoS One 9:e92799. doi: 10.1371/journal.pone.0092799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heaton BE, Barkan D, Bongiorno P, Karakousis PC, Glickman MS. 2014. Deficiency of double-strand DNA break repair does not impair Mycobacterium tuberculosis virulence in multiple animal models of infection. Infect Immun 82:3177–3185. doi: 10.1128/IAI.01540-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ford CB, Lin PL, Chase MR, Shah RR, Iartchouk O, Galagan J, Mohaideen N, Ioerger TR, Sacchettini JC, Lipsitch M, Flynn JL, Fortune SM. 2011. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet 43:482–486. doi: 10.1038/ng.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colangeli R, Arcus VL, Cursons RT, Ruthe A, Karalus N, Coley K, Manning SD, Kim S, Marchiano E, Alland D. 2014. Whole genome sequencing of Mycobacterium tuberculosis reveals slow growth and low mutation rates during latent infections in humans. PLoS One 9:e91024. doi: 10.1371/journal.pone.0091024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosas-Magallanes V, Deschavanne P, Quintana-Murci L, Brosch R, Gicquel B, Neyrolles O. 2006. Horizontal transfer of a virulence operon to the ancestor of Mycobacterium tuberculosis. Mol Biol Evol 23:1129–1135. doi: 10.1093/molbev/msj120. [DOI] [PubMed] [Google Scholar]
- 36.Veyrier F, Pletzer D, Turenne C, Behr MA. 2009. Phylogenetic detection of horizontal gene transfer during the step-wise genesis of Mycobacterium tuberculosis. BMC Evol Biol 9:196. doi: 10.1186/1471-2148-9-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jucker MT, Falkinham JO. 1990. Epidemiology of infection by nontuberculous mycobacteria. IX. Evidence for two DNA homology groups among small plasmids in Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum. Am Rev Respir Dis 142:858–862. [DOI] [PubMed] [Google Scholar]
- 38.Kirby C, Waring A, Griffin TJ, Falkinham JO, Grindley NDF, Derbyshire KM. 2002. Cryptic plasmids of Mycobacterium avium: Tn552 to the rescue. Mol Microbiol 43:173–186. doi: 10.1046/j.1365-2958.2002.02729.x. [DOI] [PubMed] [Google Scholar]
- 39.Stinear TP, Mve-Obiang A, Small PLC, Frigui W, Pryor MJ, Brosch R, Jenkin GA, Johnson PDR, Davies JK, Lee RE, Adusumilli S, Garnier T, Haydock SF, Leadlay PF, Cole ST. 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci U S A 101:1345–1349. doi: 10.1073/pnas.0305877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ummels R, Abdallah AM, Kuiper V, Aâjoud A, Sparrius M, Naeem R, Spaink HP, van Soolingen D, Pain A, Bitter W. 2014. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. mBio 5:e01744-14. doi: 10.1128/mBio.01744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bachrach G, Colston MJ, Bercovier H, Bar-Nir D, Anderson C, Papavinasasundaram KG. 2000. A new single-copy mycobacterial plasmid, pMF1, from Mycobacterium fortuitum which is compatible with the pAL5000 replicon. Microbiology 146(Part 2):297–303. doi: 10.1099/00221287-146-2-297. [DOI] [PubMed] [Google Scholar]
- 42.Gormley EP, Davies J. 1991. Transfer of plasmid RSF1010 by conjugation from Escherichia coli to Streptomyces lividans and Mycobacterium smegmatis. J Bacteriol 173:6705–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coros A, Callahan B, Battaglioli E, Derbyshire KM. 2008. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol Microbiol 69:794–808. doi: 10.1111/j.1365-2958.2008.06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gray TA, Krywy JA, Harold J, Palumbo MJ, Derbyshire KM. 2013. Distributive conjugal transfer in mycobacteria generates progeny with meiotic-like genome-wide mosaicism, allowing mapping of a mating identity locus. PLoS Biol 11:e1001602. doi: 10.1371/journal.pbio.1001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leão SC, Matsumoto CK, Carneiro A, Ramos RT, Nogueira CL, Lima JD, Lima KV, Lopes ML, Schneider H, Azevedo VA, da Costa da Silva A. 2013. The detection and sequencing of a broad-host-range conjugative IncP-1β plasmid in an epidemic strain of Mycobacterium abscessus subsp. bolletii. PLoS One 8:e60746. doi: 10.1371/journal.pone.0060746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabello MC, Matsumoto CK, Almeida LGP, Menendez MC, Oliveira RS, Silva RM, Garcia MJ, Leão SC. 2012. First description of natural and experimental conjugation between mycobacteria mediated by a linear plasmid. PLoS One 7:e29884. doi: 10.1371/journal.pone.0029884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhatt A, Kieser HM, Melton RE, Kieser T. 2002. Plasmid transfer from Streptomyces to Mycobacterium smegmatis by spontaneous transformation. Mol Microbiol 43:135–146. doi: 10.1046/j.1365-2958.2002.02722.x. [DOI] [PubMed] [Google Scholar]
- 48.Bardarov S, Bardarov S Jr, Pavelka MS Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- 49.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR, Hendrix RW, Hatfull GF. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171–182. doi: 10.1016/S0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 50.Tsolaki AG, Hirsh AE, DeRiemer K, Enciso JA, Wong MZ, Hannan M, Goguet de la Salmoniere Y-OL, Aman K, Kato-Maeda M, Small PM. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc Natl Acad Sci U S A 101:4865–4870. doi: 10.1073/pnas.0305634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isaza JP, Duque C, Gomez V, Robledo J, Barrera LF, Alzate JF. 2012. Whole genome shotgun sequencing of one Colombian clinical isolate of Mycobacterium tuberculosis reveals DosR regulon gene deletions. FEMS Microbiol Lett 330:113–120. doi: 10.1111/j.1574-6968.2012.02540.x. [DOI] [PubMed] [Google Scholar]
- 52.Brosch R, Gordon SV, Buchrieser C, Pym AS, Garnier T, Cole ST. 2000. Comparative genomics uncovers large tandem chromosomal duplications in Mycobacterium bovis BCG Pasteur. Yeast 17:111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, Parsons LM, Pym AS, Samper S, van Soolingen D, Cole ST. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A 99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, DeBoy R, Dodson R, Gwinn M, Haft D, Hickey E, Kolonay JF, Nelson WC, Umayam LA, Ermolaeva M, Salzberg SL, Delcher A, Utterback T, Weidman J, Khouri H, Gill J, Mikula A, Bishai W, Jacobs WR Jr, Venter JC, Fraser CM. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol 184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, Blom J, Wirth T, Jaenicke S, Schuback S, Rüsch-Gerdes S, Supply P, Kalinowski J, Niemann S. 2013. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med 10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, Eyre DW, Wilson DJ, Hawkey PM, Crook DW, Parkhill J, Harris D, Walker AS, Bowden R, Monk P, Smith EG, Peto TE. 2013. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kato-Maeda M, Ho C, Passarelli B, Banaei N, Grinsdale J, Flores L, Anderson J, Murray M, Rose G, Kawamura LM, Pourmand N, Tariq MA, Gagneux S, Hopewell PC. 2013. Use of whole genome sequencing to determine the microevolution of Mycobacterium tuberculosis during an outbreak. PLoS One 8:e58235. doi: 10.1371/journal.pone.0058235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Viratyosin W, Kulawonganunchai S, Smittipat N, Juthayothin T, Penpassakarn P, Pasomsub E, Chantratita W, Chaiprasert A, Palittapongarnpim P. 2013. Draft genome sequence of the Mycobacterium tuberculosis strain 43-16836, belonging to the Indo-Oceanic lineage, isolated from tuberculous meningitis in Thailand. Genome Announc 1(5):e00801-13. doi: 10.1128/genomeA.00801-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Juhlin I. 1960. Methods for the grouping and typing of mycobacteria. 2. Differentiation of Mycobacterium tuberculosis into typus humanus and typus bovinus. Acta Pathol Microbiol Scand 50:188–194. [DOI] [PubMed] [Google Scholar]
- 60.Lucchesi M, Zubiani M, Maldarizzi B. 1960. On the typing of Mycobacterium tuberculosis by means of the bacteriostatic action of thiophenecarboxylic acid hydrazide (TCH) and pyromucic acid hydrazide (BSH). Ann Ist Carlo Forlanini 20:514–522. (In Italian.) [PubMed] [Google Scholar]
- 61.Lebek G. 1954. The usefulness of the Wagener's bromocresol purple for typing of Mycobacterium tuberculosis. Zentralbl Bakteriol Orig 161:324–327. (In German.) [PubMed] [Google Scholar]
- 62.Van Marwyck C. 1954. Measurement of respiration and glycolysis in Mycobacterium tuberculosis for typing and for determination of chemotherapeutic effect. Arch Hyg Bakteriol 138:561–568. [PubMed] [Google Scholar]
- 63.Stur D. 1955. Bromo-cresol-purple culture medium for typing of Mycobacterium tuberculosis. Acta Pathol Microbiol Scand 37:279–285. [PubMed] [Google Scholar]
- 64.Hnatko SI. 1953. The isolation of bacteriophages for mycobacteria with reference to phage typing of the genus. Can J Med Sci 31:462–473. [DOI] [PubMed] [Google Scholar]
- 65.Froman S, Will DW, Bogen E. 1954. Bacteriophage active against virulent Mycobacterium tuberculosis. I. Isolation and activity. Am J Public Health Nations Health 44:1326–1333. doi: 10.2105/AJPH.44.10.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takeya K, Yoshimura T. 1957. Isolation of bacteriophage active against all types of Mycobacterium tuberculosis. J Bacteriol 74:540–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ribon W. 7 March 2012. Biochemical isolation and identification of mycobacteria. Biochemical testing. InTech doi: 10.5772/34309. [DOI] [Google Scholar]
- 68.Horne DJ, Pinto LM, Arentz M, Lin S-YG, Desmond E, Flores LL, Steingart KR, Minion J. 2013. Diagnostic accuracy and reproducibility of WHO-endorsed phenotypic drug susceptibility testing methods for first-line and second-line antituberculosis drugs. J Clin Microbiol 51:393–401. doi: 10.1128/JCM.02724-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Minion J, Leung E, Menzies D, Pai M. 2010. Microscopic-observation drug susceptibility and thin layer agar assays for the detection of drug resistant tuberculosis: a systematic review and meta-analysis. Lancet Infect Dis 10:688–698. doi: 10.1016/S1473-3099(10)70165-1. [DOI] [PubMed] [Google Scholar]
- 70.Martin A, Portaels F, Palomino JC. 2007. Colorimetric redox-indicator methods for the rapid detection of multidrug resistance in Mycobacterium tuberculosis: a systematic review and meta-analysis. J Antimicrob Chemother 59:175–183. [DOI] [PubMed] [Google Scholar]
- 71.Martin A, Panaiotov S, Portaels F, Hoffner S, Palomino JC, Angeby K. 2008. The nitrate reductase assay for the rapid detection of isoniazid and rifampicin resistance in Mycobacterium tuberculosis: a systematic review and meta-analysis. J Antimicrob Chemother 62:56–64. doi: 10.1093/jac/dkn139. [DOI] [PubMed] [Google Scholar]
- 72.Snider DE, Jones WD, Good RC. 1984. The usefulness of phage typing Mycobacterium tuberculosis isolates. Am Rev Respir Dis 130:1095–1099. [DOI] [PubMed] [Google Scholar]
- 73.Jones WD. 1988. Bacteriophage typing of Mycobacterium tuberculosis cultures from incidents of suspected laboratory cross-contamination. Tubercle 69:43–46. doi: 10.1016/0041-3879(88)90039-6. [DOI] [PubMed] [Google Scholar]
- 74.Rado TA, Bates JH, Engel HW, Mankiewicz E, Murohashi T, Mizuguchi Y, Sula L. 1975. World Health Organization studies on bacteriophage typing of mycobacteria. Subdivision of the species Mycobacterium tuberculosis. Am Rev Respir Dis 111:459–468. [DOI] [PubMed] [Google Scholar]
- 75.Gruft H, Johnson R, Claflin R, Loder A. 1984. Phage-typing and drug-resistance patterns as tools in mycobacterial epidemiology. Am Rev Respir Dis 130:96–97. [DOI] [PubMed] [Google Scholar]
- 76.Leite CQF, da Silva Rocha A, de Andrade Leite SR, Ferreira RMC, Suffys PN, de Souza Fonseca L, Saad MHF. 2005. A comparison of mycolic acid analysis for nontuberculous mycobacteria identification by thin-layer chromatography and molecular methods. Microbiol Immunol 49:571–578. doi: 10.1111/j.1348-0421.2005.tb03642.x. [DOI] [PubMed] [Google Scholar]
- 77.Huet G, Constant P, Malaga W, Lanéelle M-A, Kremer K, van Soolingen D, Daffé M, Guilhot C. 2009. A lipid profile typifies the Beijing strains of Mycobacterium tuberculosis. J Biol Chem 284:27101–27113. doi: 10.1074/jbc.M109.041939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Secanella-Fandos S, Luquin M, Pérez-Trujillo M, Julián E. 2011. Revisited mycolic acid pattern of Mycobacterium confluentis using thin-layer chromatography. J Chromatogr B Analyt Technol Biomed Life Sci 879:2821–2826. doi: 10.1016/j.jchromb.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 79.Butler WR, Thibert L, Kilburn JO. 1992. Identification of Mycobacterium avium complex strains and some similar species by high-performance liquid chromatography. J Clin Microbiol 30:2698–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Glickman SE, Kilburn JO, Butler WR, Ramos LS. 1994. Rapid identification of mycolic acid patterns of mycobacteria by high-performance liquid chromatography using pattern recognition software and a Mycobacterium library. J Clin Microbiol 32:740–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Slosárek M, Alugupalli S, Kaustova J, Larsson L. 1996. Rapid detection of Mycobacterium kansasii in water by gas chromatography/mass spectrometry. J Microbiol Methods 27:229–232. doi: 10.1016/S0167-7012(96)00954-2. [DOI] [Google Scholar]
- 82.O'Sullivan DM, Nicoara SC, Mutetwa R, Mungofa S, Lee OY-C, Minnikin DE, Bardwell MW, Corbett EL, McNerney R, Morgan GH. 2012. Detection of Mycobacterium tuberculosis in sputum by gas chromatography-mass spectrometry of methyl mycocerosates released by thermochemolysis. PLoS One 7:e32836. doi: 10.1371/journal.pone.0032836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pignone M, Greth KM, Cooper J, Emerson D, Tang J. 2006. Identification of mycobacteria by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry. J Clin Microbiol 44:1963–1970. doi: 10.1128/JCM.01959-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shitikov E, Ilina E, Chernousova L, Borovskaya A, Rukin I, Afanas'ev M, Smirnova T, Vorobyeva A, Larionova E, Andreevskaya S, Kostrzewa M, Govorun V. 2012. Mass spectrometry based methods for the discrimination and typing of mycobacteria. Infect Genet Evol 12:838–845. doi: 10.1016/j.meegid.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 85.Szewczyk R, Kowalski K, Janiszewska-Drobinska B, Druszczyńska M. 2013. Rapid method for Mycobacterium tuberculosis identification using electrospray ionization tandem mass spectrometry analysis of mycolic acids. Diagn Microbiol Infect Dis 76:298–305. doi: 10.1016/j.diagmicrobio.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 86.Saleeb PG, Drake SK, Murray PR, Zelazny AM. 2011. Identification of mycobacteria in solid-culture media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 49:1790–1794. doi: 10.1128/JCM.02135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen JHK, Yam W-C, Ngan AHY, Fung AMY, Woo W-L, Yan M-K, Choi GKY, Ho P-L, Cheng VCC, Yuen K-Y. 2013. Advantages of using matrix-assisted laser desorption ionization–time of flight mass spectrometry as a rapid diagnostic tool for identification of yeasts and mycobacteria in the clinical microbiological laboratory. J Clin Microbiol 51:3981–3987. doi: 10.1128/JCM.01437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mathema B, Kurepina N, Yang G, Shashkina E, Manca C, Mehaffy C, Bielefeldt-Ohmann H, Ahuja S, Fallows DA, Izzo A, Bifani P, Dobos K, Kaplan G, Kreiswirth BN. 2012. Epidemiologic consequences of microvariation in Mycobacterium tuberculosis. J Infect Dis 205:964–974. doi: 10.1093/infdis/jir876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Toney N, Adekambi T, Toney S, Yakrus M, Butler WR. 2010. Revival and emended description of “Mycobacterium paraffinicum” Davis, Chase and Raymond 1956 as Mycobacterium paraffinicum sp. nov., nom. rev. Int J Syst Evol Microbiol 60:2307–2313. doi: 10.1099/ijs.0.016972-0. [DOI] [PubMed] [Google Scholar]
- 90.Wasem CF, McCarthy CM, Murray LW. 1991. Multilocus enzyme electrophoresis analysis of the Mycobacterium avium complex and other mycobacteria. J Clin Microbiol 29:264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Feizabadi MM, Robertson ID, Cousins DV, Dawson D, Chew W, Gilbert GL, Hampson DJ. 1996. Genetic characterization of Mycobacterium avium isolates recovered from humans and animals in Australia. Epidemiol Infect 116:41–49. doi: 10.1017/S0950268800058945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Y, Yakrus MA, Graviss EA, Williams-Bouyer N, Turenne C, Kabani A, Wallace RJ. 2004. Pulsed-field gel electrophoresis study of Mycobacterium abscessus isolates previously affected by DNA degradation. J Clin Microbiol 42:5582–5587. doi: 10.1128/JCM.42.12.5582-5587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.O'Shea B, Khare S, Klein P, Roussel A, Adams LG, Ficht TA, Rice-Ficht AC. 2011. Amplified fragment length polymorphism reveals specific epigenetic distinctions between Mycobacterium avium subspecies paratuberculosis isolates of various isolation types. J Clin Microbiol 49:2222–2229. doi: 10.1128/JCM.01123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shell SS, Prestwich EG, Baek S-H, Shah RR, Sassetti CM, Dedon PC, Fortune SM. 2013. DNA methylation impacts gene expression and ensures hypoxic survival of Mycobacterium tuberculosis. PLoS Pathog 9:e1003419. doi: 10.1371/journal.ppat.1003419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Han XY, Pham AS, Tarrand JJ, Sood PK, Luthra R. 2002. Rapid and accurate identification of mycobacteria by sequencing hypervariable regions of the 16S ribosomal RNA gene. Am J Clin Pathol 118:796–801. doi: 10.1309/HN44-XQYM-JMAQ-2EDL. [DOI] [PubMed] [Google Scholar]
- 96.Rogall T, Flohr T, Böttger EC. 1990. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J Gen Microbiol 136:1915–1920. doi: 10.1099/00221287-136-9-1915. [DOI] [PubMed] [Google Scholar]
- 97.Hernandez SM, Morlock GP, Butler WR, Crawford JT, Cooksey RC. 1999. Identification of Mycobacterium species by PCR-restriction fragment length polymorphism analyses using fluorescence capillary electrophoresis. J Clin Microbiol 37:3688–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ong C-S, Ngeow Y-F, Yap S-F, Tay S-T. 2010. Evaluation of PCR-RFLP analysis targeting hsp65 and rpoB genes for the typing of mycobacterial isolates in Malaysia. J Med Microbiol 59:1311–1316. doi: 10.1099/jmm.0.021139-0. [DOI] [PubMed] [Google Scholar]
- 99.Saifi M, Jabbarzadeh E, Bahrmand AR, Karimi A, Pourazar S, Fateh A, Masoumi M, Vahidi E. 2013. hsp65-PRA identification of non-tuberculosis mycobacteria from 4892 samples suspicious for mycobacterial infections. Clin Microbiol Infect 19:723–728. doi: 10.1111/j.1469-0691.2012.04005.x. [DOI] [PubMed] [Google Scholar]
- 100.Dvorska L, Bartos M, Pavlik I, Martin G, Erler W. 2001. Strategies for differentiation, identification and typing of medically important species of mycobacteria by molecular methods. Vet Med 11–12:309–328. [Google Scholar]
- 101.Dalovisio JR, Montenegro-James S, Kemmerly SA, Genre CF, Chambers R, Greer D, Pankey GA, Failla DM, Haydel KG, Hutchinson L, Lindley MF, Nunez BM, Praba A, Eisenach KD, Cooper ES. 1996. Comparison of the amplified Mycobacterium tuberculosis (MTB) direct test, Amplicor MTB PCR, and IS6110-PCR for detection of MTB in respiratory specimens Clin Infect Dis 23:1099–1106; discussion 1107–1108. [DOI] [PubMed] [Google Scholar]
- 102.Bloemberg GV, Voit A, Ritter C, Deggim V, Böttger EC. 2013. Evaluation of Cobas TaqMan MTB for direct detection of the Mycobacterium tuberculosis complex in comparison with Cobas Amplicor MTB. J Clin Microbiol 51:2112–2117. doi: 10.1128/JCM.00142-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rodríguez-Lázaro D, Lloyd J, Herrewegh A, Ikonomopoulos J, D'Agostino M, Pla M, Cook N. 2004. A molecular beacon-based real-time NASBA assay for detection of Mycobacterium avium subsp. paratuberculosis in water and milk. FEMS Microbiol Lett 237:119–126. doi: 10.1111/j.1574-6968.2004.tb09686.x. [DOI] [PubMed] [Google Scholar]
- 104.Barrett A, Magee JG, Freeman R. 2002. An evaluation of the BD ProbeTec ET system for the direct detection of Mycobacterium tuberculosis in respiratory samples. J Med Microbiol 51:895–898. doi: 10.1099/0022-1317-51-10-895. [DOI] [PubMed] [Google Scholar]
- 105.Ichiyama S, Iinuma Y, Tawada Y, Yamori S, Hasegawa Y, Shimokata K, Nakashima N. 1996. Evaluation of Gen-Probe Amplified Mycobacterium Tuberculosis Direct Test and Roche PCR-microwell plate hybridization method (AMPLICOR MYCOBACTERIUM) for direct detection of mycobacteria. J Clin Microbiol 34:130–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kauppinen J, Pelkonen J, Katila M-L. 1994. RFLP analysis of Mycobacterium malmoense strains using ribosomal RNA gene probes: an addition tool to examine intraspecies variation. J Microbiol Methods 19:261–267. doi: 10.1016/0167-7012(94)90029-9. [DOI] [Google Scholar]
- 107.Pourahmad F, Thompson KD, Taggart JB, Adams A, Richards RH. 2008. Evaluation of the INNO-LiPA mycobacteria v2 assay for identification of aquatic mycobacteria. J Fish Dis 31:931–940. doi: 10.1111/j.1365-2761.2008.00968.x. [DOI] [PubMed] [Google Scholar]
- 108.Kusunoki S, Ezaki T, Tamesada M, Hatanaka Y, Asano K, Hashimoto Y, Yabuuchi E. 1991. Application of colorimetric microdilution plate hybridization for rapid genetic identification of 22 Mycobacterium species. J Clin Microbiol 29:1596–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yam WC, Cheng VCC, Hui WT, Wang LN, Seto WH, Yuen KY. 2004. Direct detection of Mycobacterium tuberculosis in clinical specimens using single-tube biotinylated nested polymerase chain reaction-enzyme linked immunoassay (PCR-ELISA). Diagn Microbiol Infect Dis 48:271–275. doi: 10.1016/j.diagmicrobio.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 110.Kim JH, Kim YJ, Ki C-S, Kim J-Y, Lee NY. 2011. Evaluation of Cobas TaqMan MTB PCR for detection of Mycobacterium tuberculosis. J Clin Microbiol 49:173–176. doi: 10.1128/JCM.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Picardeau M, Prod'Hom G, Raskine L, LePennec MP, Vincent V. 1997. Genotypic characterization of five subspecies of Mycobacterium kansasii. J Clin Microbiol 35:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pai M, Flores LL, Pai N, Hubbard A, Riley LW, Colford JM. 2003. Diagnostic accuracy of nucleic acid amplification tests for tuberculous meningitis: a systematic review and meta-analysis. Lancet Infect Dis 3:633–643. doi: 10.1016/S1473-3099(03)00772-2. [DOI] [PubMed] [Google Scholar]
- 113.Pai M. 2004. The accuracy and reliability of nucleic acid amplification tests in the diagnosis of tuberculosis. Natl Med J India 17:233–236. [PubMed] [Google Scholar]
- 114.Pai M, Flores LL, Hubbard A, Riley LW, Colford JM. 2004. Nucleic acid amplification tests in the diagnosis of tuberculous pleuritis: a systematic review and meta-analysis. BMC Infect Dis 4:6. doi: 10.1186/1471-2334-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Flores LL, Pai M, Colford JM, Riley LW. 2005. In-house nucleic acid amplification tests for the detection of Mycobacterium tuberculosis in sputum specimens: meta-analysis and meta-regression. BMC Microbiol 5:55. doi: 10.1186/1471-2180-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Menzies D. 2004. What is the current and potential role of diagnostic tests other than sputum microscopy and culture?, p 87–91. In Frieden T. (ed), Toman's tuberculosis. Case detection, treatment, and monitoring: questions and answers, 2nd ed, World Health Organization, Geneva, Switzerland. [Google Scholar]
- 117.Ahmad S, Mokaddas E, Jaber A-A. 2004. Rapid detection of ethambutol-resistant Mycobacterium tuberculosis strains by PCR-RFLP targeting embB codons 306 and 497 and iniA codon 501 mutations. Mol Cell Probes 18:299–306. doi: 10.1016/j.mcp.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 118.Röltgen K, Assan-Ampah K, Danso E, Yeboah-Manu D, Pluschke G. 2012. Development of a temperature-switch PCR-based SNP typing method for Mycobacterium ulcerans. PLoS Negl Trop Dis 6:e1904. doi: 10.1371/journal.pntd.0001904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Herrera-León L, Molina T, Saíz P, Sáez-Nieto JA, Jiménez MS. 2005. New multiplex PCR for rapid detection of isoniazid-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother 49:144–147. doi: 10.1128/AAC.49.1.144-147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Piatek AS, Tyagi S, Pol AC, Telenti A, Miller LP, Kramer FR, Alland D. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat Biotechnol 16:359–363. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- 121.Piatek AS, Telenti A, Murray MR, El-Hajj H, Jacobs WR, Kramer FR, Alland D. 2000. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob Agents Chemother 44:103–110. doi: 10.1128/AAC.44.1.103-110.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chakravorty S, Kothari H, Aladegbami B, Cho EJ, Lee JS, Roh SS, Kim H, Kwak H, Lee EG, Hwang SH, Banada PP, Safi H, Via LE, Cho S-N, Barry CE, Alland D. 2012. Rapid, high-throughput detection of rifampin resistance and heteroresistance in Mycobacterium tuberculosis by use of sloppy molecular beacon melting temperature coding. J Clin Microbiol 50:2194–2202. doi: 10.1128/JCM.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Torres MJ, Criado A, Palomares JC, Aznar J. 2000. Use of real-time PCR and fluorimetry for rapid detection of rifampin and isoniazid resistance-associated mutations in Mycobacterium tuberculosis. J Clin Microbiol 38:3194–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hristea A, Otelea D, Paraschiv S, Macri A, Baicus C, Moldovan O, Tinischi M, Arama V, Streinu-Cercel A. 2010. Detection of Mycobacterium tuberculosis resistance mutations to rifampin and isoniazid by real-time PCR. Indian J Med Microbiol 28:211–216. doi: 10.4103/0255-0857.66474. [DOI] [PubMed] [Google Scholar]
- 125.Ramirez MV, Cowart KC, Campbell PJ, Morlock GP, Sikes D, Winchell JM, Posey JE. 2010. Rapid detection of multidrug-resistant Mycobacterium tuberculosis by use of real-time PCR and high-resolution melt analysis. J Clin Microbiol 48:4003–4009. doi: 10.1128/JCM.00812-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hu S, Li G, Li H, Liu X, Niu J, Quan S, Wang F, Wen H, Xu Y, Li Q. 2014. Rapid detection of isoniazid resistance in Mycobacterium tuberculosis isolates by use of real-time-PCR-based melting curve analysis. J Clin Microbiol 52:1644–1652. doi: 10.1128/JCM.03395-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Argac D, Bulbul O, Shahzad MS, Acar E, Altuncul H, Filoglu G. 2009. Optimization and validation of 10 mitochondrial DNA SNPs using SNaPshot kit. Forensic Sci Int Genet Suppl Ser 2:99–101. doi: 10.1016/j.fsigss.2009.08.194. [DOI] [Google Scholar]
- 128.Bouakaze C, Keyser C, de Martino SJ, Sougakoff W, Veziris N, Dabernat H, Ludes B. 2010. Identification and genotyping of Mycobacterium tuberculosis complex species by use of a SNaPshot minisequencing-based assay. J Clin Microbiol 48:1758–1766. doi: 10.1128/JCM.02255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang H, Yue J, Han M, Yang J, Zhao Y. 2010. Rapid method for identification of six common species of mycobacteria based on multiplex SNP analysis. J Clin Microbiol 48:247–250. doi: 10.1128/JCM.01084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bouakaze C, Keyser C, Gonzalez A, Sougakoff W, Veziris N, Dabernat H, Jaulhac B, Ludes B. 2011. Matrix-assisted laser desorption ionization–time of flight mass spectrometry-based single nucleotide polymorphism genotyping assay using iPLEX Gold technology for identification of Mycobacterium tuberculosis complex species and lineages. J Clin Microbiol 49:3292–3299. doi: 10.1128/JCM.00744-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yao C, Zhu T, Li Y, Zhang L, Zhang B, Huang J, Fu W. 2010. Detection of rpoB, katG and inhA gene mutations in Mycobacterium tuberculosis clinical isolates from Chongqing as determined by microarray. Clin Microbiol Infect 16:1639–1643. doi: 10.1111/j.1469-0691.2010.03267.x. [DOI] [PubMed] [Google Scholar]
- 132.Linger Y, Kukhtin A, Golova J, Perov A, Lambarqui A, Bryant L, Rudy GB, Dionne K, Fisher SL, Parrish N, Chandler DP. 2014. Simplified microarray system for simultaneously detecting rifampin, isoniazid, ethambutol, and streptomycin resistance markers in Mycobacterium tuberculosis. J Clin Microbiol 52:2100–2107. doi: 10.1128/JCM.00238-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bergval IL, Vijzelaar RNCP, Dalla Costa ER, Schuitema ARJ, Oskam L, Kritski AL, Klatser PR, Anthony RM. 2008. Development of multiplex assay for rapid characterization of Mycobacterium tuberculosis. J Clin Microbiol 46:689–699. doi: 10.1128/JCM.01821-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bergval I, Sengstake S, Brankova N, Levterova V, Abadía E, Tadumaze N, Bablishvili N, Akhalaia M, Tuin K, Schuitema A, Panaiotov S, Bachiyska E, Kantardjiev T, de Zwaan R, Schürch A, van Soolingen D, van't Hoog A, Cobelens F, Aspindzelashvili R, Sola C, Klatser P, Anthony R. 2012. Combined species identification, genotyping, and drug resistance detection of Mycobacterium tuberculosis cultures by MLPA on a bead-based array. PLoS One 7:e43240. doi: 10.1371/journal.pone.0043240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sengstake S, Bablishvili N, Schuitema A, Bzekalava N, Abadia E, de Beer J, Tadumadze N, Akhalaia M, Tuin K, Tukvadze N, Aspindzelashvili R, Bachiyska E, Panaiotov S, Sola C, van Soolingen D, Klatser P, Anthony R, Bergval I. 2014. Optimizing multiplex SNP-based data analysis for genotyping of Mycobacterium tuberculosis isolates. BMC Genomics 15:572. doi: 10.1186/1471-2164-15-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stucki D, Malla B, Hostettler S, Huna T, Feldmann J, Yeboah-Manu D, Borrell S, Fenner L, Comas I, Coscollà M, Gagneux S. 2012. Two new rapid SNP-typing methods for classifying Mycobacterium tuberculosis complex into the main phylogenetic lineages. PLoS One 7:e41253. doi: 10.1371/journal.pone.0041253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Engström A, Zardán Gómez de la Torre T, Strømme M, Nilsson M, Herthnek D. 2013. Detection of rifampicin resistance in Mycobacterium tuberculosis by padlock probes and magnetic nanobead-based readout. PLoS One 8:e62015. doi: 10.1371/journal.pone.0062015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen X, Wang B, Yang W, Kong F, Li C, Sun Z, Jelfs P, Gilbert GL. 2014. Rolling circle amplification for direct detection of rpoB gene mutations in Mycobacterium tuberculosis isolates from clinical specimens. J Clin Microbiol 52:1540–1548. doi: 10.1128/JCM.00065-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.World Health Organization. 2008. Policy statement: molecular line probe assays for rapid screening of patients at risk of multidrug-resistant tuberculosis (MDR-TB). World Health Organization, Geneva, Switzerland. [Google Scholar]
- 140.Gazi MA, Islam MR, Kibria MG, Mahmud Z. 2015. General and advanced diagnostic tools to detect Mycobacterium tuberculosis and their drug susceptibility: a review. Eur J Clin Microbiol Infect Dis 34:851–861. doi: 10.1007/s10096-014-2306-5. [DOI] [PubMed] [Google Scholar]
- 141.Yüksel P, Tansel O. 2009. Characterization of pncA mutations of pyrazinamide-resistant Mycobacterium tuberculosis in Turkey. New Microbiol 32:153–158. [PubMed] [Google Scholar]
- 142.Lefmann M, Honisch C, Böcker S, Storm N, von Wintzingerode F, Schlötelburg C, Moter A, van den Boom D, Göbel UB. 2004. Novel mass spectrometry-based tool for genotypic identification of mycobacteria. J Clin Microbiol 42:339–346. doi: 10.1128/JCM.42.1.339-346.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Simner PJ, Buckwalter SP, Uhl JR, Wengenack NL. 2013. Identification of Mycobacterium species and Mycobacterium tuberculosis complex resistance determinants by use of PCR-electrospray ionization mass spectrometry. J Clin Microbiol 51:3492–3498. doi: 10.1128/JCM.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kolb J, Hillemann D, Möbius P, Reetz J, Lahiri A, Lewin A, Rüsch-Gerdes S, Richter E. 2014. Genetic characterization of German Mycobacterium avium strains isolated from different hosts and specimens by multilocus sequence typing. Int J Med Microbiol 304:941–948. doi: 10.1016/j.ijmm.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 145.Jeon S, Lim N, Kwon S, Shim T, Park M, Kim B-J, Kim S. 2014. Molecular typing of Mycobacterium intracellulare using pulsed-field gel electrophoresis, variable-number tandem-repeat analysis, mycobacteria interspersed repetitive-unit-variable-number tandem repeat typing, and multilocus sequence typing: molecular characterization and comparison of each typing methods. Osong Public Health Res Perspect 5:119–130. doi: 10.1016/j.phrp.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Adékambi T, Drancourt M. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int J Syst Evol Microbiol 54:2095–2105. doi: 10.1099/ijs.0.63094-0. [DOI] [PubMed] [Google Scholar]
- 147.Kim SY, Kang YA, Bae IK, Yim J-J, Park MS, Kim YS, Kim SK, Chang J, Jeong SH. 2013. Standardization of multilocus sequence typing scheme for Mycobacterium abscessus and Mycobacterium massiliense. Diagn Microbiol Infect Dis 77:143–149. doi: 10.1016/j.diagmicrobio.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 148.Machado GE, Matsumoto CK, Chimara E, da Silva Duarte R, de Freitas D, Palaci M, Hadad DJ, Lima KVB, Lopes ML, Ramos JP, Campos CE, Caldas PC, Heym B, Leão SC. 2014. Multilocus sequence typing scheme versus pulsed-field gel electrophoresis for typing Mycobacterium abscessus isolates. J Clin Microbiol 52:2881–2891. doi: 10.1128/JCM.00688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Macheras E, Konjek J, Roux A-L, Thiberge J-M, Bastian S, Leão SC, Palaci M, Sivadon-Tardy V, Gutierrez C, Richter E, Rüsch-Gerdes S, Pfyffer GE, Bodmer T, Jarlier V, Cambau E, Brisse S, Caro V, Rastogi N, Gaillard J-L, Heym B. 2014. Multilocus sequence typing scheme for the Mycobacterium abscessus complex. Res Microbiol 165:82–90. doi: 10.1016/j.resmic.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 150.Fangous M-S, Mougari F, Gouriou S, Calvez E, Raskine L, Cambau E, Payan C, Héry-Arnaud G. 2014. Classification algorithm for subspecies identification within the Mycobacterium abscessus species, based on matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 52:3362–3369. doi: 10.1128/JCM.00788-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, Roach JC, Kremer K, Petrov DA, Feldman MW, Gagneux S. 2008. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol 6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mestre O, Luo T, Dos Vultos T, Kremer K, Murray A, Namouchi A, Jackson C, Rauzier J, Bifani P, Warren R, Rasolofo V, Mei J, Gao Q, Gicquel B. 2011. Phylogeny of Mycobacterium tuberculosis Beijing strains constructed from polymorphisms in genes involved in DNA replication, recombination and repair. PLoS One 6:e16020. doi: 10.1371/journal.pone.0016020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Copin R, Coscollá M, Seiffert SN, Bothamley G, Sutherland J, Mbayo G, Gagneux S, Ernst JD. 2014. Sequence diversity in the pe_pgrs genes of Mycobacterium tuberculosis is independent of human T cell recognition. mBio 5:e00960-13. doi: 10.1128/mBio.00960-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Baker L, Brown T, Maiden MC, Drobniewski F. 2004. Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis. Emerg Infect Dis 10:1568–1577. doi: 10.3201/eid1009.040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Stackebrandt E, Van de Peer Y, Vandamme P, Thompson FL, Swings J. 2005. Opinion: re-evaluating prokaryotic species. Nat Rev Microbiol 3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 156.Collins DM, De Lisle GW. 1984. DNA restriction endonuclease analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG. J Gen Microbiol 130:1019–1021. [DOI] [PubMed] [Google Scholar]
- 157.Schwartz DC, Cantor CR. 1984. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37:67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 158.Wang M, Lai E. 1995. Pulsed field separation of large supercoiled and open-circular DNAs and its application to bacterial artificial chromosome cloning. Electrophoresis 16:1–7. doi: 10.1002/elps.1150160102. [DOI] [PubMed] [Google Scholar]
- 159.Feizabadi MM, Robertson ID, Edwards R, Cousins DV, Hampson DJ. 1997. Genetic differentiation of Australian isolates of Mycobacterium tuberculosis by pulsed-field gel electrophoresis. J Med Microbiol 46:501–505. doi: 10.1099/00222615-46-6-501. [DOI] [PubMed] [Google Scholar]
- 160.Feizabadi MM, Robertson ID, Cousins DV, Hampson DJ. 1996. Genomic analysis of Mycobacterium bovis and other members of the Mycobacterium tuberculosis complex by isoenzyme analysis and pulsed-field gel electrophoresis. J Clin Microbiol 34:1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang Y, Wallace RJ, Mazurek GH. 1995. Genetic differences between BCG substrains. Tuber Lung Dis 76:43–50. [DOI] [PubMed] [Google Scholar]
- 162.Zhang Y, Mazurek GH, Cave MD, Eisenach KD, Pang Y, Murphy DT, Wallace RJ. 1992. DNA polymorphisms in strains of Mycobacterium tuberculosis analyzed by pulsed-field gel electrophoresis: a tool for epidemiology. J Clin Microbiol 30:1551–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Singh SP, Salamon H, Lahti CJ, Farid-Moyer M, Small PM. 1999. Use of pulsed-field gel electrophoresis for molecular epidemiologic and population genetic studies of Mycobacterium tuberculosis. J Clin Microbiol 37:1927–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Iinuma Y, Ichiyama S, Hasegawa Y, Shimokata K, Kawahara S, Matsushima T. 1997. Large-restriction-fragment analysis of Mycobacterium kansasii genomic DNA and its application in molecular typing. J Clin Microbiol 35:596–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang Y, Rajagopalan M, Brown BA, Wallace RJ. 1997. Randomly amplified polymorphic DNA PCR for comparison of Mycobacterium abscessus strains from nosocomial outbreaks. J Clin Microbiol 35:3132–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang Y, Mann LB, Wilson RW, Brown-Elliott BA, Vincent V, Iinuma Y, Wallace RJ. 2004. Molecular analysis of Mycobacterium kansasii isolates from the United States. J Clin Microbiol 42:119–125. doi: 10.1128/JCM.42.1.119-125.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mazurek GH, Hartman S, Zhang Y, Brown BA, Hector JS, Murphy D, Wallace RJ. 1993. Large DNA restriction fragment polymorphism in the Mycobacterium avium-M. intracellulare complex: a potential epidemiologic tool. J Clin Microbiol 31:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stevenson K, Alvarez J, Bakker D, Biet F, de Juan L, Denham S, Dimareli Z, Dohmann K, Gerlach GF, Heron I, Kopecna M, May L, Pavlik I, Sharp JM, Thibault VC, Willemsen P, Zadoks RN, Greig A. 2009. Occurrence of Mycobacterium avium subspecies paratuberculosis across host species and European countries with evidence for transmission between wildlife and domestic ruminants. BMC Microbiol 9:212. doi: 10.1186/1471-2180-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lalande V, Barbut F, Varnerot A, Febvre M, Nesa D, Wadel S, Vincent V, Petit JC. 2001. Pseudo-outbreak of Mycobacterium gordonae associated with water from refrigerated fountains. J Hosp Infect 48:76–79. doi: 10.1053/jhin.2000.0929. [DOI] [PubMed] [Google Scholar]
- 170.Yakrus MA, Straus WL. 1994. DNA polymorphisms detected in Mycobacterium haemophilum by pulsed-field gel electrophoresis. J Clin Microbiol 32:1083–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hector JS, Pang Y, Mazurek GH, Zhang Y, Brown BA, Wallace RJ. 1992. Large restriction fragment patterns of genomic Mycobacterium fortuitum DNA as strain-specific markers and their use in epidemiologic investigation of four nosocomial outbreaks. J Clin Microbiol 30:1250–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Legrand E, Radegonde N, Goh KS, Rastogi N. 2002. A pulsed-field gel electrophoresis study of Mycobacterium fortuitum in a Caribbean setting underlines high genetic diversity of the strains and excludes nosocomial outbreaks. Int J Med Microbiol 292:51–57. doi: 10.1078/1438-4221-00187. [DOI] [PubMed] [Google Scholar]
- 173.Sampaio JLM, Chimara E, Ferrazoli L, da Silva Telles MA, Del Guercio VMF, Jericó ZVN, Miyashiro K, Fortaleza CMCB, Padoveze MC, Leão SC. 2006. Application of four molecular typing methods for analysis of Mycobacterium fortuitum group strains causing post-mammaplasty infections. Clin Microbiol Infect 12:142–149. doi: 10.1111/j.1469-0691.2005.01312.x. [DOI] [PubMed] [Google Scholar]
- 174.Wallace RJ, Zhang Y, Brown BA, Fraser V, Mazurek GH, Maloney S. 1993. DNA large restriction fragment patterns of sporadic and epidemic nosocomial strains of Mycobacterium chelonae and Mycobacterium abscessus. J Clin Microbiol 31:2697–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zelazny AM, Root JM, Shea YR, Colombo RE, Shamputa IC, Stock F, Conlan S, McNulty S, Brown-Elliott BA, Wallace RJ, Olivier KN, Holland SM, Sampaio EP. 2009. Cohort study of molecular identification and typing of Mycobacterium abscessus, Mycobacterium massiliense, and Mycobacterium bolletii. J Clin Microbiol 47:1985–1995. doi: 10.1128/JCM.01688-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rodriguez JG, Mejia GA, Del Portillo P, Patarroyo ME, Murillo LA. 1995. Species-specific identification of Mycobacterium bovis by PCR. Microbiology 141(Part 9):2131–2138. [DOI] [PubMed] [Google Scholar]
- 177.Linton CJ, Jalal H, Leeming JP, Millar MR. 1994. Rapid discrimination of Mycobacterium tuberculosis strains by random amplified polymorphic DNA analysis. J Clin Microbiol 32:2169–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Linton CJ, Smart AD, Leeming JP, Jalal H, Telenti A, Bodmer T, Millar MR. 1995. Comparison of random amplified polymorphic DNA with restriction fragment length polymorphism as epidemiological typing methods for Mycobacterium tuberculosis. Clin Mol Pathol 48:M133–M135. doi: 10.1136/mp.48.3.M133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Palittapongarnpim P, Chomyc S, Fanning A, Kunimoto D. 1993. DNA fragment length polymorphism analysis of Mycobacterium tuberculosis isolates by arbitrarily primed polymerase chain reaction. J Infect Dis 167:975–978. doi: 10.1093/infdis/167.4.975. [DOI] [PubMed] [Google Scholar]
- 180.Esteban J, Zamora N, Ortiz A. 2006. Use of molecular techniques for epidemiological typing of rapidly growing mycobacteria. Clin Microbiol Infect 12:943–944. doi: 10.1111/j.1469-0691.2006.1538_1.x (Reply, 12:944.) [DOI] [PubMed] [Google Scholar]
- 181.Cooksey RC, Jhung MA, Yakrus MA, Butler WR, Adékambi T, Morlock GP, Williams M, Shams AM, Jensen BJ, Morey RE, Charles N, Toney SR, Jost KC, Dunbar DF, Bennett V, Kuan M, Srinivasan A. 2008. Multiphasic approach reveals genetic diversity of environmental and patient isolates of Mycobacterium mucogenicum and Mycobacterium phocaicum associated with an outbreak of bacteremias at a Texas hospital. Appl Environ Microbiol 74:2480–2487. doi: 10.1128/AEM.02476-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Vogiatzakis E, Stefanou S, Skroubelou A, Anagnostou S, Marinis E, Matsiota-Bernard P. 1998. Molecular markers for the investigation of Mycobacterium gordonae epidemics. J Hosp Infect 38:217–222. doi: 10.1016/S0195-6701(98)90277-8. [DOI] [PubMed] [Google Scholar]
- 183.van Ingen J, Boeree MJ, de Lange WCM, de Haas PEW, Dekhuijzen PNR, van Soolingen D. 2008. Clinical relevance of Mycobacterium szulgai in The Netherlands. Clin Infect Dis 46:1200–1205. doi: 10.1086/529443. [DOI] [PubMed] [Google Scholar]
- 184.Kauppinen J, Mäntyjärvi R, Katila ML. 1994. Random amplified polymorphic DNA genotyping of Mycobacterium malmoense. J Clin Microbiol 32:1827–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Abed Y, Davin-Regli A, Bollet C, De Micco P. 1995. Efficient discrimination of Mycobacterium tuberculosis strains by 16S-23S spacer region-based random amplified polymorphic DNA analysis. J Clin Microbiol 33:1418–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. 1996. Evaluation of the DNA fingerprinting method AFLP as an new tool in bacterial taxonomy. Microbiology 142(Part 7):1881–1893. [DOI] [PubMed] [Google Scholar]
- 187.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Krishnan MY, Radhakrishnan I, Joseph BV, Madhavi Latha GK, Ajay Kumar R, Mundayoor S. 2007. Combined use of amplified fragment length polymorphism and IS6110-RFLP in fingerprinting clinical isolates of Mycobacterium tuberculosis from Kerala, South India. BMC Infect Dis 7:86. doi: 10.1186/1471-2334-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Huys G, Rigouts L, Chemlal K, Portaels F, Swings J. 2000. Evaluation of amplified fragment length polymorphism analysis for inter- and intraspecific differentiation of Mycobacterium bovis, M. tuberculosis, and M. ulcerans. J Clin Microbiol 38:3675–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Goulding JN, Stanley J, Saunders N, Arnold C. 2000. Genome-sequence-based fluorescent amplified-fragment length polymorphism analysis of Mycobacterium tuberculosis. J Clin Microbiol 38:1121–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sims EJ, Goyal M, Arnold C. 2002. Experimental versus in silico fluorescent amplified fragment length polymorphism analysis of Mycobacterium tuberculosis: improved typing with an extended fragment range. J Clin Microbiol 40:4072–4076. doi: 10.1128/JCM.40.11.4072-4076.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Pfaller SL, Aronson TW, Holtzman AE, Covert TC. 2007. Amplified fragment length polymorphism analysis of Mycobacterium avium complex isolates recovered from southern California. J Med Microbiol 56:1152–1160. doi: 10.1099/jmm.0.47075-0. [DOI] [PubMed] [Google Scholar]
- 193.Bruijnesteijn van Coppenraet LES, Savelkoul PHM, Buffing N, van der Bijl MW, Woudenberg J, Lindeboom JA, Kiehn TE, Haverkort F, Samra Z, Kuijper EJ. 2009. Amplified fragment length polymorphism analysis of human clinical isolates of Mycobacterium haemophilum from different continents. Clin Microbiol Infect 15:924–930. doi: 10.1111/j.1469-0691.2009.02798.x. [DOI] [PubMed] [Google Scholar]
- 194.Chemlal K, Huys G, Fonteyne PA, Vincent V, Lopez AG, Rigouts L, Swings J, Meyers WM, Portaels F. 2001. Evaluation of PCR-restriction profile analysis and IS2404 restriction fragment length polymorphism and amplified fragment length polymorphism fingerprinting for identification and typing of Mycobacterium ulcerans and M. marinum. J Clin Microbiol 39:3272–3278. doi: 10.1128/JCM.39.9.3272-3278.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chemlal K, Huys G, Laval F, Vincent V, Savage C, Gutierrez C, Laneelle M-A, Swings J, Meyers WM, Daffe M, Portaels F. 2002. Characterization of an unusual mycobacterium: a possible missing link between Mycobacterium marinum and Mycobacterium ulcerans. J Clin Microbiol 40:2370–2380. doi: 10.1128/JCM.40.7.2370-2380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mostowy S, Cousins D, Brinkman J, Aranaz A, Behr MA. 2002. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J Infect Dis 186:74–80. doi: 10.1086/341068. [DOI] [PubMed] [Google Scholar]
- 197.Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, Stover CK. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol 178:1274–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Warren RM, Gey van Pittius NC, Barnard M, Hesseling A, Engelke E, de Kock M, Gutierrez MC, Chege GK, Victor TC, Hoal EG, van Helden PD. 2006. Differentiation of Mycobacterium tuberculosis complex by PCR amplification of genomic regions of difference. Int J Tuberc Lung Dis 10:818–822. [PubMed] [Google Scholar]
- 199.Halse TA, Escuyer VE, Musser KA. 2011. Evaluation of a single-tube multiplex real-time PCR for differentiation of members of the Mycobacterium tuberculosis complex in clinical specimens. J Clin Microbiol 49:2562–2567. doi: 10.1128/JCM.00467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hirsh AE, Tsolaki AG, DeRiemer K, Feldman MW, Small PM. 2004. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc Natl Acad Sci U S A 101:4871–4876. doi: 10.1073/pnas.0305627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, Nicol M, Niemann S, Kremer K, Gutierrez MC, Hilty M, Hopewell PC, Small PM. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Alland D, Lacher DW, Hazbón MH, Motiwala AS, Qi W, Fleischmann RD, Whittam TS. 2007. Role of large sequence polymorphisms (LSPs) in generating genomic diversity among clinical isolates of Mycobacterium tuberculosis and the utility of LSPs in phylogenetic analysis. J Clin Microbiol 45:39–46. doi: 10.1128/JCM.02483-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, Small PM. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 204.Paustian ML, Kapur V, Bannantine JP. 2005. Comparative genomic hybridizations reveal genetic regions within the Mycobacterium avium complex that are divergent from Mycobacterium avium subsp. paratuberculosis isolates. J Bacteriol 187:2406–2415. doi: 10.1128/JB.187.7.2406-2415.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Marsh IB, Bannantine JP, Paustian ML, Tizard ML, Kapur V, Whittington RJ. 2006. Genomic comparison of Mycobacterium avium subsp. paratuberculosis sheep and cattle strains by microarray hybridization. J Bacteriol 188:2290–2293. doi: 10.1128/JB.188.6.2290-2293.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Paustian ML, Zhu X, Sreevatsan S, Robbe-Austerman S, Kapur V, Bannantine JP. 2008. Comparative genomic analysis of Mycobacterium avium subspecies obtained from multiple host species. BMC Genomics 9:135. doi: 10.1186/1471-2164-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Rondini S, Käser M, Stinear T, Tessier M, Mangold C, Dernick G, Naegeli M, Portaels F, Certa U, Pluschke G. 2007. Ongoing genome reduction in Mycobacterium ulcerans. Emerg Infect Dis 13:1008–1015. doi: 10.3201/eid1307.060205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Käser M, Gutmann O, Hauser J, Stinear T, Cole S, Yeboah-Manu D, Dernick G, Certa U, Pluschke G. 2009. Lack of insertional-deletional polymorphism in a collection of Mycobacterium ulcerans isolates from Ghanaian Buruli ulcer patients. J Clin Microbiol 47:3640–3646. doi: 10.1128/JCM.00760-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Goguet de la Salmonière Y-OL, Kim CC, Tsolaki AG, Pym AS, Siegrist MS, Small PM. 2004. High-throughput method for detecting genomic-deletion polymorphisms. J Clin Microbiol 42:2913–2918. doi: 10.1128/JCM.42.7.2913-2918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Cardoso Oelemann M, Gomes HM, Willery E, Possuelo L, Batista Lima KV, Allix-Béguec C, Locht C, Goguet de la Salmonière Y-OL, Gutierrez MC, Suffys P, Supply P. 2011. The forest behind the tree: phylogenetic exploration of a dominant Mycobacterium tuberculosis strain lineage from a high tuberculosis burden country. PLoS One 6:e18256. doi: 10.1371/journal.pone.0018256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M. 2012. Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Loman NJ, Constantinidou C, Chan JZM, Halachev M, Sergeant M, Penn CW, Robinson ER, Pallen MJ. 2012. High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat Rev Microbiol 10:599–606. doi: 10.1038/nrmicro2850. [DOI] [PubMed] [Google Scholar]
- 213.Lee JS, Krause R, Schreiber J, Mollenkopf H-J, Kowall J, Stein R, Jeon B-Y, Kwak J-Y, Song M-K, Patron JP, Jorg S, Roh K, Cho S-N, Kaufmann SHE. 2008. Mutation in the transcriptional regulator PhoP contributes to avirulence of Mycobacterium tuberculosis H37Ra strain. Cell Host Microbe 3:97–103. doi: 10.1016/j.chom.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 214.Zheng H, Lu L, Wang B, Pu S, Zhang X, Zhu G, Shi W, Zhang L, Wang H, Wang S, Zhao G, Zhang Y. 2008. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS One 3:e2375. doi: 10.1371/journal.pone.0002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Niemann S, Köser CU, Gagneux S, Plinke C, Homolka S, Bignell H, Carter RJ, Cheetham RK, Cox A, Gormley NA, Kokko-Gonzales P, Murray LJ, Rigatti R, Smith VP, Arends FPM, Cox HS, Smith G, Archer JAC. 2009. Genomic diversity among drug sensitive and multidrug resistant isolates of Mycobacterium tuberculosis with identical DNA fingerprints. PLoS One 4:e7407. doi: 10.1371/journal.pone.0007407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Feng Y, Zhang Y, Ying C, Wang D, Du C. 2015. Nanopore-based fourth-generation DNA sequencing technology. Genomics Proteomics Bioinformatics 13:4–16. doi: 10.1016/j.gpb.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Stinear TP, Seemann T, Pidot S, Frigui W, Reysset G, Garnier T, Meurice G, Simon D, Bouchier C, Ma L, Tichit M, Porter JL, Ryan J, Johnson PDR, Davies JK, Jenkin GA, Small PLC, Jones LM, Tekaia F, Laval F, Daffé M, Parkhill J, Cole ST. 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res 17:192–200. doi: 10.1101/gr.5942807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Cho Y-J, Yi H, Chun J, Cho S-N, Daley CL, Koh W-J, Shin SJ. 2013. The genome sequence of “Mycobacterium massiliense” strain CIP 108297 suggests the independent taxonomic status of the Mycobacterium abscessus complex at the subspecies level. PLoS One 8:e81560. doi: 10.1371/journal.pone.0081560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Pepperell CS, Casto AM, Kitchen A, Granka JM, Cornejo OE, Holmes EC, Holmes EC, Birren B, Galagan J, Feldman MW. 2013. The role of selection in shaping diversity of natural M. tuberculosis populations. PLoS Pathog 9:e1003543. doi: 10.1371/journal.ppat.1003543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Choo SW, Wee WY, Ngeow YF, Mitchell W, Tan JL, Wong GJ, Zhao Y, Xiao J. 2014. Genomic reconnaissance of clinical isolates of emerging human pathogen Mycobacterium abscessus reveals high evolutionary potential. Sci Rep 4:4061. doi: 10.1038/srep04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Perdigão J, Silva H, Machado D, Macedo R, Maltez F, Silva C, Jordao L, Couto I, Mallard K, Coll F, Hill-Cawthorne GA, McNerney R, Pain A, Clark TG, Viveiros M, Portugal I. 2014. Unraveling Mycobacterium tuberculosis genomic diversity and evolution in Lisbon, Portugal, a highly drug resistant setting. BMC Genomics 15:991. doi: 10.1186/1471-2164-15-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Andries K, Verhasselt P, Guillemont J, Göhlmann HWH, Neefs J-M, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 223.Motiwala AS, Dai Y, Jones-López EC, Hwang S-H, Lee JS, Cho SN, Via LE, Barry CE, Alland D. 2010. Mutations in extensively drug-resistant Mycobacterium tuberculosis that do not code for known drug-resistance mechanisms. J Infect Dis 201:881–888. doi: 10.1086/650999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, Galagan J, Niemann S, Gagneux S. 2012. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet 44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Coll F, McNerney R, Preston MD, Guerra-Assunção JA, Warry A, Hill-Cawthorne G, Mallard K, Nair M, Miranda A, Alves A, Perdigão J, Viveiros M, Portugal I, Hasan Z, Hasan R, Glynn JR, Martin N, Pain A, Clark TG. 2015. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med 7:51. doi: 10.1186/s13073-015-0164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Coscolla M, Gagneux S. 2010. Does M. tuberculosis genomic diversity explain disease diversity? Drug Discov Today Dis Mech 7:e43–e59. doi: 10.1016/j.ddmec.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, Ernst JD, Gagneux S. 2010. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet 42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gardy JL, Johnston JC, Sui SJH, Cook VJ, Shah L, Brodkin E, Rempel S, Moore R, Zhao Y, Holt R, Varhol R, Birol I, Lem M, Sharma MK, Elwood K, Jones SJM, Brinkman FSL, Brunham RC, Tang P. 2011. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med 364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 229.Wlodarska M, Johnston JC, Gardy JL, Tang P. 2015. A microbiological revolution meets an ancient disease: improving the management of tuberculosis with genomics. Clin Microbiol Rev 28:523–539. doi: 10.1128/CMR.00124-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Walker TM, Lalor MK, Broda A, Saldana Ortega L, Morgan M, Parker L, Churchill S, Bennett K, Golubchik T, Giess AP, Del Ojo Elias C, Jeffery KJ, Bowler IC, Laurenson IF, Barrett A, Drobniewski F, McCarthy ND, Anderson LF, Abubakar I, Thomas HL, Monk P, Smith EG, Walker AS, Crook DW, Peto TE, Conlon CP. 2014. Assessment of Mycobacterium tuberculosis transmission in Oxfordshire, UK, 2007-12, with whole pathogen genome sequences: an observational study. Lancet Respir Med 2:285–292. doi: 10.1016/S2213-2600(14)70027-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Merker M, Kohl TA, Roetzer A, Truebe L, Richter E, Rüsch-Gerdes S, Fattorini L, Oggioni MR, Cox H, Varaine F, Niemann S. 2013. Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients. PLoS One 8:e82551. doi: 10.1371/journal.pone.0082551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Shamputa IC, Rigouts L, Eyongeta LA, El Aila NA, van Deun A, Salim AH, Willery E, Locht C, Supply P, Portaels F. 2004. Genotypic and phenotypic heterogeneity among Mycobacterium tuberculosis isolates from pulmonary tuberculosis patients. J Clin Microbiol 42:5528–5536. doi: 10.1128/JCM.42.12.5528-5536.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Warren RM, Victor TC, Streicher EM, Richardson M, Beyers N, Gey van Pittius NC, van Helden PD. 2004. Patients with active tuberculosis often have different strains in the same sputum specimen. Am J Respir Crit Care Med 169:610–614. doi: 10.1164/rccm.200305-714OC. [DOI] [PubMed] [Google Scholar]
- 234.Wang J-Y, Hsu H-L, Yu M-C, Chiang C-Y, Yu F-L, Yu C-J, Lee L-N, Yang P-C, TAMI Group. 2011. Mixed infection with Beijing and non-Beijing strains in pulmonary tuberculosis in Taiwan: prevalence, risk factors, and dominant strain. Clin Microbiol Infect 17:1239–1245. doi: 10.1111/j.1469-0691.2010.03401.x. [DOI] [PubMed] [Google Scholar]
- 235.Köser CU, Bryant JM, Becq J, Török ME, Ellington MJ, Marti-Renom MA, Carmichael AJ, Parkhill J, Smith GP, Peacock SJ. 2013. Whole-genome sequencing for rapid susceptibility testing of M. tuberculosis. N Engl J Med 369:290–292. doi: 10.1056/NEJMc1215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Chan JZ-M, Sergeant MJ, Lee OY-C, Minnikin DE, Besra GS, Pap I, Spigelman M, Donoghue HD, Pallen MJ. 2013. Metagenomic analysis of tuberculosis in a mummy. N Engl J Med 369:289–290. doi: 10.1056/NEJMc1302295. [DOI] [PubMed] [Google Scholar]
- 237.Doughty EL, Sergeant MJ, Adetifa I, Antonio M, Pallen MJ. 2014. Culture-independent detection and characterisation of Mycobacterium tuberculosis and M. africanum in sputum samples using shotgun metagenomics on a benchtop sequencer. PeerJ 2:e585. doi: 10.7717/peerj.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Outhred AC, Jelfs P, Suliman B, Hill-Cawthorne GA, Crawford ABH, Marais BJ, Sintchenko V. 2015. Added value of whole-genome sequencing for management of highly drug-resistant TB. J Antimicrob Chemother 70:1198–1202. doi: 10.1093/jac/dku508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Schuenemann VJ, Singh P, Mendum TA, Krause-Kyora B, Jäger G, Bos KI, Herbig A, Economou C, Benjak A, Busso P, Nebel A, Boldsen JL, Kjellström A, Wu H, Stewart GR, Taylor GM, Bauer P, Lee OY-C, Wu HHT, Minnikin DE, Besra GS, Tucker K, Roffey S, Sow SO, Cole ST, Nieselt K, Krause J. 2013. Genome-wide comparison of medieval and modern Mycobacterium leprae. Science 341:179–183. doi: 10.1126/science.1238286. [DOI] [PubMed] [Google Scholar]
- 240.Chandler M, Mahillon J. 2002. Insertion sequences revised, p 305–366. In Craig NL, Craigie R, Gellert M, Lambowitz AM (ed), Mobile DNA II. ASM Press, Washington, DC. [Google Scholar]
- 241.Safi H, Barnes PF, Lakey DL, Shams H, Samten B, Vankayalapati R, Howard ST. 2004. IS6110 functions as a mobile, monocyte-activated promoter in Mycobacterium tuberculosis. Mol Microbiol 52:999–1012. doi: 10.1111/j.1365-2958.2004.04037.x. [DOI] [PubMed] [Google Scholar]
- 242.Gordon S, Supply P. 2005. Repetitive DNA in the Mycobacterium tuberculosis complex, p 191–202. In Cole ST, Eisenach KD, McMurray DN, Jacobs WR Jr (ed), Tuberculosis and the tubercle bacillus. ASM Press, Washington, DC. [Google Scholar]
- 243.McAdam RA, Hermans PW, van Soolingen D, Zainuddin ZF, Catty D, van Embden JD, Dale JW. 1990. Characterization of a Mycobacterium tuberculosis insertion sequence belonging to the IS3 family. Mol Microbiol 4:1607–1613. doi: 10.1111/j.1365-2958.1990.tb02073.x. [DOI] [PubMed] [Google Scholar]
- 244.Thierry D, Brisson-Noël A, Vincent-Lévy-Frébault V, Nguyen S, Guesdon JL, Gicquel B. 1990. Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J Clin Microbiol 28:2668–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Thierry D, Cave MD, Eisenach KD, Crawford JT, Bates JH, Gicquel B, Guesdon JL. 1990. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res 18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Behr MA, Small PM. 1999. A historical and molecular phylogeny of BCG strains. Vaccine 17:915–922. doi: 10.1016/S0264-410X(98)00277-1. [DOI] [PubMed] [Google Scholar]
- 247.McEvoy CRE, Falmer AA, Gey van Pittius NC, Victor TC, van Helden PD, Warren RM. 2007. The role of IS6110 in the evolution of Mycobacterium tuberculosis. Tuberculosis (Edinb) 87:393–404. doi: 10.1016/j.tube.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 248.Van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol 29:2578–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Cave MD, Eisenach KD, McDermott PF, Bates JH, Crawford JT. 1991. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol Cell Probes 5:73–80. doi: 10.1016/0890-8508(91)90040-Q. [DOI] [PubMed] [Google Scholar]
- 250.Alland D, Kalkut GE, Moss AR, McAdam RA, Hahn JA, Bosworth W, Drucker E, Bloom BR. 1994. Transmission of tuberculosis in New York City—an analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med 330:1710–1716. doi: 10.1056/NEJM199406163302403. [DOI] [PubMed] [Google Scholar]
- 251.Yang ZH, de Haas PE, Wachmann CH, van Soolingen D, van Embden JD, Andersen AB. 1995. Molecular epidemiology of tuberculosis in Denmark in 1992. J Clin Microbiol 33:2077–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Jonsson J, Hoffner S, Berggren I, Bruchfeld J, Ghebremichael S, Pennhag A, Groenheit R. 2014. Comparison between RFLP and MIRU-VNTR genotyping of Mycobacterium tuberculosis strains isolated in Stockholm 2009 to 2011. PLoS One 9:e95159. doi: 10.1371/journal.pone.0095159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Joseph BV, Soman S, Radhakrishnan I, Hill V, Dhanasooraj D, Kumar RA, Rastogi N, Mundayoor S. 2013. Molecular epidemiology of Mycobacterium tuberculosis isolates from Kerala, India using IS6110-RFLP, spoligotyping and MIRU-VNTRs. Infect Genet Evol 16:157–164. doi: 10.1016/j.meegid.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 254.Zaczek A, Ziółkiewicz M, Wojtasik A, Dziadek J, Sajduda A. 2013. IS6110-based differentiation of Mycobacterium tuberculosis strains. Pol J Microbiol 62:201–204. [PubMed] [Google Scholar]
- 255.Salo WL, Aufderheide AC, Buikstra J, Holcomb TA. 1994. Identification of Mycobacterium tuberculosis DNA in a pre-Columbian Peruvian mummy. Proc Natl Acad Sci U S A 91:2091–2094. doi: 10.1073/pnas.91.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Konomi N, Lebwohl E, Mowbray K, Tattersall I, Zhang D. 2002. Detection of mycobacterial DNA in Andean mummies. J Clin Microbiol 40:4738–4740. doi: 10.1128/JCM.40.12.4738-4740.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Lalremruata A, Ball M, Bianucci R, Welte B, Nerlich AG, Kun JFJ, Pusch CM. 2013. Molecular identification of falciparum malaria and human tuberculosis co-infections in mummies from the Fayum depression (Lower Egypt). PLoS One 8:e60307. doi: 10.1371/journal.pone.0060307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Collins DM, Stephens DM. 1991. Identification of an insertion sequence, IS1081, in Mycobacterium bovis. FEMS Microbiol Lett 83:11–16. doi: 10.1111/j.1574-6968.1991.tb04380.x. [DOI] [PubMed] [Google Scholar]
- 259.Van Soolingen D, Hermans PW, de Haas PE, van Embden JD. 1992. Insertion element IS1081-associated restriction fragment length polymorphisms in Mycobacterium tuberculosis complex species: a reliable tool for recognizing Mycobacterium bovis BCG. J Clin Microbiol 30:1772–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Fang Z, Morrison N, Watt B, Doig C, Forbes KJ. 1998. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J Bacteriol 180:2102–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Van Soolingen D, Bauer J, Ritacco V, Leão SC, Pavlik I, Vincent V, Rastogi N, Gori A, Bodmer T, Garzelli C, Garcia MJ. 1998. IS1245 restriction fragment length polymorphism typing of Mycobacterium avium isolates: proposal for standardization. J Clin Microbiol 36:3051–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Johansen TB, Djønne B, Jensen MR, Olsen I. 2005. Distribution of IS1311 and IS1245 in Mycobacterium avium subspecies revisited. J Clin Microbiol 43:2500–2502. doi: 10.1128/JCM.43.5.2500-2502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Radomski N, Thibault VC, Karoui C, de Cruz K, Cochard T, Gutiérrez C, Supply P, Biet F, Boschiroli ML. 2010. Determination of genotypic diversity of Mycobacterium avium subspecies from human and animal origins by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat and IS1311 restriction fragment length polymorphism typing methods. J Clin Microbiol 48:1026–1034. doi: 10.1128/JCM.01869-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Whipple D, Kapke P, Vary C. 1990. Identification of restriction fragment length polymorphisms in DNA from Mycobacterium paratuberculosis. J Clin Microbiol 28:2561–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kunze ZM, Portaels F, McFadden JJ. 1992. Biologically distinct subtypes of Mycobacterium avium differ in possession of insertion sequence IS901. J Clin Microbiol 30:2366–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Moss MT, Malik ZP, Tizard ML, Green EP, Sanderson JD, Hermon-Taylor J. 1992. IS902, an insertion element of the chronic-enteritis-causing Mycobacterium avium subsp. silvaticum. J Gen Microbiol 138:139–145. doi: 10.1099/00221287-138-1-139. [DOI] [PubMed] [Google Scholar]
- 267.Hernandez Perez M, Fomukong NG, Hellyer T, Brown IN, Dale JW. 1994. Characterization of IS1110, a highly mobile genetic element from Mycobacterium avium. Mol Microbiol 12:717–724. doi: 10.1111/j.1365-2958.1994.tb01059.x. [DOI] [PubMed] [Google Scholar]
- 268.Puyang X, Lee K, Pawlichuk C, Kunimoto DY. 1999. IS1626, a new IS900-related Mycobacterium avium insertion sequence. Microbiology 145(Part 11):3163–3168. [DOI] [PubMed] [Google Scholar]
- 269.Sangari FJ, Bächli M, Bermudez LE, Bodmer T. 2000. Characterization of IS666, a newly described insertion element of Mycobacterium avium. Microb Comp Genomics 5:181–188. doi: 10.1089/omi.1.2000.5.181. [DOI] [PubMed] [Google Scholar]
- 270.Amonsin A, Li LL, Zhang Q, Bannantine JP, Motiwala AS, Sreevatsan S, Kapur V. 2004. Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J Clin Microbiol 42:1694–1702. doi: 10.1128/JCM.42.4.1694-1702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Thibault VC, Grayon M, Boschiroli ML, Hubbans C, Overduin P, Stevenson K, Gutierrez MC, Supply P, Biet F. 2007. New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 45:2404–2410. doi: 10.1128/JCM.00476-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Nishimori K, Eguchi M, Nakaoka Y, Onodera Y, Ito T, Tanaka K. 1995. Distribution of IS901 in strains of Mycobacterium avium complex from swine by using IS901-detecting primers that discriminate between M. avium and Mycobacterium intracellulare. J Clin Microbiol 33:2102–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Ahrens P, Giese SB, Klausen J, Inglis NF. 1995. Two markers, IS901-IS902 and p40, identified by PCR and by using monoclonal antibodies in Mycobacterium avium strains. J Clin Microbiol 33:1049–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Picardeau M, Varnerot A, Rauzier J, Gicquel B, Vincent V. 1996. Mycobacterium xenopi IS1395, a novel insertion sequence expanding the IS256 family. Microbiology 142(Part 9):2453–2461. [DOI] [PubMed] [Google Scholar]
- 275.Picardeau M, Bull TJ, Vincent V. 1997. Identification and characterization of IS-like elements in Mycobacterium gordonae. FEMS Microbiol Lett 154:95–102. doi: 10.1111/j.1574-6968.1997.tb12629.x. [DOI] [PubMed] [Google Scholar]
- 276.Picardeau M, Bull TJ, Prod'hom G, Pozniak AL, Shanson DC, Vincent V. 1997. Comparison of a new insertion element, IS1407, with established molecular markers for the characterization of Mycobacterium celatum. Int J Syst Bacteriol 47:640–644. doi: 10.1099/00207713-47-3-640. [DOI] [PubMed] [Google Scholar]
- 277.Guilhot C, Gicquel B, Davies J, Martín C. 1992. Isolation and analysis of IS6120, a new insertion sequence from Mycobacterium smegmatis. Mol Microbiol 6:107–113. doi: 10.1111/j.1365-2958.1992.tb00842.x. [DOI] [PubMed] [Google Scholar]
- 278.Stinear T, Ross BC, Davies JK, Marino L, Robins-Browne RM, Oppedisano F, Sievers A, Johnson PD. 1999. Identification and characterization of IS2404 and IS2606: two distinct repeated sequences for detection of Mycobacterium ulcerans by PCR. J Clin Microbiol 37:1018–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Palittapongarnpim P, Rienthong S, Panbangred W. 1993. Comparison of restriction fragment length polymorphism of Mycobacterium tuberculosis isolated from cerebrospinal fluid and sputum: a preliminary report. Tuber Lung Dis 74:204–207. doi: 10.1016/0962-8479(93)90013-N. [DOI] [PubMed] [Google Scholar]
- 280.Ross BC, Raios K, Jackson K, Sievers A, Dwyer B. 1991. Differentiation of Mycobacterium tuberculosis strains by use of a nonradioactive Southern blot hybridization method. J Infect Dis 163:904–907. doi: 10.1093/infdis/163.4.904. [DOI] [PubMed] [Google Scholar]
- 281.Shoemaker SA, Fisher JH, Jones WD, Scoggin CH. 1986. Restriction fragment analysis of chromosomal DNA defines different strains of Mycobacterium tuberculosis. Am Rev Respir Dis 134:210–213. [DOI] [PubMed] [Google Scholar]
- 282.Eisenach KD, Crawford JT, Bates JH. 1986. Genetic relatedness among strains of the Mycobacterium tuberculosis complex. Analysis of restriction fragment heterogeneity using cloned DNA probes. Am Rev Respir Dis 133:1065–1068. [DOI] [PubMed] [Google Scholar]
- 283.Eisenach KD, Crawford JT, Bates JH. 1988. Repetitive DNA sequences as probes for Mycobacterium tuberculosis. J Clin Microbiol 26:2240–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 31:406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Small PM, Hopewell PC, Singh SP, Paz A, Parsonnet J, Ruston DC, Schecter GF, Daley CL, Schoolnik GK. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med 330:1703–1709. [DOI] [PubMed] [Google Scholar]
- 286.Van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, Portaels F, Qing HZ, Enkhsaikan D, Nymadawa P, van Embden JD. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J Clin Microbiol 33:3234–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Van Embden JD, Van Soolingen D, Heersma HF, De Neeling AJ, Jones ME, Steiert M, Grek V, Mooi FR, Verhoef J. 1996. Establishment of a European network for the surveillance of Mycobacterium tuberculosis, MRSA and penicillin-resistant pneumococci. J Antimicrob Chemother 38:905–907. doi: 10.1093/jac/38.5.905. [DOI] [PubMed] [Google Scholar]
- 288.Van Soolingen D, Borgdorff MW, de Haas PE, Sebek MM, Veen J, Dessens M, Kremer K, van Embden JD. 1999. Molecular epidemiology of tuberculosis in the Netherlands: a nationwide study from 1993 through 1997. J Infect Dis 180:726–736. doi: 10.1086/314930. [DOI] [PubMed] [Google Scholar]
- 289.Yaganehdoost A, Graviss EA, Ross MW, Adams GJ, Ramaswamy S, Wanger A, Frothingham R, Soini H, Musser JM. 1999. Complex transmission dynamics of clonally related virulent Mycobacterium tuberculosis associated with barhopping by predominantly human immunodeficiency virus-positive gay men. J Infect Dis 180:1245–1251. doi: 10.1086/314991. [DOI] [PubMed] [Google Scholar]
- 290.Kremer K, van Soolingen D, Frothingham R, Haas WH, Hermans PW, Martín C, Palittapongarnpim P, Plikaytis BB, Riley LW, Yakrus MA, Musser JM, van Embden JD. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol 37:2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Cowan LS, Mosher L, Diem L, Massey JP, Crawford JT. 2002. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J Clin Microbiol 40:1592–1602. doi: 10.1128/JCM.40.5.1592-1602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Thong-On A, Smittipat N, Juthayothin T, Yanai H, Yamada N, Yorsangsukkamol J, Chaiprasert A, Rienthong D, Billamas P, Palittapongarnpim P. 2010. Variable-number tandem repeats typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 in Thailand. Tuberculosis (Edinb) 90:9–15. doi: 10.1016/j.tube.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 293.Varma-Basil M, Kumar S, Arora J, Angrup A, Zozio T, Banavaliker JN, Singh UB, Rastogi N, Bose M. 2011. Comparison of spoligotyping, mycobacterial interspersed repetitive units typing and IS6110-RFLP in a study of genotypic diversity of Mycobacterium tuberculosis in Delhi, North India. Mem Inst Oswaldo Cruz 106:524–535. doi: 10.1590/S0074-02762011000500002. [DOI] [PubMed] [Google Scholar]
- 294.Bauer J, Thomsen VO, Poulsen S, Andersen AB. 1997. False-positive results from cultures of Mycobacterium tuberculosis due to laboratory cross-contamination confirmed by restriction fragment length polymorphism. J Clin Microbiol 35:988–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Dale JW, Nor RM, Ramayah S, Tang TH, Zainuddin ZF. 1999. Molecular epidemiology of tuberculosis in Malaysia. J Clin Microbiol 37:1265–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Rhee JT, Tanaka MM, Behr MA, Agasino CB, Paz EA, Hopewell PC, Small PM. 2000. Use of multiple markers in population-based molecular epidemiologic studies of tuberculosis. Int J Tuberc Lung Dis 4:1111–1119. [PubMed] [Google Scholar]
- 297.Barlow RE, Gascoyne-Binzi DM, Gillespie SH, Dickens A, Qamer S, Hawkey PM. 2001. Comparison of variable number tandem repeat and IS6110-restriction fragment length polymorphism analyses for discrimination of high- and low-copy-number IS6110 Mycobacterium tuberculosis isolates. J Clin Microbiol 39:2453–2457. doi: 10.1128/JCM.39.7.2453-2457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Otal I, Isabel O, Gomez AB, Kremer K, de Haas P, García MJ, Martín C, van Soolingen D. 2008. Mapping of IS6110 insertion sites in Mycobacterium bovis isolates in relation to adaptation from the animal to human host. Vet Microbiol 129:333–341. doi: 10.1016/j.vetmic.2007.11.038. [DOI] [PubMed] [Google Scholar]
- 299.Viana-Niero C, Rodriguez CAR, Bigi F, Zanini MS, Ferreira-Neto JS, Cataldi A, Leão SC. 2006. Identification of an IS6110 insertion site in plcD, the unique phospholipase C gene of Mycobacterium bovis. J Med Microbiol 55:451–457. doi: 10.1099/jmm.0.46364-0. [DOI] [PubMed] [Google Scholar]
- 300.Mazars E, Lesjean S, Banuls AL, Gilbert M, Vincent V, Gicquel B, Tibayrenc M, Locht C, Supply P. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc Natl Acad Sci U S A 98:1901–1906. doi: 10.1073/pnas.98.4.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.McHugh TD, Newport LE, Gillespie SH. 1997. IS6110 homologs are present in multiple copies in mycobacteria other than tuberculosis-causing mycobacteria. J Clin Microbiol 35:1769–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Johansen TB, Olsen I, Jensen MR, Dahle UR, Holstad G, Djønne B. 2007. New probes used for IS1245 and IS1311 restriction fragment length polymorphism of Mycobacterium avium subsp. avium and Mycobacterium avium subsp. hominissuis isolates of human and animal origin in Norway. BMC Microbiol 7:14. doi: 10.1186/1471-2180-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Tirkkonen T, Pakarinen J, Moisander A-M, Mäkinen J, Soini H, Ali-Vehmas T. 2007. High genetic relatedness among Mycobacterium avium strains isolated from pigs and humans revealed by comparative IS1245 RFLP analysis. Vet Microbiol 125:175–181. doi: 10.1016/j.vetmic.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 304.Palittapongarnpim P, Chomyc S, Fanning A, Kunimoto D. 1993. DNA fingerprinting of Mycobacterium tuberculosis isolates by ligation-mediated polymerase chain reaction. Nucleic Acids Res 21:761–762. doi: 10.1093/nar/21.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Prod'hom G, Guilhot C, Gutierrez MC, Varnerot A, Gicquel B, Vincent V. 1997. Rapid discrimination of Mycobacterium tuberculosis complex strains by ligation-mediated PCR fingerprint analysis. J Clin Microbiol 35:3331–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Haas WH, Butler WR, Woodley CL, Crawford JT. 1993. Mixed-linker polymerase chain reaction: a new method for rapid fingerprinting of isolates of the Mycobacterium tuberculosis complex. J Clin Microbiol 31:1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Reisig F, Kremer K, Amthor B, van Soolingen D, Haas WH. 2005. Fast ligation-mediated PCR, a fast and reliable method for IS6110-based typing of Mycobacterium tuberculosis complex. J Clin Microbiol 43:5622–5627. doi: 10.1128/JCM.43.11.5622-5627.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Zaczek A, Brzostek A, Kuroń A, Wojtasik A, Sajduda A, Dziadek J. 2014. Development of a new ligation-mediated PCR method for the differentiation of Mycobacterium tuberculosis strains. Int J Tuberc Lung Dis 18:302–309. doi: 10.5588/ijtld.13.0538. [DOI] [PubMed] [Google Scholar]
- 309.Zaczek A, Brzostek A, Górna A, Sajduda A, Wojtasik A, Dziadek J. 2013. Application of FLiP method for differentiation of Mycobacterium tuberculosis strains in comparison to commonly used methods, spoligotyping and MIRU-VNTR typing. Pol J Microbiol 62:73–76. [PubMed] [Google Scholar]
- 310.Burger M, Raskin S, Brockelt SR, Amthor B, Geiss HK, Haas WH. 1998. DNA fingerprinting of Mycobacterium tuberculosis complex culture isolates collected in Brazil and spotted onto filter paper. J Clin Microbiol 36:573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Haas WH, Bretzel G, Amthor B, Schilke K, Krommes G, Rüsch-Gerdes S, Sticht-Groh V, Bremer HJ. 1997. Comparison of DNA fingerprint patterns of isolates of Mycobacterium africanum from East and West Africa. J Clin Microbiol 35:663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Butler WR, Haas WH, Crawford JT. 1996. Automated DNA fingerprinting analysis of Mycobacterium tuberculosis using fluorescent detection of PCR products. J Clin Microbiol 34:1801–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Zaczek A. 2012. Molecular methods used in discrimination of M. tuberculosis complex bacteria, p 179–189. In Kostecka J, Kaniuczak J (ed), Practical applications of environmental research. University of Rzeszów, Rzeszów, Poland. [Google Scholar]
- 314.Kremer K, Arnold C, Cataldi A, Gutiérrez MC, Haas WH, Panaiotov S, Skuce RA, Supply P, van der Zanden AGM, van Soolingen D. 2005. Discriminatory power and reproducibility of novel DNA typing methods for Mycobacterium tuberculosis complex strains. J Clin Microbiol 43:5628–5638. doi: 10.1128/JCM.43.11.5628-5638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Zaczek A, Brzostek A, Wojtasik A, Dziadek J, Sajduda A. 2013. Genotyping of clinical Mycobacterium tuberculosis isolates based on IS6110 and MIRU-VNTR polymorphisms. Biomed Res Int 2013:865197. doi: 10.1155/2013/865197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Krawczyk B, Leibner-Ciszak J, Stojowska K, Kur J. 2011. The new LM-PCR/shifter method for the genotyping of microorganisms based on the use of a class IIS restriction enzyme and ligation mediated PCR. J Microbiol Biotechnol 21:1336–1344. doi: 10.4014/jmb.1104.04019. [DOI] [PubMed] [Google Scholar]
- 317.Bonora S, Gutierrez MC, Di Perri G, Brunello F, Allegranzi B, Ligozzi M, Fontana R, Concia E, Vincent V. 1999. Comparative evaluation of ligation-mediated PCR and spoligotyping as screening methods for genotyping of Mycobacterium tuberculosis strains. J Clin Microbiol 37:3118–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Mardassi H, Namouchi A, Karbouli A, Khabouchi N, Haltiti R, Zarrouk M, Mhennii B, Kaabi S. 2005. Usefulness of ligation-mediated PCR genotyping in tracking outbreak-associated Mycobacterium tuberculosis strains. Arch Inst Pasteur Tunis 82:23–29. [PubMed] [Google Scholar]
- 319.Maes M, Kremer K, van Soolingen D, Takiff H, de Waard JH. 2008. 24-locus MIRU-VNTR genotyping is a useful tool to study the molecular epidemiology of tuberculosis among Warao Amerindians in Venezuela. Tuberculosis (Edinb) 88:490–494. doi: 10.1016/j.tube.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 320.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. 1987. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169:5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Sorek R, Kunin V, Hugenholtz P. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol 6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 322.Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 324.He L, Fan X, Xie J. 2012. Comparative genomic structures of Mycobacterium CRISPR-Cas. J Cell Biochem 113:2464–2473. doi: 10.1002/jcb.24121. [DOI] [PubMed] [Google Scholar]
- 325.Supply P, Marceau M, Mangenot S, Roche D, Rouanet C, Khanna V, Majlessi L, Criscuolo A, Tap J, Pawlik A, Fiette L, Orgeur M, Fabre M, Parmentier C, Frigui W, Simeone R, Boritsch EC, Debrie A-S, Willery E, Walker D, Quail MA, Ma L, Bouchier C, Salvignol G, Sayes F, Cascioferro A, Seemann T, Barbe V, Locht C, Gutierrez M-C, Leclerc C, Bentley SD, Stinear TP, Brisse S, Médigue C, Parkhill J, Cruveiller S, Brosch R. 2013. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet 45:172–179. doi: 10.1038/ng.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35:907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Dale JW, Brittain D, Cataldi AA, Cousins D, Crawford JT, Driscoll J, Heersma H, Lillebaek T, Quitugua T, Rastogi N, Skuce RA, Sola C, Van Soolingen D, Vincent V. 2001. Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: recommendations for standardised nomenclature. Int J Tuberc Lung Dis 5:216–219. [PubMed] [Google Scholar]
- 328.Cowan LS, Diem L, Brake MC, Crawford JT. 2004. Transfer of a Mycobacterium tuberculosis genotyping method, spoligotyping, from a reverse line-blot hybridization, membrane-based assay to the Luminex multianalyte profiling system. J Clin Microbiol 42:474–477. doi: 10.1128/JCM.42.1.474-477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Zhang J, Abadia E, Refregier G, Tafaj S, Boschiroli ML, Guillard B, Andremont A, Ruimy R, Sola C. 2010. Mycobacterium tuberculosis complex CRISPR genotyping: improving efficiency, throughput and discriminative power of “spoligotyping” with new spacers and a microbead-based hybridization assay. J Med Microbiol 59:285–294. doi: 10.1099/jmm.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 330.Honisch C, Mosko M, Arnold C, Gharbia SE, Diel R, Niemann S. 2010. Replacing reverse line blot hybridization spoligotyping of the Mycobacterium tuberculosis complex. J Clin Microbiol 48:1520–1526. doi: 10.1128/JCM.02299-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Goguet de la Salmonière YO, Li HM, Torrea G, Bunschoten A, van Embden J, Gicquel B. 1997. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J Clin Microbiol 35:2210–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Diaz R, Kremer K, de Haas PE, Gomez RI, Marrero A, Valdivia JA, van Embden JD, van Soolingen D. 1998. Molecular epidemiology of tuberculosis in Cuba outside of Havana, July 1994-June 1995: utility of spoligotyping versus IS6110 restriction fragment length polymorphism. Int J Tuberc Lung Dis 2:743–750. [PubMed] [Google Scholar]
- 333.Farnia P, Mohammadi F, Masjedi MR, Varnerot A, Zarifi AZ, Tabatabee J, Douraghei M, Ghazisaeedi K, Mansorri D, Bahadori M, Vincent V, Gutierrez C, Velayati AA. 2004. Evaluation of tuberculosis transmission in Tehran: using RFLP and spoligotyping methods. J Infect 49:94–101. doi: 10.1016/j.jinf.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 334.Gori A, Bandera A, Marchetti G, Esposti AD, Catozzi L, Nardi GP, Gazzola L, Ferrario G, van Embden JDA, van Soolingen D, Moroni M, Franzetti F. 2005. Spoligotyping and Mycobacterium tuberculosis. Emerg Infect Dis 11:1242–1248. doi: 10.3201/eid1108.040982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Gori A, Esposti AD, Bandera A, Mezzetti M, Sola C, Marchetti G, Ferrario G, Salerno F, Goyal M, Diaz R, Gazzola L, Codecasa L, Penati V, Rastogi N, Moroni M, Franzetti F. 2005. Comparison between spoligotyping and IS6110 restriction fragment length polymorphisms in molecular genotyping analysis of Mycobacterium tuberculosis strains. Mol Cell Probes 19:236–244. doi: 10.1016/j.mcp.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 336.Choi GE, Jang MH, Song EJ, Jeong SH, Kim J-S, Lee WG, Uh Y, Roh KH, Lee HS, Shin JH, Ryoo NH, Kim YR, Jeong J, Kim JH, Lee SM, Yi J, Hwang SH, Kim HH, Lee EY, Chang CL, Kim M-B, Kim YD. 2010. IS6110-restriction fragment length polymorphism and spoligotyping analysis of Mycobacterium tuberculosis clinical isolates for investigating epidemiologic distribution in Korea. J Korean Med Sci 25:1716–1721. doi: 10.3346/jkms.2010.25.12.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Rozo-Anaya JC, Ribón W. 2010. Molecular tools for Mycobacterium tuberculosis genotyping. Rev Salud Publica (Bogota) 12:510–521. [PubMed] [Google Scholar]
- 338.Van Embden JD, van Gorkom T, Kremer K, Jansen R, van Der Zeijst BA, Schouls LM. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J Bacteriol 182:2393–2401. doi: 10.1128/JB.182.9.2393-2401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Brudey K, Gutierrez MC, Vincent V, Parsons LM, Salfinger M, Rastogi N, Sola C. 2004. Mycobacterium africanum genotyping using novel spacer oligonucleotides in the direct repeat locus. J Clin Microbiol 42:5053–5057. doi: 10.1128/JCM.42.11.5053-5057.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Schürch AC, Kremer K, Kiers A, Boeree MJ, Siezen RJ, van Soolingen D. 2011. Preferential deletion events in the direct repeat locus of Mycobacterium tuberculosis. J Clin Microbiol 49:1318–1322. doi: 10.1128/JCM.01848-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Krawczyk M, Brzostek A, Gorna A, Knapska K, Ziolkiewicz M, Wojtasik A, Dziadek J. 2011. Epidemiological analysis of Mycobacterium tuberculosis strains isolated in Lodz, Poland. Int J Tuberc Lung Dis 15:1252–1258. doi: 10.5588/ijtld.10.0718. [DOI] [PubMed] [Google Scholar]
- 342.Dong H, Lu B, Zhang Y, Liu Z, Zhao X, Jiang Y, Wan K. 2009. Introduction to the standard operation program of spoligotyping on Mycobacterium tuberculosis in China. Zhonghua Liu Xing Bing Xue Za Zhi 30:384–387. [PubMed] [Google Scholar]
- 343.Kulkarni S, Sola C, Filliol I, Rastogi N, Kadival G. 2005. Spoligotyping of Mycobacterium tuberculosis isolates from patients with pulmonary tuberculosis in Mumbai, India. Res Microbiol 156:588–596. doi: 10.1016/j.resmic.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 344.Van der Zanden AG, Hoentjen AH, Heilmann FG, Weltevreden EF, Schouls LM, van Embden JD. 1998. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis complex in paraffin wax embedded tissues and in stained microscopic preparations. Mol Pathol 51:209–214. doi: 10.1136/mp.51.4.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Driscoll JR, McGarry MA, Taber HW. 1999. DNA typing of a nonviable culture of Mycobacterium tuberculosis in a homeless shelter outbreak. J Clin Microbiol 37:274–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Baron H, Hummel S, Herrmann B. 1996. Mycobacterium tuberculosis complex DNA in ancient human bones. J Archaeol Sci 23:667–671. doi: 10.1006/jasc.1996.0063. [DOI] [Google Scholar]
- 347.Donoghue HD, Spigelman M, Zias J, Gernaey-Child AM, Minnikin DE. 1998. Mycobacterium tuberculosis complex DNA in calcified pleura from remains 1400 years old. Lett Appl Microbiol 27:265–269. [PubMed] [Google Scholar]
- 348.Haas CJ, Zink A, Molńar E, Szeimies U, Reischl U, Marcsik A, Ardagna Y, Dutour O, Pálfi G, Nerlich AG. 2000. Molecular evidence for different stages of tuberculosis in ancient bone samples from Hungary. Am J Phys Anthropol 113:293–304. doi:. [DOI] [PubMed] [Google Scholar]
- 349.Spigelman M, Lemma E. 1993. The use of the polymerase chain reaction (PCR) to detect Mycobacterium tuberculosis in ancient skeletons. Int J Osteoarchaeol 3:137–143. doi: 10.1002/oa.1390030211. [DOI] [Google Scholar]
- 350.Zink AR, Sola C, Reischl U, Grabner W, Rastogi N, Wolf H, Nerlich AG. 2003. Characterization of Mycobacterium tuberculosis complex DNAs from Egyptian mummies by spoligotyping. J Clin Microbiol 41:359–367. doi: 10.1128/JCM.41.1.359-367.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Plikaytis BB, Crawford JT, Woodley CL, Butler WR, Eisenach KD, Cave MD, Shinnick TM. 1993. Rapid, amplification-based fingerprinting of Mycobacterium tuberculosis. J Gen Microbiol 139:1537–1542. doi: 10.1099/00221287-139-7-1537. [DOI] [PubMed] [Google Scholar]
- 352.Kremer K, Glynn JR, Lillebaek T, Niemann S, Kurepina NE, Kreiswirth BN, Bifani PJ, van Soolingen D. 2004. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J Clin Microbiol 42:4040–4049. doi: 10.1128/JCM.42.9.4040-4049.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Yeh RW, Ponce de Leon A, Agasino CB, Hahn JA, Daley CL, Hopewell PC, Small PM. 1998. Stability of Mycobacterium tuberculosis DNA genotypes. J Infect Dis 177:1107–1111. doi: 10.1086/517406. [DOI] [PubMed] [Google Scholar]
- 354.Coll F, Mallard K, Preston MD, Bentley S, Parkhill J, McNerney R, Martin N, Clark TG. 2012. SpolPred: rapid and accurate prediction of Mycobacterium tuberculosis spoligotypes from short genomic sequences. Bioinformatics 28:2991–2993. doi: 10.1093/bioinformatics/bts544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Frothingham R, Meeker-O'Connell WA. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144(Part 5):1189–1196. [DOI] [PubMed] [Google Scholar]
- 356.Hermans PW, van Soolingen D, van Embden JD. 1992. Characterization of a major polymorphic tandem repeat in Mycobacterium tuberculosis and its potential use in the epidemiology of Mycobacterium kansasii and Mycobacterium gordonae. J Bacteriol 174:4157–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Cole ST. 2002. Comparative and functional genomics of the Mycobacterium tuberculosis complex. Microbiology 148:2919–2928. doi: 10.1099/00221287-148-10-2919. [DOI] [PubMed] [Google Scholar]
- 358.Taillard C, Greub G, Weber R, Pfyffer GE, Bodmer T, Zimmerli S, Frei R, Bassetti S, Rohner P, Piffaretti J-C, Bernasconi E, Bille J, Telenti A, Prod'hom G. 2003. Clinical implications of Mycobacterium kansasii species heterogeneity: Swiss National Survey. J Clin Microbiol 41:1240–1244. doi: 10.1128/JCM.41.3.1240-1244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol 36:762–771. [DOI] [PubMed] [Google Scholar]
- 360.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Blackwood KS, Wolfe JN, Kabani AM. 2004. Application of mycobacterial interspersed repetitive unit typing to Manitoba tuberculosis cases: can restriction fragment length polymorphism be forgotten? J Clin Microbiol 42:5001–5006. doi: 10.1128/JCM.42.11.5001-5006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Hawkey PM, Smith EG, Evans JT, Monk P, Bryan G, Mohamed HH, Bardhan M, Pugh RN. 2003. Mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis compared to IS6110-based restriction fragment length polymorphism analysis for investigation of apparently clustered cases of tuberculosis. J Clin Microbiol 41:3514–3520. doi: 10.1128/JCM.41.8.3514-3520.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Alonso-Rodríguez N, Martínez-Lirola M, Herránz M, Sanchez-Benitez M, Barroso P, INDAL-TB Group, Bouza E, Garcia de Viedma D. 2008. Evaluation of the new advanced 15-loci MIRU-VNTR genotyping tool in Mycobacterium tuberculosis molecular epidemiology studies. BMC Microbiol 8:34. doi: 10.1186/1471-2180-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Lee ASG, Tang LLH, Lim IHK, Bellamy R, Wong S-Y. 2002. Discrimination of single-copy IS6110 DNA fingerprints of Mycobacterium tuberculosis isolates by high-resolution minisatellite-based typing. J Clin Microbiol 40:657–659. doi: 10.1128/JCM.40.2.657-659.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Asgharzadeh M, Kafil HS, Roudsary AA, Hanifi GR. 2011. Tuberculosis transmission in Northwest of Iran: using MIRU-VNTR, ETR-VNTR and IS6110-RFLP methods. Infect Genet Evol 11:124–131. doi: 10.1016/j.meegid.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 366.Cowan LS, Diem L, Monson T, Wand P, Temporado D, Oemig TV, Crawford JT. 2005. Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol 43:688–695. doi: 10.1128/JCM.43.2.688-695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Scott AN, Menzies D, Tannenbaum T-N, Thibert L, Kozak R, Joseph L, Schwartzman K, Behr MA. 2005. Sensitivities and specificities of spoligotyping and mycobacterial interspersed repetitive unit-variable-number tandem repeat typing methods for studying molecular epidemiology of tuberculosis. J Clin Microbiol 43:89–94. doi: 10.1128/JCM.43.1.89-94.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Brossier F, Sola C, Millot G, Jarlier V, Veziris N, Sougakoff W. 2014. Comparison of a semiautomated commercial repetitive-sequence-based PCR method with spoligotyping, 24-locus mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing, and restriction fragment length polymorphism-based analysis of IS6110 for Mycobacterium tuberculosis typing. J Clin Microbiol 52:4082–4086. doi: 10.1128/JCM.02226-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Wirth T, Hildebrand F, Allix-Béguec C, Wölbeling F, Kubica T, Kremer K, van Soolingen D, Rüsch-Gerdes S, Locht C, Brisse S, Meyer A, Supply P, Niemann S. 2008. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog 4:e1000160. doi: 10.1371/journal.ppat.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Vadwai V, Shetty A, Supply P, Rodrigues C. 2012. Evaluation of 24-locus MIRU-VNTR in extrapulmonary specimens: study from a tertiary centre in Mumbai. Tuberculosis (Edinb) 92:264–272. doi: 10.1016/j.tube.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 371.Oelemann MC, Diel R, Vatin V, Haas W, Rüsch-Gerdes S, Locht C, Niemann S, Supply P. 2007. Assessment of an optimized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J Clin Microbiol 45:691–697. doi: 10.1128/JCM.01393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.De Beer JL, Akkerman OW, Schürch AC, Mulder A, van der Werf TS, van der Zanden AGM, van Ingen J, van Soolingen D. 2014. Optimization of standard in-house 24-locus variable-number tandem-repeat typing for Mycobacterium tuberculosis and its direct application to clinical material. J Clin Microbiol 52:1338–1342. doi: 10.1128/JCM.03436-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Hanekom M, van der Spuy GD, van Pittius NCG, McEvoy CRE, Hoek KGP, Ndabambi SL, Jordaan AM, Victor TC, van Helden PD, Warren RM. 2008. Discordance between mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing and IS6110 restriction fragment length polymorphism genotyping for analysis of Mycobacterium tuberculosis Beijing strains in a setting of high incidence of tuberculosis. J Clin Microbiol 46:3338–3345. doi: 10.1128/JCM.00770-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Mokrousov I, Narvskaya O, Vyazovaya A, Millet J, Otten T, Vishnevsky B, Rastogi N. 2008. Mycobacterium tuberculosis Beijing genotype in Russia: in search of informative variable-number tandem-repeat loci. J Clin Microbiol 46:3576–3584. doi: 10.1128/JCM.00414-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Niemann S, Diel R, Khechinashvili G, Gegia M, Mdivani N, Tang Y-W. 2010. Mycobacterium tuberculosis Beijing lineage favors the spread of multidrug-resistant tuberculosis in the Republic of Georgia. J Clin Microbiol 48:3544–3550. doi: 10.1128/JCM.00715-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Comas I, Homolka S, Niemann S, Gagneux S. 2009. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One 4:e7815. doi: 10.1371/journal.pone.0007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Asante-Poku A, Nyaho MS, Borrell S, Comas I, Gagneux S, Yeboah-Manu D. 2014. Evaluation of customised lineage-specific sets of MIRU-VNTR loci for genotyping Mycobacterium tuberculosis complex isolates in Ghana. PLoS One 9:e92675. doi: 10.1371/journal.pone.0092675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Zheng C, Zhao Y, Zhu G, Li S, Sun H, Feng Q, Luo M, Wu F, Li X, Hill V, Rastogi N, Sun Q. 2014. Suitability of IS6110-RFLP and MIRU-VNTR for differentiating spoligotyped drug-resistant Mycobacterium tuberculosis clinical isolates from Sichuan in China. Biomed Res Int 2014:763204. doi: 10.1155/2014/763204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S. 2008. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol 46:2692–2699. doi: 10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. 2010. MIRU-VNTRplus: a Web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res 38:W326–W331. doi: 10.1093/nar/gkq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Demay C, Liens B, Burguière T, Hill V, Couvin D, Millet J, Mokrousov I, Sola C, Zozio T, Rastogi N. 2012. SITVITWEB—a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol 12:755–766. doi: 10.1016/j.meegid.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 382.De Beer JL, Ködmön C, van Ingen J, Supply P, van Soolingen D, Global Network for Molecular Surveillance of Tuberculosis 2010. 2014. Second worldwide proficiency study on variable number of tandem repeats typing of Mycobacterium tuberculosis complex. Int J Tuberc Lung Dis 18:594–600. doi: 10.5588/ijtld.13.0531. [DOI] [PubMed] [Google Scholar]
- 383.Inagaki T, Nishimori K, Yagi T, Ichikawa K, Moriyama M, Nakagawa T, Shibayama T, Uchiya K, Nikai T, Ogawa K. 2009. Comparison of a variable-number tandem-repeat (VNTR) method for typing Mycobacterium avium with mycobacterial interspersed repetitive-unit-VNTR and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 47:2156–2164. doi: 10.1128/JCM.02373-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Dauchy F-A, Dégrange S, Charron A, Dupon M, Xin Y, Bébéar C, Maugein J. 2010. Variable-number tandem-repeat markers for typing Mycobacterium intracellulare strains isolated in humans. BMC Microbiol 10:93. doi: 10.1186/1471-2180-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Lavender CJ, Stinear TP, Johnson PDR, Azuolas J, Benbow ME, Wallace JR, Fyfe JAM. 2008. Evaluation of VNTR typing for the identification of Mycobacterium ulcerans in environmental samples from Victoria, Australia. FEMS Microbiol Lett 287:250–255. doi: 10.1111/j.1574-6968.2008.01328.x. [DOI] [PubMed] [Google Scholar]
- 386.Sun G, Chen C, Li J, Gui X, Li X, Chen L, Peng F, Mei J, Gao Q. 2011. Discriminatory potential of a novel set of variable number of tandem repeats for genotyping Mycobacterium marinum. Vet Microbiol 152:200–204. doi: 10.1016/j.vetmic.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 387.Lin Z, Ni X, Zeng D, Zhou X. 2007. Research progress of ERIC (IRU). Wei Sheng Wu Xue Bao 47:370–373. [PubMed] [Google Scholar]
- 388.Sechi LA, Zanetti S, Dupré I, Delogu G, Fadda G. 1998. Enterobacterial repetitive intergenic consensus sequences as molecular targets for typing of Mycobacterium tuberculosis strains. J Clin Microbiol 36:128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Wojtasik A, Majchrzak M, Adamus-Bialek W, Augustynowicz-Kopec E, Zwolska Z, Dziadek J, Parniewski P. 2011. Trinucleotide repeat sequence-based PCR as a potential approach for genotyping Mycobacterium gordonae strains. J Microbiol Methods 85:28–32. doi: 10.1016/j.mimet.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 390.Asiimwe BB, Bagyenzi GB, Ssengooba W, Mumbowa F, Mboowa G, Wajja A, Mayanja-Kiiza H, Musoke PM, Wobudeya E, Kallenius G, Joloba ML. 2013. Species and genotypic diversity of non-tuberculous mycobacteria isolated from children investigated for pulmonary tuberculosis in rural Uganda. BMC Infect Dis 13:88. doi: 10.1186/1471-2334-13-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Sampaio JLM, Viana-Niero C, de Freitas D, Höfling-Lima AL, Leão SC. 2006. Enterobacterial repetitive intergenic consensus PCR is a useful tool for typing Mycobacterium chelonae and Mycobacterium abscessus isolates. Diagn Microbiol Infect Dis 55:107–118. doi: 10.1016/j.diagmicrobio.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 392.Englund S. 2003. IS900/ERIC-PCR as a tool to distinguish Mycobacterium avium subsp. paratuberculosis from closely related mycobacteria. Vet Microbiol 96:277–287. doi: 10.1016/j.vetmic.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 393.Otsuka Y, Parniewski P, Zwolska Z, Kai M, Fujino T, Kirikae F, Toyota E, Kudo K, Kuratsuji T, Kirikae T. 2004. Characterization of a trinucleotide repeat sequence (CGG)5 and potential use in restriction fragment length polymorphism typing of Mycobacterium tuberculosis. J Clin Microbiol 42:3538–3548. doi: 10.1128/JCM.42.8.3538-3548.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Poulet S, Cole ST. 1995. Characterization of the highly abundant polymorphic GC-rich-repetitive sequence (PGRS) present in Mycobacterium tuberculosis. Arch Microbiol 163:87–95. doi: 10.1007/BF00381781. [DOI] [PubMed] [Google Scholar]
- 395.Van Soolingen D, de Haas PE, Hermans PW, Groenen PM, van Embden JD. 1993. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol 31:1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Dela A, Sajduda A, Pawłowska I, Dziadek J. 2006. Molecular characterization of Mycobacterium tuberculosis isolates from Łódź, Poland: analysis by IS6110 restriction fragment length polymorphism and double-repetitive-element PCR. J Infect 52:346–353. doi: 10.1016/j.jinf.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 397.Flores L, Jarlsberg LG, Kim EY, Osmond D, Grinsdale J, Kawamura M, Desmond E, Hopewell PC, Kato-Maeda M. 2010. Comparison of restriction fragment length polymorphism with the polymorphic guanine-cytosine-rich sequence and spoligotyping for differentiation of Mycobacterium tuberculosis isolates with five or fewer copies of IS6110. J Clin Microbiol 48:575–578. doi: 10.1128/JCM.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Ross BC, Raios K, Jackson K, Dwyer B. 1992. Molecular cloning of a highly repeated DNA element from Mycobacterium tuberculosis and its use as an epidemiological tool. J Clin Microbiol 30:942–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Jackson K, Edwards R, Leslie DE, Hayman J. 1995. Molecular method for typing Mycobacterium ulcerans. J Clin Microbiol 33:2250–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Friedman CR, Stoeckle MY, Johnson WD, Riley LW. 1995. Double-repetitive-element PCR method for subtyping Mycobacterium tuberculosis clinical isolates. J Clin Microbiol 33:1383–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Sechi LA, Zanetti S, Delogu G, Montinaro B, Sanna A, Fadda G. 1996. Molecular epidemiology of Mycobacterium tuberculosis strains isolated from different regions of Italy and Pakistan. J Clin Microbiol 34:1825–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Warren R, Richardson M, Sampson S, Hauman JH, Beyers N, Donald PR, van Helden PD. 1996. Genotyping of Mycobacterium tuberculosis with additional markers enhances accuracy in epidemiological studies. J Clin Microbiol 34:2219–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Wiid IJ, Werely C, Beyers N, Donald P, van Helden PD. 1994. Oligonucleotide (GTG)5 as a marker for Mycobacterium tuberculosis strain identification. J Clin Microbiol 32:1318–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Cilliers FJ, Warren RM, Hauman JH, Wiid IJ, van Helden PD. 1997. Oligonucleotide (GTG)5 as an epidemiological tool in the study of nontuberculous mycobacteria. J Clin Microbiol 35:1545–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Kotłowski R, Shamputa IC, El Aila NA, Sajduda A, Rigouts L, van Deun A, Portaels F. 2004. PCR-based genotyping of Mycobacterium tuberculosis with new GC-rich repeated sequences and IS6110 inverted repeats used as primers. J Clin Microbiol 42:372–377. doi: 10.1128/JCM.42.1.372-377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Jagielski T, Augustynowicz-Kopeć E, Pawlik K, Dziadek J, Zwolska Z, Bielecki J. 2011. A two-step strategy for molecular typing of multidrug-resistant Mycobacterium tuberculosis clinical isolates from Poland. Pol J Microbiol 60:233–241. [PubMed] [Google Scholar]
- 407.Sajduda A, Dziadek J, Kotłowski R, Portaels F. 2006. Evaluation of multiple genetic markers for typing drug-resistant Mycobacterium tuberculosis strains from Poland. Diagn Microbiol Infect Dis 55:59–64. doi: 10.1016/j.diagmicrobio.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 408.Augustynowicz-Kopeć E, Jagielski T, Kozińska M, Zabost A, Klatt M, Zwolska Z. 2008. Molecular analysis of drug-resistant Mycobacterium tuberculosis isolates collected in central Poland. Clin Microbiol Infect 14:605–607. doi: 10.1111/j.1469-0691.2008.02003.x. [DOI] [PubMed] [Google Scholar]
- 409.Thomson R, Tolson C, Carter R, Coulter C, Huygens F, Hargreaves M. 2013. Isolation of nontuberculous mycobacteria (NTM) from household water and shower aerosols in patients with pulmonary disease caused by NTM. J Clin Microbiol 51:3006–3011. doi: 10.1128/JCM.00899-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Cangelosi GA, Freeman RJ, Lewis KN, Livingston-Rosanoff D, Shah KS, Milan SJ, Goldberg SV. 2004. Evaluation of a high-throughput repetitive-sequence-based PCR system for DNA fingerprinting of Mycobacterium tuberculosis and Mycobacterium avium complex strains. J Clin Microbiol 42:2685–2693. doi: 10.1128/JCM.42.6.2685-2693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Jang MH, Choi GE, Shin B-M, Lee SH, Kim S-R, Chang CL, Kim J-M. 2011. Comparison of an automated repetitive sequence-based PCR microbial typing system with IS6110-restriction fragment length polymorphism for epidemiologic investigation of clinical Mycobacterium tuberculosis isolates in Korea. Korean J Lab Med 31:282–284. doi: 10.3343/kjlm.2011.31.4.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Aitken ML, Limaye A, Pottinger P, Whimbey E, Goss CH, Tonelli MR, Cangelosi GA, Dirac MA, Olivier KN, Brown-Elliott BA, McNulty S, Wallace RJ. 2012. Respiratory outbreak of Mycobacterium abscessus subspecies massiliense in a lung transplant and cystic fibrosis center. Am J Respir Crit Care Med 185:231–232. doi: 10.1164/ajrccm.185.2.231. [DOI] [PubMed] [Google Scholar]
- 413.Thomson R, Tolson C, Sidjabat H, Huygens F, Hargreaves M. 2013. Mycobacterium abscessus isolated from municipal water—a potential source of human infection. BMC Infect Dis 13:241. doi: 10.1186/1471-2334-13-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Christianson S, Wolfe J, Soualhine H, Sharma MK. 2012. Comparison of repetitive-sequence-based polymerase chain reaction with random amplified polymorphic DNA analysis for rapid genotyping of nontuberculosis mycobacteria. Can J Microbiol 58:953–964. doi: 10.1139/w2012-068. [DOI] [PubMed] [Google Scholar]
- 415.Jagielski T, van Ingen J, Rastogi N, Dziadek J, Mazur PK, Bielecki J. 2014. Current methods in the molecular typing of Mycobacterium tuberculosis and other mycobacteria. Biomed Res Int 2014:645802. doi: 10.1155/2014/645802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Simpson E. 1949. Measurement of diversity. Nature 163:688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- 417.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Bromham L, Penny D. 2003. The modern molecular clock. Nat Rev Genet 4:216–224. doi: 10.1038/nrg1020. [DOI] [PubMed] [Google Scholar]
- 419.Bryant JM, Schürch AC, van Deutekom H, Harris SR, de Beer JL, de Jager V, Kremer K, van Hijum SAFT, Siezen RJ, Borgdorff M, Bentley SD, Parkhill J, van Soolingen D. 2013. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect Dis 13:110. doi: 10.1186/1471-2334-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.De Boer AS, Borgdorff MW, de Haas PE, Nagelkerke NJ, van Embden JD, van Soolingen D. 1999. Analysis of rate of change of IS6110 RFLP patterns of Mycobacterium tuberculosis based on serial patient isolates. J Infect Dis 180:1238–1244. doi: 10.1086/314979. [DOI] [PubMed] [Google Scholar]
- 421.Niemann S, Richter E, Rüsch-Gerdes S. 1999. Stability of Mycobacterium tuberculosis IS6110 restriction fragment length polymorphism patterns and spoligotypes determined by analyzing serial isolates from patients with drug-resistant tuberculosis. J Clin Microbiol 37:409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Warren RM, Streicher EM, Charalambous S, Churchyard G, van der Spuy GD, Grant AD, van Helden PD, Victor TC. 2002. Use of spoligotyping for accurate classification of recurrent tuberculosis. J Clin Microbiol 40:3851–3853. doi: 10.1128/JCM.40.10.3851-3853.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Pérez-Lago L, Comas I, Navarro Y, González-Candelas F, Herranz M, Bouza E, García-de-Viedma D. 2014. Whole genome sequencing analysis of intrapatient microevolution in Mycobacterium tuberculosis: potential impact on the inference of tuberculosis transmission. J Infect Dis 209:98–108. doi: 10.1093/infdis/jit439. [DOI] [PubMed] [Google Scholar]
- 424.Schürch AC, Kremer K, Kiers A, Daviena O, Boeree MJ, Siezen RJ, Smith NH, van Soolingen D. 2010. The tempo and mode of molecular evolution of Mycobacterium tuberculosis at patient-to-patient scale. Infect Genet Evol 10:108–114. doi: 10.1016/j.meegid.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 425.Navarro Y, Herranz M, Pérez-Lago L, Martínez Lirola M, INDAL-TB, Ruiz-Serrano MJ, Bouza E, García de Viedma D. 2011. Systematic survey of clonal complexity in tuberculosis at a populational level and detailed characterization of the isolates involved. J Clin Microbiol 49:4131–4137. doi: 10.1128/JCM.05203-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Seidler A, Nienhaus A, Diel R. 2004. The transmission of tuberculosis in the light of new molecular biological approaches. Occup Environ Med 61:96–102. doi: 10.1136/oem.2003.008573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Zhao S, Prenger K, Smith L, Messina T, Fan H, Jaeger E, Stephens S. 2013. Rainbow: a tool for large-scale whole-genome sequencing data analysis using cloud computing. BMC Genomics 14:425. doi: 10.1186/1471-2164-14-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Kohl TA, Diel R, Harmsen D, Rothgänger J, Walter KM, Merker M, Weniger T, Niemann S. 2014. Whole-genome-based Mycobacterium tuberculosis surveillance: a standardized, portable, and expandable approach. J Clin Microbiol 52:2479–2486. doi: 10.1128/JCM.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Qi W, Käser M, Röltgen K, Yeboah-Manu D, Pluschke G. 2009. Genomic diversity and evolution of Mycobacterium ulcerans revealed by next-generation sequencing. PLoS Pathog 5:e1000580. doi: 10.1371/journal.ppat.1000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Monot M, Honoré N, Garnier T, Zidane N, Sherafi D, Paniz-Mondolfi A, Matsuoka M, Taylor GM, Donoghue HD, Bouwman A, Mays S, Watson C, Lockwood D, Khamesipour A, Khamispour A, Dowlati Y, Jianping S, Rea TH, Vera-Cabrera L, Stefani MM, Banu S, Macdonald M, Sapkota BR, Spencer JS, Thomas J, Harshman K, Singh P, Busso P, Gattiker A, Rougemont J, Brennan PJ, Cole ST. 2009. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet 41:1282–1289. doi: 10.1038/ng.477. [DOI] [PubMed] [Google Scholar]
- 431.Sassi M, Robert C, Raoult D, Drancourt M. 2013. Noncontiguous genome sequence of Mycobacterium septicum strain DSM 44393T. Genome Announc 1(4):e00574-13. doi: 10.1128/genomeA.00574-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Reyes A, Sandoval A, Cubillos-Ruiz A, Varley KE, Hernández-Neuta I, Samper S, Martín C, García MJ, Ritacco V, López L, Robledo J, Zambrano MM, Mitra RD, Portillo PD. 2012. IS-seq: a novel high throughput survey of in vivo IS6110 transposition in multiple Mycobacterium tuberculosis genomes. BMC Genomics 13:249. doi: 10.1186/1471-2164-13-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Murray M. 2002. Determinants of cluster distribution in the molecular epidemiology of tuberculosis. Proc Natl Acad Sci U S A 99:1538–1543. doi: 10.1073/pnas.022618299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Fok A, Numata Y, Schulzer M, FitzGerald MJ. 2008. Risk factors for clustering of tuberculosis cases: a systematic review of population-based molecular epidemiology studies. Int J Tuberc Lung Dis 12:480–492. [PubMed] [Google Scholar]
- 435.Love J, Sonnenberg P, Glynn JR, Gibson A, Gopaul K, Fang Z, Le Brun F, Pitman R, Hayward AC, Innes J, Van den Bosch C, Delpech V, Drobniewski F, Watson JM. 2009. Molecular epidemiology of tuberculosis in England, 1998. Int J Tuberc Lung Dis 13:201–207. [PubMed] [Google Scholar]
- 436.Gutiérrez MC, Vincent V, Aubert D, Bizet J, Gaillot O, Lebrun L, Pendeven CL, Pennec MPL, Mathieu D, Offredo C, Pangon B, Pierre-Audigier C. 1998. Molecular fingerprinting of Mycobacterium tuberculosis and risk factors for tuberculosis transmission in Paris, France, and surrounding area. J Clin Microbiol 36:486–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Geng E, Kreiswirth B, Driver C, Li J, Burzynski J, DellaLatta P, LaPaz A, Schluger NW. 2002. Changes in the transmission of tuberculosis in New York City from 1990 to 1999. N Engl J Med 346:1453–1458. doi: 10.1056/NEJMoa012972. [DOI] [PubMed] [Google Scholar]
- 438.Ohkado A, Nagamine M, Murase Y, Uchimura K, Kaguraoka S, Tatsumi Y, Yamada N, Ohmori M, Maeda S, Maeda H, Kato S, Mori T, Ishikawa N. 2008. Molecular epidemiology of Mycobacterium tuberculosis in an urban area in Japan, 2002-2006. Int J Tuberc Lung Dis 12:548–554. [PubMed] [Google Scholar]
- 439.Franzetti F, Codecasa L, Matteelli A, Degli Esposti A, Bandera A, Lacchini C, Lombardi A, Pinsi G, Zanini F, El-Hamad I, Gori A. 2010. Genotyping analyses of tuberculosis transmission among immigrant residents in Italy. Clin Microbiol Infect 16:1149–1154. doi: 10.1111/j.1469-0691.2009.03080.x. [DOI] [PubMed] [Google Scholar]
- 440.Kempf M-C, Dunlap NE, Lok KH, Benjamin WH, Keenan NB, Kimerling ME. 2005. Long-term molecular analysis of tuberculosis strains in Alabama, a state characterized by a largely indigenous, low-risk population. J Clin Microbiol 43:870–878. doi: 10.1128/JCM.43.2.870-878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Moonan PK, Ghosh S, Oeltmann JE, Kammerer JS, Cowan LS, Navin TR. 2012. Using genotyping and geospatial scanning to estimate recent Mycobacterium tuberculosis transmission, United States. Emerg Infect Dis 18:458–465. doi: 10.3201/eid1803.111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Suwanpimolkul G, Jarlsberg LG, Grinsdale JA, Osmond D, Kawamura LM, Hopewell PC, Kato-Maeda M. 2013. Molecular epidemiology of tuberculosis in foreign-born persons living in San Francisco. Am J Respir Crit Care Med 187:998–1006. doi: 10.1164/rccm.201212-2239OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Jones-López EC, Kim S, Fregona G, Marques-Rodrigues P, Hadad DJ, Molina LPD, Vinhas S, Reilly N, Moine S, Chakravorty S, Gaeddert M, Ribeiro-Rodrigues R, Salgame P, Palaci M, Alland D, Ellner JJ, Dietze R. 2014. Importance of cough and M. tuberculosis strain type as risks for increased transmission within households. PLoS One 9:e100984. doi: 10.1371/journal.pone.0100984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Kik SV, Verver S, van Soolingen D, de Haas PEW, Cobelens FG, Kremer K, van Deutekom H, Borgdorff MW. 2008. Tuberculosis outbreaks predicted by characteristics of first patients in a DNA fingerprint cluster. Am J Respir Crit Care Med 178:96–104. doi: 10.1164/rccm.200708-1256OC. [DOI] [PubMed] [Google Scholar]
- 445.De Vries G, van Hest NAH, Baars HWM, Sebek MMGG, Richardus JH. 2010. Factors associated with the high tuberculosis case rate in an urban area. Int J Tuberc Lung Dis 14:859–865. [PubMed] [Google Scholar]
- 446.De Vries G, Aldridge RW, Cayla JA, Haas WH, Sandgren A, van Hest NA, Abubakar I, Tuberculosis in European Union Big Cities Working Group. 2014. Epidemiology of tuberculosis in big cities of the European Union and European Economic Area countries. Euro Surveill 19(9):pii=20726 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20726. [DOI] [PubMed] [Google Scholar]
- 447.Sandgren A, Sañé Schepisi M, Sotgiu G, Huitric E, Migliori GB, Manissero D, van der Werf MJ, Girardi E. 2014. Tuberculosis transmission between foreign- and native-born populations in the EU/EEA: a systematic review. Eur Respir J 43:1159–1171. doi: 10.1183/09031936.00117213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Kamper-Jørgensen Z, Andersen AB, Kok-Jensen A, Kamper-Jørgensen M, Bygbjerg IC, Andersen PH, Thomsen VO, Lillebaek T. 2012. Migrant tuberculosis: the extent of transmission in a low burden country. BMC Infect Dis 12:60. doi: 10.1186/1471-2334-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Chin DP, DeRiemer K, Small PM, de Leon AP, Steinhart R, Schecter GF, Daley CL, Moss AR, Paz EA, Jasmer RM, Agasino CB, Hopewell PC. 1998. Differences in contributing factors to tuberculosis incidence in U.S.-born and foreign-born persons. Am J Respir Crit Care Med 158:1797–1803. doi: 10.1164/ajrccm.158.6.9804029. [DOI] [PubMed] [Google Scholar]
- 450.Martínez-Lirola M, Alonso-Rodriguez N, Sánchez ML, Herranz M, Andrés S, Peñafiel T, Rogado MC, Cabezas T, Martínez J, Lucerna MA, Rodríguez M, Bonillo MDC, Bouza E, García de Viedma D. 2008. Advanced survey of tuberculosis transmission in a complex socioepidemiologic scenario with a high proportion of cases in immigrants. Clin Infect Dis 47:8–14. doi: 10.1086/588785. [DOI] [PubMed] [Google Scholar]
- 451.Borrell S, Español M, Orcau A, Tudó G, March F, Caylà JA, Jansà JM, Alcaide F, Martín-Casabona N, Salvadó M, Martinez JA, Vidal R, Sanchez F, Altet N, Rey E, Coll P, González-Martín J. 2010. Tuberculosis transmission patterns among Spanish-born and foreign-born populations in the city of Barcelona. Clin Microbiol Infect 16:568–574. doi: 10.1111/j.1469-0691.2009.02886.x. [DOI] [PubMed] [Google Scholar]
- 452.Goldblatt D, Rorman E, Chemtob D, Freidlin PJ, Cedar N, Kaidar-Shwartz H, Dveyrin Z, Mor Z. 2014. Molecular epidemiology and mapping of tuberculosis in Israel: do migrants transmit the disease to locals? Int J Tuberc Lung Dis 18:1085–1091. doi: 10.5588/ijtld.14.0186. [DOI] [PubMed] [Google Scholar]
- 453.Varghese B, Supply P, Shoukri M, Allix-Beguec C, Memish Z, Abuljadayel N, Al-Hakeem R, AlRabiah F, Al-Hajoj S. 2013. Tuberculosis transmission among immigrants and autochthonous populations of the eastern province of Saudi Arabia. PLoS One 8:e77635. doi: 10.1371/journal.pone.0077635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Braden CR, Templeton GL, Cave MD, Valway S, Onorato IM, Castro KG, Moers D, Yang Z, Stead WW, Bates JH. 1997. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. J Infect Dis 175:1446–1452. doi: 10.1086/516478. [DOI] [PubMed] [Google Scholar]
- 455.De Beer JL, Kodmon C, van der Werf MJ, van Ingen J, van Soolingen D, ECDC MDR-TB Molecular Surveillance Project Participants. 2014. Molecular surveillance of multi- and extensively drug-resistant tuberculosis transmission in the European Union from 2003 to 2011. Euro Surveill 19(11):pii=20724 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20742. [DOI] [PubMed] [Google Scholar]
- 456.López-Calleja AI, Lezcano MA, Vitoria MA, Iglesias MJ, Cebollada A, Lafoz C, Gavin P, Aristimuño L, Revillo MJ, Martin C, Samper S. 2007. Genotyping of Mycobacterium tuberculosis over two periods: a changing scenario for tuberculosis transmission. Int J Tuberc Lung Dis 11:1080–1086. [PubMed] [Google Scholar]
- 457.Bhattacharya M, Dietrich S, Mosher L, Siddiqui F, Reisberg BE, Paul WS, Warren JR. 1998. Cross-contamination of specimens with Mycobacterium tuberculosis: clinical significance, causes, and prevention. Am J Clin Pathol 109:324–330. [DOI] [PubMed] [Google Scholar]
- 458.Ribeiro FKC, Lemos EM, Hadad DJ, Leão SC, Viana-Niero C, Dietze R, Johnson JL, Eisenach KD, Palaci M. 2009. Evaluation of low-colony-number counts of Mycobacterium tuberculosis on solid media as a microbiological marker of cross-contamination. J Clin Microbiol 47:1950–1952. doi: 10.1128/JCM.00626-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Sula L, Sulová J. 1979. Accidental contamination of diagnostic cultures of mycobacteria and phage identification of the contaminating strain. Tubercle 60:159–162. doi: 10.1016/0041-3879(79)90016-3. [DOI] [PubMed] [Google Scholar]
- 460.Small PM, McClenny NB, Singh SP, Schoolnik GK, Tompkins LS, Mickelsen PA. 1993. Molecular strain typing of Mycobacterium tuberculosis to confirm cross-contamination in the mycobacteriology laboratory and modification of procedures to minimize occurrence of false-positive cultures. J Clin Microbiol 31:1677–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Burman WJ, Reves RR. 2000. Review of false-positive cultures for Mycobacterium tuberculosis and recommendations for avoiding unnecessary treatment. Clin Infect Dis 31:1390–1395. doi: 10.1086/317504. [DOI] [PubMed] [Google Scholar]
- 462.Ruddy M, McHugh TD, Dale JW, Banerjee D, Maguire H, Wilson P, Drobniewski F, Butcher P, Gillespie SH. 2002. Estimation of the rate of unrecognized cross-contamination with Mycobacterium tuberculosis in London microbiology laboratories. J Clin Microbiol 40:4100–4104. doi: 10.1128/JCM.40.11.4100-4104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Gascoyne-Binzi DM, Barlow RE, Frothingham R, Robinson G, Collyns TA, Gelletlie R, Hawkey PM. 2001. Rapid identification of laboratory contamination with Mycobacterium tuberculosis using variable number tandem repeat analysis. J Clin Microbiol 39:69–74. doi: 10.1128/JCM.39.1.69-74.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Miller AC, Sharnprapai S, Suruki R, Corkren E, Nardell EA, Driscoll JR, McGarry M, Taber H, Etkind S. 2002. Impact of genotyping of Mycobacterium tuberculosis on public health practice in Massachusetts. Emerg Infect Dis 8:1285–1289. doi: 10.3201/eid0811.020316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Jasmer RM, Roemer M, Hamilton J, Bunter J, Braden CR, Shinnick TM, Desmond EP. 2002. A prospective, multicenter study of laboratory cross-contamination of Mycobacterium tuberculosis cultures. Emerg Infect Dis 8:1260–1263. doi: 10.3201/eid0811.020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Yan J-J, Jou R, Ko W-C, Wu J-J, Yang M-L, Chen H-M. 2005. The use of variable-number tandem-repeat mycobacterial interspersed repetitive unit typing to identify laboratory cross-contamination with Mycobacterium tuberculosis. Diagn Microbiol Infect Dis 52:21–28. doi: 10.1016/j.diagmicrobio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 467.Lai C-C, Tan C-K, Lin SH, Liao C-H, Chou C-H, Huang Y-T, Hsueh P-R. 2010. Molecular evidence of false-positive cultures for Mycobacterium tuberculosis in a Taiwanese hospital with a high incidence of TB. Chest 137:1065–1070. doi: 10.1378/chest.09-1878. [DOI] [PubMed] [Google Scholar]
- 468.Thumamo BP, Asuquo AE, Abia-Bassey LN, Lawson L, Hill V, Zozio T, Emenyonu N, Eko FO, Rastogi N. 2012. Molecular epidemiology and genetic diversity of Mycobacterium tuberculosis complex in the Cross River State, Nigeria. Infect Genet Evol 12:671–677. doi: 10.1016/j.meegid.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.De Boer AS, Blommerde B, de Haas PEW, Sebek MMGG, Lambregts-van Weezenbeek KSB, Dessens M, van Soolingen D. 2002. False-positive Mycobacterium tuberculosis cultures in 44 laboratories in The Netherlands (1993 to 2000): incidence, risk factors, and consequences. J Clin Microbiol 40:4004–4009. doi: 10.1128/JCM.40.11.4004-4009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Chang CL, Kim HH, Son HC, Park SS, Lee MK, Park SK, Park WW, Jeon CH. 2001. False-positive growth of Mycobacterium tuberculosis attributable to laboratory contamination confirmed by restriction fragment length polymorphism analysis. Int J Tuberc Lung Dis 5:861–867. [PubMed] [Google Scholar]
- 471.Northrup JM, Miller AC, Nardell E, Sharnprapai S, Etkind S, Driscoll J, McGarry M, Taber HW, Elvin P, Qualls NL, Braden CR. 2002. Estimated costs of false laboratory diagnoses of tuberculosis in three patients. Emerg Infect Dis 8:1264–1270. doi: 10.3201/eid0811.020387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Agerton T, Valway S, Gore B, Pozsik C, Plikaytis B, Woodley C, Onorato I. 1997. Transmission of a highly drug-resistant strain (strain W1) of Mycobacterium tuberculosis. Community outbreak and nosocomial transmission via a contaminated bronchoscope. JAMA 278:1073–1077. [PubMed] [Google Scholar]
- 473.Larson JL, Lambert L, Stricof RL, Driscoll J, McGarry MA, Ridzon R. 2003. Potential nosocomial exposure to Mycobacterium tuberculosis from a bronchoscope. Infect Control Hosp Epidemiol 24:825–830. doi: 10.1086/502144. [DOI] [PubMed] [Google Scholar]
- 474.Kovaleva J, Peters FTM, van der Mei HC, Degener JE. 2013. Transmission of infection by flexible gastrointestinal endoscopy and bronchoscopy. Clin Microbiol Rev 26:231–254. doi: 10.1128/CMR.00085-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Bang D, Andersen AB, Thomsen VO, Lillebaek T. 2010. Recurrent tuberculosis in Denmark: relapse vs. re-infection. Int J Tuberc Lung Dis 14:447–453. [PubMed] [Google Scholar]
- 476.Verver S, Warren RM, Beyers N, Richardson M, van der Spuy GD, Borgdorff MW, Enarson DA, Behr MA, van Helden PD. 2005. Rate of reinfection tuberculosis after successful treatment is higher than rate of new tuberculosis. Am J Respir Crit Care Med 171:1430–1435. doi: 10.1164/rccm.200409-1200OC. [DOI] [PubMed] [Google Scholar]
- 477.Otal I, Martín C, Vincent-Lévy-Frebault V, Thierry D, Gicquel B. 1991. Restriction fragment length polymorphism analysis using IS6110 as an epidemiological marker in tuberculosis. J Clin Microbiol 29:1252–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Godfrey-Faussett P, Githui W, Batchelor B, Brindle R, Paul J, Hawken M, Gathua S, Odhiambo J, Ojoo S, Nunn P. 1994. Recurrence of HIV-related tuberculosis in an endemic area may be due to relapse or reinfection. Tuber Lung Dis 75:199–202. doi: 10.1016/0962-8479(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 479.Krüüner A, Pehme L, Ghebremichael S, Koivula T, Hoffner SE, Mikelsaar M. 2002. Use of molecular techniques to distinguish between treatment failure and exogenous reinfection with Mycobacterium tuberculosis. Clin Infect Dis 35:146–155. doi: 10.1086/340980. [DOI] [PubMed] [Google Scholar]
- 480.Sonnenberg P, Murray J, Glynn JR, Shearer S, Kambashi B, Godfrey-Faussett P. 2001. HIV-1 and recurrence, relapse, and reinfection of tuberculosis after cure: a cohort study in South African mineworkers. Lancet 358:1687–1693. doi: 10.1016/S0140-6736(01)06712-5. [DOI] [PubMed] [Google Scholar]
- 481.García de Viedma D, Marín M, Hernangómez S, Díaz M, Ruiz Serrano MJ, Alcalá L, Bouza E. 2002. Tuberculosis recurrences: reinfection plays a role in a population whose clinical/epidemiological characteristics do not favor reinfection. Arch Intern Med 162:1873–1879. doi: 10.1001/archinte.162.16.1873. [DOI] [PubMed] [Google Scholar]
- 482.Dobler CC, Crawford ABH, Jelfs PJ, Gilbert GL, Marks GB. 2009. Recurrence of tuberculosis in a low-incidence setting. Eur Respir J 33:160–167. doi: 10.1183/09031936.00104108. [DOI] [PubMed] [Google Scholar]
- 483.De Boer AS, Borgdorff MW, Vynnycky E, Sebek MM, van Soolingen D. 2003. Exogenous re-infection as a cause of recurrent tuberculosis in a low-incidence area. Int J Tuberc Lung Dis 7:145–152. [PubMed] [Google Scholar]
- 484.Bryant JM, Harris SR, Parkhill J, Dawson R, Diacon AH, van Helden P, Pym A, Mahayiddin AA, Chuchottaworn C, Sanne IM, Louw C, Boeree MJ, Hoelscher M, McHugh TD, Bateson ALC, Hunt RD, Mwaigwisya S, Wright L, Gillespie SH, Bentley SD. 2013. Whole-genome sequencing to establish relapse or re-infection with Mycobacterium tuberculosis: a retrospective observational study. Lancet Respir Med 1:786–792. doi: 10.1016/S2213-2600(13)70231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Meissner G. 1954. The virulence of isoniazid resistant tubercle bacilli, recovered from patients during treatment. Dis Chest 26:15–26. doi: 10.1378/chest.26.1.15. [DOI] [PubMed] [Google Scholar]
- 486.Burgos M, DeRiemer K, Small PM, Hopewell PC, Daley CL. 2003. Effect of drug resistance on the generation of secondary cases of tuberculosis. J Infect Dis 188:1878–1884. doi: 10.1086/379895. [DOI] [PubMed] [Google Scholar]
- 487.García-García ML, Ponce de León A, Jiménez-Corona ME, Jiménez-Corona A, Palacios-Martínez M, Balandrano-Campos S, Ferreyra-Reyes L, Juárez-Sandino L, Sifuentes-Osornio J, Olivera-Díaz H, Valdespino-Gómez JL, Small PM. 2000. Clinical consequences and transmissibility of drug-resistant tuberculosis in southern Mexico. Arch Intern Med 160:630–636. [DOI] [PubMed] [Google Scholar]
- 488.Godfrey-Faussett P, Sonnenberg P, Shearer SC, Bruce MC, Mee C, Morris L, Murray J. 2000. Tuberculosis control and molecular epidemiology in a South African gold-mining community. Lancet 356:1066–1071. doi: 10.1016/S0140-6736(00)02730-6. [DOI] [PubMed] [Google Scholar]
- 489.Krüüner A, Hoffner SE, Sillastu H, Danilovits M, Levina K, Svenson SB, Ghebremichael S, Koivula T, Källenius G. 2001. Spread of drug-resistant pulmonary tuberculosis in Estonia. J Clin Microbiol 39:3339–3345. doi: 10.1128/JCM.39.9.3339-3345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Gandhi NR, Weissman D, Moodley P, Ramathal M, Elson I, Kreiswirth BN, Mathema B, Shashkina E, Rothenberg R, Moll AP, Friedland G, Sturm AW, Shah NS. 2013. Nosocomial transmission of extensively drug-resistant tuberculosis in a rural hospital in South Africa. J Infect Dis 207:9–17. doi: 10.1093/infdis/jis631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Merker M, Blin C, Mona S, Duforet-Frebourg N, Lecher S, Willery E, Blum MGB, Rüsch-Gerdes S, Mokrousov I, Aleksic E, Allix-Béguec C, Antierens A, Augustynowicz-Kopeć E, Ballif M, Barletta F, Beck HP, Barry CE, Bonnet M, Borroni E, Campos-Herrero I, Cirillo D, Cox H, Crowe S, Crudu V, Diel R, Drobniewski F, Fauville-Dufaux M, Gagneux S, Ghebremichael S, Hanekom M, Hoffner S, Jiao W, Kalon S, Kohl TA, Kontsevaya I, Lillebæk T, Maeda S, Nikolayevskyy V, Rasmussen M, Rastogi N, Samper S, Sanchez-Padilla E, Savic B, Shamputa IC, Shen A, Sng L-H, Stakenas P, Toit K, Varaine F, Vukovic D, et al. 2015. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet 47:242–249. doi: 10.1038/ng.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Streicher EM, Müller B, Chihota V, Mlambo C, Tait M, Pillay M, Trollip A, Hoek KGP, Sirgel FA, Gey van Pittius NC, van Helden PD, Victor TC, Warren RM. 2012. Emergence and treatment of multidrug resistant (MDR) and extensively drug-resistant (XDR) tuberculosis in South Africa. Infect Genet Evol 12:686–694. doi: 10.1016/j.meegid.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 493.Marais BJ, Mlambo CK, Rastogi N, Zozio T, Duse AG, Victor TC, Marais E, Warren RM. 2013. Epidemic spread of multidrug-resistant tuberculosis in Johannesburg, South Africa. J Clin Microbiol 51:1818–1825. doi: 10.1128/JCM.00200-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Diriba B, Berkessa T, Mamo G, Tedla Y, Ameni G. 2013. Spoligotyping of multidrug-resistant Mycobacterium tuberculosis isolates in Ethiopia. Int J Tuberc Lung Dis 17:246–250. doi: 10.5588/ijtld.12.0195. [DOI] [PubMed] [Google Scholar]
- 495.Perdigão J, Macedo R, João I, Fernandes E, Brum L, Portugal I. 2008. Multidrug-resistant tuberculosis in Lisbon, Portugal: a molecular epidemiological perspective. Microb Drug Resist 14:133–143. doi: 10.1089/mdr.2008.0798. [DOI] [PubMed] [Google Scholar]
- 496.Gavín P, Iglesias MJ, Jiménez MS, Rodríguez-Valín E, Ibarz D, Lezcano MA, Revillo MJ, Martín C, Samper S, Spanish Working Group on MDR-TB. 2012. Long-term molecular surveillance of multidrug-resistant tuberculosis in Spain. Infect Genet Evol 12:701–710. doi: 10.1016/j.meegid.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 497.Jagielski T, Brzostek A, van Belkum A, Dziadek J, Augustynowicz-Kopeć E, Zwolska Z. 2015. A close-up on the epidemiology and transmission of multidrug-resistant tuberculosis in Poland. Eur J Clin Microbiol Infect Dis 34:41–53. doi: 10.1007/s10096-014-2202-z. [DOI] [PubMed] [Google Scholar]
- 498.Jagielski T, Augustynowicz-Kopec E, Zozio T, Rastogi N, Zwolska Z. 2010. Spoligotype-based comparative population structure analysis of multidrug-resistant and isoniazid-monoresistant Mycobacterium tuberculosis complex clinical isolates in Poland. J Clin Microbiol 48:3899–3909. doi: 10.1128/JCM.00572-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Disratthakit A, Meada S, Prammananan T, Thaipisuttikul I, Doi N, Chaiprasert A. 2015. Genotypic diversity of multidrug-, quinolone- and extensively drug-resistant Mycobacterium tuberculosis isolates in Thailand. Infect Genet Evol 32:432–439. doi: 10.1016/j.meegid.2015.03.038. [DOI] [PubMed] [Google Scholar]
- 500.Murase Y, Maeda S, Yamada H, Ohkado A, Chikamatsu K, Mizuno K, Kato S, Mitarai S. 2010. Clonal expansion of multidrug-resistant and extensively drug-resistant tuberculosis, Japan. Emerg Infect Dis 16:948–954. doi: 10.3201/eid1606.091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.He GX, Wang HY, Borgdorff MW, van Soolingen D, van der Werf MJ, Liu ZM, Li XZ, Guo H, Zhao YL, Varma JK, Tostado CP, van den Hof S. 2011. Multidrug-resistant tuberculosis, People's Republic of China, 2007-2009. Emerg Infect Dis 17:1831–1838. doi: 10.3201/eid1710.110546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Parwati I, van Crevel R, van Soolingen D. 2010. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect Dis 10:103–111. doi: 10.1016/S1473-3099(09)70330-5. [DOI] [PubMed] [Google Scholar]
- 503.Allix-Béguec C, Wahl C, Hanekom M, Nikolayevskyy V, Drobniewski F, Maeda S, Campos-Herrero I, Mokrousov I, Niemann S, Kontsevaya I, Rastogi N, Samper S, Sng L-H, Warren RM, Supply P. 2014. Proposal of a consensus set of hypervariable mycobacterial interspersed repetitive-unit-variable-number tandem-repeat loci for subtyping of Mycobacterium tuberculosis Beijing isolates. J Clin Microbiol 52:164–172. doi: 10.1128/JCM.02519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Thomson R, Tolson C, Huygens F, Hargreaves M. 2014. Strain variation amongst clinical and potable water isolates of M. kansasii using automated repetitive unit PCR. Int J Med Microbiol 304:484–489. doi: 10.1016/j.ijmm.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 505.Marshall HM, Carter R, Torbey MJ, Minion S, Tolson C, Sidjabat HE, Huygens F, Hargreaves M, Thomson RM. 2011. Mycobacterium lentiflavum in drinking water supplies, Australia. Emerg Infect Dis 17:395–402. doi: 10.3201/eid1703.090948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Thomson RM, Tolson CE, Carter R, Huygens F, Hargreaves M. 2014. Heterogeneity of clinical and environmental isolates of Mycobacterium fortuitum using repetitive element sequence-based PCR: municipal water an unlikely source of community-acquired infections. Epidemiol Infect 142:2057–2064. doi: 10.1017/S0950268813003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Feazel LM, Baumgartner LK, Peterson KL, Frank DN, Harris JK, Pace NR. 2009. Opportunistic pathogens enriched in showerhead biofilms. Proc Natl Acad Sci U S A 106:16393–16399. doi: 10.1073/pnas.0908446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Falkinham JO, Iseman MD, de Haas P, van Soolingen D. 2008. Mycobacterium avium in a shower linked to pulmonary disease. J Water Health 6:209–213. doi: 10.2166/wh.2008.032. [DOI] [PubMed] [Google Scholar]
- 509.Falkinham JO. 2011. Nontuberculous mycobacteria from household plumbing of patients with nontuberculous mycobacteria disease. Emerg Infect Dis 17:419–424. doi: 10.3201/eid1703.101510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Nishiuchi Y, Maekura R, Kitada S, Tamaru A, Taguri T, Kira Y, Hiraga T, Hirotani A, Yoshimura K, Miki M, Ito M. 2007. The recovery of Mycobacterium avium-intracellulare complex (MAC) from the residential bathrooms of patients with pulmonary MAC. Clin Infect Dis 45:347–351. doi: 10.1086/519383. [DOI] [PubMed] [Google Scholar]
- 511.Taga S, Niimi M, Kurokawa K, Nakagawa T, Ogawa K. 2012. A case of environmental infection with pulmonary Mycobacterium avium complex disease from a residential bathroom of a patient suggested by variable-number tandem-repeat typing of Mycobacterium avium tandem repeat loci. Kekkaku 87:409–414. [PubMed] [Google Scholar]
- 512.Wallace RJ, Iakhiaeva E, Williams MD, Brown-Elliott BA, Vasireddy S, Vasireddy R, Lande L, Peterson DD, Sawicki J, Kwait R, Tichenor WS, Turenne C, Falkinham JO. 2013. Absence of Mycobacterium intracellulare and presence of Mycobacterium chimaera in household water and biofilm samples of patients in the United States with Mycobacterium avium complex respiratory disease. J Clin Microbiol 51:1747–1752. doi: 10.1128/JCM.00186-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.De Groote MA, Pace NR, Fulton K, Falkinham JO. 2006. Relationships between Mycobacterium isolates from patients with pulmonary mycobacterial infection and potting soils. Appl Environ Microbiol 72:7602–7606. doi: 10.1128/AEM.00930-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Fujita K, Ito Y, Hirai T, Maekawa K, Imai S, Tatsumi S, Niimi A, Iinuma Y, Ichiyama S, Mishima M. 2013. Genetic relatedness of Mycobacterium avium-intracellulare complex isolates from patients with pulmonary MAC disease and their residential soils. Clin Microbiol Infect 19:537–541. doi: 10.1111/j.1469-0691.2012.03929.x. [DOI] [PubMed] [Google Scholar]
- 515.Van Ingen J, Blaak H, de Beer J, de Roda Husman AM, van Soolingen D. 2010. Rapidly growing nontuberculous mycobacteria cultured from home tap and shower water. Appl Environ Microbiol 76:6017–6019. doi: 10.1128/AEM.00843-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Van der Wielen PWJJ, Heijnen L, van der Kooij D. 2013. Pyrosequence analysis of the hsp65 genes of nontuberculous mycobacterium communities in unchlorinated drinking water in the Netherlands. Appl Environ Microbiol 79:6160–6166. doi: 10.1128/AEM.01591-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Van der Wielen PWJJ, van der Kooij D. 2013. Nontuberculous mycobacteria, fungi, and opportunistic pathogens in unchlorinated drinking water in The Netherlands. Appl Environ Microbiol 79:825–834. doi: 10.1128/AEM.02748-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Astagneau P, Desplaces N, Vincent V, Chicheportiche V, Botherel A, Maugat S, Lebascle K, Léonard P, Desenclos J, Grosset J, Ziza J, Brücker G. 2001. Mycobacterium xenopi spinal infections after discovertebral surgery: investigation and screening of a large outbreak. Lancet 358:747–751. doi: 10.1016/S0140-6736(01)05843-3. [DOI] [PubMed] [Google Scholar]
- 519.Zhibang Y, BiXia Z, Qishan L, Lihao C, Xiangquan L, Huaping L. 2002. Large-scale outbreak of infection with Mycobacterium chelonae subsp. abscessus after penicillin injection. J Clin Microbiol 40:2626–2628. doi: 10.1128/JCM.40.7.2626-2628.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Kim H-Y, Yun Y-J, Park CG, Lee DH, Cho YK, Park BJ, Joo S-I, Kim E-C, Hur YJ, Kim B-J, Kook Y-H. 2007. Outbreak of Mycobacterium massiliense infection associated with intramuscular injections. J Clin Microbiol 45:3127–3130. doi: 10.1128/JCM.00608-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Winthrop KL, Abrams M, Yakrus M, Schwartz I, Ely J, Gillies D, Vugia DJ. 2002. An outbreak of mycobacterial furunculosis associated with footbaths at a nail salon. N Engl J Med 346:1366–1371. doi: 10.1056/NEJMoa012643. [DOI] [PubMed] [Google Scholar]
- 522.Furuya EY, Paez A, Srinivasan A, Cooksey R, Augenbraun M, Baron M, Brudney K, Della-Latta P, Estivariz C, Fischer S, Flood M, Kellner P, Roman C, Yakrus M, Weiss D, Granowitz EV. 2008. Outbreak of Mycobacterium abscessus wound infections among “lipotourists” from the United States who underwent abdominoplasty in the Dominican Republic. Clin Infect Dis 46:1181–1188. doi: 10.1086/529191. [DOI] [PubMed] [Google Scholar]
- 523.Duarte RS, Lourenço MCS, Fonseca LS, Leão SC, Amorim ELT, Rocha ILL, Coelho FS, Viana-Niero C, Gomes KM, da Silva MG, Lorena NS, Pitombo MB, Ferreira RMC, Garcia MH, de Oliveira GP, Lupi O, Vilaça BR, Serradas LR, Chebabo A, Marques EA, Teixeira LM, Dalcolmo M, Senna SG, Sampaio JLM. 2009. Epidemic of postsurgical infections caused by Mycobacterium massiliense. J Clin Microbiol 47:2149–2155. doi: 10.1128/JCM.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Viana-Niero C, Lima KVB, Lopes ML, Rabello MCS, Marsola LR, Brilhante VCR, Durham AM, Leão SC. 2008. Molecular characterization of Mycobacterium massiliense and Mycobacterium bolletii in isolates collected from outbreaks of infections after laparoscopic surgeries and cosmetic procedures. J Clin Microbiol 46:850–855. doi: 10.1128/JCM.02052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Achermann Y, Rössle M, Hoffmann M, Deggim V, Kuster S, Zimmermann DR, Bloemberg G, Hombach M, Hasse B. 2013. Prosthetic valve endocarditis and bloodstream infection due to Mycobacterium chimaera. J Clin Microbiol 51:1769–1773. doi: 10.1128/JCM.00435-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Sax H, Bloemberg G, Hasse B, Sommerstein R, Kohler P, Achermann Y, Rössle M, Falk V, Kuster SP, Böttger EC, Weber R. 2015. Prolonged outbreak of Mycobacterium chimaera infection after open-chest heart surgery. Clin Infect Dis 61:67–75. doi: 10.1093/cid/civ198. [DOI] [PubMed] [Google Scholar]
- 527.Conger NG, O'Connell RJ, Laurel VL, Olivier KN, Graviss EA, Williams-Bouyer N, Zhang Y, Brown-Elliott BA, Wallace RJ. 2004. Mycobacterium simae outbreak associated with a hospital water supply. Infect Control Hosp Epidemiol 25:1050–1055. doi: 10.1086/502342. [DOI] [PubMed] [Google Scholar]
- 528.Esteban J, Fernández-Roblas R, Ortiz A, García-Cía J-I. 2006. Pseudo-outbreak of Mycobacterium gordonae: usefulness of randomly amplified polymorphic DNA analysis to assess the clonality of the isolates. Clin Microbiol Infect 12:677–679. doi: 10.1111/j.1469-0691.2006.01450.x. [DOI] [PubMed] [Google Scholar]
- 529.Bettiker RL, Axelrod PI, Fekete T, St John K, Truant A, Toney S, Yakrus MA. 2006. Delayed recognition of a pseudo-outbreak of Mycobacterium terrae. Am J Infect Control 34:343–347. doi: 10.1016/j.ajic.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 530.Falkinham JO. 2015. Environmental sources of nontuberculous mycobacteria. Clin Chest Med 36:35–41. doi: 10.1016/j.ccm.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 531.Wallace RJ Jr, Zhang Y, Brown-Elliott BA, Yakrus MA, Wilson RW, Mann L, Couch L, Girard WM, Griffith DE. 2002. Repeat positive cultures in Mycobacterium intracellulare lung disease after macrolide therapy represent new infections in patients with nodular bronchiectasis. J Infect Dis 186:266–273. doi: 10.1086/341207. [DOI] [PubMed] [Google Scholar]
- 532.Wallace RJ, Brown-Elliott BA, McNulty S, Philley JV, Killingley J, Wilson RW, York DS, Shepherd S, Griffith DE. 2014. Macrolide/azalide therapy for nodular/bronchiectatic Mycobacterium avium complex lung disease. Chest 146:276–282. doi: 10.1378/chest.13-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Nishimura T, Fujita-Suzuki Y, Yonemaru M, Ohkusu K, Sakagami T, Carpenter SM, Otsuka Y, Namkoong H, Yano I, Hasegawa N. 2015. Recurrence of disseminated Mycobacterium avium complex disease in a patient with anti-gamma interferon autoantibodies by reinfection. J Clin Microbiol 53:1436–1438. doi: 10.1128/JCM.03339-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Doig C, Muckersie L, Watt B, Forbes KJ. 2002. Molecular epidemiology of Mycobacterium malmoense infections in Scotland. J Clin Microbiol 40:1103–1105. doi: 10.1128/JCM.40.3.1103-1105.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Harris KA, Underwood A, Kenna DTD, Brooks A, Kavaliunaite E, Kapatai G, Tewolde R, Aurora P, Dixon G. 2015. Whole-genome sequencing and epidemiological analysis do not provide evidence for cross-transmission of Mycobacterium abscessus in a cohort of pediatric cystic fibrosis patients. Clin Infect Dis 60:1007–1016. doi: 10.1093/cid/ciu967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Ricketts WM, O'Shaughnessy TC, van Ingen J. 2014. Human-to-human transmission of Mycobacterium kansasii or victims of a shared source? Eur Respir J 44:1085–1087. doi: 10.1183/09031936.00066614. [DOI] [PubMed] [Google Scholar]
- 537.Van Ingen J, Ferro BE, Hoefsloot W, Boeree MJ, van Soolingen D. 2013. Drug treatment of pulmonary nontuberculous mycobacterial disease in HIV-negative patients: the evidence. Expert Rev Anti Infect Ther 11:1065–1077. doi: 10.1586/14787210.2013.830413. [DOI] [PubMed] [Google Scholar]
- 538.Van Ingen J, de Zwaan R, Dekhuijzen R, Boeree M, van Soolingen D. 2009. Region of difference 1 in nontuberculous Mycobacterium species adds a phylogenetic and taxonomical character. J Bacteriol 191:5865–5867. doi: 10.1128/JB.00683-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Houben D, Demangel C, van Ingen J, Perez J, Baldeón L, Abdallah AM, Caleechurn L, Bottai D, van Zon M, de Punder K, van der Laan T, Kant A, Bossers-de Vries R, Willemsen P, Bitter W, van Soolingen D, Brosch R, van der Wel N, Peters PJ. 2012. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol 14:1287–1298. doi: 10.1111/j.1462-5822.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- 540.Van Ingen J, Hoefsloot W, Buijtels PC, Tortoli E, Supply P, Dekhuijzen PNR, Boeree MJ, van Soolingen D. 2012. Characterization of a novel variant of Mycobacterium chimaera. J Med Microbiol 61:1234–1239. doi: 10.1099/jmm.0.045070-0. [DOI] [PubMed] [Google Scholar]
- 541.Park J-H, Shim T-S, Lee S-A, Lee H, Lee I-K, Kim K, Kook HY-, Kim B-J. 2010. Molecular characterization of Mycobacterium intracellulare-related strains based on the sequence analysis of hsp65, internal transcribed spacer and 16S rRNA genes. J Med Microbiol 59:1037–1043. doi: 10.1099/jmm.0.020727-0. [DOI] [PubMed] [Google Scholar]
- 542.Kim S-Y, Lee S-T, Jeong B-H, Park HY, Jeon K, Kim J-W, Shin SJ, Koh W-J. 2013. Genotyping of Mycobacterium intracellulare isolates and clinical characteristics of lung disease. Int J Tuberc Lung Dis 17:669–675. doi: 10.5588/ijtld.12.0575. [DOI] [PubMed] [Google Scholar]
- 543.Kikuchi T, Watanabe A, Gomi K, Sakakibara T, Nishimori K, Daito H, Fujimura S, Tazawa R, Inoue A, Ebina M, Tokue Y, Kaku M, Nukiwa T. 2009. Association between mycobacterial genotypes and disease progression in Mycobacterium avium pulmonary infection. Thorax 64:901–907. doi: 10.1136/thx.2009.114603. [DOI] [PubMed] [Google Scholar]
- 544.Kikuchi T, Kobashi Y, Hirano T, Tode N, Santoso A, Tamada T, Fujimura S, Mitsuhashi Y, Honda Y, Nukiwa T, Kaku M, Watanabe A, Ichinose M. 2014. Mycobacterium avium genotype is associated with the therapeutic response to lung infection. Clin Microbiol Infect 20:256–262. doi: 10.1111/1469-0691.12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Kim S-Y, Lee S-T, Jeong B-H, Jeon K, Kim J-W, Shin SJ, Koh W-J. 2012. Clinical significance of mycobacterial genotyping in Mycobacterium avium lung disease in Korea. Int J Tuberc Lung Dis 16:1393–1399. doi: 10.5588/ijtld.12.0147. [DOI] [PubMed] [Google Scholar]
- 546.Shin SJ, Choi G-E, Cho S-N, Woo SY, Jeong B-H, Jeon K, Koh W-J. 2013. Mycobacterial genotypes are associated with clinical manifestation and progression of lung disease caused by Mycobacterium abscessus and Mycobacterium massiliense. Clin Infect Dis 57:32–39. doi: 10.1093/cid/cit172. [DOI] [PubMed] [Google Scholar]
- 547.Frothingham R, Wilson KH. 1994. Molecular phylogeny of the Mycobacterium avium complex demonstrates clinically meaningful divisions. J Infect Dis 169:305–312. doi: 10.1093/infdis/169.2.305. [DOI] [PubMed] [Google Scholar]
- 548.Van Soolingen D, van Ingen J. 2012. On the road to unravelling the aetiology of non-tuberculous mycobacterial disease. Int J Tuberc Lung Dis 16:1279. doi: 10.5588/ijtld.12.0630. [DOI] [PubMed] [Google Scholar]
- 549.Simons S, van Ingen J, Hsueh P-R, Van Hung N, Dekhuijzen PNR, Boeree MJ, van Soolingen D. 2011. Nontuberculous mycobacteria in respiratory tract infections, eastern Asia. Emerg Infect Dis 17:343–349. doi: 10.3201/eid170310060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Hoefsloot W, van Ingen J, Andrejak C, Angeby K, Bauriaud R, Bemer P, Beylis N, Boeree MJ, Cacho J, Chihota V, Chimara E, Churchyard G, Cias R, Daza R, Daley CL, Dekhuijzen PNR, Domingo D, Drobniewski F, Esteban J, Fauville-Dufaux M, Folkvardsen DB, Gibbons N, Gómez-Mampaso E, Gonzalez R, Hoffmann H, Hsueh P-R, Indra A, Jagielski T, Jamieson F, Jankovic M, Jong E, Keane J, Koh W-J, Lange B, Leao S, Macedo R, Mannsåker T, Marras TK, Maugein J, Milburn HJ, Mlinkó T, Morcillo N, Morimoto K, Papaventsis D, Palenque E, Paez-Peña M, Piersimoni C, Polanová M, Rastogi N, Richter E, et al. 2013. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: an NTM-NET collaborative study. Eur Respir J 42:1604–1613. doi: 10.1183/09031936.00149212. [DOI] [PubMed] [Google Scholar]
- 551.García de Viedma D, Mokrousov I, Rastogi N. 2011. Innovations in the molecular epidemiology of tuberculosis. Enferm Infecc Microbiol Clin 29(Suppl 1):8–13. doi: 10.1016/S0213-005X(11)70012-X. [DOI] [PubMed] [Google Scholar]
- 552.Rothschild BM, Martin LD, Lev G, Bercovier H, Bar-Gal GK, Greenblatt C, Donoghue H, Spigelman M, Brittain D. 2001. Mycobacterium tuberculosis complex DNA from an extinct bison dated 17,000 years before the present. Clin Infect Dis 33:305–311. doi: 10.1086/321886. [DOI] [PubMed] [Google Scholar]
- 553.Wolfe ND, Dunavan CP, Diamond J. 2007. Origins of major human infectious diseases. Nature 447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Barry CE, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, Young D. 2009. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol 7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, Parkhill J, Malla B, Berg S, Thwaites G, Yeboah-Manu D, Bothamley G, Mei J, Wei L, Bentley S, Harris SR, Niemann S, Diel R, Aseffa A, Gao Q, Young D, Gagneux S. 2013. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet 45:1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Bos KI, Harkins KM, Herbig A, Coscolla M, Weber N, Comas I, Forrest SA, Bryant JM, Harris SR, Schuenemann VJ, Campbell TJ, Majander K, Wilbur AK, Guichon RA, Wolfe Steadman DL, Cook DC, Niemann S, Behr MA, Zumarraga M, Bastida R, Huson D, Nieselt K, Young D, Parkhill J, Buikstra JE, Gagneux S, Stone AC, Krause J. 2014. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature 514:494–497. doi: 10.1038/nature13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Daniel TM. 2005. The bioarchaeology of tuberculosis: a global view on a reemerging disease. Am J Trop Med Hyg 73:649–650. [Google Scholar]
- 558.Kay GL, Sergeant MJ, Zhou Z, Chan JZ, Millard A, Quick J, Szikossy I, Pap I, Spigelman M, Loman NJ, Achtman M, Donoghue HD, Pallen MJ. 2015. Eighteenth-century genomes show that mixed infections were common at time of peak tuberculosis in Europe. Nat Commun 6:6717. doi: 10.1038/ncomms7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, Whittam TS, Musser JM. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci U S A 94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Gordon SV, Brosch R, Billault A, Garnier T, Eiglmeier K, Cole ST. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol Microbiol 32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 561.Filliol I, Motiwala AS, Cavatore M, Qi W, Hazbón MH, Bobadilla del Valle M, Fyfe J, García-García L, Rastogi N, Sola C, Zozio T, Guerrero MI, León CI, Crabtree J, Angiuoli S, Eisenach KD, Durmaz R, Joloba ML, Rendón A, Sifuentes-Osornio J, Ponce de León A, Cave MD, Fleischmann R, Whittam TS, Alland D. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol 188:759–772. doi: 10.1128/JB.188.2.759-772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Gutacker MM, Mathema B, Soini H, Shashkina E, Kreiswirth BN, Graviss EA, Musser JM. 2006. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J Infect Dis 193:121–128. doi: 10.1086/498574. [DOI] [PubMed] [Google Scholar]
- 563.Fenner L, Malla B, Ninet B, Dubuis O, Stucki D, Borrell S, Huna T, Bodmer T, Egger M, Gagneux S. 2011. “Pseudo-Beijing”: evidence for convergent evolution in the direct repeat region of Mycobacterium tuberculosis. PLoS One 6:e24737. doi: 10.1371/journal.pone.0024737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.World Health Organization. 2015. Introducing the END TB strategy. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 565.Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, Hill DP, Kania R, Schaeffer M, St Pierre S, Twigger S, White O, Rhee SY. 2008. Big data: the future of biocuration. Nature 455:47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, Allix C, Aristimuño L, Arora J, Baumanis V, Binder L, Cafrune P, Cataldi A, Cheong S, Diel R, Ellermeier C, Evans JT, Fauville-Dufaux M, Ferdinand S, de Viedma DG, Garzelli C, Gazzola L, Gomes HM, Guttierez MC, Hawkey PM, van Helden PD, Kadival GV, Kreiswirth BN, Kremer K, Kubin M, Kulkarni SP, Liens B, Lillebaek T, Ly HM, Martin C, Martin C, Mokrousov I, Narvskaïa O, Ngeow YF, Naumann L, Niemann S, Parwati I, Rahim Z, Rasolofo-Razanamparany V, Rasolonavalona T, Rossetti ML, Rüsch-Gerdes S, Sajduda A, Samper S, Shemyakin IG, et al. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol 6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Reyes JF, Francis AR, Tanaka MM. 2008. Models of deletion for visualizing bacterial variation: an application to tuberculosis spoligotypes. BMC Bioinformatics 9:496. doi: 10.1186/1471-2105-9-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Tang C, Reyes JF, Luciani F, Francis AR, Tanaka MM. 2008. spolTools: online utilities for analyzing spoligotypes of the Mycobacterium tuberculosis complex. Bioinformatics 24:2414–2415. doi: 10.1093/bioinformatics/btn434. [DOI] [PubMed] [Google Scholar]
- 569.Ellson J, Gansner E, Koutsofios L, North SC, Woodhull G. 2002. Graphviz—open source graph drawing tools, p 483–484. In Mutzel P, Jünger M, Leipert S (ed), Graph drawing. Springer, Berlin, Germany. [Google Scholar]
- 570.Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Crooks GE, Hon G, Chandonia J-M, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Driscoll JR, Bifani PJ, Mathema B, McGarry MA, Zickas GM, Kreiswirth BN, Taber HW. 2002. Spoligologos: a bioinformatic approach to displaying and analyzing Mycobacterium tuberculosis data. Emerg Infect Dis 8:1306–1309. doi: 10.3201/eid0811.020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.CDC. 2009. TB genotyping information management system (TB GIMS). CDC, Atlanta, GA. [Google Scholar]
- 574.Smith NH, Upton P. 2012. Naming spoligotype patterns for the RD9-deleted lineage of the Mycobacterium tuberculosis complex; www.Mbovis.org. Infect Genet Evol 12:873–876. doi: 10.1016/j.meegid.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 575.Rodriguez-Campos S, González S, de Juan L, Romero B, Bezos J, Casal C, Álvarez J, Fernández-de-Mera IG, Castellanos E, Mateos A, Sáez-Llorente JL, Domínguez L, Aranaz A, Spanish Network on Surveillance Monitoring of Animal Tuberculosis. 2012. A database for animal tuberculosis (mycoDB.es) within the context of the Spanish national programme for eradication of bovine tuberculosis. Infect Genet Evol 12:877–882. doi: 10.1016/j.meegid.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 576.Shabbeer A, Cowan LS, Ozcaglar C, Rastogi N, Vandenberg SL, Yener B, Bennett KP. 2012. TB-Lineage: an online tool for classification and analysis of strains of Mycobacterium tuberculosis complex. Infect Genet Evol 12:789–797. doi: 10.1016/j.meegid.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 577.Joshi KR, Dhiman H, Scaria V. 2014. tbvar: a comprehensive genome variation resource for Mycobacterium tuberculosis. Database 2014:bat083. doi: 10.1093/database/bat083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Soares P, Alves RJ, Abecasis AB, Penha-Gonçalves C, Gomes MGM, Pereira-Leal JB. 2013. inTB—a data integration platform for molecular and clinical epidemiological analysis of tuberculosis. BMC Bioinformatics 14:264. doi: 10.1186/1471-2105-14-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. 2009. Tuberculosis drug resistance mutation database. PLoS Med 6:e1000002. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Flandrois J-P, Lina G, Dumitrescu O. 2014. MUBII-TB-DB: a database of mutations associated with antibiotic resistance in Mycobacterium tuberculosis. BMC Bioinformatics 15:107. doi: 10.1186/1471-2105-15-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581.Feuerriegel S, Schleusener V, Beckert P, Kohl TA, Miotto P, Cirillo DM, Cabibbe AM, Niemann S, Fellenberg K. 2015. PhyResSE: a Web tool delineating Mycobacterium tuberculosis antibiotic resistance and lineage from whole-genome sequencing data. J Clin Microbiol 53:1908–1914. doi: 10.1128/JCM.00025-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Starks AM, Avilés E, Cirillo DM, Denkinger CM, Dolinger DL, Emerson C, Gallarda J, Hanna D, Kim PS, Liwski R, Miotto P, Schito M, Zignol M. 2015. Collaborative effort for a centralized worldwide tuberculosis relational sequencing data platform. Clin Infect Dis 61:S141–S146. doi: 10.1093/cid/civ610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Sola C, Devallois A, Horgen L, Maïsetti J, Filliol I, Legrand E, Rastogi N. 1999. Tuberculosis in the Caribbean: using spacer oligonucleotide typing to understand strain origin and transmission. Emerg Infect Dis 5:404–414. doi: 10.3201/eid0503.990311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Sola C, Filliol I, Gutierrez MC, Mokrousov I, Vincent V, Rastogi N. 2001. Spoligotype database of Mycobacterium tuberculosis: biogeographic distribution of shared types and epidemiologic and phylogenetic perspectives. Emerg Infect Dis 7:390–396. doi: 10.3201/10.3201/eid0703.0107304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Filliol I, Driscoll JR, Van Soolingen D, Kreiswirth BN, Kremer K, Valétudie G, Anh DD, Barlow R, Banerjee D, Bifani PJ, Brudey K, Cataldi A, Cooksey RC, Cousins DV, Dale JW, Dellagostin OA, Drobniewski F, Engelmann G, Ferdinand S, Gascoyne-Binzi D, Gordon M, Gutierrez MC, Haas WH, Heersma H, Källenius G, Kassa-Kelembho E, Koivula T, Ly HM, Makristathis A, Mammina C, Martin G, Moström P, Mokrousov I, Narbonne V, Narvskaya O, Nastasi A, Niobe-Eyangoh SN, Pape JW, Rasolofo-Razanamparany V, Ridell M, Rossetti ML, Stauffer F, Suffys PN, Takiff H, Texier-Maugein J, Vincent V, De Waard JH, Sola C, Rastogi N. 2002. Global distribution of Mycobacterium tuberculosis spoligotypes. Emerg Infect Dis 8:1347–1349. doi: 10.3201/eid0811.020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Filliol I, Driscoll JR, van Soolingen D, Kreiswirth BN, Kremer K, Valétudie G, Dang DA, Barlow R, Banerjee D, Bifani PJ, Brudey K, Cataldi A, Cooksey RC, Cousins DV, Dale JW, Dellagostin OA, Drobniewski F, Engelmann G, Ferdinand S, Gascoyne-Binzi D, Gordon M, Gutierrez MC, Haas WH, Heersma H, Kassa-Kelembho E, Ho ML, Makristathis A, Mammina C, Martin G, Moström P, Mokrousov I, Narbonne V, Narvskaya O, Nastasi A, Niobe-Eyangoh SN, Pape JW, Rasolofo-Razanamparany V, Ridell M, Rossetti ML, Stauffer F, Suffys PN, Takiff H, Texier-Maugein J, Vincent V, de Waard JH, Sola C, Rastogi N. 2003. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J Clin Microbiol 41:1963–1970. doi: 10.1128/JCM.41.5.1963-1970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 587.Mokrousov I. 2012. The quiet and controversial: Ural family of Mycobacterium tuberculosis. Infect Genet Evol 12:619–629. doi: 10.1016/j.meegid.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 588.Koro Koro F, Simo YK, Piam FF, Noeske J, Gutierrez C, Kuaban C, Eyangoh SI. 2013. Population dynamics of tuberculous bacilli in Cameroon as assessed by spoligotyping. J Clin Microbiol 51:299–302. doi: 10.1128/JCM.01196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Abadia E, Zhang J, dos Vultos T, Ritacco V, Kremer K, Aktas E, Matsumoto T, Refregier G, van Soolingen D, Gicquel B, Sola C. 2010. Resolving lineage assignation on Mycobacterium tuberculosis clinical isolates classified by spoligotyping with a new high-throughput 3R SNPs based method. Infect Genet Evol 10:1066–1074. doi: 10.1016/j.meegid.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 590.Kisa O, Tarhan G, Gunal S, Albay A, Durmaz R, Saribas Z, Zozio T, Alp A, Ceyhan I, Tombak A, Rastogi N. 2012. Distribution of spoligotyping defined genotypic lineages among drug-resistant Mycobacterium tuberculosis complex clinical isolates in Ankara, Turkey. PLoS One 7:e30331. doi: 10.1371/journal.pone.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Groenheit R, Ghebremichael S, Svensson J, Rabna P, Colombatti R, Riccardi F, Couvin D, Hill V, Rastogi N, Koivula T, Källenius G. 2011. The Guinea-Bissau family of Mycobacterium tuberculosis complex revisited. PLoS One 6:e18601. doi: 10.1371/journal.pone.0018601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Groenheit R, Ghebremichael S, Pennhag A, Jonsson J, Hoffner S, Couvin D, Koivula T, Rastogi N, Källenius G. 2012. Mycobacterium tuberculosis strains potentially involved in the TB epidemic in Sweden a century ago. PLoS One 7:e46848. doi: 10.1371/journal.pone.0046848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593.Sheen P, Couvin D, Grandjean L, Zimic M, Dominguez M, Luna G, Gilman RH, Rastogi N, Moore DAJ. 2013. Genetic diversity of Mycobacterium tuberculosis in Peru and exploration of phylogenetic associations with drug resistance. PLoS One 8:e65873. doi: 10.1371/journal.pone.0065873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Smit PW, Haanperä M, Rantala P, Couvin D, Lyytikäinen O, Rastogi N, Ruutu P, Soini H. 2013. Molecular epidemiology of tuberculosis in Finland, 2008-2011. PLoS One 8:e85027. doi: 10.1371/journal.pone.0085027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Glynn JR, Whiteley J, Bifani PJ, Kremer K, van Soolingen D. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg Infect Dis 8:843–849. doi: 10.3201/eid0808.020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Couvin D, Rastogi N. 2015. Tuberculosis—a global emergency: tools and methods to monitor, understand, and control the epidemic with specific example of the Beijing lineage. Tuberculosis (Edinb) 95:S177–S189. doi: 10.1016/j.tube.2015.02.023. [DOI] [PubMed] [Google Scholar]
- 597.Mustafa Ali R, Trovato A, Couvin D, Al-Thwani AN, Borroni E, Dhaer FH, Rastogi N, Cirillo DM. 2014. Molecular epidemiology and genotyping of Mycobacterium tuberculosis isolated in Baghdad. Biomed Res Int 2014:580981. doi: 10.1155/2014/580981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Han XY, Tarrand JJ, Infante R, Jacobson KL, Truong M. 2005. Clinical significance and epidemiologic analyses of Mycobacterium avium and Mycobacterium intracellulare among patients without AIDS. J Clin Microbiol 43:4407–4412. doi: 10.1128/JCM.43.9.4407-4412.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Roth A, Fischer M, Hamid ME, Michalke S, Ludwig W, Mauch H. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J Clin Microbiol 36:139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Park H, Jang H, Kim C, Chung B, Chang CL, Park SK, Song S. 2000. Detection and identification of mycobacteria by amplification of the internal transcribed spacer regions with genus- and species-specific PCR primers. J Clin Microbiol 38:4080–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Zumarraga MJ, Resoagli EH, Cicuta ME, Martinez AR, Oritiz de Rott MI, de Millan SG, Caimi K, Gioffre A, Alito A, Bigi F, Cataldi AA, Romano MI. 2001. PCR-restriction fragment length polymorphism analysis (PRA) of Mycobacterium leprae from human lepromas and from a natural case of an armadillo of Corrientes, Argentina. Int J Lepr Other Mycobact Dis 69:21–25. [PubMed] [Google Scholar]
- 602.Kim H, Kim S-H, Shim T-S, Kim M, Bai G-H, Park Y-G, Lee S-H, Chae G-T, Cha C-Y, Kook Y-H, Kim B-J. 2005. Differentiation of Mycobacterium species by analysis of the heat-shock protein 65 gene (hsp65). Int J Syst Evol Microbiol 55:1649–1656. doi: 10.1099/ijs.0.63553-0. [DOI] [PubMed] [Google Scholar]
- 603.Chang P-L, Hsieh W-S, Chiang C-L, Yen-Liberman B, Procop GW, Chang H-T, Ho H-T. 2008. Identification of individual DNA molecule of Mycobacterium tuberculosis by nested PCR-RFLP and capillary electrophoresis. Talanta 77:182–188. doi: 10.1016/j.talanta.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 604.Chimara E, Ferrazoli L, Ueky SY, Martins MC, Durham AM, Arbeit RD, Leão SC. 2008. Reliable identification of mycobacterial species by PCR-restriction enzyme analysis (PRA)-hsp65 in a reference laboratory and elaboration of a sequence-based extended algorithm of PRA-hsp65 patterns. BMC Microbiol 8:48. doi: 10.1186/1471-2180-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Da Costa ARF, Lopes ML, Furlaneto IP, de Sousa MS, Lima KVB. 2010. Molecular identification of nontuberculous mycobacteria isolates in a Brazilian mycobacteria reference laboratory. Diagn Microbiol Infect Dis 68:390–394. doi: 10.1016/j.diagmicrobio.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 606.Shukla S, Shukla SK, Sharma R, Kumar A. 2014. Identification of Mycobacterium species from apparently healthy freshwater aquarium fish using partial sequencing and PCR-RFLP analysis of heat shock protein (hsp65) gene. J Appl Ichthyol 30:513–520. doi: 10.1111/jai.12415. [DOI] [Google Scholar]
- 607.Aravindhan V, Sulochana S, Narayanan S, Paramasivam CN, Narayanan PR. 2007. Identification & differentiation of Mycobacterium avium & M. intracellulare by PCR-RFLP assay using the groES gene. Indian J Med Res 126:575–579. [PubMed] [Google Scholar]
- 608.Blackwood KS, He C, Gunton J, Turenne CY, Wolfe J, Kabani AM. 2000. Evaluation of recA sequences for identification of Mycobacterium species. J Clin Microbiol 38:2846–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Lee H, Bang H-E, Bai G-H, Cho S-N. 2003. Novel polymorphic region of the rpoB gene containing Mycobacterium species-specific sequences and its use in identification of mycobacteria. J Clin Microbiol 41:2213–2218. doi: 10.1128/JCM.41.5.2213-2218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Kim B-J, Hong S-K, Lee K-H, Yun Y-J, Kim E-C, Park Y-G, Bai G-H, Kook Y-H. 2004. Differential identification of Mycobacterium tuberculosis complex and nontuberculous mycobacteria by duplex PCR assay using the RNA polymerase gene (rpoB). J Clin Microbiol 42:1308–1312. doi: 10.1128/JCM.42.3.1308-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611.Ben Salah I, Adékambi T, Raoult D, Drancourt M. 2008. rpoB sequence-based identification of Mycobacterium avium complex species. Microbiology 154:3715–3723. doi: 10.1099/mic.0.2008/020164-0. [DOI] [PubMed] [Google Scholar]
- 612.Arnold C, Barrett A, Cross L, Magee JG. 2012. The use of rpoB sequence analysis in the differentiation of Mycobacterium abscessus and Mycobacterium chelonae: a critical judgement in cystic fibrosis? Clin Microbiol Infect 18:E131–E133. doi: 10.1111/j.1469-0691.2012.03785.x. [DOI] [PubMed] [Google Scholar]
- 613.Takewaki S, Okuzumi K, Manabe I, Tanimura M, Miyamura K, Nakahara K, Yazaki Y, Ohkubo A, Nagai R. 1994. Nucleotide sequence comparison of the mycobacterial dnaJ gene and PCR-restriction fragment length polymorphism analysis for identification of mycobacterial species. Int J Syst Bacteriol 44:159–166. doi: 10.1099/00207713-44-1-159. [DOI] [PubMed] [Google Scholar]
- 614.Singh A, Kashyap VK. 2012. Specific and rapid detection of Mycobacterium tuberculosis complex in clinical samples by polymerase chain reaction. Interdiscip Perspect Infect Dis 2012:654694. doi: 10.1155/2012/654694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 615.Espinosa de los Monteros LE, Galán JC, Gutiérrez M, Samper S, García Marín JF, Martín C, Domínguez L, de Rafael L, Baquero F, Gómez-Mampaso E, Blázquez J. 1998. Allele-specific PCR method based on pncA and oxyR sequences for distinguishing Mycobacterium bovis from Mycobacterium tuberculosis: intraspecific M. bovis pncA sequence polymorphism. J Clin Microbiol 36:239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616.Abbadi S, El Hadidy G, Gomaa N, Cooksey R. 2009. Strain differentiation of Mycobacterium tuberculosis complex isolated from sputum of pulmonary tuberculosis patients. Int J Infect Dis 13:236–242. doi: 10.1016/j.ijid.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 617.Barouni AS, Augusto CJ, Lopes MTP, Zanini MS, Salas CE. 2004. A pncA polymorphism to differentiate between Mycobacterium bovis and Mycobacterium tuberculosis. Mol Cell Probes 18:167–170. doi: 10.1016/j.mcp.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 618.Herrmann B, Stolt P, Abdeldaim G, Rubin C-J, Kirsebom LA, Thollesson M. 2014. Differentiation and phylogenetic relationships in Mycobacterium spp with special reference to the RNase P RNA gene rnpB. Curr Microbiol 69:634–639. doi: 10.1007/s00284-014-0630-8. [DOI] [PubMed] [Google Scholar]
- 619.Khan IUH, Selvaraju SB, Yadav JS. 2005. Method for rapid identification and differentiation of the species of the Mycobacterium chelonae complex based on 16S-23S rRNA gene internal transcribed spacer PCR-restriction analysis. J Clin Microbiol 43:4466–4472. doi: 10.1128/JCM.43.9.4466-4472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Niemann S, Richter E, Rüsch-Gerdes S. 2000. Differentiation among members of the Mycobacterium tuberculosis complex by molecular and biochemical features: evidence for two pyrazinamide-susceptible subtypes of M. bovis. J Clin Microbiol 38:152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 621.Chimara E, Ferrazoli L, Leão SC. 2004. Mycobacterium tuberculosis complex differentiation using gyrB-restriction fragment length polymorphism analysis. Mem Inst Oswaldo Cruz 99:745–748. doi: 10.1590/S0074-02762004000700014. [DOI] [PubMed] [Google Scholar]
- 622.Zelazny AM, Calhoun LB, Li L, Shea YR, Fischer SH. 2005. Identification of Mycobacterium species by secA1 sequences. J Clin Microbiol 43:1051–1058. doi: 10.1128/JCM.43.3.1051-1058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Heym B, Alzari PM, Honoré N, Cole ST. 1995. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol Microbiol 15:235–245. doi: 10.1111/j.1365-2958.1995.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 624.Marttila HJ, Soini H, Huovinen P, Viljanen MK. 1996. katG mutations in isoniazid-resistant Mycobacterium tuberculosis isolates recovered from Finnish patients. Antimicrob Agents Chemother 40:2187–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Ando H, Kondo Y, Suetake T, Toyota E, Kato S, Mori T, Kirikae T. 2010. Identification of katG mutations associated with high-level isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 54:1793–1799. doi: 10.1128/AAC.01691-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 626.Jagielski T, Bakuła Z, Roeske K, Kamiński M, Napiórkowska A, Augustynowicz-Kopeć E, Zwolska Z, Bielecki J. 2014. Detection of mutations associated with isoniazid resistance in multidrug-resistant Mycobacterium tuberculosis clinical isolates. J Antimicrob Chemother 69:2369–2375. doi: 10.1093/jac/dku161. [DOI] [PubMed] [Google Scholar]
- 627.Marrakchi H, Lanéelle G, Quémard A. 2000. InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Microbiology 146:289–296. doi: 10.1099/00221287-146-2-289. [DOI] [PubMed] [Google Scholar]
- 628.Hazbón MH, Brimacombe M, Bobadilla del Valle M, Cavatore M, Guerrero MI, Varma-Basil M, Billman-Jacobe H, Lavender C, Fyfe J, García-García L, León CI, Bose M, Chaves F, Murray M, Eisenach KD, Sifuentes-Osornio J, Cave MD, Ponce de León A, Alland D. 2006. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 50:2640–2649. doi: 10.1128/AAC.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 629.Zhang M, Yue J, Yang Y, Zhang H, Lei J, Jin R, Zhang X, Wang H. 2005. Detection of mutations associated with isoniazid resistance in Mycobacterium tuberculosis isolates from China. J Clin Microbiol 43:5477–5482. doi: 10.1128/JCM.43.11.5477-5482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Lee AS, Teo AS, Wong SY. 2001. Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother 45:2157–2159. doi: 10.1128/AAC.45.7.2157-2159.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 631.Cardoso RF, Cardoso MA, Leite CQF, Sato DN, Mamizuka EM, Hirata RDC, de Mello FF, Hirata MH. 2007. Characterization of ndh gene of isoniazid resistant and susceptible Mycobacterium tuberculosis isolates from Brazil. Mem Inst Oswaldo Cruz 102:59–61. doi: 10.1590/S0074-02762007000100009. [DOI] [PubMed] [Google Scholar]
- 632.Silva MSN, Senna SG, Ribeiro MO, Valim ARM, Telles MA, Kritski A, Morlock GP, Cooksey RC, Zaha A, Rossetti MLR. 2003. Mutations in katG, inhA, and ahpC genes of Brazilian isoniazid-resistant isolates of Mycobacterium tuberculosis. J Clin Microbiol 41:4471–4474. doi: 10.1128/JCM.41.9.4471-4474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633.Kelley CL, Rouse DA, Morris SL. 1997. Analysis of ahpC gene mutations in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother 41:2057–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 634.Ahmad S, Araj GF, Akbar PK, Fares E, Chugh TD, Mustafa AS. 2000. Characterization of rpoB mutations in rifampin-resistant Mycobacterium tuberculosis isolates from the Middle East. Diagn Microbiol Infect Dis 38:227–232. doi: 10.1016/S0732-8893(00)00200-5. [DOI] [PubMed] [Google Scholar]
- 635.McCammon MT, Gillette JS, Thomas DP, Ramaswamy SV, Graviss EA, Kreiswirth BN, Vijg J, Quitugua TN. 2005. Detection of rpoB mutations associated with rifampin resistance in Mycobacterium tuberculosis using denaturing gradient gel electrophoresis. Antimicrob Agents Chemother 49:2200–2209. doi: 10.1128/AAC.49.6.2200-2209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, Zhang Y. 1997. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 41:540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637.Portugal I, Barreiro L, Moniz-Pereira J, Brum L. 2004. pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates in Portugal. Antimicrob Agents Chemother 48:2736–2738. doi: 10.1128/AAC.48.7.2736-2738.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Juréen P, Werngren J, Toro J-C, Hoffner S. 2008. Pyrazinamide resistance and pncA gene mutations in Mycobacterium tuberculosis. Antimicrob Agents Chemother 52:1852–1854. doi: 10.1128/AAC.00110-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Zhao LL, Sun Q, Liu HC, Wu XC, Xiao TY, Zhao XQ, Li GL, Jiang Y, Zeng CY, Wan K-L. 2015. Analysis of embCAB mutations associated with ethambutol resistance in multidrug-resistant Mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother 59:2045–2050. doi: 10.1128/AAC.04933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Jadaun GPS, Das R, Upadhyay P, Chauhan DS, Sharma VD, Katoch VM. 2009. Role of embCAB gene mutations in ethambutol resistance in Mycobacterium tuberculosis isolates from India. Int J Antimicrob Agents 33:483–486. doi: 10.1016/j.ijantimicag.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 641.Plinke C, Cox HS, Zarkua N, Karimovich HA, Braker K, Diel R, Rüsch-Gerdes S, Feuerriegel S, Niemann S. 2010. embCAB sequence variation among ethambutol-resistant Mycobacterium tuberculosis isolates without embB306 mutation. J Antimicrob Chemother 65:1359–1367. doi: 10.1093/jac/dkq120. [DOI] [PubMed] [Google Scholar]
- 642.Bakula Z, Napiorkowska A, Bielecki J, Augustynowicz-Kopec E, Zwolska Z, Jagielski T. 2013. Mutations in the embB gene and their association with ethambutol resistance in multidrug-resistant Mycobacterium tuberculosis clinical isolates from Poland. Biomed Res Int 2013:167954. doi: 10.1155/2013/167954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643.Cuevas-Córdoba B, Cuellar-Sánchez A, Pasissi-Crivelli A, Santana-Álvarez CA, Hernández-Illezcas J, Zenteno-Cuevas R. 2013. rrs and rpsL mutations in streptomycin-resistant isolates of Mycobacterium tuberculosis from Mexico. J Microbiol Immunol Infect 46:30–34. doi: 10.1016/j.jmii.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 644.Sreevatsan S, Pan X, Stockbauer KE, Williams DL, Kreiswirth BN, Musser JM. 1996. Characterization of rpsL and rrs mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from diverse geographic localities. Antimicrob Agents Chemother 40:1024–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 645.Jagielski T, Ignatowska H, Bakuła Z, Dziewit Ł, Napiórkowska A, Augustynowicz-Kopeć E, Zwolska Z, Bielecki J. 2014. Screening for streptomycin resistance-conferring mutations in Mycobacterium tuberculosis clinical isolates from Poland. PLoS One 9:e100078. doi: 10.1371/journal.pone.0100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 646.Wong SY, Lee JS, Kwak HK, Via LE, Boshoff HIM, Barry CE. 2011. Mutations in gidB confer low-level streptomycin resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 55:2515–2522. doi: 10.1128/AAC.01814-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 647.Verma JS, Gupta Y, Nair D, Manzoor N, Rautela RS, Rai A, Katoch VM. 2014. Evaluation of gidB alterations responsible for streptomycin resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 69:2935–2941. doi: 10.1093/jac/dku273. [DOI] [PubMed] [Google Scholar]
- 648.Du Q, Dai G, Long Q, Yu X, Dong L, Huang H, Xie J. 2013. Mycobacterium tuberculosis rrs A1401G mutation correlates with high-level resistance to kanamycin, amikacin, and capreomycin in clinical isolates from mainland China. Diagn Microbiol Infect Dis 77:138–142. doi: 10.1016/j.diagmicrobio.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 649.Suzuki Y, Katsukawa C, Tamaru A, Abe C, Makino M, Mizuguchi Y, Taniguchi H. 1998. Detection of kanamycin-resistant Mycobacterium tuberculosis by identifying mutations in the 16S rRNA gene. J Clin Microbiol 36:1220–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650.Sowajassatakul A, Prammananan T, Chaiprasert A, Phunpruch S. 2014. Molecular characterization of amikacin, kanamycin and capreomycin resistance in M/XDR-TB strains isolated in Thailand. BMC Microbiol 14:165. doi: 10.1186/1471-2180-14-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 651.Gikalo MB, Nosova EY, Krylova LY, Moroz AM. 2012. The role of eis mutations in the development of kanamycin resistance in Mycobacterium tuberculosis isolates from the Moscow region. J Antimicrob Chemother 67:2107–2109. doi: 10.1093/jac/dks178. [DOI] [PubMed] [Google Scholar]
- 652.Morlock GP, Metchock B, Sikes D, Crawford JT, Cooksey RC. 2003. ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother 47:3799–3805. doi: 10.1128/AAC.47.12.3799-3805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 653.Brossier F, Veziris N, Truffot-Pernot C, Jarlier V, Sougakoff W. 2011. Molecular investigation of resistance to the antituberculous drug ethionamide in multidrug-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother 55:355–360. doi: 10.1128/AAC.01030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.Chen J, Chen Z, Li Y, Xia W, Chen X, Chen T, Zhou L, Xu B, Xu S. 2012. Characterization of gyrA and gyrB mutations and fluoroquinolone resistance in Mycobacterium tuberculosis clinical isolates from Hubei Province, China. Braz J Infect Dis 16:136–141. doi: 10.1016/S1413-8670(12)70294-5. [DOI] [PubMed] [Google Scholar]
- 655.Li J, Gao X, Luo T, Wu J, Sun G, Liu Q, Jiang Y, Zhang Y, Mei J, Gao Q. 2014. Association of gyrA/B mutations and resistance levels to fluoroquinolones in clinical isolates of Mycobacterium tuberculosis. Emerg Microbes Infect 3:e19. doi: 10.1038/emi.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656.Mathys V, Wintjens R, Lefevre P, Bertout J, Singhal A, Kiass M, Kurepina N, Wang X-M, Mathema B, Baulard A, Kreiswirth BN, Bifani P. 2009. Molecular genetics of para-aminosalicylic acid resistance in clinical isolates and spontaneous mutants of Mycobacterium tuberculosis. Antimicrob Agents Chemother 53:2100–2109. doi: 10.1128/AAC.01197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657.Köser CU, Veerapen-Pierce RN, Summers DK, Archer JAC. 2010. Role of mutations in dihydrofolate reductase DfrA (Rv2763c) and thymidylate synthase ThyA (Rv2764c) in Mycobacterium tuberculosis drug resistance. Antimicrob Agents Chemother 54:4522–4523. doi: 10.1128/AAC.00422-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 658.Zhang X, Liu L, Zhang Y, Dai G, Huang H, Jin Q. 2015. Genetic determinants involved in p-aminosalicylic acid resistance in clinical isolates from tuberculosis patients in northern China from 2006 to 2012. Antimicrob Agents Chemother 59:1320–1324. doi: 10.1128/AAC.03695-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 659.Zhao F, Wang X-D, Erber LN, Luo M, Guo A, Yang S, Gu J, Turman BJ, Gao Y, Li D, Cui Z, Zhang Z, Bi L, Baughn AD, Zhang X-E, Deng J-Y. 2014. Binding pocket alterations in dihydrofolate synthase confer resistance to para-aminosalicylic acid in clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:1479–1487. doi: 10.1128/AAC.01775-13. [DOI] [PMC free article] [PubMed] [Google Scholar]